Supplemental Digital Content is Available in the Text.
Promoting alternative splicing of human transient receptor potential ankyrin 1 channels with splice-switching antisense oligonucleotides serves to downregulate wildtype channel expression and holds promise for treating TRPA1-related disorders.
Abstract
Activation of transient receptor potential ankyrin 1 (TRPA1) channels by both environmental irritants and endogenous inflammatory mediators leads to excitation of the nerve endings, resulting in acute sensation of pain, itch, or chronic neurogenic inflammation. As such, TRPA1 channels are actively pursued as therapeutic targets for various pathological nociception and pain disorders. We uncovered that exon 27 of human TRPA1 (hTRPA1) could be alternatively spliced into hTRPA1_27A and hTRPA1_27B splice variants. The resulting channel variants displayed reduced expression, weakened affinity to interact with WT, and suffered from complete loss of function because of disruption of the C-terminal coiled-coil domain. Using a human minigene construct, we revealed that binding of splicing factor serine/arginine-rich splicing factor 1 (SRSF1) to the exonic splicing enhancer was critical for the inclusion of intact exon 27. Knockdown of SRSF1, mutation within exonic splicing enhancer, or masking SRSF1 binding with antisense oligonucleotides promoted alternative splicing within exon 27. Finally, antisense oligonucleotides-induced alternative splicing produced transcript and protein variants that could be functionally determined as diminished endogenous TRPA1 activity in human Schwann cell-line SNF96.2 and hiPSCs-derived sensory neurons. The outcome of the work could potentially offer a novel therapeutic strategy for treating pain by targeting alternative splicing of hTRPA1.
1. Introduction
The transient receptor potential ankyrin 1 (TRPA1) channels are nonselective cation channels predominantly expressed in small–medium size nociceptive neurons.37 Influx of Na+ and Ca2+ on channel opening depolarizes nerve terminals, triggering action potential, leading to neural transmission, and sensation of pain or itch. TRPA1 channels are widely regarded as nociceptive chemosensors because of their direct activation by a plethora of exogenous and endogenous electrophilic irritants.1–5,8,21,27,42,43 Indirectly, TRPA1 current was found to be potentiated by inflammatory mediator bradykinin and pruritogens through G protein–coupled receptors.3,4,48,52 Of clinical importance, the recent discoveries of TRPA1 as a critical mediator of inflammation and pain induced by ultraviolet light,19 bacterial endotoxin,28 angiotensin II,36 and chemotherapeutic agents25,29,45 provide mechanistic basis for targeting TRPA1 channels in various pathophysiological conditions.
Activated nociceptive nerve terminals produce various neuropeptides such as substance P and calcitonin gene-related peptide (CGRP) which contribute towards vessel dilation and recruitment of immune cells in a process termed neurogenic inflammation.30,35 Secretion of cytokines such as IL-1b and tumor necrosis factor-α (TNFα) by immune cells further sensitizes nociceptors, aggravating thermal and mechanical hypersensitivity.16 Consistently, human patients with gain of function mutation in TRPA1 channels suffer from familial episodic pain syndrome, showing symptoms such as episodic attack of debilitating pain during physical stress and enhanced mechanical hyperalgesia on mustard oil treatment.22
Accumulating evidence from rodent models suggests that knockdown, genetic deletion, or pharmacological blockade of TRPA1 channels could protect against chronic pain.12,24–26,31,34,44,45,49,50,53 Most notably, it was demonstrated that intradermal injections of the TRPA1 selective agonist JT010 in human subjects was sufficient to elicit pain in a dose-response manner, which otherwise could be reversed on cotreatment with TRPA1 selective blockers.18
Although alternative splicing pattern of TRPA1 transcripts has been reported in other species such as mice and fruit flies,17,53 it is not known if the human TRPA1 transcript could also be subjected to similar regulation. Our current study revealed novel alternative splicing patterns of hTRPA1 expressed in the human dorsal root ganglion (DRG) neurons. Specifically, use of alternative splice sites within exon 27 yielded 2 alternative splice variants, hTRPA1_27A and hTRPA1_27B. The resulting channel variants displayed loss of function because of disruption within the C-terminal coiled-coil domain. By employment of a human minigene system, we uncovered serine/arginine-rich splicing factor 1 (SRSF1) as a splicing factor that interacted with the exonic splicing enhancer (ESE) within exon 27 to allow for inclusion of intact exon 27. siRNA mediated knockdown of SRSF1, mutation within ESE, or masking SRSF1 binding with antisense oligonucleotides (ASO) resulted in upregulation of 27A and 27B variants. Finally, splice-switching ASO altered the splicing pattern of native TRPA1 channels correlating with dramatically attenuated Ca2+ response to allyl isothiocyanate (AITC) treatment in both Schwann cell-line SNF96.2 and hiPSCs-derived sensory neurons. The outcome of the work could potentially offer a novel therapeutic strategy for treating neurogenic inflammation by targeting alternative splicing of hTRPA1.
2. Methods
2.1. Construction of minigenes for in vitro alternative splicing assay
The genomic DNA was extracted from HEK293FT cells with Promega Wizard SV Genomic DNA Purification System (Promega) and used a template for PCR with the primer pair Hmini_Fwd GGATCCAGTGGTGGTTGAATTTTGGCA and Hmini_Rvs GTCGACAACCATGATTTCACACGCAGC. The PCR product was subjected to molecular cloning and verified by DNA sequencing before being subcloned into the modified pRK5 vector with restriction enzyme BamHI and SalI (NEB). The mutant Minigene_E1 was generated by overlapping PCR with primer pairs HE1_Fwd GTCCTTTTAGGCTGAAttATCTTACTTTTCTCCT and HE1_Rvs AGGAGAAAAGTAAGATaaTTCAGCCTAAAAGGAC.
2.2. RT-PCR to detect an alternative splicing pattern of transient receptor potential ankyrin 1 transcript
1 μg of total RNA of human DRG (Clonetech, Cat#636150) or SNF96.2 cells was reverse transcribed into cDNA with SuperScript III (Invitrogen, Carlsbad, CA) and oligo (dT)18 primers. To perform transcript across exon 26 to exon 27 of human TRPA1 mRNA, the first round and second PCR was performed by the following primer pairs.
| 1st round PCR | hTRPA1_F | ATGAAGCGCAGCCTGAGGAAGATG |
| hTRPA1_R | CTAAGGCTCAAGATGGTGTGTTTTTGCC | |
| 2nd round PCR E24-27 | HTAE24F | AGTTGGCGACATTGCTGAG |
| HTA27R | TGTGTTTTTGCCTTGACTGC |
On the other hand, mouse DRG tissues was obtained from 3-month old C57BL/6 mice by dissection, and total RNA was extracted using the Trizol method (Invitrogen). The RNA samples were treated with RNase-free DNase (Ambion) before reverse transcription with oligo (dT)18 primers. To compare alternative splicing pattern in mouse TRPA1 (mTRPA1)mRNA, the first round and second PCR was performed by the following primer pairs.
| 1st round PCR | mTRPA1_F | ATGAAGCGCAGCCTGAGGAAGATG |
| mTRPA1_R | CTAAGGCTCAAGATGGTGTGTTTTTGCC | |
| 2nd round PCR E24-27 | MTAE24F | CTGATTGGCTTGGCGGTTGG |
| MTAE27R | CGGGTGGCTAATAGAACAATGTGTT |
To detect alternative splicing in hTRPA1 transcript expressed from minigene transfected HEK293FT cell, 0.1 μg of wildtype Minigene-WT or mutant Minigene_E1 was cotransfected with either 5 μL of 10 μM ON-TARGETplus nontargeting siRNA (NT siRNA)#1 (Dharmacon, D-001810-01-05) or ON-TARGETplus SRSF1 siRNA (Dharmacon, L-018672-01-0005) with lipofectamine 2000 (Invitrogen) according to manufacturer's protocol in 2 mL of final transfection cocktail reaching concentration of 25 nM. 48 hours after transfection, total RNA was isolated using the Trizol method (Invitrogen). The RNA samples were treated with RNase-free DNase (Ambion) before reverse transcription. For minigene RNA, first strand cDNA was synthesized with SuperScript III (Invitrogen) and reverse primers cis33 (GCGGTACCAATAAACAAGTTGGGCCATGG) specific to within SV40 polyadenylation signal in the modified pRK5 vector. Subsequently, PCR was performed with primer pair Hmini_Fwd TGCACTGGGGAAATAAGACAAGA and Hmini_Rvs GGAGCTAAGGCTCAAGATGGT.
2.3. Cloning of wildtype human transient receptor potential ankyrin 1 channel and 27A and 27B splice variants
The following primer pairs were used to clone relevant reading frames using the cDNA reverse transcribed from the human DRG sample.
| Wild type hTRPA1 | hTRPA1_cF | CTCGAGATGAAGCGCAGCCTGAGGAAGATG |
| hTRPA1_cR | CCGCGGCTAAGGCTCAAGATGGTGTGTTTTTGCC | |
| hTRPA1_27A | hTRPA1_cF | CTCGAGATGAAGCGCAGCCTGAGGAAGATG |
| 27A_cR | CCGCGGTTAAACCTGTCGGTATTTCTGCTTTA | |
| hTRPA1_27B | hTRPA1_cF | CTCGAGATGAAGCGCAGCCTGAGGAAGATG |
| 27B_cR | CCGCGGCTAAGGCTCAAGATGGTGTGTTTTTGCCTT |
The PCR products were subjected to molecular cloning and verified by DNA sequencing before cloning in pIRES2-eGFP by restriction enzyme XhoI and SaclI (NEB). In cotransfection experiment, the coding sequence of eGFP was deleted.
2.4. Surface biotinylation assay
The surface biotinylation assay was performed with Pierce Cell Surface Biotinylation and Isolation Kit (Thermofisher Scientific, A44390) with minor modification. In brief, 1.5 μg of WT, 27A, and 27B plasmids were transfected into 1.5 million HEK293FT cells cultured on 6-well plates. 48 hours post-transfection, the cells were labelled with EZ-Link Sulfo-NHS-SS-Biotin and subsequently lysed with 300 μL NP40 buffer containing 150 mM sodium chloride 1.0% Triton X-100 and 50 mM Tris, pH 8.0 supplemented with Mini, EDTA-free Protease Inhibitor Cocktail (Roche). 30 μL of the clarified lysate was saved as total lysate whereas the remaining lysate was treated to 100 μL of NeutrAvidin Agarose beads to enrich for biotin-labelled surface fraction. The beads were subsequently washed 4 times with 1 mL NP40 buffer and denatured at 95°C with 90 μL of NP40 buffer supplemented with 6x loading buffer before SDS-PAGE and Western blot. Transferrin receptor (TFR) was used a loading control for surface protein detected with TFR antibody (Thermofisher Scientific, 136800) at 1:1000 dilution.
2.5. Coimmunoprecipitation to detect binding between the wildtype and splice variant of hTRPA1 channel subunit
To facilitate coimmunoprecipitation experiment, the human influenza hemagglutinin (HA) tag sequence TACCCATACGATGTTCCAGATTACGCT and the 3xFLAG tag sequence GACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGGACTACAAAGACGATGACGACAAG were introduced before the stop codon of the hTRPA1_WT and 27A and 27B constructs by PCR.1.5 μg of HA-tagged WT, 27A, or 27B constructs were transfected along or with 1.5 μg 3xFLAG-tagged WT into HEK293FT cells cultured on 6-well plates. One day before the harvest of the cells, 50 μL of protein A/G beads (Thermofisher, Cat#20422) was washed with 1 mL NP40 buffer once and incubated with 200 μL of NP40 buffer and 1 μL of mouse (ms) anti-FLAG antibody (Sigma-Aldrich, Cat# F1804) overnight. Forty eight hours post-transfection, the cells were lysed with 200 μL NP40 supplemented with Mini, EDTA-free Protease Inhibitor Cocktail (Roche), 20 μL of the clarified lysate was used for input, and the remaining were incubated with antibody-treated beads at 4°C with rotation for 2 hours. The beads were spun down and washed 4 times with 1 mL of NP40 buffer. 20 μL of 2x loading buffer was added to the input and 70 μL into the beads and incubated at 55°C for 10 minutes.10 μL of input and 20 μL of pulled down fraction was loaded for SDS PAGE and Western blot.
2.6. Western blot to detect expression of serine/arginine-splicing factor 1 and transient receptor potential ankyrin 1
Transfected HEK293FT cells were lysed with RIPA lysis buffer at 4°C. RIPA buffer consists of 150 mM sodium chloride, 0.1% Trion X-100, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris, pH 8.0 supplemented with Mini, and EDTA-free Protease Inhibitor Cocktail (Roche). 25 μg of protein lysate was loaded for SDS-PAGE and transfer. Anti-SRSF1 (Origene, TA311077) was used in dilution of 1:1000, and β-actin was stained with anti-β-actin (Sigma-Aldrich, A2228) antibody in the dilution of 1:10,000 as the loading control. Transfected SNF96.2 cells were lysed with NP40 lysis buffer supplemented with protease Inhibitor Cocktail at 4°C. 15 μg of protein lysate was loaded for SDS-PAGE and transfer. Anti-TRPA1 (Novus Biologicals, NB110-40763) was used in dilution of 1:1000.
2.7. Electrophysiological recordings and data analysis
HEK293FT were cultured in DMEM with 10% (vol/vol) FBS and 1% penicillin/streptomycin. One day before transfection, the cells were split, and 1.5 mL of the cells was seeded in a 35 mm tissue culture dish at the density of 10,000/mL. 1.5 μg of hTRPA1_WT, 27A, or 27B constructs were singly transfected into the cells by lipofectamine 2000 (Invitrogen) according to manufacturer's protocol. In cotransfection experiment, 1.5 μg of hTRPA1_WT was cotransfected with 1.5 μg of either empty pIRES2-eGFP, hTRPA1_27A, or hTRPA1_27B. The transfected cells were incubated for 24 hours in 5% CO2 incubator at 37°C. 48 hours post-transfection, the cell was split and seeded on poly-D lysine cover slip one day before recording. The cells that fluoresced green were recorded with ACSF solution containing (in mM) 10 glucose, 125 NaCl, 25 NaHCO3, 1.25 NaH2PO4.2H20, 2.5 KCl, 1.8 CaCl2, 1 MgCl2, pH 7.4 (300-310 mOsm), and internal solution containing (in mM) 130 K-gluconate, 10 KCl, 5 EGTA, 10 HEPES, 0.5 Na3GTP, 4 MgATP, 10 Na-phosphocreatine, adjusted with KOH to pH 7.4 (290 mOsm). The cells were held at −60 mV, and TRPA1 current was evoked by 100 μM AITC (Sigma-Aldrich, 377430) and blocked with 10 μM A967079 (Tocri Biosciences, 4716).Whole cell recording are performed with MultiClamp 700B amplifier (Molecular Device), low-pass filtered at 1 kHz and the series resistance was typically <20 MΩ after >80% compensation. Data were acquired using the software pClamp9 (Molecular Devices), and analyzed using GraphPad Prism V software (San Diego, CA) and Microsoft (Seattle, WA) Excel. Data are expressed as mean values ± SEM.
2.8. Design of splice-switching antisense oligonucleotides targeting splicing of hTRPA1
Antisense oligonucleotide 5′-GGAGAAAAGUAAGAUCCUUCAGCCUAAAAG-3′ and sense oligonucleotide (SO) 5′- CUUUUAGGCUGAAGGAUCUUACUUUUCUCC-3'with 2′-O-methyl modified base and phosphorothioate linkage was generated by Integrated DNA Technologies. While phosphorothioate linkage confers endogenous nuclease resistance, 2'-O-methyl modified bases increase RNA: RNA binding affinity and stability. In some experiments, to facilitate visualization of the uptake, ASO and SO was also tagged with TYE563 at 5′ end. To test the splice-switching potential of ASO, 0.1, 1, and 10 μL of 5 μM ASO or 10 μL of 5 μM SO was transfected in 2 mL final transfection cocktail reaching concentration of 0.25, 2.5, and 25 nM for ASO or 25 nM for SO. As for SNF96.2 cells 10 μL of 10 μM ASO or SO was transfected in 4 mL final transfection cocktail reaching concentration of 25 nM. Cells were harvested 72 hours post-transfection for analysis.
2.9. Knock down of transient receptor potential ankyrin 1 in SNF96.2 cells
To knockdown TRPA1 in SNF96.2 cells, 10 μL of 10 μM ON-TARGETplus nontargeting siRNA #1 (Dharmacon, D-001810-01-05) or ON-TARGETplus TRPA1 siRNA (Dharmacon, LQ-006109-02-0010) were transfected alone or with 25 nM of ASO or SO in 4 mL final transfection cocktail in SNF96.2 cells using Lipofectamine RNAiMAX Reagent (Invitrogen) as per manufacturer's instruction. Cells were harvested 72 hours post-transfection for analysis.
2.10. Ca2+ imaging
SNF96.2cells (ATCC, CRL-2884) were cultured in DMEM with 10% (v/v) FBS and 1% penicillin/streptomycin. One day before transfection, the cells were split, and 1.5 mL of the cells was seeded on poly-D coated coverslips in a 35-mm tissue culture dish at the density of 20,000/mL. Each dish was transfected with 5 μL of 10 μM 5′-TYE563-tagged ASO or SO (Integrated DNA Technologies) using Lipofectamine RNAiMAX Reagent (Invitrogen) as per manufacturer's instruction. The medium was refreshed 8 hours after transfection, and cells were imaged 72 hours after transfection. The cells were washed with ACSF solution and incubated with 1 μM Fluo4-AM (Thermofisher Scientific, F14217) for 30 minutes and washed once with ACSF and incubated with 0.5 mL of ACSF before recording. To visualize cells transfected with 5′ TYE563-tagged oligonucleotides, before calcium imaging, the cells were excited at 588 nm and captured with 608 nm bandpass filter. To perform calcium imaging, the Fluo4 dye was excited at 480 nm, and emitted fluorescence was filtered with a 535 nm bandpass filter. The Ca2+ transient was evoked with 0.1 mM of AITC (Sigma-Aldrich) and with 2 μM of ionomycin (Sigma-Aldrich). In another set of experiment, 10 mM of A967079 (Tocris Biosciences) was coadministered with 100 μM of AITC. Time lapse images were captured by Olympus DP71 camera installed on Olympus IX81 microscope every 5 seconds in process.vsi files with cellSense software (Olympus) and subsequently exported as tiff images and analysis was performed on Image J software. ROI was selected close to the nuclei positive for oligonucleotide transfection for analysis, and Ca2+ transient was quantified as F/F0, ratio of fluorescent intensity (F) as compared with that of the first capture (F0).
2.11. Maintenance of hiPSCs culture
hiPSC(BJ-RiPS cell line) were cultured on feeder-free Matrigel-coated plate supplemented with iPSC-Brew XF prepared according to manufacturer's instruction. Passaging of iPSC were performed routinely at 1:6 split ratio every 6 to 7 days using ReLeSR (Stem Cell Technologies).
2.12. Differentiation of hiPSCs toward sensory neurons
Pluripotent stem cells were directed towards sensory neurons fate with slight modifications from a previously published protocol.10 In brief, hiPSCs were plated onto Matrigel-coated plates at a density of 1.5 × 105 cells/cm2 and inducted into anterior neuroectodermal lineage using synergistic dual SMAD inhibition by LDN-193189 (0.5 µM) and SB431542 (10 µM) on day 1. At day 3, Wnt agonist CHIR99021 (3 µM) and FGF inhibitor, SU5402 (5 µM) were used additionally to pattern neural progenitors towards the neural crest fate. At day 6, withdrawal of LDN-193189 and SB431542 coupled with addition of DAPT (2.5 µM) was used to block Notch signalling and encourage differentiation of progenitor towards neurons. Finally from day 11, neurotrophic factors including BDNF (20 ng/mL), GDNF (20 ng/mL), NGF-β (20 ng/mL), and ascorbic acids (10 ng/mL) were added to promote the maturation of sensory neurons. Sensory neurons basal media were prepared using 500 mL of DMEM/F12 supplemented with MACS Neurobrew-21 w/o Vitamin A, 0.5% GlutaMAX and 1% of N2 supplement. 6 weeks post-differentiation, the neuron cultured in 24 well were transfected with 2.5 μL of 10 μM SO or ASO complexed with 2.5 μL of 100 μM Chimeric Rabies Virus Glycoprotein Fragment (RVG - 9R) (Anaspec, Cat#AS-62565) in 500 μL medium for 72 hours before analysis by calcium imaging.
3. Results
3.1. Novel alternative splice patterns of human transient receptor potential ankyrin 1 disrupted C-terminal coiled-coil domain
On the mRNA level, post-transcriptional modification such as alternative splicing has been shown to regulate gene function by combinatorial assembly of alternative exons, yielding final protein products with different properties. Intriguingly, the transcript of mouse Trpa1 transcript in the DRG neurons was shown to be regulated by alternative splicing during inflammation.53 Exclusion of alternative exon 20 yielded a splice variant TRPA1b in addition to the full-length protein TRPA1a. Although nonfunctional on its own, coexpression of TRPA1b with the wildtype TRPA1a increased the TRPA1 current density. Notably, the increased expression level of Trpa1b transcripts correlated with the development of sensory hypersensitivity, suggesting that TRPA1 could be regulated by alternative splicing in diseased condition.
However, it is not known if human TRPA1 channels could also be subjected to similar regulation. To this end, we undertook transcript scanning, a method to comprehensively identify splice variations and compare splicing patterns of human and mouse TRPA1 transcripts expressed in DRG tissue. While human DRG RNA library was purchased from Clonetech, mouse DRG RNA was extracted from isolated DRG tissues. Both RNA samples were subjected to reverse transcription and subsequently transcript scanning was performed.41 First round PCR was performed using primer pairs spanning exon 1 to last exon 27 to amplify the full-length transcript. This was followed by amplification of small overlapping amplicon to detect alternative splicing patterns (Fig. 1A).
Figure 1.
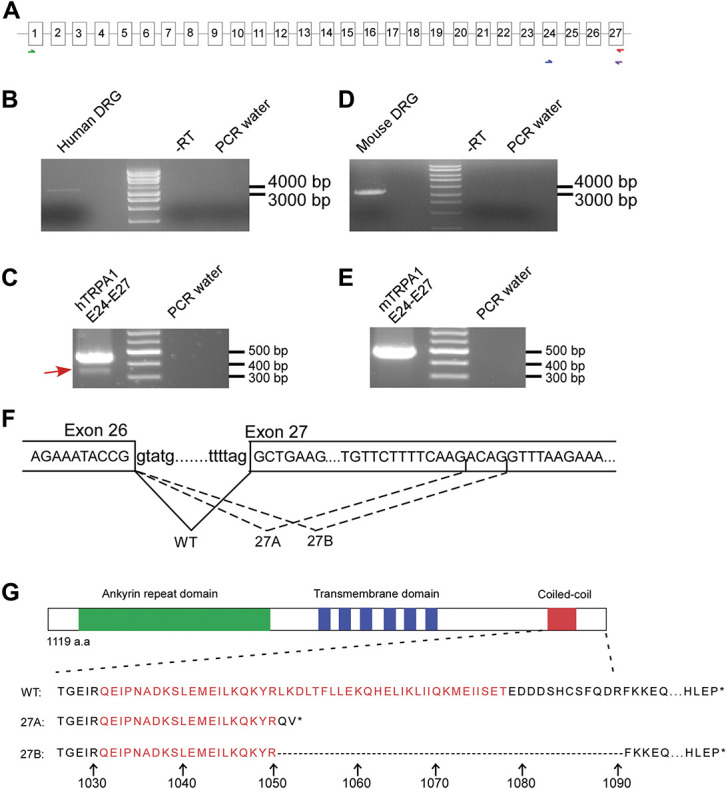
Discovery of novel alternative splicing pattern within exon 27 of hTRPA1. (A) Schematic demonstrating the strategy of performing transcript scanning across mRNA of human and mouse TRPA1. First round PCR was performed to amplify the full-length transcript as indicated by the green and red arrows. Subsequently, second round PCR spanning exon 24 to exon 27 as indicated by the blue and purple arrows was performed to screen alternative splicing pattern. (B and C) RT-PCR result of full-length PCR and transcript scanning across exon 24 to 27 of hTRPA1 transcript respectively. (D and E) RT-PCR result of transcript scanning mTrpa1 format as in B and C. (F) Top, nucleotide sequence of exon and intron boundaries across exon 26 to exon 27 of hTRPA1. Bottom, usage of 3 different splice acceptor sites of exon 27 give rise to WT or 27A and 27B splice variant respectively. (G), Top, schematic protein structure of the TRPA1 channel. The green, blue, and red boxes indicate the ankyrin repeat domain, 6 transmembrane domains and the position of the coiled-coil domain, respectively. Bottom, alignment of amino acid sequences of WT, 27A, and 27B. The arrow and number indicate position of amino acids. TRPA1, transient receptor potential ankyrin 1.
Intriguingly, amplification of human TRPA1 transcript across exon 24 to exon 27 uncovered a PCR product of lower molecular weight (Figs. 1B and C). By contrast, such an alternative splicing pattern was not observed in mouse Trpa1(mTrpa1) transcript (Figs. 1D and E). Molecular cloning followed by DNA sequencing revealed alternative usages of splice sites within exon 27 of hTRPA1 transcripts and the 2 splice variants differed by 4 nucleotides (Fig. 1F). The 2 variants were named hTRPA1_27A and hTRPA1_27B. In silico translation predicted frameshift and premature termination of hTRPA1_27A, whereas hTRPA1_27B generated deletion of 40 amino acids in the correct reading frame. Both variants are severely truncated in the C-terminal coiled-coiled domain as highlighted in red (Fig. 1G). In comparison, similar analysis across exon 17 to exon 21 detected a lower band which corresponded to the deletion of exon 20 as reported previously in mouse DRG sample.53 By contrast, such alternative splicing pattern was absent in the human DRG (Supplementary Fig. 1, available at http://links.lww.com/PAIN/B279).
3.2. Disruption of coiled-coil domain affected channel expression, assembly, and function
The crystal structure of TRPA1 channels has previously been reported33 shedding important mechanistic insight into the working of the channels. Similar to other TRP channels, the transmembrane core is a homotetramer assembled from 4 subunits of the channel through domain swap interaction. However, unique to TRPA1 channels, the coiled–coiled domains from 4 subunits form extensive stalk–like interaction structure in the centre of the channel below the permeation pore. Such stem structure contains an upper destabilizing and a lower stabilizing domain because of different positioning of the side chains of glutamine residues. It is worth to note that the stabilizing glutamine residue Gln1601 is missing in both hTRPA1_27A and 27B splice variants (Fig. 1G). On the other hand, the coiled-coil domain harbours several side chains including Lys1046 and Arg1050 from one coil, and Lys1048 and Lys1052 from an adjacent coil that form positively charged pocket to coordinate negatively charged cofactor such as inositol 6 phosphate (In6P) which sustains the activity of TRPA1 channels. Interestingly, Lys1052 is also deleted in both of the splice variants (Fig. 1G). Taken together, it was therefore tempting to hypothesize that structural disruption in both hTRPA1_27A and hTRPA1_27B variants could affect both channel assembly and function.
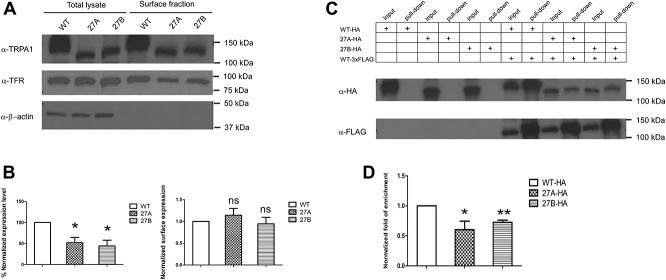
Overexpression of either the WT or splicer variants in HEK293FT cells revealed significantly lower expression of both 27A (% normalized expression level 52.1 + 12.7%) and 27B (% normalized expression level 44.3 + 13.4%) as compared with the WT. However, surface biotinylation assay showed robust surface localization of all 3 isoforms on the plasma membrane (Figs. 2A and B). Subsequently, to investigate the interaction of the 3 isoforms, WT, 27A, and 27B subunits were tagged with hemagglutinin (HA) tag, whereas the 3xFLAG tag WT subunit was used as a bait to pull down the other HA-tagged hTRPA1 subunits. Coimmunoprecipitation experiment revealed robust enrichment of the HA-tagged and 3xFLAG-tagged WT subunit in the pull down fraction over the input fraction. In comparison, both splice variants interacted with WT with significantly lower affinity. Normalized fold of enrichment for 27A and 27B subunits were 0.71 ± 0.15 and 0.62 ± 0.11, respectively. Notably, no HA-tagged channel subunits were observed in the pull down fraction when they are transfected alone, demonstrating the specificity of the system (Figs. 2C and D).
Figure 2.
hTRPA1_27A and 27B splice variants displayed the lower expression level, normal surface localization, and reduced binding affinity to the WT. (A) Surface biotinylation detected localization of WT, 27A, and 27B splice on the plasma membrane. Transferrin receptor (TFR) was used a marker for surface protein. (B) Left, densitometry analysis of the expression level of WT, 27A, and 27B in HEK293FT cells. The intensity of the WT, 27A, and 27B in total lysate fraction were first normalized against β-actin, respectively. The normalized intensity of 27A and 27B was then compared against that of the WT. * P < 0.05 (Student unpaired t test). Values shown are mean ± SEM (n = 3). Right, densitometry analysis of surface localization of WT, 27A, and 27B protein in HEK293FT cells. The ratio of surface fraction were first normalized against the input for each of the WT, 27A ,and 27B before normalization against surface over total ratio of TFR of the respective sample. The normalized intensity of 27A and 27B was then compared against the WT. ns, nonsignificant (Student unpaired t test). Values shown are mean ± SEM (n = 3). (C) Top, HEK293FT cells were transfected with HA-tagged WT, 27A or 27B variant alone or cotransfected with 3xFLAG-tagged WT. Bottom, experiment was performed by pulling down 3xFLAG-tagged WT with mouse (ms) anti-FLAG antibody, and subsequently, the HA-tagged protein subunit were detected with rabbit (rb) anti-HA antibody. The input and pull-down fractions were detected for each condition. The result is representative of 5 independent experiments. (D) Bar chart reporting the densitometry quantification of the pull-down efficiency. The band intensity of the pull-down fraction was first normalized against the input fraction for each of the HA-tagged WT, 27A, and 27B. The ratio was then normalized against the pull-down over input ratio of the 3xFLAG-tagged WT of each sample. The normalized ratio obtained for 27A and 27B variants were then compared against that of the WT * P < 0.05, ** P < 0.01 (Student unpaired t test). Values shown are mean ± SEM (n = 5). TRPA1, transient receptor potential ankyrin 1.
Overexpression of wildtype TRPA1 in HEK293FT cells produced robust AITC sensitive current (Fig. 3A). Such current was blocked by TRPA1 specific blocker A967079 and could be revived on washed out of the blocker (Fig. 3A). By contrast, 100 μM AITC evoked no current in cells transfected with either 27A or 27B variants (Figs. 3B and D). In addition, coexpression of the wildtype subunit with either 27A or 27B variant did not significantly alter the current density of the wildtype (Figs. 3E–H). Similar results were obtained when the dosage of AITC were reduced to 10 μM (Supplementary Fig. 2, available at http://links.lww.com/PAIN/B279). To ensure the same cell expresses both WT and splice variants, in the coexpression experiment, the wildtype channel subunit was expressed from a nonfluorescent vector, whereas the 27A or 27B variant was expressed within the pIRES2-eGFP vector that allowed for coexpression of eGFP protein. Subsequently, green fluorescent cells that showed AITC sensitive current were assumed to express both channel subunits, and the recordings were subjected to further analysis.
Figure 3.
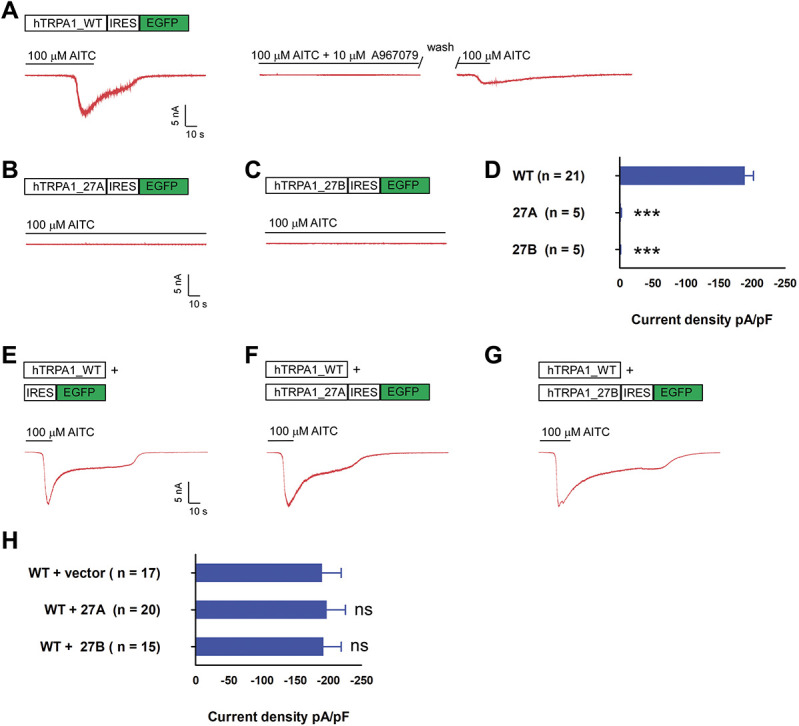
hTRPA1_27A and 27B splice variant displayed loss of function and did not affect WT current density on coexpression. (A) Top, hTRPA1 WT coding sequence was cloned into pIRES2-eGFP. On transfection, green fluorescent cells were selected for recording. Bottom, the cells were held at −60 mV, and TRPA1 current was evoked by perfusion of external solution containing 100 μM AITC as shown in the exemplar trace in red. Cotreatment of 10 μM A967079 and 100 μM AITC blocked activation of TRPA1 channels which could be reactivated on wash-out of the inhibitor. (B and C) Exemplar current of 27A and 27B on perfusion of 100 μM AITC, format as in A. (D) Bar chart reporting the peak current density of WT, 27A, and 27B singly transfected cells. *** P < 0.001 (Student unpaired t test) as compared with WT transfected cells. Values shown are mean ± SEM. (E) Exemplar current recorded from cell cotransfected with hTRPA1_WT expressed from nonfluorescent vector and empty pIRES2-eGFP vector, format as in A. (F) Exemplar current recorded from cell cotransfected with WT and 27A, format as in E. (G) Exemplar current recorded from cell cotransfected with WT and 27B, format as in E. (H) Bar chart reporting the peak current density of for conditions in F to G ns, nonsignificant (Student unpaired t test)) as compared with WT and empty vector transfected cells. Values shown are mean ± SEM. TRPA1, transient receptor potential ankyrin 1.
Transient receptor potential ankyrin 1 channels were known to be activated by strong depolarization, displaying largely outward rectifying current.6,47,54 To investigate the effect of alternative splicing on voltage-dependent gating, WT, 27A, and 27B transfected cells were stimulated with a series of voltage steps from −80 to 200 mV from holding potential of −70 mV. Although WT channels displayed strong outward rectifying current with current density of 158.0 ± 13.45 pA/pF at 200 mV, current density for 27A (17.95 ± 3.54 pA/pF) and 27B (19.55 ± 5.26 pA/pF) splice variants at 200 mV were not significantly higher than the background current observed with vector transfected cells (19.37 ± 1.76 pA/pF) (Supplementary Fig. 3, available at http://links.lww.com/PAIN/B279). In addition, cotransfection of 27A and 27B variant with WT respectively did not significantly alter the voltage-dependent current density of the WT (Supplementary Fig. 4, available at http://links.lww.com/PAIN/B279).
3.3. Serine/arginine-splicing factor 1 inhibited alternative splicing within exon 27
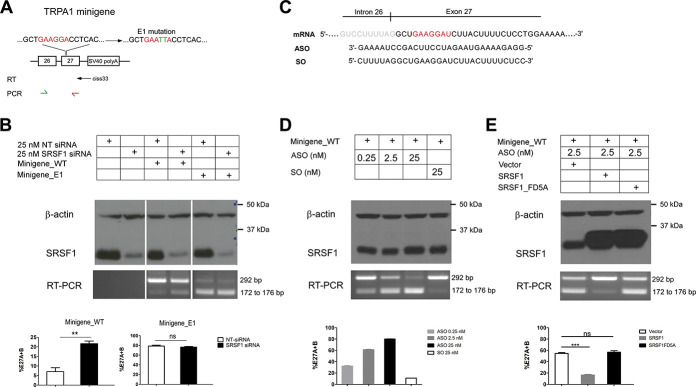
To delve further into the molecular mechanism that regulates alternative splicing of human TRPA1 channels, the genomic sequence that spans exon 26 to exon 27 was analysed in silico using the webserver RBPDB (http://rbpdb.ccbr.utoronto.ca/), and we detected a potential RNA binding motif GAAGGA in the proximal region of exon 27 that could predictably interact with splicing factor SRSF1. It has been shown that SRSF1 recognized with the purine rich ESE element to promote exon inclusion.14 To validate the regulatory role of SRSF1 in hTRPA1 alternative splicing, a minigene spanning exon 26 to exon 27 was cloned (Minigene_WT), and subsequently, the GAAGGA sequence was mutated to GAATTA (Minigene_E1) to potentially disrupt binding of SRSF1 (Fig. 4A).
Figure 4.
Identification of cis and trans element that regulates exon27 splicing. (A) Top, schematic showing the Minigene_WT structure spanning genomic sequence from exon 26 to 27 and the downstream vector coded SV40 polyA signal. In the mutant Minigene_E1 construct, the putative SRSF1 binding site “GAAGGA” was mutated to “GAATTA.” Reverse transcription was performed with ciss33 primer that is specific to SV40-polyA signal. PCR was performed from exon 26 to 27 with the primer pair as denoted by the green and red arrows. (B) Top, NT siRNA or SRSF1 siRNA was cotransfected with minigenes constructs. In negative control experiment, the minigenes were omitted in the transfection. Middle, Western blot reporting the level of β-actin and SRSF1 and RT-PCR gel reported transcript scanning of different conditions. Bottom, bar chart reporting the densitometry analysis. The percentage of alternative splicing (%E27A + B) was quantified by intensity of the lower band over the combined intensity of the top band and lower band. ** P < 0.01, ns, nonsignificant (Student unpaired t test) as compared with NT siRNA transfected cells. Values shown are mean ± SEM (n = 5). (C) Schematic denotes the intron 26-exon 27 boundary and the sequence that is targeted by ASO or SO. (D) Top, the Minigene_WT was cotransfected with SO or ASO with increasing dosage. (n = 3). Format as in B. (E) Top, the Minigene_WT was cotransfected with ASO, vector control, WT, or FD5A SRSF1. Format as in B. ** P < 0.01, ns, nonsignificant (Student unpaired t test) as compared with vector transfected cells. Values shown are mean ± SEM (n = 5). ASO, antisense oligonucleotides; SRSF1, serine/arginine-rich splicing factor 1; SO, sense sequence.
First, the Minigene_WT was transfected with either nontargeting siRNA (NT sirNA) or SRSF1 siRNA into HEK293FT cells. Knockdown of SRSF1 promoted alternative splicing as evidenced by the significant increase in the intensity of the lower band as assessed by RT-PCR experiments (Fig. 4B). Subsequent molecular cloning and DNA sequencing confirmed the identities of WT, 27A, and 27B in the PCR amplicons (data not shown). Notably, the RT-PCR system failed to detect any spurious bands without the transfection of minigenes (Fig. 4B). As hTRPA1_27A and hTRPA1_27B variants differ by only 4 nucleotides, the intensity of the lower bands was scored as a total expression level of both 27A and 27B, and the percentage of alternative splicing (%E27A + B) was therefore quantified as the intensity of the lower band over the combined intensity of the top band and lower bands. Interestingly, mutation of SRSF1 binding site in the mutant Minigene_E1 was found to dramatically promote alternative splicing (78.52 ± 2.02%) which could not be further increased by the knockdown of SRSF1 (76.86 ± 1.2%) (Fig. 4B).
To further confirm that SRSF1 is required for inclusion of exon 27 and for attenuating alternative splicing of 27A and 27B, we designed ASO which was a reverse complement to the pre-mRNA sequence to mask the binding site of SRSF1, and the sense sequence (SO) was used as a negative control (Fig. 4C). Cotransfection of ASO with Minigene_WT gave rise to robust dose-response increase in the generation of either alternative exon 27A or exon 27B as compared with SO (Fig. 4D). Transfection of either ASO or SO at all doses did not affect the expression of SRSF1 as assessed by Western blot analyses (Fig. 4D). As a further test to show that SRSF1 interacted directly with the putative sequence, wildtype SRSF1 or a mutant SRSF1 (SRSF1_FD5A) that was incapable of binding single-stranded RNA20 were cotransfected with Minigene_WT and ASO. Notably, the effect of ASO could be reversed on overexpression of wildtype SRSF1 but not the mutant SRSF1_FD5A, suggesting that the exon enhancer effect of SRSF1 required direct RNA binding.
3.4. Splice-switching antisense oligonucleotides disrupted function of endogenous hTRPA1 channels in SNF96.2 cells and hiPSCs-derived sensory neurons
As a proof-of-principle, we first tested the application of splice-switching ASO in modulating the function of TRPA1 channels in a human Schwann cell cell-line SNF96.2 that expressed endogenous TRPA1 channels.38 Although RT-PCR experiment revealed little alternative splicing on transfection with SO (%E27A + 27B, 0.68 ± 0.50%), transfection of splice-switching ASO dramatically upregulated alternative splicing events in TRPA1 mRNA (%E27A + 27B, 60.7 ± 1.6%) (Fig. 5A). Further resolution of RT-PCR amplicon revealed 2 distinct bands corresponds to 27A and 27B, respectively, and alternative splicing was not observed in other loci as checked by RT-PCR across other exonic region (Supplementary Fig. 5, available at http://links.lww.com/PAIN/B279).
Figure 5.
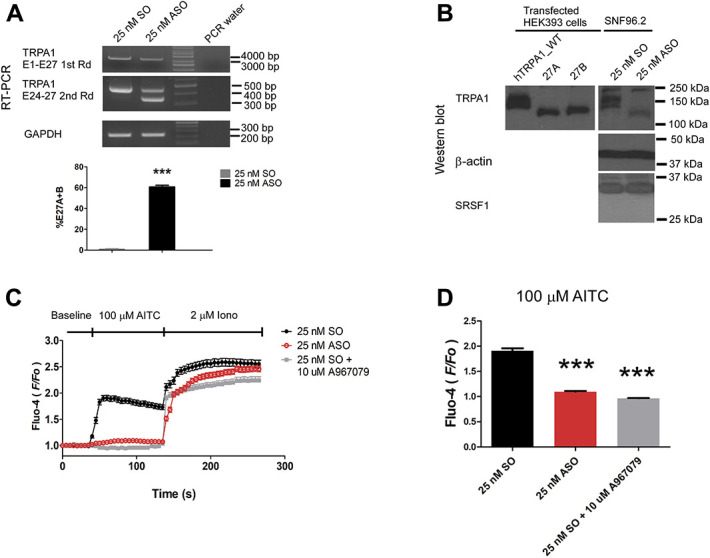
Splicing-switch ASO upregulated alternative splicing and attenuated endogenous hTRPA1 activity in SNF96.2 cells. (A) Top panel, RT-PCR result of full-length PCR (top) and subsequent transcript scanning across exon 24 to 27 of hTRPA1 transcript, respectively (middle). RT-PCR across glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control (bottom). Bottom panel, bar chart reporting the alternative splicing level of hTRPA1 in SNF96.2 cells transfected with 25 nM ASO as compared with SO. *** P < 0.01 (Student unpaired t test). Values shown are mean ± SEM (n = 3). (B) Western blot analysis of the expression of hTRPA1, β-actin, and SRSF1 in similarly transfected cells as in A. HEK293FT cells transfected with hTRPA1 WT, 27A, and 27B expression vector was included to demonstrate the change of alternative splicing pattern. (C) Diary plot reporting the calcium response quantified as the F/F0 over time for both SO (n = 259) or ASO (n = 284) transfected SNF96.2 cells or SO transfected cells cotreated with 10 μM TRPA1 specific blocker A967079 (n = 163). (D) Bar chart reporting the maximal increase in F/F0 on AITC treatment in both conditions. *** P < 0.01 (Student unpaired t test) as compared with SO-transfected cell. Values shown are mean ± SEM. ASO, antisense oligonucleotides; SRSF1, serine/arginine-rich splicing factor 1; SO, sense sequence; TRPA1, transient receptor potential ankyrin 1.
Remarkably, Western blot analyses of the similarly transfected cells detected the complete alteration of TRPA1 protein splicing pattern with ASO transfection. Two bands were observed in the SO transfected cells in the range of 150 kDa to 100 kDa. The higher band that was closer to 150 kDa that could be a result of post-translational modification of TRPA1 channels. hTRPA1 protein channels was known to undergo N-glycosylation.15 In stark contrast, on transfection with ASO, the 2 bands were replaced with a lower band closer to 100 kDa size in the protein ladder. In addition, a nonspecific band that was observed at close to 250 kDa appeared in both SO and ASO transfected cells. Juxtaposing these protein patterns against WT hTRPA1, 27A and 27B splice variants expressed from the transfected HEK293FT cells suggested that sole lower band detected in ASO transfected SNF96.2 cells could be predominantly 27A protein variants. Furthermore, in both SO and ASO transfected cells, the SRSF1 expression level was not changed significantly (Fig. 5B). Notably, knockdown of SRSF1 significantly increased the alternative splicing level (%E27A + 27B, 10.1 ± 0.9%) over the NT siRNA transfected SNF96.2 cells (%E27A + 27B, 1.0 ± 0.3%) on the RNA level. Analysis of protein lysate by Western blot revealed a weak but noticeable appearance of a band corresponded to the 27A variants similarly detected in Figure 5B and corresponding decrease of WT band intensity (Supplementary Fig. 5, available at http://links.lww.com/PAIN/B279).
To facilitate the visualization of transfected cells, ASO and SO were tagged with fluorophore TYE563 at the 5′ end of the oligonucleotides. Fluorescence microscopy imaging reported over 90% of localization of TYE563 signals in the DAPI positive nuclei, indicating efficient uptake of the oligonucleotides (Supplementary Fig. 6, available at http://links.lww.com/PAIN/B279). Intriguingly, subsequent calcium imaging assay with Fluo-4 showed that ASO strongly diminished the increase in [Ca2+]i (maximal F/F0 = 1.09 ± 0.02) as compared with SO (maximal F/F0 = 1.91 ± 0.05) in response to AITC treatment as quantified by the F/F0 (Figs. 5C and D). Cotreatment of SO-transfected cells with TRPA1-specific blocker A967079 completely blocked the AITC evoked increase in [Ca2+]i (maximal F/F0 = 0.96 ± 0.01) demonstrating the activation of TRPA1 is responsible for the AITC sensitivity in SNF96.2cells (Figs. 5C and D).
To further demonstrate the specificity of the ASO, NT siRNA or siRNA against hTRPA1 channels were cotransfected with either SO or ASO in Figure 6. Knockdown of hTRPA1 diminished expression of hTRPA1 channels not only at the mRNA level as detected by RT-PCR experiment but also totally abolished the WT protein bands. Assuredly, splice variant protein band in ASO-transfected cells was also completely abolished. However, the nonspecific bands close to 250 kDa similarly observed in Figure 5 were largely spared with knockdown of hTRPA1 (Figs. 6A and B). Expectedly, on hTRPA1 knockdown, SNF96.2 cells showed similarly diminished AITC sensitive Ca2+ signals (Figs. 6C and D).
Figure 6.
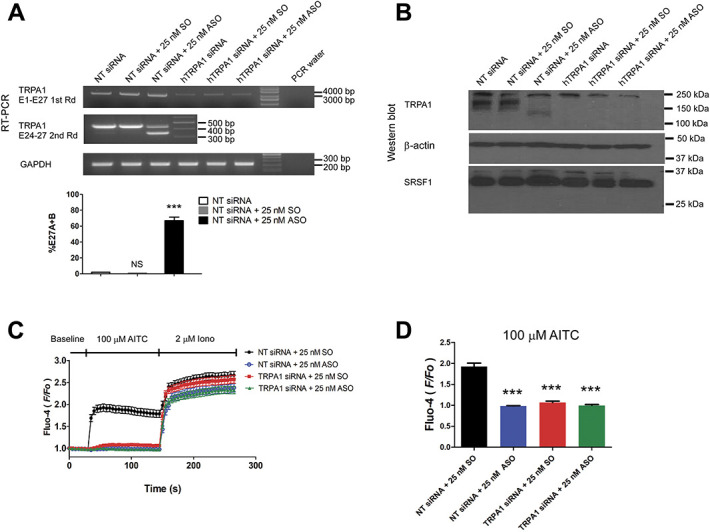
Knockdown of hTRPA1 in SNF96.2 cells verified the splice-switching effect of ASO. (A) Top panel, RT-PCR result of full-length PCR (top) of SNF96.2 cells transfected with 25 nM NT siRNA alone or transfected with 25 nM NT siRNA together with 25 nM SO or ASO, 25 nM TRPA1 siRNA alone, or transfected with 25 nM TRPA1 siRNA together with 25 nM SO or ASO. Subsequent transcript scanning across exon 24 to 27 of hTRPA1 transcript (middle). RT-PCR of GAPDH was used as the loading control (bottom). Bottom panel, bar chart reporting the alternative splicing level of hTRPA1 in SNF96.2 cells cotransfected with NT siRNA and 25 nM SO or ASO as compared with NT siRNA transfected cells. ***P < 0.01 (Student unpaired t test). Values shown are mean ± SEM (n = 3). (B). Western blot reporting the level of hTRPA1, β-actin, and SRSF1 in similarly transfected cells as in A. (C) Diary plot reporting the calcium response quantified as the F/F0 over time for NT siRNA and SO cotransfected (n = 94), NT siRNA and ASO cotransfected (n = 90), TRPA1 siRNA and SO cotransfected (n = 95), and hTRPA1 siRNA and ASO transfected (n = 86) SNF96.2 cells. (D) Bar chart reporting the maximal increase in F/F0 on AITC treatment in respective conditions. *** P < 0.01 (Student unpaired t test) as compared with SO transfected cell. Values shown are mean ± SEM. ASO, antisense oligonucleotides; SRSF1, serine/arginine-rich splicing factor 1; SO, sense sequence; TRPA1, transient receptor potential ankyrin 1.
As a relevant pain model, human IPSC–derived sensory neuronal culture was used to further investigate the effect of TRPA1-targeting splice-switch ASO. Assuredly, immunostaining of derived neurons at day 28 exhibiting confirmed the sensory neuron canonical markers, that is, peripherin (PRPH), Brn3a, and Islet1 (ISL1) whereas qPRC analysis detected increasing expression of TRPA1 beyond 28 days of differentiation (Supplementary Fig. 7, available at http://links.lww.com/PAIN/B279). Robust expression of TRPA1 protein was observed 42 days after differentiation. To overcome the low transfection efficiency usually experienced with neuronal culture and increase specificity of neuronal targeting, the fluorophore TYE563-tagged SO and ASO were complexed with commercially available RVG peptide 9 D-arginine residues (RVG-9R) peptide.23 Notably, robust red fluorescent signals were observed within most of the neuronal cell bodies 72 hours post-transfection (Figs. 7A and B). Strikingly, subsequent calcium imaging experiment revealed robust flickering calcium signals in control SO-transfected cells, indicative action potential firing (Figs. 7C and D and Supplementary Movie 1, available at http://links.lww.com/PAIN/B280) whereas a complete loss of calcium signal in response to AITC treatment was observed with ASO transfected neurons (Figs. 7C and D and Supplementary Movie 2, available at http://links.lww.com/PAIN/B281).
Figure 7.
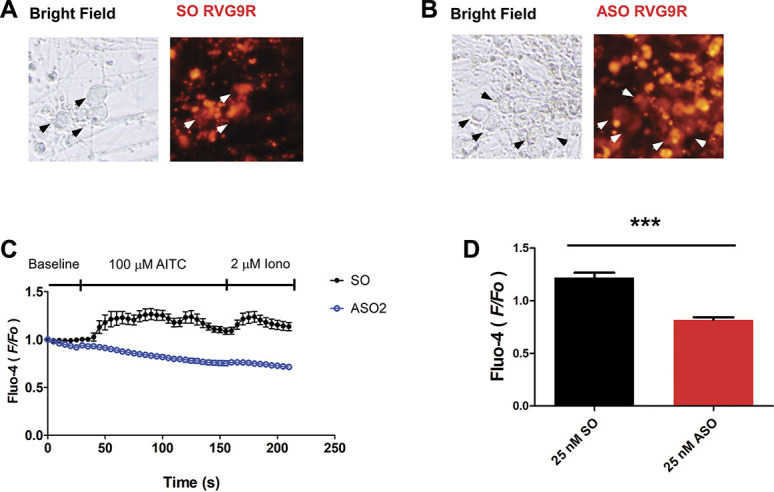
hiPSCs-derived sensory neurons transfected with ASO displayed diminished AITC-mediated activation. (A) Exemplar images of the live neurons transfected with TYE563-tagged sense oligonucleotides as control. (B) Exemplar images of the live neurons transfected with TYE563-tagged ASO. (C) Diary plot reporting the calcium response quantified as the F/F0 over time for SO transfected (n = 19) and ASO transfected (n = 22). (D) Bar chart reporting the maximal increase in F/F0 on AITC treatment in respective conditions. *** P < 0.01 (Student unpaired t test) as compared with SO-transfected cell. Values shown are mean ± SEM. ASO, antisense oligonucleotides; SO, sense sequence.
4. Discussion
Alternative splicing of pre-mRNA diversifies coding mRNA and final protein products by combinatorial assembly of exons. It is a choreographed process that is catalyzed by a multistep assembly of the spliceosome machinery. Notably, binding of splicing factors such as the SR (Ser-Arg) splicing factor to the ESE cis-element within the exon plays a crucial role in stimulating spliceosome assembly and splice site determination.13 Here, we have demonstrated that SRSF1, a member of the SR protein family, promoted inclusion of exon 27 by binding to the purine rich ESE sequence “GAAGGA” in the proximal regions of the exon. Such a conclusion was supported by the several lines of evidence. First, siRNA knockdown of SRSF1 promoted alternative splicing within exon 27. Second, mutation of the ESE sequence or blocking of SRSF1 access to the ESE sequence by ASO dramatically increased alternative splicing of exon 27 in the generation of alternative 27A and 27B variants. Finally, overexpression of wildtype SRSF1 overcame the effect of ASO. Using human Schwann cell SNF96.2 that robustly expressed the endogenous hTRPA1 mRNA and protein, we managed to recapitulated the data obtained with the minigene system in HEK293FT cells; knock down of SRSF1 caused modest but appreciable upregulation of alternative splicing observed in both mRNA and protein level which was dramatically enhanced with splice-switching ASO. Of note, alternative splicing in other exonic regions of hTRPA1 channels was not observed by transcript scanning, suggesting alternation of protein variants observed was due solely to the alternative splicing of exon 27. Strikingly, further experiment performed with native hiPSCs-derived sensory neuronal cultural confirmed the inhibitory effect of splice-switching ASO towards AITC-mediated neuronal excitation.
One reason that siRNA knockdown of SRSF1 seemed only to have modest effect in modulating alternative splicing as compared with mutation of ESE or ASO treatment could be that SRSF1 binds to ESE very effectively in mediating splicing, and the ESE sequence was therefore easily saturated with SRSF1. Alternatively, it also remains to be determined if other SR proteins could compensate for the loss of SRSF1 as different SR protein members were known to display overlapping binding specificity and therefore functional redundancy.32 Such hypotheses warrant future investigation by singly or multiplex knockout of SR proteins.
Moreover, the current study revealed that splicing pattern of TRPA1 mRNA in DRG neurons display highly contrasting species selective patterns. Although deletion of exon 20 was detected in mouse, 27A and 27B variants originated from human samples. It is puzzling given that the SRSF1 binding ESE sequence and alternative 3′ splice donor sites seem to be conserved in both human and mouse exon 27. However, it is important to note that the genomic sequence of hTRPA1 spanning exon 26 to exon 27 displayed only 69% conservation from mouse to human (Supplementary Fig. 8, available at http://links.lww.com/PAIN/B279). It could be possible that disparity in the specific sequence of other intronic or exonic splicing elements might affect splice site selection. This could be addressed in the future by extensive mutagenesis within human minigene sequence to corresponding mouse sequence. In this regard, the current study also raises a challenge to use rodent to model the regulation of alternative splicing of exon 27 in human TRPA1 channels.
Furthermore, our work highlighted the important role of coil-coiled domain in supporting intact hTRPA1 channel functions as disruption of such domain in both 27A and 27B variants reduced protein expression, channel assembly, and caused complete loss of channel function. Although the 2 splice variants 27A and 27B seems not to be functional and did not change the wildtype current on cotransfection, their roles in native sensory neurons remained to be determined. For instance, capsaicin receptor TRPV1 channels were found to associate with and suppress TRPA1channels activity in DRG neurons, whereas such inhibition was relieved in the presence of another membrane protein Tmem100 that physically interacted with both channel proteins.51 Whether the splice variants 27A and 27B play any role in such modulation and upstream regulation mechanism remains to be elucidated. It could be possible that the sensory neurons could buffer the expression level of endogenous TRPA1 by such alternative splicing mechanism under certain pathophysiological conditions.
TRPA1 channels are expressed predominantly in the nociceptive sensory neurons. Activation of TRPA1 channels specifically by both environmental irritants and endogenous inflammatory mediators excites the nerve endings, leading to both acute sensation of pain, itch and more chronic neurogenic inflammation. As such, TRPA1 channels are actively pursued as therapeutic targets for familial episodic pain syndrome patients bearing gain of function mutation in TRPA1, and patients suffering from diabetes, osteoarthritis, and chronic obstructive pulmonary disease among many chronic pathophysiological conditions.39 Beyond the nervous system, NFR-2 driven ectopic expression of TRPA1 in human lung and breast cancer cells was recently found to promote cell survival signalling pathway and confer resistance to oxidative stress and chemotherapy, highlighting other potential application of targeting TRPA1 channels. Although many small molecular blockers and pore-blocking antibodies have been proposed as therapeutic candidates for effective treatment of TRPA1-related diseases, only one such compound GRC-17536 is currently in clinical trial for the treatment of peripheral diabetic neuropathy.11,39
As a proof of principle, we have shown that modulation of TRPA1 channel splicing pattern could be a simple and yet viable approach to inhibit TRPA1 function in both sensory neuron and cancer cell line SNF96.2, supplementing the existing arsenal of channel blockers. Of significance, splice-switching ASOs such as eteplirsen and nusinersen have been used to treat Duchenne muscular dystrophy and spinal muscular atrophy, respectively.46 However, poor permeability of cell membrane to negatively charged nucleic acids could pose a major limitation to efficient delivery of ASO. Although packaging of siRNA with cationic charged polymer polyethylenimine has been shown to effectively silence NMDA receptor subunit NR1 in DRG neurons and alleviate inflammatory pain.40 Such method suffered from low specificity as polyethylenimine could expectedly distribute the payload nucleic acid nonspecifically to different cell types. Interestingly, cell-penetrating peptides derived from viral capsid proteins have emerged as promising alternatives for the efficient and specific delivery of nucleic acid into sensory neurons.9,23 Examples such as TAT peptide, which was derived from the human immunodeficiency virus or RVG-R9 peptide, which could be created by fusing RVG peptide 9 D-arginine residues can facilitate nucleotide loading and have been proposed as new strategies in this regard.7 Indeed, prior complexing of TYE563-tagged oligonucleotides with RVG-9R lead to high transfection efficiency as evidenced by the robust localization of red fluorophore within neuronal ell bodies observed in the study. In addition, our preliminary experiment using minigene cloned from Macaque monkey demonstrated that ASO could effectively promote 27A and 27B splicing (data not shown). It would be exciting to adopt and test such approaches in packaging and delivering splice-switching ASO against TRPA1 into nonhuman primates in the future.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B279, http://links.lww.com/PAIN/B280 and http://links.lww.com/PAIN/B281.
Acknowledgments
This research is supported by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education's AcRF Tier 3 grants, Grant number MOE2014-T3-1-006.
Author contribution: H. Huang conceptualized the project, designed experiment, performed the experiment, and wrote the article. S.H. Tay, W. Ng, and S.Y. Ng developed and characterized the hiPSCs-derived sensory neuronal culture, and T.W. Soong provided intellectual input, supported the project, and edited the article.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painjournalonline.com).
Contributor Information
Shermaine Huiping Tay, Email: Shermaine_Tay@imcb.a-star.edu.sg.
Winanto Ng, Email: winanto@imcb.a-star.edu.sg.
Shi Yan Ng, Email: shiyan_ng@nus.edu.sg.
Tuck Wah Soong, Email: phsstw@nus.edu.sg.
References
- [1].Andersson DA, Gentry C, Light E, Vastani N, Vallortigara J, Bierhaus A, Fleming T, Bevan S. Methylglyoxal evokes pain by stimulating TRPA1. PLoS One 2013;8:e77986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Andersson DA, Gentry C, Moss S, Bevan S. Transient receptor potential A1 is a sensory receptor for multiple products of oxidative stress. J Neurosci 2008;28:2485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bandell M, Story GM, Hwang SW, Viswanath V, Eid SR, Petrus MJ, Earley TJ, Patapoutian A. Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron 2004;41:849–57. [DOI] [PubMed] [Google Scholar]
- [4].Bautista DM, Jordt SE, Nikai T, Tsuruda PR, Read AJ, Poblete J, Yamoah EN, Basbaum AI, Julius D. TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell 2006;124:1269–82. [DOI] [PubMed] [Google Scholar]
- [5].Bautista DM, Movahed P, Hinman A, Axelsson HE, Sterner O, Hogestatt ED, Julius D, Jordt SE, Zygmunt PM. Pungent products from garlic activate the sensory ion channel TRPA1. Proc Natl Acad Sci U S A 2005;102:12248–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Benedikt J, Samad A, Ettrich R, Teisinger J, Vlachova V. Essential role for the putative S6 inner pore region in the activation gating of the human TRPA1 channel. Biochim Biophys Acta 2009;1793:1279–88. [DOI] [PubMed] [Google Scholar]
- [7].Berta T, Qadri Y, Tan PH, Ji RR. Targeting dorsal root ganglia and primary sensory neurons for the treatment of chronic pain. Expert Opin Ther Targets 2017;21:695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bessac BF, Sivula M, von Hehn CA, Caceres AI, Escalera J, Jordt SE. Transient receptor potential ankyrin 1 antagonists block the noxious effects of toxic industrial isocyanates and tear gases. Faseb J 2009;23:1102–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Borsello T, Clarke PG, Hirt L, Vercelli A, Repici M, Schorderet DF, Bogousslavsky J, Bonny C. A peptide inhibitor of c-Jun N-terminal kinase protects against excitotoxicity and cerebral ischemia. Nat Med 2003;9:1180–6. [DOI] [PubMed] [Google Scholar]
- [10].Chambers SM, Qi Y, Mica Y, Lee G, Zhang XJ, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi SH, Studer L. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol 2012;30:715–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen J, Hackos DH. TRPA1 as a drug target-promise and challenges. Naunyn Schmiedebergs Arch Pharmacol 2015;388:451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen J, Joshi SK, DiDomenico S, Perner RJ, Mikusa JP, Gauvin DM, Segreti JA, Han P, Zhang XF, Niforatos W, Bianchi BR, Baker SJ, Zhong C, Simler GH, McDonald HA, Schmidt RG, McGaraughty SP, Chu KL, Faltynek CR, Kort ME, Reilly RM, Kym PR. Selective blockade of TRPA1 channel attenuates pathological pain without altering noxious cold sensation or body temperature regulation. PAIN 2011;152:1165–72. [DOI] [PubMed] [Google Scholar]
- [13].Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 2009;10:741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cowper AE, Cáceres JF, Mayeda A, Screaton GR. Serine-arginine (SR) protein-like factors that antagonize authentic SR proteins and regulate alternative splicing. J Biol Chem 2001;276:48908–14. [DOI] [PubMed] [Google Scholar]
- [15].Egan TJ, Acuña MA, Zenobi-Wong M, Zeilhofer HU, Urech D. Effects of N-Glycosylation of the human cation channel TRPA1 on agonist-sensitivity. Biosci Rep 2016;36:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ellis A, Bennett DL. Neuroinflammation and the generation of neuropathic pain. Br J Anaesth 2013;111:26–37. [DOI] [PubMed] [Google Scholar]
- [17].Gu P, Gong J, Shang Y, Wang F, Ruppell KT, Ma Z, Sheehan AE, Freeman MR, Xiang Y. Polymodal nociception in Drosophila requires alternative splicing of TrpA1. Curr Biol 2019;29:3961–73 e3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Heber S, Gold-Binder M, Ciotu CI, Witek M, Ninidze N, Kress HG, Fischer MJM. A human TRPA1-specific pain model. J Neurosci 2019;39:3845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hill K, Schaefer M. Ultraviolet light and photosensitising agents activate TRPA1 via generation of oxidative stress. Cell Calcium 2009;45:155–64. [DOI] [PubMed] [Google Scholar]
- [20].Huang H, Kapeli K, Jin W, Wong YP, Arumugam TV, Koh JH, Srimasorn S, Mallilankaraman K, Chua JJE, Yeo GW, Soong TW. Tissue-selective restriction of RNA editing of CaV1.3 by splicing factor SRSF9. Nucleic Acids Res 2018;46:7323–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jordt SE, Bautista DM, Chuang HH, McKemy DD, Zygmunt PM, Hogestatt ED, Meng ID, Julius D. Mustard oils and cannabinoids excite sensory nerve fibres through the TRP channel ANKTM1. Nature 2004;427:260–5. [DOI] [PubMed] [Google Scholar]
- [22].Kremeyer B, Lopera F, Cox JJ, Momin A, Rugiero F, Marsh S, Woods CG, Jones NG, Paterson KJ, Fricker FR, Villegas A, Acosta N, Pineda-Trujillo NG, Ramirez JD, Zea J, Burley MW, Bedoya G, Bennett DL, Wood JN, Ruiz-Linares A. A gain-of-function mutation in TRPA1 causes familial episodic pain syndrome. Neuron 2010;66:671–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature 2007;448:39–43. [DOI] [PubMed] [Google Scholar]
- [24].Lennertz RC, Kossyreva EA, Smith AK, Stucky CL. TRPA1 mediates mechanical sensitization in nociceptors during inflammation. PLoS One 2012;7:e43597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Materazzi S, Fusi C, Benemei S, Pedretti P, Patacchini R, Nilius B, Prenen J, Creminon C, Geppetti P, Nassini R. TRPA1 and TRPV4 mediate paclitaxel-induced peripheral neuropathy in mice via a glutathione-sensitive mechanism. Pflugers Arch 2012;463:561–9. [DOI] [PubMed] [Google Scholar]
- [26].McGaraughty S, Chu KL, Perner RJ, Didomenico S, Kort ME, Kym PR. TRPA1 modulation of spontaneous and mechanically evoked firing of spinal neurons in uninjured, osteoarthritic, and inflamed rats. Mol Pain 2010;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].McNamara CR, Mandel-Brehm J, Bautista DM, Siemens J, Deranian KL, Zhao M, Hayward NJ, Chong JA, Julius D, Moran MM, Fanger CM. TRPA1 mediates formalin-induced pain. Proc Natl Acad Sci U S A 2007;104:13525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Meseguer V, Alpizar YA, Luis E, Tajada S, Denlinger B, Fajardo O, Manenschijn JA, Fernandez-Pena C, Talavera A, Kichko T, Navia B, Sanchez A, Senaris R, Reeh P, Perez-Garcia MT, Lopez-Lopez JR, Voets T, Belmonte C, Talavera K, Viana F. TRPA1 channels mediate acute neurogenic inflammation and pain produced by bacterial endotoxins. Nat Commun 2014;5:3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Miyake T, Nakamura S, Zhao M, So K, Inoue K, Numata T, Takahashi N, Shirakawa H, Mori Y, Nakagawa T, Kaneko S. Cold sensitivity of TRPA1 is unveiled by the prolyl hydroxylation blockade-induced sensitization to ROS. Nat Commun 2016;7:12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nakamura Y, Une Y, Miyano K, Abe H, Hisaoka K, Morioka N, Nakata Y. Activation of transient receptor potential ankyrin 1 evokes nociception through substance P release from primary sensory neurons. J Neurochem 2012;120:1036–47. [DOI] [PubMed] [Google Scholar]
- [31].Obata K, Katsura H, Mizushima T, Yamanaka H, Kobayashi K, Dai Y, Fukuoka T, Tokunaga A, Tominaga M, Noguchi K. TRPA1 induced in sensory neurons contributes to cold hyperalgesia after inflammation and nerve injury. J Clin Invest 2005;115:2393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M, Jr., Fu XD. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell 2013;50:223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 2015;520:511–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Petrus M, Peier AM, Bandell M, Hwang SW, Huynh T, Olney N, Jegla T, Patapoutian A. A role of TRPA1 in mechanical hyperalgesia is revealed by pharmacological inhibition. Mol Pain 2007;3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pozsgai G, Hajna Z, Bagoly T, Boros M, Kemeny A, Materazzi S, Nassini R, Helyes Z, Szolcsanyi J, Pinter E. The role of transient receptor potential ankyrin 1 (TRPA1) receptor activation in hydrogen-sulphide-induced CGRP-release and vasodilation. Eur J Pharmacol 2012;689:56–64. [DOI] [PubMed] [Google Scholar]
- [36].Shepherd AJ, Copits BA, Mickle AD, Karlsson P, Kadunganattil S, Haroutounian S, Tadinada SM, de Kloet AD, Valtcheva MV, McIlvried LA, Sheahan TD, Jain S, Ray PR, Usachev YM, Dussor G, Krause EG, Price TJ, Gereau RWt, Mohapatra DP. Angiotensin II triggers peripheral macrophage-to-sensory neuron redox crosstalk to elicit pain. J Neurosci 2018;38:7032–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stucky CL, Dubin AE, Jeske NA, Malin SA, McKemy DD, Story GM. Roles of transient receptor potential channels in pain. Brain Res Rev 2009;60:2–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Takahashi N, Chen HY, Harris IS, Stover DG, Selfors LM, Bronson RT, Deraedt T, Cichowski K, Welm AL, Mori Y, Mills GB, Brugge JS. Cancer cells Co-opt the neuronal redox-sensing channel TRPA1 to promote oxidative-stress tolerance. Cancer Cell 2018;33:985–1003 e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Talavera K, Startek JB, Alvarez-Collazo J, Boonen B, Alpizar YA, Sanchez A, Naert R, Nilius B. Mammalian transient receptor potential TRPA1 channels: from structure to disease. Physiol Rev 2020;100:725–803. [DOI] [PubMed] [Google Scholar]
- [40].Tan PH, Yu SW, Lin VC, Liu CC, Chien CF. RNA interference-mediated gene silence of the NR1 subunit of the NMDA receptor by subcutaneous injection of vector-encoding short hairpin RNA reduces formalin-induced nociception in the rat. PAIN 2011;152:573–81. [DOI] [PubMed] [Google Scholar]
- [41].Tang ZZ, Liang MC, Lu S, Yu D, Yu CY, Yue DT, Soong TW. Transcript scanning reveals novel and extensive splice variations in human l-type voltage-gated calcium channel, Cav1.2 alpha1 subunit. J Biol Chem 2004;279:44335–43. [DOI] [PubMed] [Google Scholar]
- [42].Taylor-Clark TE, Ghatta S, Bettner W, Undem BJ. Nitrooleic acid, an endogenous product of nitrative stress, activates nociceptive sensory nerves via the direct activation of TRPA1. Mol Pharmacol 2009;75:820–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Taylor-Clark TE, Undem BJ, Macglashan DW, Jr, Ghatta S, Carr MJ, McAlexander MA. Prostaglandin-induced activation of nociceptive neurons via direct interaction with transient receptor potential A1 (TRPA1). Mol Pharmacol 2008;73:274–81. [DOI] [PubMed] [Google Scholar]
- [44].Trevisan G, Hoffmeister C, Rossato MF, Oliveira SM, Silva MA, Silva CR, Fusi C, Tonello R, Minocci D, Guerra GP, Materazzi S, Nassini R, Geppetti P, Ferreira J. TRPA1 receptor stimulation by hydrogen peroxide is critical to trigger hyperalgesia and inflammation in a model of acute gout. Free Radic Biol Med 2014;72:200–9. [DOI] [PubMed] [Google Scholar]
- [45].Trevisan G, Materazzi S, Fusi C, Altomare A, Aldini G, Lodovici M, Patacchini R, Geppetti P, Nassini R. Novel therapeutic strategy to prevent chemotherapy-induced persistent sensory neuropathy by TRPA1 blockade. Cancer Res 2013;73:3120–31. [DOI] [PubMed] [Google Scholar]
- [46].Verma A. Recent advances in antisense oligonucleotide therapy in genetic neuromuscular diseases. Ann Indian Acad Neurol 2018;21:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wan X, Lu Y, Chen X, Xiong J, Zhou Y, Li P, Xia B, Li M, Zhu MX, Gao Z. Bimodal voltage dependence of TRPA1: mutations of a key pore helix residue reveal strong intrinsic voltage-dependent inactivation. Pflugers Arch 2013;466:1273–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang S, Dai Y, Fukuoka T, Yamanaka H, Kobayashi K, Obata K, Cui X, Tominaga M, Noguchi K. Phospholipase C and protein kinase A mediate bradykinin sensitization of TRPA1: a molecular mechanism of inflammatory pain. Brain 2008;131:1241–51. [DOI] [PubMed] [Google Scholar]
- [49].Wei H, Hämäläinen MM, Saarnilehto M, Koivisto A, Pertovaara A. Attenuation of mechanical hypersensitivity by an antagonist of the TRPA1 ion channel in diabetic animals. Anesthesiology 2009;111:147–54. [DOI] [PubMed] [Google Scholar]
- [50].Wei H, Karimaa M, Korjamo T, Koivisto A, Pertovaara A. Transient receptor potential ankyrin 1 ion channel contributes to guarding pain and mechanical hypersensitivity in a rat model of postoperative pain. Anesthesiology 2012;117:137–48. [DOI] [PubMed] [Google Scholar]
- [51].Weng HJ, Patel KN, Jeske NA, Bierbower SM, Zou W, Tiwari V, Zheng Q, Tang Z, Mo GC, Wang Y, Geng Y, Zhang J, Guan Y, Akopian AN, Dong X. Tmem100 is a regulator of TRPA1-TRPV1 complex and contributes to persistent pain. Neuron 2015;85:833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wilson SR, Gerhold KA, Bifolck-Fisher A, Liu Q, Patel KN, Dong X, Bautista DM. TRPA1 is required for histamine-independent, Mas-related G protein-coupled receptor-mediated itch. Nat Neurosci 2011;14:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhou Y, Suzuki Y, Uchida K, Tominaga M. Identification of a splice variant of mouse TRPA1 that regulates TRPA1 activity. Nat Commun 2013;4:2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zimova L, Sinica V, Kadkova A, Vyklicka L, Zima V, Barvik I, Vlachova V. Intracellular cavity of sensor domain controls allosteric gating of TRPA1 channel. Sci Signal 2018;11:eaan8621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B279, http://links.lww.com/PAIN/B280 and http://links.lww.com/PAIN/B281.