Abstract
Specific mutations in the RET proto-oncogene are associated with multiple endocrine neoplasia type 2A, a hereditary syndrome characterized by tumorigenesis in multiple glandular elements. In rare instances, MEN2A-associated germline RET mutations have also occurred with non-MEN2A associated cancers. One such germline mutant RET mutation occurred concomitantly in a young adult diagnosed with alveolar rhabdomyosarcoma, a pediatric and young adult soft-tissue cancer with a generally poor prognosis. Although tumor tissue samples were initially unable to provide a viable cell culture for study, tumor tissues were sequenced for molecular characteristics. Through a hierarchical clustering approach, the index case sample was matched to several genetically similar cell models, which were transformed to express the same mutant RET as the index case and used to explore potential therapeutic options for mutant RET-bearing alveolar rhabdomyosarcoma. We also determined whether the RET mutation associated with the index case caused synthetic lethality to select clinical agents. From our investigation, we did not identify synthetic lethality associated with the expression of that patient's RET variant, and overall we did not find experimental evidence for the role of RET in rhabdomyosarcoma progression.
Keywords: alveolar rhabdomyosarcoma, rhabdomyosarcoma
INTRODUCTION
Rhabdomyosarcoma (RMS) is the most prevalent pediatric soft tissue sarcoma, with frequent incidence in adolescent and young adults (Rudzinski et al. 2017). The alveolar subtype (aRMS) accounts for 20%–30% of all RMS tumors and thus represents ∼1% of all pediatric cancers. aRMS has an annual incidence within the United States of ∼350 cases/yr (Amer et al. 2019). aRMS is commonly initiated by a gene fusion event, most prevalently the PAX3:FOXO1-positive (P3F) variant, less commonly the PAX7:FOXO1-positive (P7F) variant, and rarely the atypical fusions (PAX3:NCOA1, PAX3:INO8) or even fusion-negative (Parham and Barr 2013; Shern et al. 2014). A substantial fraction of patients who develop metastatic aRMS (25%–30%) have metastatic disease at the time of diagnosis, with aRMS most frequently spreading to bone marrow, distal nodes, and bone (Ognjanovic et al. 2009; Amer et al. 2019).
Standard clinical care for aRMS is a combination of surgery, radiation, and intensive chemotherapy (Hawkins et al. 2013). Therapeutic schedules have been developed for RMS patients based on their Oberlin risk group, with the goal of reducing therapy-related toxicity in patients in low risk groups. The Oberlin risk score is determined by several factors but is strongly influenced by the number of metastatic lesions and the location of the metastatic disease. RMS patients with fusion-positive tumors are more likely to have a higher risk score than fusion-negative patients; however, fusion status has little additional prognostic value when patients are stratified by Oberlin score. The 5-yr event-free survival rates based on Oberlin score are Oberlin 0 or 1, 52.4%; Oberlin 2, 22.8%; Oberlin 3, 6.3%; and Oberlin 4, 0% (Rudzinski et al. 2017).
The research presented here complements our case report of a teenage male patient (denoted CF-00034) with a germline RET C634F mutation diagnosed with aRMS bearing the canonical PAX3:FOXO1 genomic rearrangement (Crawford et al. 2020). RET mutations occur in 1.8% of tumors across disparate disease types (Kato et al. 2017), and the PAX3:FOXO1 fusion occurs in 55% of ARMS cases (Marshall and Grosveld 2012). The RET C634F mutation is associated with Multiple Endocrine (MEN) Type 2 and has well established oncogenic potential (Schuffenecker et al. 1998; Zhou et al. 2007; Masbi et al. 2014). Prior reports demonstrate metastatic PAX3:FOXO1 aRMS in a 13-yr-old male (Agarwal et al. 1997; Jones et al. 2010) with familial MEN Type 2A induced by RET V804M mutation, a patient genotypically and phenotypically reminiscent of the index patient. MEN syndromes can also be caused by loss-of-function mutations in MEN1 or CDKN1B, such as a case of rhabdomyosarcoma with a MEN1 1280delG truncation mutation (Alpers and Jones 2010). Nonetheless, rhabdomyosarcoma bearing MEN syndromes is a rare occurrence with few published clinical cases.
RET is a transmembrane coreceptor tyrosine kinase involved in neurite guidance and outgrowth (Hezam et al. 2018). In neurons, RET stimulates a multitude of biological functions including cell survival, differentiation, proliferation, and migration/chemotaxis. In cancer cells RET is associated cell migration and proliferation, autophagy, and chemoresistance (Mulligan 2019). RET mutations occur regularly in other cancer types such as medullary thyroid cancer (43%–71% of patients) and lung carcinosarcoma (16.7%) (Kato et al. 2017), which has led to clinical trials of highly selective RET inhibitors with promising results (Ackermann et al. 2019). RET signaling requires the stepwise assembly of a signaling complex that first requires binding of a dimerized ligand of the glial cell line–derived neurotrophic factor (GDNF) family (GDNF, neurturin [NTRN], artemin [ARTN], persephin [PSPN]) by two GDNF family receptors (GFRα1–4) (Takahashi 2001), which forms a tetrameric complex that binds and induces dimerization of RET (Supplemental Fig. 1A). The active signaling complex in a 2:2:2 ratio results in trans-autophosphorylation of RET that can lead to activation of several signaling pathways including MAPK, PI3K/AKT, and Src. However, mutant RET does not have the same signaling complex requirements. Mutations in the kinase domain or RET rearrangements render the kinase constitutively active (Cranston et al. 2006), whereas mutations in the RET extracellular domain, as in the case of the index patient (C634F), render the receptor ligand-independent (Kjær et al. 2006).
Given the association of mutant RET to cancer oncogenesis and metastasis, and previous reports of mutant RET in aRMS patients (Agarwal et al. 1997; Jones et al. 2010; Crawford et al. 2020), we investigated if the germline RET mutation coincident in a subset of aRMS patients may have contributed to the clinical course of CF-00034, and if the RET C634F mutation subsequently induces atypical susceptibilities in RMS tumor cells for patients with elevated RET expression or germline RET mutations.
RESULTS
RET Signaling Components in aRMS
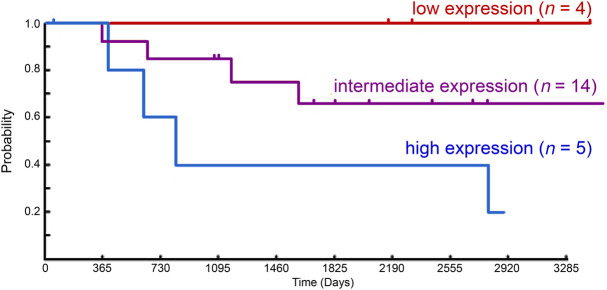
To determine the importance of RET activity in aRMS, we evaluated the survival data and paired gene expression data from sporadic cases of RMS (n = 23 cases) archived by the Children's Oncology Group (Fig. 1). RET expression demonstrated a trend associated with decreased long term survival over 10 years in an expression-dependent manner when adjusted for known clinical covariates (age, stage, sex, fusion status), although cohort sizes were small (four to 14 patients/group) and results did not reach statistical significance (P = 0.069). We next evaluated whole-transcriptome sequencing data from human aRMS biopsy samples to determine the expression of the RET signaling complex members (Supplemental Fig. 1). Although RET and GFRα were expressed in several samples, no single sample expressed both coreceptors RET and GFRα simultaneously. The evaluation of this diverse collection of RMS tumors indicates that RET is rarely expressed (∼5%), whereas coreceptors GFRA1–4 are more commonly expressed (∼34%), and all samples transcribe at least one RET ligand, with ATRN and NRTN being near-universally expressed.
Figure 1.
RET expression versus aRMS outcome. For the Intergroup Rhabdomyosarcoma Study Group 4 clinical trial (Blandford et al. 2006), increased RET gene expression in new-diagnosis biopsy samples is associated with the trend for reduced patient survival over a 10-yr period (P = 0.069, multivariate analysis adjusted for gender, age, stage, and fusion type for Pax:Foxo+ aRMS).
Endotype Analysis and Compound Screen of Endotypically Similar Cell Models
The lack of available RET-mutant aRMS cell models coupled with lack of patient-derived primary cultures at the time of our studies necessitated identifying cellular models genomically and transcriptionally similar to the index case for in vitro experiments. Similarity of cell models was quantified using an aRMS-focused dendrogram endotype computational model (Ricker et al. 2020). Multiple aRMS exome and transcriptomes sequencing data sets were obtained from internal cc-TDI CuRe-FAST samples, the Database of Genotypes and Phenotypes (dbGaP), the Gene Expression Omnibus (GEO) database, Foundation Medicine sequencing panels, the Jackson Laboratory PDX sequencing data, and Champions Oncology PDX sequencing data as well as statistical comparison of RET and RET signaling partner expression between ARMS and ERMS cohorts (Supplemental Tables 1, 2). Using DNA mutation data and RNA expression data from the accumulated aRMS sample cohort, endotype clustering analysis was performed on the aRMS sample sequencing data cohort. Hierarchical clustering identified multiple out-groups (self-clustering consistent with expectations) such as Foundation Medicine panel data (which identifies mutations in a subset of tumor-associated genes), non-PAX3 fusion aRMS samples, and fusion-negative samples.
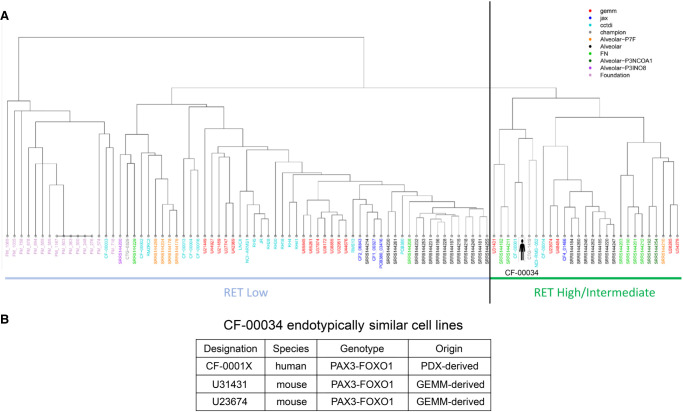
An endotype emerged which focused on RET expression as a key differentiator between hierarchical clusters (Fig. 2A). The index case was determined to be endotypically most similar to CF-00001X (a pediatric PAX3–FOXO1 aRMS with an established PDX model and PDX-derived cell line), U31431 (a PAX3–FOXO1 GEM model–derived cell culture), and four additional GEM cell models (U48484, U23674, U28285, U34279), all bearing PAX3–FOXO1 fusions (Fig. 2B). Lacking a CF-00034-originating cell culture, cell models identified through endotype analysis were used as surrogates for the index case in subsequent experiments.
Figure 2.
Index case endotype and patient avatar cell models. (A) Endotype dendrogram of PAX3–FOXO1 and PAX7–FOXO1 alveolar rhabdomyosarcoma patient samples, cell lines, patient-derived xenograft (PDX) models, and genetically engineered mouse (GEM) models. Data sets were obtained from internal cc-TDI CureFAST samples, database of genotypes and phenotypes (dbGaP) and gene expression omnibus (GEO) sequencing data, Foundation Medicine sequencing panel data, Jackson Laboratory PDX sequencing data, and Champions Oncology PDX sequencing data. Based on DNA and RNA expression of genes in both aRMS variants, multiple aRMS endotypes exist. Validation outgroups include Foundation Medicine samples (out-grouped because of focused sequencing of a subset of target genes) and PAX7–FOXO1 samples (out-grouped because of expected molecular differences from PAX3–FOXO1 samples). (B) Cell models identified as endotypically most similar to the index patient CF-00034 through endotype analysis and hierarchical clustering of aRMS samples and cell models.
The human cell model (CF-00001X) and murine cell model (U31314) endotypically most similar to index patient CF-00034 were screened in a 60-compound drug screen (Supplemental Table 3) to identify therapeutic strategies potentially applicable to the index case (e.g., synthetic lethality). U31314 supplemented with RET ligand ARTN (100 nM) was assayed a second time using the 60-compound screen; no significant difference in drug sensitivity was observed.
Although CF-00001X was sensitive to a greater number of agents than U31314, few drugs showed growth inhibition activity in both models. Of note, we observe CF-00001X was sensitive to IGFR1 inhibitors (OSI-906 and BMS-754807), HDAC inhibitors (panobinostat, CUDC101, CUDC907 and entinostat), and inhibition of several targets in the PI3K pathway (AKT, PI3K, and mTOR).
Phenotypic Analyses
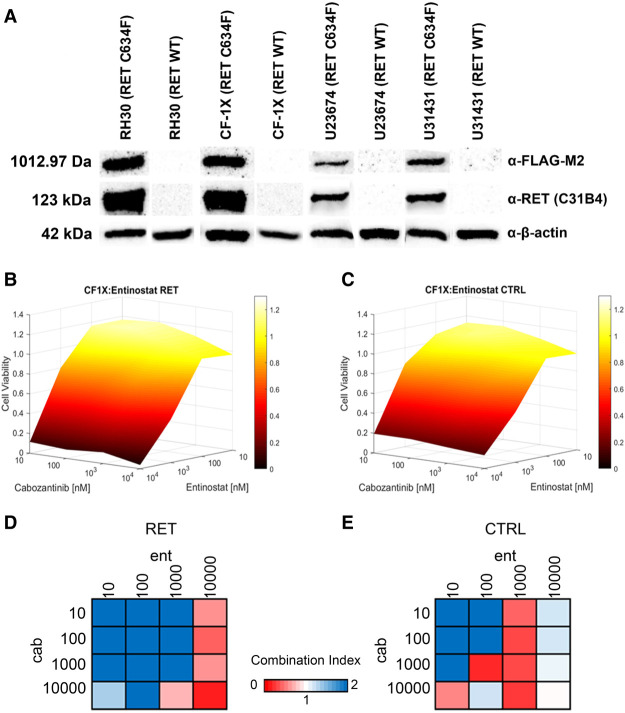
The presence of a known, characterized oncogenic germline RET mutation (Schuffenecker et al. 1998; Masbi et al. 2014) led to the hypothesis that clinical RET inhibitors, as monotherapy or in combination, would be promising treatment options for RET-mutant aRMS. To test this hypothesis, we created synthetic mutant RET cell models mimicking the genotype of the index case patient. Human and murine aRMS cells selected as representative models from the dendrogram (Fig. 2B) were transiently transfected with commercially available RET C634F expression plasmid and assayed with a panel of clinically available agents relevant to aRMS biology and the germline RET mutation (Fig. 3). Transfection of cell models was confirmed by western blot analysis (Fig. 4A), confirming expression of RET C634F and expression of MYC–DDK (FLAG) tagged protein.
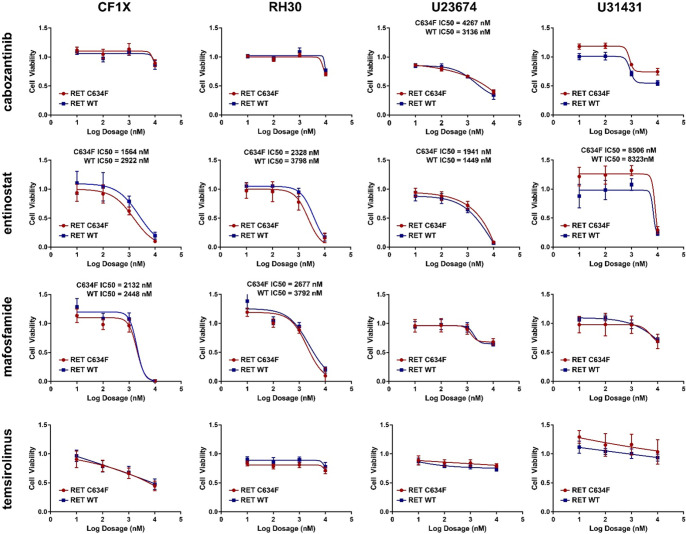
Figure 3.
Monotherapy treatment of null plasmid and RET C634F plasmid transformed endotypically similar cell models. Four cell models identified as endotypically similar to the index patient CF-00034 (CF-00001X, RH30, U23674, U31431) were treated with six different monotherapies (cabozantinib, entinostat, vincristine, cyclophosphamide prodrug, mafosfamide, temsirolimus) to determine agent response and unique sensitives induced by RET C634F protein expression.
Figure 4.
Cabozantinib-focused drug combinations in synthetic RET-mutant cell lines. (A) Western blots of null plasmid and RET C634F transformed endotypically similar cell models, confirming successful transformation and expression of RET C634F. The western blot image in this panel was generated from the grouping of two individual western blot images. No other modifications were made to the western blot image. (B) Representative 3D response manifold of RET C634F transformed RH30 cell model treated with combinations of cabozantinib and entinostat. (C) Representative 3D response manifold of null transformed RH30 cell model treated with combinations of cabozantinib and entinostat. (D) Representative Chou–Talalay Combination Index values from RET C634F cabozantinib plus entinostat combination. (E) Representative Chou–Talalay Combination Index values from null plasmid cabozantinib plus entinostat combination.
The clinical RET inhibitor cabozantinib was selected as the experimental RET inhibitor because of off-label clinical use in the previous case report (Crawford et al. 2020). Cabozantinib has been investigated clinically for RET-rearranged non-small-cell lung cancer (Drilon et al. 2016) and has shown significant clinical efficacy in RET M918T mutant medullary thyroid cancer (Krajewska et al. 2016). For combination therapies, we selected two clinical chemotherapy agents (vincristine and cyclophosphamide [tested as the prodrug mafosfamide]) not used to treat the index case patient described in the preceding manuscript (Crawford et al. 2020) and two clinical targeted inhibitor compounds (HDAC inhibitor entinostat [recently in a Phase I pediatric clinical trial, ADVL1513/NCT02780804] and mTOR inhibitor temsirolimus). Each agent was first tested as a monotherapy to determine overall single agent efficacy across RET C634F transfected and null plasmid cell models (Fig. 3). Drug response to monotherapy and cabozantinib-focused combination therapy of RET C634F transfected versus null plasmid transfected cell models were tested for statistically significant differences. For monotherapy, absolute IC50 values were used for statistical comparison when available or dose-level response vectors were used when single agents did not achieve 50% growth inhibition. In RET C634F transfected versus null cell models, no monotherapy treatment showed statistically significant change in sensitivity across all cell models (Fig. 3).
Combination therapies were tested in checkboard dosage design, with cabozantinib tested in combination with the remaining five monotherapy agents. Combination Index values and 3D dose response manifolds were generated for each cell model/drug combination (representative manifold presented in Fig. 4B,C). Several combinations exhibited synergy at specific combination doses and/or at dose regions in RET C634F synthetic cell models (representative combination index plots are presented in Fig. 4D,E). All manifold and combination index results are provided in Supplemental Table 4.
We also tested for statistically significant differences in single agent response based on IC50 values (Table 1) and overall response curve (Table 2), as well as differences in drug combination response manifolds in RET C634F transfected versus null cell models (Table 3). Although no monotherapy or combination therapy showed statistically significant change across all cell models, monotherapy entinostat showed statistically significant difference in three of four models. However, although the statistically significant human model (RH30) showed increased sensitivity to entinostat, both mouse models showed statistically decreased entinostat sensitivity. Additionally, both RH30 and U31431 show statistically significantly different response to the cabozantinib + mafosfamide combination, as well as increased overall synergy by combination index values (Supplemental Table 4). Nonetheless, the overall lack of statistically significant differences between RET C634F transfected versus null plasmid cell models across all models suggests RET C634F mutation in rhabdomyosarcoma may not induce sensitivity to RET inhibition, nor confer additional sensitivity to chemotherapy agents or targeted agents.
Table 1.
Statistical analysis of monotherapy and combination drug screens with and without synthetic RET mutation
| IC50 value comparison (P-value) | ||||
|---|---|---|---|---|
| Human | Mouse | |||
| RH30 | CF1X | U23674 | U31431 | |
| Cabozantinib | - | - | 1 × 10−04 | - |
| Entinostat | 0.7298 | 1 × 10−04 | 1 × 10−04 | 1 × 10−04 |
| Vincristine | - | - | 0.0719 | - |
| Mafosfamide | 0.9604 | 0.7746 | - | - |
Statistical comparison of RET WT and RET C634F monotherapy IC50 values. Comparison is provided as P-values, with green text indicating statistically significant differences.
Table 2.
Statistical analysis of monotherapy and combination drug screens with and without synthetic RET mutation
| Monotherapy response vector comparison (P-value) | ||||
|---|---|---|---|---|
| Human | Mouse | |||
| RH30 | CF1X | U23674 | U31431 | |
| Cabozantinib | 0.37 | 0.7706 | 0.5068 | 0.1772 |
| Entinostat | 0.1363 | 0.0215 | 0.0681 | 0.1375 |
| Vincristine | 0.0512 | 0.5773 | 0.0136 | 0.5747 |
| Mafosfamide | 0.0604 | 0.3234 | 0.3652 | 0.1196 |
Statistical comparison of RET WT and RET C634F monotherapy response vectors. Comparison is provided as P-values, with green text indicating statistically significant differences.
Table 3.
Statistical analysis of monotherapy and combination drug screens with and without synthetic RET mutation
| Combination therapy response vector comparison (P-value) | ||||
|---|---|---|---|---|
| Human | Mouse | |||
| RH30 | CF1X | U23674 | U31431 | |
| cab-ent | 0.8663 | 0.529 | 0.0252 | 0.7756 |
| cab-vnc | 0.1905 | 0.1199 | 0.4231 | 0.1532 |
| cab-maf | 0.0062 | 0.1105 | 0.3825 | 0.0319 |
Statistical comparison of RET WT and RET C634F combination therapy response vectors as P-values. Comparison is provided as P-values, with green text indicating statistically significant differences.
DISCUSSION
This research began with an unusual clinical observation, an aRMS patient with aggressive metastatic disease has an underlying germline RET C634F-activating mutation. RET activation is known to be the ligand-independent, self-activated driver mutation of MEN2A syndrome cancers and thought to be involved in lung adenocarcinoma (Wang et al. 2019), breast cancer (Paratala et al. 2018), non-small-cell lung cancer, and colorectal cancer (Gozgit et al. 2018). Given the unique genotype of the patient. the poor prognosis associated with metastatic aRMS, and previous success of RET inhibitors in RET mutant tumors (Ackermann et al. 2019), we investigated the lethality of a clinical RET inhibitor as a monotherapy or in combination with other clinically available agents when treating RET-mutant expressing cell models. Sensitivity to monotherapies and combination therapies were consistent in both null plasmid transformed and RET C634F transformed cell models, suggesting the RET mutation borne by the patient did not induce new susceptibility to RET inhibition. Although RET mutation does not necessarily guarantee complete response to a RET inhibitor (Valenciaga et al. 2018), lack of any significant susceptibility to RET inhibition is surprising and speaks to the clinical challenge aRMS presents even when a genomically relevant target is readily druggable.
Promisingly, the recently U.S. Federal Drug Administration (FDA)-approved RET inhibitor selpercatinib has been approved for the use in RET fusion-positive non-small-cell lung cancer (Drilon et al. 2020) and medullary thyroid cancer with RET mutations or RET fusions (Wirth et al. 2020), including both kinase-domain mutations and extracellular cysteine mutations. Notably, extracellular cysteine mutations show overall reduced response to selpercatinib compared to kinase domain mutations. Given the use of cabozantinib as a maintenance therapy in the index case patient, the potential impact of selpercatinib in the index case or similar patients is unclear.
We also tested a panel of multiple agents relevant to rhabdomyosarcoma biology against two of the endotype-based cell models, identifying susceptibility to IGFR1 and PI3K inhibitors in the endotypically closest human cell model, consistent with previous aRMS studies (Crosswell et al. 2006; Abraham et al. 2011; Berlow et al. 2019), again supporting RET status playing a minimal role in altering response to therapeutic agents. Although rarely occurring concomitantly, metastatic aRMS with RET C634F germline mutation presents a difficult phenotype and a dire clinical prognosis and will likely remain as intractable as classical aRMS cases.
A key challenge in the investigation of RET C634F mutant aRMS is the lack of readily available cell models and the initial biopsy material from the patient not generating a viable cell culture. Overcoming the lack of patient-derived cell model by using a cohort of sequenced aRMS samples to identify available cell models most molecularly similar to the index case is a strategy for research-level functional analysis, which may be applicable to other patients with atypical or challenging genotypes. Development of rare disease cohorts will beneficial both in understanding the disease as a whole but also in exploring the unique genotypes possessed by individual patients.
METHODS
Cell Lines
The RH30 cell line (Douglass et al. 1987) (RRID:CVCL_0041) was shared by the Children's Oncology Group Repository (https://www.cccells.org/). RH30 was cultured in RPMI-1640 (11875-093; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (26140079; Thermo Fisher Scientific) plus 1% penicillin-streptomycin (P/S) (15140-122; Thermo Fisher Scientific) maintained at 5% CO2 at 37°C.
Murine Cell Model Establishment
The mouse primary tumor cell cultures U31431 and U23674 were established from a site-of-origin conditionally induced aRMS tumor in a genetically engineered Myf6Cre,Pax3:Foxo1,p53 mouse as previously described (Abraham et al. 2014). In brief, extracted tumor was minced and digested with 10 mg/mL collagenase type IV (17104019; Thermo Fisher Scientific) overnight at 4°C. Dissociated cells were then incubated in Dulbecco's modified Eagle's medium (DMEM) (11995-073; Thermo Fisher Scientific) supplemented with 10% FBS and 1% P/S in 5% CO2 at 37°C. Cultures were maintained at low passage to minimize biological variation from the original tumor. All experiments were performed at or below passage 10.
Patient-Derived Xenograft (PDX) Model Development
All aspects of cancer tissue sharing for model development were reviewed and approved by the Children's Cancer Therapy Development Institute Institutional Review Board. The CF-00001 PDX model and subsequent cell culture (denoted CF-00001X) were described previously (Bharathy et al. 2018). Fusion status of CF-00001 PDX-derived cells (denoted CF-00001X) was confirmed by RNA sequencing for PAX3–FOXO1 fusion reads and western blotting of CF-00001X culture lysate for PAX3–FOXO1 (Bharathy et al. 2018). Cultures were maintained at low passage to minimize biological variation from the original tumor. All experiments were performed at or below passage 10. CF-00001X was cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) plus 1% P/S and maintained at 5% CO2 at 37°C. The CF-0001 PDX model is maintained at the Jackson Laboratory (http://tumor.informatics.jax.org/mtbwi/pdxDetails.do?modelID=J000099761).
Data Integration and Dendrogram Generation
To define endotypes (disease subgroups defined by combined genomic, clinical, and pathobiological feature sets unique to each patient) within aRMS, we applied hierarchical agglomerative clustering analysis to a cohort of aRMS samples including human patient samples with PAX3–FOXO1 fusions (n = 23), patient samples with PAX7–FOXO1 fusions (n = 6 samples), patient samples with PAX3–NCOA1 fusions (n = 2 samples), patient samples with PAX3–INO8 fusions (n = 1 sample), fusion-negative patient samples (n = 8), patient-derived aRMS cell lines (n = 12), Foundation Medicine panel sequencing result from human aRMS (n = 15), genetically engineered mouse models (n = 17), and PDX models of human aRMS from Champions Oncology (n = 2: CTG-0329 and CTG-1419) and the Jackson Laboratory (n = 4: CF1_00327, CF2_06843, CF4_01466, PCB380_03416).
Data from disparate human data sets were aligned to a common set of genes across DNA sequencing data and RNA sequencing data. Mouse-origin samples were converted from mouse gene identifiers to homologous human gene identifiers via BioMart (Smedley et al. 2009) and merged with the human DNA or RNA sequencing data sets, respectively. Gene expression values were normalized to read depth across samples using edgeR (Robinson et al. 2009) and then across individual genes by assigning gene-level z-scores. Zero expression values were assigned to all DNA sequencing data sets lacking RNA sequencing data (i.e., Foundation panel sequencing data sets) to allow for clustering analysis. For mutation data, samples were assigned the value 1 for each gene bearing a mutation, and 0 otherwise. The resulting quantified mutation and expression data sets were concatenated as feature vectors for downstream clustering analysis. The Euclidean distance metric was used to define distance between feature vectors, with average-linkage clustering used to merge nonsingleton feature vectors into groups. Clustering analysis was performed using RStudio Version 3.6.1 using the hdist package.
Sequencing Data Analysis
All sequencing data from patient samples, patient-derived cell lines, genetically engineered mouse models, and the Jackson Laboratory PDX models were analyzed by the previously published analysis pipeline (Kats et al. 2019). In brief, gene mutations from whole-exome sequencing experiments were identified by aligning fastq files to the GRCh38 human reference genome or GRCm38 mouse reference genome via BWA aligner, processing with GATK best practices (DePristo et al. 2011), and called with MuTect2 (Benjamin et al. 2019). Gene expression from whole transcriptome sequencing experiments was analyzed by aligning fastq files to the GRCh38 human reference genome or GRCm38 mouse reference genome using STAR aligner (Dobin et al. 2013) and quantified by RSEM (Li and Dewey 2011).
Drug Screen of Endotypically Similar Cell Models
CF-00001X and U31431, were evaluated in a proliferation assay against a 60-compound library which was previously described and reported (Kats et al. 2019). In brief, cells were cultured and harvested, then plated via BioTek MultiFlo liquid dispenser into thawed preprinted compound library 384-well opaque white wall plates (164610; Thermo Fisher Scientific). Cells in plates were incubated for 72 h then assayed for cell viability via CellTiter Glo 2.0 (G9241; Promega) luminescent assay. Drug response and IC50 values were determined using Microsoft Excel.
Transfection of PAX3–FOXO1 Alveolar Rhabdomyosarcoma Cell Models
The RET mutant plasmid RET C634F built into pCMV6 Entry. carboxy-terminal Myc/DDK FLAG Tagged cloning vector was purchased from OriGene (RC403308; OriGene) and expanded by Aldevron (http://www.aldevron.com/). The plasmid was transfected into four cell models: RH30, CF-00001X, U23674, and U31431.
Cells were plated in tissue-culture treated six well plates (140675; Thermo Fisher Scientific) at initial cell populations at 150,000 cells/well. After a 24-h acclimation period, the plated cells were transfected with DNA plasmid (lipofectamine 6 µL + 2 µg DNA) for 2 h in serum-free media (RPMI-1640 for human cells, DMEM for murine cells), and then washed and replaced with growth media. Transient transfection of RET C634F plasmid via the above methodology was confirmed for all cell models via western blot for Myc/DDK FLAG antibody (F3165; Millipore Sigma).
Immunoblotting
Following transfection, all four cell models were cultured for 24 h then harvested for protein via RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific 89900) + Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific 78441). Null plasmid cells were cultured for 48 h, and protein was isolated in the same manner. RET expression in transfected and null plasmid cell models was analyzed using ant-RET antibody (1:1000 dilution; Cell Signaling Technologies 3220). Presence of RET C634F protein was confirmed using anti-FLAG M2 antibody (1:5000 dilution; Sigma-Aldrich F1804).
Monotherapy and Combination Therapy Drug Studies of Transformed Cell Models
Cell lines transfected with either null pCMV6 Entry plasmid or RET C634F pCMV6 Entry plasmid were collected and plated in growth media at 1500 cells/well into 384-well drug plates preprinted with six agents validated as a monotherapy and five cabozantinib-focused drug combinations based on the six monotherapy agents. The following drugs were tested as monotherapy: cabozantinib (Selleck Chemicals S4001), entinostat (Selleck Chemicals S1053), vincristine (Selleck Chemicals S1241), mafosfamide (Santa Cruz Biotechnology sc-211761), and temsirolimus (Selleck Chemicals S1044). Individual agents were tested in triplicate at the following doses: [10 nM, 100 nM, 1 µM, 10 µM]. The following two-drug combinations were selected for validation: cabozantinib + entinostat, cabozantinib + vincristine, cabozantinib + mafosfamide, and cabozantinib + temsirolimus. Each combination of agents was tested at the Cartesian product of the following dosages: cabozantinib [10 nM, 100 nM, 1 µM, 10 µM] × combination agent [10 nM, 100 nM, 1 µM, 10 µM] for a total of 16 data points per combination. Three technical replicates of each monotherapy/combination screen were performed for each null plasmid– and RET C634F plasmid–transfected cell model. Cell models in growth media were plated in the preprinted plates and incubated for 72 h in 5% CO2 at 37°C. At the end of the 72-h period, cell viability was assessed by CellTiter Glo 2.0 luminescent assay per manufacturer's protocol and quantified via BioTek Synergy HT plate reader.
Drug sensitivity as IC50 value of each monotherapy was determined by Hill curve fitting using GraphPad Prism. Chou–Talalay Combination Index values (Chou 2010) for drug combinations were generated using the CompuSyn software tool. Effect values for CompuSyn monotherapy and combination therapy were determined by mean cell viability based on n = 3 independent technical replicates for each transfected cell model. Each null plasmid– and RET C634F plasmid–transfected cell model was compared for statistically significant differences of drug response to determine if RET C634F expression resulted in changes in drug sensitivity.
Sequencing Data
Sequencing data for endotype analysis and dendrogram generation were gathered from previously published data sets with the following accession IDs: GSE138269, GSM984615, GSM758578, GSE142775 (Gene Expression Omnibus database), EGAS00001004359, EGAS00001003981 (European Genome-phenome Archive), PRJNA613152 (Short Read Archive), Champions TumorGraft database, Mouse Tumor Biology Database (http://tumor.informatics.jax.org/mtbwi/pdxSearch.do), the OncoGenomics Database (https://pob.abcc.ncifcrf.gov/cgi-bin/JK), and phs001121.v1.p1 (Database of Phenotypes and Genotypes).
Statistics
Student's t-tests were used to compare IC50 values from null and RET C634F transfected aRMS cell lines treated with different monotherapy agents. Hotelling's T2 test was used to compare one-dimensional drug response vectors from null and RET C634F transfected aRMS cell lines treated with different monotherapy agents, when 50% inhibitory concentrations were not reached. Hotelling's T2 test was also used to compare two-dimensional drug response manifolds from null and RET C634F transfected aRMS cell lines treated with different combination therapy agents.
ADDITIONAL INFORMATION
Data Deposition and Access
All sequencing data used to generate results in this manuscript have been previously deposited to online sequencing databases. The following data sets under the given accession IDs were used: GSE138269, GSM984615, GSM758578, GSE142775 (Gene Expression Omnibus database), EGAS00001004359, EGAS00001003981 (European Genome-phenome Archive), PRJNA613152 (Short Read Archive), Champions TumorGraft database, Mouse Tumor Biology Database (http://tumor.informatics.jax.org/mtbwi/pdxSearch.do), the OncoGenomics Database (https://pob.abcc.ncifcrf.gov/cgi-bin/JK), and phs001121.v1.p1 (Database of Phenotypes and Genotypes).
Ethics Statement
All human tissue samples were acquired through the Cancer Registry for Familial and Sporadic Tumors (CuRe-FAST) tumor banking program. All patients enrolled in CuRe-FAST provided informed consent. All aspects of the study were reviewed and approved by the Children's Cancer Therapy Development Institute (cc-TDI) Institutional Review Board (IRB). Patient data and clinical and pathologic information are maintained in a de-identified database.
Acknowledgments
We thank Foundation Medicine for access to their pediatric tumor sequencing data. We thank Christine McCormick for graphic design assistance. We thank the parents and family of the index patient for inspiration and support of this project.
Author Contributions
N.E.B. and K.A.C. designed the experiments, interpreted results, and wrote the manuscript; C.J.B., C.N., and I.S. provided materials and edited the manuscript; E.R.R. edited the manuscript; and C.K. wrote the manuscript.
Funding
This work was funded by the Genomic Endotypes in Alveolar Rhabdomyosarcoma (GEAR) project consortium hosted on consano.org and by the Shane's Future Days foundation.
Competing Interest Statement
Author N.E.B. is a business partner to Champions Oncology (which employs author C.N.), from which PDX tissue was provided. The work in this manuscript is independent of the business relationship.
Referees
Patience Obasaju
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Abraham J, Prajapati SI, Nishijo K, Schaffer BS, Taniguchi E, Kilcoyne A, McCleish AT, Nelon LD, Giles FG, Efstratiadis A, et al. 2011. Evasion mechanisms to Igf1r inhibition in rhabdomyosarcoma. Mol Cancer Ther 10: 697–707. 10.1158/1535-7163.MCT-10-0695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham J, Nuñez-Álvarez Y, Hettmer S, Carrió E, Chen HIH, Nishijo K, Huang ET, Prajapati SI, Walker RL, Davis S, et al. 2014. Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes Dev 28: 1578–1591. 10.1101/gad.238733.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann CJ, Stock G, Tay R, Dawod M, Gomes F, Califano R. 2019. Targeted therapy for RET-rearranged non–small cell lung cancer: clinical development and future directions. Onco Targets Ther 12: 7857–7864. 10.2147/OTT.S171665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal SK, Beth Kester M, Debelenko LV, Heppner C, Emmert-Buck MR, Skarulis MC, Doppman JL, Kim YS, Lubensky IA, Zhuang Z, et al. 1997. Germline mutations of the MEN1 gene in familial multiple endocrine neoplasia type 1 and related states. Hum Mol Genet 6: 1169–1175. 10.1093/hmg/6.7.1169 [DOI] [PubMed] [Google Scholar]
- Alpers JP, Jones LK Jr. 2010. Natural history of exertional rhabdomyolysis: a population-based analysis. Muscle Nerve 42: 487–491. 10.1002/mus.21740 [DOI] [PubMed] [Google Scholar]
- Amer KM, Thomson JE, Congiusta D, Dobitsch A, Chaudhry A, Li M, Chaudhry A, Bozzo A, Siracuse B, Aytekin MN, et al. 2019. Epidemiology, incidence, and survival of rhabdomyosarcoma subtypes: SEER and ICES database analysis. J Orthop Res 37: 2226–2230. 10.1002/jor.24387 [DOI] [PubMed] [Google Scholar]
- Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L. 2019. Calling somatic SNVs and indels with Mutect2. bioRxiv 10.1101/861054 [DOI] [Google Scholar]
- Berlow NE, Rikhi R, Geltzeiler M, Abraham J, Svalina MN, Davis LE, Wise E, Mancini M, Noujaim J, Mansoor A, et al. 2019. Probabilistic modeling of personalized drug combinations from integrated chemical screen and molecular data in sarcoma. BMC Cancer 19: 593. 10.1186/s12885-019-5681-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathy N, Berlow NE, Wang E, Abraham J, Settelmeyer TP, Hooper JE, Svalina MN, Ishikawa Y, Zientek K, Bajwa Z, et al. 2018. The HDAC3–SMARCA4–miR-27a axis promotes expression of the PAX3:FOXO1 fusion oncogene in rhabdomyosarcoma. Sci Signal 11: eaau7632. 10.1126/scisignal.aau7632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandford MC, Barr FG, Lynch JC, Randall RL, Qualman SJ, Keller C. 2006. Rhabdomyosarcomas utilize developmental, myogenic growth factors for disease advantage: a report from the Children's Oncology Group. Pediatr Blood Cancer 46: 329–338. 10.1002/pbc.20466 [DOI] [PubMed] [Google Scholar]
- Chou TC. 2010. Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res 70: 440–446. 10.1158/0008-5472.CAN-09-1947 [DOI] [PubMed] [Google Scholar]
- Cranston AN, Carniti C, Oakhill K, Radzio-Andzelm E, Stone EA, McCallion AS, Hodgson S, Clarke S, Mondellini P, Leyland J, et al. 2006. RET is constitutively activated by novel tandem mutations that alter the active site resulting in multiple endocrine neoplasia type 2B. Cancer Res 66: 10179–10187. 10.1158/0008-5472.CAN-06-0884 [DOI] [PubMed] [Google Scholar]
- Crawford KA, Berlow NE, Tsay J, Lazich M, Mancini M, Noakes C, Huang T, Keller C. 2020. Case report for an adolescent with germline RET mutation and alveolar rhabdomyosarcoma. Cold Spring Harb Mol Case Stud 6: a004853. 10.1101/mcs.a004853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosswell HE, Peng Q, Prince C, De P, Durden DL. 2006. Targeting PI3K in alveolar rhabdomyosarcoma (aRMS). J Clin Oncol 24: 10025. 10.1200/jco.2006.24.18_suppl.10025 [DOI] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass EC, Valentine M, Etcubanas E, Parham D, Webber BL, Houghton PJ, Houghton JA, Green AA. 1987. A specific chromosomal abnormality in rhabdomyosarcoma. Cytogenet Cell Genet 45: 148–155. 10.1159/000132446 [DOI] [PubMed] [Google Scholar]
- Drilon A, Rekhtman N, Arcila M, Wang L, Ni A, Albano M, Van Voorthuysen M, Somwar R, Smith RS, Montecalvo J, et al. 2016. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 17: 1653–1660. 10.1016/S1470-2045(16)30562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drilon A, Oxnard GR, Tan DSW, Loong HHF, Johnson M, Gainor J, McCoach CE, Gautschi O, Besse B, Cho BC, et al. 2020. Efficacy of selpercatinib in RET fusion–positive non-small-cell lung cancer. N Engl J Med 383: 813–824. 10.1056/NEJMoa2005653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozgit JM, Chen TH, Song Y, Wardwell S, Wang F, Cai J, Li H, Edgren H, Rivera VM, Pritchard J. 2018. RET fusions observed in lung and colorectal cancers are sensitive to ponatinib. Oncotarget 9: 29654–29664. 10.18632/oncotarget.25664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins DS, Spunt SL, Skapek SX. 2013. Children's Oncology Group's 2013 blueprint for research: soft tissue sarcomas. Pediatr Blood Cancer 60: 1001–1008. 10.1002/pbc.24435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezam K, Jiang J, Sun F, Zhang X, Zhang J. 2018. Artemin promotes oncogenicity, metastasis and drug resistance in cancer cells. Rev Neurosci 29: 93–98. 10.1515/revneuro-2017-0029 [DOI] [PubMed] [Google Scholar]
- Jones AE, Albano EA, Lovell MA, Hunger SP. 2010. Metastatic alveolar rhabdomyosarcoma in multiple endocrine neoplasia type 2A. Pediatr Blood Cancer 55: 1213–1216. 10.1002/pbc.22591 [DOI] [PubMed] [Google Scholar]
- Kato S, Subbiah V, Marchlik E, Elkin SK, Carter JL, Kurzrock R. 2017. RET aberrations in diverse cancers: next-generation sequencing of 4,871 patients. Clin Cancer Res 23: 1988–1997. 10.1158/1078-0432.CCR-16-1679 [DOI] [PubMed] [Google Scholar]
- Kats D, Ricker CA, Berlow NE, Noblet B, Nicolle D, Mevel K, Branchereau S, Judde J-G, Stiverson CD, Stiverson CL, et al. 2019. Volasertib preclinical activity in high-risk hepatoblastoma. Oncotarget 10: 6403–6417. 10.18632/oncotarget.27237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjær S, Kurokawa K, Perrinjaquet M, Abrescia C, Ibáñez CF. 2006. Self-association of the transmembrane domain of RET underlies oncogenic activation by MEN2A mutations. Oncogene 25: 7086–7095. 10.1038/sj.onc.1209698 [DOI] [PubMed] [Google Scholar]
- Krajewska J, Olczyk T, Jarzab B. 2016. Cabozantinib for the treatment of progressive metastatic medullary thyroid cancer. Expert Rev Clin Pharmacol 9: 69–79. 10.1586/17512433.2016.1102052 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall AD, Grosveld GC. 2012. Alveolar rhabdomyosarcoma: the molecular drivers of PAX3/7-FOXO1-induced tumorigenesis. Skelet Muscle 2: 25. 10.1186/2044-5040-2-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masbi MH, Mohammadiasl J, Galehdari H, Ahmadzadeh A, Tabatabaiefar MA, Golchin N, Haghpanah V, Rahim F. 2014. Characterization of wild-type and mutated RET proto-oncogene associated with familial medullary thyroid cancer. Asian Pac J Cancer Prev 15: 2027–2033. 10.7314/APJCP.2014.15.5.2027 [DOI] [PubMed] [Google Scholar]
- Mulligan LM. 2019. GDNF and the RET receptor in cancer: new insights and therapeutic potential. Front Physiol 9: 1873. 10.3389/fphys.2018.01873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. 2009. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975–2005. Cancer 115: 4218–4226. 10.1002/cncr.24465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paratala BS, Chung JH, Williams CB, Yilmazel B, Petrosky W, Williams K, Schrock AB, Gay LM, Lee E, Dolfi SC, et al. 2018. RET rearrangements are actionable alterations in breast cancer. Nat Commun 9: 4821. 10.1038/s41467-018-07341-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham DM, Barr FG. 2013. Classification of rhabdomyosarcoma and its molecular basis. Adv Anat Pathol 20: 387–397. 10.1097/PAP.0b013e3182a92d0d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker C, Crawford K, Matlock K, Lathara M, Seguin B, Rudzinski E, Berlow N, Keller C. 2020. Defining an embryonal rhabdomyosarcoma endotype. Mol Case Stud 6: a005066. 10.1101/mcs.a005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2009. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudzinski ER, Anderson JR, Chi YY, Gastier-Foster JM, Astbury C, Barr FG, Skapek SX, Hawkins DS, Weigel BJ, Pappo A, et al. 2017. Histology, fusion status, and outcome in metastatic rhabdomyosarcoma: a report from the Children's Oncology Group. Pediatr Blood Cancer 64: 10.1002/pbc.26645. 10.1002/pbc.26645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuffenecker I, Virally-Monod M, Brohet R, Goldgar D, Conte-Devolx B, Leclerc L, Chabre O, Boneu A, Caron J, Houdent C, et al. 1998. Risk and penetrance of primary hyperparathyroidism in multiple endocrine neoplasia type 2A families with mutations at codon 634 of the RET proto-oncogene1. J Clin Endocrinol Metab 83: 487–491. 10.1210/jcem.83.2.4529 [DOI] [PubMed] [Google Scholar]
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. 2014. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 4: 216–231. 10.1158/2159-8290.CD-13-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. 2009. BioMart: biological queries made easy. BMC Genomics 10: 22. 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M. 2001. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev 12: 361–373. 10.1016/S1359-6101(01)00012-0 [DOI] [PubMed] [Google Scholar]
- Valenciaga A, Saji M, Yu L, Zhang X, Bumrah C, Yilmaz AS, Knippler CM, Miles W, Giordano TJ, Cote GJ, et al. 2018. Transcriptional targeting of oncogene addiction in medullary thyroid cancer. JCI Insight 3: e122225. 10.1172/jci.insight.122225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zimmermann S, Parikh K, Mansfield AS, Adjei AA. 2019. Current diagnosis and management of small-cell lung cancer. Mayo Clin Proc 94: 1599–1622. 10.1016/j.mayocp.2019.01.034 [DOI] [PubMed] [Google Scholar]
- Wirth LJ, Sherman E, Robinson B, Solomon B, Kang H, Lorch J, Worden F, Brose M, Patel J, Leboulleux S, et al. 2020. Efficacy of selpercatinib in RET-altered thyroid cancers. N Engl J Med 383: 825–835. 10.1056/NEJMoa2005651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhao Y, Cui B, Gu L, Zhu S, Li J, Liu J, Yin M, Zhao T, Yin Z, et al. 2007. RET proto-oncogene mutations are restricted to codons 634 and 918 in mainland Chinese families with MEN2A and MEN2B. Clin Endocrinol 67: 570–576. 10.1111/j.1365-2265.2007.02927.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data used to generate results in this manuscript have been previously deposited to online sequencing databases. The following data sets under the given accession IDs were used: GSE138269, GSM984615, GSM758578, GSE142775 (Gene Expression Omnibus database), EGAS00001004359, EGAS00001003981 (European Genome-phenome Archive), PRJNA613152 (Short Read Archive), Champions TumorGraft database, Mouse Tumor Biology Database (http://tumor.informatics.jax.org/mtbwi/pdxSearch.do), the OncoGenomics Database (https://pob.abcc.ncifcrf.gov/cgi-bin/JK), and phs001121.v1.p1 (Database of Phenotypes and Genotypes).