Abstract
A 52-yr-old woman presented with therapy-related acute myeloid leukemia. A bone marrow biopsy showed 21% blasts with a myeloid phenotype and no other notable features such as abnormal eosinophils. Routine nanofluidics-based reverse transcriptase polymerase chain reaction (PCR) leukemia translocation panel designed to screen for recurrent genetic abnormalities in acute leukemia detected an inversion 16 transcript variant E. This prompted rereview of karyotype and fluorescence in situ hybridization studies, which confirmed inv(16), leading to appropriate prognostication and modification of treatment. This case underscores the utility of a powerful molecular screening method for the routine detection of recurrent genetic abnormalities of acute myeloid leukemia. It was especially useful in this case because of the lack of characteristic morphologic findings seen in inversion 16 and the difficulty in its detection by conventional karyotype analysis.
Keywords: acute myeloid leukemia, hematological neoplasm
INTRODUCTION
In the World Health Organization (WHO) classification system, acute myeloid leukemia (AML) with inv(16)(p13.1q22)/CBFB–MYH11 is recognized as a separate entity within the category of AML with recurrent genetic abnormalities (Arber et al. 2016). Approximately 5%–7% of patients with de novo AML have an inv(16)(p13.1q22) or t(16;16)(p13.1;q22) abnormality. It is typically associated with distinctive morphologic findings, including a prominent monocytic component, eosinophilia, and abnormalities involving immature eosinophilic granules in late promyelocyte and myelocyte stages of eosinophil maturation. Characteristically, these abnormal eosinophilic precursors have larger than usual, dense purple-violet granulation that can obscure the cell morphologic features (Larson et al. 1986; Marlton et al. 1995; Bloomfield et al. 1997; Byrd et al. 2002; Delaunay et al. 2003; Schwind et al. 2013). The association of abnormal eosinophils and alterations in Chromosome 16 was first described by Arthur and Bloomfield in 1983 (Arthur and Bloomfield 1983). Occasionally, a small subset of cases may lack one or more of these features including eosinophilia. Rarely, AML with inv(16) can be seen in therapy-related settings.
Importantly, irrespective of morphologic findings, AML with inv(16)(p13.1q22)/CBFβ–MYH11 carries a favorable prognosis. This subtype of AML has been shown to achieve a high rate of complete remission with high-dose cytarabine during consolidation therapy (Bloomfield et al. 1998; Borthakur et al. 2008). The optimal treatment strategy for AML with inv(16) involves a regimen of intensive consolidation chemotherapy, as opposed to prolonged maintenance chemotherapy with low-dose antileukemic agents (Juliusson et al. 2009; Prebet et al. 2009). Higher remission rates are noted in elderly AML patients, as well as therapy-related settings (Larson 2006; Arahata et al. 2015). For the above reasons, it is critical to identify inv(16) in AML patients.
The breakpoints of both inv(16)(p13.1q22)/CBFB–MYH11 and t(16;16) involve the intronic regions of the core-binding factor β subunit (CBFβ) at 16q22 and the myosin heavy chain 11 smooth muscle gene (MYH11) at 16p13.1 and often occurs as a result of a pericentric inversion (Liu et al. 1993; Reilly 2005). Liu et al. showed that the first five, or rarely four, exons of CBFβ bind to variable lengths of the carboxy-terminal region of MYH11. In their study, the nucleotide numbering in the breakpoint of MYH11 ranged from 994 to 2134 (Liu et al. 1995; Reilly 2005). Multiple unique fusion transcripts, designated types A–K, can occur in AML with inv(16) as a result of different combinations in CBFβ and MYH11. Type A is the most common, accounting for >85% of inv(16) fusion transcripts. Types D and E have been reported at 5%–10% each, whereas type B, C, and F–K are predominantly single case reports (Shurtleff et al. 1995; van der Reijden et al. 1995; Schnittger et al. 2007; Schwind et al. 2013).
Detecting inv(16) is not without its challenges. By conventional karyotyping, inv(16) abnormality can be very subtle, which is a well-documented pitfall (Langabeer et al. 1997; Ritter et al. 1997; Hernández et al. 2000). Routine fluorescence in situ hybridization (FISH)/molecular studies to detect inv(16) on every AML can be expensive. For this reason, many clinical centers perform FISH or molecular testing for recurrent translocations only when there is clinical or morphologic suspicion for its presence. Furthermore, not all laboratories may have these tests available on-site and must have the sample sent to a reference center. Similarly, the detection of differentially spliced CBFB–MYH11 transcripts requires the use of multiple different reverse transcriptase–polymerase chain reaction (RT-PCR) primers (van der Reijden et al. 1995; Hernández et al. 2000). As such, discretion in ordering, based on morphological clues, is exercised in many settings.
However, on occasion, the morphologic clues may be subtle or absent. Specifically, AML cases with transcript E are more frequent in therapy-related settings and often lack abnormal eosinophil cytomorphology (Schnittger et al. 2007). In such cases, unless testing is routinely done, inv(16) can be missed, leading to suboptimal treatment approach. In addition to routine assessment of inv(16), which is expensive, screening for other recurrent genetic abnormalities such as t(8;21) and t(15;17) is also required.
Herein, we present a patient with AML with inv(16) variant E, presenting in a therapy-related setting, without any of the typical morphological features of eosinophilia or abnormal eosinophils with basophilic granules. This transcript was detected incidentally using a routine nanofluidics-based multiplex RT-PCR (leukemia translocation panel) screening assay designed to screen for the frequently identified recurrent genetic abnormalities in acute leukemia. Inv(16) abnormality was subsequently confirmed by FISH and detected by conventional karyotyping studies in retrospect. We illustrate the utility of this assay in formulating the appropriate treatment plan with the utmost benefit and highlight the need for such an assay for clinical screening of all AML patients for enabling precision medicine.
RESULTS
Case Report
A 52-yr-old woman was found to have leukocytosis, anemia, thrombocytopenia, and circulating blasts during routine follow-up. Complete blood count showed a white blood cell count of 14.2 K/µL, hemoglobin of 11.5 g/dL, and platelet count of 68 K/µL. She had a history of left breast invasive ductal carcinoma, diagnosed 5 yr prior, treated with neoadjuvant chemotherapy with fluorouracil (5-FU), doxorubicin and cyclophosphamide, and paclitaxel, followed by segmental mastectomy and radiation.
The patient underwent bone marrow aspiration and biopsy on the left posterior iliac crest.
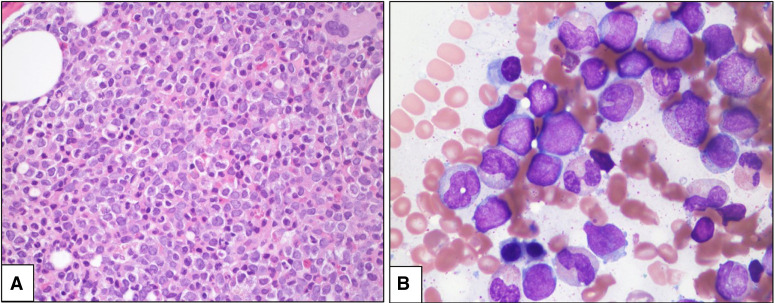
The bone marrow biopsy was hypercellular (90%) for age with trilineage hypoplasia, increased immature blasts, and marked granulocytic predominance (Fig. 1A). Bone marrow touch imprint showed ∼21% blasts, variable in size, with irregular nuclear contours, oval to folded nuclei, and some azurophilic granules (Fig. 1B). Auer rods were noted in rare blasts. The myeloid precursors showed left-shifted maturation with occasional dyspoietic changes. The erythroid precursors were decreased in proportion, with megaloblastoid maturation. Megakaryocytes were rare and showed normal morphology. There was no increase in monocytic cells. There was no eosinophilia or abnormal eosinophils with purple-violet granules. By cytochemical staining, the blasts were positive for myeloperoxidase and negative for butyrate (nonspecific) esterase. By Prussian blue stain, ring sideroblasts were not seen. A diagnosis of AML arising in a therapy-related setting was rendered. Immunohistochemical stains for pan-cytokeratin and TP53 were negative. Flow cytometry immunophenotypic analysis showed an aberrant blast population (∼20% of total analyzed events) positive for CD13, CD34, CD38, CD117, CD123, HLA-DR, and MPO. They were negative for CD2, sCD3, cCD3, CD4, CD5, CD7, CD14, CD15, CD19, CD22, CD25, CD33, CD36, CD41, CD56, CD64, and TdT. Preliminary conventional cytogenetic studies (48 h of culture) showed 46,XX,add(5p)[6]/46,XX[4].
Figure 1.
(A) Hypercellular bone marrow biopsy showing increased immature cells and granulocytic predominance (hematoxylin and eosin [H&E], 400×); (B) Bone marrow touch imprint showing increased blasts and left-shifted granulocytes with some dysplastic features (Wright–Giemsa, 1000×, oil immersion).
Inv(16) Detecting by a Leukemia Translocation Panel Screening Assay
All new patients with AML and at least 6% blasts by differential count undergo routine testing using a nanofluidics-based qualitative RT-PCR assay custom-designed for detection of eight recurring translocations including a few variants, observed in AML and acute lymphocytic leukemia (ALL) (eight translocations, nine variants). This “leukemia translocation panel” (LTP) is a novel, high-throughput single-platform nanofluidics-based approach to identify multiple leukemia-associated translocations (Table 1), which combines integrated fluidic circuit arrays and TaqMan probe–based quantitative PCR (qPCR) to simultaneously detect fusion transcripts using very little RNA.
Table 1.
Leukemia translocation panel
| t(8;21)(q22;q22); RUNX1–RUNX1T1 |
| inv(16)(p13.1q22) or t(16;16)(p13.1q22); CBFB–MYH11 variant A |
| inv(16)(p13.1q22) or t(16;16)(p13.1q22); CBFB–MYH11 variant D |
| inv(16)(p13.1q22) or t(16;16)(p13.1q22); CBFB–MYH11 variant E |
| t(15;17)(q22;q12); PML–RARA long form |
| t(15;17)(q22;q12); PML–RARA short form |
| t(15;17)(q22;q12); PML–RARA alternative form |
| t(9;22)(q34;q11.2); BCR–ABL1 b2a2 |
| t(9;22)(q34;q11.2); BCR–ABL1 b3a2 |
| t(9;22)(q34;q11.2); BCR–ABL1 e1a2 |
| t(12;21)(p13;q22); ETV6–RUNX1 |
| t(1;19)(q23;p13.3); E2A–PBX1 |
| t(4;11)(q21;q23); KMT2A–AF4 |
| t(6;9)(p23;q34); DEK–NUP214 |
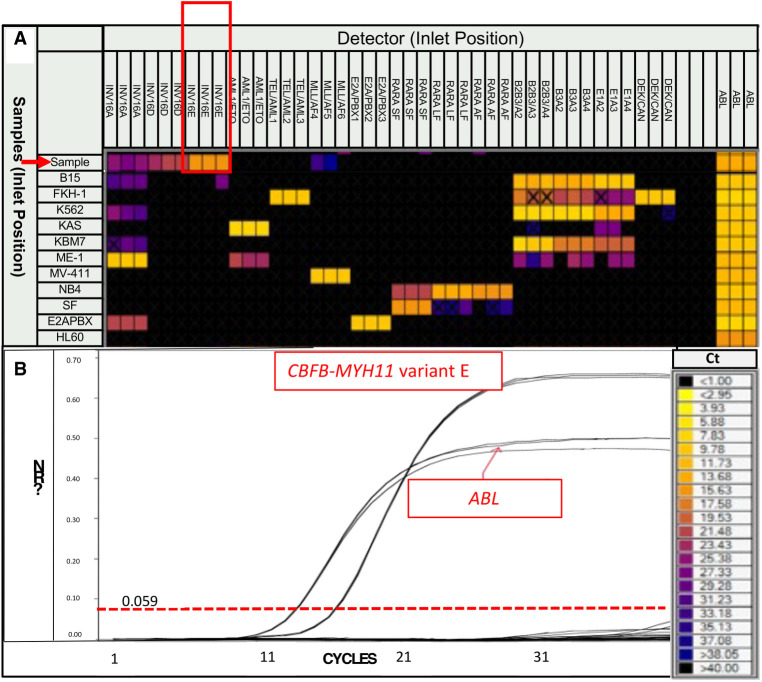
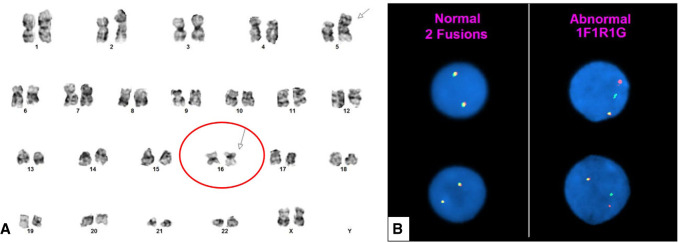
The LTP identified a CBFB–MYH11 variant E fusion transcript (Fig. 2). Based on these unexpected results, we performed FISH studies on aspirate smears for confirmation using a dual-color break-apart DNA probe (Abbott Molecular) that hybridizes to the band 16q22 (spectrum orange on the 5′ centromeric side and spectrum green on the 3′ telomeric side of the CBFB breakpoint). FISH results confirmed CBFB rearrangement in 176 of 200 (88%) interphases (Fig. 3A). Following identification of inv(16) by both FISH and molecular studies, review of the karyotype revealed 46,XX,add(5)(p15.1),inv(16)(p13.1q22)[12]/46,XX[8] (Fig. 3B).
Figure 2.
(A) Leukemia translocation panel heat map representation of cycle to threshold Ct values for each tested translocation. A result is considered positive if the Ct value is ≤25 (as indicated by the color scale) and if all samples in triplicate have a nearly identical Ct value. The patient is indicated by a red arrow, with a Ct value for inv(16) type E of ∼16, in triplicate (red box). In contrast, the Ct values for inv(16) types A and D do not meet these criteria (Ct value > 26 in at least one of the triplicate samples). Similarly, MLL(KMT2A)/AF4 also does not meet these criteria (Ct value > 30 and in only two-thirds of the triplicate samples). The positive control for each translocation is shown as an individual row (rows 31–40), and HLA-60 (row 41) serves as the negative control. (B) Amplification curve for CBFB–MYH11 variant E fusion transcript using a nanofluidics-based qualitative RT-PCR assay.
Figure 3.
(A) Conventional cytogenetics showing an inversion 16 abnormality (circled red with arrow). The karyotype was 46,XX,add(5)(p15.1),inv(16)(p13.1q22)[12]/46,XX[8]. (B) Fluorescence in situ hybridization was positive for CBFB rearrangement (176 of 200 interphases) using a CBFB inv(16) dual-color break-apart DNA probe.
Next-generation sequencing (NGS)-based mutation analysis on a Illumina MiSeq sequencer using a 28-gene myeloid panel detected low-level mutations in KRAS p.G12D (variant allele frequency, 2.7%) and FLT3 p.D835Y (variant allele frequency, 1.3%) (Table 2; Kanagal-Shamanna et al. 2016a). The latter was also confirmed using a PCR-based capillary electrophoresis assay that was positive for low-level FLT3 D835 point mutation (and negative for FLT3 internal tandem duplication [ITD]). Mutations in CEBPA (CCAAT/enhancer binding protein-alpha) gene were absent by Sanger sequencing assay.
Table 2.
Variant table
| Chromo some | Gene | Genomic coordinates | HGVS coding variant | HGVS protein | Variant(+) | Variant type | Predicted effect | Variant allele frequency | Coverage | dbSNP/dbVar ID |
|---|---|---|---|---|---|---|---|---|---|---|
| Chr 13 | FLT3 | Chr 13:28592642 | NM_004119.3(FLT3):c.2503G > T | p.D835Y | G > T | SNV/missense | Substitution | 1.3 | 9024 | rs121913488 |
| Chr 12 | KRAS | Chr 12:25398284 | NM_004985.5(KRAS): c.35G > A | p.G12D | G > A | SNV/missense | Substitution | 2.7 | 1583 | rs121913529 |
(SNV) Single-nucleotide variant.
Therapy Change and Outcome
Following the diagnosis of therapy-related AML with inv(16), the patient was treated with induction chemotherapy with CIA (cladribine, idarubicin, and cytarabine) followed by consolidation with FLAG-IDA (fludarabine, cytarabine, idarubicin). Because core-binding factor AML benefits most from high-dose Ara-C, the consolidation therapy was switched to FLAG-IDA, with a higher dose of Ara-C from CIA (the standard in the community practice would have been 3 + 7). The patient achieved complete remission, negative for minimal residual disease by flow cytometry studies, FISH studies for CBFB–MYH11, and negative for FLT3 D835. The patient could not be followed up using the in-house RT-PCR assay because it is designed for only CBFB–MYH11 transcript type A. As of last follow-up, she has been in complete molecular remission for the last 16 months.
DISCUSSION
Identification of genetic aberrations is essential for providing accurate diagnosis and prognosis, instituting appropriate treatment, and guiding subsequent follow-up testing for minimal residual disease assessment (Kanagal-Shamanna et al. 2016b). Unique management strategies in AML patients with recurrent genetic translocations such as t(15;17), t(8;21) and inv(16) are an epitome of personalized medicine.
Inv(16) abnormality is often suspected in AML with myelomonocytic maturation associated with characteristic abnormal eosinophils; however, in a therapy-related setting such as this case, morphologic clues may be absent. Furthermore, the inv(16) abnormality can be subtle, and detection by conventional karyotype can be challenging. Not surprisingly, preliminary karyotyping results from this case did not detect inv(16). For these reasons, reflex FISH testing was not ordered. However, routine use of a leukemia translocation panel to screen for recurring genetic abnormalities in all patients with AML identified inv(16) transcript E, which led to further confirmatory testing using FISH studies, guiding appropriate prognostication and treatment. This case highlights the importance of this or a similar assay in which the treatment is modified based on this cryptic finding. Detection of recurring translocations in core-binding factors, including inv(16), is essential to render a complete diagnosis, predict prognosis, and guide therapy.
The variants of fusion transcripts, including type E, are rare, but recurrent and important to recognize. Type E variant cases are associated with certain unique features. They are found more frequently in therapy-related AML, have lower white blood cell counts at diagnosis, often lack abnormal eosinophil cytomorphology, and appear to be mutually exclusive with KIT mutations. Nevertheless, in the absence of concomitant KIT mutation, there is no difference in overall survival or event-free survival compared to variant A (Schnittger et al. 2007; Schwind et al. 2013). The case under discussion showed typical features of inv(16) type E AML. The patient was initially diagnosed as therapy-related AML. Following the results of the leukemia translocation panel, inv(16) transcript variant E was detected. Because the transcript was detected incidentally in a therapy-related setting without the usual morphologic findings of AML with inv(16)(p13.1q22) or t(16;16)(13.1;q22)/CBFB–MYH11, we performed FISH studies on the bone marrow aspirate smear for confirmation. The results showed a split signal in 176 of 200 (88%) interphases consistent with CBFB rearrangement. The final diagnosis was therapy-related AML with inv(16)(p13.1q22)/CBFB–MYH11. None of the features of the clinical presentation nor morphologic findings were suggestive of an underlying core-binding factor AML. The presence of add (5p) was consistent with t-AML, without raising suspicion for the presence of inv(16), although absence of concurrent TP53 mutation (or KMT2A fusion) was unusual. In this case scenario, the only way to detect inv(16) was by routine screening for fusions, either by FISH or molecular techniques.
This case illustrates the important role of molecular diagnostic testing in making accurate diagnosis, prognostication, appropriate treatment. and guiding subsequent follow-up testing for minimal residual testing assessment. In the era of personalization of medicine, core-binding factor AML is a diagnosis that cannot be missed. This has implications on diagnostic subclassification and prognostic risk stratification. Even in a therapy-related setting, AML with inv(16) has improved outcomes compared to therapy-related AML (t-AML) without recurrent genetic abnormalities (Andersen et al. 2002; Kern et al. 2004; Schoch et al. 2004; Armand et al. 2007; Aldoss and Pullarkat 2012; Bueso-Ramos et al. 2015). AML patients with inv(16) have improved event-free survival with the FLAG regimen (Borthakur et al. 2008).
Therapy for a patient with t-AML is typically tailored to each individual based on cytogenetic risk and performance status (Dhakal et al. 2020). Intensive chemotherapy, including CPX-351 or 7 + 3, is typically used in patients with good performance status, whereas older adults or patients with poor performance status may benefit more from low-intensity regimens (hypomethylating agent or low-dose cytarabine with or without venetoclax) (Boddu et al. 2017; Lancet et al. 2018; DiNardo et al. 2019; Dhakal et al. 2020). Importantly, in our patient, the detection of inv(16) resulted in induction therapy with cladribine, idarubicin, and cytarabine and consolidation with a high-dose cytarabine based regimen, essentially FLAG (fludarabine, high-dose cytarabine, and granulocyte colony-stimulating factor). In this subset of AML, chemotherapy with a higher dose Ara-C has been shown to significantly improve patient outcomes (Bloomfield et al. 1998; Byrd et al. 2004; Schlenk et al. 2004; Marcucci et al. 2005; Lowenberg et al. 2011; Dohner et al. 2017). Performing an individual FISH probe for a specific translocation, such as for rearrangements of PML/RARA or BCR/ABL, is a fast and effective method if it is suspected clinically or pathologically. However, performing a targeted FISH test for the multiple recurrent translocations recognized by the WHO would be laborious and time-consuming. It is not practical or cost-effective to perform individual FISH or RT-PCR tests to screen for each recurrent translocation; these are more effectively used as specific confirmation methods or for monitoring once identified. In regard to cost, the expense of a FISH probe labeled with two colors (either a CBFB break-apart probe or a CBFB–MYH11 fusion probe) is usually ∼$100–$200/test. To test for the most common translocations in AML would likely cost ∼$800. In contrast, the LTP costs ∼$700 per patient. In addition to the difference in testing cost, as mentioned above, performing FISH can involve more manual labor in both the technical aspect and interpretation of the tests, which ultimately may require more personnel full-time equivalents (FTEs). The LTP is more automated and less labor-intensive and can also potentially save personnel cost. Nonetheless, a limitation of the LTP and other RNA-based assays is the need for a fresh sample from which to extract the RNA. However, this is typically not difficult to obtain from bone marrow or peripheral blood for new patients presenting with suspected leukemia. Although the LTP has proven to be a versatile assay to screen for multiple leukemia-associated recurring translocation, it is unable to detect translocations involving different positions or with different partners. As such, because KMT2A/MLL can have multiple different partners, we typically use FISH break-apart probes in order to screen for KMT2A/MLL rearrangement if suspected.
In terms of minimal residual disease (MRD) monitoring, it is important to be aware of these variant transcripts at baseline and follow up using an RT-PCR assay that is able to identify these variant transcripts or FISH if such an assay is not available. This will avoid false-negative results.
In summary, for this patient, a simple molecular screening assay identified inv(16) in an unusual setting and provided accurate prognostic information and guided the use of an appropriate therapeutic regimen. inv(16) is subtle and difficult to detect by conventional karyotype, whereas FISH testing is generally initiated based on morphologic clues that were absent in this case. Hence, a screening assay for recurrent translocations played a critical role in identification of the transcript. This case highlights the importance of a multimodal approach and underscores the important part that molecular tests can have in patient management and their growing role in personalized medicine.
METHODS
Bone Marrow Evaluation
Aspirate smears and a touch imprint were stained using Wright–Giemsa stain. The biopsy, following decalcification, and clot section were fixed in formalin and submitted for morphologic and immunohistochemical evaluation. Two hematopathologists independently reviewed the morphologic findings. Flow cytometry immunophenotypic, molecular, and cytogenetic analyses were performed on fresh material prepared from the aspiration specimen.
Flow Cytometry Immunophenotypic Studies
Immunophenotypic analysis using multicolor flow cytometry was performed on bone marrow (BM) aspirates using a FACScan instrument (Becton-Dickinson) as described previously (Kanagal-Shamanna et al. 2011). The blast population was gated using right-angle side scatter and CD45 expression. The panel of monoclonal antibodies included reagents specific for CD2, CD3, CD4, CD5, CD7, CD10, CD11a, CD13, CD14, CD15, CD19, CD22, CD33, CD34, CD36, CD38, CD41, CD45, CD49d, CD56, CD64, CD117, CD123, CD184, HLA-DR, TdT, and myeloperoxidase.
Leukemia Translocation Panel
The BioMark HD (Fluidigm Biosystems) system uses integrated fluidic circuit technology combined with real time PCR. Reverse transcription of RNA to cDNA is followed by preamplification with pooled analytes (using 1.25 µL). The preamplification is diluted 1:5, and 2.7 µL is plated on a 96-well plate with 3.3 µL of 2× TaqMan Universal PCR Master Mix (Applied Biosystems PN 4304437). The 20× targets are diluted with 2× Assay Loading Reagent (Fluidigm PN 85000736) to 10× and plated on a 96-well plate. The microfluidic plate is prepared and pressurized. The analytes and preamplified cDNA are loaded into the microfluidic plate inlets. The plate is pressurized to migrate nanoliter amounts of analyte and samples toward the center where mixing occurs. The plate is placed in the BioMark for RT-qPCR. This results in simultaneous amplification of the following translocations associated with acute leukemia: t(8;21)(q22;q22); RUNX1–RUNX1T, inv(16)(p13.1q22) or t(16;16)(p13.1;q22); CBFB–MYH11 variant A, inv(16)(p13.1q22) or t(16;16)(p13.1;q22); CBFB–MYH11 variant D, inv(16)(p13.1q22) or t(16;16)(p13.1;q22); CBFB–MYH11 variant E, t(15;17)(q22;q12); PML–RARA long form, t(15;17)(q22;q12); PML–RARA short form, t(15;17)(q22;q12); PML–RARA alternative form, t(9;22)(q34;q11.2); BCR–ABL1 b2a2, t(9;22)(q34;q11.2); BCR–ABL1 b3a2, t(9;22)(q34;q11.2); BCR–ABL1 e1a2, t(12;21)(p13.1;q22); ETV6–RUNX1, t(1;19)(q23;p13.3); E2A–PBX1, and t(4;11)(q21;q23); MLL–AF4 and t(6;9)(p23;q34); DEK–NUP214. Please refer to Supplemental Table 1 for a list of primers used.
For the sensitivity control pool, we used RNA from eight cell lines or patient samples, listed as follows: K562 (cell line positive for B3A2), KBM7 (cell line positive for B2A2), B15 (cell line positive for E1A2), NB4 (cell line positive for RARA-LF [long form]), SF (patient sample positive for RARA-SF [short form]), INV16 (purchased RNA from Invivoscribe, positive for INV16-A), and Kasumi (cell line positive for AML1-ETO). HL60 was used as the negative cell line control, and deionized DNA-free, DNase-free, RNase-free water was used as the reagent control. The analytical sensitivity of this assay was 0.1%–0.01% as determined by internal validation studies. All samples were run in triplicate. For a valid run, the cycle to threshold (Ct) values for pooled sensitivity controls should be ≤25 cycles, the Ct value for the negative control (HL-60) should be >30 cycles, and the Ct values for reagent controls should be >30 cycles. A patient result is considered positive if the Ct value was ≤25, and all samples in triplicate have a nearly identical Ct value.
NGS-Based Mutation Analysis
Targeted amplicon-based somatic mutation analysis was performed using 250 ng of genomic DNA extracted from the BM aspirate and a 28-gene NGS panel as described elsewhere (Quesada et al. 2020). This custom-designed panel detects mutations in the entire coding sequences of the following genes: ABL1, ASXL1, BRAF, DNMT3A, EGFR, EZH2, FLT3, GATA1, GATA2, HRAS, IDH1, IDH2, IKZF2, JAK2, KIT, KRAS, MDM2, MLL, MPL, MYD88, NOTCH1, NPM1, NRAS, PTPN11, RUNX1, TET2, TP53, and WT1 (coverage details provided in Supplemental Table 2). After library preparation, sequencing was performed using an Illumina MiSeq sequencer per the manufacturer's instructions. MiSeq Reporter Software was used for alignment to the reference genome (GRCh37/hg19), and variant calling was performed using OncoSeek, an in-house developed variant caller software (Routbort et al. 2012). The Integrative Genomics Viewer (IGV) was used to visualize the calls (Robinson et al. 2011). A minimum of 250× bidirectional coverage was required for variant calling, to achieve a minimum 5% allelic burden.
Fluorescence In Situ Hybridization
The in situ hybridization (ISH) technique was performed using a CBFB inv(16) dual-color break-apart DNA probe from Abbott Molecular, Inc. This probe hybridizes to the band 16q22 (spectrum orange on the 5' centromeric side and spectrum green on the 3' telomeric side of the CBFB breakpoint). Two hundred interphases were analyzed. FISH testing will not be able to identify which isoform(s) of CBFB–MYH11 transcript are involved. Our laboratory uses a CBFB break-apart probe (BAP) to detect the very rare instances in which the CBFB rearrangement involves a partner gene(s) other than MYH11. A split signal was observed in 176 interphases and intact signals in 24 interphases. These findings indicate that 88% of the cells are positive for CBFB rearrangement. The normal cutoff using a CBFB probe established at 95% (P < 0.05) confidence level in the Cytogenetics Laboratory is 4.2% for the CBFB gene rearrangement (1R1G1F), 5.0% for deletion of a CBFB gene (1F), and 3.2% for gain an extra CBFB gene (3F).
Conventional Karyotype Analysis
Conventional chromosomal analysis was performed on G-banded metaphase cells prepared from unstimulated 24- and 48-h BM aspirate cultures using standard techniques described previously (Tang et al. 2009). The median number of metaphases analyzed was 20 (range, 15–30). The karyotype was documented according to the International System for Human Cytogenetic Nomenclature (ISCN 2016) (Tang et al. 2009; Kanagal-Shamanna et al. 2011; McGowan-Jordan et al. 2016).
ADDITIONAL INFORMATION
Data Deposition and Access
Both variants reported have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession numbers VCV000012582 and VCV000016276.
Ethics Statement
The study was performed following the institutional approved protocol in accordance with the Declaration of Helsinki. Informed consent was obtained. Testing was undertaken as part of the clinical workup.
Author Contributions
R.K.-S. designed the study; A.E.Q. and R.K.-S. wrote and revised the manuscript; R.L., E.J., K.P.P., Z.T., H.A., S.M., J.D.K., G.G.-M., G.M.-B., and L.J.M. provided the study patient and related clinical and pathologic data; and all authors contributed to scientific discussions and revised and approved the final manuscript.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Linda B. Baughn
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Aldoss I, Pullarkat V. 2012. Therapy-related acute myeloid leukemia with favorable cytogenetics: still favorable? Leuk Res 36: 1547–1551. 10.1016/j.leukres.2012.09.008 [DOI] [PubMed] [Google Scholar]
- Andersen MK, Larson RA, Mauritzson N, Schnittger S, Jhanwar SC, Pedersen-Bjergaard J. 2002. Balanced chromosome abnormalities inv(16) and t(15;17) in therapy-related myelodysplastic syndromes and acute leukemia: report from an international workshop. Genes Chromosomes Cancer 33: 395–400. 10.1002/gcc.10043 [DOI] [PubMed] [Google Scholar]
- Arahata M, Shimizu Y, Asakura H, Nakao S. 2015. Persistent molecular remission of refractory acute myeloid leukemia with inv(16)(p13.1q22) in an elderly patient induced by cytarabine ocfosfate hydrate. J Hematol Oncol 8: 5. 10.1186/s13045-014-0100-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. 2016. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127: 2391–2405. 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- Armand P, Kim HT, DeAngelo DJ, Ho VT, Cutler CS, Stone RM, Ritz J, Alyea EP, Antin JH, Soiffer RJ. 2007. Impact of cytogenetics on outcome of de novo and therapy-related AML and MDS after allogeneic transplantation. Biol Blood Marrow Transplant 13: 655–664. 10.1016/j.bbmt.2007.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur DC, Bloomfield CD. 1983. Partial deletion of the long arm of chromosome 16 and bone marrow eosinophilia in acute nonlymphocytic leukemia: a new association. Blood 61: 994–998. 10.1182/blood.V61.5.994.994 [DOI] [PubMed] [Google Scholar]
- Bloomfield CD, Shuma C, Regal L, Philip PP, Hossfeld DK, Hagemeijer AM, Garson OM, Peterson BA, Sakurai M, Alimena G, et al. 1997. Long-term survival of patients with acute myeloid leukemia: a third follow-up of the Fourth International Workshop on Chromosomes in Leukemia. Cancer 80: 2191–2198. [DOI] [PubMed] [Google Scholar]
- Bloomfield CD, Lawrence D, Byrd JC, Carroll A, Pettenati MJ, Tantravahi R, Patil SR, Davey FR, Berg DT, Schiffer CA, et al. 1998. Frequency of prolonged remission duration after high-dose cytarabine intensification in acute myeloid leukemia varies by cytogenetic subtype. Cancer Res 58: 4173–4179. [PubMed] [Google Scholar]
- Boddu PC, Kantarjian HM, Ravandi F, Garcia-Manero G, Verstovsek S, Jabbour EJ, Takahashi K, Bhalla K, Konopleva M, DiNardo CD, et al. 2017. Characteristics and outcomes of older patients with secondary acute myeloid leukemia according to treatment approach. Cancer 123: 3050–3060. 10.1002/cncr.30704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthakur G, Kantarjian H, Wang X, Plunkett WK Jr, Gandhi VV, Faderl S, Garcia-Manero G, Ravandi F, Pierce S, Estey EH. 2008. Treatment of core-binding-factor in acute myelogenous leukemia with fludarabine, cytarabine, and granulocyte colony–stimulating factor results in improved event-free survival. Cancer 113: 3181–3185. 10.1002/cncr.23927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso-Ramos CE, Kanagal-Shamanna R, Routbort MJ, Hanson CA. 2015. Therapy-related myeloid neoplasms. Am J Clin Pathol 144: 207–218. 10.1309/AJCPU1JO2LYTWUAV [DOI] [PubMed] [Google Scholar]
- Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, et al. 2002. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 100: 4325–4336. 10.1182/blood-2002-03-0772 [DOI] [PubMed] [Google Scholar]
- Byrd JC, Ruppert AS, Mrózek K, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Stamberg J, Koduru PR, Moore JO, et al. 2004. Repetitive cycles of high-dose cytarabine benefit patients with acute myeloid leukemia and inv(16)(p13q22) or t(16;16)(p13;q22): results from CALGB 8461. J Clin Oncol 22: 1087–1094. 10.1200/JCO.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Delaunay J, Vey N, Leblanc T, Fenaux P, Rigal-Huguet F, Witz F, Lamy T, Auvrignon A, Blaise D, Pigneux A, et al. 2003. Prognosis of inv(16)/t(16;16) acute myeloid leukemia (AML): a survey of 110 cases from the French AML Intergroup. Blood 102: 462–469. 10.1182/blood-2002-11-3527 [DOI] [PubMed] [Google Scholar]
- Dhakal P, Pyakuryal B, Pudasainee P, Rajasurya V, Gundabolu K, Bhatt VR. 2020. Treatment strategies for therapy-related acute myeloid leukemia. Clin Lymphoma Myeloma Leuk 20: 147–155. 10.1016/j.clml.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, Frankfurt O, Konopleva M, Wei AH, Kantarjian HM, et al. 2019. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 133: 7–17. 10.1182/blood-2018-08-868752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, et al. 2017. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129: 424–447. 10.1182/blood-2016-08-733196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández JM, González MB, Granada I, Gutiérrez N, Chillón C, Ramos F, Ribera JM, González M, Feliu E, San Miguel J. 2000. Detection of inv(16) and t(16;16) by fluorescence in situ hybridization in acute myeloid leukemia M4Eo. Haematologica 85: 481–485. [PubMed] [Google Scholar]
- Juliusson G, Antunovic P, Derolf A, Lehmann S, Mollgard L, Stockelberg D, Tidefelt U, Wahlin A, Hoglund M. 2009. Age and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish Acute Leukemia Registry. Blood 113: 4179–4187. 10.1182/blood-2008-07-172007 [DOI] [PubMed] [Google Scholar]
- Kanagal-Shamanna R, Bueso-Ramos CE, Barkoh B, Lu G, Wang S, Garcia-Manero G, Vadhan-Raj S, Hoehn D, Medeiros LJ, Yin CC. 2011. Myeloid neoplasms with isolated isochromosome 17q represent a clinicopathologic entity associated with myelodysplastic/myeloproliferative features, a high risk of leukemic transformation, and wild-type TP53. Cancer 118: 2879–2888. 10.1002/cncr.26537 [DOI] [PubMed] [Google Scholar]
- Kanagal-Shamanna R, Luthra R, Yin CC, Patel KP, Takahashi K, Lu X, Lee J, Zhao C, Stingo F, Zuo Z, et al. 2016a. Myeloid neoplasms with isolated isochromosome 17q demonstrate a high frequency of mutations in SETBP1, SRSF2, ASXL1 and NRAS. Oncotarget 7: 14251–14258. 10.18632/oncotarget.7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagal-Shamanna R, Singh RR, Routbort MJ, Patel KP, Medeiros LJ, Luthra R. 2016b. Principles of analytical validation of next-generation sequencing based mutational analysis for hematologic neoplasms in a CLIA-certified laboratory. Expert Rev Mol Diagn 16: 461–472. 10.1586/14737159.2016.1142374 [DOI] [PubMed] [Google Scholar]
- Kern W, Haferlach T, Schnittger S, Hiddemann W, Schoch C. 2004. Prognosis in therapy-related acute myeloid leukemia and impact of karyotype. J Clin Oncol 22: 2510–2511. 10.1200/JCO.2004.99.301 [DOI] [PubMed] [Google Scholar]
- Lancet JE, Uy GL, Cortes JE, Newell LF, Lin TL, Ritchie EK, Stuart RK, Strickland SA, Hogge D, Solomon SR, et al. 2018. CPX-351 (cytarabine and daunorubicin) liposome for injection versus conventional cytarabine plus daunorubicin in older patients with newly diagnosed secondary acute myeloid leukemia. J Clin Oncol 36: 2684–2692. 10.1200/JCO.2017.77.6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langabeer SE, Walker H, Gale RE, Wheatley K, Burnett AK, Goldstone AH, Linch DC. 1997. Frequency of CBFβ/MYH11 fusion transcripts in patients entered into the U.K. MRC AML trials. The MRC Adult Leukaemia Working Party. Br J Haematol 96: 736–739. 10.1046/j.1365-2141.1997.d01-2096.x [DOI] [PubMed] [Google Scholar]
- Larson RA. 2006. Should therapy-related myeloid leukemia be treated like de novo acute myeloid leukemia? Haematol Rep 2: 66–70. [Google Scholar]
- Larson RA, Williams SF, Le Beau MM, Bitter MA, Vardiman JW, Rowley JD. 1986. Acute myelomonocytic leukemia with abnormal eosinophils and inv(16) or t(16;16) has a favorable prognosis. Blood 68: 1242–1249. 10.1182/blood.V68.6.1242.1242 [DOI] [PubMed] [Google Scholar]
- Liu P, Tarlé SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, Collins FS. 1993. Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science 261: 1041–1044. 10.1126/science.8351518 [DOI] [PubMed] [Google Scholar]
- Liu PP, Hajra A, Wijmenga C, Collins FS. 1995. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood 85: 2289–2302. 10.1182/blood.V85.9.2289.bloodjournal8592289 [DOI] [PubMed] [Google Scholar]
- Lowenberg B, Pabst T, Vellenga E, van Putten W, Schouten HC, Graux C, Ferrant A, Sonneveld P, Biemond BJ, Gratwohl A, et al. 2011. Cytarabine dose for acute myeloid leukemia. N Engl J Med 364: 1027–1036. 10.1056/NEJMoa1010222 [DOI] [PubMed] [Google Scholar]
- Marcucci G, Mrózek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, Mayer RJ, Pettenati MJ, Powell BL, Edwards CG, et al. 2005. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol 23: 5705–5717. 10.1200/JCO.2005.15.610 [DOI] [PubMed] [Google Scholar]
- Marlton P, Keating M, Kantarjian H, Pierce S, O'Brien S, Freireich EJ, Estey E. 1995. Cytogenetic and clinical correlates in AML patients with abnormalities of chromosome 16. Leukemia 9: 965–971. [PubMed] [Google Scholar]
- McGowan-Jordan J, Simons A, Schmid M. 2016. ISCN: an international system for human cytogenomic nomenclature (2016). Karger, Basel. [Google Scholar]
- Prebet T, Boissel N, Reutenauer S, Thomas X, Delaunay J, Cahn JY, Pigneux A, Quesnel B, Witz F, Thepot S, et al. 2009. Acute myeloid leukemia with translocation (8;21) or inversion (16) in elderly patients treated with conventional chemotherapy: a collaborative study of the French CBF-AML intergroup. J Clin Oncol 27: 4747–4753. 10.1200/JCO.2008.21.0674 [DOI] [PubMed] [Google Scholar]
- Quesada AE, Montalban-Bravo G, Luthra R, Patel KP, Sasaki K, Bueso-Ramos CE, Khoury JD, Routbort MJ, Bassett R, Hidalgo-Lopez JE, et al. 2020. Clinico-pathologic characteristics and outcomes of the World Health Organization (WHO) provisional entity de novo acute myeloid leukemia with mutated RUNX1. Mod Pathol 33: 1678–1689. 10.1038/s41379-020-0531-2 [DOI] [PubMed] [Google Scholar]
- Reilly JT. 2005. Pathogenesis of acute myeloid leukaemia and inv(16)(p13;q22): a paradigm for understanding leukaemogenesis? Br J Haematol 128: 18–34. 10.1111/j.1365-2141.2004.05236.x [DOI] [PubMed] [Google Scholar]
- Ritter M, Thiede C, Schakel U, Schmidt M, Alpen B, Pascheberg U, Mohr B, Ehninger G, Neubauer A. 1997. Underestimation of inversion (16) in acute myeloid leukaemia using standard cytogenetics as compared with polymerase chain reaction: results of a prospective investigation. Br J Haematol 98: 969–972. 10.1046/j.1365-2141.1997.2933107.x [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routbort MJ, Handal B, Patel K, Singh RR, Aldape K, Reddy N, Barkoh B, Riben M, Medeiros LJ, Luthra R, et al. 2012. OncoSeek: a versatile annotation and reporting system for next generation sequencing-based clinical mutation analysis of cancer specimens. J Mol Diagn 14: 637–748. 10.1016/S1525-1578(12)00211-5 [DOI] [Google Scholar]
- Schlenk RF, Benner A, Krauter J, Buchner T, Sauerland C, Ehninger G, Schaich M, Mohr B, Niederwieser D, Krahl R, et al. 2004. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol 22: 3741–3750. 10.1200/JCO.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Schnittger S, Bacher U, Haferlach C, Kern W, Haferlach T. 2007. Rare CBFB-MYH11 fusion transcripts in AML with inv(16)/t(16;16) are associated with therapy-related AML M4eo, atypical cytomorphology, atypical immunophenotype, atypical additional chromosomal rearrangements and low white blood cell count: a study on 162 patients. Leukemia 21: 725–731. 10.1038/sj.leu.2404531 [DOI] [PubMed] [Google Scholar]
- Schoch C, Kern W, Schnittger S, Hiddemann W, Haferlach T. 2004. Karyotype is an independent prognostic parameter in therapy-related acute myeloid leukemia (t-AML): an analysis of 93 patients with t-AML in comparison to 1091 patients with de novo AML. Leukemia 18: 120–125. 10.1038/sj.leu.2403187 [DOI] [PubMed] [Google Scholar]
- Schwind S, Edwards CG, Nicolet D, Mrózek K, Maharry K, Wu YZ, Paschka P, Eisfeld AK, Hoellerbauer P, Becker H, et al. 2013. inv(16)/t(16;16) acute myeloid leukemia with non–type A CBFB-MYH11 fusions associate with distinct clinical and genetic features and lack KIT mutations. Blood 121: 385–391. 10.1182/blood-2012-07-442772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff SA, Meyers S, Hiebert SW, Raimondi SC, Head DR, Willman CL, Wolman S, Slovak ML, Carroll AJ, Behm F, et al. 1995. Heterogeneity in CBFβ/MYH11 fusion messages encoded by the inv(16)(p13q22) and the t(16;16)(p13;q22) in acute myelogenous leukemia. Blood 85: 3695–3703. 10.1182/blood.V85.12.3695.bloodjournal85123695 [DOI] [PubMed] [Google Scholar]
- Tang JL, Hou HA, Chen CY, Liu CY, Chou WC, Tseng MH, Huang CF, Lee FY, Liu MC, Yao M, et al. 2009. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood 114: 5352–5361. 10.1182/blood-2009-05-223784 [DOI] [PubMed] [Google Scholar]
- van der Reijden BA, Lombardo M, Dauwerse HG, Giles RH, Muhlematter D, Bellomo MJ, Wessels HW, Beverstock GC, van Ommen GJ, Hagemeijer A, et al. 1995. RT-PCR diagnosis of patients with acute nonlymphocytic leukemia and inv(16)(p13q22) and identification of new alternative splicing in CBFB-MYH11 transcripts. Blood 86: 277–282. 10.1182/blood.V86.1.277.bloodjournal861277 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Both variants reported have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession numbers VCV000012582 and VCV000016276.