Abstract
Around half of the genomes in mammals are composed of transposable elements (TEs) such as DNA transposons and retrotransposons. Several mechanisms have evolved to prevent their activity and the detrimental impact of their insertional mutagenesis. Despite these potentially negative effects, TEs are essential drivers of evolution, and in certain settings, beneficial to their hosts. For instance, TEs have rewired the antiviral gene regulatory network and are required for early embryonic development. However, due to structural similarities between TE-derived and viral nucleic acids, cells can misidentify TEs as invading viruses and trigger the major antiviral innate immune pathway, the type I interferon (IFN) response. This review will focus on the different settings in which the role of TE-mediated IFN activation has been documented, including cancer and senescence. Importantly, TEs may also play a causative role in the development of complex autoimmune diseases characterized by constitutive type I IFN activation. All these observations suggest the presence of strong but opposing forces driving the coevolution of TEs and antiviral defense. A better biological understanding of the TE replicative cycle as well as of the antiviral nucleic acid sensing mechanisms will provide insights into how these two biological processes interact and will help to design better strategies to treat human diseases characterized by aberrant TE expression and/or type I IFN activation.
Keywords: mobile genetic elements, transposable elements, type I interferon, nucleic acid sensing, antiviral immunity
INTRODUCTION
Transposable elements (TEs) are a driving force in evolution but pose a potential threat to the fitness of the host by disrupting the function and/or expression of genes surrounding novel insertions or by promoting chromosomal rearrangements (Garcia-Perez et al. 2016; Bourque et al. 2018). To prevent these issues, TEs expression is generally repressed, either at the epigenetic, transcriptional, or post-transcriptional level. Intriguingly, in specific cellular contexts and developmental stages TE expression is activated and may have beneficial consequences. For instance, the RNA derived from the major mammalian retrotransposon, LINE-1, appears to be essential for maintaining the pluripotency-specific gene expression programs (Percharde et al. 2018). During viral infections the expression of TEs is also up-regulated, but it remains unclear if this is required to establish a proficient antiviral state, or is a secondary effect of the robust transcriptional response triggered upon infections (Macchietto et al. 2020; Srinivasachar Badarinarayan et al. 2020). This review will focus on the role of mammalian TE-derived nucleic acids in stimulating the antiviral innate immune response. We provide a brief overview of the major mammalian TEs and innate antiviral defence mechanisms to understand the evolutionary interactions between TEs and antiviral defence.
MOUSE AND HUMAN TRANSPOSABLE ELEMENTS: AN OVERVIEW
TEs can be defined as DNA fragments that are able to mobilize from one place to another within the genome and are divided in two major groups based on their mobilization mechanism (Finnegan 1989; Beck et al. 2011). On the one hand, DNA transposons (or class II elements) mobilize through a “cut and paste” mechanism, in which the transposon is excised from the donor site and inserted in a different genomic location. They comprise ∼3% of the human genome and are no longer competent for mobilization (Lander et al. 2001). On the other hand, retrotransposons (or class I elements) mobilize through a “copy and paste” mechanism that involves the reverse transcription of an RNA intermediate to generate a new copy of the element. Retrotransposons can be further divided based on their structure and the mechanism of reverse transcription. In the case of retrotransposons with long terminal repeats, LTR retrotransposons including endogenous retroviruses (ERVs), the double-stranded DNA (dsDNA) copy is generated in the cytoplasm using a tRNA as a primer for reverse transcription. Once synthesized, the dsDNA travels to the nucleus, where it is inserted into the genome by the integrase enzyme (Fig. 1A; Bannert and Kurth 2006). In contrast, non-LTR retrotransposons, which include long interspersed elements (LINEs) and short interspersed elements (SINEs), have developed a different strategy: A LINE-1-encoded endonuclease nicks the target DNA generating an exposed 3′-OH that serves as a primer to produce a new cDNA molecule at the site of integration. This mechanism is called target-primed reverse transcription (TPRT) (Fig. 1B; Luan et al. 1993; Cost et al. 2002).
FIGURE 1.
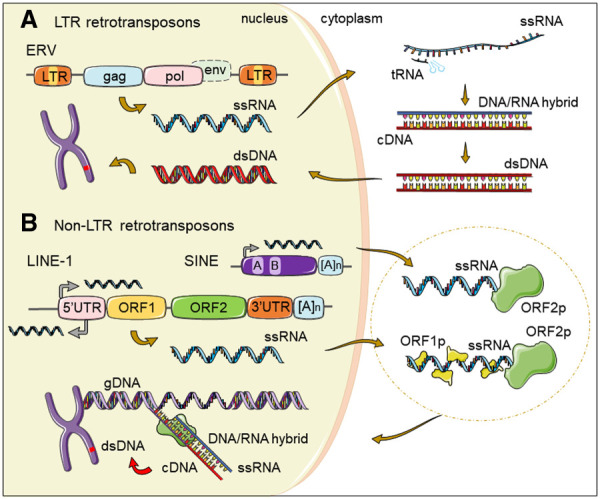
Mammalian LTR versus non-LTR retrotransposon replicative cycles. (A) ERVs are flanked by long-terminal repeats (LTRs) and encode Gag and Pol proteins while lacking a functional env gene. After transcription, the single-stranded RNA (ssRNA) from ERVs is exported to the cytoplasm. A transfer RNA (tRNA) binds the ssRNA molecule and serves as a primer for discontinuous reverse transcription to generate a dsDNA copy. The dsDNA is then imported to the nucleus where it integrates in the genome. (B) SINE elements, which include human Alu and mouse B1/B2 elements, typically contain an RNA pol III transcription start site (black arrow), sequences required for transcription (A and B boxes) and a terminal poly(A) repeat [(A)n]. Below SINEs, the structure of a full-length LINE-1 element is represented, containing a 5′ untranslated region (5′UTR), with sense and antisense RNA pol II promoter transcription start sites (black arrows), two nonoverlapping open reading frames (ORF1 and ORF2) and a short 3′UTR with a poly(A) tail. The ssRNA from LINEs and SINEs is exported to the cytoplasm and bound by LINE-1 encoded proteins, forming a ribonucleoparticle that is imported into the nucleus. ORF2p endonuclease domain nicks the genomic DNA (gDNA) at a target site, and the exposed 3′ OH group is used by the reverse transcriptase domain to prime the reverse transcription of the RNA template. This mechanism, called target-primed reverse transcription (TPRT), generates an RNA/DNA hybrid intermediate. In the next steps, which are still not completely understood, a new dsDNA copy of the SINE or LINE element is formed and integrated into a new genomic location.
In terms of genomic structure, ERVs resemble retroviruses. ERVs contain gag and pol genes which encode structural and enzymatic proteins, including the reverse transcriptase, but lack functional envelope env genes. Although human ERVs (HERVs) comprise 8% of our genome, to date, no competent HERVs have been described. However, these elements impact gene expression in many ways, acting as enhancers or via the production of long noncoding RNAs (lncRNAs) regulating pluripotency maintenance (Garcia-Perez et al. 2016). In mice, ERVs account for 10% of the genome and, in contrast to humans, several subfamilies are actively mobilized, including IAP (intracisternal A-particle) and MusD (Mus-type D related retroviruses) elements, among others (Lueders and Kuff 1977).
Around 40% of the human and mouse genomes are made of non-LTR retrotransposons and in both species, several families are active. LINEs are autonomous (i.e., they encode the proteins required for their mobilization), while SINEs require the proteins encoded by LINE elements to mobilize (Moran et al. 1996; Dewannieux et al. 2003; Hancks et al. 2011; Richardson et al. 2015). Three distant families of LINEs, LINE-1, LINE-2, and LINE-3, account for 21% of the human genome (Lander et al. 2001); however, only elements belonging to the LINE-1 family have remained active (Brouha et al. 2003; Khan et al. 2006). A competent LINE-1 is typically ∼6 kb long and contains a 5′ untranslated region (5′UTR), two nonoverlapping open reading frames (ORF1 and ORF2) and a short 3′UTR containing a poly(A) tail. ORF1 encodes an RNA binding protein with nucleic acid chaperone activity, whereas ORF2 encodes a protein with endonuclease (EN) and reverse transcriptase (RT) activities. Both ORFs are required for LINE-1 retrotransposition, but only ORF2 is essential for mobilization (for review, see Richardson et al. 2015). The LINE-1 5′UTR has an internal RNA polymerase II promoter that directs transcription of the full-length element and an antisense promoter (Swergold 1990; Speek 2001). Antisense transcription can lead to chimeric transcripts containing a portion of the LINE-1 5′UTR and locus-specific upstream genomic sequences. Furthermore, the antisense promoter drives the expression of a small peptide, ORF0, whose function is still unclear (Denli et al. 2015). It is predicted that all 5′UTRs from the 16 different primate-specific amplifying LINE-1 subfamilies (L1PA16–L1PA1) have the ability to drive transcription, despite lacking sequence homology, while the antisense promoter region is only conserved among the youngest families (L1PA6–L1PA1) (Khan et al. 2006). However, the vast majority of LINE-1 copies in mammalian genomes are 5′ truncated and therefore inactive and only ∼80–100 copies from the youngest LINE-1 subfamily L1PA1, also called L1Hs, have retained the potential to mobilize (Brouha et al. 2003; Beck et al. 2011). In mice, around 3000 copies of mouse LINE-1, from subfamilies A, TF, and GF, remain active. The 5′UTR of mouse LINEs consists of monomeric repeats and contains an RNA polymerase II sense strand promoter, while the antisense promoter is located at ORF1 (Deberardinis and Kazazian 1999; Li et al. 2014). The bidirectional transcription of opposed LINE-1 retrotransposon sequences could potentially result in the formation of double-stranded RNAs (dsRNAs), as previously suggested (Yang and Kazazian 2006). Although at the structural level mouse LINE-1s are similar to human LINEs, they differ at the 5′UTR sequence, suggesting that the transcripts derived from this region could adopt different RNA secondary structures. LINE-1 insertions within genes are biased toward the antisense orientation (Smit 1999; Flasch et al. 2019; Sultana et al. 2019), which suggests that for an important number of LINE-1 copies, full-length antisense LINE-1 RNA synthesis could be driven by the host gene promoter.
SINEs make up ∼13% and ∼8% of the human and mouse genome, respectively. Several families (e.g., human Alu and SVA elements, and mouse B1 and B2 elements) are still active and hijack the LINE-1-encoded proteins to be mobilized in trans (Dewannieux et al. 2003, 2005; Hancks et al. 2011). The most abundant SINEs elements, human Alu and mouse B1 and B2, are respectively ∼300, ∼135, and ∼200 bp-long, and are all transcribed by RNA-polymerase III (for review, see Richardson et al. 2015). However, Alu elements are highly prevalent in RNA-polymerase II transcripts, due to their preference for inserting in gene-rich regions (Lander et al. 2001). Within genes, Alus are generally located in introns and UTRs, in both sense and antisense orientations, suggesting that two different Alus present in the same transcript could form intramolecular dsRNA interactions (Deininger 2011).
DsRNA structures adopted by TEs resemble viral RNAs, which cells can aberrantly recognize as invading viruses and trigger the type I interferon (IFN) response. In the next section, the type I IFN response is discussed, and more specifically, the types of nucleic acids and receptors involved in their recognition.
THE TYPE I INTERFERON RESPONSE AND NUCLEIC ACID SENSING MECHANISMS: AN OVERVIEW
In mammals, the type I IFN response constitutes the major innate defense mechanism against viruses, and it is triggered after sensing the presence of viruses. To this end, mammalian cells have evolved a plethora of sensors that recognize typical traits of viral nucleic acids. Upon detection of viral nucleic acids, signaling pathways that culminate in the expression of type I IFNs and proinflammatory cytokines are activated. Secreted IFNs act in both a paracrine and autocrine manner. By binding the IFN receptor (IFNAR1/2) and stimulating the JAK/STAT pathway, IFNs trigger the expression of hundreds of interferon-stimulated genes (ISGs). ISGs are responsible for establishing an antiviral cellular state to prevent viral infection and replication as well as stimulating the adaptive immune response to generate long-lasting antiviral immunity (Schneider et al. 2014; Ivashkiv and Donlin 2015).
In this section, we will describe the nucleic-acid sensing mechanisms present in mammalian cells. Nucleic acid sensors localize in the cytoplasm, the cell surface, and endosomal compartments to detect the presence of viral genomes or replication intermediates. While most cytoplasmic sensors are widely expressed, the expression of endosomal receptors is generally restricted to specific cell types. Besides initiating the type I IFN response, nucleic acids sensors can also activate other antiviral pathways, such as the host translational shutoff. Here, we will summarize the different mammalian nucleic acid-sensing pathways based on the types of virus-derived nucleic acids they recognize, and that are relevant for the focus of this review.
Double- and single-stranded RNA
The presence of cytoplasmic dsRNA is the hallmark of viral infections and active viral replication (Kumar and Carmichael 1998). Virus-derived dsRNAs are sensed by the cytoplasmic retinoic acid-inducible gene I (RIG-I)-like receptors (RLR), which are composed of the three ATP-dependent DExD/H box RNA helicases, RIG-I, MDA5 (melanoma differentiation-associated gene 5), and LGP2 (laboratory of genetics and physiology 2). RIG-I preferably binds short stretches of dsRNA (<300 bp) bearing 5′-di/triphosphate groups (Hornung et al. 2006; Pichlmair et al. 2006; Myong et al. 2009; Schmidt et al. 2009; Goubau et al. 2014), while MDA5 binds long molecules of dsRNA (>1 Kbp) (Kato et al. 2008; Feng et al. 2012). Both RIG-I and MDA5 contain amino-terminal caspase activation recruitment domains (CARD) that interact with the mitochondrial antiviral signaling (MAVS) protein to activate the nuclear translocation of the transcription factors interferon regulatory factor 3 (IRF3) and IRF7 and nuclear factor kB (NF-kB) to trigger the expression of type I IFNs and proinflammatory cytokines such as interleukin 6 (IL-6), IL-8 and tumor necrosis factor α (TNF-α) (Fig. 2; Kawai et al. 2005; Seth et al. 2005). Despite the ability of LGP2 to bind dsRNA, the absence of CARD domains prevents the signal transduction for IFN production and it is assumed that the main role of LGP2 is regulating the signaling activity of both RIG-I and MDA5 (Yoneyama et al. 2005; Deddouche et al. 2014).
FIGURE 2.
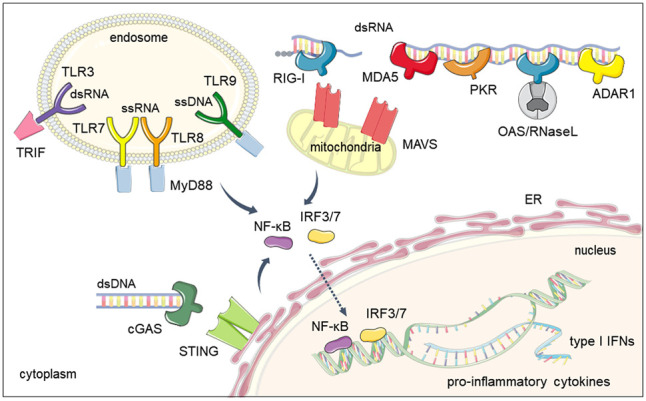
Mammalian nucleic acids sensors. Sensors are classified depending on their subcellular localization and the type of nucleic acid they recognize (DNA vs. RNA). Toll-like receptors (TLRs) usually reside at the cell membrane or the endosome and recognize several types of nucleic acids: TLR3 for dsRNA, TLR7-8 for ssRNA, and TLR9 for ssDNA and RNA/DNA hybrids. cGAS and the RIG-I-like (RLR) family of receptors are typically cytoplasmic, although cGAS can also be found in the nucleus. cGAS recognizes DNA, while RLRs, MDA5 and RIG-I recognize long or short dsRNA molecules, respectively. Upon nucleic acid recognition, these receptors trigger a signalling cascade that promotes nuclear translocation of the IRF3/7 and NF-kB transcription factors and expression of type I IFNs and proinflammatory cytokines. Other cytoplasmic proteins recognize virus-derived dsRNA to induce the host translational shutoff response (PKR), widespread cytoplasmic RNA degradation (OAS/RNase L) and deamination of dsRNA molecules (ADAR1).
Type I IFN expression can also be triggered by toll-like receptor 3 (TLR3)-mediated dsRNA recognition. This sensor belongs to the toll-like family receptor (TLR) and, depending on the cell-type, is localized at the cell membrane or endosomal compartment. TLR3 binding to dsRNA activates a TRIF-dependent signaling pathway that culminates in expression of type I IFNs and proinflammatory cytokines (Alexopoulou et al. 2001; Oshiumi et al. 2003). Other members of the TLR family recognize viral RNA of a single-stranded conformation (ssRNA). Human TLR7 senses ssRNA and TLR8 binds ssRNA in conjunction with products of RNase degradation to trigger type I IFN expression in a myeloid differentiation primary response 88 (MyD88)-dependent pathway (Fig. 2; Diebold et al. 2004; Heil et al. 2004; Greulich et al. 2019; Ostendorf et al. 2020).
Besides IFN production, the presence of dsRNA in the cytoplasm of cells can activate other pathways that are important for antiviral defense. These include the activation of the inflammasome, the host translational shutoff, the formation of stress granules and widespread RNA degradation. Recognition of dsRNA by the DEAH-box helicase 33 (DHX33) activates the NLR family pyrin domain containing 3 (NLRP3) inflammasome, a proinflammatory pathway characterized by the production of IL-1β and IL-18 (Mitoma et al. 2013). Upon dsRNA binding, the interferon-inducible protein kinase R (PKR) phosphorylates the initiation factor of translation eIF2α to block cap-dependent mRNA translation of viral and host mRNAs in a nonselective manner (Levin and London 1978). The inhibition of translation also results in the formation of stress granules in response to viral dsRNA accumulation (Kedersha et al. 1999; Okonski and Samuel 2013). These cytoplasmic foci are composed of untranslated mRNAs and translational incompetent complexes (Miller 2011). Finally, the IFN-stimulated oligoadenylate synthetase (OAS)/RNase L system, whose activity is also triggered by the presence of virus-derived dsRNA, causes widespread degradation of cytoplasmic RNAs (Kumar and Carmichael 1998). Activation of OAS enzymatic activity by dsRNA results in the generation of 2′–5′ oligoadenylates. These oligomers promote the dimerization and activation of RNase L, an endonuclease that causes widespread degradation of cytoplasmic ssRNA, thus also promoting the translational arrest characteristic of infected cells (Floyd-Smith et al. 1981; Kodym et al. 2009; Burke et al. 2019; Rath et al. 2019). Interestingly, the OAS/RNase L system was also shown to be involved in controlling LINE-1 and IAP retrotransposition levels (Zhang et al. 2014).
Other IFN-stimulated dsRNA binding proteins that are important in regulating type I IFN activation include the adenosine deaminase acting on RNA 1 (ADAR1), which converts adenosines to inosines in dsRNA molecules (George et al. 2014). ADAR1 activity results in the destabilization of dsRNA structures, thus preventing RLR-dependent activation of the type I IFN response (Mannion et al. 2014). This activity has been shown to be important for destabilizing dsRNA-structures formed by Alu elements to avoid their recognition as virus-derived dsRNAs (Chung et al. 2018).
Double- and single-stranded DNA
The major pathway for cytoplasmic viral DNA sensing is initiated by the cyclic GMP-AMP synthase (cGAS) (Sun et al. 2013). Stimulation of cGAS activity by dsDNA results in the formation of cyclic-GMP-AMP (cGAMP) and signaling through the endoplasmic reticulum-associated stimulator of interferon genes (STING) protein. Analogous to RLR-signaling, the cGAS/STING pathway promotes IRF3 and NF-kB nuclear translocation to induce type I IFN and proinflammatory cytokines expression (Wu et al. 2013). Although activation of cGAS is dependent on the length of dsDNA, this sensor can also bind stem–loop structures formed within ssDNA or DNA/RNA hybrids (Fig. 2; Mankan et al. 2014; Herzner et al. 2015; Luecke et al. 2017). cGAS has been classically considered a cytoplasmic sensor, but it can also localize in the nucleus and recognize viral-derived DNA in this compartment (Lahaye et al. 2018).
The endosomal TLR9 receptor preferentially recognizes unmethylated ssDNA (Hemmi et al. 2000) but is also stimulated by DNA/RNA hybrids (Rigby et al. 2014). Similarly to TLR7 and 8, TLR9 signals in a MyD88-dependent manner leading to the production of type I IFNs and proinflammatory cytokines through the activation of transcription factors IRF3/-7 and NF-kB (Fig. 2; Kawai et al. 2004; Hoshino et al. 2006).
Viral dsDNA can also trigger the absent in melanoma 2 (AIM2)-inflammasome resulting in the production of IL-1β and IL-18, as well as pyroptosis, a caspase-1 dependent cell death program associated with inflammation (Fernandes-Alnemri et al. 2009; Rathinam et al. 2010). In mice, all AIM2-like receptors are dispensable for the type I IFN response to intracellular DNA (Gray et al. 2016).
Besides IFN production and inflammasome activation, the presence of cytoplasmic DNA is sensed by the family of cytidine-deaminases known as apolipoprotein B mRNA-editing enzyme or APOBEC (Salter et al. 2016). Their enzymatic activity catalyzes the conversion of cytidines to uridines in ssDNA, as well as in RNA. This activity has been shown to hypermutate the cDNA of both HIV-1 and retrotransposons (LINEs, SINEs, and ERVs), suggesting that APOBECs have a role in defending against both endogenous and exogenous genomic parasites (Harris et al. 2003; Esnault et al. 2005; Chiu et al. 2006; Muckenfuss et al. 2006; Carmi et al. 2011; Richardson et al. 2014).
The APOBEC, ADAR, and RNase L examples illustrate the substantial overlap between the mechanisms guarding defence against viruses and TEs. In addition, TE-derived nucleic acids can be sensed by cellular nucleic acid sensors and trigger the type I IFN response, pointing out that TE activity needs to be tightly controlled to improve the accuracy of antiviral defence.
TE-DERIVED NUCLEIC ACIDS AS TRIGGERS OF THE TYPE I IFN RESPONSE
There is mounting evidence of TEs acting as substrates for the classical viral-nucleic acid sensors in fields ranging from autoimmune diseases to cancer. Here we will summarize the literature reporting TE-mediated IFN activation and discuss some of the questions that these findings raise considering the current knowledge in TE biology and nucleic acid sensing.
Cancer
Several types of cancer show reactivation of ERVs and LINEs expression, a feature that is considered to contribute to cellular transformation (Burns 2017; Bannert et al. 2018). The transcriptional silencing of both types of TEs in healthy cells is controlled by DNA methylation, KRAB-zinc finger proteins and histone repressive marks (Rowe and Trono 2011). Tumors are generally characterized by low DNA CpG methylation levels compared to healthy tissues. Considering that most methylated CpG sites are concentrated in the highly repetitive sequences of the genome, the expression of ERVs and LINEs is, as a consequence, reactivated in cancers (Ehrlich 2009; Burns 2017). Counterintuitively, DNA-methylation inhibitors are used as therapeutic agents in specific types of cancer, such as hematological malignancies, raising the question of what the molecular consequences of further demethylation of cancerous cells are. Two independent reports showed that treatment of colorectal and ovarian carcinoma cells with low doses of the demethylating agent 5-aza-2′-deoxycytidine (5-aza) led to the accumulation of dsRNAs and subsequent activation of the IFN response (Chiappinelli et al. 2015; Roulois et al. 2015). In colorectal cancer, 5-aza treatment resulted in activation of the IFN response in a MDA5/MAVS-dependent manner with a concomitant up-regulation of ERV expression (Roulois et al. 2015). In ovarian carcinoma cells, treatment with 5-aza resulted in activation of IFN and ISG expression through the TLR3/MAVS pathway, an effect that was partially rescued by inhibiting the expression of two specific ERVs. Although the relationship between IFNs and cancer is complex, IFNs are positive players in stimulating antitumor immunity (Budhwani et al. 2018; Borden 2019). For instance, treatment with demethylating agents slowed the population doubling time of colorectal cancer cells in vitro and their ability to develop tumors after injection in nude mice, and importantly, these effects were abrogated by preventing IFN activation. In a mouse model of melanoma, 5-aza sensitized cells to anti-CTLA4 immune checkpoint therapy (Chiappinelli et al. 2015). All these results suggest that dsRNA-mediated immune activation of the RLR pathway could constitute a novel therapeutic target to control tumor growth. Other chemotherapeutic agents, such as doxorubicin and anthracyclines have also been shown to trigger a TLR3-dependent IFN activation (Sistigu et al. 2014). The chemotherapeutic agent RRx-001 was also reported to induce an IFN and ISG response in a human colon carcinoma cell line. Treatment with RRx-001 caused accumulation of dsRNA, as revealed by anti-dsRNA immunofluorescence (J2 antibody), and a concomitant reactivation of the expression of endogenous retroviral genes (Zhao et al. 2017). Similarly, loss of proteins responsible for ERV silencing, such as SETDB1, Trim24, and KAP1, resulted in IFN activation in cancerous and noncancerous settings upon accumulation of ERV and LINE transcripts (Herquel et al. 2013; Cuellar et al. 2017; Tie et al. 2018).
Only recently, failure to epigenetically silence LINE-1 retrotransposon expression has been associated with IFN activation. The absence of p53, the most commonly mutated tumor suppressor gene in cancer, resulted in increased expression of locus-specific copies of LINE-1 and was accompanied by an inflammatory signature. This activation of the immune response was reverted by treatment with the reverse transcriptase inhibitor (RTi) lamivudine (3TC) (Tiwari et al. 2020). RTis are compounds that can disrupt the replication cycle of both exogenous retroviruses and endogenous retrotransposons by interfering with the reverse transcription step. Some RTis, such as lamivudine and emtricitabine, are selective and only inhibit LINE-1 RT (Banuelos-Sanchez et al. 2019). These findings suggest that the nucleic acid responsible for IFN activation in the absence of p53 is the product of LINE-1 retrotranscription (Tiwari et al. 2020). Previously, p53-deficient mouse embryonic fibroblasts were found to accumulate mitochondrial-derived dsRNAs capable of triggering IFN expression (Wiatrek et al. 2019), and SINE B1 and B2 elements, which led to a “suicidal” IFN response (Leonova et al. 2013). These findings suggest that several types of TEs and other endogenous nucleic acids could be responsible for triggering the IFN response in the context of p53 deficiency. An alternative epigenetic pathway controlling LINE-1 expression is orchestrated by the human silencing hub (HUSH) complex. The expression of the central subunit, MPP8, was found to be decreased in cancer and correlated with tumors displaying an immune signature. Loss of the HUSH complex resulted in increased LINE-1 expression and IFN activation in a RLR/MAVS-dependent manner, suggesting that LINE-1 dsRNA can also act as a trigger of the IFN response (Tunbak et al. 2020).
It is becoming increasingly evident that the expression of dsRNA-forming loci is activated after treatment with DNA demethylating agents or inactivation of epigenetic repressors (Fig. 3A). However, the exact origin of the nucleic acids responsible for the immune activation is still unclear. A recent attempt to answer this question found that the innate immune sensor MDA5 recognizes Alu elements from colorectal cancer cells in vitro, despite Alu expression not being increased after low-dosage 5-aza treatment (Mehdipour et al. 2020). Therefore, a more targeted search for in vivo RNA ligands of the RLR receptors, for instance using CLIP-seq (cross-linking and immunoprecipitation coupled to high-throughput sequencing) or similar techniques, will help to elucidate the identity of the specific substrates for activating the IFN response in cancer after demethylating treatment.
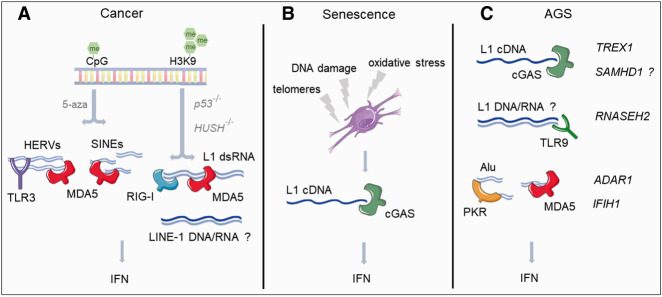
FIGURE 3.
Recognition of TEs as viral nucleic acids in cancer, senescence, and Aicardi–Goutières syndrome (AGS). (A) Treatment of cancer cells with demethylating agents (5-aza) or genetic manipulation of factors involved in epigenetic repression (HUSH complex) or genetic stability (p53) leads to increased expression of different types of retrotransposons, including HERVs, LINEs, and SINEs. The RNA molecules derived from these elements are recognized by nucleic acid sensors, triggering the type I IFN response. (B) Changes in the genomic architecture of senescent cells are responsible for increased expression of LINE-1 elements. In senescence, the sensor cGAS recognizes the retrotranscription product (cDNA) of LINE-1 triggering IFN expression. (C) Mutations in genes involved in nucleic acid metabolism (TREX1, SAMHD1, RNASEH2A/B/C, and ADAR1) and nucleic acid sensing (IFIH1, which encodes MDA5) are associated with AGS, a disease characterized by IFNs expression in the absence of infections. LINE-1 cDNA accumulation in the absence of functional TREX1 has been suggested to trigger IFN expression in a cGAS-dependent manner, this mechanism could also operate in the context of SAMHD1 mutations. The accumulation of unedited dsRNA Alu molecules in the absence of functional ADAR1 triggers IFN expression in a MDA5-dependent manner and the host translational shutoff by PKR. Gain-of-function mutations of IFIH1 lead to recognition of dsRNA-Alu molecules as nonself and consequent IFN activation. The role of TE in the context of RNASEH2 mutations is still unclear, but, hypothetically, accumulation of LINE-1 RNA/DNA hybrids could be triggering the IFN response in this context.
Cellular senescence
Cellular senescence is an irreversible state of cell cycle arrest associated with aging, but also wound healing and development, typically triggered in response to stress or damaging agents. For instance, telomere shortening, mitochondrial dysfunction, oxidative stress and DNA damage can trigger senescence (Hernandez-Segura et al. 2018). Apart from the permanent cell cycle arrest, the senescent state is characterized by the “senescence-associated secretory phenotype” (SASP), where cells release cytokines, chemokines, extracellular matrix components, and growth factors (Novakova et al. 2010; Lopes-Paciencia et al. 2019). Senescence is also accompanied by changes in the genome architecture, resulting in the loss of repressive epigenetic marks and heterochromatin formation and consequent increased expression of the human retrotransposons Alu, SVA, and LINE-1 (Wang et al. 2011; De Cecco et al. 2013; Van Meter et al. 2014). The increase in retrotransposon expression has been observed in different models of in vitro-induced cellular senescence. High cell-passage numbers, oxidative stress, hypomethylation (5-aza) and treatment with chemotherapy agents (adriamycin) resulted in dysregulation of TE expression, with LINE-1 and ERV-1 elements being the predominant up-regulated biotypes. Interestingly, TE up-regulation was found to be accompanied by activation of the proinflammatory phenotype (SASP) and the type I IFN response (Colombo et al. 2018). Using a model of late senescence, LINE-1 was found to be responsible for activation of the type I IFN response in a cGAS/STING-dependent manner (Fig. 3B). Treatment of cells with shRNAs against LINE-1 or the RTi lamivudine reverted the activation of the IFN response, suggesting that the LINE-1 cDNA was responsible for triggering IFN expression (De Cecco et al. 2019). Similar results were reported in a mouse model of premature aging. SIRT6 knockout mice display increased genomic instability and a premature aging phenotype which is accompanied by increased LINE-1 expression and a cGAS-dependent activation of the type I IFN response (Mostoslavsky et al. 2006; Van Meter et al. 2014; Simon et al. 2019). Many of the phenotypes associated with SIRT6 deficiency, such as premature aging, colitis and increased IFN expression, were reverted after treatment with RTis, suggesting a central role for the LINE-1/IFN interaction in the disease-associated traits of SIRT6 KO mice (Simon et al. 2019).
An additional exciting observation is the requirement of an active IFN response for establishing a full senescent phenotype. Blocking IFN signaling by knocking out the IFN receptor (IFNAR) abolished the establishment of a fully mature SASP response (De Cecco et al. 2019). Interestingly, the cGAS/STING pathway has also been shown to be necessary to establish senescence (Glück et al. 2017; Yang et al. 2017). The exact role of the IFN response in promoting a full-senescent phenotype remains an open question.
One common observation from these reports is the proposed role for cytoplasmic-localised LINE-1 cDNA as the molecule responsible for triggering IFN expression in a cGAS-dependent manner. Considering that the accepted model for the LINE-1 lifecycle suggests that the retrotranscription step occurs in the nucleus, further experimentation is necessary to reconcile these findings. It is possible that senescent cells localize both LINE-1 retrotranscription and/or cGAS in different subcellular compartments than actively cycling cells. For instance, senescent cells could be retrotranscribing LINE-1 in the cytoplasm, if cytoplasmic RNA/DNA molecules could act as primer. Only recently, Alu elements have been shown to be retrotranscribed in the cytoplasm by the LINE-1-encoded machinery independently of its retrotransposition, bringing closer the possibility that LINE-1 RNA could be also used as template to generate LINE-1 cDNA in the cytoplasm (Fukuda et al. 2021). Alternatively, LINE-1 cDNA could also be recognized by cGAS in the nucleus, as a significant proportion of this sensor has been found in the nuclear compartment of specific cell types (Xia et al. 2018; Gentili et al. 2019; Volkman et al. 2019). It is important to note that nuclear cGAS does not react to genomic DNA because its binding to nucleosomes maintains this sensor in an “inactive” state and incompetent for signaling (Zierhut et al. 2019; Boyer et al. 2020; Cao et al. 2020; Kujirai et al. 2020; Pathare et al. 2020). In addition, BAF, a chromatin-associated protein, has also been recently shown to compete cGAS for dsDNA binding, thus preventing recognition of self-DNA (Guey et al. 2020). Considering that newly retrotranscribed LINE-1 cDNA will lack chromatinization during de novo insertion, we hypothesize that this could provide a short window of opportunity for nuclear cGAS to sense the LINE-1 cDNA as an invading pathogen. It will be interesting to measure BAF levels in senescent cells as well as the nuclear envelope integrity, but also test whether nuclear/cytoplasmic cGAS is responsible for recognizing the LINE-1 cDNA or DNA/RNA intermediate during the reverse transcription step. This will help to clarify how LINE-1 cDNA can trigger the cGAS–STING sensing pathway.
Autoimmune diseases
In recent years, TEs-derived nucleic acids have attracted much attention as molecules that may be responsible for driving inflammation in the context of autoimmune diseases. In this section, we will summarize the literature reporting TE-mediated immune activation in disease, both in the context of the rare autosomal recessive autoinflammatory disorder Aicardi–Goutières syndrome and the much more common autoimmune disease, systemic lupus erythematosus.
Aicardi–Goutières syndrome
Aicardi–Goutières syndrome (AGS) is a heritable form of inflammatory encephalopathy with symptoms resembling congenital viral infections of the brain (Crow et al. 2014), including a characteristic elevation of type I IFN levels both in serum and cerebrospinal fluid of patients (Goutières et al. 1998). Mutations in different genes involved in the metabolism or sensing of nucleic acids can lead to AGS (Crow and Manel 2015; Rice et al. 2020). Therefore, AGS has become the paradigm in the study of the role of aberrant accumulation/sensing of endogenous nucleic acids as triggers of the IFN response. Although the precise origin of the nucleic acids driving IFN activation is still unclear, here, we will summarize the current evidence supporting the hypothesis that retrotransposition-intermediates (RNA and DNA) can act as the immunostimulatory molecules triggering the type I IFN response in AGS.
Gain-of-function mutations in the sensor IFIH1 and partial loss of function mutations in TREX1, SAMHD1, and ADAR1 genes, as well as the genes encoding for the three subunits of the RNase H2 complex can lead to AGS. Besides the classical function of most of these genes in regulating different aspects of nucleic acids metabolism, TREX1, SAMHD1, and ADAR1 have also been implicated in restricting the mobilization of LINE-1 retrotransposons (Stetson et al. 2008; Zhao et al. 2013; Orecchini et al. 2016; Li et al. 2017a). Intriguingly, RNase H2 has been described both as a promoter as well as an inhibitor of LINE-1 retrotransposition (Bartsch et al. 2017; Benitez-Guijarro et al. 2018). The role of these genes in controlling retrotransposition suggests that their loss of function in AGS may lead to increased accumulation of TE-intermediates that could trigger innate immune activation in an RNA (ADAR1) or DNA-dependent (TREX1, RNase H2, and SAMHD1) manner (Fig. 3C). Below, we summarize the proposed roles for proteins mutated in AGS on the control of TEs.
TREX1
This gene encodes a 3′ to 5′ DNA exonuclease whose expression is controlled by IFNs (ISG) (Mazur and Perrino 2001; Grieves et al. 2015). TREX1 loss of function or dominant negative mutations have been reported in AGS, familial chilblain lupus and systemic lupus erythematosus patients (Crow et al. 2006a; Lee-Kirsch et al. 2007; Rodero and Crow 2016). One of the first animal models for Trex1 deficiency revealed that this enzyme is required to prevent a lethal inflammatory phenotype in mice (Morita et al. 2004). The cGAS/STING-dependent autoinflammatory phenotype of Trex1-deficient mice correlated with the accumulation of ssDNA from LINE-1, LTR endogenous retroviruses and SINE elements (Stetson et al. 2008; Gall et al. 2012; Gray et al. 2015). Supporting the notion that Trex1 may regulate the levels of retrotransposition byproducts, in vitro retrotransposition assays showed reduced mobilization of human LINE-1 element and the mouse IAP LTR retrotransposon during Trex1 overexpression (Stetson et al. 2008). All this led to the hypothesis that mouse Trex1 metabolizes the ssDNA products of TE retrotranscription, preventing their accumulation and consequent activation of the innate immune response.
In humans, neurons, neuronal progenitor cells (NPCs) and astrocytes derived from TREX1-deficient human stem cells also displayed an increase in intracellular DNA species, including LINE-1 elements and elevated type I IFN levels. The accumulation of LINE-1 cDNA and consequent IFN activation was proposed to be responsible for the neurotoxic phenotype, as this was reverted after treatment with the RTis or after antagonizing IFN signaling (Thomas et al. 2017). These results suggest that in both humans and mice, the absence of TREX1 can lead to LINE-1 cDNA accumulation triggering the IFN response through the cGAS–STING pathway (Fig. 3C). However, this hypothesis has been challenged by the observation that treatment of Trex1−/− mice with RTis did not reduce the spontaneous ISG signature, suggesting that retrotransposon-independent mechanisms may also contribute to the inflammatory phenotype (Achleitner et al. 2017).
RNase H2
The RNase H2 complex is composed of the three different subunits: RNASEH2A, RNASEH2B, and RNASEH2C. RNase H2 activity is essential for genome stability and mammalian embryonic development (Hiller et al. 2012; Reijns et al. 2012). This ribonuclease initiates the removal of ribonucleotides incorporated in genomic DNA during replication, and it is also thought to be involved in resolving R-loops formed during transcription, by catalyzing the degradation of RNA in RNA/DNA hybrids. Biallelic mutations in the genes encoding the three subunits are the most common cause of AGS, and monoallelic mutations have been associated with increased risk of systemic lupus erythematosus (Crow et al. 2006b, 2015; Günther et al. 2015). Analysis of mice with partial-loss-of-function AGS disease mutations in Rnaseh2a and Rnaseh2b, as well as experiments using Rnaseh2b knockout cells established that RNase H2 function is necessary to prevent activation of the IFN response and ISG expression in a cGAS/STING-dependent manner (Mackenzie et al. 2016; Pokatayev et al. 2016). For instance, mutation in the highly conserved glycine residue (G37S), near the catalytic center of the RNASEH2A subunit, causes a severe early onset presentation of AGS. In mice, this mutation was shown to activate ISGs expression, with a concomitant accumulation of cytoplasmic LINE-1 DNA. This led to the hypothesis that accumulation of LINE-1 DNA in the absence of functional RNase H2 could lead to activation of the IFN response (Pokatayev et al. 2016). Similarly, a mouse model for the most common missense mutation found in AGS patients (RNASEH2B-A177T), led to ISG activation in a cGAS/STING-dependent manner (Mackenzie et al. 2016). However, the role of RNase H2 in the control of retrotransposons is still controversial, as it has been suggested to be necessary for both restricting and facilitating LINE-1 activity. Restriction of LINE-1 retrotransposition was suggested to involve a physical interaction with the well-known LINE-1 restricting factor MOV10 (Choi et al. 2018), while two other reports contradicted these findings and showed that RNase H2 function is necessary for LINE-1 mobilization (Bartsch et al. 2017; Benitez-Guijarro et al. 2018). The proposed model is that LINE-1 elements rely on the endogenous RNase H activity of cellular RNase H2 to degrade the RNA moiety after reverse transcription and successfully synthesize their second strand cDNA to complete their retrotransposition cycle (Benitez-Guijarro et al. 2018). Considering this model, the absence of functional RNase H2 could lead to the accumulation of unprocessed LINE-1 RNA/DNA hybrids and potentially trigger the IFN response in a cGAS/STING-dependent manner (Fig. 3C; Mankan et al. 2014; Rigby et al. 2014).
Importantly, the absence of RNase H2 also causes a significant increase in DNA damage, widespread DNA hypomethylation, and micronuclei formation, which can trigger the IFN response independent of retrotransposon intermediates (Reijns et al. 2012; Lim et al. 2015; MacKenzie et al. 2017). Therefore, further investigation is necessary to clarify the specific nature of the nucleic acids that trigger IFN expression in the context of RNase H2 deficiency.
SAMHD1
Mutations in this gene are found in AGS, as well as in patients with solid tumors and leukemia (Rice et al. 2009; Clifford et al. 2014; Rentoft et al. 2016). This gene encodes a deoxynucleoside triphosphate triphosphohydrolase (dNTPase) that controls dNTP levels in cells and whose expression is controlled by IFNs (ISG). Ensuring optimal dNTPs levels is required for efficient cellular DNA replication and cDNA synthesis of retrovirus and retrotransposons. In the absence of functional SAMHD1, ssDNA fragments from stalled replication forks are released to the cytoplasm where they are thought to activate IFN expression in a cGAS/STING-dependent manner (Coquel et al. 2018). Considering the impact of SAMHD1 on dNTP levels, this enzyme also acts as a restriction factor against the HIV-1 retrovirus and the LINE-1 retrotransposon (Fig. 3C; Lahouassa et al. 2012; Zhao et al. 2013). SAMHD1-mediated restriction of HIV-1 can be ameliorated by dNTP supplementation, but the exact role of SAMHD1 enzymatic activity in the context of LINE-1s remains unclear, as its role in restricting LINE-1 mobilization has been shown to be both dependent and independent of its enzymatic activity (Zhao et al. 2013; Herrmann et al. 2018). For instance, AGS-associated SAMHD1 mutations outside the catalytical sites also fail to restrict LINE-1 mobilization (Zhao et al. 2013). The current proposed model is that SAMHD1 restricts LINE-1 mobilization by both reducing ORF2p levels and LINE-1 reverse transcription, as a consequence. Alternatively, SAMHD1 has been shown to directly interact with ORF2p and locally deplete dNTP levels to prevent LINE-1 retrotranscription at the site of integration (Zhao et al. 2013; Herrmann et al. 2018). In addition to these models, SAMHD1 has also been proposed to inhibit LINE-1 retrotransposition by promoting the sequestration of the LINE-1 RNP in cytoplasmic stress granules (Hu et al. 2015). The exact role of increased retrotransposon intermediates in activating the IFN response in the absence of functional SAMHD1 remains unknown.
ADAR1
This gene encodes two different isoforms of the RNA-editing enzyme ADAR1, p110, and p150, and partial loss-of-function mutations have been shown to cause AGS (Rice et al. 2012). While ADAR1 p110 is constitutively expressed and mostly nuclear, the p150 isoform is IFN-inducible and both nuclear and cytoplasmic (Mannion et al. 2015). Both ADAR1 isoforms catalyze the hydrolytic deamination of adenosine to inosine in dsRNA molecules. Editing can result in amino acid recoding when in protein-coding sequences or disrupt important base-pairing interactions in dsRNA secondary structures. High-throughput studies of the human transcriptome demonstrated that the vast majority of edited residues correspond to noncoding regions, with around 90% residing in the primate-specific Alu element (Athanasiadis et al. 2004; Levanon et al. 2004). The high abundance of sense and antisense Alu elements suggest that they are prone to forming dsRNA molecules, which can be deaminated by ADARs. Loss of Adar1 in mice leads to early embryonic lethality (E11.5–12.5), a phenotype that can be rescued to birth by preventing IFN activation of the RLR-sensing pathway by interfering with MDA5 or MAVS expression (Wang et al. 2000; Hartner et al. 2009; Mannion et al. 2014; Liddicoat et al. 2015; Pestal et al. 2015). These results suggest that, in the absence of ADAR1, unedited dsRNAs accumulate and act as a substrate for the MDA5/MAVS signaling pathway for IFN production (Fig. 3C). Supporting these observations, the accumulation of endogenous immunogenic dsRNAs in the absence of ADAR1 also stimulated the translational shutoff by stimulating PKR activity (Chung et al. 2018) and the OAS/RNase L system (Li et al. 2017b).
ADAR1 has also been shown to control LINE-1 retrotransposition independent of its deaminating activity by binding to the LINE-1 RNP complex and restricting its retrotransposition through direct interaction with the LINE-1 RNA (Orecchini et al. 2016). In other settings, the accumulation of LINE-1 RNA in the absence of ADAR1 was also shown to trigger the IFN response in an RLR-dependent manner (Zhao et al. 2018). These results suggest that both LINE-1 RNA and unedited Alu elements could act as immunostimulatory molecules in the absence of functional ADAR1.
IFIH1
This interferon-stimulated gene is the only known factor mutated in AGS that is not directly involved in the metabolism of nucleic acids, as it encodes the dsRNA sensor MDA5. IFIH1 mutations are also associated with systemic lupus and type I diabetes (Smyth et al. 2006; Gateva et al. 2009; Oda et al. 2014; Rice et al. 2014). Gain-of-function mutations in IFIH1 have been reported to lead to IFN activation by preventing discrimination of self versus nonself RNA. DsRNAs derived from sense and antisense Alu elements have been proposed to be the preferred endogenous substrate for gain-of-function mutations of MDA5 (Fig. 3C; Ahmad et al. 2018). Mutated MDA5 was shown to bind and oligomerize on inverted Alu repeats, with a preference for inverted repeats that are in close genomic proximity (< 1Kb) and derived from a single transcript, while wild-type MDA5 could only oligomerize on unedited or perfectly paired Alu-dsRNA structures, suggesting that ADAR1-mediated editing of Alu-dsRNAs could act as a protective mechanism to avoid their erroneous recognition as nonself molecules (Ahmad et al. 2018; Mehdipour et al. 2020).
Retrotransposon-independent IFN activation in AGS
Despite the increasing evidence of retrotransposon intermediates as drivers of the IFN response in AGS, an alternative model exists which suggests that IFN expression is activated by DNA damage. Constitutive DNA damage signaling is associated with cell cycle delay, cellular senescence, and up-regulation of IFNs and ISGs. Importantly, DNA damage and genomic instability are phenotypes widely reported upon mutation of several AGS-genes, including TREX1 (Yang et al. 2007), RNASEH2A, RNASEH2B, RNASEH2C (Hiller et al. 2012; Reijns et al. 2012; Uehara et al. 2018), and SAMHD1 (Günther et al. 2015; Kretschmer et al. 2015). For instance, cells lacking SAMHD1 showed accumulation of DNA damage and ssDNA fragment in the cytosol, where they activated the cGAS/STING pathway to induce expression of type I IFNs (Coquel et al. 2018). Interestingly, RNase H2-deficient cells accumulated cytosolic DNA aggregates virtually indistinguishable from micronuclei (Bartsch et al. 2017), which colocalized with the nucleic acid sensors cGAS inducing a cGAS/STING-dependent innate immune response (Mackenzie et al. 2016, 2017). These findings point toward a link between AGS-related genes and the DNA-damage response through self-DNA becoming exposed in the cytosol and consequent IFN activation.
The hypothesis that unrestricted retrotransposon activity contributes to AGS is supported by the observation that RTis can treat and even prevent disease-associated phenotypes of AGS. However, treatment of Trex1-null mice with both specific and nonspecific LINE-1 RTis led to confounding results. Beck-Engeser et al. reported an improved survival and reduced inflammatory infiltrates in the heart after combinatorial RTi treatment (Beck-Engeser et al. 2011), but Achleitner et al. failed to reproduce these results (Achleitner et al. 2017). In the human context, RTis rescued the neurotoxicity phenotype of AGS-associated TREX1 mutations (Thomas et al. 2017). Recently, stavudine (d4T) was shown to effectively block LINE-1 activity and decrease STING activation after hypoxia-ischemia, reducing neurodegeneration (Gamdzyk et al. 2020). Importantly, a clinical trial using a similar strategy (ClinicalTrials.gov identifier: NCT02363452) reported positive effects of a therapy consisting of a combination of three RTis (lamivudine, abacavir, and zidovudine) in patients with AGS. Treatment resulted in a reduction in the IFN score and IFN-α protein levels. Interestingly, this effect was greatest among the four patients with mutations in components of the RNase H2 complex (Rice et al. 2018). These results support the hypothesis that RTi therapy can prevent IFN signaling in AGS patients by inhibiting reverse transcription driven by endogenous retrotransposons. However, RTis have also been reported to provide an intrinsic anti-inflammatory effect independent of TLRs, the cGAS/STING pathway and its reverse transcriptase inhibition activity (Tarallo et al. 2012; Kerur et al. 2013; Fowler et al. 2014). Taken together, the available data suggest that accumulation of reverse-transcribed DNA species results in an inflammatory phenotype mediated by the cGAS/STING pathway which can be reversed by RTis; however, the exact contribution of their intrinsic anti-inflammatory effect remains unclear.
There are other unanswered questions regarding the mechanisms underlying the induction of the IFN response by retrotransposon-intermediates in AGS and senescence. For instance, it is unclear how the LINE-1 cDNA accumulates in the cytoplasm of AGS cellular models to engage with cGAS for IFN activation. As previously suggested, one possibility is that the LINE-1 cDNA is synthesized directly in this cellular compartment. For this, the LINE-1 RNA and encoded proteins might perform reverse transcription in situ using a nonstandard primer in the cytoplasm.
We propose a model in which by-products of active endogenous retrotransposons in addition to the DNA damage associated with some AGS-mutations are drivers of the IFN response. Clearly, further work is needed to determine the importance of retroelement activity and the detection of retroelement-derived nucleic acids by innate immune sensors in AGS.
Systemic lupus erythematosus
The first noninfectious human disease to be associated with an increase in type I IFN activity was systemic lupus erythematosus (SLE) (Hooks et al. 1979). SLE is an autoimmune disease characterized by the production of autoantibodies targeting nucleic acids and nuclear-associated proteins. A link between AGS and lupus was highlighted by the discovery of TREX1, SAMHD1, RNASEH2A/B/C, and IFIH1 mutations in SLE patients (Lee-Kirsch et al. 2007; Cunninghame Graham et al. 2011; Ravenscroft et al. 2011; Günther et al. 2015). Crow and Rehwinkel were the first to report the significant overlap between AGS and lupus and highlighted the interplay between nucleic acid metabolism and autoimmune disease (Crow and Rehwinkel 2009). Shortly after, Crow put forward LINE-1 retrotransposons as potential endogenous stimuli triggering the immune response and promoting autoimmunity (Crow 2010). Indeed, TEs are dysregulated in SLE and LINE-1 mRNA transcripts were found to be increased, inducing type I IFN production in vitro through both TLR-dependent and TLR-independent pathways (Mavragani et al. 2016; Kelly et al. 2018). In addition, the relationship between autoimmunity and TEs also involves the production of autoantibodies. Autoantibodies against the RNA binding protein Ro60 are present in individuals with SLE and other autoimmune disorders. Alu elements were found associated with the Ro60 immune complexes from the blood of individuals with lupus (Hung et al. 2015), and autoantibodies against LINE-1 ORF1 are characteristic of a population of SLE patients with severe and active disease and higher type I IFN score (Carter et al. 2020).
Unlike non-LTRs, the potential role of HERV in autoimmunity has been more widely explored. HERV proteins can be recognized by the host as exogenous and trigger an immune response and antibody production. Patients suffering from autoimmune disorders present autoantibodies against the gag and env regions of retroviruses that can cross-react with endogenous factors. In SLE for example, patients produce autoantibodies against the p30 gag protein from the endogenous retrovirus HRES-1 that can cross-react with the common autoantigen U1-70K (Perl et al. 1995). Similar findings in other autoimmune diseases have led to the formulation of a model in which HERV antigens stimulate immunity due to their similarity to exogenous viral proteins, a process termed “molecular mimicry.” Several reviews have extensively discussed the evidence for HERV in the aetiopathogenesis of autoimmune disorders, including lupus, diabetes mellitus, multiple sclerosis, Sjögren's syndrome, and rheumatoid arthritis, among others (Balada et al. 2009, 2010; Brodziak et al. 2012; Morandi et al. 2017; Grandi and Tramontano 2018a,b; Greenig 2019; Morris et al. 2019; Talotta et al. 2020).
CONCLUDING REMARKS: IS THE RELATIONSHIP BETWEEN TEs AND IFNs RELEVANT FOR EARLY DEVELOPMENT?
The expression of ERVs and non-LTR retrotransposons is generally silenced in mammalian adult tissues; however, expression of retrotransposons is characteristic of germ cells, early embryos and embryonic stem cells (ESCs) (Peaston et al. 2004; Garcia-Perez et al. 2007; Macia et al. 2011; Schumann et al. 2019). The epigenetic reprogramming of cells during early development to erase germline-specific marks and acquire totipotent and pluripotent capacity involves a general demethylated state resulting in increased TE expression. After fertilization, the zygote undergoes several divisions despite being transcriptionally inactive. The embryonic genome becomes active during the two-cell stage in mouse, and eight-cell stage in humans. During these stages, cells are defined as totipotent, as they have the ability to generate a full organism, including the embryonic and extraembryonic tissues. Later in development, during the blastocyst stage (day 3.5 post-fertilization in mice and day 5 in humans), embryonic stem cells, which can be derived from the inner cell mass, are only pluripotent, which means that they can give rise to the three germ layers (ectoderm, endoderm, and mesoderm) that will form the embryo, but not to the extra-embryonic tissue, such as the placenta. Although many mechanisms have evolved to prevent the deleterious effects of TE activity during these stages, the expression of TEs may also be beneficial during early development. Interestingly, the waves of expression pattern of the different classes of transposable elements differs, suggesting that they could contribute to modulating the cellular fate during early development (for reviews, see Gerdes et al. 2016; Torres-Padilla 2020). For example, mouse MERVL is highly transcribed during the two-cell (2C)-stage, and sequences derived from this TE function as promoters to drive the expression of 2C-stage specific transcripts (Macfarlan et al. 2012). The HERVK is expressed during the human embryonic genome activation stage (eight-cell) and sustained during the blastocyst stage. During these stages, the HERVK-encoded protein Rec can bind the 3′UTR of cellular mRNAs promoting their association with polyribosomes (Grow et al. 2015). The expression of another primate-specific transposon, HERVH, is only activated later in development, during the blastocyst stage (Göke et al. 2015). Interestingly, the lncRNA derived from HERVH acts as a scaffold to maintain the pluripotency transcriptional network typical of stem cells through physical interaction with OCT4 and coactivators (Lu et al. 2014). More recently, the RNA from LINE-1 has been suggested to be required for the maintenance of embryonic stem cell self-renewal by silencing the 2C-stage transcriptional program (Percharde et al. 2018).
Despite the high expression of TEs during early development, TE-nucleic acids cannot trigger the IFN response since this pathway is known to be inactive during this stage. ESCs, both from human and mouse origin, are unable to synthesize IFNs upon infections with viruses or when challenged with viral DNA or RNA mimics, and the ability to produce IFNs only becomes active after differentiation (Burke et al. 1978; Chen et al. 2010; Wang et al. 2013; Witteveldt et al. 2019). In agreement, in vivo studies of mouse development confirmed that the ability to produce IFNs only becomes active after early postimplantation (Barlow et al. 1984). The inability of pluripotent cells to produce IFNs has been attributed to both the low expression of the RIG-I-like receptors and the action of miRNAs silencing on MAVS, a central factor for RNA immunity (Wang et al. 2013; Witteveldt et al. 2019). ESCs also fail to produce IFNs in response to cytoplasmic DNA, suggesting that the cGAS/STING pathway is also inactive during pluripotency (Witteveldt et al. 2019). Why the pluripotent stage of development needs to silence the IFN response is unknown, but we hypothesize it may be a requirement to prevent the aberrant production of IFNs upon recognition of TE-derived nucleic acids. In addition, this may be required to avoid the deleterious effects that IFN production has on ESC biology. For instance, constitutive activation of the type I IFN transcription factor IRF7 in ESCs has been shown to drive the expression of a subset of ISGs as well as dysregulation of pluripotency and lineage-specific genes (Eggenberger et al. 2019). All these findings together lead us to hypothesize that TEs have exerted a strong evolutionary pressure to supress the main antiviral response during early mammalian development.
ACKNOWLEDGMENTS
We thank Dr. Francisco Sanchez-Luque, Dr. Martin Reijns, and Priscilla Chin for helpful discussions and comments on the manuscript. This work was funded by grants from the Wellcome Trust 107665/Z/15/Z and Royal Society grant RGS\R1\191368 to S.M., and from MINECO SAF2015-71589-P and Ramón y Cajal grant RYC-2016-21395 to S.R.H. Figures were designed using Smart Servier Medical Art (https://smart.servier.com).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078721.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Achleitner M, Kleefisch M, Hennig A, Peschke K, Polikarpova A, Oertel R, Gabriel B, Schulze L, Lindeman D, Gerbaulet A, et al. 2017. Lack of Trex1 causes systemic autoimmunity despite the presence of antiretroviral drugs. J Immunol 199: 2261–2269. 10.4049/jimmunol.1700714 [DOI] [PubMed] [Google Scholar]
- Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang C-Z, Hur S. 2018. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172: 797–810.e13. 10.1016/j.cell.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413: 732–738. 10.1038/35099560 [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balada E, Ordi-Ros J, Vilardell-Tarrés M. 2009. Molecular mechanisms mediated by human endogenous retroviruses (HERVs) in autoimmunity. Rev Med Virol 19: 273–286. 10.1002/rmv.622 [DOI] [PubMed] [Google Scholar]
- Balada E, Vilardell-Tarrés M, Ordi-Ros J. 2010. Implication of human endogenous retroviruses in the development of autoimmune diseases. Int Rev Immunol 29: 351–370. 10.3109/08830185.2010.485333 [DOI] [PubMed] [Google Scholar]
- Bannert N, Kurth R. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet 7: 149–173. 10.1146/annurev.genom.7.080505.115700 [DOI] [PubMed] [Google Scholar]
- Bannert N, Hofmann H, Block A, Hohn O. 2018. HERVs new role in cancer: from accused perpetrators to cheerful protectors. Front Microbiol 9: 178. 10.3389/fmicb.2018.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuelos-Sanchez G, Sanchez L, Benitez-Guijarro M, Franco-Montalban F, Tamayo JA, Garcia-Perez JL. 2019. Synthesis and characterization of specific reverse transcriptase inhibitors for mammalian LINE-1 retrotransposons. Cell Chem Biol 26: 1095–1109.e14. 10.1016/j.chembiol.2019.04.010 [DOI] [PubMed] [Google Scholar]
- Barlow DP, Randle BJ, Burke DC. 1984. Interferon synthesis in the early post-implantation mouse embryo. Differentiation 27: 229–235. 10.1111/j.1432-0436.1984.tb01433.x [DOI] [PubMed] [Google Scholar]
- Bartsch K, Knittler K, Borowski C, Rudnik S, Damme M, Aden K, Spehlmann ME, Frey N, Saftig P, Chalaris A, et al. 2017. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum Mol Genet 26: 3960–3972. 10.1093/hmg/ddx283 [DOI] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran J V. 2011. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12: 187–215. 10.1146/annurev-genom-082509-141802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck-Engeser GB, Eilat D, Wabl M. 2011. An autoimmune disease prevented by anti-retroviral drugs. Retrovirology 8: 91. 10.1186/1742-4690-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Guijarro M, Lopez-Ruiz C, Tarnauskaite˙ Ž, Murina O, Mian Mohammad M, Williams TC, Fluteau A, Sanchez L, Vilar-Astasio R, Garcia-Canadas M, et al. 2018. RNase H2, mutated in Aicardi-Goutières syndrome, promotes LINE-1 retrotransposition. EMBO J 37: e98506. 10.15252/embj.201798506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden EC. 2019. Interferons α and β in cancer: therapeutic opportunities from new insights. Nat Rev Drug Discov 18: 219–234. 10.1038/s41573-018-0011-2 [DOI] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, et al. 2018. Ten things you should know about transposable elements. Genome Biol 19: 199. 10.1186/s13059-018-1577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JA, Spangler CJ, Strauss JD, Cesmat AP, Liu P, McGinty RK, Zhang Q. 2020. Structural basis of nucleosome-dependent cGAS inhibition. Science 370: eabd0609. 10.1126/science.abd0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodziak A, Ziółko E, Muc-Wierzgoń M, Nowakowska-Zajdel E, Kokot T, Klakla K. 2012. The role of human endogenous retroviruses in the pathogenesis of autoimmune diseases. Med Sci Monit 18: RA80. 10.12659/MSM.882892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Morant J V, Kazazian HH. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285. 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhwani M, Mazzieri R, Dolcetti R. 2018. Plasticity of type I interferon-mediated responses in cancer therapy: from anti-tumor immunity to resistance. Front Oncol 8: 322. 10.3389/fonc.2018.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DC, Graham CF, Lehman JM. 1978. Appearance of interferon inducibility and sensitivity during differentiation of murine teratocarcinoma cells in vitro. Cell 13: 243–248. 10.1016/0092-8674(78)90193-9 [DOI] [PubMed] [Google Scholar]
- Burke JM, Moon SL, Matheny T, Parker R. 2019. RNase L reprograms translation by widespread mRNA turnover escaped by antiviral mRNAs. Mol Cell 75: 1203–1217.e5. 10.1016/j.molcel.2019.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH. 2017. Transposable elements in cancer. Nat Rev Cancer 17: 415–424. 10.1038/nrc.2017.35 [DOI] [PubMed] [Google Scholar]
- Cao D, Han X, Fan X, Xu RM, Zhang X. 2020. Structural basis for nucleosome-mediated inhibition of cGAS activity. Cell Res 30: 1088–1097. 10.1038/s41422-020-00422-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi S, Church GM, Levanon EY. 2011. Large-scale DNA editing of retrotransposons accelerates mammalian genome evolution. Nat Commun 2: 519. 10.1038/ncomms1525 [DOI] [PubMed] [Google Scholar]
- Carter V, LaCava J, Taylor MS, Liang SY, Mustelin C, Ukadike KC, Bengtsson A, Lood C, Mustelin T. 2020. High prevalence and disease correlation of autoantibodies against p40 encoded by long interspersed nuclear elements in systemic lupus erythematosus. Arthritis Rheumatol 72: 89–99. 10.1002/art.41054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Yang L, Carmichael GG. 2010. Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle 9: 3552–3564. 10.4161/cc.9.17.12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci 103: 15588–15593. 10.1073/pnas.0604524103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Hwang S-Y, Ahn K. 2018. Interplay between RNASEH2 and MOV10 controls LINE-1 retrotransposition. Nucleic Acids Res 46: 1912–1926. 10.1093/nar/gkx1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, et al. 2018. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172: 811–824.e14. 10.1016/j.cell.2017.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Louis T, Robbe P, Ackroyd S, Burns A, Timbs AT, Wright Colopy G, Dreau H, Sigaux F, Judde JG, et al. 2014. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 123: 1021–1031. 10.1182/blood-2013-04-490847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AR, Elias HK, Ramsingh G. 2018. Senescence induction universally activates transposable element expression. Cell Cycle 17: 1846–1857. 10.1080/15384101.2018.1502576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel F, Silva MJ, Técher H, Zadorozhny K, Sharma S, Nieminuszczy J, Mettling C, Dardillac E, Barthe A, Schmitz AL, et al. 2018. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557: 57–61. 10.1038/s41586-018-0050-1 [DOI] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J 21: 5899–5910. 10.1093/emboj/cdf592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MK. 2010. Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease. Autoimmunity 43: 7–16. 10.3109/08916930903374865 [DOI] [PubMed] [Google Scholar]
- Crow YJ, Manel N. 2015. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol 15: 429–440. 10.1038/nri3850 [DOI] [PubMed] [Google Scholar]
- Crow YJ, Rehwinkel J. 2009. Aicardi-Goutières syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet 18: R130. 10.1093/hmg/ddp293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, Van Bokhoven H, Brunner HG, Hamel BC, et al. 2006a. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat Genet 38: 917–920. 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. 2006b. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet 38: 910–916. 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- Crow YJ, Vanderver A, Orcesi S, Kuijpers TW, Rice GI. 2014. Therapies in Aicardi-Goutières syndrome. Clin Exp Immunol 175: 1–8. 10.1111/cei.12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GMA, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, et al. 2015. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet Part A 167: 296–312. 10.1002/ajmg.a.36887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar L, Herzner AM, Zhang X, Goyal Y, Watanabe C, Friedman BA, Janakiraman V, Durinck S, Stinson J, Arnott D, et al. 2017. Silencing of retrotransposons by SET DB1 inhibits the interferon response in acute myeloid leukemia. J Cell Biol 216: 3535–3549. 10.1083/jcb.201612160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunninghame Graham DS, Morris DL, Bhangale TR, Criswell LA, Syvänen A-C, Rönnblom L, Behrens TW, Graham RR, Vyse TJ. 2011. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet 7: e1002341. 10.1371/journal.pgen.1002341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Kazazian HH. 1999. Analysis of the promoter from an expanding mouse retrotransposon subfamily. Genomics 56: 317–323. 10.1006/geno.1998.5729 [DOI] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM. 2013. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12: 247–256. 10.1111/acel.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, Caligiana A, Brocculi G, Adney EM, Boeke JD, et al. 2019. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566: 73–78. 10.1038/s41586-018-0784-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard P V, Borg A, Matthews N, Feng Q, van Kuppeveld FJM, et al. 2014. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife 3: e01535. 10.7554/eLife.01535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. 2011. Alu elements: know the SINEs. Genome Biol 12: 1–12. 10.1186/gb-2011-12-12-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MCN, Diedrich JK, Aslanian A, Ma J, Moresco JJ, et al. 2015. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163: 583–593. 10.1016/j.cell.2015.09.025 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48. 10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T, Yaniv M. 2005. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol 349: 241–247. 10.1016/j.jmb.2005.03.068 [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis E Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303: 1529–1531. 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- Eggenberger J, Blanco-Melo D, Panis M, Brennand KJ, Ten Oever BR. 2019. Type I interferon response impairs differentiation potential of pluripotent stem cells. Proc Natl Acad Sci 116: 1384–1393. 10.1073/pnas.1812449116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. 2009. DNA hypomethylation in cancer cells. Epigenomics 1: 239–259. 10.2217/epi.09.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433: 430–433. 10.1038/nature03238 [DOI] [PubMed] [Google Scholar]
- Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJM. 2012. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep 2: 1187–1196. 10.1016/j.celrep.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458: 509–513. 10.1038/nature07710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan DJ. 1989. Eukaryotic transposable elements and genome evolution. Trends Genet 5: 103–107. 10.1016/0168-9525(89)90039-5 [DOI] [PubMed] [Google Scholar]
- Flasch DA, Macia Á, Sánchez L, Ljungman M, Heras SR, García-Pérez JL, Wilson TE, Moran J V. 2019. Genome-wide de novo L1 retrotransposition connects endonuclease activity with replication. Cell 177: 837–851.e28. 10.1016/j.cell.2019.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd-Smith G, Slattery E, Lengyel P. 1981. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate–dependent endonuclease. Science 212: 1030–1032. 10.1126/science.6165080 [DOI] [PubMed] [Google Scholar]
- Fowler BJ, Gelfand BD, Kim Y, Kerur N, Tarallo V, Hirano Y, Amarnath S, Fowler DH, Radwan M, Young MT, et al. 2014. Nucleoside reverse transcriptase inhibitors possess intrinsic anti-inflammatory activity. Science 346: 1000–1003. 10.1126/science.1261754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Varshney A, Fowler BJ, Wang S, Narendran S, Ambati K, Yasuma T, Magagnoli J, Leung H, Hirahara S, et al. 2021. Cytoplasmic synthesis of endogenous Alu complementary DNA via reverse transcription and implications in age-related macular degeneration. Proc Natl Acad Sci 118: e2022751118. 10.1073/pnas.2022751118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Barber GN, Stetson DB. 2012. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36: 120–131. 10.1016/j.immuni.2011.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamdzyk M, Doycheva DM, Araujo C, Ocak U, Luo Y, Tang J, Zhang JH. 2020. cGAS/STING pathway activation contributes to delayed neurodegeneration in neonatal hypoxia-ischemia rat model: possible involvement of LINE-1. Mol Neurobiol 57: 2600–2619. 10.1007/s12035-020-01904-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Marchetto MCN, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran J V. 2007. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet 16: 1569–1577. 10.1093/hmg/ddm105 [DOI] [PubMed] [Google Scholar]
- Garcia-Perez JL, Widmann TJ, Adams IR. 2016. The impact of transposable elements on mammalian development. Dev 143: 4101–4114. 10.1242/dev.132639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, et al. 2009. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 41: 1228–1233. 10.1038/ng.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili M, Lahaye X, Nadalin F, Nader GFP, Puig Lombardi E, Herve S, De Silva NS, Rookhuizen DC, Zueva E, Goudot C, et al. 2019. The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep 26: 2377–2393.e13. 10.1016/j.celrep.2019.01.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, John L, Samuel CE. 2014. An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1). J Interf Cytokine Res 34: 437–446. 10.1089/jir.2014.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes P, Richardson SR, Mager DL, Faulkner GJ. 2016. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol 17: 100. 10.1186/s13059-016-0965-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glück S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, Bridgeman A, Rehwinkel J, Zender L, Ablasser A. 2017. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol 19: 1061–1070. 10.1038/ncb3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, Lu X, Chan YS, Ng HH, Ly LH, Sachs F, Szczerbinska I. 2015. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16: 135–141. 10.1016/j.stem.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, et al. 2014. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514: 372–375. 10.1038/nature13590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutières F, Aicardi J, Barth PG, Lebon P. 1998. Aicardi-Goutières syndrome: an update and results of interferon-α studies. Ann Neurol 44: 900–907. 10.1002/ana.410440608 [DOI] [PubMed] [Google Scholar]
- Grandi N, Tramontano E. 2018a. HERV envelope proteins: physiological role and pathogenic potential in cancer and autoimmunity. Front Microbiol 9: 462. 10.3389/fmicb.2018.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N, Tramontano E. 2018b. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol 9: 2039. 10.3389/fimmu.2018.02039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Treuting PM, Woodward JJ, Stetson DB. 2015. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi–Goutières syndrome. J Immunol 195: 1939–1943. 10.4049/jimmunol.1500969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Winship D, Snyder JM, Child SJ, Geballe AP, Stetson DB. 2016. The AIM2-like receptors are dispensable for the interferon response to intracellular DNA. Immunity 45: 255–266. 10.1016/j.immuni.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenig M. 2019. HERVs, immunity, and autoimmunity: understanding the connection. PeerJ 7: e6711. 10.7717/peerj.6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich W, Wagner M, Gaidt MM, Stafford C, Cheng Y, Linder A, Carell T, Hornung V. 2019. TLR8 is a sensor of RNase T2 degradation products. Cell 179: 1264–1275.e13. 10.1016/j.cell.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieves JL, Fye JM, Harvey S, Grayson JM, Hollis T, Perrino FW. 2015. Exonuclease TREX1 degrades double-stranded DNA to prevent spontaneous lupus-like inflammatory disease. Proc Natl Acad Sci 112: 5117–5122. 10.1073/pnas.1423804112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. 2015. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522: 221–246. 10.1038/nature14308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guey B, Wischnewski M, Decout A, Makasheva K, Kaynak M, Saka MS, Fierz B, Ablasser A. 2020. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 369: 823–828. 10.1126/science.aaw6421 [DOI] [PubMed] [Google Scholar]
- Günther C, Kind B, Reijns MAM, Berndt N, Martinez-Bueno M, Wolf C, Tüngler V, Chara O, Lee YA, Hübner N, et al. 2015. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J Clin Invest 125: 413–424. 10.1172/JCI78001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH. 2011. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet 20: 3386–3400. 10.1093/hmg/ddr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113: 803–809. 10.1016/S0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 10: 109–115. 10.1038/ni.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. 2004. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303: 1526–1529. 10.1126/science.1093620 [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408: 740–745. 10.1038/35047123 [DOI] [PubMed] [Google Scholar]
- Hernandez-Segura A, Nehme J, Demaria M. 2018. Hallmarks of cellular senescence. Trends Cell Biol 28: 436–453. 10.1016/j.tcb.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Herquel B, Ouararhni K, Martianov I, Le Gras S, Ye T, Keime C, Lerouge T, Jost B, Cammas F, Losson R, et al. 2013. Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nat Struct Mol Biol 20: 339–346. 10.1038/nsmb.2496 [DOI] [PubMed] [Google Scholar]
- Herrmann A, Wittmann S, Thomas D, Shepard CN, Kim B, Ferreirós N, Gramberg T. 2018. The SAMHD1-mediated block of LINE-1 retroelements is regulated by phosphorylation. Mob DNA 9: 11. 10.1186/s13100-018-0116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzner A-M, Hagmann CA, Goldeck M, Wolter S, Kübler K, Wittmann S, Gramberg T, Andreeva L, Hopfner K-P, Mertens C, et al. 2015. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol 16: 1025–1033. 10.1038/ni.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A. 2012. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med 209: 1419–1426. 10.1084/jem.20120876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. 1979. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med 301: 5–8. 10.1056/NEJM197907053010102 [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- Hoshino K, Sugiyama T, Matsumoto M, Tanaka T, Saito M, Hemmi H, Ohara O, Akira S, Kaisho T. 2006. IκB kinase-α is critical for interferon-α production induced by Toll-like receptors 7 and 9. Nature 440: 949–953. 10.1038/nature04641 [DOI] [PubMed] [Google Scholar]
- Hu S, Li J, Xu F, Mei S, Le Duff Y, Yin L, Pang X, Cen S, Jin Q, Liang C, et al. 2015. SAMHD1 inhibits LINE-1 retrotransposition by promoting stress granule formation. PLoS Genet 11: e1005367. 10.1371/journal.pgen.1005367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Pratt GA, Sundararaman B, Towsend MJ, Chaivorapol C, Bhangale T, Graham RR, Ortmann W, Criswell LA, Yeo GW, et al. 2015. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science 350: 455–459. 10.1126/science.aac7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. 2015. Regulation of type I interferon responses. Nat Rev Immunol 14: 36–49. 10.1038/nri3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-I and melanoma differentiation–associated gene 5. J Exp Med 205: 1601–1610. 10.1084/jem.20080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, Terai K, Matsuda M, Inoue JI, Uematsu S, et al. 2004. Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol 5: 1061–1068. 10.1038/ni1118 [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1441. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M, Lihua S, Zhe Z, Li S, Yoselin P, Michelle P, Sullivan Kathleen E. 2018. Transposable element dysregulation in systemic lupus erythematosus and regulation by histone conformation and Hsp90. Clin Immunol 197: 6–18. 10.1016/j.clim.2018.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Hirano Y, Tarallo V, Fowler BJ, Bastos-Carvalho A, Yasuma T, Yasuma R, Kim Y, Hinton DR, Kirschning CJ, et al. 2013. TLR-independent and P2X7-dependent signaling mediate Alu RNA-induced NLRP3 inflammasome activation in geographic atrophy. Investig Ophthalmol Vis Sci 54: 7395–7401. 10.1167/iovs.13-12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 16: 78–87. 10.1101/gr.4001406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodym R, Kodym E, Story MD. 2009. 2′–5′-Oligoadenylate synthetase is activated by a specific RNA sequence motif. Biochem Biophys Res Commun 388: 317–322. 10.1016/j.bbrc.2009.07.167 [DOI] [PubMed] [Google Scholar]
- Kretschmer S, Wolf C, König N, Staroske W, Guck J, Häusler M, Luksch H, Nguyen LA, Kim B, Alexopoulou D, et al. 2015. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann Rheum Dis 74: e17. 10.1136/annrheumdis-2013-204845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujirai T, Zierhut C, Takizawa Y, Kim R, Negishi L, Uruma N, Hirai S, Funabiki H, Kurumizaka H. 2020. Structural basis for the inhibition of cGAS by nucleosomes. Science 370: 455–458. 10.1126/science.abd0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Carmichael GG. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol Mol Biol Rev 62: 1415–1434. 10.1128/MMBR.62.4.1415-1434.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye X, Gentili M, Silvin A, Conrad C, Picard L, Jouve M, Zueva E, Maurin M, Nadalin F, Knott GJ, et al. 2018. NONO detects the nuclear HIV capsid to promote cGAS-mediated innate immune activation. Cell 175: 488–501.e22. 10.1016/j.cell.2018.08.062 [DOI] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, et al. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13: 223–228. 10.1038/ni.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, et al. 2001. Initial sequencing and analysis of the human genome: International Human Genome Sequencing Consortium. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, De Silva U, Bailey SL, Witte T, Vyse TJ, et al. 2007. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet 39: 1065–1067. 10.1038/ng2091 [DOI] [PubMed] [Google Scholar]
- Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, Sen GC, Komarova EA, Gudkov A V. 2013. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci 110: E89–E98. 10.1073/pnas.1216922110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005. 10.1038/nbt996 [DOI] [PubMed] [Google Scholar]
- Levin D, London IM. 1978. Regulation of protein synthesis: activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc Natl Acad Sci 75: 1121–1125. 10.1073/pnas.75.3.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kannan M, Trivett AL, Liao H, Wu X, Akagi K, Symer DE. 2014. An antisense promoter in mouse L1 retrotransposon open reading frame-1 initiates expression of diverse fusion transcripts and limits retrotransposition. Nucleic Acids Res 42: 4546–4562. 10.1093/nar/gku091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Du J, Goodier JL, Hou J, Kang J, Kazazian HH, Zhao K, Yu X-F. 2017a. Aicardi-Goutìeres syndrome protein TREX1 suppresses L1 and maintains genome integrity through exonuclease-independent ORF1p depletion. Nucleic Acids Res 45: 4619–4631. 10.1093/nar/gkx178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Banerjee S, Goldstein SA, Dong B, Gaughan C, Rath S, Donovan J, Korennykh A, Silverman RH, Weiss SR. 2017b. Ribonuclease l mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. Elife 6: e25687. 10.7554/eLife.25687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349: 1115–1120. 10.1126/science.aac7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YW, Sanz LA, Xu X, Hartono SR, Chédin F. 2015. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi–Goutières syndrome. Elife 4: e08007. 10.7554/eLife.08007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Paciencia S, Saint-Germain E, Rowell MC, Ruiz AF, Kalegari P, Ferbeyre G. 2019. The senescence-associated secretory phenotype and its regulation. Cytokine 117: 15–22. 10.1016/j.cyto.2019.01.013 [DOI] [PubMed] [Google Scholar]
- Lu X, Sachs F, Ramsay LA, Jacques PÉ, Göke J, Bourque G, Ng HH. 2014. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol 21: 423–425. 10.1038/nsmb.2799 [DOI] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605. 10.1016/0092-8674(93)90078-5 [DOI] [PubMed] [Google Scholar]
- Luecke S, Holleufer A, Christensen MH, Jønsson KL, Boni GA, Sørensen LK, Johannsen M, Jakobsen MR, Hartmann R, Paludan SR. 2017. cGAS is activated by DNA in a length-dependent manner. EMBO Rep 18: 1707–1715. 10.15252/embr.201744017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders KK, Kuff EL. 1977. Sequences associated with intracisternal A particles are reiterated in the mouse genome. Cell 12: 963–972. 10.1016/0092-8674(77)90161-1 [DOI] [PubMed] [Google Scholar]
- Macchietto MG, Langlois RA, Shen SS. 2020. Virus-induced transposable element expression up-regulation in human and mouse host cells. Life Sci Alliance 3: 201900536. 10.26508/lsa.201900536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, Marchal JA, Badge RM, Garcia-Perez JL. 2011. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol 31: 300–316. 10.1128/MCB.00561-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Lettice L, Tarnauskaite˙ Ž, Reddy K, Dix F, Revuelta A, Abbondati E, Rigby RE, Rabe B, et al. 2016. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J 35: 831–844. 10.15252/embj.201593339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. 2017. CGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548: 461–465. 10.1038/nature23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Schmidt T, Chauhan D, Goldeck M, Höning K, Gaidt M, Kubarenko A V, Andreeva L, Hopfner K, Hornung V. 2014. Cytosolic RNA:DNA hybrids activate the cGAS–STING axis. EMBO J 33: 2937–2946. 10.15252/embj.201488726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 9: 1482–1494. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion N, Arieti F, Gallo A, Keegan L, O'Connell M. 2015. New insights into the biological role of mammalian ADARs; the RNA editing proteins. Biomolecules 5: 2338–2362. 10.3390/biom5042338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavragani CP, Sagalovskiy I, Guo Q, Nezos A, Kapsogeorgou EK, Lu P, Liang Zhou J, Kirou KA, Seshan S V, Moutsopoulos HM, et al. 2016. Expression of long interspersed nuclear element 1 retroelements and induction of type I interferon in patients with systemic autoimmune disease. Arthritis Rheumatol 68: 2686–2696. 10.1002/art.39795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. 2001. Excision of 3′ termini by the Trex1 and TREX2 3′→5′ exonucleases. Characterization of the recombinant proteins. J Biol Chem 276: 17022–17029. 10.1074/jbc.M100623200 [DOI] [PubMed] [Google Scholar]
- Mehdipour P, Marhon SA, Ettayebi I, Chakravarthy A, Hosseini A, Wang Y, de Castro FA, Loo Yau H, Ishak C, Abelson S, et al. 2020. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 588: 169–173. 10.1038/s41586-020-2844-1 [DOI] [PubMed] [Google Scholar]
- Miller CL. 2011. Stress granules and virus replication. Future Virol 6: 1329–1338. 10.2217/fvl.11.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma H, Hanabuchi S, Kim T, Bao M, Zhang Z, Sugimoto N, Liu YJ. 2013. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity 39: 123–135. 10.1016/j.immuni.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927. 10.1016/S0092-8674(00)81998-4 [DOI] [PubMed] [Google Scholar]
- Morandi E, Tanasescu R, Tarlinton RE, Constantinescu CS, Zhang W, Tench C, Gran B. 2017. The association between human endogenous retroviruses and multiple sclerosis: a systematic review and meta-analysis. PLoS One 12: e0172415. 10.1371/journal.pone.0172415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. 2004. Gene-targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol 24: 6719–6727. 10.1128/MCB.24.15.6719-6727.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G, Maes M, Murdjeva M, Puri BK. 2019. Do human endogenous retroviruses contribute to multiple sclerosis, and if so, how? Mol Neurobiol 56: 2590–2605. 10.1007/s12035-018-1255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. 2006. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329. 10.1016/j.cell.2005.11.044 [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 281: 22161–22172. 10.1074/jbc.M601716200 [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish P V, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. 2009. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323: 1070–1074. 10.1126/science.1168352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova Z, Hubackova S, Kosar M, Janderova-Rossmeislova L, Dobrovolna J, Vasicova P, Vancurova M, Horejsi Z, Hozak P, Bartek J, et al. 2010. Cytokine expression and signaling in drug-induced cellular senescence. Oncogene 29: 273–284. 10.1038/onc.2009.318 [DOI] [PubMed] [Google Scholar]
- Oda H, Nakagawa K, Abe J, Awaya T, Funabiki M, Hijikata A, Nishikomori R, Funatsuka M, Ohshima Y, Sugawara Y, et al. 2014. Aicardi-Goutières syndrome is caused by IFIH1 mutations. Am J Hum Genet 95: 121–125. 10.1016/j.ajhg.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonski KM, Samuel CE. 2013. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J Virol 87: 756–766. 10.1128/JVI.02270-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orecchini E, Doria M, Antonioni A, Galardi S, Ciafrè SA, Ciafrè C, Frassinelli L, Mancone C, Montaldo C, Tripodi M, et al. 2016. ADAR1 restricts LINE-1 retrotransposition. Nucleic Acids Res 45: 155–168. 10.1093/nar/gkw834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-β induction. Nat Immunol 4: 161–167. 10.1038/ni886 [DOI] [PubMed] [Google Scholar]
- Ostendorf T, Zillinger T, Andryka K, Schlee-Guimaraes TM, Schmitz S, Marx S, Bayrak K, Linke R, Salgert S, Wegner J, et al. 2020. Immune sensing of synthetic, bacterial, and protozoan RNA by toll-like receptor 8 requires coordinated processing by RNase T2 and RNase 2. Immunity 52: 591–605.e6. 10.1016/j.immuni.2020.03.009 [DOI] [PubMed] [Google Scholar]
- Pathare GR, Decout A, Glück S, Cavadini S, Makasheva K, Hovius R, Kempf G, Weiss J, Kozicka Z, Guey B, et al. 2020. Structural mechanism of cGAS inhibition by the nucleosome. Nature 587: 668–672. 10.1038/s41586-020-2750-6 [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov A V, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606. 10.1016/j.devcel.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M. 2018. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174: 391–405.e19. 10.1016/j.cell.2018.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A, Colombo E, Dai H, Agarwal R, Mark KA, Banki K, Poiesz BJ, Phillips PE, Hoch SO, Reveille JD, et al. 1995. Antibody reactivity to the HRES-1 endogenous retroviral element identifies a subset of patients with systemic lupus erythematosus and overlap syndromes. Arthritis Rheum 38: 1660–1671. 10.1002/art.1780381119 [DOI] [PubMed] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. 2015. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43: 933–944. 10.1016/j.immuni.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis E Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- Pokatayev V, Hasin N, Chon H, Cerritelli SM, Sakhuja K, Ward JM, Douglas Morris H, Yan N, Crouch RJ. 2016. RNase H2 catalytic core Aicardi-Goutières syndrome-related mutant invokes cGAS-STING innate immunesensing pathway in mice. J Exp Med 213: 329–336. 10.1084/jem.20151464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S, Prangley E, Donovan J, Demarest K, Wingreen NS, Meir Y, Korennykh A. 2019. Concerted 2-5A-mediated mRNA decay and transcription reprogram protein synthesis in the dsRNA response. Mol Cell 75: 1218–1228.e6. 10.1016/j.molcel.2019.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VAK, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 11: 395–402. 10.1038/ni.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft JC, Suri M, Rice GI, Szynkiewicz M, Crow YJ. 2011. Autosomal dominant inheritance of a heterozygous mutation in SAMHD1 causing familial chilblain lupus. Am J Med Genet Part A 155: 235–237. 10.1002/ajmg.a.33778 [DOI] [PubMed] [Google Scholar]
- Reijns MAM, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. 2012. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149: 1008–1022. 10.1016/j.cell.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentoft M, Lindell K, Tran P, Chabes AL, Buckland RJ, Watt DL, Marjavaara L, Nilsson AK, Melin B, Trygg J, et al. 2016. Heterozygous colon cancer-associated mutations of SAMHD1 have functional significance. Proc Natl Acad Sci 113: 4723–4728. 10.1073/pnas.1519128113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, et al. 2009. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 41: 829–832. 10.1038/ng.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GMA, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. 2012. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet 44: 1243–1248. 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Del Toro Duany Y, Jenkinson EM, Forte GMA, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, et al. 2014. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 46: 503–509. 10.1038/ng.2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Meyzer C, Bouazza N, Hully M, Boddaert N, Semeraro M, Zeef LAH, Rozenberg F, Bondet V, Duffy D, et al. 2018. Reverse-transcriptase inhibitors in the Aicardi–Goutières syndrome. N Engl J Med 379: 2275–2277. 10.1056/NEJMc1810983 [DOI] [PubMed] [Google Scholar]
- Rice GI, Park S, Gavazzi F, Adang LA, Ayuk LA, Van Eyck L, Seabra L, Barrea C, Battini R, Belot A, et al. 2020. Genetic and phenotypic spectrum associated with IFIH1 gain-of-function. Hum Mutat 41: 837–849. 10.1002/humu.23975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran J V. 2014. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife 3: e02008. 10.7554/eLife.02008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran J V. 2015. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. In Mobile DNA III, pp. 1165–1208. ASM Press, Washington, DC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby RE, Webb LM, Mackenzie KJ, Li Y, Leitch A, Reijns MAM, Lundie RJ, Revuelta A, Davidson DJ, Diebold S, et al. 2014. RNA:DNA hybrids are a novel molecular pattern sensed by TLR9. EMBO J 33: 542–558. 10.1002/embj.201386117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero MP, Crow YJ. 2016. Type I interferon-mediated monogenic autoinflammation: the type I interferonopathies, a conceptual overview. J Exp Med 213: 2527–2538. 10.1084/jem.20161596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, et al. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162: 961–973. 10.1016/j.cell.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D. 2011. Dynamic control of endogenous retroviruses during development. Virology 411: 273–287. 10.1016/j.virol.2010.12.007 [DOI] [PubMed] [Google Scholar]
- Salter JD, Bennett RP, Smith HC. 2016. The APOBEC protein family: united by structure, divergent in function. Trends Biochem Sci 41: 578–594. 10.1016/j.tibs.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 2009. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci 106: 12067–12072. 10.1073/pnas.0900971106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32: 513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann GG, Fuchs N V, Tristán-Ramos P, Sebe A, Ivics Z, Heras SR. 2019. The impact of transposable element activity on therapeutically relevant human stem cells. Mob DNA 10: 1–23. 10.1186/s13100-019-0151-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122: 669–682. 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Simon M, Van Meter M, Ablaeva J, Ke Z, Gonzalez RS, Taguchi T, De Cecco M, Leonova KI, Kogan V, Helfand SL, et al. 2019. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab 29: 871–885.e5. 10.1016/j.cmet.2019.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remédios C, et al. 2014. Cancer cell–autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med 20: 1301–1309. 10.1038/nm.3708 [DOI] [PubMed] [Google Scholar]
- Smit AF. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev 9: 657–663. 10.1016/S0959-437X(99)00031-3 [DOI] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, et al. 2006. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet 38: 617–619. 10.1038/ng1800 [DOI] [PubMed] [Google Scholar]
- Speek M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 21: 1973–1985. 10.1128/MCB.21.6.1973-1985.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasachar Badarinarayan S, Shcherbakova I, Langer S, Koepke L, Preising A, Hotter D, Kirchhoff F, Sparrer KM, Schotta G, Sauter D. 2020. HIV-1 infection activates endogenous retroviral promoters regulating antiviral gene expression. Nucleic Acids Res 48: 10890–10908. 10.1093/nar/gkaa832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134: 587–598. 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana T, van Essen D, Siol O, Bailly-Bechet M, Philippe C, Zine El Aabidine A, Pioger L, Nigumann P, Saccani S, Andrau JC, et al. 2019. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol Cell 74: 555–570.e7. 10.1016/j.molcel.2019.02.036 [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold GD. 1990. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol 10: 6718–6729. 10.1128/MCB.10.12.6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talotta R, Atzeni F, Laska MJ. 2020. The contribution of HERV-E clone 4-1 and other HERV-E members to the pathogenesis of rheumatic autoimmune diseases. APMIS 128: 367–377. 10.1111/apm.13039 [DOI] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al. 2012. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149: 847–859. 10.1016/j.cell.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CA, Tejwani L, Trujillo CA, Negraes PD, Herai RH, Mesci P, Macia A, Crow YJ, Muotri AR. 2017. Modeling of TREX1-dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell 21: 319–331.e8. 10.1016/j.stem.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie CH, Fernandes L, Conde L, Robbez-Masson L, Sumner RP, Peacock T, Rodriguez-Plata MT, Mickute G, Gifford R, Towers GJ, et al. 2018. KAP1 regulates endogenous retroviruses in adult human cells and contributes to innate immune control. EMBO Rep 19: e45000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari B, Jones AE, Caillet CJ, Das S, Royer SK, Abrams JM. 2020. p53 directly represses human LINE1 transposons. Genes Dev 34: 1439–1451. 10.1101/gad.343186.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla ME. 2020. On transposons and totipotency. Philos Trans R Soc Lond B Biol Sci 375: 20190339. 10.1098/rstb.2019.0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunbak H, Enriquez-Gasca R, Tie CH, Gould PA, Mlcochova P, Gupta RK, Fernandes L, Holt J, van der Veen AG, Giampazolias E, et al. 2020. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat Commun 11: 5387. 10.1038/s41467-020-19170-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Cerritelli SM, Hasin N, Sakhuja K, London M, Iranzo J, Chon H, Grinberg A, Crouch RJ. 2018. Two RNase H2 mutants with differential rNMP processing activity reveal a threshold of ribonucleotide tolerance for embryonic development. Cell Rep 25: 1135–1145.e5. 10.1016/j.celrep.2018.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. 2014. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 5: 5011. 10.1038/ncomms6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Cambier S, Gray EE, Stetson DB. 2019. Tight nuclear tethering of cGAS is essential for preventing autoreactivity. Elife 8: e47491. 10.7554/eLife.47491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K. 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290: 1765–1768. 10.1126/science.290.5497.1765 [DOI] [PubMed] [Google Scholar]
- Wang J, Geesman GJ, Hostikka SL, Atallah M, Blackwell B, Lee E, Cook PJ, Pasaniuc B, Shariat G, Halperin E, et al. 2011. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 10: 3016–3030. 10.4161/cc.10.17.17543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Wang J, Paul AM, Acharya D, Bai F, Huang F, Guo YL. 2013. Mouse embryonic stem cells are deficient in type I interferon expression in response to viral infections and double-stranded RNA. J Biol Chem 288: 15926–15936. 10.1074/jbc.M112.421438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiatrek DM, Candela ME, Sedmík J, Oppelt J, Keegan LP, O'Connell MA. 2019. Activation of innate immunity by mitochondrial dsRNA in mouse cells lacking p53 protein. RNA 25: 713–726. 10.1261/rna.069625.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witteveldt J, Knol LI, Macias S. 2019. MicroRNA-deficient mouse embryonic stem cells acquire a functional interferon response. Elife 8: e44171. 10.7554/eLife.44171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339: 826–830. 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, Qu Y, Fan Z. 2018. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48: 688–701. 10.1016/j.immuni.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HH. 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol 13: 763–771. 10.1038/nsmb1141 [DOI] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE. 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 131: 873–886. 10.1016/j.cell.2007.10.017 [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Ren J, Chen Q, Chen ZJ. 2017. cGAS is essential for cellular senescence. Proc Natl Acad Sci 114: E4612–E4620. 10.1073/pnas.1705499114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo Y-M, Gale M, Akira S, et al. 2005. Shared and unique functions of the DExD/H-Box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175: 2851–2858. 10.4049/jimmunol.175.5.2851 [DOI] [PubMed] [Google Scholar]
- Zhang A, Dong B, Doucet AJ, Moldovan JB, Moran J V, Silverman RH. 2014. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res 42: 3803–3820. 10.1093/nar/gkt1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, et al. 2013. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutières syndrome-related SAMHD1. Cell Rep 4: 1108–1115. 10.1016/j.celrep.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ning S, Nolley R, Scicinski J, Oronsky B, Knox SJ, Peehl DM. 2017. The immunomodulatory anticancer agent, RRx-001, induces an interferon response through epigenetic induction of viral mimicry. Clin Epigenetics 9: 4. 10.1186/s13148-017-0312-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Du J, Peng Y, Li P, Wang S, Wang Y, Hou J, Kang J, Zheng W, Hua S, et al. 2018. LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways. J Autoimmun 90: 105–115. 10.1016/j.jaut.2018.02.007 [DOI] [PubMed] [Google Scholar]
- Zierhut C, Yamaguchi N, Paredes M, Luo JD, Carroll T, Funabiki H. 2019. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178: 302–315.e23. 10.1016/j.cell.2019.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]