Abstract
Variants in the X-linked gene AIFM1 (apoptosis-inducing factor mitochondria-associated 1) are associated with a highly variable clinical presentation that encompasses motor neuropathy, ataxia, encephalopathies, deafness, and cognitive impairment. AIFM1 encodes a mitochondrial flavin adenine dinucleotide (FAD)-dependent nicotinamide adenine dinucleotide (NADH) oxidoreductase, with roles in the regulation of respiratory complex assembly and function, production of reactive oxygen species, and the coordination of a caspase-independent type of apoptosis known as parthanatos. In this report, we describe a missense AIFM1 variant (absent in reference population databases; c.506C > T, p.Pro169Leu) identified in the proband and sibling of a family with three affected males. The proband, his brother, and their maternal uncle all exhibited severe multisystem pathology, metabolic acidosis, and early demise. Metabolic testing on the proband revealed normal activity of the pyruvate dehydrogenase complex in skin fibroblasts. Absent or partial deficiency of cytochrome c oxidase was found in muscle fibers, however, supporting a Complex IV mitochondrial deficiency. Functional studies carried out on fibroblasts from the proband demonstrated reduced steady state levels of the AIFM1 protein, decreased Complex I subunit abundance, elevated sensitivity to the apoptosis inducer staurosporine, and increased nuclear condensation when grown in galactose-containing media. The reduced abundance of AIFM1 in the patient cells could not be stabilized with riboflavin or protease inhibitor treatment. Together, these findings suggest that the normal function of the AIFM1 gene product within mitochondria, and its response to apoptotic stimuli, are impaired by this variant, likely accounting for the severity of the phenotype seen in these patients. These findings also imply tissue-specific effects of this variant on different mitochondrial complexes. This study expands the genetic and phenotypic spectrum associated with AIFM1 variants, with the combination of exome sequencing and functional studies allowing a diagnosis to finally be confirmed for this family.
Keywords: congenital lactic acidosis, infantile encephalopathy, lethal infantile mitochondrial myopathy, mitochondrial encephalopathy
INTRODUCTION
The X-linked gene AIFM1 (apoptosis-inducing factor mitochondria-associated 1) encodes a 67-kDa flavin adenine dinucleotide (FAD)-dependent nicotinamide adenine dinucleotide (NADH) oxidase that localizes to mitochondria (Susin et al. 1999; Bano and Prehn 2018). Under normal conditions, AIFM1 facilitates the assembly and/or import of complexes within the respiratory chain in this organelle (McKenzie and Ryan 2010; Bano and Prehn 2018; Herrmann and Riemer 2020). In response to apoptotic stimuli, however, AIFM1 is released into the cytosol following proteolytic cleavage of its amino-terminal sequence. This form of the enzyme translocates into the nucleus where it participates in a caspase-independent form of DNA fragmentation and programmed cell death referred to as parthanatos (Candé et al. 2002; Lipton and Bossy-Wetzel 2002; Cregan et al. 2004; Wang and Ge 2020). Its capacity to trigger cell death is controlled in part by its association with other proteins including HSP70 and cyclophilin A (Candé et al. 2004). The protein contains multiple conserved domains including two FAD domains and an NADH binding domain, and its function is controlled in part by redox-dependent conformational changes (Herrmann and Riemer 2020; Novo et al. 2020).
AIFM1 missense mutations are associated with a range of pathologies in humans, including progressive encephalopathies, auditory neuropathy, and intellectual disability (Cowchock et al. 1985; Ghezzi et al. 2010; Rinaldi et al. 2012; Ardissone et al. 2015; Diodato et al. 2016; Mierzewska et al. 2017; Miyake et al. 2017; Morton et al. 2017; Heimer et al. 2018; Bogdanova-Mihaylova et al. 2019; Kawarai et al. 2020; Pandolfo et al. 2020). Nearly 40 different variants have been reported to date with some identified in patients with Charcot–Marie–Tooth (CMT) type 4 or auditory neuropathy. No frameshift or nonsense variants have been reported in the Human Gene Mutation Database (HGMD), and the gene is highly intolerant to loss of function (pLI = 1 in gnomAD). Correlations drawn from reported cases suggest that the phenotypic severity and tissues affected may relate to the different cell functions (mitochondrial respiration, regulation of cell death) (Bano and Prehn 2018). Although a complete appreciation of how different AIFM1 functions are balanced in different organ systems is still lacking, functional assessment of new variants can offer important clues.
Here we report the characterization of an AIFM1 variant (c.506C > T, p.Pro169Leu) in a family with three affected males whose clinical presentation lies within the most severe that have been described. All three exhibited multisystem pathology, metabolic acidosis, and early demise. Our functional characterization of fibroblasts from the proband revealed reduced steady state levels of the AIFM1 protein, Complex I deficiency, and an increase in staurosporine- and galactose-induced nuclear condensation. Combined with metabolic findings in the proband, these data implicate that the normal function of AIFM1 within mitochondria and its roles in the regulation of cell death are both likely affected. The impact of this variant in relationship to other known AIFM1 variants is discussed.
RESULTS
Human Subjects Ethics approval was obtained from the Duke University Medical Center Institutional Review Board (Pro00014158). Informed consent was obtained by the family of the affected individuals in this study.
Clinical Presentation and Family History
The proband (P1) presented 12 yr ago. Prenatal ultrasound at 25 wk gestation found bilateral mildly dilated cerebral lateral ventricles with “teardrop” conformation consistent with colpocephaly, partial agenesis of the corpus callosum, pyelectasis, asymmetric growth restriction (<3%), and polyhydraminios. Fetal heart echocardiogram identified biventricular myocardial hypertrophy.
Delivery by cesarean section occurred at 37 wk. Apgar scores were 2 (1 min) and 7 (5 min). Reflexes could not be elicited. His tone and respiratory and heart rates remained depressed. He was small for gestational age, at 1370 g, length 39.5 cm, and head circumference (HC) 29 cm (all <3%), and was intubated immediately after birth and transferred to neonatal intensive care on a ventilator. His whole blood lactic acid at delivery was 16.0 mmol/L, pH 7.12 mmol/L, CO2 13 mmol/L, and glucose 301 mg/dL. He had severe metabolic acidosis, lactic acidosis, and an elevated anion gap. Neurologic examination was notable for poor tone and slow reflexes and magnetic resonance imaging (MRI) could not be performed. On exam, he was noted to have a high arched palate, frontal bossing, hypoplastic left fifth digit distal phalange, hypospadias, and undescended testes. He became bradycardic and continued to deteriorate on maximum ventilation and died on day 8 of life.
Muscle Biopsy
On muscle, the hematoxylin and eosin (H&E) staining revealed normal fiber size and shape. Moreover, trichrome staining failed to show any mitochondrial aggregates, nemaline rods, or fibrosis. Phosphorylase activity and alkaline phosphatase staining were normal but staining for cytochrome c oxidase was negative. Visualization by electron microscopy showed mitochondria with normal ultrastructural features and no indication of glycogen or lipid storage.
Autopsy
The proband was confirmed to have teardrop formation of the lateral ventricles with cavum septum pellucidum with intraventricular fusion of the ependyma and the fornix. There was abnormal development and migration of the periventricular germinal matrix with agenesis of the posterior half of the corpus callosum. He had severe type 2 astrocyte hyperplasia with focal cystic degeneration of the occipital white matter and intraventricular germinal matrix hemorrhage, left greater than right. He had a coarctation of the aorta distal to the left subclavian artery, preductal, large patent ductus arteriosus (PDA), secundum atrial septal defect (ASD), and small membranous ventricular septal defect (VSD) with right ventricular hypertrophy and dilation, and right atrial dilation. There was bilateral moderate to severe hydronephrosis and an acute hepatic infarct noted with severe micro- and macrovesicular steatosis of the liver.
Metabolic Evaluation
Analysis of plasma amino acids and organic acids in the proband showed grossly elevated lactate; marked increases in ketones, branched chain ketoacids, pyruvate, and tyrosine metabolites; and modest elevations in glutamine, alanine, and valine, all supporting a diagnosis of primary lactic acidemia. The pyruvate dehydrogenase complex (PDC) in cultured skin fibroblasts was normal in relation to concurrent and prior controls and shows normal activation/inactivation after preincubation with dichloroacetate/fluoride. The activity of dihydrolipoamide dehydrogenase (the E3 component of PDC and another mitochondrial reference enzyme) was normal and the ratio of PDC/E3 was within the control range. The PDC assay in skeletal muscle was also normal. Follow-up studies revealed Complex IV cytochrome c oxidase was reduced at 2% of the mean (0.40 [range 24.4 ± 9.9 nmol/min/mg protein]) in muscle, further suggestive of a mitochondrial defect.
Genetic Testing
Sequencing of mitochondrial DNA (mtDNA) COX subunits, SURF1, SCO1, SCO2, COX, and ATP7A, as well as the X-linked COX subunit (COX7B), did not show any clinically significant findings. Mitochondrial genome sequencing identified a homoplasmic rare variant, m.9909T > C (p.Phe235Leu in COIII), which was also homoplasmic in the asymptomatic mother. Chromosome microarray was normal.
Family History
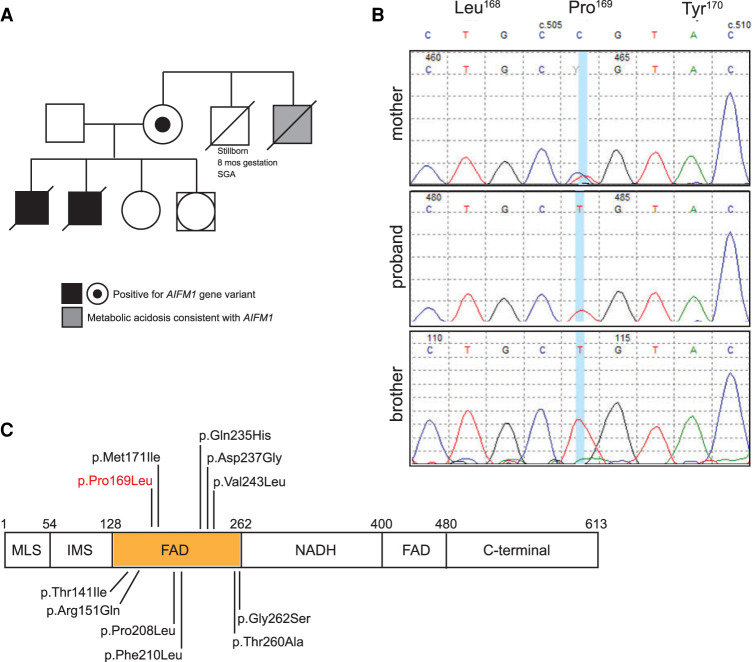
The family history was positive for a full brother and a maternal uncle, both presented with severe metabolic acidosis, lactic acidosis, and elevated anion gap (see family history in Fig. 1A). The maternal grandmother also experienced a stillborn small for gestational age (SGA) male born at 8 mo gestation, with no additional information known. Clinical information on the sibling and maternal uncle was available only through medical record review.
Figure 1.
Family history and confirmation of AIFM1 sequence variants. (A) Pedigree of the family showing the two affected brothers, the carrier mother, and the affected maternal uncle. (B) Sequence traces from the Sanger confirmation of the two affected brothers and the carrier mother. (C) Depiction of the different conserved domains in the AIFM1 protein and the location of different missense variants in the first flavin adenine dinucleotide (FAD) domain. (MLS) mitochondrial localization sequence, (IMS) intermembrane space, (NADH) nicotinamide adenine dinucleotide.
The proband's brother (P2), born 14 yr ago, 2 yr earlier than the proband, was identified on prenatal ultrasound (∼27–30 wk) with symmetric intrauterine growth restriction (IUGR) (<5%), ventriculomegaly, abnormal cerebellum, and oligohydramnios. He was delivered at 37 wk gestation by cesarean section for nonreassuring heart rate, with a birth weight of 2.17 kg (7.5%), length at 46 cm (29.5%), and HC at 32.0 cm (30.9%). Apgars were 3 (1 min), 6 (5 min), and 9 (10 min). His cord blood pH was 7.28 mmol/L and pCO2 was 45. He was hypotonic, apneic, and bradycardic and was intubated and ventilated within the first 5 min of life. On physical examination, he was noted to have low-set ears, bilateral enoph-thalmos and a prominent philtrum, undescended tests, and contracture deformity of the first interphalangeal joint of both fourth fingers and wrists. Echo identified a PDA with prominent left to right shunt, mild to moderate tricuspid regurgitation, and retroesophageal subclavian artery. His status deteriorated, ventilator support was withdrawn, and he died on day 2 of life. Muscle biopsy was not done. On autopsy the brain had an abnormal and poorly defined gray-white junction with lack of myelination of the white matter, ventricles were dilated, corpus callosum was present, and there was evidence of remote necrosis. The PDHA1 was normal on sequencing. Skin fibroblast assay of pyruvate dehydrogenase complex was normal.
The maternal uncle of the proband (P3), born 38 yr ago, 26 yr before the proband, was delivered by cesarean section at 35 wk gestation, had severe IUGR, birth weight 1.34 kg, HC 22 cm, both <3%. Apgar scores were 8 (1 min) and 9 (5 min). He was noted at 18 h of life to have severe metabolic acidosis with an anion gap. He became apneic and was intubated and ventilated. He developed seizures and had minimal spontaneous movement, and no reflexes could be elicited. Head ultrasound noted symmetrical mild prominence of the lateral ventricles. Ventilation became increasing difficult on maximum support and he died day 39 of life. His pyruvate carboxylase and pyruvate dehydrogenase were normal in skin fibroblasts, but cytochrome oxidase was deficient. The lactate to pyruvate ratio was elevated. Muscle biopsy was not done. Limited autopsy was performed, but no gross brain or liver abnormalities noted. The heart was enlarged as a result of biventricular hypertrophy with slightly patent foramen ovale and ductus arteriosus.
Genomic Analysis
In 2010, because of suspicion of an X-linked disorder, the unaffected mother of the two siblings had an X-inactivation assay completed at the Greenwood Genetic Center. The resulting X-inactivation ratio was 100:0, which is consistent with a highly skewed inactivation pattern and indicative of a potential X-linked disorder. In 2014, following up on this finding, an X-linked intellectual disability panel was also performed at the Greenwood Genetic Center on the unaffected mother (Supplemental Table 1). This analysis identified a heterozygous c.506C > T (p.Pro169Leu) alteration in the AIFM1 gene (Table 1). The change was subsequently confirmed with Sanger sequencing (Fig. 1B). DNA was available from cultured fibroblasts on the proband and his brother, who were both found to have the AIFM1 gene variant. No tissue was available on the maternal uncle to allow for investigation of his status for this gene variant. Maternal grandmother was also unavailable. Exome sequencing was performed on the proband to look for any variants of interest but did not detect any other clinically significant findings besides the AIFM1 change.
Table 1.
Variant table
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | ClinVar submission ID | Genotype | Parent of origin | Comments |
|---|---|---|---|---|---|---|---|---|---|
| AIFM1 | Xq26.1 | c.506C > T | p.Pro169Leu | Missense | Substitution | SUB9454895 | Hemizygous | Maternal | Likely pathogenic (PS3, PM1, PM2, PP3) |
At the time of identification, the c.506C > T (p.Pro169Leu) variant was absent from the 6500 samples of the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project Exome Variant Server, so the alteration was originally reported as a variant of uncertain significance. Currently, the p.Pro169Leu variant is also absent from the gnomAD population database but has been uploaded into the ClinVar database (accession number RCV001262516.1). The clinical phenotype of the patient in the ClinVar database is combined oxidative phosphorylation deficiency type 6 (COXPD6). This missense change is located in the FAD-dependent oxidoreductase domain of the AIFM1 protein, suggesting this amino acid substitution could have a negative impact on the enzymatic function. The location of this variant and others within the first FAD domain of the AIFM1 protein is shown in Figure 1C. The variant is also predicted to be damaging by SIFT, PolyPhen, and MutationTaster. Targeted testing of DNA from cultured fibroblasts and muscle biopsy tissue from the two deceased sons revealed both samples to be hemizygous for the AIFM1 c.506C > T variant. This segregation suggested the p.Pro169Leu was causative for the observed clinical picture of this family, but, without any further analysis, the alteration still remained a variant of uncertain significance.
Functional Analyses
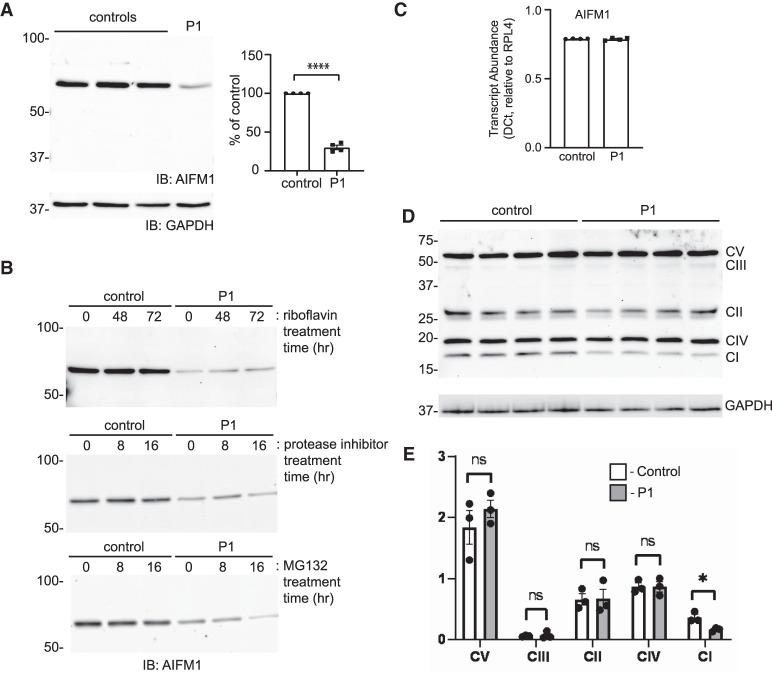
With a likely defect in mitochondrial function established from the biochemical testing, functional studies in fibroblasts from the proband were focused on delineating how the p.Pro169Leu variant impacts the AIFM1 protein, its localization, and its response to apoptotic stimuli. Western blot analysis for AIFM1 revealed an ∼70% decrease in the steady state level of the protein in the patient fibroblasts (Fig. 2A). Based on the location of the variant within the first FAD domain and prior reports that riboflavin supplementation may be beneficial for patients with AIFM1 variants (Ghezzi et al. 2010; Heimer et al. 2018), we asked whether riboflavin addition to the culture media would be able to restore stability of the AIFM1 protein. As shown in Figure 2B, we did not see any effect of 50 µM riboflavin supplementation after 48 or 72 h of treatment. To further explore the basis for the reduced level of the AIFM1 protein, cells were also treated with an EDTA-free protease inhibitor cocktail and with the proteasome inhibitor MG132. Neither inhibitor was capable of restoring the decreased steady state abundance of AIFM1 in the patient cells. Furthermore, the effect of the variant on AIFM1 levels does not appear to be acting at the level of transcript abundance or stability as quantitative polymerase chain reaction (qPCR) analysis did not reveal any differences in transcript levels between control and patient fibroblasts (Fig. 2C). Taken together, these data argue that the p.Pro169Leu variant may cause inherent instability in the AIFM1 protein that is evident when cells are analyzed biochemically.
Figure 2.
Reduced AIFM1 levels and variable decreases in respiratory chain complexes in patient fibroblasts. (A) Representative western blot of control and P1 fibroblasts and quantification of relative abundance by densitometry (n = 4). Error bar represents standard error of the mean. (B) Western blot for AIFM1 protein in control and patient cells treated with either 50 µM riboflavin for 48 and 72 h, 10 mM protease inhibitor cocktail for 8 and 16 h, or 10 mM MG132 for 8 and 16 h. (C) Quantitative polymerase chain reaction (PCR) analysis of AIFM1 transcripts in wild-type (WT) and patient fibroblasts. (D) Western blot of different electron transport chain subunits in four replicates of control and P1 fibroblast lysates. The position of each Complex subunit is marked. (E) Quantification by densitometry of the relative abundance of each subunit (n = 3). Error bars represent standard error of the mean.
Western blot analysis to look at the steady state abundance of different respiratory chain complex subunits was performed to ask how the reduction in AIFM1 protein impacts the stability/abundance of these complexes. The use of an antibody cocktail for this experiment allowed the simultaneous detection of subunits within the five complexes. As show in Figure 2D (and quantitatively in Fig. 2E), the patient fibroblasts show a reproducible decrease in the Complex I subunit NDUFB8, whereas the Complex II-V subunits were largely unchanged. In light of the striking absence of the Complex IV component cytochrome c oxidase in muscle, these data from fibroblasts indicate that the variant AIFM1 protein impacts different respiratory complex subunits in different tissues.
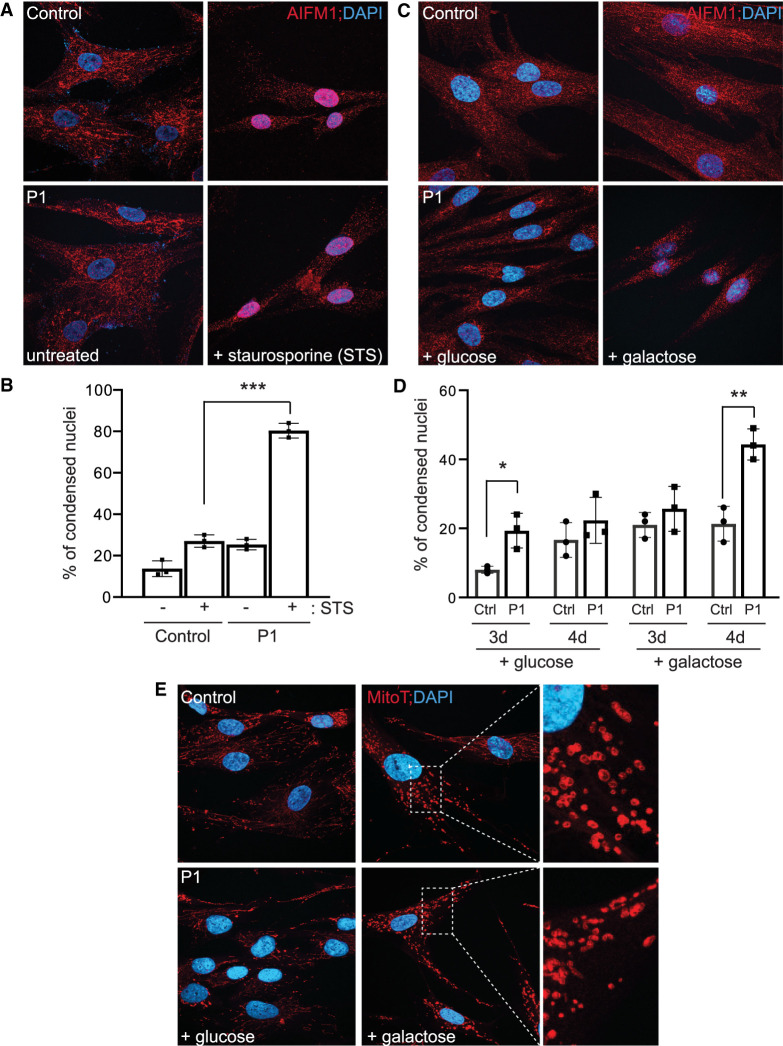
We next asked whether the p.Pro169Leu variant sensitizes the response to the apoptotic inducer, staurosporine. Control and patient fibroblasts were treated with 1 µM staurosporine for 75 min and the localization of AIFM1 was monitored, along with nuclear morphology. Under normal, untreated conditions, the variant protein localizes to the mitochondria as expected (Fig. 3A). Consistent with its demonstrated role in the caspase-independent apoptosis (e.g., parthanatos), the AIFM1 protein is lost from the mitochondria after staurosporine treatment and eventually accumulates in the nucleus, where it triggers cell death. In control cells, this nuclear localization corresponds with a change in overall cell morphology that is in line with the apoptotic phenotype. We quantified the percentage of cells with condensed nuclei in both untreated and treated control and patient cells (shown in Fig. 3B), revealing a significant increase in the percentage of patient cells with a condensed nuclei upon treatment. Despite the reduced steady state level of the protein, these results suggest that the lower amount of the AIFM1 variant protein is associated with a greater sensitivity and/or more rapid response to this apoptotic trigger. This observation aligns with similar experiments performed on other AIFM1 variants (Ghezzi et al. 2010; Rinaldi et al. 2012). In an effort to characterize both the mitochondrial dysfunction and effects on cell death caused by the variant, control and patient fibroblasts were grown in media containing either glucose- or galactose-containing media for several days followed by immunostaining for AIFM1 or MitoTracker CMX ROS staining (Fig. 3C–E). The replacement of glucose with galactose in the growth media causes the cells to rely on oxidative phosphorylation for ATP production instead of glycolysis, which may help uncover an underlying mitochondrial dysfunction under this stress. The effects of galactose might also trigger cell death albeit at a much slower rate than staurosporine treatment, allowing AIFM1 translocation to potentially be monitored. Using AIFM1 staining, we observed increased collapse of the mitochondrial network and a qualitative increase in AIFM1 protein translocation to the nucleus in patient cells after 4 d in galactose (Fig. 3C). These phenotypes corresponded with a significant elevation in the percentage of patient cells with condensed nuclei after 4 d in galactose (Fig. 3D). Substantial fragmentation of the mitochondrial network (as assessed by MitoTracker staining) was apparent in both control and patients cells after culture in galactose for 3 and 4 d. Although difficult to quantify the extent of fragmentation in these cells without the use of advanced software, MitoTracker staining did uncover an unusual morphology to the mitochondrial membrane in patient cells grown in galactose (Fig. 3E). The fragmented mitochondria were more diffuse compared to control cells, with poorly defined outer membrane morphology (see inset in Fig. 3E). The significance of this phenotype is not clear but it could reflect a defect in AIFM1-dependent assembly of mitochondrial complexes within the membrane.
Figure 3.
Increased sensitivity of patient fibroblasts to the apoptotic inducer staurosporine. (A) Immunostaining of control and P1 fibroblasts using an antibody to AIFM1 before and after treatment with staurosporine. DAPI is used as a nuclear marker. (B) Quantification of the number of condensed nuclei in untreated and staurosporine-treated control and P1 fibroblasts. *** P < 0.001. (C) Representative images of AIFM1 immunostaining in wild-type (WT) and patient fibroblasts cultured in glucose- or galactose-containing media for 96 h. (D) Quantification of the percentage of condensed nuclei under these growth conditions for both 72 and 96 h. (E) Representative images of MitoTracker CMX ROS Red staining in cells incubated with glucose or galactose for 96 h. Enlarged inset is provided to highlight the morphology of the fragmented mitochondria.
DISCUSSION
In this report, we describe a family with three affected males, two of whom were confirmed to bear an AIFM1 variant (c.506C > T; p.Pro169Leu). All died early in life, placing their course among the most severe cases associated with this gene that have been reported. Initial diagnostic efforts for this family focused on finding the cause for their severe lactic acidosis. Biochemical analysis of the skeletal muscle and skin fibroblasts from the first patient in this study demonstrated normal pyruvate dehydrogenase complex activity but deficiency in Complex IV or cytochrome c oxidase. Ultimately, targeted NGS testing was performed, revealing the hemizygous variant in P1 and his brother, and subsequent functional studies were carried out to add further insight into pathogenicity. Collectively, these efforts have confirmed a formal diagnosis for this family 38 yr after the first case (the maternal uncle of the two siblings) was identified.
Prior studies on AIFM1 variants have demonstrated variable effects on Complex I, III, or IV level/activity that likely arise from AIFM1's proposed role in assembly of the electron transport chain complexes (Calvo et al. 2010; Kettwig et al. 2015; Diodato et al. 2016; Pronicka et al. 2016; Morton et al. 2017). In this case, effects appear to be limited to Complex IV in muscle and Complex I in fibroblasts, revealing possible tissue specificity with regard to the affected complex. Despite the reduction in Complex I in the patient fibroblasts, we were unable to specifically link this decrease to any obvious impairment in mitochondrial respiration or function. However, an unusual abnormality in mitochondrial morphology was observed in the patient fibroblasts following culture in galactose-containing media. The connection between these mitochondrial phenotypes and the increased sensitivity to inducers of cell death is still not fully defined. Based on the current data in fibroblasts and available metabolic information from muscle, we hypothesize that this AIFM1 variant leads to tissue-specific effects on different mitochondrial complexes, with the profound loss of Complex IV in muscle (and perhaps other tissues) providing the most relevant cause for the severe course and early demise. The development of new cell models will be required to formally test this hypothesis in future studies.
Nearly 40 AIFM1 missense variants have been reported in HGMD, spanning every conserved domain in the protein, but no robust genotype/phenotype correlations have been emerged, and only a small fraction of these variants have been studied at a functional level in patient cells. There are other reported variants in the first FAD domain with close proximity to the proline residue at 169, although most appear to be associated with more slowly progressive and attenuated forms of disease. One example is a Chinese family bearing a p.Met171Ile change in AIFM1 that presents with X-linked Charcot–Marie–Tooth type 4, a condition with childhood onset and slowly progressive motor and sensory neuropathy (Wang et al. 2018). Other missense variants with close proximity to each other, such as those at positions 492 and 493 within the second FAD domain, have been reported with substantial differences in phenotypic severity. The p.Tyr492His variant results in a severe form of the disease resembling our family whereas the p.Glu493Val is found in a patient with Cowchock syndrome (Rinaldi et al. 2012; Pronicka et al. 2016). Thus, location of the variants within different conserved domains of AIFM1 does not alone predict severity. Rather, we speculate that it is likely the extent to which the different functions that this protein carries out are affected. One consistent feature in the most severe cases where functional work was performed is a Complex IV or Complex I/IV deficiency (noted in patients with the p.Pro169Leu and p.Tyr492His variants). Whether this involvement of these complexes triggers a more rapid deterioration of mitochondrial function or simply reflects a broader impact in affected tissues is not known. A systematic analysis of these different mitochondrial complexes is warranted for future severe cases to help strengthen this correlation.
The biochemical and functional studies implicate pathogenic roles for this variant in both mitochondrial function and the regulation of cell death. As noted by other groups, our AIFM1 variant leads to a more pronounced effect on cell death following treatment with the apoptotic inducer staurosporine. As AIFM1 is required for the process of parthanatos, this finding is somewhat counterintuitive with the lower steady state levels of the protein in patient fibroblasts. Nonetheless, it is possible that variants in AIFM1 might either result in more efficient release of the protein from mitochondria or lead to enhanced binding to DNA once in the nucleus, as suggested by earlier work. We provide qualitative evidence for more efficient translocation of AIFM1 in the patient cells following galactose treatment, a slower inducer of this translocation than staurosporine. AIFM1 is proteolytically processed as part of its release from the mitochondria so it is possible that the variant could also facilitate this cleavage. Our immunostaining experiments did not reveal any obvious mitochondrial morphology defects when cells are grown in glucose-containing media (Fig. 3A). We were unable to detect the cleaved form of AIFM1 in our Western blot experiments.
Combined with the reduction in the steady state level of AIFM1 protein in patient fibroblasts, it is likely that the involvement of all these processes contributed to the early demise of the patients. Although we do not know the impact of the variant on the structure of the AIFM1 protein, at least some effect on the secondary or tertiary structure is likely evident based on the replacement of the proline residue at this position. Such conformational changes could impact the ability of the protein to bind flavins and might also cause it to be released from the mitochondria more easily following staurosporine or galactose treatment. The systematic functional analysis of these variants in patient cells provides the opportunity to better understand how the effects of different variants converge on the phenotype in patients.
METHODS
Genomic Analysis
Genomic DNA from the unaffected female was extracted from peripheral blood using conventional DNA isolation methodology. The DNA was then enriched for genes on the Greenwood Genetic Center X-linked intellectual disability panel using RainDance microdroplet technology (RainDance Technologies). Sequencing of these 90 genes was performed on an Applied Biosystems SOLiD 3 Plus Sequencer with coverage at 400× (Supplemental Table 1). Traditional Sanger sequencing was used to confirm the AIFM1 c.506C > T variant identified by next-generation sequencing (NGS). Targeted Sanger sequencing was performed on DNA from affected male fibroblast and muscle biopsy samples to detect the previously identified AIFM1 c.506C > T change. X-inactivation analysis of the androgen receptor (AR) locus was performed on genomic DNA extracted from peripheral blood of the unaffected female. The X-inactivation pattern was determined by PCR analysis of a polymorphic CAG repeat in the first exon of the AR gene. Amplification of the AR gene both before and after digestion with the methylation-sensitive HpaII restriction enzyme was used to determine the methylation status of both X chromosomes (Allen et al. 1992). pLI (probability of being loss-of-function intolerant) score is a constraint metric to define genes with a high probability of intolerance to heterozygous loss-of-function variation. pLI scores range from 0 to 1, with scores closer to 1 indicating greater intolerance to protein-truncating variation.
For exome sequencing, DNA libraries were prepared from genomic DNA isolated from the proband fibroblasts, using the Agilent SureSelectXT Clinical Research Exome v2 capture kit (Agilent Technologies). Briefly, DNA was fragmented using the Covaris ME220 system (Covaris), and fragments of 150–200 bp were selected using AMPure XP beads (Beckman Coulter). Fragments were subsequently end-repaired, adenylated at the 3′ end, ligated to sequencing adaptors, and then PCR-amplified using the SureSelectXT Library Preparation kit (Agilent Technologies) with DNA being purified using AMPure XP beads after each of these steps. The concentration of the enriched libraries was determined using Quant-IT fluorometry on the Victor Nivo Fluorometer (PerkinElmer). An amount of 750 ng of each DNA library was used for hybridization and capture with the SureSelectXT Clinical Research Exome v2 probes (Agilent Technologies). After hybridization, the RNA-bound DNA was retained, and unhybridized material was washed away. Captured fragments were amplified by PCR and purified. The quality of the enriched libraries was evaluated using a D1000 Tape on the TapeStation 4200 (Agilent Technologies). Libraries were quantified on a Victor Nivo Fluorometer (PerkinElmer) using a Quant-IT Broad Range kit (Life Technologies), and separate libraries were pooled and sequenced using an Illumina NovaSeq 6000° Sequencing System at 157× coverage (Illumina Inc.) per the manufacturer's protocol.
Cell Culture and Lysate Preparation
Fibroblast cells were cultured in DMEM (ThermoFisher 10-017-CV) supplemented with 10% fetal bovine serum (VWR 97068-085) and 1% penicillin/streptomycin (ThermoFisher 30-002-CI) in a 5% CO2 humidified tissue culture incubator. For lysate preparation, cells were released by trypsinization, and then lysed on ice in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with protease inhibitor mixture. After incubation for 30 min on ice, the lysate was vortexed, and centrifuged at 20,000g for 10 min at 4°C. Protein concentrations were quantified by Micro BCA Protein Assay (ThermoFisher #23235).
Protein Biochemistry and Western Blotting
For western blot analysis, 20 µg of cell lysate was separated on a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel. Protein was transferred to 0.45 µm pore nitrocellulose at 110 V for 2 h at 4°C. The membrane was rinsed and blocked membranes were blocked with 5% milk/TBST for 1 h at room temperature. Blots were incubated overnight with a rabbit polyclonal anti-AIF antibody (Sigma-Aldrich AB16501) at 1:1000 at 4°C, washed, and then incubated with horseradish peroxidase (HRP) conjugated anti-rabbit (1:2000) for 1 h before washing and developing with SuperSignal West Pico PLUS ECL reagent (ThermoFisher 34577). All images were captured on the Bio-Rad ChemiDoc MP Imaging System (Bio-Rad 12003154). Analysis was done with Image Lab Software (Bio-Rad 1709690, ver5.2.1). For the riboflavin supplementation and protease inhibition experiments, control and P1 fibroblasts were treated for various times prior to lysis and western blotting. To analyze the stability of Complex I-V subunits, an antibody cocktail kit (Total OXPHOS Human WB Antibody Cocktail; AbCam ab110411) was used, allowing all subunits to be resolved and detected in a single blot. Lysates (without dithiothreitol [DTT] or denaturation) were resolved on a 13.5% gel, transferred to PVDF, and incubated overnight with the primary antibodies (1:1000) followed by anti-mouse secondary (1:1000) for 2 h. This analysis was performed in triplicate and the relative abundance of each subunit determined by densitometry, using GAPDH abundance as a normalizing factor.
Immunostaining and Imaging
Fibroblasts were seeded on coverslips and cultured overnight in a 12-well plate. Seeded cells were grown in glucose- or galactose-containing media (5 mM final concentration) for different times. In other experiments, staurosporine was added at a final concentration of 1 µM to cells grown in glucose and cells were incubated for 75 min before washing and fixation in 3.7% formaldehyde for 10 min. Following incubation with primary anti-AIFM1 antibody and secondary anti-rabbit-conjugated AlexaFluor 568, the coverslips were mounted on slides using DAPI-containing ProLong Gold mounting media. For the MitoTracker staining, cells were incubated with 100 nM MitoTracker CMX ROS dye (Cell Signaling Technology 9082S) for 30 min followed by fixation using ice cold methanol and mounting using DAPI-containing ProLong Gold mounting media. Slides were imaged using an Olympus FV3000 confocal microscope. The number of condensed nuclei were quantified in three independent experiments by determining the percentage of cells with condensed or apoptotic nuclei relative to the total number of nuclei counted. For each experiment/slide, a minimum of 100 nuclei was counted.
ADDITIONAL INFORMATON
Data Deposition and Access
The variant has been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) under accession number VCV000982823.2.
Ethics Statement
Informed consent was obtained from the family of the affected individuals involved in this study. Human Subjects Ethics approval was obtained from the Duke University Medical Center Institutional Review Board (Pro00014158).
Acknowledgments
We are thankful to the family for their participation in this study.
Author Contributions
R.S., A.M.-R., and M.F. conceptualized the project; R.S., H.F.-S., R.C., A.M.-R., and M.F. curated the data; R.C. and H.F.-S. performed the formal analysis; R.S., A.M.-R., and M.F. acquired funding; T.M., M.Ma., H.F.-S., and Y.-H.J. investigated; A.M.-R., M.F., and H.F.-S. were responsible for methodology, R.S., A.M.-R., and M.F. administered the project; M.Mc., Y.-H.J., and A.M.-R. provided resources; R.S., H.F.-S., M.F., and A.M.-R. supervised the project; H.F.-S. visualized the project; R.S., A.M.-R., and R.C. wrote the original draft; and R.S. and A.M.-R. wrote, reviewed, and edited the manuscript.
Funding
This work was supported by the Greenwood Genetic Center.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. 1992. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51: 1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Ardissone A, Piscosquito G, Legati A, Langella T, Lamantea E, Garavaglia B, Salsano E, Farina L, Moroni I, Pareyson D, et al. 2015. A slowly progressive mitochondrial encephalomyopathy widens the spectrum of AIFM1 disorders. Neurology 84: 2193–2195. 10.1212/WNL.0000000000001613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano D, Prehn JHM. 2018. Apoptosis-inducing factor (AIF) in physiology and disease: the tale of a repented natural born killer. EBioMedicine 30: 29–37. 10.1016/j.ebiom.2018.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanova-Mihaylova P, Alexander MD, Murphy RP, Chen H, Healy DG, Walsh RA, Murphy SM. 2019. Clinical spectrum of AIFM1-associated disease in an Irish family, from mild neuropathy to severe cerebellar ataxia with colour blindness. J Peripher Nerv Syst 24: 348–353. 10.1111/jns.12348 [DOI] [PubMed] [Google Scholar]
- Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, Rivas M, Guiducci C, Bruno DL, Goldberger OA, et al. 2010. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet 42: 851–858. 10.1038/ng.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candé C, Cohen I, Daugas E, Ravagnan L, Larochette N, Zamzami N, Kroemer G. 2002. Apoptosis-inducing factor (AIF): a novel caspase-independent death effector released from mitochondria. Biochimie 84: 215–222. 10.1016/S0300-9084(02)01374-3 [DOI] [PubMed] [Google Scholar]
- Candé C, Vahsen N, Kouranti I, Schmitt E, Daugas E, Spahr C, Luban J, Kroemer RT, Giordanetto F, Garrido C, et al. 2004. AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene 23: 1514–1521. 10.1038/sj.onc.1207279 [DOI] [PubMed] [Google Scholar]
- Cowchock FS, Duckett SW, Streletz LJ, Graziani LJ, Jackson LG. 1985. X-linked motor-sensory neuropathy type-II with deafness and mental retardation: a new disorder. Am J Med Genet 20: 307–315. 10.1002/ajmg.1320200214 [DOI] [PubMed] [Google Scholar]
- Cregan SP, Dawson VL, Slack RS. 2004. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene 23: 2785–2796. 10.1038/sj.onc.1207517 [DOI] [PubMed] [Google Scholar]
- Diodato D, Tasca G, Verrigni D, D'Amico A, Rizza T, Tozzi G, Martinelli D, Verardo M, Invernizzi F, Nasca A, et al. 2016. A novel AIFM1 mutation expands the phenotype to an infantile motor neuron disease. Eur J Hum Genet 24: 463–466. 10.1038/ejhg.2015.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi D, Sevrioukova I, Invernizzi F, Lamperti C, Mora M, D'Adamo P, Novara F, Zuffardi O, Uziel G, Zeviani M. 2010. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am J Hum Genet 86: 639–649. 10.1016/j.ajhg.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimer G, Eyal E, Zhu X, Ruzzo EK, Marek-Yagel D, Sagiv D, Anikster Y, Reznik-Wolf H, Pras E, Oz Levi D, et al. 2018. Mutations in AIFM1 cause an X-linked childhood cerebellar ataxia partially responsive to riboflavin. Eur J Paediatr Neurol 22: 93–101. 10.1016/j.ejpn.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. 2020. Apoptosis inducing factor and mitochondrial NADH dehydrogenases: redox-controlled gear boxes to switch between mitochondrial biogenesis and cell death. Biol Chem 402: 289–297. 10.1515/hsz-2020-0254. [DOI] [PubMed] [Google Scholar]
- Kawarai T, Yamazaki H, Yamakami K, Tsukamoto-Miyashiro A, Kodama M, Rumore R, Caltagirone C, Nishino I, Orlacchio A. 2020. A novel AIFM1 missense mutation in a Japanese patient with ataxic sensory neuronopathy and hearing impairment. J Neurol Sci 409: 116584. 10.1016/j.jns.2019.116584 [DOI] [PubMed] [Google Scholar]
- Kettwig M, Schubach M, Zimmermann FA, Klinge L, Mayr JA, Biskup S, Sperl W, Gartner J, Huppke P. 2015. From ventriculomegaly to severe muscular atrophy: expansion of the clinical spectrum related to mutations in AIFM1. Mitochondrion 21: 12–18. 10.1016/j.mito.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Lipton SA, Bossy-Wetzel E. 2002. Dueling activities of AIF in cell death versus survival: DNA binding and redox activity. Cell 111: 147–150. 10.1016/S0092-8674(02)01046-2 [DOI] [PubMed] [Google Scholar]
- McKenzie M, Ryan MT. 2010. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life 62: 497–502. 10.1002/iub.335 [DOI] [PubMed] [Google Scholar]
- Mierzewska H, Rydzanicz M, Bieganski T, Kosinska J, Mierzewska-Schmidt M, Lugowska A, Pollak A, Stawinski P, Walczak A, Kedra A, et al. 2017. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with AIFM1 mutation: a novel phenotype of the mitochondrial disease. Clin Genet 91: 30–37. 10.1111/cge.12792 [DOI] [PubMed] [Google Scholar]
- Miyake N, Wolf NI, Cayami FK, Crawford J, Bley A, Bulas D, Conant A, Bent SJ, Gripp KW, Hahn A, et al. 2017. X-linked hypomyelination with spondylometaphyseal dysplasia (H-SMD) associated with mutations in AIFM1. Neurogenetics 18: 185–194. 10.1007/s10048-017-0520-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton SU, Prabhu SP, Lidov HGW, Shi J, Anselm I, Brownstein CA, Bainbridge MN, Beggs AH, Vargas SO, Agrawal PB. 2017. AIFM1 mutation presenting with fatal encephalomyopathy and mitochondrial disease in an infant. Cold Spring Harb Mol Case Stud 3: a001560. 10.1101/mcs.a001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo N, Ferreira P, Medina M. 2020. The apoptosis-inducing factor family: moonlighting proteins in the crosstalk between mitochondria and nuclei. IUBMB Life 73: 568–581. 10.1002/iub.2390. [DOI] [PubMed] [Google Scholar]
- Pandolfo M, Rai M, Remiche G, Desmyter L, Vandernoot I. 2020. Cerebellar ataxia, neuropathy, hearing loss, and intellectual disability due to AIFM1 mutation. Neurol Genet 6: e420. 10.1212/NXG.0000000000000420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronicka E, Piekutowska-Abramczuk D, Ciara E, Trubicka J, Rokicki D, Karkucinska-Wieckowska A, Pajdowska M, Jurkiewicz E, Halat P, Kosinska J, et al. 2016. New perspective in diagnostics of mitochondrial disorders: two years’ experience with whole-exome sequencing at a national paediatric centre. J Transl Med 14: 174. 10.1186/s12967-016-0930-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi C, Grunseich C, Sevrioukova IF, Schindler A, Horkayne-Szakaly I, Lamperti C, Landoure G, Kennerson ML, Burnett BG, Bonnemann C, et al. 2012. Cowchock syndrome is associated with a mutation in apoptosis-inducing factor. Am J Hum Genet 91: 1095–1102. 10.1016/j.ajhg.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, et al. 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397: 441–446. 10.1038/17135 [DOI] [PubMed] [Google Scholar]
- Wang X, Ge P. 2020. Parthanatos in the pathogenesis of nervous system diseases. Neuroscience 449: 241–250. 10.1016/j.neuroscience.2020.09.049 [DOI] [PubMed] [Google Scholar]
- Wang B, Li X, Wang J, Liu L, Xie Y, Huang S, Pakhrin PS, Jin Q, Zhu C, Tang B, et al. 2018. A novel AIFM1mutation in a Chinese family with X-linked Charcot–Marie–Tooth disease type 4. Neuromuscul Disord 28: 652–659. 10.1016/j.nmd.2018.05.008 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variant has been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) under accession number VCV000982823.2.