Abstract
The stem cell–specific RNA-binding protein TRIM71/LIN-41 was the first identified target of the prodifferentiation and tumor suppressor miRNA let-7. TRIM71 has essential functions in embryonic development and a proposed oncogenic role in several cancer types, such as hepatocellular carcinoma. Here, we show that TRIM71 regulates let-7 expression and activity via two independent mechanisms. On the one hand, TRIM71 enhances pre-let-7 degradation through its direct interaction with LIN28 and TUT4, thereby inhibiting let-7 maturation and indirectly promoting the stabilization of let-7 targets. On the other hand, TRIM71 represses the activity of mature let-7 via its RNA-dependent interaction with the RNA-induced silencing complex (RISC) effector protein AGO2. We found that TRIM71 directly binds and stabilizes let-7 targets, suggesting that let-7 activity inhibition occurs on active RISCs. MiRNA enrichment analysis of several transcriptomic data sets from mouse embryonic stem cells and human hepatocellular carcinoma cells suggests that these let-7 regulatory mechanisms shape transcriptomic changes during developmental and oncogenic processes. Altogether, our work reveals a novel role for TRIM71 as a miRNA repressor and sheds light on a dual mechanism of let-7 regulation, uncovering a bistable switch between TRIM71 and let-7 miRNAs that regulates the balance between proliferation and differentiation.
Keywords: TRIM71/LIN-41, RNA-binding protein (RBP), E3 ubiquitin ligase, miRNAs, let-7 family, AGO2, LIN28, TUT4, embryonic development, hepatocellular carcinoma, stem cells and cancer biology
INTRODUCTION
TRIM71/LIN-41 was first discovered as a heterochronic gene in the nematode C. elegans, where it was identified as a major target of the prodifferentiation lethal 7 (let-7) miRNA family (Reinhart et al. 2000; Slack et al. 2000). Later studies confirmed the conservation of TRIM71/LIN-41 and its repression by let-7 miRNAs in other species (Pasquinelli et al. 2000; Slack et al. 2000; Kloosterman et al. 2004; Lancman et al. 2005; Schulman et al. 2005; Lin et al. 2007), highlighting the importance of this TRIM-NHL protein in the control of developmental processes. In mice, Trim71 is highly expressed during early embryonic development and its expression starts to decrease at around E10.5 due to an increase of let-7 and miR-125 miRNAs in the course of differentiation (Schulman et al. 2005). Despite its short expression window, TRIM71 is essential for embryonic development in several vertebrate and invertebrate species (Ecsedi and Grosshans 2013). Embryonic lethality in mice occurs at E12.5 and is accompanied by neural tube closure defects (Schulman et al. 2008; Cuevas et al. 2015; Mitschka et al. 2015), underscoring a fundamental role of TRIM71 in the development of the nervous system. Furthermore, recurrent TRIM71 mutations have been identified in congenital hydrocephalus (CH) patients (Furey et al. 2018), a brain developmental disease which is associated with neural tube closure defects (Kahle et al. 2016) and is characterized by an abnormal accumulation of cerebrospinal fluid (CSF) within the brain ventricles. Interestingly, TRIM71 expression has been observed in CSF-producing ependymal cells in the adult brain (Cuevas et al. 2015), revealing yet-unknown postnatal TRIM71 functions in the nervous system. Last, TRIM71 is known to be expressed in adult mouse testes (Rybak et al. 2009), and has been recently found to play an essential role in the embryonic development of the germline (Torres-Fernández et al. 2021) as well as in adult spermatogenesis (Du et al. 2020).
On the molecular level, TRIM71 has a set of specialized domains enabling its function as an E3 ubiquitin ligase (Chen et al. 2012; Nguyen et al. 2017; Ren et al. 2018; Hu et al. 2019) and an mRNA-binding and repressor protein (Chang et al. 2012; Loedige et al. 2013; Worringer et al. 2014; Mitschka et al. 2015; Torres-Fernández et al. 2019; Welte et al. 2019). A number of protein and mRNA targets regulated by TRIM71 have been linked to the developmental phenotypes observed in TRIM71-deficient mice (Chang et al. 2012; Chen et al. 2012; Loedige et al. 2013; Worringer et al. 2014; Mitschka et al. 2015; Nguyen et al. 2017; Li et al. 2019; Torres-Fernández et al. 2019). TRIM71 has also been described as a miRNA-binding protein (Treiber et al. 2017). Furthermore, several proteins involved in the miRNA pathway, namely AGO1, AGO2, AGO4, DICER, and LIN28B, are confirmed protein interactors of TRIM71 (Rybak et al. 2009; Chang et al. 2012; Loedige et al. 2013; Lee et al. 2014; Torres-Fernández et al. 2019). However, the role of TRIM71 in the regulation of miRNAs remains poorly understood. Previous studies in our laboratory showed that TRIM71-deficient murine embryonic stem cells (ESCs) had an altered miRNA expression landscape as compared to wild-type ESCs (Mitschka et al. 2015), but it is yet unclear how TRIM71 regulates miRNA expression. Furthermore, TRIM71 was reported to decrease global miRNA activity via ubiquitination and proteasomal degradation of AGO2 (Rybak et al. 2009). However, TRIM71-mediated changes in AGO2 protein stability could not be confirmed by several other studies (Chang et al. 2012; Chen et al. 2012; Loedige et al. 2013; Mitschka et al. 2015; Torres-Fernández et al. 2019). Thus, the functional significance of the interaction between TRIM71 and AGO2 remains elusive.
We therefore aimed to investigate the role of TRIM71 in the regulation of miRNA expression and activity. Our results show that TRIM71 is not only a let-7 target, but also a let-7 repressor which uses two independent mechanisms to regulate let-7 expression and activity. For the regulation of let-7 expression, TRIM71 interacts with the let-7 repressor complex formed by LIN28 and TUT4, which intercepts let-7 miRNAs at their precursor stage and labels them for degradation. Furthermore, the interaction between TRIM71 and AGO2 results in a specific repression of let-7 activity, without affecting AGO2 stability or global miRNA activity. Together, these TRIM71-mediated miRNA repression pathways alter the transcriptome of ESCs and hepatocellular carcinoma cells, resulting in the stabilization of multiple let-7 targets during developmental and oncogenic processes.
RESULTS
TRIM71 interferes with the last processing step of let-7 biogenesis in ESCs
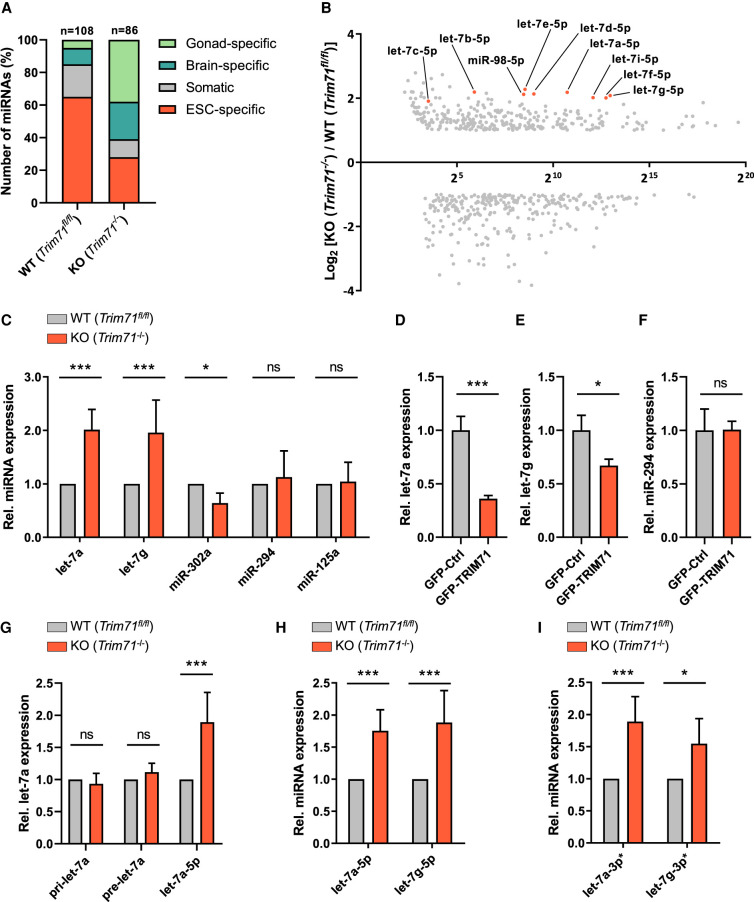
TRIM-NHL proteins have been connected to the miRNA pathway in several species (Wulczyn et al. 2010). In a previous study, we analyzed the impact of TRIM71 deficiency on the transcriptome of murine ESCs by performing RNA sequencing in undifferentiated wild-type (WT, Trim71fl/fl) and Trim71 knockout (KO, Trim71−/−) ESCs (Mitschka et al. 2015). Reanalysis of these data focusing on miRNA expression shows that the miRNome of TRIM71-deficient ESCs is substantially altered, with increased expression of differentiation-promoting brain-specific and gonad-specific miRNAs and reduced expression of several ESC-specific miRNAs (Fig. 1A; Mitschka et al. 2015). Interestingly, all members of the let-7 miRNA family were up-regulated in TRIM71-deficient ESCs (Fig. 1B). qRT-PCR analysis confirmed a significant twofold up-regulation of let-7a and let-7g species and the down-regulation of the stem cell–specific miR-302 in TRIM71-deficient ESCs (Fig. 1C; Supplemental Fig. 1A–D). The expression of other miRNAs such as miR-125, which targets Trim71 mRNA for degradation (Schulman et al. 2005, 2008), or the stem cell–specific miR-294, which functionally counteracts let-7 miRNAs (Melton et al. 2010), remained unaltered upon TRIM71 depletion (Fig. 1C; Supplemental Fig. 1E–H).
FIGURE 1.
TRIM71 interferes with the last processing step of let-7 biogenesis in ESCs. (A) Representation of relative expression differences in the miRNome of wild-type (WT, Trim71fl/fl) and Trim71 knockout (KO, Trim71−/−) ESCs (GSE62509). MiRNAs were classified as previously described into four categories (gonad-specific, brain-specific, ESC-specific, or somatic miRNAs) and displayed as percentages (n = 590). (B) Scatter plot showing the relative fold change (FC, with −1 ≤ FC > +1) of differentially expressed miRNAs in Trim71 KO cells plotted against their baseline expression level in WT ESCs (GSE62509). Guide strands (5p) of all let-7 family members are marked. (C) RT-qPCR showing relative expression of the indicated miRNAs in WT and Trim71 KO ESCs (n = 3–10). (D) RT-qPCR showing relative expression of let-7a, (E) let-7g and (F) miR-294 miRNAs in WT ESCs transiently overexpressing GFP-Ctrl or GFP-TRIM71 48 hpt (n = 3). (G) RT-qPCR showing relative pri-, pre- and mature let-7a expression in WT and Trim71 KO ESCs (n = 3–5). (H) RT-qPCR showing relative guide (5p) and (I) passenger (3p*) strand levels of mature let-7a and let-7g miRNAs in in WT and Trim71 KO ESCs (n = 3–5). RT-qPCR quantification of all miRNAs was normalized to the levels of the housekeeping U6 snRNA. Error bars represent SD. (***) P < 0.005, (**) P < 0.01, (*) P < 0.05, (ns) nonsignificant (unpaired Student's t-test). See also Supplemental Figures 1, 2.
Trim71 mRNA is a well-known let-7 target (Reinhart et al. 2000; Slack et al. 2000; Kloosterman et al. 2004; Schulman et al. 2005, 2008; Kanamoto et al. 2006; Lin et al. 2007; O'Farrell et al. 2008) but the fact that let-7 members are up-regulated upon TRIM71 depletion could suggest a double negative feedback loop between TRIM71 and let-7 miRNAs. To confirm that let-7 up-regulation was not an artefact of clonal selection but rather a direct consequence of TRIM71 depletion, we induced the deletion of the floxed Trim71 alleles in WT (Trim71fl/fl/Rosa26-CerERT2) ESCs via 4-hydroxytamoxifen (4-OHT) treatment and measured let-7a levels over time. We detected a gradual increase of let-7a miRNA expression that coincided with the loss of Trim71 mRNA and protein upon 4-OHT treatment (Supplemental Fig. 2A,B). Conversely, TRIM71 overexpression in WT ESCs resulted in a significant down-regulation of let-7a and let-7g, but not miR-294 (Fig. 1D–F). These results revealed a specific and direct involvement of TRIM71 in the regulation of let-7 expression in ESCs.
To determine at which stage of let-7 biogenesis this regulation occurs, we measured primary (pri-let-7a), precursor (pre-let-7a), and mature let-7a levels in WT and Trim71 KO ESCs. We found that mature let-7a levels were significantly up-regulated in Trim71 KO ESCs, whereas pri- and pre-miRNAs remained unaltered (Fig. 1G). This indicated that TRIM71 affects either the last step of let-7 maturation (i.e., pre-let-7 processing to mature let-7 miRNA duplex), or the stability of the mature miRNA (i.e., half-life of the let-7 guide strand). In order to distinguish between these two possibilities, we analyzed the relative abundance of guide (5p) and passenger (3p*) strands of let-7 miRNAs in WT and Trim71 KO ESCs and found both strands equally increased in Trim71 KO ESCs (Fig. 1H,I; Supplemental Fig. 2C,D). This suggested that TRIM71 interferes with the last processing step of let-7 biogenesis, resulting in the down-regulation of the mature miRNA duplex.
TRIM71 depends on LIN28A for the down-regulation of let-7 expression in ESCs
LIN28 proteins have an established role as inhibitors of let-7 biogenesis in undifferentiated stem cells (Heo et al. 2008, 2009). In vertebrates, two LIN28 paralogs exist—LIN28A and LIN28B—and both proteins are able to directly interact with pre-let-7 to promote its degradation (Nam et al. 2011; Mayr and Heinemann 2013). Additionally, LIN28B sequesters pri-let-7 and pre-let-7 inside the nucleus, preventing its processing and nuclear export (Piskounova et al. 2011). Notably, TRIM71 and LIN28 proteins have highly similar expression patterns during development in C. elegans, zebrafish and mouse (Ouchi et al. 2014). Furthermore, their expression is highly correlated in healthy adult human tissues (Supplemental Fig. 3A,B), as well as in tumor samples of different cancer types (Supplemental Fig. 3C,D; Tang et al. 2017).
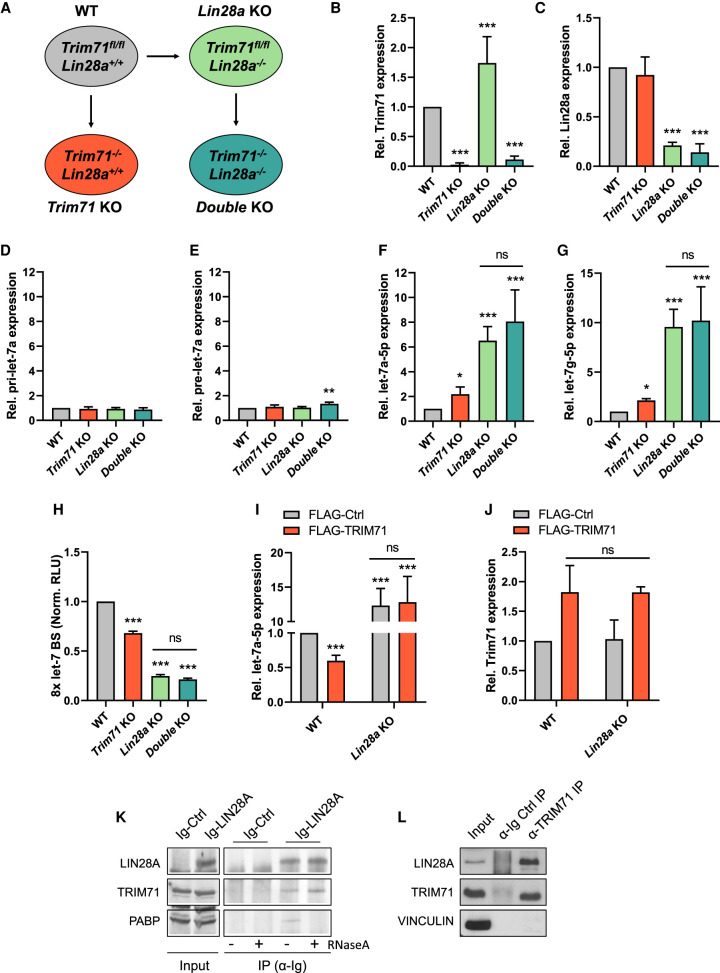
In order to investigate whether TRIM71-mediated let-7 regulation is linked to LIN28 function, we generated Lin28a KO ESCs (Trim71fl/fl; Lin28−/−) as well as double Trim71/Lin28a KO ESCs (Trim71−/−; Lin28−/−) (Fig. 2A–C). Similar to what we previously reported for Trim71 KO ESCs (Mitschka et al. 2015) and agreeing with previous studies in Lin28a KO ESCs (Zhang et al. 2016), we found that stemness was unimpaired in all KO cell lines. All cell lines had an ESC-characteristic morphology (Supplemental Fig. 4A,B) and a robust expression of stemness markers such as Nanog, Myc, and Pou5f1/Oct4 (Supplemental Fig. 4C–F). We then measured the relative levels of pri-let-7a, pre-let-7a and mature let-7a-5p and let-7g-5p miRNAs in WT, Trim71 KO, Lin28a KO, and double KO ESCs. Again, we found no changes in either primary or precursor miRNA expression (Fig. 2D,E), while the levels of mature let-7a-5p and let-7g-5p—but not miR-294 (Supplemental Fig. 4G)—were significantly increased in all KO cell lines (Fig. 2F,G). We also confirmed that the up-regulation of let-7 was not indirectly caused by changes in the expression of Lin28b or Zchcc11/tut4 (Supplemental Fig. 4C,H,I), which are also known to participate in the repression of let-7 expression (Heo et al. 2009; Thornton et al. 2012; Mayr and Heinemann 2013).
FIGURE 2.
TRIM71 depends on LIN28A for the down-regulation of let-7 expression in ESCs. (A) Schematic representation for the generation of the indicated ESC lines. Lin28a KO ESCs were generated from WT (Trim71fl/fl) ESCs via TALENs. Trim71 KO and Trim71–Lin28a double KO were generated by addition of 4-OHT to WT and Lin28a KO ESCs, respectively (see Materials and Methods for details). (B) RT-qPCR showing relative mRNA expression of Trim71 and (C) Lin28a in the generated ESC lines (n = 6–8). (D) RT-qPCR showing relative expression of pri-let-7a, (E) pre-let-7a, (F) mature let-7a-5p and (G) mature let-7g-5p miRNAs in the different ESC lines (n = 4–8). (H) Let-7 reporter assay after transient transfection of a Renilla luciferase reporter under the control of a 3′UTR containing 8x Let-7 binding sites (BS) in the different ESC lines (n = 3). (Norm. RLU) Normalized relative light units. (I) RT-qPCR showing relative expression of mature let-7a-5p and (J) Trim71 mRNA in WT and Lin28a KO ESCs transiently overexpressing FLAG-Ctrl or FLAG-TRIM71 48 hpt (n = 3–6). RT-qPCR quantification of miRNAs and mRNAs was normalized to the levels of the housekeeping U6 snRNA and Hprt mRNA, respectively. Error bars represent SD. (***) P < 0.005, (**) P < 0.01, (*) P < 0.05, (ns) nonsignificant (unpaired Student's t-test between WT and each KO condition, unless indicated by a line joining the two compared conditions). (K) Immunoblot showing the RNA-independent coprecipitation of endogenous TRIM71 with Ig-tagged LIN28A overexpressed in wild-type ESCs. PABP was used as a control for an RNA-dependent interaction with LIN28A. (L) Immunoblot showing the coprecipitation of endogenous LIN28A with endogenous TRIM71 in wild-type ESCs. See also Supplemental Figures 3, 4.
Importantly, although the up-regulation of let-7 was stronger in Lin28a KO ESCs than in Trim71 KO ESCs, no additional increase of mature let-7a/g was observed for the double KO compared to the single Lin28a KO (Fig. 2F,G). These results were confirmed by let-7 reporter assays in ESCs, in which a luciferase reporter containing 8x let-7 binding sites (BS) (Iwasaki et al. 2009) was significantly repressed in all KO lines, but no additional repression was observed in the double KO as compared to Lin28a KO ESCs (Fig. 2H). This suggested that the suppressive effect of TRIM71 on let-7 expression in ESCs is dependent on LIN28A. Indeed, while overexpression of TRIM71 in WT ESCs reduced let-7a levels by ∼40% (Fig. 2I), a comparable overexpression of TRIM71 had no effect on let-7a expression in Lin28a KO ESCs (Fig. 2I,J). These results collectively showed that TRIM71-mediated let-7 down-regulation in ESCs depends on LIN28A. We also confirmed a direct interaction between TRIM71 and LIN28A in WT ESCs (Fig. 2K,L).
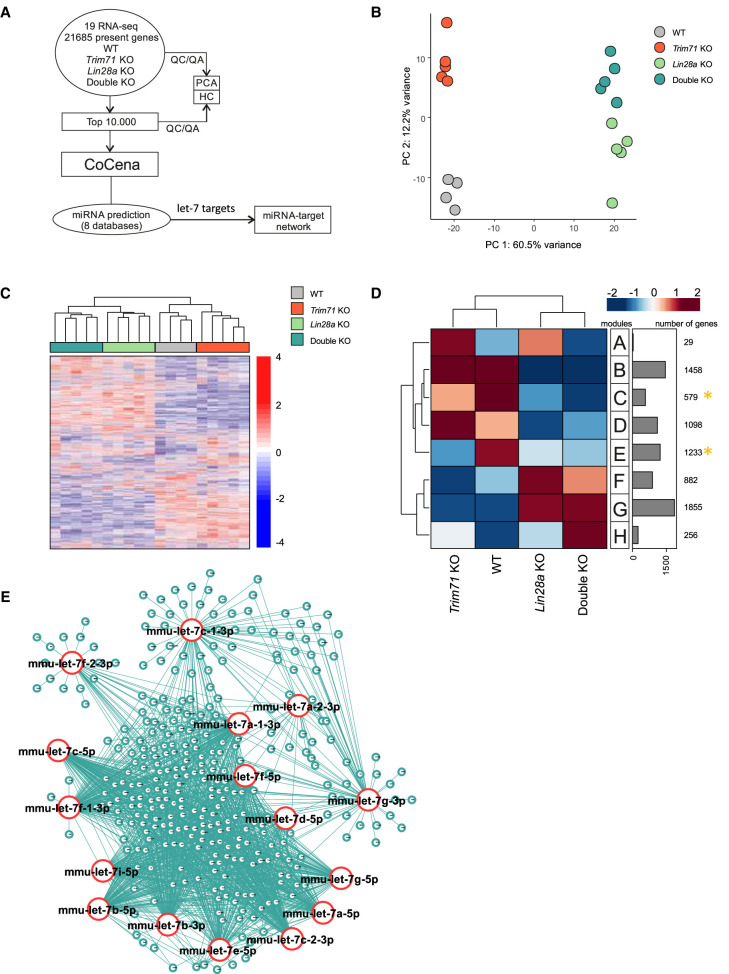
Our let-7 reporter assay (Fig. 2H) suggested that the changes in let-7 expression observed in Trim71 KO, Lin28a KO and double KO ESCs may have an impact on the expression of let-7 mRNA targets. In order to evaluate this effect on physiological targets endogenously expressed in ESCs, we conducted RNA-seq in WT, Trim71 KO, Lin28a KO and double KO ESCs (Fig. 3A). Principal component analysis (PCA) showed close clustering of sample replicates, but distinct clusters corresponding to each cell line, with Lin28a KO and double KO ESCs clustering close to each other (Fig. 3B,C). We then conducted coexpression network analysis (Fig. 3D) and found two groups of genes which were consistently down-regulated in all KO cell lines compared to WT ESCs (modules C and E, marked with a yellow asterisk in Fig. 3D). To evaluate the possibility that some of those down-regulated genes had been repressed by let-7, we conducted an unbiased miRNA enrichment analysis in silico using the online software ShinyGO v0.61 (Ge et al. 2020). This software identifies miRNA targets present in a list of genes and returns the top 100 miRNAs whose targets are significantly enriched in that list. Using the 1812 genes of modules C and E as input, we found the let-7 family among the top enriched miRNA families (P-value [FDR] = 2.7 × 10−07), with a total of 118 let-7 targets (≈6.5%) found within modules C and E (Supplemental Table 1A). However, a limitation of this analysis is that ShinyGO only considers data from a single chosen database. In order to increase the robustness of our analysis, we then integrated data from eight different miRNA databases (see details in Materials and Methods) and found a total of 329 (≈18.2%) predicted targets for the different let-7 family members present in the analyzed modules (Fig. 3E; Supplemental Table 1B). Collectively, our analysis indicated that a substantial part of the changes observed in the down-regulated transcriptome of each KO ESC line could be caused by the common up-regulation of let-7 miRNAs.
FIGURE 3.
Analysis of transcriptomic profiles of Trim71 KO, Lin28a KO and double KO ESCs reveals a common signature of let-7 targets down-regulation. (A) Schematic representation of the workflow for the transcriptomic analysis of ESCs. (B) Principal component analysis (PCA) showing individual sequencing experiments for each genotype (n = 4–5). (C) Hierarchical clustering of the 10,000 most variable genes among the different ESC lines. (D) Coexpression network analysis (CoCena) for the transcriptomes of the different ESC lines. Modules with commonly down-regulated genes among all KO ESC lines are marked with a yellow asterisk and were used for E. (E) miRNA network showing targets for the different let-7 family members found in modules C and E (marked with a yellow asterisk in D). See also Supplemental Figures 5, 6 and Supplemental Table 1.
Gene ontology (GO) term enrichment analysis showed that these let-7 mRNA targets mostly participate in functions related to cell cycle control (Supplemental Fig. 5A,B). However, in line with an unaltered expression of stemness markers (see again Supplemental Fig. 4), we did not detect proliferation defects in Trim71 KO ESCs under steady state conditions (Supplemental Fig. 6A–C). However, the regulation of let-7 targets involved in cell cycle functions might impact proliferation once ESC differentiation starts. We had previously reported coexistence of intact stemness and priming of neural differentiation in Trim71 KO ESCs, which translated into an accelerated differentiation into neural progenitor cells (NPCs) upon addition of the proper stimuli (Mitschka et al. 2015). Hence, we investigated cell proliferation in the course of ESC differentiation into NPCs. Indeed, Trim71 KO ESCs showed decreased proliferation rates in the course of N2B27-induced neural differentiation (Supplemental Fig. 6D–H).
TRIM71 represses let-7 maturation through its interaction with the TUT4/LIN28 complex
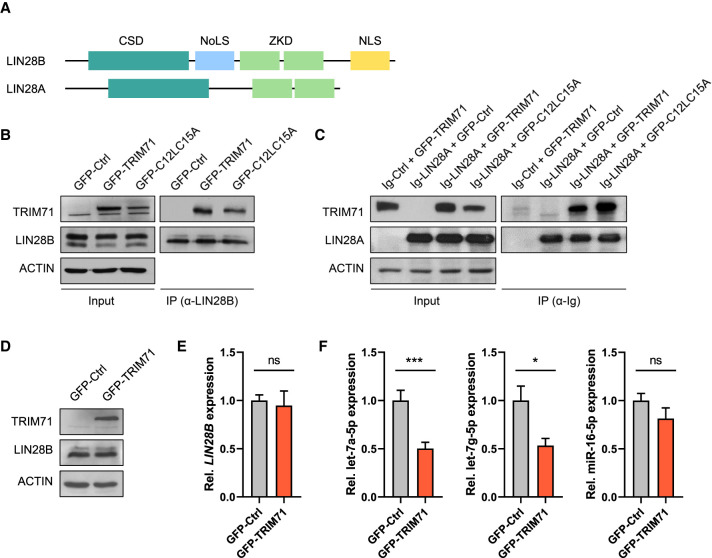
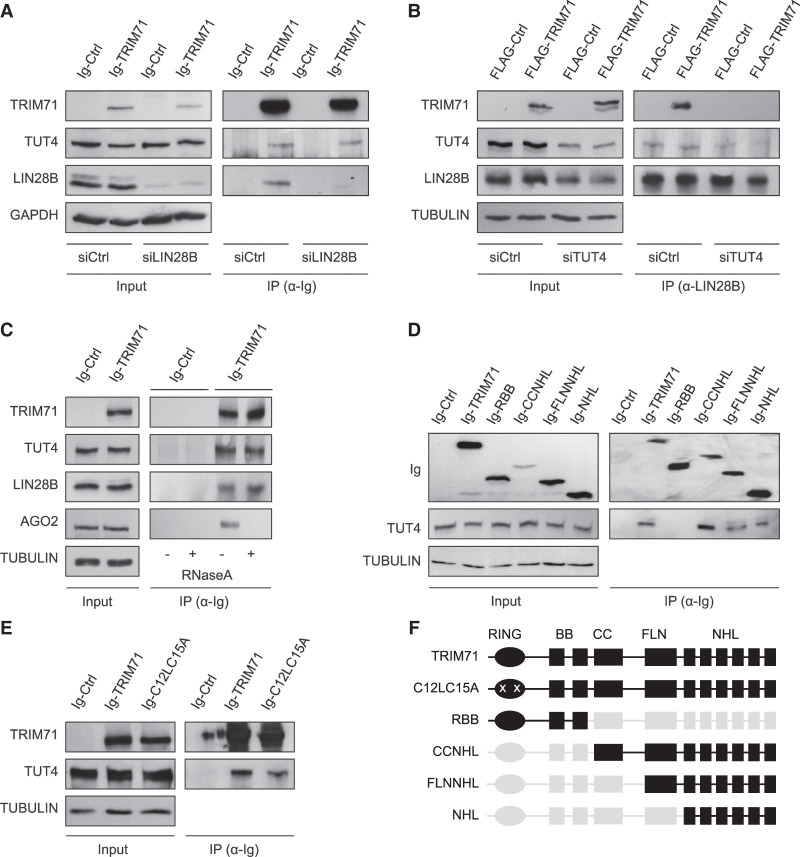
After finding that TRIM71 cooperates with LIN28A in the repression of let-7 expression, we next aimed at elucidating the underlying molecular mechanism. A previous study reported an interaction between TRIM71 and the paralog protein LIN28B in HEK293T cells (Lee et al. 2014). It was suggested that TRIM71 mediates the ubiquitination and proteasomal degradation of LIN28B, thereby inducing let-7 up-regulation (Lee et al. 2014). We found that both wild-type TRIM71 and the RING ubiquitination mutant C12LC15A (Rybak et al. 2009; Lee et al. 2014) coprecipitated with endogenous LIN28B in HEK293T cells (Fig. 4A,B). Both TRIM71 variants also interacted with ectopically expressed LIN28A (Fig. 4C). However, we observed that overexpression of TRIM71 in HEK293T cells did not affect endogenous LIN28B protein or mRNA levels (Fig. 4D,E), and instead resulted in the specific down-regulation of let-7a/g-5p miRNAs—but not of the housekeeping miR-16—(Fig. 4F), similar to our observations in ESCs. Thus, our results showed that TRIM71 does not promote LIN28B degradation, and instead suggested that TRIM71 can functionally cooperate with both LIN28 proteins to repress the expression of let-7 miRNAs.
FIGURE 4.
TRIM71 interacts with LIN28 proteins and specifically regulates let-7 miRNAs in HEK293T cells. (A) Schematic representation of both LIN28 paralog proteins (LIN28A and LIN28B) domain organization. (CSD) Cold shock domain, (ZKD) zinc knuckle domain, (NoLS) putative nucleolar localization sequence, (NLS) nuclear localization signal. (B) Representative immunoblot showing GFP-tagged TRIM71 and C12LC15A coprecipitation with endogenous LIN28B in HEK293T cells. (C) Representative immunoblot showing GFP-tagged TRIM71 and C12LC15A coprecipitation with Ig-tagged LIN28A overexpressed in HEK293T cells. Overexpression of an Ig-empty control vector together with wild-type GFP-TRIM71 (first lane) excluded an Ig-mediated TRIM71–LIN28A interaction. (D) Representative immunoblot showing ectopic TRIM71 and endogenous LIN28B protein levels in HEK293 cells stably overexpressing GFP-Ctrl or GFP-TRIM71. (E) RT-qPCR showing relative expression of LIN28B mRNA (n = 3) and (F) the indicated mature miRNAs (n = 6) in HEK293 cells stably overexpressing GFP-Ctrl or GFP-TRIM71. RT-qPCR quantification of miRNAs and mRNAs was normalized to the levels of the housekeeping U6 snRNA and HPRT1 mRNA, respectively. Error bars represent SD. (***) P < 0.005, (*) P < 0.05, (ns) nonsignificant (unpaired Student's t-test). See also Supplemental Figure 7.
Next, we designed several truncated constructs to map the interaction between TRIM71 and LIN28 proteins via coprecipitation experiments in HEK293T cells. We found that the cold-shock domain (CSD) of LIN28 proteins (Supplemental Fig. 7A,B) and the NHL domain of TRIM71 (Supplemental Fig. 7C–E), respectively, are required and sufficient to establish their interaction in cell culture. We then produced a recombinant human TRIM71 NHL domain and evaluated its direct interaction with the LIN28A-pre-let7 complex in vitro via electrophoretic mobility shift assays (EMSA). As expected, we found LIN28A to specifically interact with pre-let-7 (KD ≈ 55.5), but not with pre-miR-16 (Supplemental Fig. 8A,B). Strikingly, the addition of TRIM71's NHL domain did neither result in a detectable shift of the LIN28A-pre-let-7 complex (PR) nor the pre-let-7 alone (R) at any of the tested NHL protein concentrations (Supplemental Fig. 8C,D). Based on these findings, we reasoned that other cellular components are required to mediate or stabilize the interaction between TRIM71 and LIN28.
LIN28 proteins are known to induce pre-let-7 degradation through the recruitment of the terminal uridyl-transferase enzymes TUT4/7 (Heo et al. 2009; Thornton et al. 2012). Thus, we first evaluated the ability of TRIM71 to independently interact with TUT4. Indeed, we found endogenous TUT4 coprecipitated with TRIM71 in HEK293T cells, which lack LIN28A, as well as in LIN28B knockdown HEK293T cells (Fig. 5A). This indicated that LIN28 proteins are not indirectly mediating the interaction between TRIM71 and TUT4. In contrast, the binding between TRIM71 and endogenous LIN28B was abrogated upon TUT4 knockdown, suggesting that TUT4 facilitates the interaction between TRIM71 and LIN28B (Fig. 5B). The interaction between TRIM71 and TUT4/LIN28B was resistant to RNase treatment (Fig. 5C), demonstrating that these three partners form a stable protein complex. Consistent with these results, the NHL domain of TRIM71 was sufficient for TUT4 binding in HEK293T cells (Fig. 5D,F). Furthermore, the RING ubiquitination mutant C12LC15A, which we had previously shown to also bind LIN28 proteins, was also coprecipitated with TUT4 (Fig. 5E,F). Collectively, our data strongly support the formation of a tripartite TRIM71/TUT4/LIN28 complex which seems to mediate the degradation of pre-let-7 miRNAs more efficiently than the TUT4/LIN28 complex alone.
FIGURE 5.
The interaction between TRIM71 and LIN28 is mediated by the uridylating enzyme TUT4. (A) Representative immunoblot showing the coprecipitation of endogenous TUT4 with ectopically expressed Ig-TRIM71 in control HEK293T cells (siCtrl)—which lack LIN28A expression—and upon LIN28B knockdown (siLIN28B). (B) Representative immunoblot showing the coprecipitation of ectopically expressed FLAG-TRIM71 with endogenous LIN28B in control (siCtrl) and TUT4 knockdown (siTUT4) HEK293T cells. (C) Representative immunoblot showing the RNA-independent interaction between TRIM71 and TUT4 or LIN28B. AGO2 was used as a control of an RNA-dependent interaction. (D) Representative immunoblot showing the coprecipitation of endogenous TUT4 with different Ig-tagged TRIM71 constructs in HEK293T cells, depicted in F. (E) Representative immunoblot showing the coprecipitation of endogenous TUT4 with Ig-tagged TRIM71 and C12LC15A overexpressed in HEK293T cells. (F) Schematic representation of TRIM71 constructs used for IP assays in D and E. For each construct, present domains are depicted in black, deleted domains are depicted in gray and mutations are marked with a white “x.” See also Supplemental Figure 8.
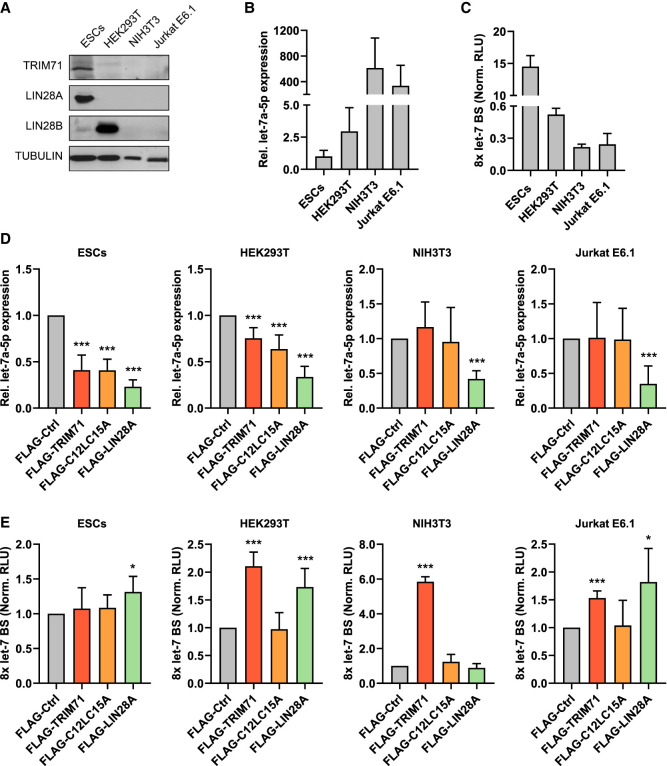
Since both TRIM71 and the ubiquitin ligase mutant C12LC15A were able to interact with TUT4/LIN28, we then asked whether C12LC15A could also mediate let-7 down-regulation. To this end, we evaluated the ability of wild-type TRIM71 and ubiquitin ligase mutant C12LC15A to regulate let-7 expression and let-7 activity in several cell lines with different expression levels of LIN28 proteins: (i) ESCs, which mostly express LIN28A, (ii) HEK293T cells which exclusively express LIN28B, (iii) the mouse embryonic fibroblast cell line NIH3T3 lacking expression of both LIN28 proteins, and (iv) the human malignant T cell line Jurkat E6.1, in which both LIN28 proteins are lacking as well (Fig. 6A). As expected, cell lines expressing either LIN28A or LIN28B (ESCs and HEK293T) showed lower basal let-7a expression levels than cell lines lacking LIN28 proteins (NIH3T3 and Jurkat E6.1) (Fig. 6B). Accordingly, let-7 activity was also lower in LIN28-expressing cells, as shown by a higher de-repression of the let-7 luciferase reporter (Fig. 6C). Overexpression of both TRIM71 and C12LC15A resulted in a 60% down-regulation of let-7 expression in ESCs, and a 25%–30% down-regulation of let-7 expression in HEK293T cells, proving that the RING domain (E3 ubiquitin ligase activity) of TRIM71 is not required for this function (Fig. 6D; Supplemental Fig. 9). In contrast, overexpression of TRIM71 or C12LC15A in NIH3T3 and Jurkat E6.1 cells did not affect let-7 levels, which were only down-regulated upon LIN28A overexpression (Fig. 6D; Supplemental Fig. 9). So far, our results have shown that TRIM71 establishes an RNA-independent interaction with the TUT4/LIN28 complex via its NHL domain, and enhances the repression of pre-let-7 maturation in an ubiquitination-independent manner.
FIGURE 6.
TRIM71 represses let-7 expression and activity via two independent mechanisms. (A) Representative immunoblot showing the levels of endogenously expressed TRIM71 and LIN28 proteins in the indicated cell lines, used for experiments depicted in B–E. (B) RT-qPCR showing the basal expression levels of mature let-7a miRNA in the indicated cell lines (n = 3 6). (C) Let-7 reporter assay upon transient transfection of a Renilla luciferase reporter under the control of a 3′UTR containing 8x Let-7 binding sites (BS) in the indicated cell lines (n = 3). (Norm. RLU) Normalized relative light units. (D) RT-qPCR showing the expression of mature let-7a miRNA in the indicated cell lines upon overexpression of FLAG-tagged TRIM71, C12L15A and LIN28A (n = 3–8). (E) Let-7 reporter assays in the indicated cell lines upon overexpression of FLAG-tagged TRIM71, C12L15A, and LIN28A (n = 3–8). (Norm. RLU) Normalized relative light units. RT-qPCR quantification of let-7a was normalized to the levels of the housekeeping U6 snRNA. Error bars represent SD. (***) P < 0.005, (*) P < 0.05 (unpaired Student's t-test between FLAG-Ctrl and each other condition). See also Supplemental Figure 9.
TRIM71 regulates let-7 activity via a TUT4/LIN28-independent mechanism
When we analyzed let-7 activity in ESCs, HEK293T, NIH3T3, and JurkatE6.1 cell lines via luciferase reporter assays, we noticed striking discrepancies with the parallel let-7 expression measurements. Whereas both wild-type TRIM71 and RING ubiquitination mutant C12LC15A were able to down-regulate let-7 expression to the same extent (Fig. 6D), only wild-type TRIM71 induced a significant de-repression of the let-7 reporter (Fig. 6E; Supplemental Fig. 9)—from now on referred to as repression of let-7 activity. Furthermore, although a TRIM71-mediated down-regulation of let-7 expression was only observed in LIN28-expressing cells (Fig. 6D), a TRIM71-dependent repression of let-7 activity was also found in cells lacking LIN28 expression (Fig. 6E). These results strongly suggested that TRIM71 uses distinct and independent mechanisms for the regulation of let-7 expression and activity.
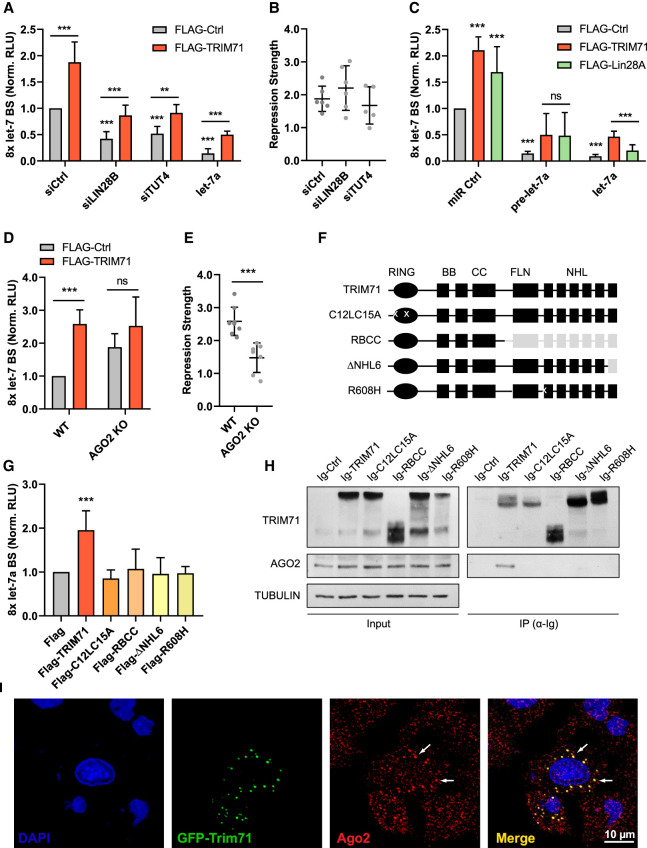
In line with this hypothesis, TRIM71 continued to repress let-7 activity after knockdown of either LIN28B or TUT4 in HEK293T cells (Fig. 7A,B; Supplemental Fig. 10A,B). Interestingly, TRIM71 was able to repress let-7 activity even upon overexpression of a mature let-7a miRNA duplex (Fig. 7A). These data indicated that the mechanism enabling TRIM71-dependent let-7 activity regulation occurs downstream from the processes regulating let-7 expression levels. To confirm this, we evaluated the repression of let-7 activity by TRIM71 and LIN28A upon overexpression of a pre-let-7a miRNA stem–loop and a mature let-7a miRNA duplex (Fig. 7C). Both TRIM71 and LIN28 were able to relieve the repression of the let-7 reporter to a similar extent upon pre-let-7 overexpression. However, only TRIM71 was able to significantly relieve the repression of the let-7 reporter upon mature let-7a overexpression (Fig. 7C). These data showed that TRIM71-mediated let-7 activity regulation acts downstream from the TUT4/LIN28-dependent let-7 expression regulation, which operates at the precursor miRNA stage. Thus, our results show that TRIM71 regulates let-7 expression and activity via two independent, mechanistically discernable functions: On the one hand, TRIM71 relies on its interaction with the TUT4/LIN28/pre-let 7 complex to repress miRNA maturation, and thus down-regulates let-7 expression; on the other hand, TRIM71 relies on its E3 ubiquitin ligase function to repress the activity of the mature let-7 miRNA in a TUT4/LIN28-independent manner.
FIGURE 7.
TRIM71 relies on AGO2 binding for the specific repression of let-7 activity. (A) Let-7 reporter assay upon overexpression of FLAG-Ctrl or FLAG-TRIM71 in control (siCtrl), LIN28B knockdown (siLIN28B) and TUT4 knockdown (siTUT4) HEK293T cells. Ectopic let-7 miRNA duplex expression was used as a control for efficient let-7 reporter repression, but revealed a regulation of mature let-7 activity (n = 4–6). (B) TRIM71-mediated let-7 activity repression strength, calculated from values in A, as Norm. RLU (FLAG-TRIM71)/Norm. RLU (FLAG-Ctrl) for each condition. (C) Let-7 reporter assay in HEK293T cells upon FLAG-Ctrl, FLAG-TRIM71 or FLAG-LIN28A coexpression with either a control miRNA duplex (miR Ctrl), a let-7a precursor stem–loop (pre-let-7a), or a mature let-7a duplex (let-7a) (n = 3–8). (D) Let-7 reporter assay after overexpression of FLAG-Ctrl or FLAG-TRIM71 in wild-type (WT) and AGO2 knockout (KO) HEK293T cells (n = 6–8). (E) TRIM71-mediated let-7 activity repression strength, calculated from values in D, as Norm. RLU (FLAG-TRIM71)/Norm. RLU (FLAG-Ctrl) for each condition. (F) Schematic representation of TRIM71 constructs used in G and H. For each construct, present domains are depicted in black, deleted domains are depicted in gray and mutations are marked with a white “x.” (G) Let-7 reporter assay in HEK293T cells upon overexpression of different FLAG-tagged TRIM71 constructs depicted in F (n = 3–10). (H) Representative immunoblot showing the coprecipitation of endogenous AGO2 with different Ig-tagged TRIM71 constructs—depicted in F—overexpressed in HEK293T cells. (I) Representative confocal microscopy images showing the partial colocalization of GFP-tagged TRIM71 and AGO2 in perinuclear foci. (Norm. RLU) Normalized relative light units. Error bars represent SD. (***) P < 0.005, (**) P < 0.01, (ns) nonsignificant (unpaired Student's t-test between the main control condition and each other condition, unless indicated by a line joining the two compared conditions). See also Supplemental Figure 10.
TRIM71 relies on AGO2 binding for the specific repression of let-7 activity
We next aimed to investigate how TRIM71 could repress let-7 miRNA activity. The activity of mature miRNAs is assisted by the RNA-induced silencing complex (RISC). Argonaute (AGO) proteins—particularly the well-characterized AGO2—are the major effectors of the RISC, since they bind the miRNA duplex, identify the guide strand, and find the complementary mRNA to repress its translation and often induce its degradation (Hutvagner and Simard 2008). Several TRIM-NHL proteins, including TRIM71, are known to interact with AGO proteins in various species (Rybak et al. 2009; Chang et al. 2012; Chen et al. 2013; Loedige et al. 2013; Mitschka et al. 2015; Torres-Fernández et al. 2019). To evaluate whether TRIM71-mediated let-7 activity repression is linked to AGO2 function, we conducted let-7 reporter assays in wild-type (WT, AGO+/+) and AGO2 knockout (KO, AGO−/−, Torres-Fernández et al. 2019) HEK293T cells (Supplemental Fig. 10C). Indeed, we found a diminished TRIM71-mediated repression of let-7 activity in AGO2 knockout HEK293T cells (Fig. 7D,E). Although this effect could be attributed to a general diminished miRNA activity in AGO2 KO cells, these cells continued to down-regulate the let-7 target HMGA2 upon overexpression of mature let-7 (Supplemental Fig. 10D). This suggests that in the absence of AGO2, AGO1/3/4 proteins may also enable miRNA-mediated silencing as it was previously described (Su et al. 2009).
The interaction between TRIM71 and AGO2 has been mapped to the NHL domain and is known to be RNA-dependent (Chang et al. 2012; Loedige et al. 2013; Torres-Fernández et al. 2019), as we have also shown in Figure 5C. We therefore evaluated the ability of several TRIM71 NHL domain mutants with impaired RNA binding ability, namely RBCC (Loedige et al. 2013), ΔNHL6 (Torres-Fernández et al. 2019), and R608H (Furey et al. 2018; Welte et al. 2019), as well as of the RING ubiquitination mutant C12LC15A, to bind AGO2 and to repress let-7 activity. We found that all investigated TRIM71 mutants failed to bind AGO2 and regulate let-7 activity (Fig. 7F–H). This correlation strongly suggested that TRIM71 depends on AGO2 for the inhibition of let-7 activity.
A previous study postulated that TRIM71 inhibits miRNA activity through a reduction of AGO2 protein via ubiquitination (Rybak et al. 2009). However, later publications have repeatedly shown in various systems that TRIM71 does not induce changes in AGO2 stability (Chang et al. 2012; Chen et al. 2012; Loedige et al. 2013; Mitschka et al. 2015; Torres-Fernández et al. 2019), as also shown in Figures 5C and 7H. Accordingly, luciferase reporter assays for other miRNAs showed that TRIM71 does not repress miRNA activity globally, as the activity of miR-16, miR-19b or miR-122 remained unaffected upon TRIM71 overexpression in HEK293T cells (Supplemental Fig. 10E). In contrast, TRIM71 repressed the activity of several let-7 family members, namely let-7a, let-7c, let-7e, and let-7g (Supplemental Fig. 10E). We also found that TRIM71 colocalized with only a subset of AGO2 foci in HEK293T cells (Fig. 7I), suggesting that TRIM71 may be associated with specific miRNA-containing AGO2/RISCs.
So far, we had observed an inhibition of let-7 activity in HEK293T cells, NIH3T3 cells and Jurkat E6.1 cells upon TRIM71 overexpression. We then confirmed a TRIM71-mediated let-7 activity repression also in murine Trim71 KO ESCs and NE4C cells (Supplemental Fig. 10F,G; Welte et al. 2019) as well as human TRIM71 knockdown and TRIM71-overexpressing NCCIT cells (Supplemental Fig. 10H,I). Thus, this mechanism is functional in several developmental and oncogenic cell line models. Collectively, our data indicate that TRIM71 represses the activity of several let-7 miRNAs in an AGO2-dependent manner. To interact with AGO2, TRIM71 requires both an intact NHL domain and a functional RING domain. This interaction does not result in decreased AGO2 protein stability or a global repression of miRNA activity.
TRIM71 binds and stabilizes let-7 mRNA targets in ESCs
TRIM71 is known to interact or colocalize with several RISC-associated factors, namely DICER (Rybak et al. 2009), MOV10 and TNRC6B (Loedige et al. 2013). Furthermore, TRIM71 is an mRNA-binding protein (Loedige et al. 2013; Torres-Fernández et al. 2019; Welte et al. 2019), and we have shown the interaction between TRIM71 and AGO2 to be RNA-dependent. Thus, we hypothesized that TRIM71 interacts with AGO2/RISC via its binding to specific mRNAs. To test this hypothesis, we asked whether TRIM71 binds and stabilizes let-7 targets in ESCs. To this end, we analyzed a published data set which includes RNA-seq data from TRIM71 RNA-IP experiments in ESCs, accompanied by transcriptomics on several Trim71 mutant ESCs, including Trim71 KO ESCs, Trim71 RING mutant ESCs, and Trim71 NHL mutant ESCs (Welte et al. 2019). RING mutant ESCs harbor the E3 ligase-impairing mutation C12LC15A (Rybak et al. 2009; Lee et al. 2014), and NHL mutant ESCs contain the R738A point mutation which was previously shown to impair TRIM71's ability to bind RNA (Kumari et al. 2018).
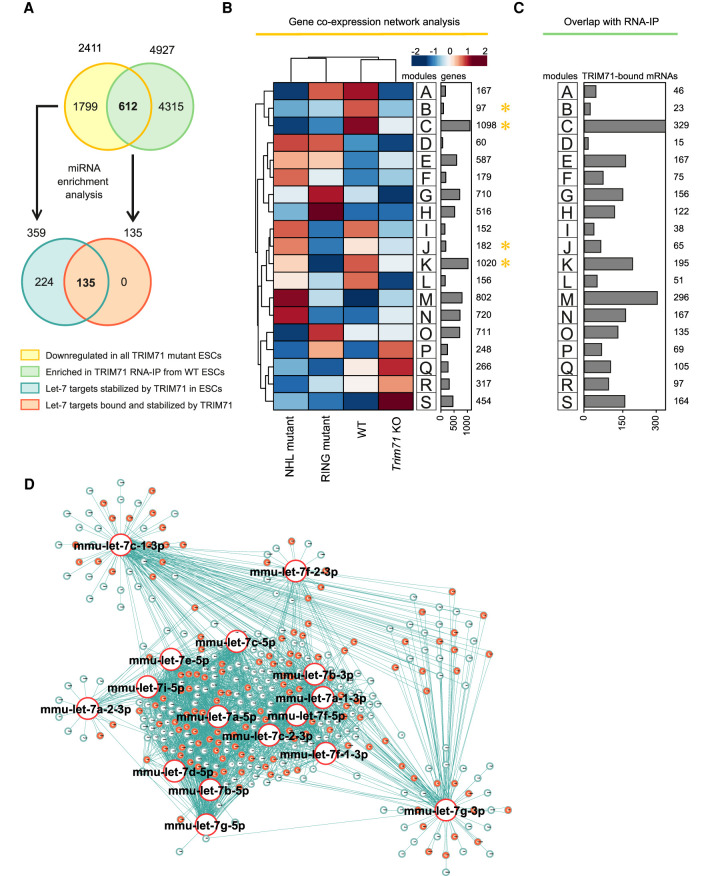
Since we have shown that both the RING and the NHL domains are required for AGO2 binding and repression of let-7 activity, we expected let-7 targets regulated by the TRIM71/AGO2 axis to be commonly down-regulated in all mutant ESCs. Coexpression network analysis of transcripts derived from WT, Trim71 KO, RING mutant, and NHL mutant ESCs showed four groups of genes commonly down-regulated in all mutant ESCs (modules B, C, J, and K, marked with a yellow asterisk) (Fig. 8A,B). We then used the 2411 genes corresponding to all four modules for miRNA enrichment analysis via ShinyGO and found the let-7 family to be among the top enriched miRNA families (P-value [FDR] = 2.54 × 10−04), with a total of 131 let-7 targets (≈ 5.4%) identified within modules B, C, J, and K (Supplemental Table 2A). Integrating information from eight different databases increased this number to 359 predicted let-7 targets (≈15%) (Fig. 8A,D; Supplemental Table 2B). These results recapitulated our previous findings with a new data set, showing that TRIM71 depletion results in the down-regulation of multiple let-7 targets in ESCs. We then overlapped the gene expression data with RNA-IP data to find mRNAs that are bound and stabilized by TRIM71 (Fig. 8A–C). This approach identified 612 mRNAs which were again enriched for let-7 miRNA targets (P-value [FDR] = 1.20 × 10−05), with a total of 48 let-7 targets (≈7.8%) within all mRNAs bound and stabilized by TRIM71 (Supplemental Table 3A). Integrating information from eight different databases increased this number to 135 predicted let-7 targets (≈ 22%) (Fig. 8A,D; Supplemental Table 3B). This revealed that 37.6% (135) of all let-7 targets that are stabilized in the presence of TRIM71 (359) are also directly bound by TRIM71.
FIGURE 8.
TRIM71 directly binds and stabilizes let-7 targets in ESCs. (A) Schematic representation for the analysis conducted in B–D. Genes commonly down-regulated in all Trim71 mutant ESCs (yellow)—obtained from B—are enriched for let-7 targets (blue) (Supplemental Table 2). Genes found enriched (greater than twofold) in TRIM71 RNA-IPs in ESCs (green) were overlapped with genes commonly down-regulated in all Trim71 mutant ESCs (yellow)—overlap depicted in C—to identify genes bound and stabilized by TRIM71 in ESCs. For those genes, miRNA enrichment analysis was conducted (Supplemental Table 3) to identify let-7 targets bound and stabilized by TRIM71 (orange). (B) Coexpression network analysis for the indicated ESC lines using an available data set (GSE134125) revealed genes commonly down-regulated in all Trim71 mutant ESCs (modules B, C, J, and K, marked with a yellow asterisk). (C) Overlap of TRIM71 RNA-IP data in ESCs (GSE134125) with gene expression data, showing the number of TRIM71-bound mRNAs in ESCs within each module from B. (D) miRNA network showing targets for the different let-7 family members found among commonly down-regulated genes in all Trim71 mutant ESCs (modules B, C, J, and K from B), with let-7 targets directly bound by TRIM71 in ESCs marked in orange. See also Supplemental Tables 2, 3.
Altogether, our analysis shows that TRIM71 interacts with specific let-7 mRNA targets, resulting in their stabilization, and that both the RING and the NHL domains of TRIM71 are required for the regulation of this subset of mRNAs. Having previously shown that both the RING and the NHL domains are required for AGO2 binding and let-7 activity regulation, our results strongly suggest that TRIM71 represses let-7 activity by inhibiting AGO2 function on active RISCs, to which TRIM71 is recruited via the interaction with specific mRNAs, that is, let-7 targets.
TRIM71 binds and stabilizes let-7 mRNA targets in HCC cells
TRIM71 is up-regulated in patients with hepatocellular carcinoma (HCC) and its expression is correlated with advanced tumor stages and poor prognosis (Chen et al. 2013; Torres-Fernández et al. 2019). Several studies have shown that TRIM71 promotes proliferation in several HCC cell lines, including HepG2, Hep3B, and Huh7 (Chen et al. 2013; Torres-Fernández et al. 2019; Foster et al. 2020). In one of these studies, an increase of let-7 activity was observed in TRIM71 knockdown cells (Chen et al. 2013). This effect was attributed to the previously reported global inhibition of miRNA activity caused by TRIM71-mediated ubiquitination and proteasomal degradation of AGO2 (Rybak et al. 2009). However, we did not find any evidence supporting a role for TRIM71 in the regulation of AGO2 protein stability.
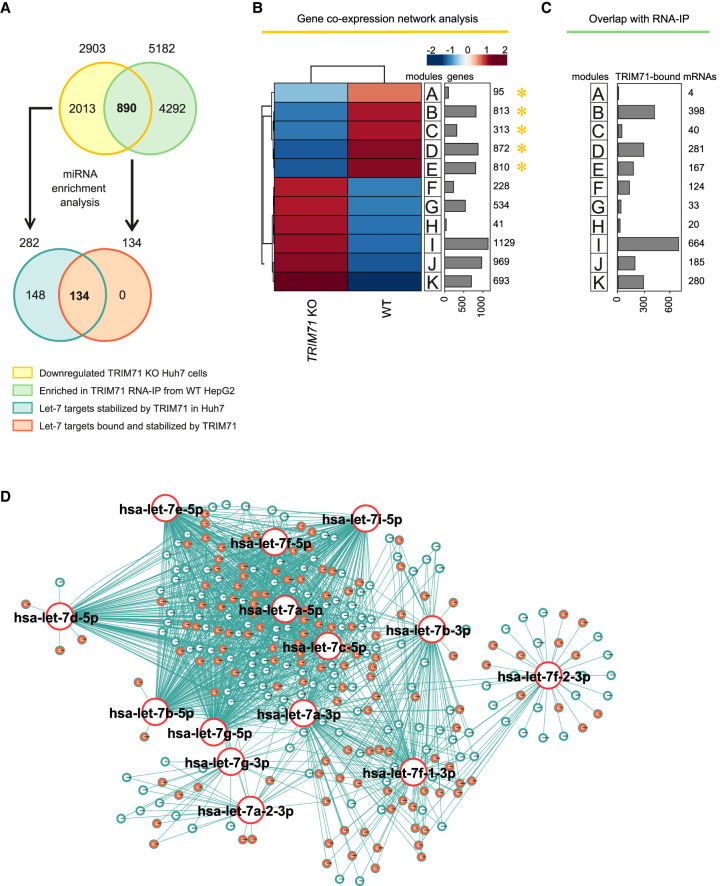
Thus, we first evaluated AGO2-TRIM71 interaction in wild-type HepG2 cells (Supplemental Fig. 8J) together with AGO2 expression levels (Supplemental Fig. 8K) and let-7 activity (Supplemental Fig. 8L) upon TRIM71 knockdown in HepG2 cells. Our results showed that TRIM71 binds AGO2 and regulates let-7 activity in HepG2 cells without affecting AGO2 protein levels (Supplemental Fig. 8J–L). We then used an available transcriptomic data set of TRIM71 KO Huh7 cells (Welte et al. 2019), and conducted coexpression network analysis (Fig. 9A,B). We selected significantly down-regulated mRNAs in TRIM71 KO cells (2903 genes from modules A–E, marked with a yellow asterisk in Fig. 9B) and conducted miRNA enrichment analysis via ShinyGO. Again, we found the let-7 family to be among the top enriched miRNA families (P-value [FDR] = 9.89 × 10−04), with a total of 216 down-regulated let-7 targets (≈7.4%) (Supplemental Table 4A). Integrating information from eight different miRNA data bases increased this number to 282 predicted targets (≈9.7%) for the members of the let-7 family (Fig. 9A,D; Supplemental Table 4B).
FIGURE 9.
TRIM71 directly binds and stabilizes let-7 targets in HCC cells. (A) Schematic representation for the analysis conducted in B–D. Genes down-regulated in TRIM71 KO Huh7 cells (yellow)—obtained from B—are enriched for let-7 targets (blue) (Supplemental Table 4). Genes found enriched (greater than twofold) in TRIM71 RNA-IPs in HepG2 cells (green) were overlapped with genes down-regulated in TRIM71 KO Huh7 cells (yellow)—overlap depicted in C—to identify genes bound and stabilized by TRIM71. For those genes, miRNA enrichment analysis was conducted (Supplemental Table 5) to identify let-7 targets bound and stabilized by TRIM71 in HCC cells (orange). (B) Coexpression network analysis for WT and TRIM71 KO Huh7 cells using an available data set (GSE134125) revealed genes down-regulated in TRIM71 KO Huh7 cells (modules A–E marked with a yellow asterisk). (C) Overlap of TRIM71 RNA-IP data in HepG2 cells with gene expression data, showing the number of TRIM71-bound mRNAs in HCC cells within each module from B. (D) miRNA network showing targets for the different let-7 family members found among the down-regulated genes in TRIM71 KO Huh7 cells (modules A–E), with let-7 targets directly bound by TRIM71 in HepG2 cells marked in orange. See also Supplemental Tables 4, 5.
To evaluate whether TRIM71 was also able to interact with some of these let-7 targets in HCC cells, we used another available TRIM71 RNA-IP data set from HepG2 cells (Foster et al. 2020) and overlapped it with genes found to be down-regulated in TRIM71 KO Huh7 cells (Fig. 9A–C). We found a total of 890 genes bound and stabilized by TRIM71 in HCC cells, 87 of which (≈9.8%) were found to be let-7 targets (P-value [FDR] = 1.30 × 10−05) (Supplemental Table 5A). Integrating information from eight different miRNA data bases increased this number to a total of 134 predicted targets (≈15%) (Fig. 9A,D; Supplemental Table 5B). This shows that 47.5% (134) of all let-7 targets stabilized in the presence of TRIM71 (282) were also directly bound by TRIM71. While we propose that these targets are regulated by TRIM71 via the AGO2-dependent mechanism, let-7 targets stabilized but not bound by TRIM71 may be presumably still indirectly affected by TRIM71-dependent control of let-7 expression via the TUT4/LIN28 axis. Altogether, these results indicate that the TRIM71-mediated let-7 regulatory mechanisms described in our study are not restricted to ESCs, but are also active in hepatocellular carcinoma cells.
Let-7 targets bound and stabilized by TRIM71 are heavily enriched in TRIM71-binding hairpins
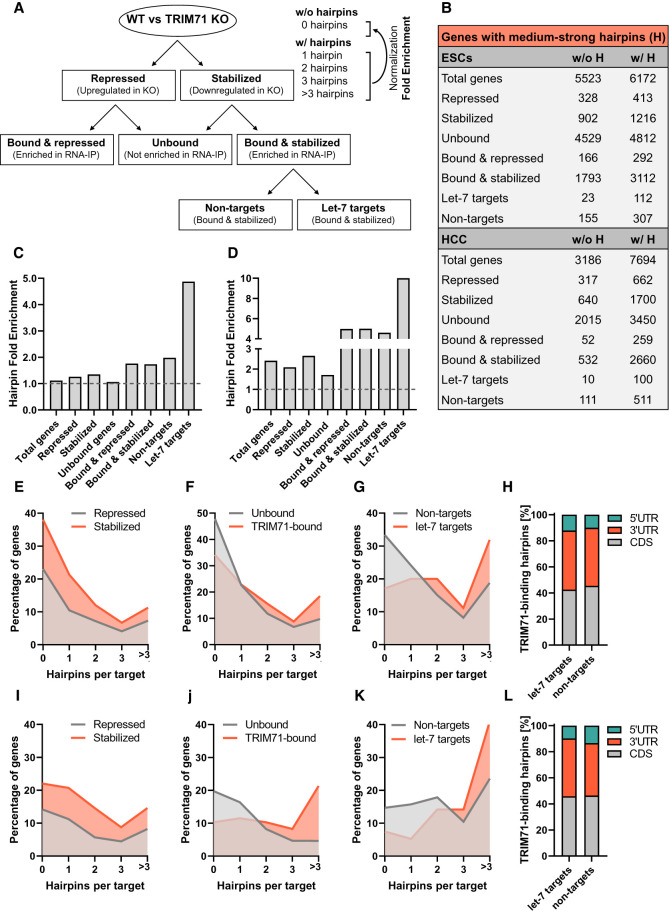
So far, TRIM71 has been described as an mRNA-binding and repressor protein. TRIM71 directly interacts with conserved RNA structural elements (TRIM71-binding hairpins) mostly present in the 3′UTR and coding sequence (CDS) of its target mRNAs and is reported to trigger mRNA degradation following mRNA recognition (Loedige et al. 2013; Kumari et al. 2018; Torres-Fernández et al. 2019; Welte et al. 2019). The present work, however, uncovers a novel role for TRIM71 as a positive regulator of mRNAs by modulating the impact of let-7 miRNAs on these targets. We wondered whether mRNA target recognition by TRIM71 underlies different principles for mRNAs that are either stabilized or degraded. For this purpose, we analyzed whether different groups of targets derived from our previous analysis (Fig. 10A) are enriched for TRIM71-binding hairpins by using an available data set with global predictions for the presence, number, and position of these hairpins across mouse and human genomes (Welte et al. 2019).
FIGURE 10.
let-7 targets bound and stabilized by TRIM71 in ESCs and HCC cells are heavily enriched in TRIM71-binding hairpins. (A) Schematic representation for the target groups analyzed for the identification of RNA structural elements well-characterized as TRIM71-binding hairpins. (B) Number of genes with (w/ H) and without (w/o H) medium or strong hairpins (hairpin strength classification according to Welte et al. 2019) identified in the different target groups depicted in A. (C) Hairpin fold change enrichment in ESCs and (D) HCC cells, calculated from values specified in B by dividing the number of “targets w/ H,” in which one or more middle or strong hairpins were identified, by the number of “targets w/o H” (represented by the baseline), in which neither medium nor strong hairpins were identified. (E–G) Percentage of genes with 0, 1, 2, 3, or >3 hairpins identified in the specified target groups in ESCs. (H) Number of TRIM71-binding hairpins identified within the 5′UTR, coding sequence (CDS) and 3′UTR of mRNAs found to be bound and stabilized by TRIM71 in ESCs, depicted as percentages. (I–K) Percentage of genes with 0, 1, 2, 3, or >3 hairpins identified in the specified target groups in HCC cells. (L) Number of TRIM71-binding hairpins identified within the 5′UTR, CDS and 3′UTR of mRNAs found to be bound and stabilized by TRIM71 in HCC cells.
We first evaluated the number of mRNAs with (w/ H) and without (w/o H) hairpins within each group for ESCs and HCC cells (Fig. 10A,B) in order to calculate their enrichment in each group (Fig. 10C,D). Then, we also analyzed the number and position of these hairpins (Fig. 10E–L). TRIM71-binding hairpins were identified in about half of the mRNAs expressed in ESCs and about two thirds of the mRNAs expressed in HCC cells (group: total). A similar proportion was found for targets that were either up-regulated (group: repressed by TRIM71) or down-regulated (group: stabilized by TRIM71) in Trim71 KO ESCs (Fig. 10B,C,E) or TRIM71 KO HCC cells (Fig. 10B,D,I). No enrichment was expected for these groups since not all expression changes occurring upon TRIM71 depletion are directly driven by TRIM71 target binding. In contrast, directly bound targets (groups: bound & repressed and bound & stabilized) indeed showed an enrichment of hairpins (Fig. 10B–D), with ∼20% of bound mRNAs containing more than three hairpins in ESCs (Fig. 10F) and HCC cells (Fig. 10J). Last, we divided the target group bound & stabilized into let-7 targets and non-targets (Fig. 10A) in order to evaluate possible differences in hairpin fold enrichment as well as hairpin number and position (5′UTR, CDS or 3′UTR) between these two groups. Strikingly, let-7 targets that were bound and stabilized by TRIM71 showed the strongest hairpin enrichment among all groups (Fig. 10A–D). Less than 20% and 10% of let-7 targets lacked hairpins and more than 40% and 30% of let-7 targets contained >3 hairpins in ESCs and HCC, respectively (Fig. 10G,K). No differences were found between let-7 targets and non-targets concerning the position of these hairpins, which were mostly located along the CDS and the 3′UTR (Fig. 10H,L). These results suggest that cooperative binding of TRIM71 to these mRNAs may directly compete with or indirectly modulate let-7 binding to such mRNAs. Last, our results challenge the idea that TRIM71 is a default repressor of its mRNA targets and indicate that the fate of TRIM71-bound mRNAs is not determined by different binding modes.
DISCUSSION
Regulation of miRNAs is one of the major mechanisms to fine-tune protein expression and function. TRIM-NHL proteins have been linked to the regulation of the miRNA pathway in several species (Wulczyn et al. 2010). In the present work we performed a comprehensive analysis of TRIM71-dependent regulation of miRNA expression and activity, specifically focusing on the regulation of the let-7 miRNA family.
By analyzing the miRNome of TRIM71-deficient murine ESCs (Mitschka et al. 2015), we identified let-7 family members as some of the strongest up-regulated miRNAs. Mature miRNA duplexes of several let-7 members were found up-regulated in Trim71 KO ESCs, while expression of pri- and pre-let-7 was unaltered, suggesting that TRIM71 interferes with pre-let-7 maturation. Since LIN28 proteins are known to assist pre-let-7 degradation (Heo et al. 2008, 2009), we generated Lin28a KO and Trim71–Lin28a double KO cells, and found an up-regulation of mature let-7 miRNAs—but not of pri- and pre-miRNA molecules—in all KO cell lines. Accordingly, all KO cell lines shared the down-regulation of multiple let-7 targets in their transcriptomic profiles, many of which participate in cell cycle regulation. However, an effect of TRIM71 deficiency on ESCs proliferation was only apparent upon induction of neural differentiation. This suggests that TRIM71 is not essential for ESCs maintenance but may rather control the balance between proliferation and differentiation during early embryonic development in order to ensure a sufficient supply of cells before they terminally differentiate. Therefore, the role of TRIM71 might be more accurately described as an inhibitor of premature differentiation rather than a bona fide stemness factor. Thus, the role of TRIM71 in the regulation of let-7 expression may be especially relevant during differentiation, when LIN28 amounts start to be limiting. In other words, once ESC differentiation is triggered and let-7 starts down-regulating its targets, including Trim71 and Lin28, the cooperation between TRIM71 and LIN28 may be essential to fine-tune a progressive up-regulation of let-7 in order to prevent premature differentiation.
Such a cooperation was apparent in the Trim71–Lin28a double KO ESC line, which did not show an additional let-7 up-regulation to that observed in single Lin28a KO ESCs, suggesting that TRIM71 depends on LIN28A for the regulation of let-7 expression. Confirming TRIM71's dependency on LIN28A for this function, overexpression of TRIM71 resulted in a significant down-regulation of let-7 only in WT, but not Lin28a KO ESCs. We found that TRIM71 can likewise functionally cooperate with the paralog protein LIN28B for the repression of let-7 expression in HEK293T cells. We demonstrated that TRIM71 is able to interact with both LIN28 proteins, and found the TRIM71-LIN28 interaction to be RNA-independent, E3 ligase-independent, and mediated by TRIM71's NHL domain and LIN28 protein's CSD, respectively. Further analysis revealed that the uridylating enzyme TUT4 facilitates the interaction between TRIM71 and LIN28 proteins. Accordingly, the interaction between TRIM71 and TUT4 was also RNA-independent, E3 ligase-independent and mediated by the TRIM71's NHL domain. Altogether, we showed that TRIM71 represses let-7 expression in cooperation with the TUT4/LIN28 complex, a well-established inhibitor of pre-let-7 maturation (Heo et al. 2009). It remains to be investigated how exactly TRIM71 enhances the function of the LIN28/TUT4 protein complex. Interestingly, a similar effect on pre-let-7 regulation was previously described for TRIM25, another member of the TRIM protein family that lacks an NHL domain (Choudhury et al. 2014). In this case, TRIM25 was found to directly interact via its coiled-coil domain with the pre-let-7 stem–loop and to enhance TUT4-mediated pre-let-7 uridylation. The role of TRIM25 as an E3 ligase in this context was not investigated and the exact mechanism of uridylation enhancement remains to be elucidated (Choudhury et al. 2014). Nevertheless, this mechanism revealed the existence of miRNA-specific cofactors that stimulate TUT4-mediated uridylation. Thus, TRIM71 could also enhance pre-let-degradation by stimulating TUT4-mediated uridylation, but in order to confirm this hypothesis, pre-let7 uridylation levels should be investigated in the presence and absence of TRIM71 in the future.
Our EMSA experiments showed that, unlike TRIM25 (Choudhury et al. 2014), TRIM71 is not able to directly interact with pre-let-7 miRNAs. This result is in line with a previous study that investigated ESC-specific miRNA-binding proteins and identified TRIM71 as one of them for its binding to pre-miR-1 and pre-miR-29a, but not pre-let-7 species (Treiber et al. 2017). Thus, an interaction between TRIM71 and pre-let-7 is only indirectly mediated by the TUT4/LIN28 complex. Therefore, TUT4 and LIN28, but not TRIM71, are responsible for pre-miRNA target specificity. Hence, we propose that TRIM71 may be able to enhance the degradation of other pre-miRNAs reported to be regulated by the TUT4/LIN28 axis. Indeed, TRIM71-deficient ESCs showed an up-regulation of miR-9 and miR-200 species (Mitschka et al. 2015), both of which are prodifferentiation and tumor suppressor miRNAs whose precursors are targeted for degradation via the TUT4/LIN28 complex (Heo et al. 2009; Peter 2009; Nowak et al. 2014). Notably, both miR-9 and miR-200 families were also present among the top enriched miRNAs in all of our miRNA enrichment analysis. Thus, our study also provides other miRNA candidates which might be regulated in the same fashion as let-7 miRNAs.
Further supporting a TRIM71/TUT4/LIN28-mediated let-7 regulation mechanism, the overexpression of TRIM71 resulted in decreased let-7 expression in ESCs and HEK293T cells, which express LIN28A and LIN28B, respectively, but not in NIH3T3 or Jurkat E6.1 cells, which lack LIN28A/B expression. Notably, TRIM71-mediated let-7 down-regulation was stronger in ESCs than in HEK293T cells. This difference might be explained by the distinct LIN28 proteins expressed in either of these cell lines. While both LIN28 proteins can regulate let-7 in the cytoplasm in a TUT4-dependent manner (Heo et al. 2008, 2009; Suzuki et al. 2015), LIN28B can additionally block let-7 biogenesis in a TUT4-independent manner within the nucleus, where it binds pri- and pre-let-7 molecules preventing further processing and cytoplasmic export (Heo et al. 2008; Piskounova et al. 2011). Given the cytoplasmic localization of TRIM71 (Rybak et al. 2009; Chang et al. 2012; Torres-Fernández et al. 2019), the efficiency of TRIM71-mediated let-7 regulation in LIN28B-expressing cells may be limited by the amount of LIN28B available in the cytoplasm, which is known to vary among cell types (Guo et al. 2006; Piskounova et al. 2011; Wang et al. 2016).
Importantly, we provided evidence that the E3 ligase function of TRIM71 is dispensable for the regulation of let-7 expression, as the RING ubiquitination mutant C12LC15A could interact with TUT4/LIN28 and repress let-7 expression to a similar extent as the wild-type TRIM71. In contrast, only wild-type TRIM71, but not C12LC15A, was able to repress let-7 activity in luciferase reporter assays. Furthermore, TRIM71-mediated repression of let-7 activity was also observed in NIH3T3 and Jurkat E6.1 cells in which let-7 expression was unaltered. These results revealed that TRIM71 uses two independent molecular mechanisms for the regulation of let-7 expression and activity. Further analysis confirmed that the regulation of let-7 activity occurred independently and downstream from the TUT4/LIN28 function.
An earlier study had identified TRIM71 as a global inhibitor of miRNA activity, a function that was attributed to TRIM71-mediated AGO2 protein ubiquitination and proteasomal degradation (Rybak et al. 2009). However, we and others (Chang et al. 2012; Chen et al. 2012; Loedige et al. 2013; Mitschka et al. 2015; Torres-Fernández et al. 2019) did not detect TRIM71-mediated changes of AGO2 expression. Accordingly, we observed a miRNA-specific rather than a global inhibition of miRNA activity upon TRIM71 overexpression, as well as a partial but not total TRIM71–AGO2 colocalization. Furthermore, we found that TRIM71-mediated repression of let-7 activity was impaired in AGO2 knockout HEK293T cells, and observed an impaired ability to repress let-7 activity in all TRIM71 mutants which failed to interact with AGO2. These results strongly suggested that TRIM71-mediated let-7 activity repression is linked to the functional inhibition of AGO2 rather than to AGO2 degradation. It remains to be elucidated if this mechanism applies to all different let-7 members, as well as to other specific miRNA families, but it certainly does not apply to all cellular miRNAs.
Other TRIM-NHL proteins, namely drosophila orthologues Brat, Mei-P26 and Wech/Dappled, C. elegans NHL-2 and mammalian TRIM32, were all found to interact with AGO proteins, but in none of these cases did the interaction result in reduced AGO protein stability (Neumüller et al. 2008; Hammell et al. 2009; Schwamborn et al. 2009; Loedige et al. 2013). Interestingly, C. elegans NHL-2 and mammalian TRIM32 were also found to affect miRNA activity (Hammell et al. 2009; Schwamborn et al. 2009). Specifically, both NHL-2 and TRIM32 were found to enhance the activity of let-7 miRNAs and to promote cell differentiation (Hammell et al. 2009; Schwamborn et al. 2009). Although TRIM71- and TRIM32-mediated miRNA activity regulation results in opposite functional outcomes, their molecular mechanisms seem to be highly similar: Both proteins interact with AGO proteins via their NHL domain (Schwamborn et al. 2009; Chang et al. 2012; Loedige et al. 2013) and affect the activity of specific miRNAs, including let-7. Furthermore, both TRIM32 and TRIM71 were found to induce AGO2 ubiquitination without leading to their degradation (Loedige et al. 2013), suggesting that ubiquitination-dependent events may regulate AGO2 function without affecting its stability.
Several post-translational modifications are known to alter AGO2 function (Martinez and Gregory 2013; Jee and Lai 2014). For instance, AGO2 phosphorylation was shown to reduce let-7 activity by causing the dissociation of let-7 from the RISC complex, resulting in the stabilization of let-7 targets (Patranabis and Bhattacharyya 2016). Similarly, TRIM71-mediated AGO2 ubiquitination could lead to allosteric inhibition of AGO2 binding to specific miRNAs, resulting in the repression of miRNA activity. Alternatively, TRIM71 autoubiquitination could be required for AGO2 binding. This type of autoregulation has been observed for TRIM32, where autoubiquitination is required for its localization to cytoplasmic processing bodies (Ichimura et al. 2013). In support of this hypothesis, our results showed that the TRIM71 ubiquitination mutant C12LC15A not only failed to repress let-7 activity, but also to interact with AGO2.
The interaction of TRIM71 with AGO2 required not only a functional RING domain, but also an intact NHL domain. We showed the interaction between TRIM71 and AGO2 to be RNA-sensitive, and TRIM71 NHL mutants with impaired RNA-binding capacity failed to bind AGO2. Thus, we hypothesized that TRIM71 binds AGO2 on active RISCs through its interaction with specific let-7 target mRNAs. This may also explain why TRIM71 does not affect global miRNA activity, as its miRNA specificity would be determined by the TRIM71-bound mRNA target. Indeed, we found that TRIM71 interacts with multiple let-7 targets in mouse ESCs and human HCC cells, resulting in their stabilization. Importantly, TRIM71 required both the RING and NHL intact domains for the stabilization of those targets, again suggesting that the TRIM71–AGO2 interaction is required for the repression of let-7 activity. Thus, our data collectively support a role for TRIM71 as an inhibitor of let7-associated RISCs.
Together with another recent study (Foster et al. 2020), our work reports a novel role for TRIM71 as a positive post-transcriptional regulator of mRNA expression. We have found that many let-7 targets which are directly bound and stabilized by TRIM71 in ESCs and HCC cells, display an enrichment in conserved RNA structural elements (hairpins), previously defined as TRIM71-binding motifs (Kumari et al. 2018; Torres-Fernández et al. 2019; Welte et al. 2019). Like most functional miRNA binding sites, many of these hairpins are found in the 3′UTR of mRNA targets. Thus, the interaction of TRIM71 with these hairpins may impede let-7 binding to the 3′UTR of target mRNAs on active RISCs, resulting in a decreased AGO2/RISC function. Our results show that the fate (degradation or stabilization) of the mRNA target is not controlled by differential TRIM71-binding modes and thus may depend on other events occurring downstream from target mRNA recognition, or the presence of different protein cofactors.
In summary, TRIM71 was first discovered more than 20 years ago as an mRNA target of the miRNA let-7. In the present study, we have uncovered a role for TRIM71 as a let-7 repressor, revealing a bistable switch between these two highly conserved developmental regulators. Our study shows that TRIM71 modulates the let-7 miRNA pathway via specific interactions with two distinct protein complexes, TUT4/LIN28 and AGO2/mRNAs (RISC), in order to regulate let-7 expression and let-7 activity, respectively. These mechanisms induce significant transcriptomic changes in ESCs and HCC cells, suggesting that TRIM71-mediated miRNA regulatory mechanisms play important roles in both developmental and oncogenic processes. Future studies may shed further light on the precise in vivo implications of TRIM71-mediated miRNA regulation during embryogenesis and tumorigenesis.
MATERIALS AND METHODS
Cell lines and cell culture
The generation of wild-type mouse ESCs (WT, Trim71fl/fl) from conditional Trim71 full knockout mouse (Trim71fl/fl; Rosa26-CreERT2) was described in our previous work (Mitschka et al. 2015). These cells were then used to generate Lin28a KO ESCs (Trim71fl/fl; Lin28a−/−) via gene editing using transcription activator-like effectors nucleases (TALENs) as previously described (Sanjana et al. 2012). The TALEs were engineered to bind specific DNA sequences in the Lin28a locus (5′-GGGGCCCGGGGCCACGGGC-3′ and 5′-GCTGGTTGGACACCGAGCC-3′) and were fused to the unspecific FokI endonuclease catalytic domain to generate a double strand break close downstream from the Lin28a start codon. A donor DNA with Lin28a homology arms was designed to insert a G-418 resistance gene within the Lin28a locus by homologous recombination to allow preselection of edited clones, while disrupting Lin28a coding sequence.
Trim71 knockout ESCs (KO, Trim71−/−) and Trim71–Lin28a double KO ESCs (Trim71−/−; Lin28a−/−) were generated from WT (Trim71fl/fl) and Lin28a KO ESCs (Trim71fl/fl; Lin28a−/−), respectively, by addition of 500 nM of 4-hydroxytamoxifen (4-OHT) in their culture media for 48 h, followed by further culture for 72 h to achieve full protein depletion. The generation of HEK293 cells stably overexpressing GFP or GFP-TRIM71, and of AGO2 KO HEK293T cells was described in our previous work (Torres-Fernández et al. 2019).
ESCs were cultured in 0.1% gelatin-coated dishes and maintained in 2i + LIF media (DMEM knockout media supplemented with 15% FCS, 1% Penicillin-Streptomycin, 0.1 mM NEAA, 2 mM L-GlutaMAX, 100 µM β- mercaptoethanol, 0.2% in-house produced LIF [supernatant from L929 cells], 1 µM of MEK/ERK inhibitor PD0325091 and 3 µM of GSK-3 inhibitor CHIR99021). HEK293(T) cells, NIH3T3 cells and NCCIT cells were maintained in DMEM media supplemented with 10% FBS and 600 µg/mL of G418 antibiotic solution (HEK293) or 1% Penicillin-Streptomycin antibiotic solution (HEK293T, NIH3T3, and NCCIT). HepG2 and Jurkat E6.1 cells were maintained in RPMI 1640 media supplemented with 10% FBS and 1% Penicillin-Streptomycin antibiotic solution. WT and Trim71 KO NE4C cells were a kind gift of Professor Helge Großhans and were generated and characterized in a previous study (Welte et al. 2019). NE4C cells were cultured in Poly-L-Lysine coated plates (200 µg/mL) and maintained in MEM media supplemented with 2 mM L-Glutamine, 10% FBS and 1% Penicillin-Streptomycin antibiotic solution.
DNA and RNA transient transfections
DNA transfection in HEK293 and HEK293T cells was conducted by the well-established calcium phosphate method, using 25 µg DNA/mL of calcium phosphate solution. DNA transfection in NIH3T3, NCCIT and HepG2 cells was conducted with Lipofectamine 2000 reagent following the manufacturer's instructions (Invitrogen) and using a ratio 1 µg:2 µL DNA:Lipofectamine 2000. DNA transfection in ESCs was conducted with PANfect reagent following the manufacturer's instructions (PAN biotech) and using a ratio 1 µg:2 µL DNA:PANfect. Jurkat E6.1 cells were transfected by electroporation in 1:1 RPMI:FBS media using an exponential wave program in a Bio-Rad electroporation system and providing a single pulse of 240 V. All DNA-transfected cells were harvested 48 h post-transfection (hpt) for further analysis. siRNA/miRNA transfection for all cell lines was conducted with Lipofectamine RNAiMAX reagent following the manufacturer's instructions (Invitrogen) and using a ratio 10 pmol:2 µL RNA:Lipofectamine RNAiMAX. Cells transfected with siRNAs/miRNAs were harvested 72 hpt for further analysis. For experiments requiring siRNA/miRNA and DNA transfections, DNA was transfected 24 h after siRNA/miRNA transfection. MiRNAs and siRNAs used can be found in Supplemental Tables 1 and 2, respectively.
RNA isolation, generation of cDNA libraries, and RNA-seq
For RNA isolation, 5 × 106–2 × 107 murine ESCs were lysed in TRIzol, and total RNA was extracted according to the manufactures’ protocol. The precipitated RNA was solved in RNase-free water. RNA quality was assessed by measuring the ratio of absorbance at 260 and 280 nm using a Nanodrop 2000 Spectrometer as well as by visualization of the integrity of the 28S and 18S bands on agarose gels. Total RNA was then converted into double-stranded cDNA libraries following the manufacturer's recommendations using the Illumina TruSeq RNA Sample Preparation Kit v2. Shortly, mRNA was purified from 100 ng of total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in Illumina proprietary fragmentation buffer. First-strand cDNA was synthesized using random oligonucleotides and SuperScript II. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities and enzymes were removed. After adenylation of 3′ ends of DNA fragments, Illumina PE adapter oligonucleotides were ligated to prepare for hybridization. DNA fragments with ligated adapter molecules were selectively enriched using Illumina PCR primer PE1.0 and PE2.0 in a 15 cycle PCR reaction. Size-selection and purification of cDNA fragments with preferentially 200 bp in length was performed using SPRIBeads (Beckman-Coulter). Size-distribution of cDNA libraries was measured using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent). cDNA libraries were quantified using KAPA Library Quantification Kits (Kapa Biosystems) and were used as a template for high-throughput sequencing. To this end, cluster generation was conducted on a cBot, and a 2 × 100 bp paired-end run was performed on a HiSeq1500.
RNA-seq preprocessing
Sequenced reads were aligned and quantified using STAR software (v2.7.3a) (Dobin et al. 2013) and the murine reference genome (GRCm38) from the Genome Reference Consortium. Raw counts were imported using the DESeqDataSetFromHTSeqCount function from DEseq2 (v1.26.0) (Love et al. 2014) and were rlog transformed according to the DEseq2 pipeline. DESeq2 was used for the calculation of normalized counts for each transcript using default parameters. All normalized transcripts with a maximum overall row mean lower than 10 were excluded, resulting in 21,685 present transcripts. RNA-seq data can be accessed under GSE163635.
Construction of Coexpression networks analysis (CoCena)
To define differences and similarities in transcript expression patterns among the different groups, CoCena was performed based on Pearson correlation. CoCena is a network-based approach to identify clusters of genes that are coexpressed in a series of observed conditions based on data retrieved from RNA-sequencing. The tool offers a variety of functions that allow subsequent in-depth analysis of the biological context associated with the found clusters. The 10,000 most variable genes were selected as input for the analysis. A Pearson correlation coefficient cutoff of 0.915 (7713 nodes and 410,684 edges) was chosen to construct scale-free networks.
MiRNA enrichment analysis
Each input list of genes used for miRNA enrichment analysis consisted of CoCena gene modules found to be down-regulated in a given mutant genotype as compared to the wild-type counterpart (also called negative modules). Specifically, we used genes commonly down-regulated in Trim71 KO ESCs, Lin28a KO ESCs, and double KO ESCs, derived from our own analysis (GSE163635). We additionally used genes commonly down-regulated in Trim71 KO ESCs, Trim71 RING mutant (C12LC15A) ESCs, and Trim71 NHL mutant (R738A) ESCs, derived from a previous study which also included mRNAs enriched in TRIM71 RNA-IPs of ESCs (GSE134125) (Welte et al. 2019). Last, we used two published data sets derived from previous studies in HCC cells (Welte et al. 2019; Foster et al. 2020), and focused on genes significantly down-regulated in TRIM71 KO Huh7 cells (GSE134125) (Welte et al. 2019) and genes significantly enriched in TRIM71 RNA-IPs of HepG2 cells (Foster et al. 2020). In general, miRNA enrichment analysis was first conducted on negative modules to identify let-7 targets among the mRNAs found stabilized in the presence of TRIM71. Then, negative modules were overlapped with RNA-IP data to identify mRNAs bound and stabilized by TRIM71 (intersection). Last, miRNA enrichment analysis was conducted on the intersection to identify let-7 targets bound and stabilized by TRIM71.
miRNA enrichment analysis was performed with ShinyGo v0.61 online software (Ge et al. 2020). This software identifies miRNA targets present in a list of genes and returns the top X miRNAs whose targets are significantly enriched in that list based on hypergeometric distribution followed by false discovery rate (FDR) correction (Ge et al. 2020). For this analysis, we selected a FDR cutoff of 0.05 for the top 100 enriched miRNAs. The GRIMSON (mouse) and TargetScan (human) miRNA databases were selected for the analysis of ESCs and HCC cells, respectively, as these two databases provide the enrichment for each given miRNA family. An additional analysis was then conducted to identify specific targets of individual miRNA members within each family. To this end, we used the multiMiR R package, which scans an input list of genes for predicted miRNA targets by integrating information from eight different databases (DIANA-microT, ElMMo, MicroCosm, miRanda, miRDB, PicTar, PITA, and TargetScan) (Ru et al. 2014). For miRNA network visualization, individual members of the let-7 miRNA family were depicted connected to each of their respective identified targets. Only genes predicted to be let-7 targets by at least four different databases were used for visualization. Networks were visualized using the software Cytoscape (v3.8.2).
RNA extraction and qRT-PCR quantification
RNA was extracted from cell pellets using the TRIzol-containing reagent TriFAST (peqGold) according to the manufacturer's instructions. RNA pellets were resuspended in RNase-free water, and DNA digestion was performed prior to RNA quantification. 0.5–1 µg of RNA were reverse-transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions. The cDNA was then diluted 1:5, and a relative quantification of specific genes was performed in a Bio-Rad qCycler using TaqMan probes in iTaq Universal Probes Supermix (Bio-Rad). TaqMan probes used can be found in Supplemental Table 3.
Luciferase reporter assays
For luciferase assays, cells were cotransfected with the required psiCHECK2 dual luciferase plasmid and the specified pRK5-FLAG construct in a 1:4 ratio, and cells were harvested for further analysis 48 hpt. If miRNA overexpression or siRNA-mediated knockdown was required, miRNA/siRNA transfection was conducted 24 h before DNA transfection. psiCHECK2 plasmids contained a 3′UTR sequence with several miRNA binding sites (BS) in tandem downstream from Renilla luciferase coding sequence, while the firefly luciferase, expressed under the control of a different promoter and not subjected to any altered 3′UTR regulation, was used as normalization control. To eliminate the possible 3′UTR-independent impact that each condition/construct could have on the psiCHECK2 vector backbone, a psiCHECK2 vector with no insert downstream from the Renilla sequence (Renilla-empty) was generated. Then, the Renilla-3′UTR/Firefly ratio was calculated and then normalized to the Renilla-empty/Firefly ratio calculated for the same condition. The resultant values were specified as normalized relative light units (norm. RLU). The repression strength (norm. RLU [FLAG-TRIM71]/norm. RLU [FLAG-Empty]) was calculated as an additional parameter representing the degree of TRIM71-mediated repression for each miRNA. Repression strength values higher than 1 were observed upon repression of a specific miRNA, while values equal or below 1 implied that no repression was observed. Renilla and firefly luciferase signals were read in a MicroLumat Plus LB96V luminometer by using the Dual Luciferase Reporter Assay kit (Promega) following the manufacturers’ instructions. The 8x let-7 BS psiCHECK2 plasmid was acquired from Addgene (#20931), and the 3x miRNA BS psiCHECK2 plasmids (used in Supplemental Fig. 10E) were generated by inserting artificial sequences containing three miRNA-binding sites in tandem via XhoI/NotI directional cloning.
Protein extraction and western blotting (WB)
Cell pellets were lysed in RIPA Buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1 mM Na3VO4, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM glycerophosphate) supplemented with protease inhibitors and protein lysates were precleared by centrifugation and quantified using the BCA assay kit (Pierce) according to the manufacturer's instructions. Protein lysates were then denatured by incubation with SDS Buffer (12% glycerol, 60 mM Na2EDTA pH 8, 0.6% SDS, 0.003% bromophenol blue) for 10 min at 95°C and separated by PAGE-SDS in Laemmli Buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). Proteins were then wet transferred to a nitrocellulose membrane in transfer buffer (25 mM Tris–HCl pH 7.6, 192 mM glycine, 20% methanol, 0.03% SDS) and membranes were then blocked with 3% BSA in 1× TBST (50 mM Tris–HCl pH 7.6, 150 mM NaCl, 0.05% Tween-20) prior to overnight incubation at 4°C with the required primary antibodies. After washing the membrane three times with 1× TBST, they were incubated with a suitable HRP-coupled secondary antibody for 1 h at RT followed by three washing steps with 1× TBST. Membranes were developed with ECL substrate kit (Pierce) according to the manufacturer's instructions. Antibodies used can be found in Supplemental Table 4.
Protein immunoprecipitation (IP)
For protein interaction studies, proteins were extracted on IP buffer (50 mM Tris–HCl pH 7.5, 1 mM EGTA, 1 mM EDTA, 270 mM Sucrose, 1% Triton X-100) supplemented with phosphatase inhibitors (10 mM Glycerophosphate, 50 mM Sodium Fluoride, 5 mM Sodium Pyrophosphate, 1 mM Sodium Vanadate) and protease inhibitors. Protein lysates were precleared by centrifugation at 12,000 rpm and 4°C for 5 min, quantified using the BCA assay kit (Pierce), and at least 500 µg of protein were incubated with 10 µL of magnetic beads at 4°C in a rotating wheel for 5 h or overnight. A portion of each whole-cell lysate was retained as input control. Sigma ANTI-FLAG M2 magnetic beads were used for the IP of Flag-tagged proteins and Protein A/G Dynabeads for the IP of Ig-tagged proteins or endogenous proteins by precoupling the beads to the desired primary antibody. Precoupling was performed in 300 µL of IP buffer with the suitable antibodies diluted 1:50 at 4°C for 2 h in a rotating wheel. After IP, beads were washed five times with 500 µL IP buffer and bound proteins were eluted from the beads by boiling in SDS buffer, followed by PAGE-SDS separation and WB as described above. For RNase treatment after IP, washed IP fractions were incubated in 200 µL IP buffer containing 200 µg/mL RNase A at 37°C for 30 min and washed three more times with IP buffer before protein elution. Antibodies used can be found in Supplemental Table 4.
Electrophoretic mobility-shift assays (EMSA)
The generation of a recombinant TRIM71 NHL protein (FLAG-NHL) was previously described (Torres-Fernández et al. 2019). The capability of MYC-LIN28A (OriGene #TP303397) and FLAG-NHL recombinant proteins to directly bind pre-miRNAs was evaluated by EMSA. pre-let-7 and pre-miR-16 molecules (miRVana) were radioactively 5′-labeled with 32P isotopes by incubation with 32P-γATP and the phosphorylating enzyme T4 PNK (Roche) for 30 min at 37°C, followed by enzyme inactivation for 10 min at 65°C. 32P-Labeled pre-miRNAs were then purified with ProbeQuant G-50 MicroColumns (Merck) according to the manufacturer's instructions in order to eliminate the T4 PNK enzyme and the excess of 32P-γATP. Radioactivity levels (cpm = counts per minute) of the purified samples were quantified using a liquid scintillation counter. A fixed amount of each 32P-labeled pre-miRNA (60 fmol) was incubated with increasing concentrations of MYC-LIN28A and/or FLAG-NHL recombinant proteins in a range of 10–1000 nM, and samples were brought to the equilibrium for 30 min at 37°C in binding buffer (10 mM MOPS pH 7.5, 50 mM KCl, 5 mM MgCl2, 3 mM EDTA pH 8, 30 µg/mL heparin, 5% glycerol) supplemented with 1 mM DTT and competitor yeast tRNA in a 1000-fold excess over the 32P-pre-miRNA to prevent unspecific binding. Control samples containing only each of the 32P-labeled pre-miRNAs were included to evaluate the RNA migration in the absence of protein(s). Samples were then loaded on a 10% native PAGE gel and run for 4 h at 200 V. Gels were then dehydrated for 30–45 min at 80°C in a vacuum dryer and exposed to autoradiography films for the visualization of 32P-Labeled pre-miRNAs–protein complexes, detected as up-shifted bands as compared to the bands observed in control samples. Densitometry quantification of up-shifted bands was conducted with the LI-COR Image Studio Lite software and binding-saturation curves were built and adjusted by nonlinear regression using GraphPad Prism7 to estimate the equilibrium dissociation constant (KD) of pre-miRNAs–protein complexes.
Confocal microscopy
eGFP-Trim71-transfected cells were seeded on glass coverslips coated with Poly-L-lysine (200 µg/mL), fixed with 4% PFA and permeabilized with 0.2% Triton X-100/PBS. Samples were then incubated in a dark humid chamber with anti-AGO2 antibody (1:200) for 1 h at RT followed by incubation with Alexa Fluor-647-coupled anti-rabbit secondary antibody (Jackson ImmunoResearch) for 1 h at RT. Images were taken using an Olympus Fluoview 1000 confocal microscope equipped with a Plan Apochromat 60×, NA 1.4 oil immersion objective (Olympus) and DIC.
eFluo670 proliferation assays
Cells were stained with the proliferation dye eFluor670 (eBiosciences) diluted to 5 µM in PBS (1 mL of PBS per 106 cells) for 10 min at 37°C in the dark. The labeling was stopped by addition of 4–5 volumes of cold FCS and incubation for 5 min on ice. Cells were washed once with complete growth medium and once with PBS before seeding them at different densities per condition in multiwell plates. A fraction of the cells was used to measure the initial fluorescence intensity (day 0) via flow cytometry. The loss of fluorescence intensity overtime was monitored by flow cytometry for 4 d after staining. Flow cytometry data was then analyzed with FlowJo. The number of cell divisions at the end of the experiment was calculated assuming a decrease of the median fluorescence intensity (MFI) by half upon each cell division with the following formula: Division Number = log2 [(MFIday0–MFIunstained)/(MFIday4–MFIunstained)]. The average cell cycle duration was estimated at the end of the experiment from the number of cell divisions. For the investigation of proliferation in the course of neural differentiation, ESCs were stained as described above and directly seeded in N2B27 differentiation media (Neurobasal medium mixed with DMEM/F12 medium 1:1, supplemented with 1% Penicillin-Streptomycin, 50 mg/mL BSA, 0.5% GlutaMAX, 1% N2 Supplement and 2% B27 Supplement), which induces differentiation into neural progenitor cells within 4 days, as previously described (Abranches et al. 2009; Mitschka et al. 2015).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Veit Hornung and Dr. Thomas S. Ebert (LMU, Munich) for kindly providing us with the AGO2 KO HEK293T cells and Professor Helge Großhans (FMI, Basel) for the Trim71 KO NE4C cells. We also thank Professor Shinya Yamanaka (CiRA, Kyoto) and the J. David Gladstone Institutes for providing us with the TRIM71 antibody that we used for endogenous protein immunoprecipitation. We furthermore thank all members of our laboratory and the LIMES Institute for general advice and discussion. L.T.F. held a stipend from Bayer AG and Immunosensation Cluster of Excellence. W.K. and J.L.S. are funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy EXC2151—390873048.
Author contributions: L.T.F. and S.M. designed and performed all experiments and wrote the manuscript. T.U., K.D., M. Becker, K.H., and J.L.S. conducted RNA-seq and bioinformatic analysis. M. Beyer designed the TALENs cloning strategy. J.W. conducted several experiments under the supervision of L.T. S.W. conducted Co-IPs in ESCs during the review process. W.K. supervised experimental design and performance and cowrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078696.121.
REFERENCES
- Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, Bekman E. 2009. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS One 4: e6286. 10.1371/journal.pone.0006286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H-M, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. 2012. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun 3: 923. 10.1038/ncomms1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lai F, Niswander L. 2012. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev 26: 803–815. 10.1101/gad.187641.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Yuan RH, Yang WC, Hsu HC, Jeng YM. 2013. The stem cell E3-ligase Lin-41 promotes liver cancer progression through inhibition of microRNA-mediated gene silencing. J Pathol 229: 486–496. 10.1002/path.4130 [DOI] [PubMed] [Google Scholar]
- Choudhury NR, Nowak JS, Zuo J, Rappsilber J, Spoel SH, Michlewski G. 2014. Trim25 is an RNA-specific activator of Lin28a/TuT4-mediated uridylation. Cell Rep 9: 1265–1272. 10.1016/j.celrep.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas E, Rybak-Wolf A, Rohde AM, Nguyen DTT, Wulczyn FG. 2015. Lin41/Trim71 is essential for mouse development and specifically expressed in postnatal ependymal cells of the brain. Front Cell Dev Biol 3: 20. 10.3389/fcell.2015.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Wang X, Luo M, Xu W, Zhou T, Wang M, Yu L, Li L, Cai L, Wang PJ, et al. 2020. mRBPome capture identifies the RNA binding protein TRIM71, an essential regulator of spermatogonial differentiation. Development 147: 184655. 10.1242/dev.184655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M, Grosshans H. 2013. LIN-41/TRIM71: emancipation of a miRNA target. Genes Dev 27: 581–589. 10.1101/gad.207266.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DJ, Chang HM, Haswell JR, Gregory RI, Slack FJ. 2020. TRIM71 binds to IMP1 and is capable of positive and negative regulation of target RNAs. Cell Cycle 19: 2314–2326. 10.1080/15384101.2020.1804232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey CG, Choi J, Jin SC, Zeng X, Timberlake AT, Nelson-Williams C, Mansuri MS, Lu Q, Duran D, Panchagnula S, et al. 2018. De novo mutation in genes regulating neural stem cell fate in human congenital hydrocephalus. Neuron 99: 302–314.e4. 10.1016/j.neuron.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge SX, Jung D, Jung D, Yao R. 2020. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36: 2628–2629. 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. 2006. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 384: 51–61. 10.1016/j.gene.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. 2009. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938. 10.1016/j.cell.2009.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284. 10.1016/j.molcel.2008.09.014 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708. 10.1016/j.cell.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, Egranov SD, Zhang Y, Xia W, Gong J, et al. 2019. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol 20: 835–851. 10.1038/s41590-019-0400-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. 2008. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32. 10.1038/nrm2321 [DOI] [PubMed] [Google Scholar]
- Ichimura T, Taoka M, Shoji I, Kato H, Sato T, Hatakeyama S, Isobe T, Hachiya N. 2013. 14-3-3 proteins sequester a pool of soluble TRIM32 ubiquitin ligase to repress autoubiquitination and cytoplasmic body formation. J Cell Sci 126: 2014–2026. 10.1242/jcs.122069 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y. 2009. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell 34: 58–67. 10.1016/j.molcel.2009.02.010 [DOI] [PubMed] [Google Scholar]
- Jee D, Lai EC. 2014. Alteration of miRNA activity via context-specific modifications of Argonaute proteins. Trends Cell Biol 24: 546–553. 10.1016/j.tcb.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle KT, Kulkarni AV, Limbrick DD Jr, Warf BC. 2016. Hydrocephalus in children. Lancet 387: 788–799. 10.1016/S0140-6736(15)60694-8 [DOI] [PubMed] [Google Scholar]
- Kanamoto T, Terada K, Yoshikawa H, Furukawa T. 2006. Cloning and regulation of the vertebrate homologue oflin-41 that functions as a heterochronic gene in Caenorhabditis elegans. Dev Dyn 235: 1142–1149. 10.1002/dvdy.20712 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, Ketting RF, Plasterk RHA. 2004. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res 32: 6284–6291. 10.1093/nar/gkh968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P, Aeschimann F, Gaidatzis D, Keusch JJ, Ghosh P, Neagu A, Pachulska-Wieczorek K, Bujnicki JM, Gut H, Großhans H, et al. 2018. Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition. Nat Commun 9: 1549. 10.1038/s41467-018-03920-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancman JJ, Caruccio NC, Harfe BD, Pasquinelli AE, Schageman JJ, Pertsemlidis A, Fallon JF. 2005. Analysis of the regulation of lin-41 during chick and mouse limb development. Dev Dyn 234: 948–960. 10.1002/dvdy.20591 [DOI] [PubMed] [Google Scholar]
- Lee SH, Cho S, Kim MS, Choi K, Cho JY, Gwak HS, Kim YJ, Yoo H, Lee SH, Park JB, et al. 2014. The ubiquitin ligase human TRIM71 regulates let-7 microRNA biogenesis via modulation of Lin28B protein. Biochim Biophys Acta 1839: 374–386. 10.1016/j.bbagrm.2014.02.017 [DOI] [PubMed] [Google Scholar]
- Li YP, Duan FF, Zhao YT, Gu KL, Liao LQ, Su HB, Hao J, Zhang K, Yang N, Wang Y. 2019. A TRIM71 binding long noncoding RNA Trincr1 represses FGF/ERK signaling in embryonic stem cells. Nat Commun 10: 1368. 10.1038/s41467-019-08911-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-C, Hsieh L-C, Kuo M-W, Yu J, Kuo H-H, Lo W-L, Lin R-J, Yu AL, Li W-H. 2007. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evol 24: 2525–2534. 10.1093/molbev/msm195 [DOI] [PubMed] [Google Scholar]
- Loedige I, Gaidatzis D, Sack R, Meister G, Filipowicz W. 2013. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res 41: 518–532. 10.1093/nar/gks1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Gregory RI. 2013. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA 19: 605–612. 10.1261/rna.036434.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr F, Heinemann U. 2013. Mechanisms of Lin28-mediated miRNA and mRNA regulation—a structural and functional perspective. Int J Mol Sci 14: 16532–16553. 10.3390/ijms140816532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626. 10.1038/nature08725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschka S, Ulas T, Goller T, Schneider K, Egert A, Mertens J, Brüstle O, Schorle H, Beyer M, Klee K, et al. 2015. Co-existence of intact stemness and priming of neural differentiation programs in mES cells lacking Trim71. Sci Rep 5: 11126. 10.1038/srep11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. 2011. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147: 1080–1091. 10.1016/j.cell.2011.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. 2008. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454: 241–245. 10.1038/nature07014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DTT, Richter D, Michel G, Mitschka S, Kolanus W, Cuevas E, Wulczyn FG. 2017. The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ 24: 1063–1078. 10.1038/cdd.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JS, Choudhury NR, De Lima Alves F, Rappsilber J, Michlewski G. 2014. Lin28a regulates neuronal differentiation and controls miR-9 production. Nat Commun 5: 3687. 10.1038/ncomms4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell F, Esfahani SS, Engström Y, Kylsten P. 2008. Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev Dyn 237: 196–208. 10.1002/dvdy.21396 [DOI] [PubMed] [Google Scholar]
- Ouchi Y, Yamamoto J, Iwamoto T. 2014. The heterochronic genes lin-28a and lin-28b play an essential and evolutionarily conserved role in early zebrafish development. PLoS One 9: e88086. 10.1371/journal.pone.0088086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89. 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Patranabis S, Bhattacharyya SN. 2016. Phosphorylation of Ago2 and subsequent inactivation of let-7a RNP-specific microRNAs control differentiation of mammalian sympathetic neurons. Mol Cell Biol 36: 1260–1271. 10.1128/MCB.00054-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter ME. 2009. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 8: 843–852. 10.4161/cc.8.6.7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. 2011. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147: 1066–1079. 10.1016/j.cell.2011.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Ren H, Xu Y, Wang Q, Jiang J, Wudumuli HL, Zhang Q, Zhang X, Wang E, Sun L, et al. 2018. E3 ubiquitin ligase tripartite motif-containing 71 promotes the proliferation of non-small cell lung cancer through the inhibitor of kappaB-α/nuclear factor kappaB pathway. Oncotarget 9: 10880–10890. 10.18632/oncotarget.19075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, Mahaffey S, Rossi S, Calin GA, Bemis L, et al. 2014. The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res 42: e133. 10.1093/nar/gku631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG. 2009. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 11: 1411–1420. 10.1038/ncb1987 [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. 2012. A transcription activator-like effector toolbox for genome engineering. Nat Protoc 7: 171–192. 10.1038/nprot.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BRM, Esquela-Kerscher A, Slack FJ. 2005. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn 234: 1046–1054. 10.1002/dvdy.20599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BRM, Liang X, Stahlhut C, Delconte C, Stefani G, Slack FJ. 2008. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 7: 3935–3942. 10.4161/cc.7.24.7397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA. 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925. 10.1016/j.cell.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5: 659–669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. 2009. Essential and overlapping functions for mammalian argonautes in microRNA silencing. Genes Dev 23: 304–317. 10.1101/gad.1749809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Katsura A, Miyazono K. 2015. A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B. Cancer Sci 106: 1174–1181. 10.1111/cas.12721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. 2017. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45: W98–W102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. 2012. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18: 1875–1885. 10.1261/rna.034538.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Fernández LA, Jux B, Bille M, Port Y, Schneider K, Geyer M, Mayer G, Kolanus W. 2019. The mRNA repressor TRIM71 cooperates with Nonsense-Mediated Decay factors to destabilize the mRNA of CDKN1A/p21. Nucleic Acids Res 47: 11861–11879. 10.1093/nar/gkz1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Fernández LA, Emich J, Port Y, Mitschka S, Wöste M, Schneider S, Fietz D, Oud MS, Di Persio S, Neuhaus N, et al. 2021. TRIM71 deficiency causes germ cell loss during mouse embryogenesis and is associated with human male infertility. Front Cell Dev Bio 9: 658966. 10.3389/fcell.2021.658966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Plessmann U, Harlander S, Daiß JL, Eichner N, Lehmann G, Schall K, Urlaub H, Meister G. 2017. A compendium of RNA-binding proteins that regulate microRNA biogenesis. Mol Cell 66: 270–284.e13. 10.1016/j.molcel.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Wang T, He Y, Zhu Y, Chen M, Weng M, Yang C, Zhang Y, Ning N, Zhao R, Yang W, et al. 2016. Comparison of the expression and function of Lin28A and Lin28B in colon cancer. Oncotarget 7: 79605–79616. 10.18632/oncotarget.12869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte T, Tuck AC, Papasaikas P, Carl SH, Flemr M, Knuckles P, Rankova A, Bühler M, Großhans H. 2019. The RNA hairpin binder TRIM71 modulates alternative splicing by repressing MBNL1. Genes Dev 33: 1221–1235. 10.1101/gad.328492.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K, Narita M, Srivastava D, Yamanaka S. 2014. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14: 40–52. 10.1016/j.stem.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Cuevas E, Franzoni E, Rybak A. 2010. MiRNAs need a trim: regulation of miRNA activity by trim-NHL proteins. Adv Exp Med Biol 700: 85–105. 10.1007/978-1-4419-7823-3_9 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M, et al. 2016. LIN28 regulates stem cell metabolism and conversion to primed pluripotency accession numbers GSE67568. Cell Stem Cell 19: 66–80. 10.1016/j.stem.2016.05.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.