Supplemental Digital Content is Available in the Text.
An integrated workflow combining behavioral testing, mass-spectrometry–based proteomics, and network activity predictions revealed a hitherto unknown dorsal root ganglia protein signature relevant for incisional pain in mice.
Keywords: Incision; Postoperative pain, Proteomics; DRG; Non-evoked pain; Protein networks, Mass spectrometry
Abstract
After surgery, acute pain is still managed insufficiently and may lead to short-term and long-term complications including chronic postsurgical pain and an increased prescription of opioids. Thus, identifying new targets specifically implicated in postoperative pain is of utmost importance to develop effective and nonaddictive analgesics. Here, we used an integrated and multimethod workflow to reveal unprecedented insights into proteome dynamics in dorsal root ganglia (DRG) of mice after plantar incision (INC). Based on a detailed characterization of INC-associated pain-related behavior profiles, including a novel paradigm for nonevoked pain, we performed quantitative mass-spectrometry–based proteomics in DRG 1 day after INC. Our data revealed a hitherto unknown INC-regulated protein signature in DRG with changes in distinct proteins and cellular signaling pathways. In particular, we show the differential regulation of 44 protein candidates, many of which are annotated with pathways related to immune and inflammatory responses such as MAPK/extracellular signal-regulated kinases signaling. Subsequent orthogonal assays comprised multiplex Western blotting, bioinformatic protein network analysis, and immunolabeling in independent mouse cohorts to validate (1) the INC-induced regulation of immune/inflammatory pathways and (2) the high priority candidate Annexin A1. Taken together, our results propose novel potential targets in the context of incision and, therefore, represent a highly valuable resource for further mechanistic and translational studies of postoperative pain.
1. Introduction
Pain management after surgery is still far from being optimal. Early after surgery, severe pain occurs in a very high number of patients, contributes to (sub)acute complications, and impedes the acute healing process.22,26,71 Furthermore, severe acute pain after surgery may facilitate chronic pain and long-term opioid intake after surgery.20,28,68 Thus, novel non–opioid-based treatment options targeting pathway-specific mechanisms relevant for acute postoperative pain are urgently needed.
To investigate the pathophysiology of acute incisional pain, specific rodent “postoperative” pain models have been developed.54 Incision injury, as a surrogate for postoperative pain, is known to induce the activation of diverse nociceptive signaling pathways and inflammatory responses in peripheral tissues and dorsal root ganglia (DRG) innervating the injured region.55,64 In particular, alterations of proteins involved in aforementioned processes may drive activity-dependent plasticity, alter the excitability in peripheral nociceptors and, consequently, in the central nervous system.64 These changes are distinct to those after other types of tissue injuries and contribute to pain-related behaviors after incision.15,54 However, our knowledge of the mechanistic underpinnings, specific cellular protein networks, and possible interactions within major signaling pathways governing these changes in DRG after incision injury is still very limited. Therefore, innovative and unbiased approaches are necessary to expand our knowledge, identify new targets, and ultimately design new therapeutic options.
Current pain research increasingly exploits the potential of genomics and transcriptomics to globally characterize changes at multiple levels of the nociceptive system.5,25,60 Recent hypothesis-driven studies investigated mRNA levels in the skin, DRG, and spinal dorsal horn after incision in rodents and discovered multiple mediators to be regulated.13,48,57,65 However, transcript levels correlate only up to 50% with factual levels of corresponding proteins, which renders the interpretation of RNA-based results and identification of novel targets are challenging.44 Hence, a thorough understanding of incision-induced dynamics warrants the inclusion of comprehensive studies at the proteome level. Proteome-based approaches allow monitoring changes of functionally relevant proteins. In addition, comprehensive protein analyses enable their quantitative and qualitative interrogation in terms of associated protein networks and their predicted activity as a prerequisite to develop combinatorial therapeutic interventions in the future.30,62,77,81
Here, we combined the established mouse plantar incision model53 and the assessment of pain-associated behavior profiles with quantitative and comprehensive mass spectrometry–based proteomics27 to reveal proteome dynamics in lumbar DRG in an unbiased manner on postoperative day 1 (POD 1). Postoperative day 1 represents a time point at which mice show prominent pain-associated behaviors. Based on these proteome analyses, we identified molecular changes in distinct proteins and cellular signaling pathways, which we then orthogonally validated by protein assays combined with activity predictions on the cellular and network level. Taken together, our data provide unprecedented insights into DRG protein network dynamics and modulation of immune/inflammatory pathways on plantar incision. Importantly, our results propose distinct protein networks as potential novel targets in the context of incisional pain and, therefore, represent a highly valuable resource for further mechanistic and potentially therapeutic studies.
2. Material and methods
2.1. Animal care and general information
All experiments (Figs. 1A–E) were approved by the Animal Ethics Committee of the State Agency for Nature, Environment and Consumer Protection North Rhine-Westphalia, Recklinghausen, Germany, and are in accordance with the ethical guidelines for investigation of experimental pain in conscious animals.82 Adult male C57BL/6 J mice (age: 10 weeks; 26 ± 2.1 g, mean ± SEM local breeding; total n = 84) were kept in a 12/12 hours day/night cycle with access to food and water ad libitum under standardized specific pathogen-free conditions. All mice were euthanized by carbon dioxide. Incision injury and evoked mechanical and heat stimuli were performed on the right hind paw of mice.
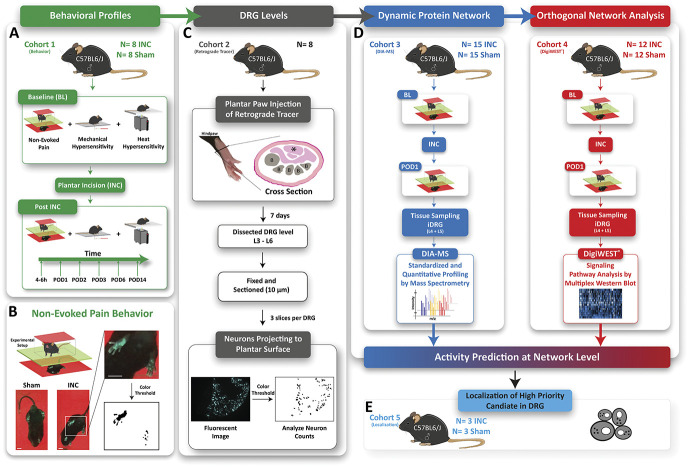
Figure 1.
Study design. (A) Characterization of the time course of nonevoked pain (NEP) and evoked (mechanical hypersenstivity, heat hypersensitivity) pain-related behavior profiles after plantar incision. (B) Assessment of nonevoked pain was performed by comparing weight-bearing (print area) of the paw at the incision site (ipsilateral) with the nonincised paw (contralateral) (cohort 1, green, INC N = 8, Sham N = 8). Scale bar = 1 cm. (C) The level of DRG projecting to the plantar surface of the hind paw was determined in our C57Bl6/J mouse colony (cohort 2, grey, naïve N = 8). B = bone, asterisk = musculus flexor digitorum brevis. (D) Mice (N = 54) were assessed for NEP at baseline (BL, 24 hours prior) and 24 hours after (POD 1) plantar incision (INC) or Sham. After determining NEP, mice were sacrificed, and ipsilateral dorsal root ganglia (iDRG) were isolated. iDRG were used for quantitative proteome profiling (DIA-MS; cohort 3, blue, INC N = 15, Sham N = 15) or orthogonal validation by DigiWEST multiplex Western blots (DigiWEST; cohort 4, red, INC N = 12, and Sham N = 12). Obtained results from DIA-MS and DigiWEST analysis were integrated by bioinformatic activity prediction at the network level. (E) Orthogonal validation of our data by immunohistochemistry in ipsilateral L4 DRG at the cell level (cohort 5, light blue, INC N = 3, Sham N = 3).
2.2. Plantar incision
Mice were initially anesthetized with 5% isoflurane in 100% oxygen and maintained with 1,5% to 2% isoflurane through a nose cone during the entire procedure. Paw incision (INC) was performed based on Brennan et al.8 and adapted to mice, as described previously.16,53 In brief, the entire right hind paw was disinfected by 100% ethanol and Betadine (povidone-iodine). The glabrous epidermis, dermis, fascia, and musculus flexor digitorum brevis were incised longitudinally with a scalpel (No 11, incision: start 2 mm from the proximal edge of the heel, 5 mm length). In addition, the muscle was elevated and shortly retracted with a forceps (Dumount #5). A mattress suture with 6-0 prolene closed the skin. The incision was covered by Betadine using a cotton swab. Sham-treated mice (only anesthesia, same exposure duration, no suture, and no incision) were used as a control for the surgical incision. On postoperative day 3 (POD 3), the sutures were removed from mice of cohort 1 under short-term isoflurane anesthesia under sterile conditions. Mice were randomly assigned to the groups (INC and Sham) using the Microsoft Excel randomization function.
2.3. Pain-related behavior profiles—general
Pain-related assays were conducted randomly by experimenters of both genders in the morning (8-12 am). Blinding regarding incision vs sham is not possible because the animals in the INC group have surgical suture until POD 3 (see section “plantar incision”) increased hind paw thickness and show apparent changes in pain-related behavior. However, data analysis was performed in a blinded manner. The order of behavioral testing in cohort 1 (Fig. 1A, cohort 1, green, INC N = 8, and Sham N = 8) was nonevoked pain assessment (NEP), mechanical hypersensitivity, and heat hypersensitivity with a resting interval of 1 hour between tests. During the behavioral tests, mice were not handled.
2.4. Nonevoked pain assessment (guarding pain at rest)
Nonevoked pain was determined by comparing the weight-bearing (print area) of the paw at the incision site (ipsilateral) with the nonincised paw (contralateral). Typically, plantar incision resulted in an unbalanced distribution of weight-bearing caused by guarding the ipsilateral paw to ambulatory activities.2 Mice were separately placed in a transparent box (7 × 5 × 5 cm), which is covered by an LED-panel (illumined in red) on a green light illuminated glass plate (Fig. 1B). With viewing direction towards the ventral side of the mouse footprints, a camera captures an image every 30 seconds for a total period of 10 minutes; before (baseline, BL) and at 4∼6 hours POD 1, POD 2, POD 3, POD 6, and POD 14 for cohort 1 (Fig. 1A), BL and POD 1 for cohort 3 and 4 (Fig. 1D). The print areas of both hind paws were determined with the software ImageJ by color thresholding from 10 different images for each mouse blinded with respect to the experimental group. The ratio was calculated by the print area of the ipsilateral to the contralateral hind paw for every image. Image analysis (in total 10 images per mouse and time point) was based on predefined exclusion criteria such as visible grooming during capturing, rearing, or an unsharp image caused by the movement of the hind paw. A change in the print area ratio ipsilateral/contralateral represents the degree of guarding pain at rest in the ipsilateral hind paw (Fig. 1B). Statistical analysis showed that INC and Sham groups (at POD 1, cohort 3 and 4, Fig. 1D) did not exhibit overlapping minimum or maximum values, and no outliers were identified by the ROUT method with a Q = 5%. All mice exhibited a clearly defined behavioral phenotype (ie, with prominent nonevoked pain on INC); thus, all mice were included for further proteome analysis.
2.5. Mechanical hypersensitivity
Mechanical hypersensitivity was assessed by using calibrated von Frey filaments (Bioseb Danmic Global, LLC, San Jose, CA) with a logarithmic force range from 0.07 g to 2 g. After a 15-minute habituation in a transparent box (7 × 5 × 5 cm) on mesh (7 mm × 7 mm), the von Frey filaments were applied 5 times (1 Hz) to the plantar aspect of the right hind paw. The filaments were used in an ascending order until a withdrawal response occurred or the cut-off limit of 2 g was reached. The median force of 3 trials (5-10 minutes break between trials) was determined as 60% paw withdrawal threshold.
2.6. Heat hypersensitivity
Heat hypersensitivity was measured as paw withdrawal latency (PWL) to radiant heat using the Hargreaves device (IITC Life Science, Halogen lamp intensity 17%, temperature of the test platform, 30°C). The radiant heat intensity (17%) was adjusted to produce withdrawal latencies around 10 to 12 seconds at the baseline level17). Here, mice were placed on a prewarmed glass plate (30°C). After 15 minutes of habituation, a radiant heat source was applied to the plantar aspect of the hind paw. The PWL was measured with a cut-off time set at 20 seconds. Five trials at intervals of 5 to 10 minutes were taken to estimate the average PWL to heat stimuli.
2.7. Retrograde tracing of projection neurons into the incised area of mice hind paw
The structural organization of the sciatic nerve across species, strains, and substrains has been indicated to be heterogeneous.40,59 Therefore, it is essential to identify the peripheral sensory neurons (PSNs) of DRG that project into the incised area in the individual mouse colony. Retrograde labeling allowed identifying PSNs (and corresponding DRG) that project into the incised area of the hind paw in our local C57BL/6 J colony. The method was adopted by Rigaud et al.59 In brief, retrograde tracing was performed by injection (subcutaneous + intramuscular, 29 G needle) of True Blue dye (20 µl, 1% solution in phosphate-buffered saline (PBS), Sigma-Aldrich, Germany) under isoflurane anesthesia in 8 naïve male mice (Fig. 1C, cohort 2, grey, N = 8). The dye was injected into the skin of the plantar aspect and the musculus flexor digitorum brevis, which represented the region where the incision in cohorts 1, 3, 4, and 5 was made. Ipsilateral and contralateral lumbar DRG were dissected immediately after euthanasia 7 days after injection. Dorsal root ganglia levels were identified using anatomical images and bony landmarks according to the previous report.59 Dorsal root ganglia were separately fixed in 4% paraformaldehyde (PFA) at 4°C overnight. After washing in 0.1 M PBS (3 × 15 minutes), the tissue was cryopreserved in 0.4 M followed by 0.8 M sucrose until it had sagged. The tissues were embedded in Tissue-Tek O.C.T. compound and stored at −80°C. 10 µm thin slices of DRG tissue were prepared on a cryostat and mounted on SUPERFROST PLUS microspore slides (Thermo). For each DRG, 3 randomized slices from 2 independent experimenters (D.S. and K.K.) were examined in a blinded manner regarding the number of total DRG neurons and True Blue–positive neurons by ImageJ.61 The autofluorescent threshold was determined by measuring the brightest pixel in corresponding contralateral DRG.
2.8. Protein isolation for quantitative data-independent acquisition-mass spectrometry
Tissues for proteome analysis were isolated from mice of the incision group (INC, N = 15) and the sham group (Sham, N = 15) (Fig. 1D, cohort 3, blue). Ipsilateral lumbar DRG (iDRG; L4, and L5) were isolated immediately after CO2 euthanasia, snap frozen in liquid nitrogen, and stored at −80°C. Ipsilateral dorsal root ganglia from 5 mice were pooled to obtain 3 replicates per condition (INC vs Sham). Protein isolation was performed as described previously.5,9 In brief, the frozen tissue was homogenized in 4% sodium dodecyl sulfate (SDS) lysis buffer (4% SDS in 100 mM Tris, 10 mM DTT, 5% glycerol, complete protease inhibitor cocktail (Roche, Basel, Switzerland), pH 7.5). After, the homogenate was incubated at 70°C for 10 minutes and centrifuged at 10,000 × g for 5 minutes at room temperature to remove cell debris. Proteins were precipitated by the addition of 5 × volume prechilled acetone (Roth, Germany) and incubated for 2 hours at −20°C. The protein precipitate was centrifuged at 14,000 × g for 30 minutes, the pellet washed with ice cold 80% ethanol (AppliedChem, Germany), and centrifuged again at 14,000 × g for 30 minutes. The proteins were air dried, resuspended in 2% SDS lysis buffer, and analyzed by quantitative data-independent acquisition (DIA) mass spectrometry (MS).
2.9. Data-independent acquisition mass spectrometry and data analysis
Sample preparation, DIA-MS (performed by Biognosys AG, Switzerland), and data analysis were performed as previously reported60 with the modification that data were analyzed with Spectronaut Pulsar (Biognosys AG, Switzerland) with precursor and protein false discovery rate set at 0.01. For data analysis, our previously described pan-mouse spectral library was used.60 To calculate expression changes in the INC group, we used very stringent criteria, that is, peptides were only considered if they were detected across all 3 Sham replicates and all 3 INC replicates (q-complete analysis in Spectronaut).5 Mean log2 ratios were calculated for each protein ID and statistically analyzed using the Benjamini–Hochberg (BH) procedure, as previously described.5 Regulated proteins were defined as having a BH-adjusted P-value < 0.05 (Q < 0.05). Any potential keratin contaminations were removed. For comparison of DIA-MS data with relevant previously published datasets, we used reviewed mouse UniProt identifiers essentially as outlined previously.5
2.10. DigiWEST multiplex western blot
Protein isolation for DigiWEST analysis was performed as described before.69 Lysates of iDRG (L4 and L5) were obtained from a second independent mouse cohort with the same inclusion criteria regarding nonevoked pain behavior (Fig. 1D, cohort 4, red, INC N = 12, Sham N = 12) as for DIA-MS analysis (please see workflow and behavior analysis in Fig. 1A; iDRG from 3 mice were pooled/replicate, 4 biological replicates in total). As analytes are independent of each other, they were individually analyzed for statistical significance using multiple t-tests without assuming a consistent standard deviation (GraphPad Prism 8.0.2, San Diego, CA).
2.11. Ingenuity Pathway Analysis
The list of significantly regulated proteins (BH-adjusted P-value, Q-value < 0.05) was uploaded to Ingenuity Pathway Analysis (IPA) (Qiagen, 2000-2017; upload date: 07/2017), and network activity prediction analysis was performed.
2.12. Gene ontology analysis
The list of significantly regulated proteins (BH-adjusted P-value, Q-value < 0.05) was uploaded to the web interface STRING (string-db.org).21 For gene ontology (GO) analysis, only significant enrichments ( false discovery rate < 0.05) in biological process (BP) and REACTOME pathways are reported. STRING settings for network visualization: confidence view, confidence level 0.7, and clustering algorithm MCL set to 3.
2.13. Immunohistochemistry in mouse dorsal root ganglia
The localization of the high priority candidate Annexin A1 (Anxa1) in DRG was realized with immunohistochemistry (IHC) in a separate cohort of mice (Fig. 1E, cohort 5, light blue; INC N = 3, Sham N = 3). Dorsal root ganglia tissue sections (for tissue preparation, cryopreservation, and slicing, see “Retrograde tracing experiments”) were thawed and subsequently surrounded by Pap Pen (Dako, Glostrup, Denmark). After, incubation steps were performed in a humid chamber (Thermo Scientific, Marietta, OH). After washing with 0.1 PBS (1 × 15 minutes, 3 × 5 minutes), slices were incubated for 10 minutes in TrueBlack IF Background Suppressor (Biotium, San Francisco, CA) and for 1 hour in TrueBlack IF Blocking Buffer at room temperature.
Sections were incubated in primary antibody solution overnight at 4°C, followed by washing steps and incubation with secondary antibodies at room temperature for 1 hour. After the final washing steps, slides were covered up with Confocal matrix (Micro-Tech-Lab, Austria). Isolectin B4 (IB4)-positive PSNs were identified by biotin-conjugated IB4 (1:50) and Pacific Blue–conjugated streptavidin (Invitrogen, Marietta, OH) at 1:50 dilution. Image acquisition was performed at a Zeiss Apotome 2 in a blinded manner concerning the experimental group and antibody combinations in L4 iDRG. Exposure times for the single channels were used: ultraviolet (352 nm): 1200 ms, green fluorescent protein (488 nm): 900 ms, and red fluorescent protein (594 nm): 900 ms. Only for presentation purposes, equal adjustments of brightness and contrast were made evenly for all images with the Zen software (Zeiss, Germany). Three randomized slices from iDRG L4 and L5 per mouse of both groups (Fig. 1E, cohort 5, light blue, INC N = 3, Sham N = 3) were examined in a blinded manner (2 independent experimenters, D.S. and K.K.) regarding the number of total DRG neurons and Anxa1-positive neurons by ImageJ.61 An increase in Anxa1-positive neurons was detected in each slide of each mouse of the INC group (N = 3, 3 slides per mouse).
2.14. Antibodies used in this study
Primary antibodies: mouse Anxa1, sc-12740 AF546, Santa Cruz Biotechnology, Dallas, TX (IHC, 1:100); rabbit neurofilament protein 200, N4142, Sigma-Aldrich, Germany (IHC, 1:250); rabbit peripherin AB1530, Chemicon, US (1:200); rabbit calcitonin gene-related peptide (CGRP), C8198, Sigma-Aldrich, Germany (IHC, 1:500); rabbit glial fibrillary acidic protein (GFAP), Z0334, Dako, US (1:400 in 1%BSA/PBS); Fuji Film, Japan (1:500). Secondary antibodies were purchased from Abcam and used at 1:1000 dilution; goat antirabbit AlexaFluor (AF) 488 (ab150077).
2.15. Systematic literature search
We performed a systematic literature search in Medline to assess whether regulated proteins and predicted signaling pathways (extracellular signal-regulated kinases [ERK], serine/threonine kinase Akt [Akt], and protein kinase C [PKC]) have already been investigated in the context of incisional pain in preclinical studies. For this, the “animal filter” for systematic search in MEDLINE was used,34 combined with a search string to identify all studies that used the “plantar incision model” in rodents, restricted by a custom timespan from 1996 (first plantar incision model was published) until the present (see Table, Supplement Digital Content 1, available at http://links.lww.com/PAIN/B274). This string was combined with the full protein name and the UniProt number. The identified publications were manually reviewed by 2 independent reviewers for the following criteria for relevance: (1) use of the plantar incision model according to Brennan et al.8 for rats and Pogatzki and Raja53/Banik et al.2 for mice and (2) investigation of protein expression by quantitative or qualitative methods along the pain axis (eg, peripheral tissue, DRG, spinal cord, or brain) after plantar incision (see Table, Supplement Digital Content 2, available at http://links.lww.com/PAIN/B274). Only publications, which fulfilled the inclusion criteria, are reported (see Tables, Supplement Digital Content 3-5, available at http://links.lww.com/PAIN/B274; publications, which report expression analyses in DRG are highlighted in red).
2.16. Statistical analysis
Data were analyzed using GraphPad Prism 8.0.2 (San Diego). All data are represented as mean ± SEM unless indicated otherwise. Sample sizes were a priori determined in dependence on the amount of tissue required for each analytical method (DIA-MS and DigiWEST multiplex Western blot) and are in line with standards in the field. All replicates were biological. All statistical tests are indicated in the respective figure legend and are 2-sided. For proteome data and protein network analysis, we used the Benjamini–Hochberg (BH)-adjusted P-value, Q-value < 0.05. There were no missing data.
2.17. Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium by the PRIDE52 partner repository with the data set identifier PXD022706.
3. Results
We used an integrated workflow to study proteome dynamics on incisional pain in male mice (Fig. 1). Incisional pain was induced using the plantar incision model (and Sham-treated mice as controls) and assessed by measuring diverse pain-related behaviors, including our novel assay for nonevoked pain behavior (Figs. 1A, B, D). The level of DRG projecting to the plantar surface of the hind paw was determined in our local C57Bl6/J mouse colony (Fig. 1C). Ipsilateral lumbar dorsal root ganglia (iDRG) of the respective level were isolated for quantitative data-independent acquisition mass spectrometry (DIA-MS)-based proteomics at POD 1 (Fig. 1D, cohort 3). Data-independent acquisition mass spectrometry enabled us to reveal a differential protein signature on incision, further evaluated by bioinformatic pathway analysis. Our results were orthogonally validated by experimental pathway analysis using quantitative multiplex Western blot analysis (DigiWEST) (Fig. 1D, cohort 4) and by immunohistochemistry in DRG (Fig. 1E, cohort 5). In this way, we revealed unprecedented insights into protein network dynamics for acute incisional pain.
3.1. Study rationale—behavior profiles, dorsal root ganglia levels
At the beginning of the study, 2 rationales had to be determined: (1) identification of a time point suitable for analysis of proteome dynamics associated with incisional pain and (2) characterization of DRG levels that correspond to peripheral sensory neurons (PSNs) that project to the incised area of the hind paw in our C57Bl6/J mouse colony.
Pain-related behavioral profiles were determined in an independent cohort (Figs. 1A and 2A, cohort 1, green, INC N = 8, Sham N = 8, and see RAW data, Supplemental Digital Content 6, available at http://links.lww.com/PAIN/B274). Nonevoked pain behavior was observed up to POD 6, represented by a significant decrease of change (%) compared with baseline (BL) and Sham condition. In addition, mice exhibited significant hypersensitivity to mechanical and heat stimuli, starting with the acute phase (4∼6 hours) and lasting up to POD 3 after incision (Fig. 2A). For all pain modalities, no significant difference was determined between (4∼6 hours) and POD 1. Mechanical and heat hypersensitivity characteristics regarding intensity and time are consistent with previous results in this model.63 Taken together, our behavior data suggest that POD 1 represents an appropriate time point to study proteome dynamics in incisional pain.
Figure 2.
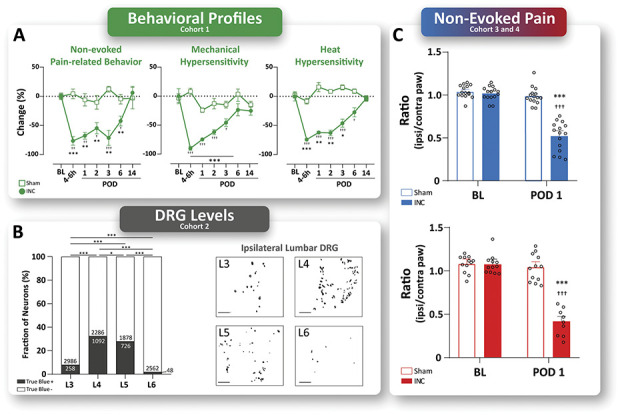
Study rationales—behavior profiles and DRG levels. (A) On plantar incision (INC), pain-related behavior profiles of nonevoked (NEP) and evoked (mechanical and heat) pain-related behaviors were evaluated in an independent cohort (cohort 1, green, INC N = 8, Sham N = 8). INC-induced NEP, as well as mechanical and heat hypersensitivity started in the acute phase (4∼6 hours) after surgery and lasted up to postoperative day (POD) 3 (mechanical and heat hypersensitivity) and up to POD 6 (NEP), respectively. At POD 1, 2, and 3, no significant differences were found for either pain-related behavior. Mice were randomly assigned to experimental groups (INC and Sham), and experiments were performed in a balanced study design. Data are expressed as change in % (mean ± SEM). Statistics were performed by 2-way ANOVA followed by Dunnett's post-hoc test. P-Values: ***P < 0.001 vs baseline (BL), †††P < 0.001 vs Sham. (B) The contribution of DRG levels to the sciatic nerve was determined by retrograde tracing (True Blue dye) in a separate cohort of male naïve mice of our local C57BL6/J colony (cohort 2, grey, naïve N = 8). Representative images of the ipsilateral lumbar DRG of levels L3 to L6. True Blue–positive neurons were counted 7 days after tracer injection and are significantly increased in ipsilateral DRG L4, and L5 compared with L3 and L6. The results are expressed as the absolute number of True Blue–positive and negative neurons, and the percentage of neurons in total. Scale bar = 200 µm. Statistics: Fisher's exact test (contingency). P-Values: ***P < 0.001. (C) Determination of behavior-based inclusion criterion for cohorts 3 and 4 (cohort 3, blue, INC N = 15, Sham N = 15; cohort 4, red, INC N = 12, and Sham N = 12). At POD 1, INC-induced significant NEP in both cohorts. The results are expressed as mean (ratio incised/nonincised paw) ± SEM and single values for each mice (open circles). P-Values: ***P < 0.001 vs baseline (BL), †††P < 0.001 vs Sham by 2-way ANOVA followed by Dunnett's post-hoc test. ANOVA, analysis of variance; DRG, dorsal root ganglia.
The anatomical organization of the sciatic nerve is heterogeneous, and the extent of the DRG level (lumbar levels L3 to L6) contribution has been shown to be dependent on the species, strain, substrain, and genetics.59 Therefore, before starting the study, we assessed which DRG levels correspond to PSNs that project to the incised area of the hind paw in our local C57Bl6/J mouse colony to ensure adequate selection of DRG for the proteome analysis. The distributions of True Blue–labeled PSNs were determined in a separate naïve male cohort (Fig. 1B, cohort 2, grey, naïve N = 8). We identified 258 True Blue–positive cells (Fig. 2B) in L3 iDRG (8.13%, 258/3244 in total, P < 0.001 vs L4, L5, L6, Fisher's exact test), 1092 in the L4 iDRG (30.62%, 1092/3378 in total, P < 0.05 vs L5, P < 0.001 vs L3, L6, Fisher's exact test), 726 in L5 iDRG (26.4%, 726/2604 in total, P < 0.05 vs L4, P < 0.001 vs L3, L6, Fisher's exact test), and 48 in L6 iDRG (1.91%, 48/2610 in total, P < 0.001 vs L3, L4, L5, Fisher's exact test). Compared with the number of positive cells in the L4 iDRG, significantly fewer marked cells were found in other levels (P < 0.05, Fisher's exact test). Based on these data, which characterize the nerve projections to the plantar surface of the hind paw in male mice of our local mouse colony (yet are not universally valid), L4 and L5 DRG were harvested and pooled for subsequent proteome analysis.
3.2. Mouse behavior combined with unbiased quantitative proteomics reveals the regulation of 44 proteins at POD 1 on plantar incision
Two independent cohorts of mice were used (total n = 54), one for DIA-MS (Fig. 1D, cohort 3, blue, INC N = 15, Sham N = 15), the other for follow up by DigiWEST (Fig. 1D, cohort 4, red, INC N = 12, Sham N = 12). Behavior analysis was performed before (baseline, BL) and 24 hours after incision (POD 1, Fig. 2C). Specifically, we assessed nonevoked pain at rest by comparing the print area of ipsilateral and contralateral hind paws in incised (INC) and (Sham) (sham) mice (please see Methods for details). The selection of POD 1 and nonevoked pain at rest as a paradigm is based on our a priori characterization of the time course of diverse pain-related behaviors in an independent cohort beforehand (Fig. 2A). In cohort 3 (Fig. 2C blue), the print area decreased from 1.02 ± 0.02 (Mean ± SEM) at BL before incision to 0.52 ± 0.04 at POD 1 after incision. By contrast, Sham mice exhibited stable print areas with 1.04 ± 0.02 at BL and 0.99 ± 0.03 at POD 1 as expected (***P < 0.001 vs BL, †††P < 0.001 vs Sham Fig. 2C, blue). In cohort 4 (Fig. 2C red), the print area decreased from 1.08 ± 0.03 at BL before incision to 0.42 ± 0.05 at POD 1 after incision; again, sham mice displayed unchanged print areas with 1.08 ± 0.03 at BL and 1.03 ± 0.16 at POD 1 (Fig. 2C, red). Thus, plantar incision caused nonevoked pain behavior at POD 1 in both cohorts, as indicated by a significant decrease in the ratio of the print area between the ipsi/contralateral paws compared with baseline (BL).
After the behavioral assessment, cohort 3 was sacrificed for dissection of iDRG (L4 and L5, Fig. 1D) for the purpose of MS-based proteomics. In total, iDRG from 15 INC and 15 Sham animals were pooled in 3 biological replicates each (ie, iDRG from 5 mice/replicate) and prepared for DIA-MS. We then followed our established proteomics workflow using liquid chromatography coupled mass spectrometry in DIA mode. For comprehensive data analysis, we used our pan-mouse spectral library harboring proteins detected across the mouse pain axis, from the sciatic nerve, lumbar DRG, and spinal cord to different brain regions as previously reported5,60 (http://painproteome.em.mpg.de/). In this way, we reproducibly identified 4618 proteins across all biological replicates (Fig. 3A).
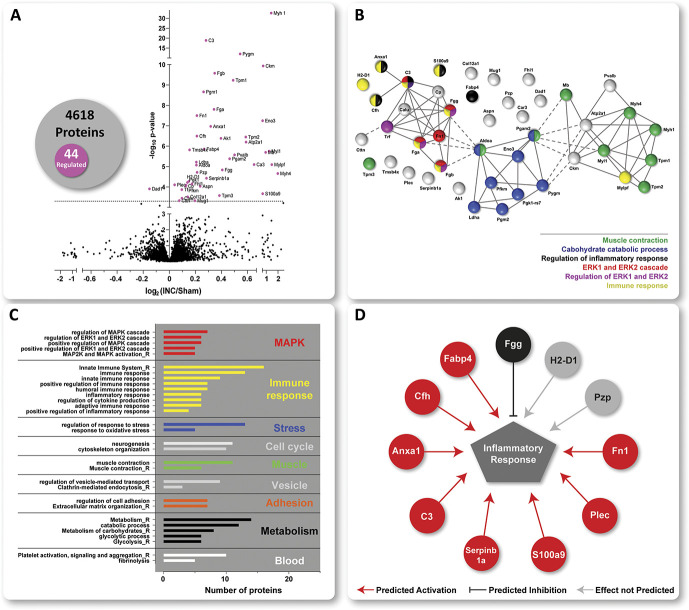
Figure 3.
Protein signature of incisional pain in iDRG. (A) The Volcano plot displays the mean log2 fold change (log2 INC/Sham) of detected proteins and corresponding -log10 of Q values on comparing protein intensities between INC and Sham. 44 of a total of 4618 quantified proteins were significantly regulated (magenta-colored circles above the dotted horizontal line, which represents Q < 0.05, ie, BH-adjusted P-value) on INC compared with Sham. The numbers are summarized in the Venn diagram. STRING-based predicted, relationship across the 44 regulated proteins (B). Three major clusters were identified with functionally distinct gene ontology (GO) annotations (see grey inlay) such as muscle contraction, carbohydrate catabolic process, immune and inflammatory responses, including regulation of ERK1/2. Settings for network visualization with STRING were as follows: confidence view, confidence level 0.7, and clustering algorithm MCL set to 3. GO-analysis for biological process (BP) and REACTOME (R) pathways (C). Regulated proteins are associated with major cellular pathways of which selected ones are shown, for example, stress response, cell cycle, vesicle-mediated transport, adhesion, and blood platelets). For full data sets, please see Table, Supplemental Digital Content 7, available at http://links.lww.com/PAIN/B275. Only annotated pathways, which exhibit significant enrichment ( false discovery rate < 0.05; assessed via the web-based interface STRING) are reported. Several regulated candidates are implicated in inflammatory responses (D) as assessed by Ingenuity Pathway Analysis (IPA)-based activity prediction analysis (IPA, Qiagen, 2000-2017). Red lines predicted activation; black lines represent predicted inhibition; grey lines show predictions, which are not defined. DRG, dorsal root ganglia.
On statistical comparison (corrected for multiple testing using the Benjamini–Hochberg (BH) Q-value,5,60,67 for complete data set, please see table, Supplemental Digital Content 7, available at http://links.lww.com/PAIN/B275), we observed little overall proteome changes as visualized in the volcano plot (Fig. 3A). Specifically, 44 of 4618 quantified proteins (ie, 0.95% of detected proteins, list in Table 1) were significantly regulated (Fig. 3A; magenta-colored candidates) on INC compared with Sham with all but one candidate being upregulated. Interestingly, thus, biological replicates of each condition only displayed limited coclustering (see figure, Supplemental Digital Content 8, available at http://links.lww.com/PAIN/B274) despite highly reproducible DIA–MS-based quantification across samples. Once more, these data highlight the importance of including sufficient numbers of mice in each study (in this study: 15 mice/condition) to allow the robust identification of significant protein alterations beyond the variable biological background.
Table 1.
INC-regulated candidate proteins identified by proteomics.
| Uniprot Id | Gene name | LOG2 (INC/Sham) | N (no. of identified peptides) | BH (q-value) | Name |
|---|---|---|---|---|---|
| Q5SX40 | Myh 1 | 1.416 | 42 | 1.07E-29 | Myosin-1 |
| P01027 | C3 | 0.281 | 62 | 3.09E-16 | Complement C3 |
| Q9WUB3 | Pygm | 0.543 | 39 | 1.37E-09 | Glycogen phosphorylase, muscle form |
| P07310 | Ckm | 0.735 | 27 | 1.35E-07 | Creatine kinase M-type |
| Q8K0E8 | Fgb | 0.348 | 18 | 2.44E-07 | Fibrinogen beta chain |
| P58771 | Tpm1 | 0.489 | 25 | 4.53E-07 | Tropomyosin alpha-1 chain |
| Q9D0F9 | Pgm1 | 0.263 | 33 | 1.44E-06 | Phosphoglucomutase-1 |
| E9PV24 | Fga | 0.343 | 18 | 9.03E-06 | Fibrinogen alpha chain |
| P11276 | Fn1 | 0.211 | 33 | 1.62E-05 | Fibronectin |
| P21550 | Eno3 | 0.690 | 17 | 2.61E-05 | Beta-enolase |
| P10107 | Anxa1 | 0.317 | 16 | 4.48E-05 | Annexin A1 |
| P06909 | Cfh | 0.211 | 12 | 0.0001227 | Complement factor H |
| P58774 | Tpm2 | 0.586 | 17 | 0.0001266 | Tropomyosin beta chain |
| Q9R0Y5 | Ak1 | 0.396 | 22 | 0.0001366 | Adenylate kinase isoenzyme 1 |
| Q8R429 | Atp2a1 | 0.588 | 22 | 0.0001941 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 |
| P04117 | Fabp4 | 0.266 | 10 | 0.0004020 | Fatty acid–binding protein, adipocyte |
| P20065 | Tmsb4x | 0.155 | 15 | 0.0004121 | Thymosin beta-4 |
| P05977 | Myl1 | 1.236 | 8 | 0.000450 | Myosin light chain 1/3, skeletal muscle isoform |
| P04247 | Mb | 0.949 | 12 | 0.0004906 | Myoglobin |
| P32848 | Pvalb | 0.500 | 18 | 0.0006162 | Parvalbumin alpha |
| O70250 | Pgam2 | 0.462 | 16 | 0.0009156 | Phosphoglycerate mutase 2 |
| P06151 | Ldha | 0.207 | 28 | 0.0012775 | L-lactate dehydrogenase A chain |
| P16015 | Ca3 | 0.650 | 15 | 0.0015511 | Carbonic anhydrase 3 |
| P97457 | Mylpf | 1.472 | 5 | 0.0015511 | Myosin regulatory light chain 2, skeletal muscle isoform |
| P05064 | Aldoa | 0.208 | 52 | 0.0015620 | Fructose–bisphosphate aldolase A |
| Q8VCM7 | Fgg | 0.406 | 19 | 0.002596 | Fibrinogen gamma chain |
| Q61838 | Pzp | 0.213 | 38 | 0.003306 | Pregnancy zone protein |
| Q5SX39 | Myh4 | 1.977 | 4 | 0.003611 | Myosin-4 |
| Q9D154 | Serpinb1a | 0.285 | 21 | 0.0059320 | Leukocyte elastase inhibitor A |
| P01899 | H2-D1 | 0.150 | 11 | 0.0081110 | H-2 class I histocompatibility antigen, D-B alpha chain |
| P09411 | Pgk1 | 0.139 | 42 | 0.0090205 | Phosphoglycerate kinase 1 |
| P97447 | Fhl1 | 0.174 | 15 | 0.0101767 | Four and a half LIM domains protein 1 |
| Q9QXS1 | Plec | 0.041 | 353 | 0.0109909 | Plectin |
| P47857 | Pfkm | 0.129 | 33 | 0.0113854 | ATP-dependent 6-phosphofructokinase, muscle type |
| Q61147 | Cp | 0.125 | 35 | 0.0119051 | Ceruloplasmin |
| Q99MQ4 | Aspn | 0.236 | 8 | 0.0119197 | Asporin |
| P61804 | Dad1 | −0.152 | 5 | 0.0155192 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit |
| Q921I1 | Tf | 0.095 | 74 | 0.0164564 | Serotransferrin |
| P31725 | S100a9 | 0.700 | 7 | 0.0254775 | Protein S100-A9 |
| P21107 | Tpm3 | 0.384 | 11 | 0.0304592 | Tropomyosin alpha-3 chain |
| Q60847 | Col12a1 | 0.141 | 47 | 0.0346081 | Collagen alpha-1(XII) chain |
| O35887 | Calu | 0.097 | 15 | 0.0396033 | Calumenin |
| P28665 | Mug1 | 0.196 | 37 | 0.0496802 | Murinoglobulin-1 |
| Q60598 | cttn | 0.077 | 23 | 0.0496802 | Src substrate cortactin |
BH, Benjamini–Hochberg.
3.3. Plantar incision alters major biological pathways in ipsilateral dorsal root ganglia
We then used the web-based STRING interface (www.string-db.org,21) to visualize predicted relationships across the 44 regulated proteins. This analysis revealed 3 major clusters with functionally distinct GO annotations, that is, proteins involved in muscle contractions, proteins involved in carbohydrate catabolic processes, and proteins involved in immune and inflammatory responses, including ERK1/2 signaling (Fig. 3B). Overall, GO-analysis for biological process (BP) and Reactome pathways (R) suggested that regulated proteins were associated with major cellular pathways such as the ones mentioned above and, additionally, stress response, cell cycle, vesicle-mediated transport, adhesion, and blood platelets (Fig. 3C; for detailed lists, please see table, Supplemental Digital Content 7, available at http://links.lww.com/PAIN/B275). In particular, the association with pathways implicated in immune and inflammatory responses was very pronounced. Indeed, IPA-based activity prediction highlighted several significantly regulated candidates, which are known to modulate inflammatory responses such as Annexin A1 (Anxa1), S100-calcium-binding protein A9 (S100a9), fibronectin 1 (Fn1), and fatty acid–binding protein 4 (Fabp4) to name a few (Fig. 3D).
3.4. Orthogonal quantitative and network analysis of inflammatory signaling pathways on incision
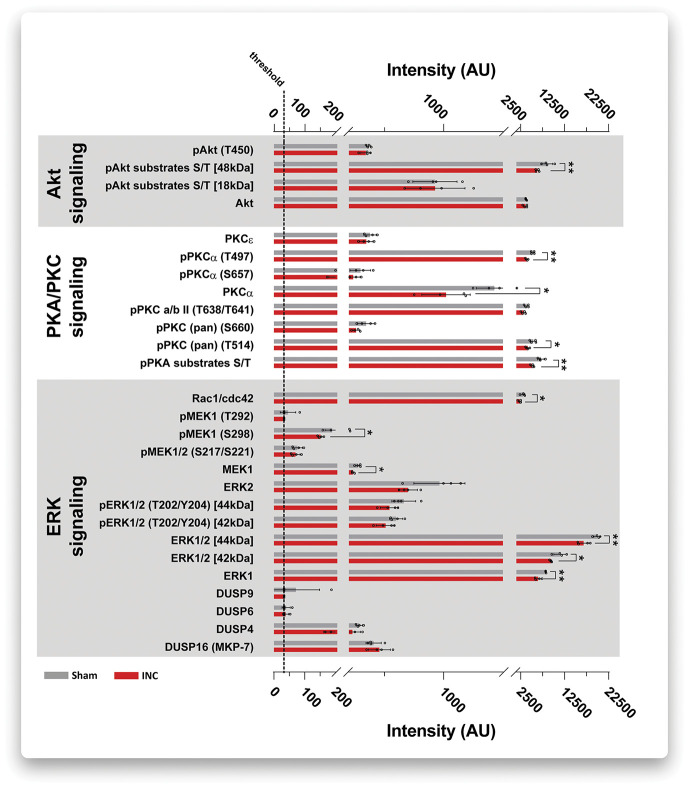
Functional categorization and network analysis based on previously annotated information (Fig. 3B-D) exhibit a certain degree of ambiguity. Therefore, we aimed at validating and extending our data on the modulation of immune/inflammatory signaling pathways by an orthogonal experimental approach. To this end, we used quantitative multiplex Western blot analysis (DigiWest). Lysates of iDRG (4 biological replicates) obtained from an independent mouse cohort (cohort 4, red, INC N = 12, Sham N = 12, please see study design and pain-related behavior analysis in Fig. 1D and Fig, 2C; 3 mice were pooled/replicate) were subjected to multiplex profiling using 87 analytes. Analytes were selected based on their previously described or predicted association with inflammatory/immune signaling, such as highly interconnected signaling cascades involving ERK1/2, PKA/PKC, and Akt (for the full list of analytes and results, please see Table, Supplemental Digital Content 9, available at http://links.lww.com/PAIN/B274). Even more, we included, whenever possible (depending on the availability of specific antibodies), several phosphorylated protein forms of analytes given the functional relevance of the phosphorylation status for the regulation of cellular signaling cascades.38,43,49 For example, a hallmark of ERK1/2 signaling is its activation by phosphorylation, which has already been reported to be functionally relevant in the mouse incision model.13 Unfortunately, direct validation of here identified candidates regulated on INC (Table 1) was not possible because antibodies did not pass quality criteria for multiplex Western blotting.
Nonetheless, and in line with our DIA-MS results and network analysis (Fig. 3B,C), we observed prominent dysregulation of ERK1/2 signaling and, additionally, altered PKA/PKC and Akt signaling (Fig. 4 and see table, Supplemental Digital Content 9, available at http://links.lww.com/PAIN/B274). Given the known crosstalk among these pathways and their involvement in diverse cellular functions,47,76 a straightforward interpretation of the net effects of here measured alterations is not possible. Importantly, thus, the decrease in the total abundance of ERK1/2 on INC (Fig. 4, data on ERK1/2 and on ERK1) paired with stable levels of phosphorylated ERK1/2 (Fig. 4 data on pERK1/2) results in a higher pERK1/2 vs total ERK1/2 ratio indicative of activation of the ERK1/2 pathway.23,80 These data are in accordance with our DIA-MS results and network analysis (Fig. 3C).
Figure 4.
DigiWEST multiplex Western blot validated alterations in distinct signaling pathways. Plantar incision (INC, red bars) induced significant dysregulation of multiple members of ERK1/2 signaling, PKA/PKC signaling, and Akt signaling (***P < 0.001 vs Sham, grey bars; 3 mice were pooled/replicate, 4 biological replicates in total). Results are expressed as scatter bar plots showing the mean ± SEM and single values for each biological replicate (open circles). P-values: ***P < 0.0001, multiple t-tests without assuming a consistent SE. Legend: T, threonine; S, serine; P, denotes the phosphorylated form of the respective analyte; numbers behind T/S indicate the respective amino acid residue; kDa, kilo Dalton (as ERK1/2 isoforms are known to run at different molecular weights).69
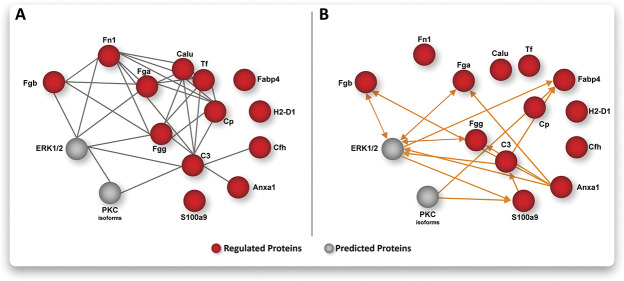
Emerging network medicine holds the promise to increase treatment specificity by combinatorial targeting of multiple central nodes involved in pathological processes.3,29,32 Thus, we aimed at interrogating our results on the relevance of ERK1/2 and PKC signaling from a network point of view. To this end, we tested for predictions of activity between ERK1/2 and PKC with those proteins that are annotated (shown in Fig. 3D) to be involved in immune/inflammatory signaling using IPA (Fig. 5A; and see figure, Supplemental Digital Content 10 for original IPA-network output, available at http://links.lww.com/PAIN/B274). This analysis revealed a protein network of INC-regulated candidates, which is predicted to be implicated in ERK1/2 signaling cascades (Fig. 5B; see figure, Supplemental Digital Content 10, available at http://links.lww.com/PAIN/B274) in line with our GO-BP and Reactome results (Fig. 3C). Similarly, PKC signaling is predicted to be activated by some members of this protein network, albeit to a lesser extent (Fig. 5B). Importantly, these predictions are consistent with our multiplex Western blot analysis endorsing our results (Fig. 4).
Figure 5.
Activity prediction on plantar incision in iDRG on the network-level. Bioinformatic network analysis integrating DIA-MS data and multiplex Western blots (A) complemented with activity prediction (B). Note the prominent role of ERK signaling as a central hub among proteins associated with immune and inflammatory responses (from Fig. 3D). Node color code: red, regulated proteins based on DIA-MS; grey, predicted proteins based on multiplex Western blots; orange lines indicate predicted activation among nodes. All graphs are based on IPA analysis (for original IPA results, please see Figure, Supplement Digital Content 10, available at http://links.lww.com/PAIN/B274). DRG, dorsal root ganglia; DIA-MS, data-independent acquisition mass spectrometry; IPA, ingenuity pathway analysis.
3.5. Comprehensive and systematic literature search on INC-regulated proteins
Although our data propose the implication of several INC-regulated candidate proteins in ERK1/2 signaling (Fig. 5), ERK1/2 per se is unlikely to serve as a new analgetic target given its ubiquitous functions.7,11 Therefore, we instead focused our attention on predicted INC-regulated modulators of ERK1/2 signaling (Fig. 5).
We first performed an extensive and systematic literature search to assess the novelty of here identified INC-regulated candidates, that is, and we asked whether candidates have previously been mentioned in the context of INC (Supplemental Digital Content 1: search string, available at http://links.lww.com/PAIN/B274). Remarkably, our literature search suggests that all here identified candidates seem to represent novel INC-regulated proteins (Supplemental Digital Content 2: results of literature search, available at http://links.lww.com/PAIN/B274). Interestingly, a systematic literature search on aforementioned signaling pathways altered on INC, that is, those involving ERK1/2, AKT, or PKC signaling (Fig. 5), suggested their implication in incisional pain by other studies (Supplemental Digital Content 3-5: results of literature search, available at http://links.lww.com/PAIN/B274) endorsing our findings.
Next, we queried select DIA-MS proteomics and transcriptomics data sets to assess (1) whether our candidate proteins have been reported in the context of neuropathic pain in mice5,36 (comparable DIA-MS proteomics datasets on other pain models are unfortunately not available, preventing further comparisons) and (2) in which cell types of DRG their transcripts are expressed (Table 2). Interestingly, 31 of all INC-regulated candidates identified in our study were reported to be also regulated in the DRG proteome on neuropathic pain induced by spared-nerve injury (SNI)5 (Table 2, first column). Among these 31 overlapping candidates 11 (highlighted by an X in bold font; Table 2, first column) were also shown above to modulate ERK1/2 signaling (Fig. 5).
Table 2.
Comparisons of INC-regulated candidate proteins with selected published data sets.
| Regulated proteins POD 1 after incision | Gene name | Overlap | |||
|---|---|---|---|---|---|
| Neuropathic Pain (POD 28) (Barry et al. 2018) | mRNA expressed in sorted nociceptors (Jager et al. 2020) | Top 300 mRNA (Usoskin et al. 2014) | mRNA expressed in SCG (Jager et al. 2020) | ||
| Myosin-1 | Myh 1 | ||||
| Complement C3 | C3 | X | |||
| Glycogen phosphorylase, muscle form | Pygm | X | |||
| Creatine kinase M-type | Ckm | X | |||
| Fibrinogen beta chain | Fgb | X | |||
| Tropomyosin alpha-1 chain | Tpm1 | X | X | X | |
| Phosphoglucomutase-1 | Pgm1 | X | X | X | |
| Fibrinogen alpha chain | Fga | X | |||
| Fibronectin | Fn1 | X | |||
| Beta-enolase | Eno3 | X | X | ||
| Annexin A1 | Anxa1 | X | X | X | |
| Complement factor H | Cfh | X | X | ||
| Tropomyosin beta chain | Tpm2 | X | X | ||
| Adenylate kinase isoenzyme 1 | Ak1 | X | X | X | X |
| Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | Atp2a1 | ||||
| Fatty acid–binding protein, adipocyte | Fabp4 | ||||
| Thymosin beta-4 | Tmsb4x | X | X | ||
| Myosin light chain 1/3, skeletal muscle isoform | Myl1 | ||||
| Myoglobin | Mb | X | |||
| Parvalbumin alpha | Pvalb | X | X | ||
| Phosphoglycerate mutase 2 | Pgam2 | ||||
| L-lactate dehydrogenase A chain | Ldha | X | X | ||
| Carbonic anhydrase 3 | Ca3 | X | |||
| Myosin regulatory light chain 2, skeletal muscle isoform | Mylpf | X | |||
| Fructose–bisphosphate aldolase A | Aldoa | X | X | ||
| Fibrinogen gamma chain | Fgg | X | X | ||
| Pregnancy zone protein | Pzp | X | |||
| Myosin-4 | Myh4 | ||||
| Leukocyte elastase inhibitor A | Serpinb1a | X | |||
| H-2 class I histocompatibility antigen, D-B alpha chain | H2-D1 | X | X | X | |
| Phosphoglycerate kinase 1 | Pgk1 | X | X | X | X |
| Four and a half LIM domains protein 1 | Fhl1 | X | X | X | |
| Plectin | Plec | X | X | X | |
| ATP-dependent 6-phosphofructokinase, muscle type | Pfkm | X | X | X | |
| Ceruloplasmin | Cp | X | X | ||
| Asporin | Aspn | X | |||
| Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit | Dad1 | X | X | X | |
| Serotransferrin | Tf | X | |||
| Protein S100-A9 | S100a9 | X | |||
| Tropomyosin alpha-3 chain | Tpm3 | X | X | X | |
| Collagen alpha-1(XII) chain | Col12a1 | X | X | ||
| Calumenin | Calu | X | X | X | |
| Murinoglobulin-1 | Mug1 | X | |||
| Src substrate cortactin | cttn | X | X | X | |
Comparison of INC-regulated proteins with other selected datasets. Data sets are given on top of each column with respective reference. Proteins, which have been shown to be regulated during neuropathic pain at POD 28 after spared-nerve injury5 and are predicted to be involved in the ERK signaling network (Fig. 5), are highlighted by an X in bold font. Candidates, which are expressed (on the transcript level) in SCG (right column) and have been shown to be regulated at POD 3 after full sciatic nerve ligation36 are highlighted by an X in bold font in the right column. SCG, satellite glia cells.
In addition, transcripts of 21 INC-regulated candidate proteins were previously detected to be expressed in nociceptors,36 and transcripts of 4 candidate proteins are included in the TOP300 transcripts across neuronal DRG subpopulations reported by Usoskin and colleagues.70 A comparison to the overall transcriptome of satellite glia cells (SCG) revealed an overlap with 20 INC-regulated proteins of which 2 (Anxa1 and Col12a1, highlighted by an X in bold font; Table 2, last column) appeared to be regulated in SCG on the transcript level at POD 3 after full sciatic nerve ligation.36 Taken together, these comparisons suggest that we have identified INC-regulated candidate proteins with expression across DRG cell types, for example, neurons and SCG.
3.6. Anxa1 is expressed in sensory neurons on incision
Among INC-regulated candidate proteins, Anxa1 represents a high priority candidate given (1) the fact that it has previously been implicated in other pain models (Table 2), (2) its novelty in the context of incisional pain (Supplemental Digital Content 2: results of literature search, available at http://links.lww.com/PAIN/B274), and (3) its predicted involvement in incisional pain as a modulator of inflammatory/immune signaling (Fig. 3B-D) and ERK1/2 (Fig. 5). Therefore, we used immunohistochemistry to investigate Anxa1 expression (and potential changes thereof) in DRG cell types on Sham- vs INC-treated mice. We used established cell population markers to determine the localization of Anxa1 in ipsilateral L4 DRG (Fig. 6). Anxa1-positive cells were largely colocalized with glial fibrillary acidic protein (GFAP) positive satellite glial cells (SGC) surrounding DRG neurons in Sham-treated mice (Fig. 6A). By contrast, on INC, Anxa1 appeared to be additionally expressed across neuronal subpopulations (please see the arrows in Fig. 6B, Inset), including Peripherin-positive neurons (representing unmyelinated C-fiber and thinly myelinated Aδ fiber neurons), calcitonin gene-related peptide (CGRP)-positive peptidergic nociceptors (representing neurons that give rise to C- or Aδ-fibers), and Isolectin B4 (IB4)-positive nociceptors (Fig. 6B, Inset). Of note, we used retrograde tracing (with True Blue, Fig. 6) to highlight those neurons that primarily innervate the plantar surface, several of which were also Anxa1 positive (please see asterisks in Fig. 6B, Inset). In total, we identified 40 Anxa1-positive neurons in L4 and L5 DRG (1.79%, 40/2223 in total) under sham conditions. On INC, a significantly higher number of Anxa1-positive neurons (13.59%, 288/2118 in total, P < 0.05, Fisher's exact test) were detected (see representative images in Fig. 6).
Figure 6.
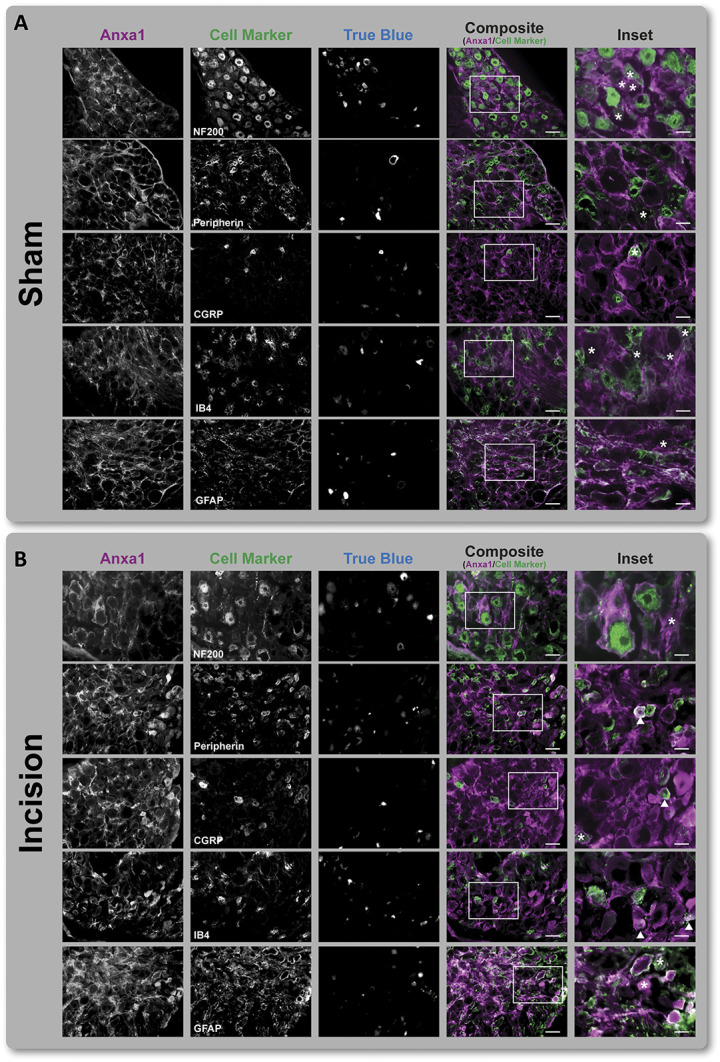
Localization of Anxa1 in L4 ipsilateral DRG. (A) Under Sham conditions, Anxa1 is colocalized with GFAP-positive cells (ie, satellite glia cells) but not with neuronal subpopulations (labeled by indicated marker proteins) or retrogradely labeled neurons (True Blue positive) in ipsilateral L4 DRG. (B) By contrast, plantar incision (postoperative day 1) induced neuronal expression of Anxa1 in Peripherin-, CGRP- and IB4-positive neurons (indicated by arrows, Inset). Retrogradely labeled neurons are marked with an asterisk (Inset). Scale bar = 50 µm (composite), 20 µm (Inset). DRG, dorsal root ganglia.
4. Discussion
To characterize proteome changes associated with plantar incision in male mice, we used here, for the first time, a multimethod integrated workflow consisting of a novel behavior assay to determine nonevoked pain, unbiased and quantitative proteome profiling, followed by orthogonal validation based on quantitative multiplex Western blot analysis, network pathway analysis, and immunolabeling. In this way, we revealed a hitherto unknown protein signature in DRG at POD 1 after incision (INC). In particular, we show the differential regulation of 44 proteins in INC samples compared with sham controls. Among these INC-regulated candidates, many seem to be implicated in pathways associated with immune and inflammatory responses such as mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) signaling. Subsequent orthogonal assays in independent mouse cohorts validated the INC-induced regulation of distinct signaling pathways and the high priority candidate Anxa1 in line with our bioinformatic network analysis.
We performed proteomic analysis at POD 1 after INC because at this time point, pain-associated behaviors are fully established in the mouse incision model, translating well to pain in patients 1 day after surgery.6,55,64 By unbiased proteome profiling, we identified 44 INC-regulated candidate proteins. This relatively low number of modulated proteins in DRG after INC contrasts the 715 regulated proteins identified in our previous data set on neuropathic pain after spared-nerve injury (SNI).5 This finding likely reflects overt differences between these 2 pain models concerning the extent of tissue trauma (SNI > INC), potential nerve damage (SNI > INC), and peripheral inflammation (SNI < INC).42,45,52 Although time points, at which pain-related behaviors are fully established were chosen for tissue collection in both pain models (INC: POD 1; SNI: POD 28), this might additionally contribute to observed differences in the number of regulated proteins. Future studies designed to investigate proteome dynamics along the complete time course of each pain model will undoubtedly provide additional and highly valuable insights into the development and resolution of distinct pain states across pain models. Interestingly, 31 of the 44 INC-regulated proteins reported here also seem to be altered during neuropathic pain (in aforementioned SNI-model) in the peripheral nervous system5 (Table 2). This overlap may be indicative of proteins, which are generally implicated in pain independently of the pain entity, and may therefore contribute to common pain-related behaviors.4 By contrast, 13 INC-regulated candidates were uniquely regulated on incision (Table 2) and might represent a specific peripheral protein signature on INC. Note, however, that “unique” only refers to the comparison with this specific DRG proteome data set on SNI;5 thus, similarities to other pain models are possible, albeit not analyzed here. To fully exploit the potential of our DIA-MS results, we performed extensive bioinformatics network analysis, which revealed the association (may it be direct or indirect) of INC-regulated candidates with diverse biological pathways (Figs. 3C, D). Interestingly, pathways related to mitochondrial function were unaltered on INC. This finding not only represents another major difference to our previous work in the SNI model5 but may also suggest a certain degree of specificity of here identified INC-regulated candidates and associated pathways.
Many INC-regulated candidates exhibited prominent association with immune and inflammatory response pathways (eg, Fgg, Fga, Fgb, Fn1, and S100a9) (Fig. 3D), in particular those that involve MAPK/ERK and PKC signaling. Changes in MAPK/ERK58,72,79 and PKC signaling74 and their contributions to pain-related behavior have previously been reported in the context of plantar incision (supplement content 3-5, available at http://links.lww.com/PAIN/B274). However, targeting MAPK/ERK directly (eg, by inhibitors) in a clinical setting is likely to be limited, given their ubiquitous expression and function.7,11,75 Our results (Figs. 3 and 4) not only support the involvement of these signaling pathways in incisional pain. Rather, IPA-based network analysis (Fig. 5) identified potential upstream modulators of ERK signaling such as complement component 3 (C3), fibrinogen A/B/G (Fga, Fgb, Fgg), and Annexin1 (Anxa1),33 none of which have so far been described in the context of incisional pain (Supplemental Digital Content 2: results of our literature search, available at http://links.lww.com/PAIN/B274). Using immunolabeling, we focused specifically on Anxa1. We could show its localization to DRG neurons on INC, which contrasted its nearly exclusive expression in SCG of Sham-treated mice (Fig. 6). Certainly, we cannot exclude very low level—beyond the detection threshold in our experiments—expression of Anxa1 in DRG of Sham-treated mice.51 Even so, the immunolabeling results correlate with the increase of Anxa1 protein levels as measured by DIA-MS (Fig. 3A). Anxa1 (formerly known as lipocortin-1) has originally been identified as a mediator of anti-inflammatory actions of glucocorticoids.10,18,33,42 It has further been shown to act through MAPK/ERK signaling endorsing our network prediction analysis (Fig. 5). Anxa1 is reported to be involved in (anti) inflammation18,19,24,42 and as an analgesic modulator of inflammation-associated pain.1,14,19,45,51 Our results suggest that Anxa1 might act upstream of ERK1/2 signaling, possibly in DRG neurons and SCG. Whether Anxa1 might be involved in analgesic/anti-inflammatory processing after incision injury remains an attractive hypothesis to be investigated in future studies. Among potential targets/effectors of ERK signaling identified by IPA-pathway analysis were the S100 calcium-binding protein A9 (S100a9) and the fatty acid–binding protein 4 (Fabp4) (Fig. 5). S100a9 is mainly expressed in immune cells,73 and its upregulation is a crucial factor in inflammatory/immune processes, in part, by cytokine secretion and MAPK/ERK signaling.41,73 Results from preclinical studies suggested antinociceptive effects of the C-terminus of S100a9 on neuropathic pain.50 Likewise, Fabp4 is expressed in immune cells35,46 and has been implicated in endothelial dysfunction, inflammation,12 and associated diseases such as diabetes.35 Although beyond the scope of this study, it will be fascinating to exploit our hypothesis-generating results in future work aimed at determining the functional and potential therapeutic relevance of here identified candidate proteins for incisional pain.
From a technical point of view, certain aspects need to be considered for interpreting our results. Whole DRG lysates contain proteins from several cell types, that is, neurons, fibroblasts, SCG, and immune cells. Furthermore, vascular permeability in DRG is high compared with the central nervous system,37 facilitating the recruitment of humoral proteins such as fibrinogens or fibronectin.39 Because of this cellular complexity and heterogeneity, we cannot assign the detected changes in protein abundance/phosphorylation to specific DRG cell types after incision—an issue common to all “-omics” approaches not performed on the single-cell level. To address this in part, we determined the cellular localization of Anxa1 in DRG by immunolabeling (Fig. 6) and compared our data sets with selected published resources on the DRG transcriptome/proteome (Table 2).5,36,70 In essence, these comparisons suggest that we have profiled proteins expressed in DRG sensory neurons alongside other DRG cell types such as SCG. Another technical aspect relates to the pooling of DRG from different levels to obtain sufficient tissue per DIA-MS replicate. To minimize heterogeneity (selection bias), we limited this pooling to DRG from levels L4 and L5 because retrograde tracing enabled us to determine DRG L4 and L5 as major contributors to the plantar hind paw innervation in our in-house mouse colony (Fig. 2B). Given known differences in hind paw innervation between and within mouse strains,59 we advocate that future studies should implement similar methods to characterize respective mouse cohorts.
Taken together, our data represent a valuable resource for exploring and testing novel therapeutical targets for incisional pain. This aspect is of high relevance for advancing pain treatments considering that severe side effects hamper not only existing treatment regimens but also stall clinical trials.56
Network medicine nourishes the hope of identifying and correcting dysfunctional networks. Along these lines, INC-regulated candidate proteins associated with distinct pathways may conceptually be exploited to correct dysfunctional signaling by combinatorial targeting. This strategy would harbor enormous advantages compared with traditional pharmacotherapies aimed at single targets: increased efficacy, specificity, and safety.31,66,78 For example, we have identified potential effectors and modulators of MAPK/ERK signaling (Fig. 5), which hold the promise to represent novel combinatorial targets to more specifically alter incision-induced pathological changes in DRG with likely reduced side effects compared with those targeting MAPK/ERK signaling directly.
5. Conclusion
Combining a mouse model of incisional pain including a novel behavioral test paradigm with quantitative proteome profiling, we identified 44 regulated proteins in DRG at POD 1. Our orthogonal assays and bioinformatic network analysis validated these results and revealed novel candidates, such as Anxa1, as attractive targets in the context of incision-induced pain. Thus, the here identified incision-associated protein signature in DRG opens new avenues for hypothesis-driven functional and translational investigations into incisional pain.
Conflict of interest statement
During the last 5 years, E. Pogatzki-Zahn received financial support from Mundipharma GmbH and Grunenthal for research activities and from Grünenthal, MSD Sharp & DOHME GmbH, Mundipharma GmbH, Mundipharma International, Janssen-Cilag GmbH, Fresenius Kabi and AcelRx for advisory board activities and/or lecture fees. None of this research support/funds was used for or influenced this article, and E. Pogatzki-Zahn declares no conflict of interest. D. Gomez-Varela received a research award by Biognosys AG (Zurich, Switzerland). This research award has not been used or influenced this article, and D. Gomez-Varela declares that there is no conflict of interest. G. Erdmann is an employee of NMI and NMI TT Pharma services, which provides DigiWEST analysis. M. Schmidt received research awards and travel support by the German Pain Society (DGSS), both of which were sponsored by Astellas Pharma GmbH (Germany). In addition, MS received one-time consulting honoraria by Grünenthal GmbH, (Germany). None of the funding sources influenced the content of this study. The remaining authors have no conflicts of interest to declare.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B274 and http://links.lww.com/PAIN/B275.
Acknowledgments
The authors thank Julia Sondermann (PhD) (formerly at MPI of Experimental Medicine, Goettingen; now: Department of Pharmacology and Toxicology, University of Vienna, Vienna) for technical help with some experiments, and Alison Barry (MSc) (formerly at MPI of Experimental Medicine, Goettingen; now University of Oxford, United Kingdom) for preliminary data analysis. The authors thank Sergej Zeiter (formerly MPI of Experimental Medicine, Goettingen) Tanja Nielsson (MPI of Experimental Medicine, Goettingen), Mirjam Augustin, and Dagmar Evers (Department for Anesthesiology, operative Intensive Care and Pain Medicine, University Hospital Muenster) for excellent technical assistance. The authors are grateful to the Schmidt laboratory at the MPI for Experimental Medicine for thoughtful comments on the study. Daniel Segelcke and Manuela Schmidt contributed equally to this article.
The study was supported by the Max Planck Society and the German Research Foundation (D.F.G.) (PO 1319/3-1 to E.P.Z., SCHM2533/6-1, and SCHM2533/4-1 to M.S. and GO2481/3-1 to D.G.V.).
Author contributions: E. Pogatzki-Zahn and M. Schmidt conceived, designed, and cosupervised the study. E. Pogatzki-Zahn contributed to the development of behavioral experiments and systematic search, analyzed behavioral experiments, systematic search, and immunolabeling data. D. Gomez-Varela performed DIA-MS analysis and provided guidance for the proteomics part of the study. G. Erdmann performed the DigiWEST analysis and supported the study with technical advice. K. Kaschube assisted in retrograde tracing, behavioral experiments, tissue preparation, and immunolabeling. D. Segelcke contributed to study design, developed and performed retrograde tracing, behavioral experiments, immunolabeling, and systematic search, prepared the tissue for DIA-MS, and created figures and tables. M. Schmidt analyzed proteomics data and DigiWEST data, performed bioinformatics network analysis, and prepared figures and tables. E. Pogatzki-Zahn, D. Segelcke, and M. Schmidt wrote the article. All authors reviewed the article. Previous presentation: Protein signature of postoperative pain in mice, M. Schmidt, D. Segelcke, Allison Barry, Mirjam Augustin, E.M. Pogatzki-Zahn, IASP World Pain Congress 12 Sept 2018 to 16 Sept 2018, Boston, MA, United States.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painjournalonline.com).
Contributor Information
David Gomez-Varela, Email: gomez@em.mpg.de.
Gerrit Erdmann, Email: erdmann@nmi-tt.de.
Katharina Kaschube, Email: K.Kaschube@web.de.
Daniel Segelcke, Email: segelcke@anit.uni-muenster.de.
Manuela Schmidt, Email: mschmidt@em.mpg.de.
References
- [1].Ayoub SS, Yazid S, Flower RJ. Increased susceptibility of annexin-A1 null mice to nociceptive pain is indicative of a spinal antinociceptive action of annexin-A1. Br J Pharmacol 2008;154:1135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Banik RK, Woo YC, Park SS, Brennan TJ. Strain and sex influence on pain sensitivity after plantar incision in the mouse. Anesthesiology 2006;105:1246–53. [DOI] [PubMed] [Google Scholar]
- [3].Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Baron R, Maier C, Attal N, Binder A, Bouhassira D, Cruccu G, Finnerup NB, Haanpää M, Hansson P, Jensen TS, Freynhagen R, Kennedy JD, Magerl W, Mainka T, Reimer M, Rice ASC, Segerdahl M, Serra J, Sindrup S, Sommer C, Tölle T, Vollert J, Treede R-D. Peripheral neuropathic pain: a mechanism-related organizing principle based on sensory profiles. PAIN 2017;158:261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barry AM, Sondermann JR, Sondermann J-H, Gomez-Varela D, Schmidt M. Region-resolved quantitative proteome profiling reveals molecular dynamics associated with chronic pain in the PNS and spinal cord. Front Mol Neurosci 2018;11:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bisgaard T, Klarskov B, Rosenberg J, Kehlet H. Characteristics and prediction of early pain after laparoscopic cholecystectomy. PAIN 2001;90:261–9. [DOI] [PubMed] [Google Scholar]
- [7].Braicu C, Buse M, Busuioc C, Drula R, Gulei D, Raduly L, Rusu A, Irimie A, Atanasov AG, Slaby O, Ionescu C, Berindan-Neagoe I. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers (Basel) 2019;11. doi: 10.3390/cancers11101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brennan TJ, Vandermeulen EP, Gebhart GF. Characterization of a rat model of incisional pain. PAIN 1996;64:493–501. [DOI] [PubMed] [Google Scholar]
- [9].Bruderer R, Sondermann J, Tsou C-C, Barrantes-Freer A, Stadelmann C, Nesvizhskii AI, Schmidt M, Reiter L, Gomez-Varela D. New targeted approaches for the quantification of dataindependent acquisition mass spectrometry. Proteomics 2017;17:1700021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Buckingham JC, Flower RJ. Lipocortin 1: a second messenger of glucocorticoid action in the hypothalamo-pituitary-adrenocortical axis. Mol Med Today 1997;3:296–302. [DOI] [PubMed] [Google Scholar]
- [11].Buscà R, Pouysségur J, Lenormand P. ERK1 and ERK2 map kinases: specific roles or functional redundancy? Front Cell Dev Biol 2016;4:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cai H, Liu Q, Gao D, Wang T, Chen T, Yan G, Chen K, Xu Y, Wang H, Li Y, Zhu W. Novel fatty acid binding protein 4 (FABP4) inhibitors: virtual screening, synthesis and crystal structure determination. Eur J Med Chem 2015;90:241–50. [DOI] [PubMed] [Google Scholar]
- [13].Chaumette T, Delay L, Barbier J, Boudieu L, Aissouni Y, Meleine M, Lashermes A, Legha W, Antraigue S, Carvalho FA, Eschalier A, Ardid D, Moqrich A, Marchand F. c-Jun/p38MAPK/ASIC3 pathways specifically activated by nerve growth factor through TrkA are crucial for mechanical allodynia development. PAIN 2020;161:1109–23. [DOI] [PubMed] [Google Scholar]
- [14].Chen L, Lv F, Pei L. Annexin 1: a glucocorticoid-inducible protein that modulates inflammatory pain. Eur J Pain 2014;18:338–47. [DOI] [PubMed] [Google Scholar]
- [15].Cowie AM, Moehring F, O'Hara C, Stucky CL. Optogenetic inhibition of CGRPα sensory neurons reveals their distinct roles in neuropathic and incisional pain. J Neurosci 2018;38:5807–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cowie AM, Stucky CL. A mouse model of postoperative pain. Bio Protoc 2019;9:e3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Deuis JR, Dvorakova LS, Vetter I. Methods used to evaluate pain behaviors in rodents. Front Mol Neurosci 2017;10:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ding Y, Flores J, Klebe D, Li P, McBride DW, Tang J, Zhang JH. Annexin A1 attenuates neuroinflammation through FPR2/p38/COX-2 pathway after intracerebral hemorrhage in male mice. J Neurosci Res 2020;98:168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ferreira SH, Cunha FQ, Lorenzetti BB, Michelin MA, Perretti M, Flower RJ, Poole S. Role of lipocortin-1 in the anti-hyperalgesic actions of dexamethasone. Br J Pharmacol 1997;121:883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fletcher D, Stamer UM, Pogatzki-Zahn E, Zaslansky R, Tanase NV, Perruchoud C, Kranke P, Komann M, Lehman T, Meissner W. Chronic postsurgical pain in Europe: an observational study. Eur J Anaesthesiol 2015;32:725–34. [DOI] [PubMed] [Google Scholar]
- [21].Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, Mering Cvon, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41:D808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gan TJ, Habib AS, Miller TE, White W, Apfelbaum JL. Incidence, patient satisfaction, and perceptions of post-surgical pain: results from a US national survey. Curr Med Res Opin 2014;30:149–60. [DOI] [PubMed] [Google Scholar]
- [23].Gao Y-J, Ji R-R. c-Fos and pERK, which is a better marker for neuronal activation and central sensitization after noxious stimulation and tissue injury? Open Pain J 2009;2:11–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gavins FNE, Hickey MJ. Annexin A1 and the regulation of innate and adaptive immunity. Front Immun 2012;3:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gazerani P, Vinterhøj HSH. “Omics”: an emerging field in pain research and management. Future Neurol 2016;11:255–65. [Google Scholar]
- [26].Gerbershagen HJ, Aduckathil S, van Wijck, Albert JM, Peelen LM, Kalkman CJ, Meissner W. Pain intensity on the first day after surgery: a prospective cohort study comparing 179 surgical procedures. Anesthesiology 2013;118:934–44. [DOI] [PubMed] [Google Scholar]
- [27].Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012;11:O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Glare P, Aubrey KR, Myles PS. Transition from acute to chronic pain after surgery. Lancet 2019;393:1537–46. [DOI] [PubMed] [Google Scholar]
- [29].Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabási A-L. The human disease network. PNAS 2007;104:8685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Grabowski P, Hesse S, Hollizeck S, Rohlfs M, Behrends U, Sherkat R, Tamary H, Ünal E, Somech R, Patıroğlu T, Canzar S, van der Werff Ten Bosch J, Klein C, Rappsilber J. Proteome analysis of human neutrophil granulocytes from patients with monogenic disease using dataindependent acquisition. Mol Cell Proteomics 2019;18:760–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Grenald SA, Young MA, Wang Y, Ossipov MH, Ibrahim MM, Largent-Milnes TM, Vanderah TW. Synergistic attenuation of chronic pain using mu opioid and cannabinoid receptor 2 agonists. Neuropharmacology 2017;116:59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Guney E, Menche J, Vidal M, Barábasi A-L. Network-based in silico drug efficacy screening. Nat Commun 2016;7:10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hebeda CB, Sandri S, Benis CM, Paula-Silva Md, Loiola RA, Reutelingsperger C, Perretti M, Farsky SHP. Annexin A1/formyl peptide receptor pathway controls uterine receptivity to the blastocyst. Cells 2020;9:1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hooijmans CR, Tillema A, Leenaars M, Ritskes-Hoitinga M. Enhancing search efficiency by means of a search filter for finding all studies on animal experimentation in PubMed. Lab Anim 2010;44:170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hui X, Li H, Zhou Z, Lam KSL, Xiao Y, Wu D, Ding K, Wang Y, Vanhoutte PM, Xu A. Adipocyte fatty acid-binding protein modulates inflammatory responses in macrophages through a positive feedback loop involving c-jun NH2-terminal kinases and activator protein-1*. J Biol Chem 2010;285:10273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jager SE, Pallesen LT, Richner M, Harley P, Hore Z, McMahon S, Denk F, Vaegter CB. Changes in the transcriptional fingerprint of satellite glial cells following peripheral nerve injury. Glia 2020;68:1375–95. [DOI] [PubMed] [Google Scholar]
- [37].Jimenez-Andrade JM, Herrera MB, Ghilardi JR, Vardanyan M, Melemedjian OK, Mantyh PW. Vascularization of the dorsal root ganglia and peripheral nerve of the mouse: implications for chemical-induced peripheral sensory neuropathies. Mol Pain 2008;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jünger MA, Aebersold R. Mass spectrometry-driven phosphoproteomics: patterning the systems biology mosaic. Wiley Interdiscip Rev Dev Biol 2014;3:83–112. [DOI] [PubMed] [Google Scholar]
- [39].Komori N, Takemori N, Kim HK, Singh A, Hwang S-H, Foreman RD, Chung K, Chung JM, Matsumoto H. Proteomics study of neuropathic and nonneuropathic dorsal root ganglia: altered protein regulation following segmental spinal nerve ligation injury. Physiol Genomics 2007;29:215–30. [DOI] [PubMed] [Google Scholar]
- [40].Laedermann CJ, Pertin M, Suter MR, Decosterd I. Voltage-gated sodium channel expression in mouse DRG after SNI leads to re-evaluation of projections of injured fibers. Mol Pain 2014;10:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lee D-G, Woo J-W, Kwok S-K, Cho M-L, Park S-H. MRP8 promotes Th17 differentiation via upregulation of IL-6 production by fibroblast-like synoviocytes in rheumatoid arthritis. Exp Mol Med 2013;45:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lima KM, Vago JP, Caux TR, Negreiros-Lima GL, Sugimoto MA, Tavares LP, Arribada RG, Carmo AAF, Galvão I, Costa BRC, Soriani FM, Pinho V, Solito E, Perretti M, Teixeira MM, Sousa LP. The resolution of acute inflammation induced by cyclic AMP is dependent on annexin A1. J Biol Chem 2017;292:13758–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu JJ, Sharma K, Zangrandi L, Chen C, Humphrey SJ, Chiu Y-T, Spetea M, Liu-Chen L-Y, Schwarzer C, Mann M. In vivo brain GPCR signaling elucidated by phosphoproteomics. Science 2018;360:eaao4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016;165:535–50. [DOI] [PubMed] [Google Scholar]
- [45].Luo Z, Wang H, Fang S, Li L, Li X, Shi J, Zhu M, Tan Z, Lu Z. Annexin-1 mimetic peptide Ac2-26 suppresses inflammatory mediators in LPS-induced astrocytes and ameliorates pain hypersensitivity in a rat model of inflammatory pain. Cell Mol Neurobiol 2020;40:569–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Makowski L, Brittingham KC, Reynolds JM, Suttles J, Hotamisligil GS. The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity: MACROPHAGE expression OF aP2 impacts peroxisome PROLIFERATOR-ACTIVATED receptor γ and IκB kinase activities*. J Biol Chem 2005;280:12888–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci 2011;36:320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mueller A, Starobova H, Morgan M, Dekan Z, Cheneval O, Schroeder CI, Alewood PF, Deuis JR, Vetter I. Antiallodynic effects of the selective NaV1.7 inhibitor Pn3a in a mouse model of acute postsurgical pain: evidence for analgesic synergy with opioids and baclofen. PAIN 2019;160:1766–80. [DOI] [PubMed] [Google Scholar]
- [49].Needham EJ, Parker BL, Burykin T, James DE, Humphrey SJ. Illuminating the dark phosphoproteome. Sci Signal 2019;12:eaau8645 [DOI] [PubMed] [Google Scholar]
- [50].Paccola CC, Gutierrez VP, Longo I, Juliano L, Juliano MA, Giorgi R. Antinociceptive effect of the Cterminus of murine S100A9 protein on experimental neuropathic pain. Peptides 2008;29:1806–14. [DOI] [PubMed] [Google Scholar]
- [51].Pei L, Zhang J, ZHAO F, Su T, Wei H, Tian J, Li M, Shi J. Annexin 1 exerts anti-nociceptive effects after peripheral inflammatory pain through formyl-peptide-receptor-like 1 in rat dorsal root ganglion. Br J Anaesth 2011;107:948–58. [DOI] [PubMed] [Google Scholar]
- [52].Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019;47:D442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pogatzki EM, Raja SN. A mouse model of incisional pain. Anesthesiology 2003;99:1023–7. [DOI] [PubMed] [Google Scholar]
- [54].Pogatzki-Zahn E, Segelcke D, Zahn P. Mechanisms of acute and chronic pain after surgery: update from findings in experimental animal models. Curr Opin Anaesthesiol 2018;31:575–85. [DOI] [PubMed] [Google Scholar]
- [55].Pogatzki-Zahn EM, Segelcke D, Schug SA. Postoperative pain-from mechanisms to treatment. Pain Rep 2017;2:e588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Price TJ, Gold MS. From mechanism to cure: renewing the goal to eliminate the disease of pain. Pain Med 2018;19:1525–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Raithel SJ, Sapio MR, LaPaglia DM, Iadarola MJ, Mannes AJ. Transcriptional changes in dorsal spinal cord persist after surgical incision despite preemptive analgesia with peripheral resiniferatoxin. Anesthesiology 2017;128:620–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Reichl S, Segelcke D, Keller V, Jonas R, Boecker A, Wenk M, Evers D, Zahn PK, Pogatzki-Zahn EM. Activation of glial glutamate transporter via MAPK p38 prevents enhanced and long-lasting nonevoked resting pain after surgical incision in rats. Neuropharmacology 2016;105:607–17. [DOI] [PubMed] [Google Scholar]
- [59].Rigaud M, Gemes G, Barabas M-E, Chernoff DI, Abram SE, Stucky CL, Hogan QH. Species and strain differences in rodent sciatic nerve anatomy: implications for studies of neuropathic pain. PAIN 2008;136:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rouwette T, Sondermann J, Avenali L, Gomez-Varela D, Schmidt M. Standardized profiling of the membrane-enriched proteome of mouse dorsal root ganglia (DRG) provides novel insights into chronic pain. Mol Cell Proteomics 2016;15:2152–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature 2011;473:337–42. [DOI] [PubMed] [Google Scholar]
- [63].Segelcke D, Pogatzki-Zahn EM. Pathophysiology of postoperative pain. In: The Senses: A Comprehensive Reference. Amsterdam, the Netherlands: Elsevier Academic Press, 2021. pp. 604-27. [Google Scholar]
- [64].Segelcke D, Pradier B, Pogatzki-Zahn E. Advances in assessment of pain behaviors and mechanisms of post-operative pain models. Curr Opin Physiol 2019;11:85–92. [Google Scholar]
- [65].Spofford CM, Brennan TJ. Gene expression in skin, muscle, and dorsal root ganglion after plantar incision in the rat. Anesthesiology 2012;117:161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Stone A, Grol MW, Ruan MZC, Dawson B, Chen Y, Jiang M-M, Song I-W, Jayaram P, Cela R, Gannon F, Lee BHL. Combinatorial Prg4 and il-1ra gene therapy protects against hyperalgesia and cartilage degeneration in post-traumatic osteoarthritis. Hum Gene Ther 2019;30:225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B (Statistical Methodology) 2002;64:479–98. [Google Scholar]
- [68].Sun EC, Darnall BD, Baker LC, Mackey S. Incidence of and risk factors for chronic opioid use among opioid-naive patients in the postoperative period. JAMA Intern Med 2016;176:1286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Treindl F, Ruprecht B, Beiter Y, Schultz S, Döttinger A, Staebler A, Joos TO, Kling S, Poetz O, Fehm T, Neubauer H, Kuster B, Templin MF. A bead-based western for high-throughput cellular signal transduction analyses. Nat Commun 2016;7:12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2014;18:145 EP. [DOI] [PubMed] [Google Scholar]
- [71].van Boekel RLM, Warlé MC, Nielen RGC, Vissers KCP, van der Sande R, Bronkhorst EM, Lerou JGC, Steegers MAH. Relationship between postoperative pain and overall 30-day complications in a broad surgical population: an observational study. Ann Surgery 2017;269:856–65. [DOI] [PubMed] [Google Scholar]
- [72].van den Heuvel I, Reichl S, Segelcke D, Zahn PK, Pogatzki-Zahn EM. Selective prevention of mechanical hyperalgesia after incision by spinal ERK1/2 inhibition. Eur J Pain 2015;19:225–35. [DOI] [PubMed] [Google Scholar]
- [73].Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immun 2018;9:1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wang Y, Wu J, Guo R, Zhao Y, Zhang M, Chen Z, Wu A, Yue Y. Surgical incision induces phosphorylation of AMPA receptor GluR1 subunits at Serine-831 sites and GluR1 trafficking in spinal cord dorsal horn via a protein kinase Cγ-dependent mechanism. Neuroscience 2013;240:361–70. [DOI] [PubMed] [Google Scholar]
- [75].Wellbrock C, Arozarena I. The complexity of the ERK/MAP-Kinase pathway and the treatment of melanoma skin cancer. Front Cell Dev Biol 2016;4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Willemen HLDM, Eijkelkamp N. Sensory signaling pathways in inflammatory and neuropathic pain. In: Wood JN, editor. The Oxford handbook of the neurobiology of pain. New York, NY: Oxford University Press, 2020. [Google Scholar]
- [77].Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, Houten SM, Amariuta T, Wolski W, Zamboni N, Aebersold R, Auwerx J. Systems proteomics of liver mitochondria function. Science 2016;352:aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Woolf CJ. Capturing novel non-opioid pain targets. Biol Psychiatry 2020;87:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Yamakita S, Horii Y, Takemura H, Matsuoka Y, Yamashita A, Yamaguchi Y, Matsuda M, Sawa T, Amaya F. Synergistic activation of ERK1/2 between A-fiber neurons and glial cells in the DRG contributes to pain hypersensitivity after tissue injury. Mol Pain 2018;14:1744806918767508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yang P, Humphrey SJ, Cinghu S, Pathania R, Oldfield AJ, Kumar D, Perera D, Yang JYH, James DE, Mann M, Jothi R. Multi-omic profiling reveals dynamics of the phased progression of pluripotency. Cell Syst 2019;8:427–45.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhan X, Cheng J, Huang Z, Han Z, Helm B, Liu X, Zhang J, Wang T-F, Ni D, Huang K. Correlation analysis of histopathology and proteogenomics data for breast cancer. Mol Cell Proteomics 2019;18(8 suppl 1):S37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zimmermann M. Ethical guidelines for investigations of experimental pain in conscious animals. PAIN 1983;16:109–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B274 and http://links.lww.com/PAIN/B275.