Abstract
The current ecosystem of single-cell RNA-seq platforms is rapidly expanding, but robust solutions for single-cell and single-molecule full-length RNA sequencing are virtually absent. A high-throughput solution that covers all aspects is necessary to study the complex life of mRNA on the single-cell level. The Nanopore platform offers long read sequencing and can be integrated with the popular single-cell sequencing method on the 10× Chromium platform. However, the high error-rate of Nanopore reads poses a challenge in downstream processing (e.g., for cell barcode assignment). We propose a solution to this particular problem by using a hybrid sequencing approach on Nanopore and Illumina platforms. Our software ScNapBar enables cell barcode assignment with high accuracy, especially if sequencing saturation is low. ScNapBar uses unique molecular identifier (UMI) or Naïve Bayes probabilistic approaches in the barcode assignment, depending on the available Illumina sequencing depth. We have benchmarked the two approaches on simulated and real Nanopore data sets. We further applied ScNapBar to pools of cells with an active or a silenced nonsense-mediated RNA decay pathway. Our Nanopore read assignment distinguishes the respective cell populations and reveals characteristic nonsense-mediated mRNA decay events depending on cell status.
Keywords: Bayesian, 10× genomics, cell barcode assignment, nonsense-mediated mRNA decay (NMD)
INTRODUCTION
Full-length cDNA sequencing allows us to investigate the differential isoforms of transcripts, which is especially useful in studying the complex life of mRNA. Compared to the Illumina sequencing approaches, third-generation sequencing generates much longer reads and thus avoids artifacts from transcriptome assembly, but often has limitations such as low throughput and poor base-calling accuracy. Two principal third-generation sequencing platforms exist: Oxford Nanopore Technologies (ONT) and Pacific Biosciences (PacBio) (Volden et al. 2018). Others and we chose the ONT platform to study full-length mRNA transcripts due to its better scalability and flexibility (Lebrigand et al. 2020). Full-length transcriptome sequencing can be taken to the single level by sequencing barcoded 10× Genomics cDNA libraries. However, this brings about certain challenges, which we address in our work.
First, the native error rate of Nanopore DNA sequencing is <5% on the latest R10.3 platform (http://nanoporetech.com) as opposed to the typical Illumina error rate of 0.1%. Due to its high error rate, barcode identification and assignment are challenging for single-cell sequencing. In the 10× Genomics single-cell protocol, about 99% barcode sequences from Illumina sequencing can be exactly matched to the 16-bp cell barcodes, while with Nanopore sequencing, the exact matches are <50% (0.99916 vs. 0.9516). Many experimental and computational approaches have been developed to correct Nanopore data. For example, the rolling circle to concatemeric consensus (R2C2) approach can produce two million full-length cDNA sequences per MinION flow cell and achieved 98% accuracy (Volden et al. 2018; Cole et al. 2020; Volden and Vollmers 2020). Single-cell Nanopore sequencing with UMIs (ScNaUmi-seq) can assign cellular barcodes with 99.8% accuracy (Lebrigand et al. 2020). However, R2C2 requires sufficient sequencing coverage to call consensus reads, and ScNaUmi-seq requires high sequencing depth to guarantee an adequate overlap of UMI sequences between Illumina and Nanopore libraries.
On the other hand, end-to-end solutions for barcode demultiplexing and read quality filtering on the ONT platform are still in its infancy. For example, Mandalorion uses BLAT (Kent 2002) for barcode demultiplexing (Byrne et al. 2017). Porechop (https://github.com/rrwick/Porechop) uses SeqAn (Döring et al. 2008) for adapter removal and barcode demultiplexing in Nanopore sequencing, but it is based on the best alignment, which could be error-prone. Minibar (Krehenwinkel et al. 2019), Deep-binner (Wick et al. 2018), and DeePlexiCon (Smith et al. 2020) are only suitable for multiplexing a few barcoded samples rather than the single-cell library which contains several thousands of barcodes.
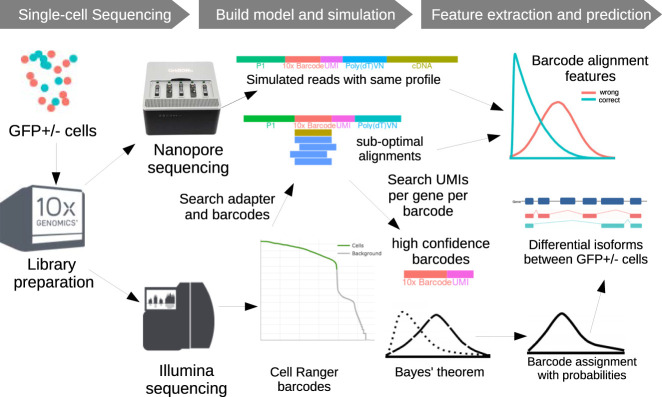
Therefore, we developed a software tool called ScNapBar (single-cell Nanopore barcode demultiplexer) that demultiplexes Nanopore barcodes and is particularly suited for low depth Illumina and Nanopore sequencing. We evaluated the performance of ScNapBar and demonstrated its high accuracy in cell barcode assignment for simulated and real Nanopore data. Our workflow is presented in Figure 1.
FIGURE 1.
Combined single-cell Illumina and Nanopore sequencing strategy. GFP+/− cells are pooled and sequenced on the Illumina and Nanopore platform. The Nanopore platform generates long cDNA sequencing reads that are used in barcode calling and estimating read error parameters. The Illumina data are used to estimate the total number of cells in sequencing and the represented cell barcodes. The simulated data are then used to parameterize a Bayesian model of barcode alignment features to discriminate correct versus false barcode assignments. This model is then used on the real data to assign cell barcodes to Nanopore reads. The GFP label and known NMD transcripts can be used to validate this assignment.
RESULTS
Benchmarking the two ScNapBar run modes
ScNapBar offers two run modes. The first one uses cell barcode and UMI information without any additional modeling aspect. The second one introduces a probabilistic model, which performs very well in cases of low sequencing saturation (i.e., UMI coverage in Illumina data).
The UMI approach of ScNapBar
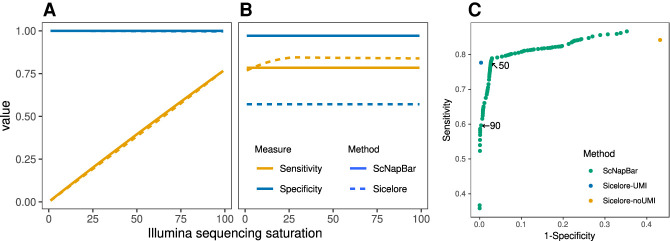
The UMI approach requires a matching cell barcode and UMI tag and was first developed in Sicelore (Lebrigand et al. 2020). Any cell barcode predictions that are supported by the presence of both, barcode and UMI alignment, are very reliable. We performed an in silico benchmark of cell barcode assignment when both, cell barcode and UMI, are found in the Nanopore read. We observed an average specificity of 99.9% (ScNapBar) and 99.8% (Sicelore) over 100 averaged simulation runs (Fig. 2A). As expected, sensitivity heavily depends on Illumina sequencing saturation (Fig. 2A). As the UMI approach relies on consistent genomic mappings for the Illumina and Nanopore reads, other challenges include insufficient or inaccurate genome annotations causing wrong gene assignment; chimeric or super-long Nanopore reads assigned to multiple genes increase the risk of assigning a false UMI.
FIGURE 2.
Sensitivity and specificity of ScNapBar and Sicelore on 100 Illumina libraries with different levels of saturation. (A) Barcode assignment with UMI matches. (B) Barcode assignment without UMI matches (ScNapBar score >50). (C) Benchmark of the specificity and sensitivity of the Illumina library with 100% saturation. We compared the barcode assignments with ScNapBar score >1–99, and the assignments from Sicelore with UMI support are roughly equivalent to the ScNapBar score >90.
The probabilistic approach of ScNapBar
Complementary to the UMI approach, we implemented a Bayesian approach in ScNapBar, which covers the situation of low Illumina sequencing saturation. In our second approach, UMI alignments are no longer used. ScNapBar evaluates probability scores for each barcode alignment instead. Illumina sequencing saturation measures the uniqueness of the transcripts detected in the Illumina library. Given that we have performed Illumina and Nanopore sequencing in our approach, the Illumina sequencing saturation limits the overlap of cell barcodes and UMIs with the low depth Nanopore libraries. To explore more realistic saturation scenarios, we estimated the Illumina sequencing saturation for our pilot data set with the Cell Ranger software. Herein, sequencing saturation is calculated as
| (1) |
where ndeduped reads is the number of unique (valid cell-barcode, valid UMI, gene) combinations among confidently mapped reads and nreads is the total number of confidently mapped, valid cell-barcode, valid UMI reads. For example, we have observed a saturation of 11.3% for our pilot data set.
We have simulated one million Nanopore reads with an error model, which was estimated from our reference Nanopore libraries (see Materials and Methods) using the same gene-barcode-UMI composition as given by the Illumina library and a sequencing saturation of 100%. We trained a Naïve Bayes classifier (see Materials and Methods) from barcode and adapter alignments of one Nanopore library and applied the model for computing the likelihood of the matched barcodes P (r|bi) on the other library. Then we used the frequencies of the given barcodes in the Illumina library as prior probabilities P (bi) and calculated the posterior probability P (bi|r) from the likelihood and prior probabilities. We scored each barcode alignment by multiplying the P (bi|r) by 100 and assigned the best matching barcode with the highest score (>50) as the predicted barcode assignment. Using the probability scores as mentioned, ScNapBar correctly assigned 65.8% barcodes from one million simulated Nanopore reads, of which 26.5% contains at least one mismatch or indel (Supplemental Fig. S1).
We estimate a user data specific error model, simulate data from which users pick the Bayes score cutoff, which meets their requirements on sensitivity and specificity, respectively. We inspected the densities of the probability scores by examining the ground-truth barcodes and confirmed that the correct barcode assignments are enriched in high scoring barcodes (Supplemental Fig. S2b).
Our probabilistic model outperforms Sicelore for cases where UMI information is sparse and cannot be used to assign cell barcodes. In the absence of UMIs, ScNapBar reaches 97.1% specificity while Sicelore reaches only 57.1% (Fig. 2B).
We examined performance metrics of cell barcode assignment over a range of score cutoffs (from 1 to 99), and the specificity increases while the sensitivity decreases along with the increased thresholds (Supplemental Fig. S3). We pooled the simulated results from FC1 and FC2 together and use the Sicelore assignments as baselines. At some cutoff thresholds, ScNapBar has better F1 scores than Sicelore (e.g., cutoff = 50), and ScNapBar score >90 is as accurate as Sicelore with UMI from the receiver-operating characteristic (ROC) graph (Fig. 2C).
The runtime performance of ScNapBar
ScNapBar is based on the Needleman–Wunsch algorithm (gap-end free, semiglobal sequence alignment) of FLEXBAR (Dodt et al. 2012; Roehr et al. 2017), and Sicelore is based on the “brute force approach,” which hashes all possible sequence tag variants (including indels) up to a certain edit distance (2 or 3) of the given barcode sequences. The time complexity of ScNapBar and Sicelore can be represented as Equations 2a and 2b, respectively:
| (2a) |
| (2b) |
where npos is the number of nucleotides downstream from the adapter, and lpos = 2npos + 1 as Sicelore typically searches the same number of nucleotides upstream and downstream from the ending position of the adapter. ncb stands for the number of barcodes in the whitelist from Illumina sequencing. ned is typically two or three as larger edit distances increase runtime drastically and are not necessarily due to the increasing error rate. lcb is the length of the barcode and is 16 in this study.
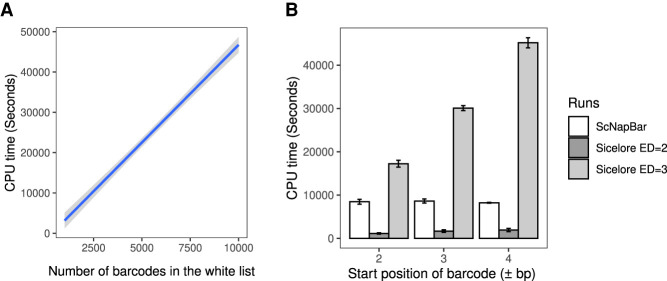
We compared the runtime between ScNapBar and Sicelore with regards to start positions of barcodes (number of nucleotides between adapter and barcode). We discovered that Sicelore may be orders of magnitude slower than ScNapBar given the same search space (2052 cellular barcodes, edit distance = 3), but also its runtime increases exponentially as the barcode start position increases (Fig. 3B). Sincelore by default searches ±1-nt from the end of the adapter, which may limit the nucleotides search space and could cause false assignments. Importantly, we did observe that 7.7% of all ground-truth barcodes have offsets >1-nt in the simulated read set, and 7.8% of all barcode assignments with ScNapBar score ≥50 in real Nanopore reads. We assessed the impact of various factors (e.g., indels ≥3 against <3) on cell barcode assignment accuracy using Fisher's test (see Supplemental Table S1). Our findings imply that larger offsets could effectively reduce the false-positive rate, which is feasible in less time with ScNapBar.
FIGURE 3.
Sicelore and ScNapBar CPU time comparison. (A) ScNapBar CPU time depends on the number of whitelist barcodes (allowing an edit distance of >2 and and offset of up to 4 bp between adapter and barcode). Gray area represents the standard deviation for 10 runs. (B) Comparison of ScNapBar and Sicelore CPU times. Benchmark was measured using one million barcode sequences and 2052 barcodes in the whitelist.
We also performed real runtime comparison on barcode assignment on the previously simulated one million Nanopore reads. In this test, we provided ScNapBar with ten barcode white lists, which contain from 1000 to 10,000 of the most abundant barcodes, and ScNapBar's runtime is only dependent on the number of barcodes to search given the other factors are fixed in this study (Fig. 3A). Then we tested Sicelore with searching parameters of barcode edit distance between two and three, barcode start position from ±2 bp to ±4 bp, and UMI edit distance of 0. ScNapBar requires only one-fifth CPU time than Sicelore when barcode start position = ±4 bp and barcode edit distance = 3 are considered in both programs (Fig. 3B).
The performance of ScNapBar on the real data
The performance of ScNapBar on an Illumina library with high sequencing saturations
We tested our ScNapBar software with the UMI approach (option 1) on the data set from the Sicelore paper (NCBI GEO GSE130708). Herein, Illumina sequencing saturation reaches 90.5%. Similar to Sicelore, we extracted the UMI whitelists for each gene or genomic window (500 bp) from the Illumina library. The 500 bp threshold is useful when a matched UMI is found in Illumina data for the same genomic region but not for the same gene. This situation may arise from incomplete gene annotations or mapping ambiguities. We observed that 99% of the Nanopore reads are within this 500 bp window when comparing the mapping positions of the Illumina and Nanopore reads that have the same UMIs in this data set. We set the minimum length of an UMI match to 7 in ScNapBar. In summary, Sicelore and ScNapBar assigned barcodes to 84.3% and 77.2% of the 9,743,819 Nanopore reads (Supplemental Fig. S4), respectively. 88.4% of the assigned barcodes are identical.
The performance of ScNapBar on an Illumina library with low sequencing saturations
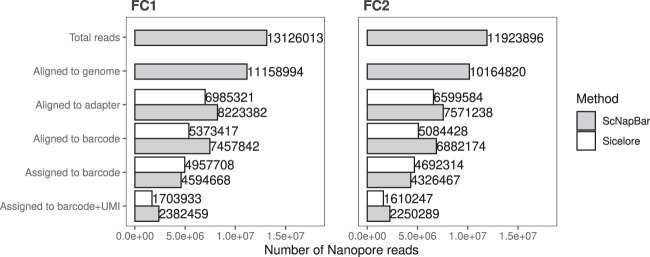
We ran ScNapBar with the Bayesian approach (option 2) on our NMD data set, which only has an Illumina saturation of 11.3%. ScNapBar assigns 35.0% and 36.3% of the Nanopore reads to cell barcodes with probability score >50, while Sicelore assigns 40.8% and 42.5% without using UMIs (“Assigned to barcode” in Fig. 4) and only assigns 4.0% and 4.2% of the Nanopore reads using the UMI approach for FC1 and FC2, respectively. The correlations of the number of gene/UMIs for each cell between Illumina and Nanopore imply good matches from the ScNapBar assignments (Supplemental Fig. S5).
FIGURE 4.
Number of the Nanopore reads identified by ScNapBar and Sicelore at each processing step. We inspected each processing step on real data (low lllumina saturation of 11.3%). The first two steps are identical for both workflows. Total Reads: Number of input reads, aligned to genome: Number of reads aligned to genome. The next three steps are workflow-specific: Aligned to adapter: Number of reads with identified adapter sequence, aligned to barcode: Number of reads with aligned barcode sequence, Assigned to barcode: Number of predictions by each workflow. The last step is a validation of the previous assignment step after additional Illumina sequencing, which increases the Illumina saturation to 52%, and using UMI matches, see main text.
In order to validate the correctness of our cell barcode assignments, we have sequenced ≈485 million additional Illumina reads from the same library. The saturation reaches 52% after combining both Illumina sequencing data (80.8 million reads from primary run and 485 million from secondary run). We searched UMIs in the Nanopore reads from the combined Illumina runs using both ScNapBar and Sicelore. We ran ScNapBar by allowing up to three edit distances between the UMIs of the Nanopore and Illumina reads, and ran Sicelore with the default parameters (–maxUMIfalseMatchPercent 1 –maxBCfalseMatchPercent 5). CBCs and UMIs must be matched for the same genomic region within a window of 500 nt as the Illumina library. We observed ≈50% of the barcode assignments by ScNapBar have matched UMIs (“Assigned to barcode + UMI” in Fig. 4).
Single-cell clustering and splicing in a pool of wild-type and NMD mutant cells
Although alternative splicing increases the coding potential of the human genome, aberrant isoforms are frequently generated that contain premature termination codons (PTCs) (Lewis et al. 2003). Regular stop codons are normally located in the last exon of a transcript or at most 50 nt upstream of the last exon–exon junction (Lindeboom et al. 2019). Alternative splicing can result in PTCs by exon inclusion/exclusion events or can convert normal stop codons into PTCs by splicing in the 3′-UTR. Transcripts harboring PTCs are rapidly degraded by the nonsense-mediated mRNA decay (NMD) machinery, not only to remove faulty mRNAs, but also to fine-tune and regulate the transcriptome. 5%–40% of all expressed human genes are directly or indirectly altered in expression levels, splicing pattern, or isoform composition by the NMD pathway (Boehm et al. 2020). We have sequenced a pool of NMD active and inactive cells and expect to see an enrichment of transcripts with PTCs in GFP− cells.
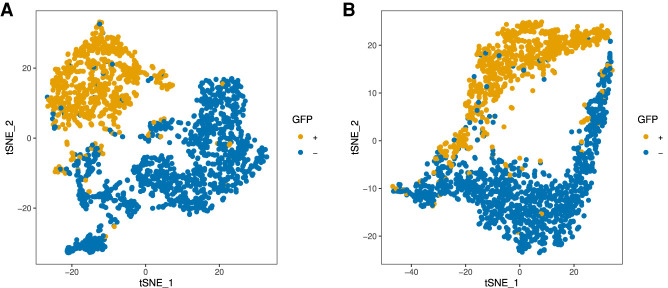
We use the GFP label as an independent confirmation of cellular NMD status and pooled data from both experiments (FC1 and FC2). For the Nanopore data, Seurat identifies 13,807 expressed genes across 1850 cells. We extracted the GFP+ barcodes from the Illumina reads mapping and rendered the corresponding cells in different colors in the t-SNE plots (Fig. 5). The locations of the GFP+ cells appear in distinct subclusters in the Illumina and Nanopore t-SNE plots.
FIGURE 5.
The t-SNE plots of gene-cell count matrices. (A) Illumina. (B) Nanopore.
We characterized the structural changes of the assembled Nanopore transcripts based on our customized transcriptome annotations using NMD Classifier (Hsu et al. 2017). The pool of SMG7-KO/SMG6-KD (GFP−) cells harbors almost twice as many inclusion/exclusion events, which lead to the formation of a PTC (Supplemental Fig. S6a). We quantified the expression level of 14,185 known NMD transcripts annotated by Ensembl release 101. After removing the nonexpressed transcripts from both flow cell runs, the remaining 6423 NMD transcripts have shown significantly higher NMD transcript expression in the SMG7-KO/SMG6-KD (GFP−) cells than the WT (GFP+) cells (Supplemental Fig. S6b). We reason that the lowered NMD response is clearly visible by the enrichment of PTC-containing transcripts in the pool of SMG7-KO/SMG6-KD (GFP−) cells. Consequently, the cell barcode assignments meet our “biological” expectations.
We investigated a well-established NMD target SRSF2 in detail (Sureau et al. 2001). The wild-type isoforms are present in both GFP+/− cells, while in the GFP− cells, the PTC-containing isoforms are more abundant in the GFP− cells (Supplemental Fig. S7a). The view on the SRSF2 genome locus confirmed the different splicing junctions between two cell types (Supplemental Fig. S7b). The inclusion of exon 3 (middle) is clearly favored GFP− cells.
DISCUSSION
The current ecosystem of single-cell RNA-seq platforms is rapidly expanding, but robust solutions for single-cell and single-molecule full-length RNA sequencing are virtually absent. In our manuscript, we combined Oxford Nanopore single-molecule sequencing of 10× Genomics cDNA libraries and developed a novel software tool to arrive at single-cell, single-molecule, full cDNA length resolution. In contrast to Lebrigand et al. (2020), our Bayesian method for cell barcode assignment performs superiorly in situations of low sequencing saturation. Even in the light of expected improvements of ONT sequencing error rates, ScNapBar offers improved performance in the aforementioned use case (see Supplemental Fig. S8). In summary, we could track in a well-controlled setting, that is, by using GFP labeled cells and strong transcriptome perturbations, full-length transcript information at a single-cell level. We have identified differential RNA splicing linked to NMD pathway activity across our cell population. Our high-throughput full-length RNA sequencing solution is a necessary step forward toward studying the complex life of mRNA on a single-cell level. This opens up unprecedented opportunities in low saturation settings such as multiplexed CRISPR-based screens.
MATERIALS AND METHODS
Single-cell samples preparation and experiment
We performed an experiment using two different Flp-In-T-REx-293 cell lines: the wild-type cell line with stably integrated FLAG-emGFP and a SMG7 knockout (KO) cell line (generated and established in Boehm et al. 2020). Wild-type cells (GFP+) were transfected with siRNA against Luciferase and the SMG7 KO cells (GFP−) were transfected with an siRNA against SMG6. Two days after siRNA transfection, we mixed both cell types at a 1:1 ratio with a target of 2000 cells in total. cDNA was prepared according to the 10× Genomics Chromium Single Cell 3′ Reagent Kit User Guide (v3 Chemistry) from the pool of ≈2000 cells with a yield of 1.68 µg cDNA (42 ng/µL concentration, 40 µL volume) and a fragment size of 1.5 kbp. At this point, all cDNA fragments carry the 16 nt cellular barcode and 12 nt UMI at the Poly (dT) end. 25% (10 µL) of this cDNA solution continued with the original 10× Genomics protocol to create an Illumina 3′ mRNA library with P5 and P7 Illumina adapters, and paired-end reads of this library will present cellular barcodes and UMI with the first read and a 90 nt second read containing 3′ mRNA sequence. Further on, we produced two ONT libraries with 200 ng each of the same cDNA (0.2 pmoles) with the ONT Direct cDNA Sequencing Kit SQK-DCS109 protocol according to the manufactures’ standard procedures. We sequenced each library on one GridIon flowcell (FLO-MIN106D R9 Version/R9.4.1) creating reads of full cDNA length that contained the same compositions of ≈2000 cellular barcodes as the Illumina data (based on the same cells) but a different composition of UMIs (different transcripts).
Illumina reads processing and identification of cellular barcodes
We used 10× Genomics Cell Ranger 3.1 (https://github.com/10XGenomics/cellranger) to map the Illumina reads onto the reference genome. In our NMD data set, the DNA sequences of luciferase were appended to the reference genome, and therefore the GFP+ cells can be called from Cell Ranger. Cell Ranger also corrects the sequencing errors in the barcode and unique molecular identifier (UMI) sequences. Cell Ranger estimates the number of cells using a Good-Turing frequency estimation model (https://support.10xgenomics.com) and characterized the identified barcodes into the cell-associated and background-associated barcodes. We used the cell-associated barcode sequences as the cellular barcode whitelist in the following analyses. Our Cell Ranger analysis estimated 2052 sequenced cells (Supplemental Table S2). The read counts per cell of the estimated cell barcodes are shown in Supplemental Figure S9.
Nanopore reads processing, mapping, and gene assignment
We sequenced the two independently prepared Nanopore libraries from the same cDNA on two Nanopore R9.4 GridION flow cells (FC1 and FC2). The base-calling of Nanopore reads was done using Guppy v3.3.3, resulting in 13,126,013 and 11,923,896 reads, respectively. We aligned the Nanopore reads onto the corresponding reference genome using minimap2 v2.17 (Li 2018) in the spliced alignment mode (-ax splice). The two Nanopore runs yielded 11,158,994 and 10,164,820 mappable reads, respectively. We further assigned gene names to Nanopore reads using the “TagReadWithGeneExon” program from the Drop-seq tools (Macosko et al. 2015). We assembled all the Nanopore reads and extended transcriptome annotations using StringTie v2.1.1 (Pertea et al. 2015). The FPKM level of the assembled transcripts were quantified using Ballgown v2.14.1 (Frazee et al. 2015).
Identification of the adapter, barcode, UMI, and poly(A)-tail sequences from Nanopore reads
We removed the cDNA sequences from Nanopore reads and extracted up to 100 bp from both ends. We developed a modified version of FLEXBAR (Dodt et al. 2012; Roehr et al. 2017) to align P1 primer adapter sequence with the following parameters (“-ao 10 -ae 0.3 -ag -2 -hr T -hi 10 -he 0.3 -be 0.2 -bg -2 -bo 5 -ul 26 -kb 3 -fl 100”). Then we aligned the Nanopore reads that have valid adapters to the cellular barcodes which have been previously identified by Cell Ranger. We scanned the poly(A) sequences using the homopolymer-trimming function of FLEXBAR downstream from the cell barcode. Once the poly(A) sequences were found, the UMI sequences between the poly(A) and barcode were searched using MUMmer 4.0 (Marçais et al. 2018) (with parameters “-maxmatch -b -c -l 7 -F”) and in-house scripts against the Illumina UMIs of the same cell and the same gene or genomic regions (±500 bp from each end of the reads). In the end, ScNapBar output the alignment score of the adapter, the number of mismatches and indel from the barcode alignment, the length of poly(A) and UMI sequences, as well as the length of the gap between the barcode and adapter. We use these features to estimate the likelihood of the barcode assignment in the steps shown in Figure 1.
Simulation and engineering of discriminative features from the barcode and adapter alignments
We characterized the correct and false barcode assignment by simulating Nanopore reads. We created some artificial template sequences which contain only the P1 primer, cellular barcode, and UMI sequences at the same frequencies as the Illumina library, followed by 20 bp oligo(dT) and 32 bp cDNA sequences. In the next step, we first used NanoSim (Yang et al. 2017) to estimate the error profile of our Nanopore library, then we generated one million Nanopore reads from the artificial template using the NanoSim simulator with the previously estimated error profile. We aligned the simulated Nanopore reads to the adapter and barcode sequences using ScNapBar. We compared the sequences in the simulated Nanopore reads and the sequences from the artificial template and labeled the assigned barcode as correct or false accordingly. By comparing sequence and alignment features [adaptor score, poly(T) length, barcode indel, barcode mismatch, barcode start] of correct and false assignments, we found that the two categories (false, true) could be discriminated by these features (Supplemental Fig. S10c). We then assessed the importance of each feature toward the correctness of the assignment (Supplemental Fig. S10a). As these features are uncorrelated (Supplemental Fig. S10b), we train a Naïve Bayes model from these features to predict the likelihood of the correctness of a barcode assignment.
Calculate cell barcode posterior probability using prior probabilities from the Illumina data set
We denote b1, b2, …, bn as barcodes that match to read r and define P (b1|r) as the probability that barcode b1 was sequenced given r is observed. Following Bayes’ theorem, P (b1|r) could be computed as in Equation 3a, and further computed as in Equation 3b according to the total probability theorem:
| (3a) |
| (3b) |
where P (r|b1) and P (r|bn) are computed by the Naïve Bayes predictor, and priors P (b1) and P (bn) can be estimated from the observed barcode counts in Illumina sequencing. For practical reasons, as the probabilities for the unaligned barcodes that contain a lot of mismatches are pretty low, we add a pseudocount of one to the denominator to represent them. Because we have sequenced the same library twice using the Nanopore and Illumina sequencer, we assume prior probabilities P (b) are the same for the Nanopore and the Illumina platform (Supplemental Fig. S2a).
Quality assessment and clustering of the single-cell libraries
A metagene body coverage analysis confirmed the near full-length character of the Nanopore approach (Supplemental Fig. S11a). After assigning gene names and cell barcodes to the Nanopore reads, we processed the gene-barcode expression matrix using Seurat v3.1.1 (Butler et al. 2018) by keeping all genes that are expressed in at least three cells, and cells with more than 200 genes expressed. We then scaled the expression matrix by a factor of 10,000 and log-normalized and performed the t-SNE analysis.
DATA DEPOSITION
All sequencing data were deposited in NBCI's BioProject database (BioProject ID PRJNA722142). ScNapBar workflow (code and tutorial) is available at https://github.com/dieterich-lab/single-cell-nanopore.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Etienne Boileau, Thiago Britto Borges, Tobias Jakobi for proofreading and comments. The authors are grateful to Marek Franitza for running the experiments on the 10× platform and to Christian Becker for running ONT sequencing. This work was supported by Informatics for Life funded by the Klaus Tschira Foundation.
Author contributions: Q.W. implemented the ScNapBar workflow, performed data analyses, and wrote the manuscript. S.B. modified the FLEXBAR implementation. V.B. performed cell culture experiments and helped to draft the manuscript. N.G. helped to draft the manuscript and acquired funding. J.A. performed all sequencing experiments and helped to draft the manuscript. C.D. supervised the project, performed data analyses, acquired funding, and wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078154.120.
Freely available online through the RNA Open Access option.
REFERENCES
- Boehm V, Kueckelmann S, Gerbracht JV, Britto-Borges T, Altmüller J, Dieterich C, Gehring NH. 2020. Nonsense-mediated mRNA decay relies on “two-factor authentication” by SMG5-SMG7. bioRxiv 10.1101/2020.07.07.191437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A, Beaudin AE, Olsen HE, Jain M, Cole C, Palmer T, DuBois RM, Forsberg EC, Akeson M, Vollmers C. 2017. Nanopore long-read RNA-seq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun 8: 16027. 10.1038/ncomms16027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Byrne A, Adams M, Volden R, Vollmers C. 2020. Complete characterization of the human immune cell transcriptome using accurate full-length cDNA sequencing. Genome Res 30: 589–601. 10.1101/gr.257188.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M, Roehr JT, Ahmed R, Dieterich C. 2012. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 1: 895–905. 10.3390/biology1030895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring A, Weese D, Rausch T, Reinert K. 2008. SeqAn: an efficient, generic C++ library for sequence analysis. BMC Bioinformatics 9: 11. 10.1186/1471-2105-9-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. 2015. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol 33: 243–246. 10.1038/nbt.3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MK, Lin HY, Chen FC. 2017. NMD Classifier: a reliable and systematic classification tool for nonsense-mediated decay events. PLoS ONE 12: e0174798. 10.1371/journal.pone.0174798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—The BLAST-like alignment tool. Genome Res 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krehenwinkel H, Pomerantz A, Henderson JB, Kennedy SR, Lim JY, Swamy V, Shoobridge JD, Graham N, Patel NH, Gillespie RG, et al. 2019. Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. Gigascience 8: giz006. 10.1093/gigascience/giz006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrigand K, Magnone V, Barbry P, Waldmann R. 2020. High throughput error corrected Nanopore single cell transcriptome sequencing. Nat commun 11: 4025. 10.1038/s41467-020-17800-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci 100: 189–192. 10.1073/pnas.0136770100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom RGH, Vermeulen M, Lehner B, Supek F. 2019. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat Genet 51: 1645–1651. 10.1038/s41588-019-0517-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol 14: e1005944. 10.1371/journal.pcbi.1005944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehr JT, Dieterich C, Reinert K. 2017. Flexbar 3.0: SIMD and multicore parallelization. Bioinformatics 33: 2941–2942. 10.1093/bioinformatics/btx330 [DOI] [PubMed] [Google Scholar]
- Smith MA, Ersavas T, Ferguson JM, Liu H, Lucas MC, Begik O, Bojarski L, Barton K, Novoa EM. 2020. Molecular barcoding of native RNAs using nanopore sequencing and deep learning. Genome Res 30: 1345–1353. 10.1101/gr.260836.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stévenin J, Soret J. 2001. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J 20: 1785–1796. 10.1093/emboj/20.7.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volden R, Vollmers C. 2020. Highly multiplexed single-cell full-length cDNA sequencing of human immune cells with 10X Genomics and R2C2. bioRxiv 10.1101/2020.01.10.902361 [DOI] [Google Scholar]
- Volden R, Palmer T, Byrne A, Cole C, Schmitz RJ, Green RE, Vollmers C. 2018. Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc Natl Acad Sci 115: 9726–9731. 10.1073/pnas.1806447115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Holt KE. 2018. Deepbinner: demultiplexing barcoded Oxford Nanopore reads with deep convolutional neural networks. PLoS Comput Biol 14: e1006583. 10.1371/journal.pcbi.1006583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Chu J, Warren RL, Birol I. 2017. NanoSim: nanopore sequence read simulator based on statistical characterization. Gigascience 6: 1–6. 10.1093/gigascience/gix089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.