Sap-sucking aphids are insects of huge agricultural concern, not only because of direct damage caused by feeding but also because of their ability to transmit various plant pathogens. Some bacteria that grow on leaf surfaces, such as Pseudomonas syringae, can infect and kill aphids, making them potentially useful in the biological control of pest aphids.
KEYWORDS: Pseudomonas syringae, aphids, biocontrol, insect-microbe interactions, phyllosphere
ABSTRACT
Interactions between epiphytic bacteria and herbivorous insects are ubiquitous on plants, but little is known about their ecological implications. Aphids are devastating crop pests worldwide, so understanding how epiphytic bacteria impact aphid populations is critically important. Recent evidence demonstrates that plant-associated bacteria, such as Pseudomonas syringae, can be highly virulent to one species of aphid, the pea aphid (Acyrthosiphon pisum). Currently, we have no knowledge on how broad this phenomenon is across diverse aphid species that are of high agricultural concern. In controlled experiments using oral exposure in an artificial diet, we challenged five aphid species of agricultural importance with three strains of P. syringae that vary in virulence to the pea aphid. These strains also vary in epiphytic ability and comprise two phytopathogens and one non-plant-pathogenic strain. In general, differences in virulence to aphids remained relatively constant across strains regardless of the aphid species, except for the bird cherry-oat aphid (Rhopalosiphum padi), which is significantly less susceptible to two P. syringae strains. We demonstrate that lower infection incidence likely plays a role in the reduced susceptibility. Importantly, these data support previous results showing that interactions with epiphytic bacteria are important for aphids and may play a large, but underappreciated, role in insect population dynamics. Our study illustrates a potential role of epiphytic bacteria in the biological control of aphid pests broadly but suggests the need for more research encompassing a greater diversity of pest species.
IMPORTANCE Sap-sucking aphids are insects of huge agricultural concern, not only because of direct damage caused by feeding but also because of their ability to transmit various plant pathogens. Some bacteria that grow on leaf surfaces, such as Pseudomonas syringae, can infect and kill aphids, making them potentially useful in the biological control of pest aphids. However, only one aphid species, the pea aphid (Acyrthosiphon pisum), has been tested for infection by P. syringae. Here, we challenged five aphid species of agricultural importance with three strains of P. syringae that vary in virulence to the pea aphid. We found that four of these aphid species were susceptible to infection and death, suggesting that these bacteria are broadly useful for biological control. However, one aphid species was much more resistant to infection, indicating that more testing on diverse aphid species is needed.
INTRODUCTION
Transient interactions between microbes growing on plant surfaces and herbivorous insect species are ubiquitous in the phyllosphere, the aboveground parts of plants, but are complex and dynamic. Some interactions are mutualistic, some are commensal, where the insect benefits, others where the microbe benefits, and many others are parasitic (1–3). In the case of interactions between epiphytic bacteria and insects, insects may serve as vectors for dispersal (4, 5), but there is also evidence that some epiphytes, including several phytopathogens, such as Pseudomonas syringae, can infect and kill agricultural pest insects (6–8). Consequently, ecological interactions between microbes and insects on plants could be harnessed to prevent agricultural losses from pest insects and also protect native plants threatened by invasive pests, but they must first be better understood in order to predict potential outcomes.
Pest insect interactions with potential epiphytic pathogens such as P. syringae have been described in detail for only one species, the pea aphid (Acyrthosiphon pisum) (7–10). A. pisum appears to be exploited by several phytopathogens, including Dickeya dadantii, Pantoea stewartii, Erwinia aphidicola, and P. syringae (6–9). In all of these studies, the bacteria were able to infect and grow to high titers in the insect (usually in the gut) and to also cause death to the host. For P. syringae specifically, we have shown that there is significant variation across strains in their virulence to A. pisum, with 23% to 83% of aphids dead 72 h after oral exposure (11, 12). The frequency with which strains infect aphids from the leaf surface also varies significantly and increases over time, with 12 to 45% of aphids infected at 48 h and 23 to 100% infected after 5 days on experimental plants sprayed with P. syringae strains (12). These interactions with A. pisum are likely to be reflected broadly across other aphid species, as we know that the more distantly related sap-sucking insect, the sweet potato whitefly (Bemisia tabaci), shows similar death rates after ingesting P. syringae strains (11, 13).
P. syringae is a well-studied, diverse, and highly successful environmental bacterium (14, 15). A species complex in the Gammaproteobacteria, P. syringae can be isolated from plant surfaces, soils, leaf litter, and throughout the water cycle (16–18). Although well studied as a plant pathogen, much is still unknown regarding the breadth of host plant use or mechanisms of virulence of most P. syringae strains to insects. Strains are often very phenotypically distinct even though they may be very phylogenetically close (19, 20), and current pathovar designations are not predictive of the potential host plant range of a strain (21). Some strains also have been shown to use aphids as vectors and hosts, with variable levels of virulence and infection (7, 12). However, it is unclear how these interactions affect diverse aphid species and how broadly this phenomenon occurs across different agricultural systems.
There are more than 5,000 species of aphid worldwide, and many are devastating crop pests. Individual species of aphid cause huge economic losses worldwide across crops such as soybean, cruciferous vegetables, and various grains (22). As sap-sucking insects that reproduce rapidly, aphids can cause damage through direct feeding on the phloem of plants (23). However, many species of aphid are also efficient vectors of plant pathogens, particularly viruses and fungi (24, 25). In most agricultural systems, rapidly growing aphid populations are extremely hard to control, and many species of aphid have a high propensity for evolving resistance to chemical pesticides (26, 27). Consequently, in recent decades attention has turned to developing alternative control methods, such as biocontrol techniques employing natural predators and pathogens of aphids. These include parasitoid wasps, predacious larvae, and fungal pathogens to which many species of aphid have known susceptibilities (28–30). However, comparatively little is known about bacterial pathogens of aphids that may have the potential for biocontrol. Epiphytic bacteria in particular may be suitable, as aphids would frequently interact with them on leaf surfaces. Many strains of the bacterium P. syringae grow epiphytically on economically important plants such as fruits, vegetables, and ornamental plants, making it a widely applicable and useful system for exploring biocontrol possibilities (16).
It is almost certain that the vast majority of, if not all, species of aphid encounter epiphytic bacteria while feeding, as some bacterial strains can reach population densities of 105 to 107 cells/cm2 on plant surfaces (16, 31–33). For A. pisum, the relatively low minimum infectious dose of less than 10 cells of either P. syringae or D. dadantii (9, 34) suggests that infection likelihood in the field is high, as such low densities of epiphytic bacteria would be common on plant surfaces. The more highly virulent strains of P. syringae also tend to be strains that grow better epiphytically and, therefore, are likely to be found more frequently on plants (11, 12). This suggests that aphid pathogens like P. syringae do not need to be at high densities on leaves to impact the population dynamics of aphid populations. However, to be able to make useful predictions, data across a larger number of agriculturally important species is needed.
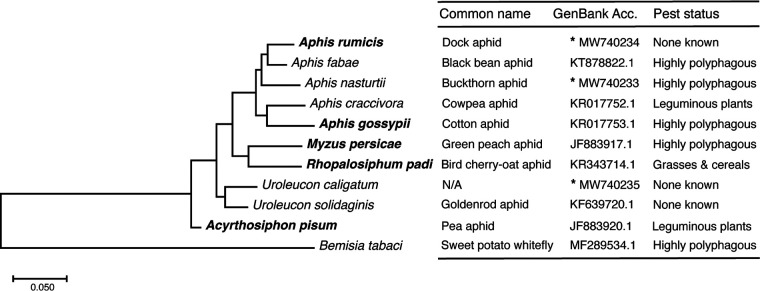
Here, we investigated whether the absolute and relative virulence of three strains of P. syringae (P. syringae pv. aptata DSM50252, pv. syringae B728a, and Cit7) were consistent across five aphid species, including four important pest species. We define virulence as the proportion of aphids dead at the final time point of the assays. Expectations of the virulence of each strain were based on previous results of the virulence of the strains when orally ingested by the pea aphid (11, 12). We also explored whether the likelihood of infection by one strain across different aphid species could impact observed differences in virulence. Of the five aphid species compared here, four are in the top 15 aphid species of most agricultural importance (35): the pea aphid, A. pisum; the cotton aphid, Aphis gossypii; the bird cherry-oat aphid, Rhopalosiphum padi; and the green peach aphid, Myzus persicae. In addition, the dock aphid, Aphis rumicis, was included as an additional species in the genus Aphis, which includes a high density of pest species (Fig. 1). If commonalities could be found across these phylogenetically distant and phenotypically different aphid species, we could better predict the dynamics of these infections in the field.
FIG 1.
Phylogeny and pest capabilities of diverse aphid species. The phylogenetic tree is based on mitochondrial COI sequences, with GenBank accession numbers given in the table. Aphids included in the study are in boldface. An asterisk indicates COI sequences that were generated in the current study (MW740233 to MW740235).
RESULTS
Three bacterial strains were chosen for these assays based on previous results from Smee et al. (11) and also based on their plant pathogenicity and collection source (Table 1). All three were originally isolated from plant surfaces, although this is not necessarily indicative of their potential host range. Two are known phytopathogens yet differed in their expected virulence to aphids: Ptt50252 showed low levels of virulence to A. pisum, whereas PsyB728a showed high levels of virulence (11). The third strain, Cit7, is considered a non-plant pathogen and has been used as a biocontrol strain against phytopathogenic strains (36), but it previously showed high virulence to A. pisum.
TABLE 1.
Summary of Pseudomonas syringae strains included in the assays
Virulence assays.
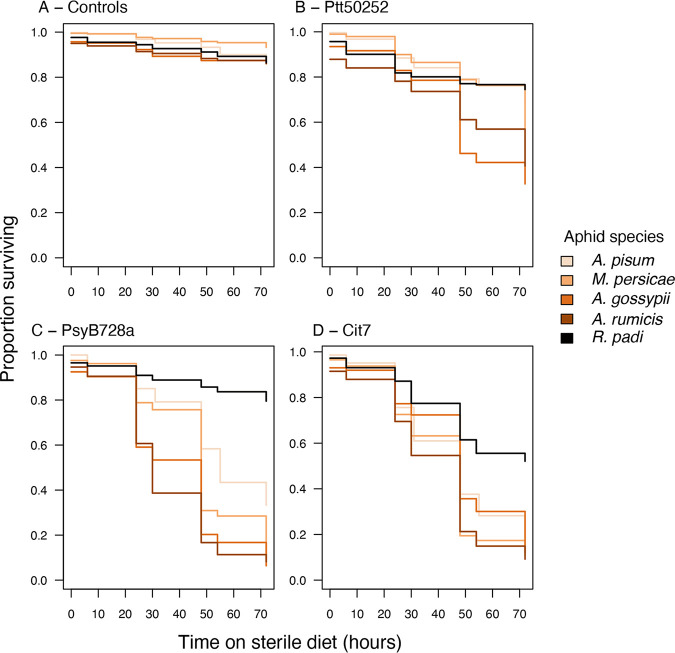
Using in vitro assays with bacterial suspensions in artificial diet, aphid death was recorded at multiple times up to 72 h post-exposure. At the final time point (72 h), there was considerable variation in virulence both for each bacterial strain across aphid species and within each aphid species across the three strains (generalized linear mixed model [GLMM] for interaction between aphid species and bacterial strain: χ212 = 281.41, P < 0.001) (Fig. 2 and Tables 2 and 3). There were no significant differences in mortality for the control treatments, where aphids were fed buffer mixed with diet; more than 85% of aphids survived for all five species (Fig. 2).
FIG 2.
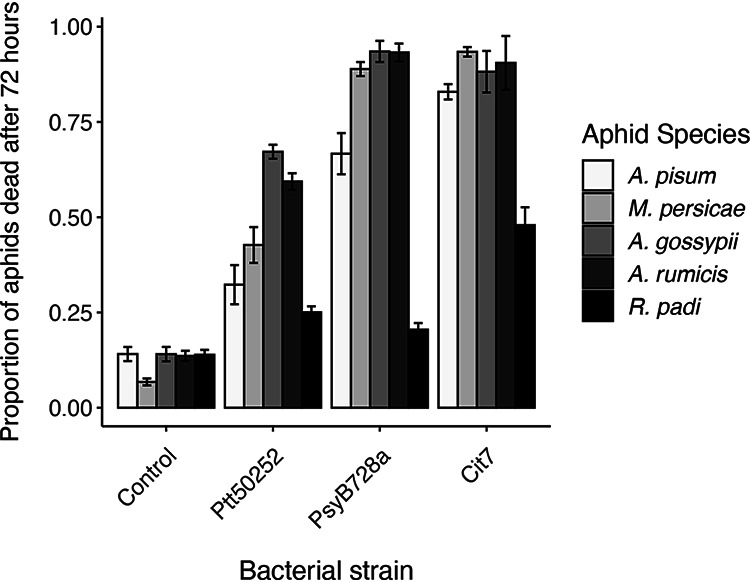
Comparison of the virulence of three P. syringae strains to five aphid species and a control buffer-only treatment. Mean values are plotted ± standard errors. In a GLMM analysis, the interaction between bacterial strain and aphid species was highly significant (χ212 = 281.41, P < 0.001). Post hoc pairwise comparisons are presented in Tables 2 and 3.
TABLE 2.
Post hoc comparisons of virulence between aphid species within a bacterial strain treatment
| Comparison |
P value for: |
|||
|---|---|---|---|---|
| Control | Ptt50252 | PsyB728a | Cit7 | |
| A. pisum-A. gossypii | 1 | <0.0001 | <0.0001 | 1 |
| A. pisum-A. rumicis | 1 | <0.0001 | <0.0001 | 0.167 |
| A. pisum-M. persicae | 0.054 | 0.842 | 0.004 | 0.004 |
| A. pisum-R. padi | 1 | 0.551 | <0.0001 | <0.0001 |
| A. gossypii-A. rumicis | 1 | 1 | 1 | 1 |
| A. gossypii-M. persicae | 0.145 | 0.004 | 0.5763 | 0.441 |
| A. gossypii-R. padi | 1 | <0.0001 | <0.0001 | <0.0001 |
| A. rumicis-M. persicae | 0.11 | 0.089 | 1 | 1 |
| A. rumicis-R. padi | 1 | <0.0001 | <0.0001 | <0.0001 |
| M. persicae-R. padi | 0.113 | 0.029 | <0.0001 | <0.0001 |
TABLE 3.
Post hoc comparisons of virulence between bacterial strains within a single aphid species
| Comparison |
P value for: |
||||
|---|---|---|---|---|---|
| A. pisum | A. gossypii | A. rumicis | M. persicae | R. padi | |
| Control-Ptt50252 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.034 |
| Control-PsyB728a | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.309 |
| Control-Cit7 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Ptt50252-PsyB728a | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 1 |
| Ptt50252-Cit7 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| PsyB728a-Cit7 | 0.318 | 0.061 | 1 | 0.206 | <0.0001 |
The relative virulence of each bacterial strain was generally consistent across all aphid species, with Ptt50252 being the least virulent (25 to 67% mortality) and Cit7 being the most virulent (48 to 93% mortality). Notably, R. padi was more resistant to all P. syringae strains, with no strain causing over 50% mortality to this species. Strain Cit7 showed the highest virulence to R. padi but conferred significantly lower mortality than the other aphid species (Table 2). Responses of R. padi were also unique in that it was the only species in which death rates after feeding on either Ptt50252 or PsyB728a did not differ significantly from death rates in the control treatment or each other (Table 3).
In general, both Aphis species were more susceptible to all three bacterial strains than the other aphid species, with a minimum death rate of 59% of A. rumicis when exposed to Ptt50252. Over 88% of both aphid species died when exposed to either PsyB728a or Cit7. M. persicae was similarly susceptible to PsyB728a and Cit7 but less so to Ptt50252. In general, A. pisum showed lower death rates than the other susceptible species (apart from R. padi) across all bacterial strains, although not always significantly so (Table 2).
Across all time points of the assay, there are evident differences in death rate for aphids exposed to the different bacterial strains (Fig. 3). In particular, R. padi shows different temporal responses to Ptt50252 and PsyB728a despite having very similar final death rates at 72 h of 25% and 21%, respectively. After exposure to PsyB728a, R. padi shows a very slow cumulative death toll compared to the other aphid species, but in response to Ptt50252 there is an initial sharp decline, in line with the other aphid species, which then levels off at later time points (Fig. 3B and C). Despite Cox proportional hazards analysis being quite conservative, the survival rate of R. padi across the 72-h period is significantly higher than those of all the other aphid species for all three bacterial strains, except pea aphids infected with strain Ptt50252 (Table 4).
FIG 3.
Kaplan-Meier survival curves showing the proportion of aphids surviving over a 72-h period after ingesting strains of P. syringae. Data are separated by bacterial treatment: controls (fed buffer mixed with artificial diet) (A), Ptt50252 (B), PsyB728a (C), and Cit7 (D).
TABLE 4.
Survival ranges across bacterial treatments for each aphid speciesa
| Bacterial treatment and aphid species | Zdf=4b | HRc | Survival ranged |
|---|---|---|---|
| Controls | |||
| R. padi | 0.83–0.90 | ||
| A. pisum | 0.23 | 1.03 | 0.84–0.88 |
| M. persicae | −3.97 | 0.48*** | 0.91–0.96 |
| A. gossypii | 0.25 | 1.05 | 0.83–0.90 |
| A. rumicis | 0.26 | 1.05 | 0.83–0.90 |
| Ptt50252 | |||
| R. padi | 0.69–0.80 | ||
| A. pisum | 1.01 | 1.21 | 0.65–0.72 |
| M. persicae | 3.94 | 1.66*** | 0.52–0.63 |
| A. gossypii | 13.40 | 3.44*** | 0.28–0.39 |
| A. rumicis | 8.29 | 2.84*** | 0.35–0.47 |
| PsyB728a | |||
| R. padi | 0.75–0.84 | ||
| A. pisum | 9.39 | 4.48*** | 0.28–0.39 |
| M. persicae | 18.43 | 7.94*** | 0.08–0.15 |
| A. gossypii | 18.35 | 11.39*** | 0.04–0.10 |
| A. rumicis | 20.89 | 12.80*** | 0.06–0.12 |
| Cit7 | |||
| R. padi | 0.47–0.58 | ||
| A. pisum | 5.86 | 2.44*** | 0.13–0.22 |
| M. persicae | 8.28 | 3.23*** | 0.07–0.15 |
| A. gossypii | 3.93 | 2.58*** | 0.09–0.16 |
| A. rumicis | 6.27 | 3.53*** | 0.06–0.13 |
Statistics are based on Cox proportional hazard analysis, and the hazard ratio (HR) is included to indicate effect size compared to R. padi (HR of 1, no effect; <1, a reduction in the hazard; >1, an increase in the hazard).
The Z value is the Wald statistic with 4 degrees of freedom.
Three asterisks indicate the value shows significantly different survival compared to R. padi at P < 0.001.
95% confidence intervals for survival probability at 72 h postexposure.
Infection assay.
Low virulence could be attributed to low incidence of infection if aphids do not easily become infected in a given treatment. We define infection here by our ability to detect a bacterial cell density that we know is sufficient to cause aphid death (34) within aphids that have been orally exposed to bacteria. When this level of infection is detected, we assume these aphids will eventually die, although they do not display symptoms of infection at this time point. We investigated the incidence of infection after ingestion of bacterial suspensions in artificial diet for three of the aphid species. These species (A. pisum, A. rumicis, and R. padi) are phylogenetically diverse yet also showed various levels of susceptibility. Susceptibility to strain PsyB728a was tested, as it displayed the most variation in virulence across the five aphid species.
All three aphid species became infected by PsyB728a, although the mean proportion of infected aphids within a species varied greatly, from 0.15 ± 0.04 (R. padi) to 0.94 ± 0.03 (A. rumicis) (Fig. 4). Also echoing the same pattern as that for virulence, PsyB728a infected A. pisum less frequently and more variably than A. rumicis (0.81 ± 0.10) (Fig. 4). Overall, there was a significant difference in infection frequency across aphid species (analysis of variance [ANOVA]; F2,6 = 47.31, P < 0.001), with only the pairwise comparison between A. pisum and A. rumicis not differing significantly.
FIG 4.
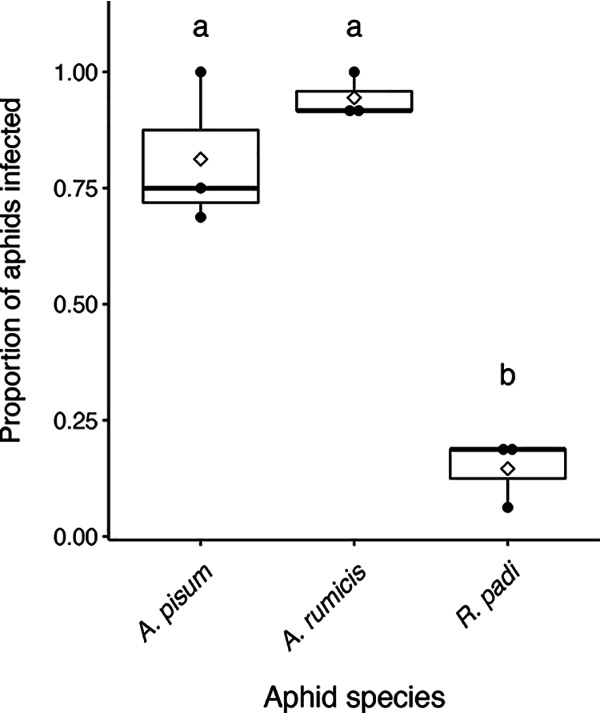
Proportion of aphids infected 24 h after being moved to sterile diet, following 24 h of feeding on artificial diet mixed with PsyB728a. Solid black points show independent data from the three experimental replicates. The solid horizontal line in each boxplot denotes the median, and empty diamonds indicate the means. Post hoc lettering denotes significant differences between aphid species (ANOVA; F2,6 = 47.31, P < 0.001).
DISCUSSION
Three diverse strains of P. syringae that vary in plant pathogenicity and epiphytic growth ability exhibited significant and generally consistent virulence toward five phylogenetically distant aphid species (Fig. 2). Over 20 strains of P. syringae have already been characterized for their virulence to the pea aphid (12), but this is the first study to explore the potential effects on other important aphid species. Given this breadth of pathogenicity across aphid species, we speculate that P. syringae also infects other sap-sucking hemipterans, as previous results found that the sweet potato whitefly (B. tabaci) suffers mortality very similar to that of pea aphids when exposed to some strains of P. syringae (11). Our data further erode the paradigm that bacterial phytopathogens only exploit plant hosts, as more evidence of epiphytic bacteria utilizing insect hosts continues to accumulate.
One species of aphid, R. padi, showed significantly lower susceptibility to the two most virulent strains tested, PsyB728a and Cit7, than the other aphid species. Even more intriguingly, R. padi was not susceptible at all to either PsyB728a or the low-virulence strain Ptt50252. The large difference in susceptibility between R. padi and the other aphid species tested prompts various hypotheses. Compared to aphids such as A. pisum, R. padi is a very small aphid, and the reduced size of its stylet and digestive tract may prevent the ingestion of certain bacteria. Studies on stylet size in whiteflies and aphids have found this to be a factor that influences the insect’s ability to acquire vectored pathogens (37, 38). However, other aphids used in the assay that were highly susceptible to P. syringae (such as A. gossypii and M. persicae) were also small. Therefore, overall body size may not matter for infection here, or it may not correlate well with stylet size. Additionally, R. padi did display a moderate level of susceptibility to the non-plant pathogen Cit7, indicating that some bacterial cells could be picked up orally.
We subsequently determined that the propensity of an aphid species to become infected by different strains of P. syringae could be a determining factor in how virulent a strain is. We show that three aphid species, including R. padi, can ingest bacterial cells and become infected by strain PsyB728a (Fig. 4). However, the proportion of R. padi individuals infected was much lower than that for A. pisum and A. rumicis, which both displayed much higher death rates after oral exposure to PsyB728a. Understanding why different aphid species have more or less propensity to acquire bacterial cells may be pivotal to discovering attributes of successful microbial biocontrol candidates. As strains of P. syringae can vary in cell size and particle size is known to be an important factor for movement through stylets, it is possible that cell size influences infection incidence. We speculate that the higher death rate of R. padi exposed to Cit7 is caused by higher infection incidence, which, in turn, may be due to the size of cells or other properties of Cit7.
Differences in aphid susceptibility may vary across aphid genera, but our data suggest that phylogeny plays an integral role. Two Aphis species were included in the current study to have multiple representatives from this particular genus, which is comprised of many important pest species. Both A. gossypii and A. rumicis showed very similar death rates after exposure to all three bacterial strains (Fig. 2, Table 1), suggesting that susceptibility is common within this genus. Our screening for endosymbionts found that our line of A. rumicis harbored the bacterial symbiont Rickettsiella, which has been demonstrated to provide some protection against fungal pathogens (39) and is involved in pea aphid coloration, changing red aphids to green (40). Due to the very similar mortality obtained for the two Aphis species, we conclude that Rickettsiella had no impact on the interaction of A. rumicis with P. syringae. However, the current study used only one aphid genotype per species. Aphid phenotypes, such as resistance to fungal pathogens and parasitoid wasps, can vary substantially across genotypes within a species (41, 42). Although these results present a general pattern of consistent susceptibility within a genus and even across species, more variation in response would be likely across more aphid genetic diversity.
Other immunological and nonimmunological mechanisms may affect the proportion of aphids succumbing to P. syringae infection and virulence. In assays utilizing the plant pathogen D. dadantii, it seemed that some aphids could regularly resist infection, indicating a partial immune response (43). It is possible that R. padi is less susceptible to P. syringae due to an immune response. Pea aphids have also been shown to avoid strains of P. syringae that are fluorescent (44); however, the current study conducted all feeding assays under UV-blocking plastic in a controlled setting using an artificial diet, so visual detection of bacteria was unlikely. However, it would be of interest to determine if R. padi had some mechanism for avoiding virulent strains, as both vision and olfactory cues have the potential to alert them to the presence of entomopathogenic bacteria.
Aphid species such as M. persicae, shown here to be susceptible to all three P. syringae strains, are phenomenally well adapted to be successful pests. They are extremely polyphagous, efficient virus vectors, and they exhibit great genetic variability across many traits, including the ability to develop resistance to pesticides. Fungal pathogens are already employed in some systems as biocontrol measures against aphid infestation (29), but bacterial pathogens have been largely overlooked until now. Given that some strains of P. syringae are not plant pathogenic but grow well epiphytically and also cause a high incidence of death to many aphid species, there is potential for their use as effective microbial biocontrol agents. Testing a wider phylogenetic range of aphids, and other sap-feeding or chewing herbivores, will allow us to more easily predict the role of epiphytic bacteria on important crop pests and their potential as applied microbial pest control agents.
MATERIALS AND METHODS
Bacterial cultures.
Three bacterial strains were used based on knowledge from previous work by Smee et al. (11, 12): an expected low-virulence phytopathogenic strain (P. syringae pv. aptata DSM50252), a higher virulence phytopathogenic strain (P. syringae pv. syringae B728a), and a non-plant-pathogenic strain (P. syringae Cit7) (Table 1). Cultures were grown and kept on King’s B medium with rifampin (50 ng/μl), as the strains all display resistance to this antibiotic. Culture plates and all experimental plates were kept incubated at 27°C, and overnight cultures were grown in an incubator shaker set at 28°C and 250 rpm. For each experimental assay, bacterial solutions were prepared from an overnight culture that was pelleted and washed in 10 mM MgCl2 twice and then resuspended and diluted to an optical density at 600 nm (OD600) of 0.8.
Aphid collection, identification, and maintenance.
Five species of aphid from the subfamily Aphidinae were used in the study. The pea aphid, Acyrthosiphon pisum, clone CWR09/18, was collected by Angela Douglas in Freeville, NY, in 2009. The cotton aphid, Aphis gossypii, had been maintained on cucumber for more than 20 years and was obtained from Keith Perry (45). The bird cherry-oat aphid, Rhopalosiphum padi, was originally collected by W. F. Rochow (46) and was obtained from Stewart Gray, along with the green peach aphid, Myzus persicae, which was originally collected from a greenhouse infestation approximately 15 to 20 years ago (47). The dock aphid, Aphis rumicis, was collected by Tory Hendry on Rumex sp. in Dryden, NY, in August 2018; a single female was used to establish a colony. Identification of A. rumicis was confirmed by phenotypic traits and cytochrome c oxidase subunit I (COI) barcode sequencing using primers LepF (5′-ATTCAACCAATCATAAAGATATTGG-3′) and LepR (5′-TAAACTTCTGGATGTCCAAAAAATCA-3′). The COI sequence was 100% similar to A. rumicis sequence HQ970784.1 in GenBank. Additionally, in the field and laboratory, these aphids caused curling of Rumex leaves, a feature of A. rumicis.
The A. pisum clone was known to harbor no endosymbionts besides the obligate Buchnera aphidicola. As well as confirming this, the other four aphid species were also screened for secondary symbionts by amplification of the bacterial 16S rRNA gene using the universal primers 10F (5′-AGTTTGATCATGGCTCAGATTG-3′) and 35R (5′-CCTTCATCGCCTCTGACTGC-3′), which detect a wide range of Eubacteria, with the notable exception of the obligate symbiont Buchnera (48, 49). For positive samples, specific diagnostic primers were then used to determine which facultative symbionts were harbored (50). The only occurrence of secondary symbionts was Rickettsiella sp. harbored by A. rumicis.
Clonal populations of each aphid species were kept as follows. A. rumicis on Rumex sp. was grown from seeds obtained from their collection site; A. gossypii was grown on cucumber (Cucumis sativus); M. persicae was grown on turnip (Brassica rapa); A. pisum was grown on fava bean (Vicia faba); and R. padi was grown on barley (Hordeum vulgare). All reproduce parthenogenetically under summer conditions, so all laboratory populations were kept at a light:dark cycle of 16:8 h at 20°C. For experiments, all aphids were age controlled to be 5 to 6 days old.
Virulence assays.
To test the virulence of each strain to the five aphid species, we used oral infection assays with an artificial diet (51). Bacterial solutions at an OD600 of 0.8 were added at a ratio of 1:5 to artificial diet, with 10 mM MgCl2 buffer used for the negative-control treatment. A feeding sachet was created by filling a 96-well plate with the diet, 200 μl per well, and covering the plate with Parafilm. One aphid was placed in each well of a second 96-well plate, and the feeding sachet was inverted and placed above it, so each aphid had access to the mixture of diet and bacteria. The plates were kept at 20°C, and after 24 h the feeding sachet was replaced with a new sachet of just sterile diet. The diet was replaced again after an additional 24 and 48 h. Mortality was recorded at 0, 6, 24, 30, 48, 54, and 72 h after replacing the bacterium-diet mixture with sterile diet. Aphids were assumed dead if they were observed at the bottom of the well and did not move when the plate was agitated or if they had turned brown. Assays were replicated a minimum of three times for each combination of aphid species and bacterial strain, using fresh overnight bacterial cultures each time.
Infection assays.
Infection assays were used to determine if differences in virulence to aphids were a result of how successfully the bacteria were infecting aphids. Pathovar PsyB728a was used, as it displayed the greatest variation in virulence across the five aphid species and had already been shown to infect aphids successfully (7, 12). Three of the aphid species were used, A. rumicis, A. pisum, and R. padi, to which PsyB728a had demonstrated high, medium, and low virulence, respectively. Aphids were fed artificial diet mixed with PsyB728a for 24 h as before and were then transferred to feeding sachets of just sterile diet. After another 24 h, 12 to 16 live aphids were randomly selected and homogenized in 100 μl 10 mM MgCl2 buffer. A 5-μl droplet from each sample was plated, and the spots with a CFU count higher than 5 for A. pisum and A. rumicis were marked as infected to be consistent with previously published data (11, 12). Due to lower infection rates, we adapted our cutoff to 2 CFU for R. padi to be more permissible, although the majority of the positive samples had greater than 10 CFU. Assays were replicated three times for each aphid species to give a total of 36 to 48 aphids tested per species.
Statistical methods.
All statistical analyses were conducted in R version 4.0.0 (52). Infection data were analyzed using an ANOVA of the proportion of aphids infected in each of the three replicates per aphid species. The model was checked for normality of residuals and heteroscedasticity by visual inspection of histograms and plots of residuals against predicted values. Significant differences in strain virulence at 72 h postinfection were assessed using a generalized linear mixed model (GLMM) with a binomial distribution, using the R packages lme4 (53) and car (54). Fixed effects included an interaction between bacterial strain and aphid species, and experimental block was included as a random factor. To determine significant differences in the above-described analyses between aphid species or between bacterial strains, post hoc tests were assessed using the R package emmeans (55), and pairwise comparisons were adjusted with a Bonferroni correction.
Survival analyses were conducted using the R package survival (56). Kaplan-Meier curves were drawn to visualize survival data over the whole period of the assays for each aphid species per bacterial strain and to estimate survival probabilities (57). Cox proportional hazard survival models were carried out for each bacterial treatment, with aphid species being the only fixed variable tested, and an experimental block was included as a clustering factor to adjust standard errors appropriately.
Data availability.
Mitochondrial COI sequence data generated during the current study are available under the following GenBank accession numbers: A. nasturtii, MW740233; A. rumicis, MW740234; and Uroleucon caligatum, MW740235.
ACKNOWLEDGMENTS
We thank Kathryn Herr and Alexandra Glasgow for their help in the Hendry laboratory throughout this project.
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project no. 1010544.
REFERENCES
- 1.Stout MJ, Thaler JS, Thomma BPHJ. 2006. Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. AnnuRev Entomol 51:663–689. 10.1146/annurev.ento.51.110104.151117. [DOI] [PubMed] [Google Scholar]
- 2.Biere A, Bennett AE. 2013. Three-way interactions between plants, microbes and insects. Funct Ecol 27:567–573. 10.1111/1365-2435.12100. [DOI] [Google Scholar]
- 3.Tack AJM, Dicke M. 2013. Plant pathogens structure arthropod communities across multiple spatial and temporal scales. Funct Ecol 27:633–645. 10.1111/1365-2435.12087. [DOI] [Google Scholar]
- 4.Emmett BJ, Baker LAE. 1971. Insect transmission of fireblight. Plant Pathol 20:41–45. 10.1111/j.1365-3059.1971.tb00507.x. [DOI] [Google Scholar]
- 5.Esker PD, Nutter FW. 2002. Assessing the risk of Stewart’s disease of cornthrough improved knowledge of the role of the corn flea beetle vector. Phytopathology 92:668–670. 10.1094/PHYTO.2002.92.6.668. [DOI] [PubMed] [Google Scholar]
- 6.Harada H, Ishikawa H. 1997. Experimental pathogenicity of Erwinia aphidicola to pea aphid, Acyrthosiphon pisum. J Gen Appl Microbiol 43:363–367. 10.2323/jgam.43.363. [DOI] [PubMed] [Google Scholar]
- 7.Stavrinides J, McCloskey JK, Ochman H. 2009. Pea aphid as both host and vector for the phytopathogenic bacterium Pseudomonas syringae. Appl Environ Microbiol 75:2230–2235. 10.1128/AEM.02860-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stavrinides J, No A, Ochman H. 2010. A single genetic locus in the phytopathogen Pantoea stewartii enables gut colonization and pathogenicity in an insect host. Environ Microbiol 12:147–155. 10.1111/j.1462-2920.2009.02056.x. [DOI] [PubMed] [Google Scholar]
- 9.Grenier A-M, Duport G, Pagès S, Condemine G, Rahbé Y. 2006. The phytopathogen Dickeya dadantii (Erwinia chrysanthemi 3937) is a pathogen of the pea aphid. Appl Environ Microbiol 72:1956–1965. 10.1128/AEM.72.3.1956-1965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadarasah G, Stavrinides J. 2011. Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol Rev 35:555–575. 10.1111/j.1574-6976.2011.00264.x. [DOI] [PubMed] [Google Scholar]
- 11.Smee MR, Baltrus DA, Hendry TA. 2017. Entomopathogenicity to two hemipteran insects is common but variable across epiphytic Pseudomonas syringae strains. Front Plant Sci 8:2149. 10.3389/fpls.2017.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smee MR, Real-Ramirez I, Hendry TA. 2019. Insects as phyllosphere microbiome engineers: effects of aphids on a plant pathogen. bioRxiv 10.1101/797738. [DOI]
- 13.Hendry TA, Hunter MS, Baltrus DA. 2014. The facultative symbiont Rickettsia protects an invasive whitefly against entomopathogenic Pseudomonas syringae strains. Appl Environ Microbiol 80:7161–7168. 10.1128/AEM.02447-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baltrus DA, McCann HC, Guttman DS. 2017. Evolution, genomics and epidemiology of Pseudomonas syringae. Mol Plant Pathol 18:152–168. 10.1111/mpp.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin X-F, Kvitko B, He SY. 2018. Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16:316–328. 10.1038/nrmicro.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirano SS, Upper CD. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev 64:624–653. 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris CE, Kinkel LL, Xiao K, Prior P, Sands DC. 2007. Surprising niche for the plant pathogen Pseudomonas syringae. Infect Genet Evol 7:84–92. 10.1016/j.meegid.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Monteil CL, Guilbaud C, Glaux C, Lafolie F, Soubeyrand S, Morris CE. 2012. Emigration of the plant pathogen Pseudomonas syringae from leaf littercontributes to its population dynamics in alpine snowpack. EnvironMicrobiol 14:2099–2112. 10.1111/j.1462-2920.2011.02680.x. [DOI] [PubMed] [Google Scholar]
- 19.Hwang MSH, Morgan RL, Sarkar SF, Wang PW, Guttman DS. 2005. Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl Environ Microbiol 71:5182–5191. 10.1128/AEM.71.9.5182-5191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baltrus DA, Nishimura MT, Dougherty KM, Biswas S, Mukhtar MS, Vicente J, Holub EB, Dangl JL. 2012. The molecular basis of host specialization in bean pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 25:877–888. 10.1094/MPMI-08-11-0218. [DOI] [PubMed] [Google Scholar]
- 21.Morris CE, Lamichhane JR, Nikolić I, Stanković S, Moury B. 2019. The overlapping continuum of host range among strains in the Pseudomonas syringae complex. Phytopathol Res 1:4. 10.1186/s42483-018-0010-6. [DOI] [Google Scholar]
- 22.Emden HV, Harrington R. 2017. Aphids as crop pests, 2nd ed. CABI Publishing, Oxfordshire, UK. [Google Scholar]
- 23.Rabbinge R, Drees EM, Graaf M, Verberne FCM, Wesselo A. 1981. Damage effects of cereal aphids in wheat. Neth J Plant Pathol 87:217–232. 10.1007/BF02084437. [DOI] [Google Scholar]
- 24.Nault LR. 1997. Arthropod transmission of plant viruses: a new synthesis. Ann Entomol Soc Am 90:521–541. 10.1093/aesa/90.5.521. [DOI] [Google Scholar]
- 25.Ng JCK, Perry KL. 2004. Transmission of plant viruses by aphid vectors. Mol Plant Pathol 5:505–511. 10.1111/j.1364-3703.2004.00240.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang K-Y, Guo Q-L, Xia X-M, Wang H-Y, Liu T-X. 2007. Resistance of Aphis gossypii (Homoptera: Aphididae) to selected insecticides on cotton from five cotton production regions in Shandong. J Pestic Sci 32:372–378. 10.1584/jpestics.G06-51. [DOI] [Google Scholar]
- 27.Bass C, Puinean AM, Zimmer CT, Denholm I, Field LM, Foster SP, Gutbrod O, Nauen R, Slater R, Williamson MS. 2014. The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem Mol Biol 51:41–51. 10.1016/j.ibmb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Akmal M, Freed S, Malik MN, Gul HT. 2013. Efficacy of Beauveria bassiana (Deuteromycotina: Hypomycetes) against different aphid species under laboratory conditions. Pak J Zool 45:71–78. [Google Scholar]
- 29.Hance T, Kohandani-Tafresh F, Munaut F. 2017. Biological control, p 448–493. In Emden HV, Harrington R (ed), Aphids as crop pests, 2nd ed. CABI Publishing, Oxfordshire, UK. [Google Scholar]
- 30.Mukherjee A, Debnath P, Ghosh SK, Medda PK. 2020. Biological control of papaya aphid (Aphis gossypii Glover) using entomopathogenic fungi. Vegetos 33:1–10. 10.1007/s42535-019-00072-x. [DOI] [Google Scholar]
- 31.Andrews JH, Harris RF. 2000. The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol 38:145–180. 10.1146/annurev.phyto.38.1.145. [DOI] [PubMed] [Google Scholar]
- 32.Beattie GA, Lindow SE. 1995. The secret life of foliar bacterial pathogens on leaves. Annu Rev Phytopathol 33:145–172. 10.1146/annurev.py.33.090195.001045. [DOI] [PubMed] [Google Scholar]
- 33.Monier J-M, Lindow SE. 2004. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl Environ Microbiol 70:346–355. 10.1128/aem.70.1.346-355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendry TA, Clark KJ, Baltrus DA. 2016. A highly infective plant-associated bacterium influences reproductive rates in pea aphids. R Soc Open Sci 3:150478. 10.1098/rsos.150478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackman RL, Eastop VF. 2017. Taxonomic issues, p 1–36. In Emden HV, Harrington R (ed), Aphids as crop pests, 2nd ed. CABI Publishing, Oxfordshire, UK. [Google Scholar]
- 36.Byrne JM, Dianese AC, Ji P, Campbell HL, Cuppels DA, Louws FJ, Miller SA, Jones JB, Wilson M. 2005. Biological control of bacterial spot of tomato under field conditions at several locations in North America. Biol Control 32:408–418. 10.1016/j.biocontrol.2004.12.001. [DOI] [Google Scholar]
- 37.Rosell RC, Davidson EW, Jancovich JK, Hendrix DL, Brown JK. 2003. Size limitations in the filter chamber and digestive tract of nymphal and adult Bemisia tabaci whiteflies (Hemiptera: Aleyrodidae). Ann Entomol SocAm 96:544–552. 10.1603/0013-8746(2003)096[0544:SLITFC]2.0.CO;2. [DOI] [Google Scholar]
- 38.Steinkraus DC. 2006. Factors affecting transmission of fungal pathogens of aphids. J Invertebr Pathol 92:125–131. 10.1016/j.jip.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Łukasik P, van Asch M, Guo H, Ferrari J, Godfray HCJ. 2013. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett 16:214–218. 10.1111/ele.12031. [DOI] [PubMed] [Google Scholar]
- 40.Tsuchida T, Koga R, Horikawa M, Tsunoda T, Maoka T, Matsumoto S, Simon J-C, Fukatsu T. 2010. Symbiotic bacterium modifies aphid body color. Science 330:1102–1104. 10.1126/science.1195463. [DOI] [PubMed] [Google Scholar]
- 41.Parker BJ, Hrček J, McLean AHC, Godfray HCJ. 2017. Genotype specificity among hosts, pathogens, and beneficial microbes influences the strength of symbiont-mediated protection. Evolution 71:1222–1231. 10.1111/evo.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez AJ, Ritter SG, Doremus MR, Russell JA, Oliver KM. 2014. Aphid-encoded variability in susceptibility to a parasitoid. BMC Evol Biol 14:127. 10.1186/1471-2148-14-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costechareyre D, Balmand S, Condemine G, Rahbé Y. 2012. Dickeya dadantii, a plant pathogenic bacterium producing Cyt-Like entomotoxins, causes septicemia in the pea aphid Acyrthosiphon pisum. PLoS One 7:e30702. 10.1371/journal.pone.0030702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendry TA, Ligon RA, Besler KR, Fay RL, Smee MR. 2018. Visual detection and avoidance of pathogenic bacteria by aphids. Curr Biol 28:3158–3164. 10.1016/j.cub.2018.07.073. [DOI] [PubMed] [Google Scholar]
- 45.Perry KL, Zhang L, Shintaku MH, Palukaitis P. 1994. Mapping determinants in cucumber mosaic virus for transmission by Aphis gossypii. Virology 205:591–595. 10.1006/viro.1994.1686. [DOI] [PubMed] [Google Scholar]
- 46.Rochow WF. 1969. Biological properties of four isolates of barley yellow dwarf virus. Phytopathology 59:1580–1589. [PubMed] [Google Scholar]
- 47.Mondal S, Lin Y-H, Carroll JE, Wenninger EJ, Bosque-Pérez NA, Whitworth JL, Hutchinson P, Eigenbrode S, Gray SM. 2017. Potato virus Y transmission efficiency from potato infected with single or multiple virus strains.Phytopathology 107:491–498. 10.1094/PHYTO-09-16-0322-R. [DOI] [PubMed] [Google Scholar]
- 48.Sandstrom JP, Russell JA, White JP, Moran NA. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10:217–228. 10.1046/j.1365-294x.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 49.Russell JA, Moran NA. 2005. Horizontal transfer of bacterial symbionts: heritability and fitness effects in a novel aphid host. Appl Environ Microbiol 71:7987–7994. 10.1128/AEM.71.12.7987-7994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McLean AHC, van Asch M, Ferrari J, Godfray HCJ. 2011. Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc Biol Sci 278:760–766. 10.1098/rspb.2010.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prosser WA, Douglas AE. 1992. A test of the hypotheses that nitrogen is upgraded and recycled in an aphid (Acyrthosiphon pisum) symbiosis. J Insect Physiol 38:93–99. 10.1016/0022-1910(92)90037-E. [DOI] [Google Scholar]
- 52.R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/. [Google Scholar]
- 53.Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 54.Fox J, Weisberg S. 2019. An R companion to applied regression, 3rd ed. Sage, Thousand Oaks, CA. [Google Scholar]
- 55.Lenth R. 2020. emmeans: estimated marginal means, aka least-squares means. R package version 1.4.6. https://CRAN.R-project.org/package=emmeans.
- 56.Therneau TM. 2020. A package for survival analysis in R. R package version 3.1–12. https://CRAN.R-project.org/package=survival.
- 57.Kaplan EL, Meier P. 1958. Nonparametric estimation from incomplete observations. J Am Stat Assoc 53:457–481. 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 58.Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 7:e1002132. 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berge O, Monteil CL, Bartoli C, Chandeysson C, Guilbaud C, Sands DC, Morris CE. 2014. A user’s guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS One 9:e105547. 10.1371/journal.pone.0105547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loper JE, Lindow SE. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449. 10.1094/Phyto-77-1449. [DOI] [Google Scholar]
- 61.Hirano SS, Upper CD. 1990. Population biology and epidemiology of Pseudomonas syringae. Annu Rev Phytopathol 28:155–177. 10.1146/annurev.py.28.090190.001103. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Mitochondrial COI sequence data generated during the current study are available under the following GenBank accession numbers: A. nasturtii, MW740233; A. rumicis, MW740234; and Uroleucon caligatum, MW740235.