ABSTRACT
The ongoing COVID-19 pandemic has led to more than 159 million confirmed cases with over 3.3 million deaths worldwide, but it remains mystery why most infected individuals (∼98%) were asymptomatic or only experienced mild illness. The same mystery applies to the deadly 1918 H1N1 influenza pandemic, which has puzzled the field for a century. Here we discuss dual potential properties of the 1918 H1N1 pandemic viruses that led to the high fatality rate in the small portion of severe cases, while about 98% infected persons in the United States were self-limited with mild symptoms, or even asymptomatic. These variations now have been postulated to be impacted by polymorphisms of the sialic acid receptors in the general population. Since coronaviruses (CoVs) also recognize sialic acid receptors and cause severe acute respiratory syndrome epidemics and pandemics, similar principles of influenza virus evolution and pandemicity may also apply to CoVs. A potential common principle of pathogen/host co-evolution of influenza and CoVs under selection of host sialic acids in parallel with different epidemic and pandemic influenza and coronaviruses is discussed.
KEYWORDS: Influenza and coronavirus, 1918 H1N1 pandemic, sialic acid receptor, polymorphism, co-evolution
Introduction
Influenza A viruses (IAVs) are widespread pathogens that cause annual seasonal epidemics and irregular, unpredictable pandemics with severe pathogenic effects and high fatality rates. Six IAV pandemics with three HA subtypes (H1, H2, and H3) have been documented in the past century, including the deadliest 1918 H1N1 pandemic that claimed ∼50 million lives globally. IAVs also infect wild and domestic animals with wild waterfowl as the natural reservoir. All known pandemic and other human, mammalian and poultry IAVs are believed to be descended from wild-bird viruses. It is also known that all native avian and many animal IAVs prefer binding sialic acids (Sias) linked to galactoses with an α2,3-linkage (Siaα2,3Gal), whereas human-adapted IAVs prefer the α2,6 linkage (Siaα2,6Gal). However, the application of these rules derived from research of influenza disease and epidemiology to date has been unable to explain many unusual epidemics, the high fatality pattern of the 1918 H1N1 pandemic [1–6], and other influenza pandemics in the past. This knowledge gap in understanding of major pandemics, such as the 1918 H1N1 pandemic, as well as the current COVID-19 pandemic, complicates development of strategies to control and prevent future influenza epidemics and pandemics.
Here we discuss a dual mutational mechanism of influenza virus evolution in humans that has resulted in two human H1 subtypes, a human-like H1 D225 subtype and an avian-like H1 225G subtype, each with distinct transmissibility and pathogenicity. We argue that the H1 D225 subtype caused the widespread transmission, while the H1 225G subtype caused high fatality in a small subpopulation of the 1918 H1N1 pandemic, and both subtypes also caused H1N1 seasonal epidemics but with much lower epidemic scales. Specifically, we postulate that the well-adapted human-like H1 D225 subtype originated from an avian virus [7] that adapted to humans by recognizing the 2,6-Sias in the upper respiratory tracts of the general population, which consequently led to high transmissibility and is therefore responsible for the widespread nature of both seasonal epidemics and pandemics with mild illness. On the other hand, the avian-like 225G subtype is descended from the human D225 subtype, which infected a small subset of the human population with a dominant 2,3-Sia in their lower respiratory tracts that resulted in limited transmissibility but led to severe cases with high fatality rates due to lower respiratory tract infections, resulting in pneumonia, which is more common in pandemics. Indeed, quasispecies with D225 and 225G subtypes of the hemagglutinin (HA) genes have been identified in archived specimens of the 1918 H1N1 pandemic victims [7–9] representing a valuable discovery.
Elucidation of the dual subtypes explains both the widespread but mild epidemic cases that occur in the majority of the human population that express 2,6-Sias and the severe cases that occurs in the minority of the same human population with polymorphic 2,3-Sias. This would also help explain pandemic threats posed by other emerging avian HA subtype viruses, such as the H5N1 and H7N9 subtypes. These emerging avian viruses cause usually sporadic epidemics with high fatality rates but with limited human transmission likely only involving in the small subpopulation with the 2,3-Sias in their lower respiratory tracts and thus may represent low pandemic risks. Since coronaviruses (CoVs) also recognize Sias as host receptors [10–12], this new knowledge regarding influenza viruses may help the understanding of CoV-caused epidemics and pandemics under selection of polymorphic human Sia receptors, such as the ongoing COVID-19 pandemic.
The H1 D225 subtype may be responsible for the widespread nature and mild cases of the 1918 H1N1 pandemic
This hypothesis is based on a bidirectional concerted evolution of humans with an upregulation of the 2,6-Sia expression in the upper respiratory tracts of humans as shown by Sia signals in different cell types and body parts of humans compared with the great apes (chimpanzee, bonobo, gorilla, and orangutan) [13]. Considering the facts that the great apes are closest relatives of humans, sharing 97–99% of DNA sequence identity, and that chimpanzees are naturally resistant to the human 1918 H1N1 subtype recognizing the 2,6-Sias [13, 14], it is assumed that the dominant 2,6-Sias in the upper respiratory tracts of modern human populations could be a result of human co-evolution in an arms-race of our ancient hominid ancestors against the deadly avian IAVs or other 2,3-Sia binding pathogens over the past 100–200 thousand years [13], which possibly experienced multiple selective swipes driven by Sia-binding pathogens.
On the other hand, the emergence of the H1 D225 subtype in humans could also have been a countermeasure of the deadly avian viruses that switched from “avian-like” to a “human-like” Sia-binding properties under selection of the 2,6-Sias in humans in the upper respiratory tracts, resulting in widespread mild cases in both seasonal epidemics and pandemics. Phylogenetic analyses of the 1918 H1 pandemic viruses showed wide variations of their HA sequences compared with their closest avian relatives [9]. This indicates that complicated mutational mechanisms were involved in evolution and adaptation required for a native avian influenza strain to infect humans [7,15]. This process may have involved complex gene constellations, explaining why only three HA subtype (H1, H2 and H3) from the 18 known avian HA subtypes had successfully adapted to humans and became human endemic. This scenario could also explain why many avian HA subtypes that emerge in humans cause severe epidemics with high fatality rates mainly in the small subpopulation of humans who express 2,3-Sias in their lower respiratory tract. Avian H5N1 and H7N9 subtypes are such examples, usually with limited human to human transmissibility and therefore they represent low risks of pandemic threats (see below). Finally, since a few animal species, such as domestic pigs and ferrets, also express 2,6-Sias in their upper respiratory tracts, it would not be surprising to detect 1918 H1 viruses in these animal species, suggesting that the 2009 H1N1 pandemic could have originated in humans without a need of an animal intermediator.
The H1 225G subtype viruses are responsible for the severe pathogenicity and high fatality of the 1918 pandemic
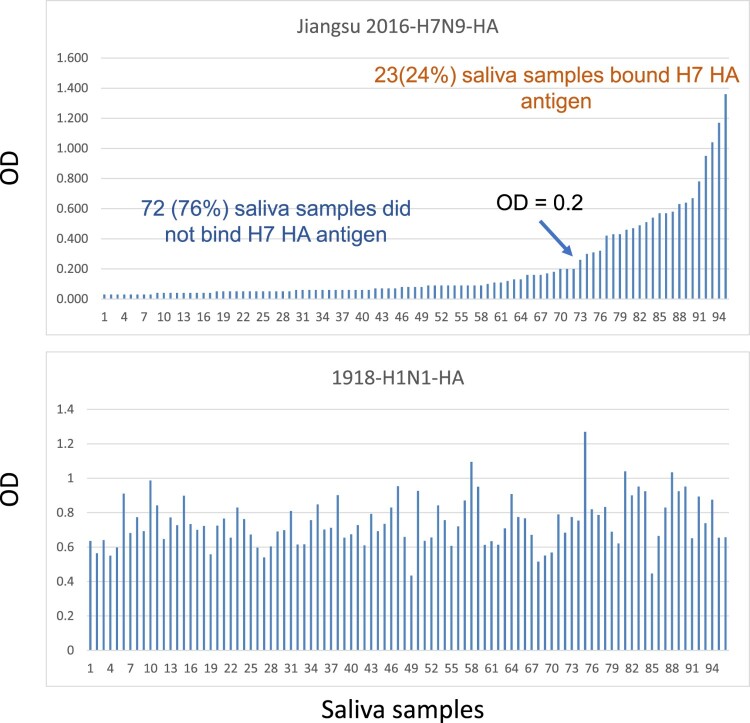
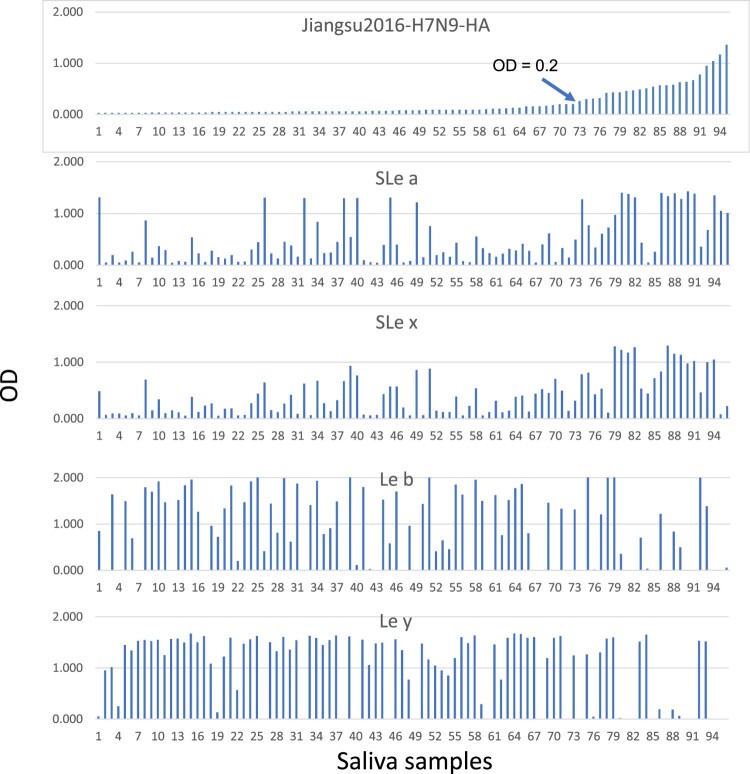
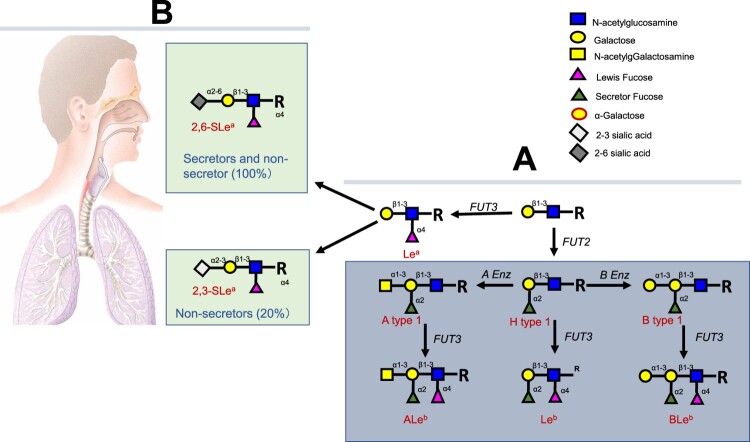
We first formulated the hypothesis of polymorphic 2,3-Sias in humans after finding that only ∼30% of human subjects revealed ELISA binding signals in their saliva specimens by a recombinant HA of the avian H7N9 viruses that bind 2,3- but not 2,6-linked Sias [16]. Our further studies of saliva specimens from a panel of healthy adults confirmed the results with ∼24% of the saliva panel binding to the H7 HA antigens but, by comparison, 100% of the saliva panel revealed binding signals with a recombinant 1918 H1 antigen that is known to recognize 2,6-Sias (Figure 1). The saliva H7 binding profiles correlated with the secretor status of the saliva sample donors and reciprocal correlations were observed between secretors and non-secretor donors (Figure 2). Around 80% of the general population are secretors and ∼20% are non-secretors in the North America and European countries. The saliva donors’ secretor types are controlled by the FUT2 gene encoding a fucose-transferase responsible for synthesis of fucose containing oligosaccharides in the ABO, Lewis and secretor (H) histo-blood group antigen (HBGA) families, in which the FUT2 fucose transferase catalysing the addition of a fucose with a 1,2-linkage to the galactose (Figure 3). Thus, lack of saliva binding by the H7 HA antigen in the majority secretor donors is a result of the preference of the H7 HA antigen to 2,3-Sias. The synthesis of the 2,3-Sias is blocked in the majority secretor population due to steric interference by the 1,2-linked fucose, and this leads to the sterically allowed synthesis of the 2,6-Sias in the majority secretor population. This observation leads to a hypothesis of dual causes of influenza pandemics with a human-like H1 subtype infecting most of the human population with dominant 2,6-Sias in their upper respiratory tracts responsible for the high transmissibility and widespread nature and another avian-like H1 subtype infecting a small subset of human population with a dominant 2,3-Sias in the lower respiratory tracts that were responsible for the severe cases and high fatality rates of the 1918 pandemic.
Figure 1.
Binding assays of H7N9 and 1918 H1N1 HAs with saliva samples from a panel of health adults. Saliva samples from healthy employees in the Cincinnati Children’s Hospital Medical Center collected for studies of norovirus and rotavirus binding profiles of host histo blood group antigens in our previous studies [17,18] were used. Recombinant H7N9 and 1918 H1N1 HAs were constructed and expressed as described previously [16]. The 95 tested saliva sample donors were sorted based on the OD binding signals of H7N9 HA (top panel).
Figure 2.
Saliva binding profiles of H7N9 HA in association with the sialic acid types of saliva donors. The binding signals of sialic Lewis a (SLe a), sialic Lewis x (SLe x), Lewis b (Le b) and Lewis y (Le y) in the saliva samples were performed ELISA using commercial monoclonal antibodies described previously [13]. The 95 tested saliva sample donors were sorted based on the OD binding signals of H7N9 HA (top panel).
Figure 3.
Synthesis of the ABH and Lewis histo-blood group antigens and deduced blocking or masking of synthesis of 2,3-sialic acid antigens by preoccupied 1,2-linked fucose by the H fucose-transferase encoded by the FUT 2 gene in humans. (A) Biosynthesis of type 1 based HBGAs. Synthesis proceeds by stepwise addition of monosaccharide units from a precursor disaccharide present at the terminus of glycan chains from either O-linked or N-linked glycans of glycoproteins, or from glycolipids (R) [19]. (B) Deduced blocking or masking of synthesis of the 2,3-sialic acid antigens by preoccupied 1,2-linked fucose synthesized by the H fucose-transferase encoded by the FUT 2 gene in humans following a study of H7N9-infected patients involved in an outbreak in China in season 2016/17. Around 80% of the general populations are secretor positive and the expression of the 2,3-linked sialic acids are blocked or masked by the preoccupied 1,2-linked fucose, suggesting that secretors may be naturally resistant to H7N9 IAVs because they mainly express the 2, 6-linked sialic acids in the upper respiratory tracts and may not express or express low amounts of 2,3-linked sialic acids in their lower respiratory tracts. The 20% non-secretors may also express 2,6-linked sialic acids in the upper respiratory tracts but mainly express the 2,3-linked sialic acids in the lower respiratory tracts.
The scenario of dual causes of influenza epidemics and pandemics also explains the unusual epidemic patterns observed during the 1918 H1N1 pandemic. For example, three epidemic waves occurred in the 1918 H1N1 pandemic with a much more severe fall of 1918 wave compared to the 1918 spring wave that could have been due to a low start of the circulating 225G subtype in the spring wave because the 225G viruses may have suffered low survival during the off-season and needed time to build up circulation by continual accumulation with the new emerging 225G variants over the course of the pandemic. On the other hand, the high mortality rates of young adults in the 1918 pandemic could have been due to a lack of pre-existing antibodies against the D225 H1 and 225G subtypes due to dominant H3N2 pandemics and epidemics [20] during their childhood resulting in the W-shaped age mortality compared with other age groups [1,2,20,21], although increased mortality rates of young adult soldiers in World War I could also have contributed to the W shape of age-specific fatalities. Furthermore, the reason why the elderly populations had a relative lower mortality rate than expected could also be due to their exposures in early life and therefore they may have obtained immunity against the H1N1 subtype viruses. This was demonstrated by the detection of protective antibodies against the 1918 H1N1 subtype viruses in seniors who survived the 1918 pandemic [22, 23].
Mutant H1 D225G subtype viruses also have been identified in clinical specimens from the 2009 H1N1 pandemic [24, 25]. Such H1 225G variants were detected more frequently in specimens collected from the lower respiratory tracts than in the upper respiratory tracts in severe cases and in cases with serious clinical outcomes and death [24]. Mutant 225G HAs bind a broader range of 2,3-linked secretor sequences of a type expressed on ciliated bronchial epithelial cells and on epithelia within the lung [26]. Evidence of D225G mutant viruses arisen de novo in vivo has also been suggested by a study of an immunocompromised human case by comparison of 225G variants that appeared in specimens harvested at different stages of clinical illness and in ferret models [27] following challenge of mixed D225/225G inoculum or cloned 225G isolates [27]. These findings support the idea that the 2009 H1N1 pandemic subtype is a descendent of the D225 subtype of the 1918 pandemic with a newly emerging H1 225G subtype arisen de novo in humans and triggered or strengthened the 2009 pandemic with or without an animal intermediator. Thus, while the D225 subtype is well-adapted to humans and causes upper respiratory tract infections with mild illness in the general human population expressing the 2,6-Sias, the 225G subtype mainly causes severe lower respiratory tract infections and pneumonia in the subpopulation with dominant 2,3-Sias in their lower respiratory tracts and is mainly responsible for the observed high fatality rates. This principle may apply to all H1N1 caused pandemics that have occurred in the past century and similar cycles of seasonal epidemics and occasional pandemics that are expected to continue.
Impact on disease control and prevention of influenza epidemics and pandemics
The new understanding of influenza virus evolution under selection of polymorphic Sia receptors in humans presented here helps explain many heretofore unanswered questions concerning the unusual epidemic patterns and high fatality rates of influenza pandemics. This new knowledge may impact future vaccine development against influenza epidemics and pandemics. For example, since the well-adapted 1918 human H1N1 subtype is the mainstream of viruses causing both seasonal infections and pandemics and is under strong positive selection by the dominant 2,6-Sias in the general human population, a vaccine targeting the antigenic determinants in the HA receptor binding domains may provide long lasting protection to avoid annual booster vaccinations. On the other hand, immune responses to conserved antigenic determinants on HA heads outside the receptor binding domains may also occur and lead to lifelong positive or negative immune effects, known as “antigen imprinting” or “antigenic seniority” [28–30], which also need to be considered in new vaccine development. Future studies to develop highly sensitive and specific assays to measure Sia receptor-specific blocking or neutralizing antibody responses for vaccine efficacy assessment are important and necessary.
The elucidation of the origins and evolutionary paths of different seasonal/pandemic viruses and the new emerging zoonotic avian HA subtypes should also help guide research and strategy development to predict and prepare against future influenza pandemics. For example, the human H1N1 subtype is well adapted possibly by one entrance and became human endemic, thus, the annual seasonal epidemics and occasional pandemic cycles could continue into the future, which may or may not need an animal intermediator. On the other hand, cross-species transmission of many newly emerging avian influenza HA subtypes into humans was permissible to only a small subpopulation expressing the 2,3-Sias, such as the emerging H5N1 and H7N9 subtypes, and these viruses may not pose pandemic threats. Thus, while an avian vaccine is important for protection of the poultry industry, a human vaccine may not be necessary; instead a screening for 2,3-Sia positive individuals and prohibit them to work with poultry may be an effective strategy. Establishment of Sia-type identity in the general population may also help disease control and prevention against many other Sia-recognized viral pathogens.
The same principle of influenza virus evolution under selection of polymorphic 2,3-Sias in humans may also apply to coronavirus-caused epidemics and pandemics
Based on the new understanding of influenza virus evolution under control of the polymorphism of 2,3-linked Sias in humans, we propose that the same principle may also apply to CoV-caused epidemics and pandemics because CoVs also recognize Sia receptors [11, 12, 31–33]. CoVs also infect wild and domestic animals with wild bats as the natural reservoir and known human and many mammalian CoVs are believed to be descended from bats. It is also known that some animal CoVs prefer binding Sias linked to galactose with a α2,3-linkage (Siaα2,3Gal), however, at least one human-adapted CoV subtype, the human-adapted HCoV-OC43, prefers the α2,6 linkage (Siaα2,6Gal) [12]. By comparison of the seven CoV strains that emerged in humans in the past century (Table 1), we propose a convergent evolutionary relationship between CoVs and influenza viruses under selection of Sia receptors as a common niche, with individual sub-lineages emerging in parallel between the two viral species and causing subtype-specific seasonal epidemics and pandemics.
Table 1.
Deduced sialic acids and their glycan linkages as receptors for selected human and zoonotic IAVs and CoVs.
| Virus | Receptor | Glycan linkage | Clinical infections and epidemics |
|---|---|---|---|
| IAVs | |||
| H1N1 | Neu5Ac-Sias | α2,6-linkage* | Seasonal and pandemic flu, 1918, 2009 |
| H2N2 | Neu5Ac-Sias | α2,6-linkage | Seasonal and pandemic flu 1957 |
| H3N2 | Neu5Ac-Sias | α2,6-linkage | Seasonal and Pandemic flu 1968 |
| H5N1 | Neu5Ac-Sias | α2,3-linkage? | Avian flu 1997– |
| H7N9 | Neu5Ac-Sias | α2,3-linkage | Avian flu 2013– |
| CoVs | |||
| HCoV-OC43 | 9-O-AC-Sias | α2,6-linkage | Endemic in human, mild respiratory tract infections |
| HCoV-HKU1 | 9-O-AC-Sias | α2,6-linkage? | Endemic in human, mild respiratory tract infections |
| HCoV-NL63 | 9-O-AC-Sias | α2,6-linkage? | Endemic in human, mild respiratory tract infections |
| HCoV-229E | 9-O-AC-Sias | α2,6-linkage? | Endemic in human, mild respiratory tract infections |
| SARS-CoV | Neu5Ac-Sias | α2,3-linkage? | Severe acute respiratory syndromes 2002–2003 |
| SARS-CoV2 | Neu5Ac-Sias | α2,6-linkage?* | Severe acute respiratory syndromes 2019– |
| MERS-CoV | Neu5Ac-Sias | α2,3-linkage | Middle East respiratory syndromes 2012–2014 |
Notes: ? Indicating deduced sialic acids and types of glycan linkages based on our preliminary studies and rationale literatures. * Indicating also recognizes α2,3-linkage.
For example, the four strains of human CoVs, HCoV-NL63, HCoV-229E (α-coronaviruses) and HCoV-OC43 and HCoV-HKU1 (β-coronaviruses) that are endemic, i.e. well-tolerated in humans, and account for up to 30% of mild respiratory tract infections, recognize 9-O-AC modified sialic acids (9-O-AC-Sias) with 2,6-linkages (Table 1) [12]. This situation is similar to the seasonal influenza C viruses (ICVs) that also recognize 9-O-AC-Sias with 2,6-linkages and only cause mild infections associated with the common cold. In addition, the Middle East respiratory syndrome coronaviruses (MERS-CoVs) that emerged in the Arabian Peninsula in 2012, which caused recurrent sporadic outbreaks in humans with a fatality rate of 35%, were introduced to humans via domestic dromedary camels and caused only sporadic cases until 2017 with limited human-to-human transmission [11, 12]. This situation is very similar to the zoonotic avian H7N9 IAVs that were transmitted to humans through domestic poultry and caused sporadic cases in outbreaks for five seasons in China since 2012 before being contained in 2017 [34]. Interestingly, both MERS and H7N9 recognize unmodified sialic acids (Neu5Ac-Sias) with 2,3-glycan linkages [11] (Table 1) and are of animal origins. This may explain their limited human-to-human transmission and sporadic cases with severe illness and high human fatality rates, which may again be due to recognition of the less common 2,3-linked Sias in lower respiratory tracts that occur in just ∼20% of the human population. Similarly, the SARS-CoV that emerged in 2002 and was responsible for an epidemic that spread to five continents with a fatality rate of ∼10% before being contained in 2003, could also be explained by the same mechanism of cross-species transmission as occurred in the MERS CoVs.
On the other hand, the continuation of the COVID-19 pandemic caused by SARS-CoV-2 is very similar to the 1918 H1N1 pandemic, and therefore may share a similar principle of a zoonotic source and a cross-species pathway for transmission as occurred in the 1918 H1N1 influenza pandemic that was widespread due to high transmissibility and had a high fatality rate. Limited evidence supporting this hypothesis includes the finding that the SARS-CoV-2 recognizes non-modified Sias [35, 36] consistent with their close genetic relatedness with the SARS- and MERS-CoVs [36–38] that also recognize non-modified Sias (for MERS-CoVs, [39]). Thus, we propose that these three CoVs may have a common ancestor and share similar evolutionary pathways of serial mutations and adaptations that occurred with the influenza seasonal and pandemic viruses, but with two new CoV subtypes; one with a gained human-like 2,6-Sia binding property with high transmissibility responsible for the asymptomatic and mild cases occurring in the majority secretor general population, and the other subtype with an animal 2,3-Sia binding property and mainly circulating in the small non-secretor subpopulation with dominant 2,3-Sias in their lower respiratory tracts, but that has been responsible for the severe cases and high fatality rate seen in the COVID-19 pandemic. Like the 1918 H1N1 pandemic, the deduced new CoV subtypes also may have existed in humans for some time, and possibly was responsible for several low-level seasonal epidemics before the current pandemic surfaced in the fall of 2019. An estimated 2–15 year pre-pandemic emergence period has been suggested for the occurrence of the pandemic in 1918 [7, 15], suggesting that COVID-19 pandemic viruses could also have existed in humans after the initial cross-species introduction to humans and underwent multiple low-level seasonal epidemics before the occurrence of the pandemic in the fall of 2019. Thus, due to the expected timeline required for pre-pandemic emergence, it is highly unlikely that the SARS-CoV-2 viruses were originated from laboratory manipulation, rather than by natural selection in humans following zoonotic transfer.
Finally, CoVs also recognize protein receptors, with the SARS-CoV-2 recognizing the host angiotensin-converting enzyme 2 (ACE2) via the viral spike proteins and employ the cellular serine protease TMPRSS2 for priming as the major cellular entry point. Variations in expression of these host proteins could influence the entry of SARS-CoV-2 viruses into the host cells and thus affect the transmissibility and even severity of the disease. In addition, variability in innate immune system components among humans also may contribute to the heterogenous disease courses of COVID-19 patients [40] which also need to be considered in clinical and epidemiological assessments for strategy development in control of the COVID-19 and prevention of future CoV epidemics and pandemics.
Conclusion and perspective
The elucidation of the polymorphic Sias in the modern human population significantly improved our understanding on the widespread natures and high fatality in only a small subpopulation of influenza epidemics and pandemics which will impact future strategy for influenza disease and epidemic control and prevention. Vaccines targeting the highly conserved antigenic determinants in the receptor binding domains on the HA head to the unanimous 2,6-Sias in the general population would likely induce long-lasting immune protection without the need of the annual booster. The deduced long-term immune protection may help the reassessment of the “original antigen sin” or “antigen imprinting” phenomenon for better understanding of adaptive immunity against influenza viruses for better vaccine designs. New understanding of the emerging avian influenza H5N1 and H7N9 subtype-caused epidemics in only a small subpopulation with dominant 2,3-Sias may provide the means to develop a rational strategy for epidemic control and prevention in the poultry industry and targeted human protection of the susceptible individuals. The same principles of evolution, disease patterns, and epidemiology of influenza viruses may also apply to CoVs with possible similar strategies between the SARS-CoV-2 and influenza H1N1 and between SARS-/MERS-CoVs and H5N1/H7N9, respectively.
Our hypothesis to explain influenza and coronavirus evolution and pandemicity remains preliminary particularly for the CoV-caused epidemics and pandemics and further studies are required to collect solid evidence to support our hypothesis. The most important study is to verify the association of susceptibility and severity of illness in CoV infected patients with their genetic and phenotypic makeup of the 2,6- vs. 2,3-Sias in their upper vs. lower respiratory tracts through large population studies of CoV-infected patients with different clinical outcomes in different countries and ethnic populations. In addition, determination of the distributions of the 2,3- and 2,6-Sias in different ethnic populations and age groups in different countries of the world population will be important to guide strategy and policy-making decisions made to control and prevent influenza and coronavirus-caused epidemics and pandemics in the future. These studies are also important for understanding many other viral and microorganism pathogens that also recognize Sia receptors [41–44].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Taubenberger JK. The origin and virulence of the 1918 “Spanish” influenza virus. Proc Am Philos Soc. 2006;150(1):86–112. [PMC free article] [PubMed] [Google Scholar]
- 2.Taubenberger JK, Morens DM.. Influenza viruses: breaking all the rules. mBio. 2013;4(4):e00365-13. Doi: 10.1128/mBio.00365-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM, Fauci AS.. The next influenza pandemic: can it be predicted? JAMA. 2007;297(18):2025–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morens DM, Fauci AS.. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis. 2007;195(7):1018–1028. [DOI] [PubMed] [Google Scholar]
- 5.Morens DM, Taubenberger JK, Fauci AS.. Pandemic influenza viruses–hoping for the road not taken. N Engl J Med. 2013;368(25):2345–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morens DM, Taubenberger JK.. Making universal influenza vaccines: lessons from the 1918 pandemic. J Infect Dis. 2019;219(Suppl_1):S5–S13. Doi: 10.1093/infdis/jiy728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reid AH, Fanning TG, Hultin JV, et al. . Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci USA. 1999;96(4):1651–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheng Z-M, Chertow DS, Ambroggio X, et al. . Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc Natl Acad Sci USA. 2011;108(39):16416–16421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid AH, Janczewski TA, Lourens RM, et al. . 1918 influenza pandemic caused by highly conserved viruses with two receptor-binding variants. Emerging Infect Dis. 2003;9(10):1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunkel F, Herrler G.. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology. 1993;195(1):195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Hulswit RJG, Widjaja I, et al. . Identification of sialic acid-binding function for the middle east respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci USA. 2017;114(40):E8508–E8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tortorici MA, Walls AC, Lang Y, et al. . Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019;26(6):481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagneux P, Cheriyan M, Hurtado-Ziola N, et al. . Human-specific regulation of alpha 2-6-linked sialic acids. J Biol Chem. 2003;278(48):48245–48250. [DOI] [PubMed] [Google Scholar]
- 14.Snyder MH, Stephenson EH, Young H, et al. . Infectivity and antigenicity of live avian-human influenza A reassortant virus: comparison of intranasal and aerosol routes in squirrel monkeys. J Infect Dis. 1986;154(4):709–711. [DOI] [PubMed] [Google Scholar]
- 15.Smith GJD, Bahl J, Vijaykrishna D, et al. . Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci USA. 2009;106(28):11709–11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan M, Cui L, Huo X, et al. . Saliva as a source of reagent to study human susceptibility to avian influenza H7N9 virus infection. Emerg Microbes Infect. 2018;7(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang P, Farkas T, Zhong W, et al. . Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol. 2005;79(11):6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang P, Farkas T, Marionneau S, et al. . Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J Infect Dis. 2003;188(1):19–31. [DOI] [PubMed] [Google Scholar]
- 19.Marionneau S, Cailleau-Thomas A, Rocher J, et al. . ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie. 2001;83(7):565–573. [DOI] [PubMed] [Google Scholar]
- 20.Taubenberger JK, Hultin JV, Morens DM.. Discovery and characterization of the 1918 pandemic influenza virus in historical context. Antivir Ther. 2007;12(4 Pt B):581–591. [PMC free article] [PubMed] [Google Scholar]
- 21.Taubenberger JK, Morens DM.. 1918 influenza: the mother of all pandemics. Emerging Infect Dis. 2006;12(1):15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viboud C, Eisenstein J, Reid AH, et al. . Age- and sex-specific mortality associated with the 1918–1919 influenza pandemic in Kentucky. J Infect Dis. 2013;207(5):721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worobey M, Han GZ, Rambaut A.. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci USA. 2014;111(22):8107–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan PKS, Lee N, Joynt GM, et al. . Clinical and virological course of infection with haemagglutinin D222G mutant strain of 2009 pandemic influenza A (H1N1) virus. J Clin Virol. 2011;50(4):320–324. [DOI] [PubMed] [Google Scholar]
- 25.Piralla A, Pariani E, Giardina F, et al. . Molecular characterization of influenza strains in patients admitted to intensive care units during the 2017–2018 season. Int J Mol Sci. 2019;20(11):2664. Doi: 10.3390/ijms20112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Childs RA, Matrosovich T, et al. . Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J Virol. 2010;84(22):12069–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Memoli MJ, Bristol T, Proudfoot KE, et al. . In vivo evaluation of pathogenicity and transmissibility of influenza A(H1N1)pdm09 hemagglutinin receptor binding domain 222 intrahost variants isolated from a single immunocompromised patient. Virology. 2012;428(1):21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry C, Palm AKE, Krammer F, et al. . From original antigenic sin to the universal influenza virus vaccine. Trends Immunol. 2018;39(1):70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ranjeva S, Subramanian R, Fang VJ, et al. . Age-specific differences in the dynamics of protective immunity to influenza. Nat Commun. 2019;10(1):1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang A, Stacey HD, Mullarkey CE, et al. . Original antigenic sin: how first exposure shapes lifelong anti-influenza virus immune responses. J Immunol. 2019;202(2):335–340. [DOI] [PubMed] [Google Scholar]
- 31.Huang X, Dong W, Milewska A, et al. . Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol. 2015;89(14):7202–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hulswit RJG, Lang Y, Bakkers MJG, et al. . Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci USA. 2019;116(7):2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlasak R, Luytjes W, Spaan W, et al. . Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA. 1988;85(12):4526–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun X, Belser JA, Pappas C, et al. . Risk assessment of fifth-wave H7N9 influenza A viruses in mammalian models. J Virol. 2018;93(1):e01740-18. Doi: 10.1128/JVI.01740-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker AN, Richards S-J, Guy CS, et al. . The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent Sci. 2020;6(11):2046–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Awasthi M, Gulati S, Sarkar DP, et al. . The sialoside-binding pocket of SARS-CoV-2 spike glycoprotein structurally resembles MERS-CoV. Viruses. 2020;12(9):909. Doi: 10.3390/v12090909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrosillo N, Viceconte G, Ergonul O, et al. . COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020;26(6):729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benvenuto D, Giovanetti M, Ciccozzi A, et al. . The 2019-new coronavirus epidemic: evidence for virus evolution. J Med Virol. 2020;92(4):455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park Y-J, Walls AC, Wang Z, et al. . Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat Struct Mol Biol. 2019;26(12):1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultze JL, Aschenbrenner AC.. COVID-19 and the human innate immune system. Cell. 2021;184(7):1671–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14(8):351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varki A. Nothing in glycobiology makes sense, except in the light of evolution. Cell. 2006;126(5):841–845. [DOI] [PubMed] [Google Scholar]
- 43.Kim CH. SARS-CoV-2 evolutionary adaptation toward host entry and recognition of receptor O-Acetyl sialylation in virus-host interaction. Int J Mol Sci. 2020;21(12):4549. Doi: 10.3390/ijms21124549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schauer R. Sialic acids: fascinating sugars in higher animals and man. Zoology. 2004;107(1):49–64. [DOI] [PubMed] [Google Scholar]