Abstract
Background
Immune landscape of cancer has been increasingly recognized as a key feature affecting disease progression, prognosis and therapeutic response. Here, we sought to comprehensively characterize the patterns of tumor-infiltrating immune cells (TIIs) in primary oral squamous cell carcinoma (OSCC) and develop immune features-derived models for prognostication and therapeutic prediction.
Methods
A total number of 392 patients with OSCC receiving ablative surgery at three independent centers were retrospectively enrolled and defined as training, testing and validation cohorts. Detailed features of 12 types of TIIs at center of tumor and invasive margin were assessed by immunohistochemistry coupled with digital quantification. TIIs abundance in OSCC was also estimated by bioinformatics approaches using multiple publicly available data sets. Prognostic models based on selected immune features were trained via machine learning approach, validated in independent cohorts and evaluated by time-dependent area under the curves and concordance index (C-index). Immune types of OSCC were further identified by consensus clustering and their associations with genetic, molecular features and patient survival were clarified.
Results
Patterns of TIIs infiltration varied among patients and dynamically evolved along with tumor progression. Prognostic models based on selected TIIs were identified as efficient and sensitive biomarkers to stratify patients into subgroups with favorable or inferior survival as well as responders or non-responders to postoperative radiotherapy or immunotherapy. These models outperformed multiple conventional biomarkers and immune-related scores in prognostic prediction. Furthermore, we identified two main immune subtypes of OSCC (immune-hot and immune-cold) which harbored characteristic TIIs infiltrations and genomic and molecular features, and associated with patient survival.
Conclusions
Our results delineated immune landscape and subtypes in OSCC, consolidated their clinical values as robust biomarkers to predict patient survival and therapeutic benefits and reinforced key roles of TIIs and tumor-immune interactions underlying oral tumorigenesis, ultimately facilitating development of tailed immunotherapeutic strategies.
Keywords: immunotherapy, head and neck neoplasms
Background
Intricate crosstalk between cancer cells and host immune system fuels tumor immune evasion and ultimately results in tumor overgrowth, metastatic dissemination and therapeutic failure.1 Tumor-infiltrating immune cells (TIIs) exert multifaceted functions either protumor or antitumor roles during diverse stages of tumorigenesis, highlighting their functional diversity and plasticity.2 Therefore, in-depth characterization of immune landscape in cancer will facilitate to unveil the key roles of immune surveillance and evasion driving tumor initiation and progression, and also provide essential information to inform rational design of therapy, predict therapeutic response and patient prognosis.3–5
Prognostic impacts of various TIIs have been demonstrated in a broad spectrum of human cancer.5 6 Densities, locations and functional status of TIIs subsets such as CD8+ cytolytic T cells have been found to be significantly associated with better survival, while M2 tumor-associated macrophages and myeloid-derived suppressor cells (MDSCs) correlated with inferior survival.6–10 Moreover, integrative analyses of multiple TIIs subsets usually outperformed single type of TIIs in prognostic prediction as exemplified by the well-established consensus Immunoscore.11–13
Oral squamous cell carcinoma (OSCC) represents one of the leading cancers worldwide with substantial mortality and morbidity.14 Current prognostic markers for OSCC largely rely on TNM staging system which is not sufficient and accurate to predict patient survival as the prognosis of patients within the same categories vary considerably.15 Although the efficiencies of immune checkpoint blockade (ICB) to improve outcomes have documented in patients with recurrent or metastatic OSCC, reliable biomarkers for patient stratification and response prediction are still lacking.16 Until now, immune context and its clinical significance have just begun to be explored in OSCC.17–19 However, large knowledge gaps still remain concerning comprehensive spatiotemporal characterization of TIIs, their clinical significance as well as the associated molecular determinants in OSCC.
In the present study, we sought to comprehensively characterize the immune landscape in OSCC via standardized immunohistochemical staining (IHC) coupled with digital quantification or in silicon deconvolution analyses of publicly available RNA-seq data sets in multiple independent cohorts, respectively. Our results revealed the compositions, heterogeneity and patterns of TIIs infiltration in OSCC samples, reinforced their values in predicting prognosis and therapeutic response.
Methods
Detailed descriptions of all materials and methods are presented in the online supplemental materials available online.
jitc-2021-002434supp001.pdf (210.5KB, pdf)
Results
Demographic and clinicopathological characteristics of patients enrolled
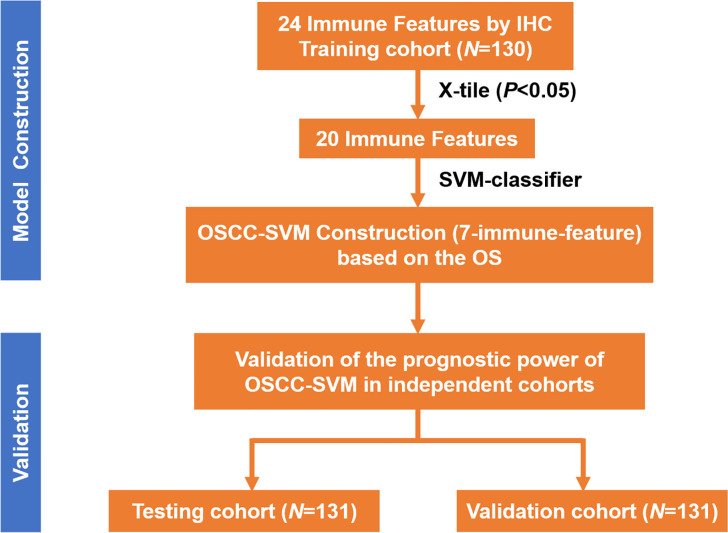
As illustrated in figure 1, we exploited a training-testing-validation cohort design to explore the immune landscape and its clinical significance in primary OSCC. Detailed demographic and clinicopathological features of patients from these three independent cohorts were described in online supplemental table S1. The distributions of gender, age, clinicopathological parameters as well as durations of follow-up were comparable. Until last follow-up, 197 patients (67 in the training cohort, 64 in the testing cohort, 66 in the validation cohort; 197/392, 50.26%) died due to disease recurrence, metastasis or unknown reasons.
Figure 1.
Study design of immune landscape in our OSCC cohorts. IHC, immunohistochemical staining; OSCC, oral squamous cell carcinoma; OS, overall survival; SVM, support vector machine.
jitc-2021-002434supp002.pdf (25KB, pdf)
Locations, densities and prognostic significance of 12 subtypes of TIIs in OSCC
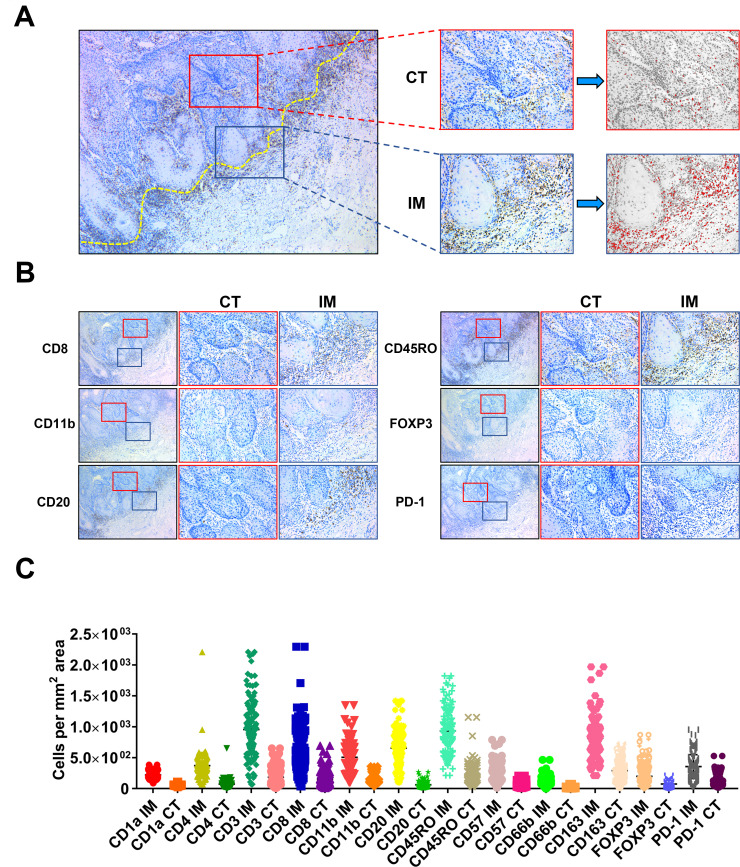
To comprehensively delineate the immune landscape of OSCC, we utilized the conventional IHC for labeling various TIIs using representative markers, followed by digital quantifications to capture their characteristics of locations (center of tumor (CT) and invasive margin (IM)) and densities in whole slides (figure 2A). These IHC markers for TIIs subsets were selected based on previous reports and PD-1 was chosen as a surrogate marker for functional status of those infiltrating lymphocytes.3 5 8 9 Representative IHC staining and quantification pipeline of TIIs subsets of interest were shown in figure 2B. To measure the reproducibility and reliability of our quantification approach, 30 cases in each cohort were randomly selected and independently assessed by two examiners. As shown in online supplemental figure S1, highly consistent results were achieved between two examiners in terms of CD8+ T-cell densities, thus in part supporting the reproducibility of our digital quantification. The densities of TIIs subsets (cells/mm2) in samples from the training cohort varied considerably among samples and in diverse areas (CT, IM) in individual samples (figure 2C). Noticeably, all these TIIs examined were more frequently infiltrated in IM region than CT compartment in OSCC. The patterns of TIIs infiltration differed among samples as evidenced by reduced amounts of CD3+ T cells, CD8+ T cells, CD20+ B cells and increased CD11b+ myeloid cells and CD163+ macrophages either in CT or IM compartment in advanced diseases as compared with diseases within early stages (online supplemental figure S2). This implied that immune landscape was not static but spatiotemporally dynamic as diseases progressed, similar as previous reports indicated.8
Figure 2.
Schematic illustration of methods to quantify TIIs at CT and IM in OSCC sample. (A, B) Illustration of TIIs quantification method in conventional IHC image in OSCC. The dot yellow line marked the border between tumor and the adjacent non-tumor stroma. The area labeled in red box was defined as center of tumor (CT), while the area labeled in blue box was defined as invasive margin (IM). Numbers of TIIs in tumor CT and IM were automatically counted by ImageJ software. (C) The abundance of 24 TIIs subsets in OSSC in the training cohort (n=130) was shown. IH, immunohistochemical staining; OSCC, oral squamous cell carcinoma; TIIs, tumor-infiltrating immune cells.
jitc-2021-002434supp003.pdf (213.8KB, pdf)
jitc-2021-002434supp004.pdf (196.2KB, pdf)
Next, we employed the X-tile plots to generate optimal cut-off values of all 24 immune features based on the overall survival (OS) data in the training cohort. Our results indicated that higher amounts of CD1a IM, CD1a CT, CD3 IM, etc., associated with improved OS (p<0.05), while lower amounts of CD11b IM and CD11b CT associated with improved OS (p<0.05). However, we failed to identify significant associations between CD4 IM, CD4 CT, CD66b IM or FOXP3 IM and OS in the training cohort. Subsequently, we performed univariate regression analyses and revealed that high densities of CD1a IM, CD1a CT, CD3 IM, etc., were significantly associated with better OS and disease-free survival (DFS), while lower densities of CD11b IM and CD11b CT associated with improved OS and DFS (p<0.05; online supplemental table S2). Moreover, we performed consensus clustering based on 24 immune features and survival, and separated the training cohort into two patient subgroups (clusters 1 and 2, online supplemental figure S3A). As shown in online supplemental figure S3B, C, samples in cluster 2 were significantly enriched with CD3+, CD8+, CD20+, CD45RO+ TIIs subsets but fewer CD11b+ myeloid cells as compared with cluster 1. In addition, patients in cluster 2 had fewer deaths, improved OS (p=0.0297) and DFS (p=0.0199) than those in cluster 1. Thus, we defined the cluster 1 as immune-cold subtype and cluster 2 as immune-hot subtype based on the immune landscape and patient survival.
jitc-2021-002434supp005.pdf (31.4KB, pdf)
jitc-2021-002434supp006.pdf (919KB, pdf)
Development and validation of a prognostic immune signature using SVM classifier for OSCC
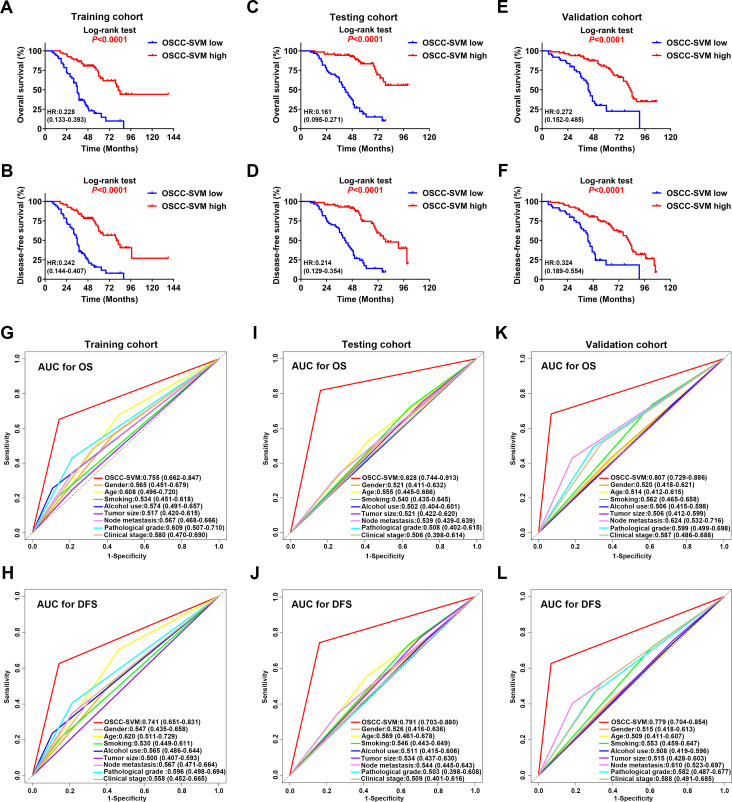
Next, we employed the support vector machine (SVM) algorithm to develop a novel OSCC-SVM immune signature. Leveraging these data from the training cohort, our OSCC-SVM signature integrated seven immune features from the 20 candidate features with prognostic significance as mentioned above including CD8 IM, CD45RO IM, CD11b IM, CD11b CT, CD20 CT, FOXP3 CT and PD-1 CT as pivotal prognostic factors. Tumors with high OSCC-SVM generally exhibited increased CD8 IM, CD20CT, CD45RO IM and PD-1 CT and reduced CD11b IM and CD11b CT (online supplemental figure S4). The Kaplan-Meier curves and univariate analyses revealed that these seven immune features significantly associated with OS and DFS in all three cohorts (online supplemental figure S5-S10, online supplemental tables S3 and S4). As shown in figure 3A–F, higher OSCC-SVM significantly associated with improved OS and DFS (Kaplan-Meier analyses, p<0.0001). To further confirm the prognostic values of this OSCC-SVM signature, we applied it to the testing and validation cohorts and found similar results. Complementarily, results from univariate analyses were in line with this notion that high OSCC-SVM was significantly associated with improved survival (online supplemental tables S3 and S4). Furthermore, multivariate Cox proportional regression analyses after adjustment for other clinicopathological variables replicated that OSCC-SVM was an independent prognostic factor affecting patient survival (p<0.001; online supplemental tables S5-S7).
Figure 3.
Prognostic significance and predictive ability of SVM classifier for OSCC. (A–F) Kaplan-Meier analyses of overall survival (OS, upper panel) and disease-free survival (DFS, lower panel) based on high or low OSCC-SVM classifier in patients from the training cohort (A, B); n=130), testing cohort (C, D); n=131) and validation cohort (E, F); n=131). (G–L) The sensitivity and specificity of OSCC-SVM and other clinicopathological parameters in prognostic prediction (OS, the upper panel; DFS, the lower panel) were estimated by 5-year ROC curves in training (G, H), testing (I, J) and validation cohorts (K, L). AUC, area under the curves; OSCC, oral squamous cell carcinoma; ROC, receiver operating characteristics; SVM, support vector machine.
jitc-2021-002434supp007.pdf (513.6KB, pdf)
jitc-2021-002434supp008.pdf (560KB, pdf)
jitc-2021-002434supp009.pdf (787KB, pdf)
jitc-2021-002434supp010.pdf (704.9KB, pdf)
jitc-2021-002434supp011.pdf (816.5KB, pdf)
jitc-2021-002434supp012.pdf (706.2KB, pdf)
jitc-2021-002434supp013.pdf (744.1KB, pdf)
jitc-2021-002434supp014.pdf (29.5KB, pdf)
jitc-2021-002434supp015.pdf (28.8KB, pdf)
jitc-2021-002434supp016.pdf (15.2KB, pdf)
jitc-2021-002434supp017.pdf (15.2KB, pdf)
jitc-2021-002434supp018.pdf (15.1KB, pdf)
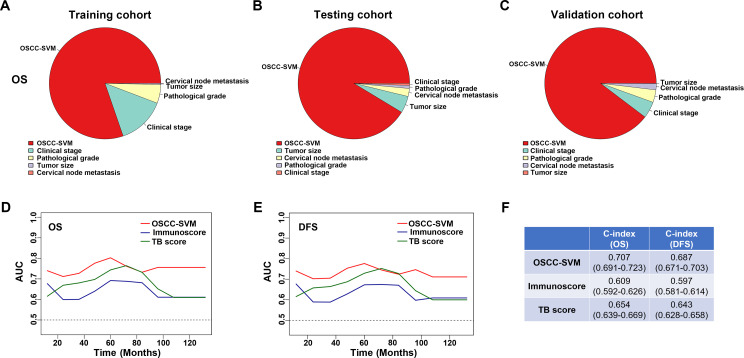
We sought to compare the predictive abilities of OSCC-SVM with other clinicopathological parameters including tumor size, cervical nodal metastasis, pathological grade and clinical stage by the area under the curves (AUC) for OS and DFS. When predicted OS and DFS for patients in three cohorts were estimated by 5-year receiver operating characteristics (ROC) curves, the AUCs for OSCC-SVM were 0.755, 0.741 in training cohort, better than other clinicopathological parameters (<0.7, figure 3G, H). Similar findings were observed in both testing and validation cohorts (figure 3I–L). Similar findings were also replicated when we plotted 3-year ROC curves in all cohorts (online supplemental figure S11). Moreover, to assess the relative importance of OSCC-SVM and these relevant clinicopathological parameters to survival risk, we performed the χ2 proportion test and found that this OSCC-SVM contributed the largest part of risk than other parameters (figure 4A–C and online supplemental figure S12). To compare the predictive performance of our OSCC-SVM with two well-established immune-related scores, we calculated the Immunoscore and T and B cell (TB) score in all patients. As shown in online supplemental figure S13, as expected, higher Immunoscore and TB score associated with improved OS and DFS (Kaplan-Meier curves, p<0.0001). Noticeably, time-independent ROC curves estimated that OSCC-SVM was superior to Immunoscore and TB score as evidenced by the highest sensitivity and specificity (figure 4D, E). Moreover, concordance index (C-index) in prognostic prediction also supported the better performance of OSCC-SVM (figure 4F). Collectively, these data established that OSCC-SVM represented as a novel effective immune-based biomarker for OSCC prognosis.
Figure 4.
Contributions of OSCC-SVM to risk of patient survival and its performance in prognostic prediction. (A–C) Relative importance of each parameter to OS risk using the χ2 proportion test for OSCC-SVM and other clinical parameters in training (A), testing (B) and validation cohorts (C). (D, E) The sensitivity and specificity of OSCC-SVM, Immunoscore and TB score in prognostic prediction were estimated by time-dependent ROC curves. (F) The concordance index (C-index) of OSCC-SVM, Immunoscore and TB score in prognostic prediction was shown. AUC, area under the curves; DFS, disease-free survival; OS, overall survival; OSCC, oral squamous cell carcinoma; ROC, receiver operating characteristics; SVM, support vector machine; TB, T and B cell.
jitc-2021-002434supp019.pdf (886.8KB, pdf)
jitc-2021-002434supp020.pdf (471.9KB, pdf)
jitc-2021-002434supp021.pdf (757.8KB, pdf)
In silicon estimates of TIIs in OSCC from TCGA data sets
Given the increasingly accessible RNA-seq data and the burgeoning concept of personalized oncology in the clinic, bioinformatics assessments of bulk RNA-seq have allowed for an unprecedented view about the compositions, locations and functions of diverse TIIs within cancer tumor microenvironment (TME).4 13 We first assessed the spectrum of immune cell infiltrations in OSCC via single sample Gene Set Enrichment Analysis (ssGSEA) approach which deconvolved the relative abundance of each cell type based on the expression of marker gene sets. In total, 328 OSCC samples from The Cancer Genome Atlas (TCGA)-OSCC data set with transcriptional profiling data and clinical characteristics available were included. As shown in figure 5A, significant heterogeneity in terms of TIIs infiltrations in OSCC were found. The optimal cut-off values for each TIIs subsets assessed by X-tile were utilized to stratify patients into the high-infiltration subgroup and low-infiltration subgroup. Then, we exploited Least absolute shrinkage and selection operator (LASSO) regression model to extract 14 features from these abovementioned 24 TIIs and construct a prognostic prediction model by calculating risk score (figure 5B). Subsequently, patients were dichotomized into low-risk or high-risk subgroups based on median of risk scores. Kaplan-Meier analyses revealed that this prognostic model could robustly stratify patients with distinct prognosis as evidenced by patients in the high-risk group had significantly reduced OS than those in the low-risk group (p<0.0001, figure 5C). To assess the predictive abilities of this prognostic model, the ROC analyses at different follow-up times were applied and revealed that our model had satisfactory sensitivity and specificity for OS (AUC at 3 years: 0.731; AUC at 5 years: 0.711, figure 5D). Similar findings were replicated in pooled data from two independent OSCC cohorts (GSE41613 and GSE42743, figure 5E, F).
Figure 5.
Prognostic models constructed based on TIIs estimated by ssGSEA. (A) The heatmap of 328 OSCC patients from TCGA-OSCC data set using ssGSEA scores from 24 immune cell types was shown. Tcm cells, central memory T cells; Tem cells, effector memory T cells; Tgd cells, gamma delta T cells; Th1 cells, T helper 1 cells; Th2 cells, T helper 2 cells; DC cells, dendritic cells; pDC, plasmacytoid dendritic cells; iDC, interstitial dendritic cells; aDC cells, activated dendritic cells; NK cells, natural killer cells; TFH cells, T follicular helper cells; TReg cells, regulatory T cells. (B) The coefficient profile plot of 24 immune cells was produced against the log lambda sequence. Tuning parameters (lambda) in the LASSO model were selected via tenfold cross-validation and minimum criteria. Dotted vertical lines were drawn at the optimal values using the minimum criteria and the 1 SE of the minimum criteria (the 1-SE criteria). (C) The Kaplan-Meier analyses revealed significant associations between 14-immune-features and OS in patients from the training cohort (TCGA-OSCC). (D) The time-dependent ROC curve analyses with 3, 5 years as the defining point were performed to evaluate the predictive values of the 14-immune-features based risk score in the training cohort (TCGA-OSCC). (E) The Kaplan-Meier analyses revealed significant associations between 14-immune-features based risk score and OS in patients from the validation cohort (GSE41613 and GSE42743). (F) The time-dependent ROC curve analyses with 3, 5 years as the defining point were performed to evaluate the predictive values of the 14-immune-features based risk score in the validation cohort (GSE41613 and GSE42743). (G, H) The Kaplan-Meier curves of OS for patients with high-risk score (G) and low-risk score (H) in predicting the benefits of postoperative radiotherapy in the TCGA-OSCC cohort. (I–N) The immunotherapeutic benefit of the 14 immune cell signature in IMvigor210 (I, J), GSE91061 (K, L) and GSE78220 (M, N) cohorts. AUC, area under the curves; CR, complete response; LASSO, least absolute shrinkage and selection operator; OS, overall survival; OSCC, oral squamous cell carcinoma; PD, progressive disease; PR, partial response; ROC, receiver operating characteristics; RT, radiotherapy; SD, stable disease; ssGSEA, single sample Gene Set Enrichment Analysis; TIIs, tumor-infiltrating immune cells; TCGA, The Cancer Genome Atlas.
To avoid the bias of single deconvolution algorithm and further strengthen our findings, we utilized CIBERSORT algorithm to calculate 22 types of TIIs in both tumor and normal samples using the TCGA-OSCC data set.20 As shown in online supplemental figure S14A, B, the fractions of TIIs varied significantly among individuals. As compared with their presence in normal counterparts, the proportions of memory B cells, resting natural killer (NK) cells, M0 macrophages, M1 macrophages, activated dendritic cells and eosinophils in OSCC were much higher, while the others were lower (p<0.05, (online supplemental figure S14C). When patterns of TIIs infiltration were compared among samples with diverse clinical stages, we found that reduced CD8+ T cells, activated NK cells and increased M2 macrophages were frequently observed in samples with advanced stages relative to those in early stages, although densities of some TIIs subsets remained unchanged or inconsistently varied among each stage (online supplemental figure S14D).
jitc-2021-002434supp022.pdf (795KB, pdf)
Next, we employed LASSO regression analyses and selected 13 features from 22 TIIs to build another independent prognostic model (online supplemental figure S15A, B). Similar with our ssGSEA-derived model, patients in the high-risk subgroup had significantly shorter OS than those in the low-risk subgroup (p<0.0001, online supplemental figure S15C). Performance of this model in prognostic prediction was satisfactory (AUC at 3 years: 0.698; AUC at 5 years: 0.707, online supplemental figure S15D). These findings were further validated in pooled samples from GSE41613 and GSE42743 data sets (online supplemental figure S15E, F).
jitc-2021-002434supp023.pdf (645.4KB, pdf)
Clinical values of TIIs-based risk score in predicting response to therapy
To further extend the clinical significance of our TIIs-based risk score beyond prognostic prediction, we utilized OSCC data set from TCGA and revealed that patients from high-risk subgroup had survival benefits from postsurgical radiotherapy as evidenced by patients without radiotherapy had significantly reduced OS than those with radiotherapy (p=0.0035, figure 5G), although no survival benefits from radiotherapy were obtained in patients from low-risk subgroup (figure 5H). Given that currently no data sets about OSCC receiving immunotherapy were publicly available, as a complement, we selected the data sets from melanoma and urothelial cancer cohorts (IMvigor210, GSE91061 and GSE78220) to explore the values of our TIIs-base risk score to predict benefits from ICB immunotherapy. We calculated the TIIs-derived risk scores for each patient from these cohorts via ssGSEA and found that these scores robustly separated patients into subgroups with high or low OS (p=0.016; 0.0079; 0.036, figure 5I, K and M). As expected, these scores were significantly increased in patients with response to immunotherapy as compared with those with no response to immunotherapy (figure 5J, L and N). Importantly, the performance of our TIIs-based risk score was superior to selected predictive biomarkers for cancer immunotherapy like PD-L1, PD-1 expression and cytolytic activity (online supplemental figure S16), indicative of the robustness and translational potentials of our model.
jitc-2021-002434supp024.pdf (157KB, pdf)
Consensus clustering for patients based on immune landscape in OSCC
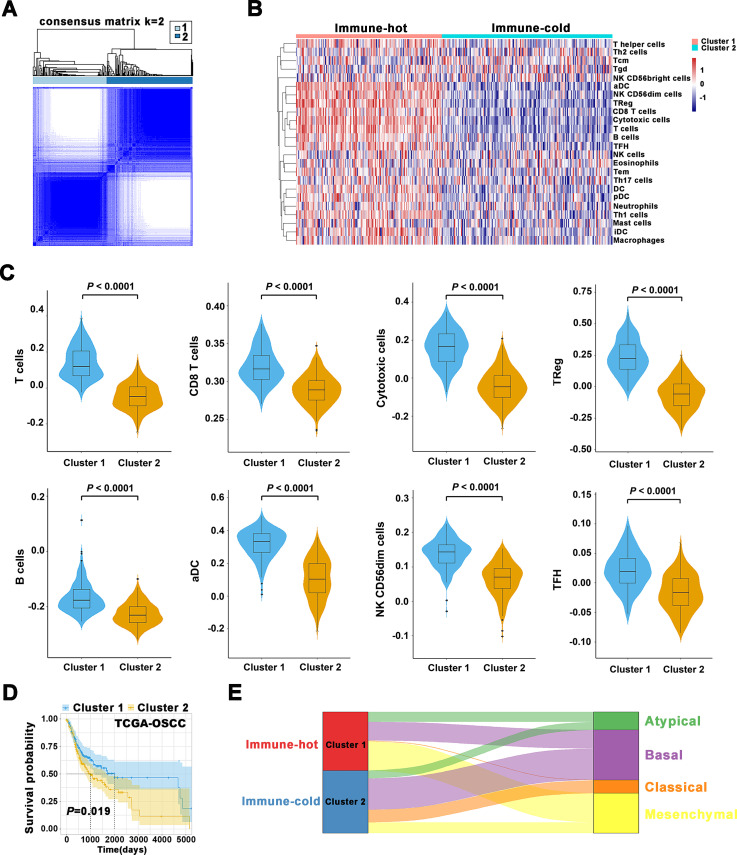
Based on 24 TIIs estimated by ssGSEA algorithm in OSCC samples from TCGA data set, the k=2 was identified with optimal clustering stability by consensus clustering (figure 6A, B). A total of 328 patients with OSCC were clustered into two subgroups, namely, cluster 1 (n=151) and cluster 2 (n=177). As shown in figure 6B, C, most of these TIIs such as B cells, CD8 T cells, cytotoxic cells, TReg, NK were significantly enriched in the cluster 1 over cluster 2. Importantly, patients in cluster 1 had improved OS relative to those in cluster 2 (Kaplan-Meier curves, p=0.019, figure 6D). Thus, we refined the cluster 1 as immune-hot subtype and cluster 2 as immune-cold subtype. Importantly, immune clustering of OSCC into immune-hot and immune-cold subtypes were reproducibly observed using data sets from GSE41613 and GSE42743 (online supplemental figure S17). In addition, such immune subtypes were also fitted in another independent head neck squamous cell carcinoma (HNSCC) data set (GSE65858). Although the statistical analyses failed to achieve significance (p=0.15, 0.12 for OS, progression-free survival), this strategy tended to stratify patients into subgroups with high or low survival (online supplemental figure S18). Together, these results supported the robustness and reproducibility of this immune subtyping in OSCC.
Figure 6.
Immune subtypes in OSCC and their associations with molecular features and patient survival. (A) Consensus clustering matrix for k=2. (B) The heatmap showed detailed distributions of 24 types of immune cells in patients from TCGA-OSCC cohort and separated samples into two clusters (immune-hot, immune-cold). Tcm cells, central memory T cells; Tem cells, effector memory T cells; Tgd cells, gamma delta T cells; Th1 cells, T helper 1 cells; Th2 cells, T helper 2 cells; DC cells, dendritic cells; pDC, plasmacytoid dendritic cells; iDC, interstitial dendritic cells; aDC cells, activated dendritic cells; NK cells, natural killer cells; TFH cells, T follicular helper cells; TReg cells, regulatory T cells. (C) The abundance of T cells, CD8 T cells, cytotoxic cells, TReg, B cells, aDC, NK CD56dim cells, TFH in two clusters was compared. (D) The Kaplan-Meier analyses revealed significant associations between immune subtypes and OS in patients from TCGA-OSCC cohort. (E) Alluvial diagram revealed associations between two immune subtypes and four molecular subtypes in TCGA-OSCC patients. OS, overall survival; OSCC, oral squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
jitc-2021-002434supp025.pdf (257.5KB, pdf)
jitc-2021-002434supp026.pdf (270.1KB, pdf)
To explore potential correlations between our immune clusters and molecular subtypes in OSCC, we adopted previously reported four molecular types of HNSCC21 and found 17.1% of atypical, 31.8% of basal, 1.1% of classical and 50% of mesenchymal subtype in cluster 1 (immune-hot) in contrast to 12.8% of atypical, 50% of basal, 20.2% of classical and 17% of mesenchymal subtype in cluster 2 (immune-cold, figure 6E). In addition, we adopted the six immune subtypes of pan-SCC22 or pan-cancer4 and found no obvious enrichments and remarkable difference of distribution between two clusters (online supplemental figure S19). To gain more molecular insights underlying immune clustering, we performed Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses of the differentially expressed genes (DEGs) between samples in both clusters. As expected, these DEGs were significantly enriched in functional categories involved in T-cell receptor signaling pathway and chemical carcinogenesis (online supplemental figure S20A, C), as well as T-cell activation and response to xenobiotic stimulus (online supplemental figure S20B, D). Moreover, the results from GSEA further revealed that IL6-JAK-STAT3 signaling pathway and adaptive immune response were significantly enriched (online supplemental figure S21). In addition, protein-protein interaction network for these DEGs by weighted correlation network analysis revealed that several modules enriched in PD-L1/PD-1 checkpoint pathway in cancer and T-cell receptor signaling pathway, etc (online supplemental figure S22).
jitc-2021-002434supp027.pdf (27.8KB, pdf)
jitc-2021-002434supp028.pdf (819KB, pdf)
jitc-2021-002434supp029.pdf (670.7KB, pdf)
jitc-2021-002434supp030.pdf (246.3KB, pdf)
Next, we compared genomic variations, mutation burden, multiple immune-related signatures/biomarkers and expression of selected immune modulatory genes which had been established with clinical significance across human cancers. As displayed in online supplemental figure S23, the spectrum of gene mutations identified in both clusters was generally similar, but the incidence of P53 mutations in immune-cold cluster prevailed over immune-hot cluster. Of note, copy number variations of several immune-related genes such as CD274, CD72 and PDCD1LG2 in immune-cold cluster shown amplifications and overexpression as compared with immune-hot cluster (online supplemental figure S24). We also calculated multiple immune-related signatures/biomarkers including tumor mutation burden (TMB), cytolytic score, T-cell receptor diversity, T-cell inflamed gene expression profile signature, tumor inflammation signature, chemokine gene expression signature, IFN-γ signature as well as tertiary lymphoid structures signature23–25 and revealed that the majority of these markers or signatures were significantly enriched in cluster 1 over cluster 2, except the TMB (p<0.0001, online supplemental figure S25). Additionally, transcriptional profiling of immune-modulatory molecules revealed that most genes were overrepresented in cluster 1 (online supplemental figure S26).
jitc-2021-002434supp031.pdf (728.8KB, pdf)
jitc-2021-002434supp032.pdf (1.1MB, pdf)
jitc-2021-002434supp033.pdf (518KB, pdf)
jitc-2021-002434supp034.pdf (951.4KB, pdf)
Discussion
Cancer immunotherapy has shown unprecedented success but yielded substantial benefits only in a small proportion of patients and selected cancer types, highlighting the need to characterize immune landscape of individual cancer, identify potential responders and design rational therapeutic strategies.1 5 26 Herein, we comprehensively delineated immune landscape and subtypes of OSCC and developed robust TIIs-derived prognostic models by machine learning approaches to predict patient survival and response of therapy.
Immune landscape in OSCC and prognostic significance of individual TIIs subtypes
Mounting evidence has established that the densities, locations and functional status of TIIs have remarkable impacts on patient survival and therapeutic response.3 6 7 27 In line with these findings, our data from both IHC staining/quantification and bioinformatics estimate revealed diverse patterns of TIIs infiltration and varied amounts of individual TIIs fractions among OSCC samples, likely reflecting the heterogeneities of both cancers and hosts. Moreover, spatial enrichment of TIIs subsets at the IM compartments indicated their recruitments from local environment or systemic circulation in hosts, and also suggested compartment-specific functions of specific TIIs subset.3 12 28 Noticeably, the numbers of TIIs subsets with putative antitumor immunity such as CD8+ T cells tended to diminish, whereas those with protumorigenic functions such as MDSCs displayed enrichments in diseases with advanced stages when compared with early-staged diseases. These results reinforced the critical involvements of TIIs during OSCC progression and strongly supported further longitudinal and functional dissections of individual TIIs subsets underlying oral tumorigenesis.
With regard to prognostic significance of individual TIIs, intratumoral infiltrations of TIIs labeled by CD8 or CD45RO were significantly associated with favorable prognosis, whereas those subsets stained by CD11b correlated with inferior prognosis among cancers.7 9 27 29 Consistent with these, our data from IHC/computational enumeration revealed similar trends for individual TIIs subsets in OSCC. However, the prognostic impacts of neutrophils and regulatory T cells seemed contradictory to some previous thoughts regarding their expected roles in cancer immunity.30 31 We reasoned that these discrepancies might be due to diverse genetic backgrounds, etiologies and cellular origins of cancer, diverse subtypes or phenotypes among these TIIs subsets as well as varied quantification approaches.3 6 32 Indeed, these phenomena regarding heterogeneous prognostic impacts from these TIIs fractions were frequently observed in multiple cancers including gastric, colorectal and oral cancers.10 32–34
Integrative TIIs-derived prognostic models constructed by machine learning for OSCC
Prognostic models integrating multiple parameters usually outperform those from single parameter.5 12 Multiple prognostic scores or signatures based on TIIs and immune-associated gene signatures have been established for patient prognostication and prediction of therapeutic response.11 13 23 34 35 Consistent with this notion, we applied SVM and LASSO approaches to extract essential immune features using IHC data from our patient cohorts or bulk transcriptomic data from publicly available cohorts, and constructed robust prognostic models. These integrative prognostic models efficiently stratified patients into subgroups with favorable or dismal survival with satisfactory sensibility and specificity. Importantly, these models were superior to some clinicopathological parameters such as TNM staging as well as two novel immune-related scores (Immunoscore and TB score).
Moreover, besides remarkable prognostic significance, we found that these risk scores from our prognostic models can identify patients who had survival benefits from postoperative radiotherapy in patient subgroup with high risk scores. Moreover, our results revealed that patients with high risk scores had significant lower survival as compared with those with low risk scores in three independent patient cohorts including urothelial cancer and melanoma receiving ICB immunotherapies. Although our risk scores derived from TIIs worked well in predicting response of ICB in a few types of cancer, much work is needed to determine their utilities and performances in predicting ICB response in patients with OSCC. Collectively, our models derived from TIIs as novel and robust biomarkers hold translational potentials, which deserve further prospective validations.
Immune subtypes and their prognostic significance and associated genetic and molecular features in OSCC
From the perspective of immune landscape, two main clusters/subtypes harboring diverse characteristics of TIIs infiltrations were identified from our cohorts. Although subtle differences existed in detailed distributions of TIIs among two subtypes identified from two different analytic pipelines (experimental or bioinformatics), significant enrichments of antitumor TIIs subsets (CD8+ T cell, CD20+ B cells, etc.) were commonly found in immune-hot subtype as compared with immune-cold subtype. As expected, these patient stratifications were well consistent with differential survival. Further explorations of genetic and molecular features found that the frequency of TP53 mutation, the most common gene mutated in HNSCC, was found higher in immune-cold cluster with inhibitory cancer immunity, which was supported by the findings regarding the loss of antitumor immunity following p53 mutation.36 37 Transcriptional profiles of key genes involved in cancer immunity further revealed overrepresentations of molecules associated with cytolytic activity, antigen-presentation and immune stimulation in immune-hot cluster, indicative of strong immunity to avoid tumor escape. However, we found that the abundance of some immune checkpoint molecules like PD-L1, PD-1 and LAG3 appeared to be upregulated in immune-hot samples with better prognosis, which seemed contrast to their expected suppressive roles in cancer immunity and tumorigenesis. Similar with our results, several lines of evidence have shown that expression of these immune checkpoints usually positively correlated with densities with CD4+, CD8+ TIIs or total tumor-infiltrating lymphocytes as well as favorable prognosis across multiple tumors including HNSCC.38–42 Indeed, the prognostic impacts of these immune checkpoints in HNSCC and OSCC remained inconsistent and sometimes contradictory.43 44 We reasoned that this discrepancy might associate with several factors including methodological difference, diverse etiology and genetic background of tumor as well as treatments. Moreover, these immune checkpoints, although originally defined as markers for immune suppression and exhaustion, might be also viewed as surrogate markers and witness of T-cell receptor activation and endogenous antitumor immune response, reflecting enrichment of activated tumor-associated antigen-specific T cells.38 39 45 46 This idea was supported by the facts that their abundance positively correlated with TIIs densities, favorable prognosis and markers of T-cell activation in multiple cancers.38 45 46 Finally, the abundance of these molecules was calculated from bulk-sample transcriptomic data rather than single TIIs fractions, which complicated accurate explaining of their significance. Together, these findings further substantiate high complexity and heterogeneity involved in tumor-stroma crosstalk and intricate interactions between immune-regulatory molecules and individual TII subsets in TME.
With regard to molecular subtypes corresponding to these two immune subtypes, significant proportions of patients in both clusters were classified with mesenchymal and basal, respectively. This was well consistent with previous findings that most OSCCs were categorized as basal and mesenchymal subtypes and the basal subtype was characterized by epidermal growth factor receptor (EGFR) activation, more aggressiveness and poorer outcome while the mesenchymal subtype was shown enrichment in immune markers, activated immune phenotype and likely to respond to immunotherapy.47 However, when we linked two immune subtypes of OSCC to two previous reported immune subtypes for cancer, no significant enrichments were detected, thus suggesting a cancer-type-specific pattern of immune infiltration for OSCC.4 22
Limitations of study
The main advantages of our study lie in complementary and integrative analytical pipelines to characterize TIIs infiltrations in OSCC from multiple independent cohorts. These two independent approaches achieved similar results, thus highlighting the reliability and robustness of our findings. Given the advantages and weakness of these two approaches, their applications might depend on research purpose and experimental design.8 10 12 However, our study has certain limitations including its retrospective nature and potential biases for patient enrollment. Accurate dissections of specific TIIs subsets especially their functional diversities and detailed characterization of spatiotemporal dynamics of immune landscape during OSCC progression and therapy will offer key insights into OSCC development.8 48 In addition, genomic and molecular networks dictating TIIs functions during OSCC initiation and progression still deserve further explorations.19 49 Moreover, various etiological factors including tobacco abuse and human papillomavirus (HPV) status might be better included to explore their contributions to dysregulated tumor immunity in OSCC. Although the prevalence of HPV infection in OSCC is quite low, its contributions to altered immune infiltration and dysfunctional immunity should not be overlooked.50
Conclusions
In summary, we characterized immune landscape in OSCC by conventional pathological approaches and bioinformatics analytic pipelines, developed robust prognostic models derived from TIIs by machine learning to stratify patients into subgroups with different survival as well as benefits from post-radiotherapy and immunotherapy. We proposed two immune clusters harboring characteristic patterns of TIIs infiltration, genomic and molecular features, and significant prognostic impacts in OSCC. These findings offered insights and clues in novel OSCC classification regimes and facilitated identifications of biomarkers and targets for immunotherapeutic strategies, which warrants further perspective investigations in larger cohorts receiving immunotherapies.
jitc-2021-002434supp035.pdf (13.7KB, pdf)
Footnotes
PD, YJ and YL contributed equally.
Contributors: Experimental analysis, data collection and analysis and manuscript writing: DP, JY, LY. Data collection and statistical analyses: WX, LJ, ZC, JL, ZW, YE, ZP, DX, WH, YH, YJ, SX, WL, WYu, JH, WYa. Study concept and supervision of the whole project: CJ. Read and approved the final version of the manuscript: all authors.
Funding: This work is financially supported, in whole or in part, by National Natural Science Foundation of China (82072991), Key Research Program in Jiangsu Province-Social Developmental Project (BE2020706), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (2018-87), Qing-Lan Project and Jiangsu Young Medical Talent Project (QNRC2016851).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Author note: PD, YJ and YL contributed equally.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. The data sets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The whole study was reviewed and approved by the Ethic Committee of Affiliated School of Stomatology, Nanjing Medical University, and performed in accordance with the ethical standards of the 1964 Declaration of Helsinki.
References
- 1.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541–50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galon J, Bruni D. Tumor immunology and tumor evolution: intertwined histories. Immunity 2020;52:55–81. 10.1016/j.immuni.2019.12.018 [DOI] [PubMed] [Google Scholar]
- 3.Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 4.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2019;51:411–2. 10.1016/j.immuni.2019.08.004 [DOI] [PubMed] [Google Scholar]
- 5.Bruni D, Angell HK, Galon J. The immune contexture and immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer 2020;20:662–80. 10.1038/s41568-020-0285-7 [DOI] [PubMed] [Google Scholar]
- 6.Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938–45. 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahmoud SMA, Paish EC, Powe DG, et al. Tumor-Infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 2011;29:1949–55. 10.1200/JCO.2010.30.5037 [DOI] [PubMed] [Google Scholar]
- 8.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 9.Pagès F, Kirilovsky A, Mlecnik B, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol 2009;27:5944–51. 10.1200/JCO.2008.19.6147 [DOI] [PubMed] [Google Scholar]
- 10.Challoner BR, von Loga K, Woolston A, et al. Computational image analysis of T-cell infiltrates in resectable gastric cancer: association with survival and molecular subtypes. J Natl Cancer Inst 2021;113:88–98. 10.1093/jnci/djaa051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Y, Zhang Q, Hu Y, et al. Immunoscore signature: a prognostic and predictive tool in gastric cancer. Ann Surg 2018;267:504–13. 10.1097/SLA.0000000000002116 [DOI] [PubMed] [Google Scholar]
- 12.Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018;391:2128–39. 10.1016/S0140-6736(18)30789-X [DOI] [PubMed] [Google Scholar]
- 13.Chen Y-P, Wang Y-Q, Lv J-W, et al. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann Oncol 2019;30:68–75. 10.1093/annonc/mdy470 [DOI] [PubMed] [Google Scholar]
- 14.Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma--an update. CA Cancer J Clin 2015;65:401–21. 10.3322/caac.21293 [DOI] [PubMed] [Google Scholar]
- 15.Lydiatt WM, Patel SG, O'Sullivan B, et al. Head and neck cancers-major changes in the American joint Committee on cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67:122–37. 10.3322/caac.21389 [DOI] [PubMed] [Google Scholar]
- 16.Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol 2016;17:956–65. 10.1016/S1470-2045(16)30066-3 [DOI] [PubMed] [Google Scholar]
- 17.Wolf GT, Chepeha DB, Bellile E, et al. Tumor infiltrating lymphocytes (TIL) and prognosis in oral cavity squamous carcinoma: a preliminary study. Oral Oncol 2015;51:90–5. 10.1016/j.oraloncology.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boxberg M, Leising L, Steiger K, et al. Composition and clinical impact of the immunologic tumor microenvironment in oral squamous cell carcinoma. J Immunol 2019;202:278–91. 10.4049/jimmunol.1800242 [DOI] [PubMed] [Google Scholar]
- 19.Feng Z, Bethmann D, Kappler M, et al. Multiparametric immune profiling in HPV- oral squamous cell cancer. JCI Insight 2017;2:e93652. 10.1172/jci.insight.93652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Network . Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Cui Y, Nambiar DK, et al. The immune subtypes and landscape of squamous cell carcinoma. Clin Cancer Res 2019;25:clincanres.4085.2018–3537. 10.1158/1078-0432.CCR-18-4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danaher P, Warren S, Lu R, et al. Pan-Cancer adaptive immune resistance as defined by the tumor inflammation signature (TIS): results from the cancer genome atlas (TCGA). J Immunother Cancer 2018;6:63. 10.1186/s40425-018-0367-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabrita R, Lauss M, Sanna A, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020;577:561–5. 10.1038/s41586-019-1914-8 [DOI] [PubMed] [Google Scholar]
- 25.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity 2020;52:17–35. 10.1016/j.immuni.2019.12.011 [DOI] [PubMed] [Google Scholar]
- 27.Hu Y, He M-Y, Zhu L-F, et al. Tumor-Associated macrophages correlate with the clinicopathological features and poor outcomes via inducing epithelial to mesenchymal transition in oral squamous cell carcinoma. J Exp Clin Cancer Res 2016;35:12. 10.1186/s13046-015-0281-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peled M, Onn A, Herbst RS. Tumor-Infiltrating Lymphocytes-Location for prognostic evaluation. Clin Cancer Res 2019;25:1449–51. 10.1158/1078-0432.CCR-18-3803 [DOI] [PubMed] [Google Scholar]
- 29.Gabrilovich DI. Myeloid-Derived suppressor cells. Cancer Immunol Res 2017;5:3–8. 10.1158/2326-6066.CIR-16-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.deLeeuw RJ, Kost SE, Kakal JA, et al. The prognostic value of Foxp3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res 2012;18:3022–9. 10.1158/1078-0432.CCR-11-3216 [DOI] [PubMed] [Google Scholar]
- 31.Donskov F. Immunomonitoring and prognostic relevance of neutrophils in clinical trials. Semin Cancer Biol 2013;23:200–7. 10.1016/j.semcancer.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 32.Fridlender ZG, Sun J, Kim S, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell 2009;16:183–94. 10.1016/j.ccr.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Liu H, Shen Z, et al. Tumor-Infiltrating neutrophils is prognostic and predictive for postoperative adjuvant chemotherapy benefit in patients with gastric cancer. Ann Surg 2018;267:311–8. 10.1097/SLA.0000000000002058 [DOI] [PubMed] [Google Scholar]
- 34.Zhou C, Diao P, Wu Y, et al. Development and validation of a seven-immune-feature-based prognostic score for oral squamous cell carcinoma after curative resection. Int J Cancer 2020;146:1152–63. 10.1002/ijc.32571 [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Li Z-Y, Zhou G-Q, et al. An immune-related gene prognostic index for head and neck squamous cell carcinoma. Clin Cancer Res 2021;27:330–41. 10.1158/1078-0432.CCR-20-2166 [DOI] [PubMed] [Google Scholar]
- 36.Jiang Z, Liu Z, Li M, et al. Immunogenomics analysis reveals that TP53 mutations inhibit tumor immunity in gastric cancer. Transl Oncol 2018;11:1171–87. 10.1016/j.tranon.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muñoz-Fontela C, Mandinova A, Aaronson SA, et al. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat Rev Immunol 2016;16:741–50. 10.1038/nri.2016.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badoual C, Hans S, Merillon N, et al. PD-1-expressing tumor-infiltrating T cells are a favorable prognostic biomarker in HPV-associated head and neck cancer. Cancer Res 2013;73:128–38. 10.1158/0008-5472.CAN-12-2606 [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Canteli M, Granda-Díaz R, Del Rio-Ibisate N, et al. PD-L1 expression correlates with tumor-infiltrating lymphocytes and better prognosis in patients with HPV-negative head and neck squamous cell carcinomas. Cancer Immunol Immunother 2020;69:2089–100. 10.1007/s00262-020-02604-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carreras J, Lopez-Guillermo A, Roncador G, et al. High numbers of tumor-infiltrating programmed cell death 1-positive regulatory lymphocytes are associated with improved overall survival in follicular lymphoma. J Clin Oncol 2009;27:1470–6. 10.1200/JCO.2008.18.0513 [DOI] [PubMed] [Google Scholar]
- 41.Schneider S, Kadletz L, Wiebringhaus R, et al. PD-1 and PD-L1 expression in HNSCC primary cancer and related lymph node metastasis - impact on clinical outcome. Histopathology 2018;73:573–84. 10.1111/his.13646 [DOI] [PubMed] [Google Scholar]
- 42.Eckstein M, Strissel P, Strick R, et al. Cytotoxic T-cell-related gene expression signature predicts improved survival in muscle-invasive urothelial bladder cancer patients after radical cystectomy and adjuvant chemotherapy. J Immunother Cancer 2020;8:e000162. 10.1136/jitc-2019-000162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghanizada M, Jakobsen KK, Grønhøj C, et al. The effects of checkpoint inhibition on head and neck squamous cell carcinoma: a systematic review. Oral Oncol 2019;90:67–73. 10.1016/j.oraloncology.2019.01.018 [DOI] [PubMed] [Google Scholar]
- 44.Lenouvel D, González-Moles Miguel Ángel, Ruiz-Ávila I, et al. Prognostic and clinicopathological significance of PD-L1 overexpression in oral squamous cell carcinoma: a systematic review and comprehensive meta-analysis. Oral Oncol 2020;106:104722. 10.1016/j.oraloncology.2020.104722 [DOI] [PubMed] [Google Scholar]
- 45.Gros A, Robbins PF, Yao X, et al. PD-1 identifies the patient-specific CD8⁺ tumor-reactive repertoire infiltrating human tumors. J Clin Invest 2014;124:2246–59. 10.1172/JCI73639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganesan A-P, Clarke J, Wood O, et al. Tissue-Resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol 2017;18:940–50. 10.1038/ni.3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chai AWY, Lim KP, Cheong SC. Translational genomics and recent advances in oral squamous cell carcinoma. Semin Cancer Biol 2020;61:71–83. 10.1016/j.semcancer.2019.09.011 [DOI] [PubMed] [Google Scholar]
- 48.Woolston A, Khan K, Spain G, et al. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during anti-EGFR treatment in colorectal cancer. Cancer Cell 2019;36:35–50. 10.1016/j.ccell.2019.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desrichard A, Kuo F, Chowell D, et al. Tobacco smoking-associated alterations in the immune microenvironment of squamous cell carcinomas. J Natl Cancer Inst 2018;110:1386–92. 10.1093/jnci/djy060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nauta IH, Heideman DAM, Brink A, et al. The unveiled reality of human papillomavirus as risk factor for oral cavity squamous cell carcinoma. Int J Cancer 2021;149:420–30. 10.1002/ijc.33514 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-002434supp001.pdf (210.5KB, pdf)
jitc-2021-002434supp002.pdf (25KB, pdf)
jitc-2021-002434supp003.pdf (213.8KB, pdf)
jitc-2021-002434supp004.pdf (196.2KB, pdf)
jitc-2021-002434supp005.pdf (31.4KB, pdf)
jitc-2021-002434supp006.pdf (919KB, pdf)
jitc-2021-002434supp007.pdf (513.6KB, pdf)
jitc-2021-002434supp008.pdf (560KB, pdf)
jitc-2021-002434supp009.pdf (787KB, pdf)
jitc-2021-002434supp010.pdf (704.9KB, pdf)
jitc-2021-002434supp011.pdf (816.5KB, pdf)
jitc-2021-002434supp012.pdf (706.2KB, pdf)
jitc-2021-002434supp013.pdf (744.1KB, pdf)
jitc-2021-002434supp014.pdf (29.5KB, pdf)
jitc-2021-002434supp015.pdf (28.8KB, pdf)
jitc-2021-002434supp016.pdf (15.2KB, pdf)
jitc-2021-002434supp017.pdf (15.2KB, pdf)
jitc-2021-002434supp018.pdf (15.1KB, pdf)
jitc-2021-002434supp019.pdf (886.8KB, pdf)
jitc-2021-002434supp020.pdf (471.9KB, pdf)
jitc-2021-002434supp021.pdf (757.8KB, pdf)
jitc-2021-002434supp022.pdf (795KB, pdf)
jitc-2021-002434supp023.pdf (645.4KB, pdf)
jitc-2021-002434supp024.pdf (157KB, pdf)
jitc-2021-002434supp025.pdf (257.5KB, pdf)
jitc-2021-002434supp026.pdf (270.1KB, pdf)
jitc-2021-002434supp027.pdf (27.8KB, pdf)
jitc-2021-002434supp028.pdf (819KB, pdf)
jitc-2021-002434supp029.pdf (670.7KB, pdf)
jitc-2021-002434supp030.pdf (246.3KB, pdf)
jitc-2021-002434supp031.pdf (728.8KB, pdf)
jitc-2021-002434supp032.pdf (1.1MB, pdf)
jitc-2021-002434supp033.pdf (518KB, pdf)
jitc-2021-002434supp034.pdf (951.4KB, pdf)
jitc-2021-002434supp035.pdf (13.7KB, pdf)
Data Availability Statement
Data are available upon reasonable request. The data sets used and/or analysed during the current study are available from the corresponding author on reasonable request.