Our research revealed the microbial network of the LCE reactor microbiome for n-caproate production at high concentrations, which will provide a foundation for designing or engineering the LCE reactor microbiome to recover n-caproate from organic waste streams in the future. In addition, the hypothetical model of the reactor microbiome that we proposed may offer guidance for researchers to find the underlying microbial mechanism when they encounter low-efficiency n-caproate production from the LCE process.
KEYWORDS: valuable biochemical, n-caproate, microbial ecology, lactate-driven chain elongation, organic waste streams
ABSTRACT
Lactate-driven chain elongation (LCE) has emerged as a new biotechnology to upgrade organic waste streams into a valuable biochemical and fuel precursor, medium-chain carboxylate, n-caproate. Considering that a low cost of downstream extraction is critical for biorefinery technology, a high concentration of n-caproate production is very important to improve the scale-up of the LCE process. We report here that in a nonsterile open environment, the n-caproate concentration was increased from the previous record of 25.7 g·liter−1 to a new high level of 33.7 g·liter−1 (76.8 g chemical oxygen demand [COD]·liter−1), with the highest production rate being 11.5 g·liter−1·day−1 (26.2 g COD·liter−1·day−1). In addition, the LCE process remained stable, with an average concentration of n-caproate production of 20.2 ± 5.62 g·liter−1 (46.1 ± 12.8 g COD·liter−1) for 780 days. Dynamic changes in taxonomic composition integrated with metagenomic data reveal the microbial ecology for long-term production of high concentrations of n-caproate: (i) the core microbiome is related to efficient functional groups, such as Ruminococcaceae (with functional strain CPB6); (ii) the core bacteria can maintain stability for long-term operation; (iii) the microbial network has relatively low microbe-microbe interaction strength; and (iv) low relative abundance and variety of competitors. The network structure could be shaped by hydraulic retention time (HRT) over time, and long-term operation at an HRT of 8 days displayed higher efficacy.
IMPORTANCE Our research revealed the microbial network of the LCE reactor microbiome for n-caproate production at high concentrations, which will provide a foundation for designing or engineering the LCE reactor microbiome to recover n-caproate from organic waste streams in the future. In addition, the hypothetical model of the reactor microbiome that we proposed may offer guidance for researchers to find the underlying microbial mechanism when they encounter low-efficiency n-caproate production from the LCE process. We anticipate that our research will rapidly advance LCE biotechnology with the goal of promoting the sustainable development of human society.
INTRODUCTION
With the increasing generation of organic waste streams coinciding with the threat of climate change, humans have an urgent need to pursue sustainable development. The carboxylate platform is a fast-growing platform for the transformation of organic waste into renewable resources (1–3).
At present, n-caproate, with six carbon atoms, is considered one of the most important products of carboxylate platforms due to its special characteristics: (i) easy extraction for its low solubility; (ii) precursor of drop-in fuels and full-performance jet or diesel fuel (4, 5); and (iii) high added value for its potential industrial utilization as a precursor for green insecticides and antifungal agents, flavoring agent in food and cigarettes, and parenteral nutrition in pharmacology, which is being more widely used in foods, drugs, and cosmetics (1, 6–8).
Driven by an electron donor (ethanol, lactate, etc.), functional microorganisms gradually synthesize medium-chain carboxylates (MCCs; with 6 to ∼12 carbon atoms) from acetyl-coenzyme A (CoA) via chain elongation (reverse β oxidation cycle or fatty acid biosynthesis) (9–15). The first-generation chain elongation biotechnology is ethanol-driven chain elongation (ECE), which can efficiently upgrade ethanol and organic waste into n-caproate (16–19). Generally, the organic waste for high concentrations of ethanol production required targeted regulation (acid/enzyme pretreatment, simultaneous saccharification, and fermentation using specific yeasts/fungi/bacteria); therefore, if we consider the perspective of resource recycling rather than bioethanol upgrading, the ECE process usually requires exogenous ethanol addition (1, 20–23), resulting in two defects: increased costs and risks of environmental pollution from the manufacture and transportation of ethanol and residue ethanol treatment (24).
A new process, lactate-to-n-caproate conversion (11, 13, 25, 26), may minimize the two problems for the electron donor lactate, which can be easily fermented to a high concentration (18.6 to 96.5 g liter−1) (13, 27) from organic wastes (especially those rich in carbohydrates), providing a clever way to circumvent exogenous lactate supplementation (8, 27–29). Previously, n-caproate was found to be a by-product of lactate fermentation in the pure culture of Megasphaera elsdenii (30), a typical chain-elongating species, which can form high concentrations of n-caproate from glucose (15); however, the n-caproate concentration was too low to attract the attention of researchers. In 2015, we first demonstrated that lactate can serve as an electron donor in the same way as ethanol to synthesize n-caproate as the main product (13). Thereafter, Kucek et al. took the first step to produce n-caproate from a continuously fed bioreactor (31). Researchers successively confirmed continuous n-caproate production from many organic waste streams, mainly via lactate-to-n-caproate conversion without exogenous electron donor supplementation, e.g., lignocellulosic biomass (grass) (28), acid whey wastewater (8, 32), diluted food waste (27, 29), and liquor brewing wastewater (33). Therefore, LCE biotechnology can provide a dual benefit of reducing cost and environmental impact compared with ECE biotechnology.
However, as a waste stream recovery biotechnology, the long-term operation efficacy is more meaningful for industrial implementation. Although batch experiments once achieved as high as 23.4 g liter−1 n-caproate production from lactate (13), long-term studies only reported an n-caproate production level of approximately 10.0 g·liter−1 (Table 1), raising a question regarding low concentrations of n-caproate production. Researchers have explored several ways to improve the efficiency of the LCE process, such as in-line extraction (8), granular sludge (33), and parameter optimization (e.g., pH [34] and HRT [8, 35]), which could greatly accelerate the production rate of n-caproate but had little effect on the enhancement of n-caproate accumulation, which seems to have reached a maximum. However, for industrial application, it is not economically feasible to extract n-caproate at low concentrations, even though n-caproate is much easier to extract than ethanol and lactate. Extraction usually involves membrane filtration, pH adjustment, liquid extraction, and product recovery, and each extraction will wear out facilities and generate resource consumption (e.g., energy and solvent) (35). Indeed, a low concentration of n-caproate in the LCE process will significantly increase the downstream extraction cost, limiting the further translation of LCE technology into industrial reality.
TABLE 1.
Comparison of n-caproate production via lactate chain elongation by continuously fed reactor microbiome
| Substrate | Reactor type | No. of days | pH | Dominant microbiome | RA (%) | Max concn (g liter−1) | Max production (g liter−1 day−1) | Reference or source |
|---|---|---|---|---|---|---|---|---|
| Thin stillage + H2 | 1-Liter reactor (with electro chemical cells) | 42 | 5.4∼5.7 | Megasphaera elsdenii | 57 | 2.08 | 0.58 | 39 |
| l-Lactate | 0.7-Liter AFa (with in-line extraction) | 220 | 5.0 | Acinetobacter sp. | 62.9 | —e | 3.03 | 31 |
| Grass | 100-ml reactor (with electrochemical cells) | 30 | 5.5∼6.2 | Clostridium IV (related to strain BS-1) | 28 | 4.09 | —e | 28 |
| Acid whey | 0.7-liter CSTRb (with in-line extraction) | 90 | 5.0 | Bacteroidales | 21.7 | —e | 1.68 | 8 |
| Clostridiales | 12.6 | |||||||
| 15× diluted food waste leachate + H2 | 6.0-liter leach-bed | 100 | Initial, 7.0d | Clostridium | 42 | 9.59 | —e | 29 |
| Acid whey | 1.0-liter UASBc | 400 | 5.5 | Ruminococcaceae | 20.5 | 10.4 | 3.20 | 32 |
| Prevotellaceae | 17.9 | |||||||
| Liquor brewing wastewater | 5.0-liter AF | 485 | 6.5 | Clostridium IV | 25.5 | 3.59 | 10.9 | 33 |
| Xylan + lactate | 1.0-liter CSTR | 148 | 5.5 | Ruminiclostridium 5 | 42.3 | —e | 3.60 | 49 |
| 10–15% (vol/vol) food waste | 1.0-liter stirred tank reactor | 45 | 6.0 | Caproiciproducensf | 32–72 | 5.4 | 1.81 | 27 |
| Lactate | 0.9-liter CSTR | 194 | 5.0–6.5 | Caproiciproducens | 2.0–67.8 | 8.58 | —e | 34 |
| dl-Lactate | 3.0-liter CSTR | 780 | 6.0 | Caproiciproducensg | 53.9 | 33.7 | 11.5 | This study |
AF, anaerobic filter.
CSTR, continuously stirred tank reactor.
UASB, upflow anaerobic sludge blanket.
Only the initial pH of each cycle was controlled.
No related data given.
Caproiciproducens in this source, formerly Clostridium IV, showed 91 to 95% similarity to Caproiciproducens galactitolivorans.
Caproiciproducens related to Ruminococcaceae strain CPB6.
Microbial ecology has been widely considered to strongly constrain or accelerate the efficacy of functional taxa and further determine the concentration of products (36–38). We also noticed analogous phenomena in the field of chain elongation. For example, a Megasphaera elsdenii-dominated reactor microbiome produced 2.08 g liter−1 n-caproate (39), while a Ruminococcaceae-occupied reactor microbiome produced 10.4 g liter−1 n-caproate (32). Mesophilic Ruminococcaceae strain CPB6-containing ecosystems with distinct microbial compositions produced different concentrations of n-caproate (4.39 g liter−1 [29] versus 12.9 g liter−1 [13]) from the same amount of lactate (∼30 g liter−1) under mild acid conditions (note that strain CPB6 is affiliated with Caproiciproducens in the SILVA database). However, at present, knowledge of the reactor microbiome of the LCE process is still in its infancy. Researchers could find the dominant microbiome in a mixed-culture biosystem, but it is usually difficult to identify the most important microbe for n-caproate production (8, 31, 40). Through omics technology, studies have distinguished many functional microbiomes, which expands our knowledge of chain elongation (12, 41). Nevertheless, these omics-based understandings were restricted in the chain elongation biosystem, which mainly produces n-butyrate rather than n-caproate as the dominant product. Moreover, the connections between microbial ecology and n-caproate yield remain unknown. These knowledge gaps in the LCE microbiome make it a long and arduous journey to improve the performance of the LCE process for practical industrial applications.
The purpose of this study was to explore the microbial ecological mechanism for the production of high concentrations of n-caproate via the LCE process. First, we inoculated the chain elongation mixed microbiome into semicontinuously operating reactors fed dl-lactate containing artificial wastewater. We then operated the bioprocessor for more than 2 years (780 days) to examine the performance of the microbial system under nonsterile open conditions over time. We tracked the dynamic changes in taxonomic composition and integrated metagenomic analyses to illustrate variations in the ecological profile of the mixed microbiome and predict the metabolic network, yielding important insights into the microbial mechanism (including microbial structure, stability, and competitive and functional microbiomes) for the production of high concentrations of n-caproate from lactate. Finally, we propose a microbial model hypothesis, laying a foundation to improve the design or control LCE processes.
RESULTS
Performance of a Ruminococcaceae-dominated reactor microbiome under nonsterile open conditions.
During the long-term operating period, we achieved 735 days of n-caproate production from lactate (Tables 2 and 3). The average n-caproate concentration in the effluent was maintained at 20.2 ± 5.62 g·liter−1 (1,044.8 ± 289.6 mmol C·liter−1), and the maximum concentration reached 33.7 g·liter−1 (1,743.1 mmol C·liter−1) on day 690 (Tables 2 and 3 and Fig. 1). Moreover, the highest n-caproate to total carboxylate ratio was 93.4 mmol C %, with an average value of 68.1 ± 10.6 mmol C %. The bioreactor produced 10.5 ± 2.56 liters of total gas (mean value) every day, and hydrogen and methane presented 12.9% ± 3.79% and 12.3% ± 5.81%, respectively (Tables 2 and 3).
TABLE 2.
Dominant carboxylates produced in lactate-fed reactor during 780 days of operation
| Parameter | Value for: |
|||||
|---|---|---|---|---|---|---|
| Acetate | n-Butyrate |
n-Caproate |
Propionate | n-Valerate | ||
| Avg | Maximum | |||||
| Avg concn (g liter−1) | 2.14 ± 1.53 | 2.88 ± 1.55 | 20.2 ± 5.62 | 33.7 | 1.26 ± 1.35 | 4.77 ± 2.03 |
| Specificity (mM C, %) | 4.48 ± 2.56 | 8.66 ± 4.63 | 68.1 ± 10.6 | 93.4 | 3.20 ± 3.19 | 15.6 ± 6.30 |
TABLE 3.
Dominant gases produced in lactate-fed reactor during 780 days of operation
| Avg gas component (%) for: | |||
|---|---|---|---|
| H2 | CO2 | CH4 | N2 |
| 12.9 ± 3.79 | 63.4 ± 4.68 | 12.3 ± 5.81 | 10.9 ± 2.26 |
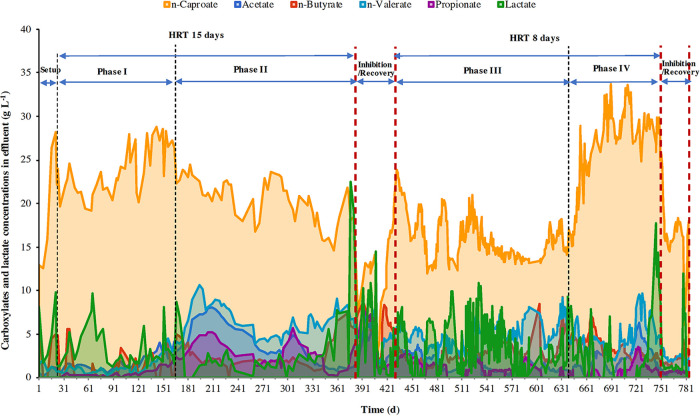
FIG 1.
Reactor performance over the operating period of 780 days. Effluent C2 to C6 carboxylate concentrations were used at 30°C, pH 6.0.
A graphical summary of reactor performance can be seen in Fig. 1. In the 15-day setup stage, we continuously fed lactate into the bioreactor without drainage for n-caproate production and accumulation. The reactor was then started in continuous operation mode with a hydraulic retention time (HRT) of 15 days. During the first phase (16 to 180 days), the reactor maintained a steady state with an average concentration of 23.9 ± 2.74 g·liter−1. In the next 220 days, the n-caproate concentration declined gradually while short-chain carboxylates (SCCs) increased, especially n-valerate. Therefore, we decreased the HRT to 8 days. During the following 200 days, the n-caproate concentration declined gradually and fluctuated between 13.0 g·liter−1 and 23.0 g·liter−1, with a mean value of 16.1 ± 2.53 g·liter−1. Thereafter, from day 600 on, the concentration of n-caproate started to increase gradually to 33.7 g·liter−1 on day 690 and stabilized at a high level (mean value, 28.4 ± 2.4 g·liter−1) for 96 days.
In addition, the chain elongation reactor showed severe inhibition twice, from day 386 to 407 and day 753 to 777. In the beginning of inhibition, no n-caproate production was observed, and then the concentration dropped to a low level. During the inhibition periods, the inactive reactor microbiome showed little lactate utilization and carboxylate production or degradation. To recover the activity of the reactor microbiome, we centrifuged the whole fermentation broth, discarded the supernatant, and fed fresh artificial wastewater. Thereafter, the reactor microbiome soon recovered, and the n-caproate concentration exceeded 10 g liter−1 within 3 to 10 days.
To our knowledge, this is the first report that long-term n-caproate production at higher concentrations can be achieved, and the maximum n-caproate concentration from lactate can exceed 30 g liter−1. We previously obtained a high n-caproate concentration of 23.4 g liter−1 in batch experiments (13), and Duber et al. (32) presumed that the high concentration of n-caproate production was due to excessive lactate supplementation. In contrast, Kucek et al. (31) suggested that excessive lactate will induce additional electron flow toward pyruvate and odd-numbered chain elongation products (e.g., propionate), significantly lowering the n-caproate production rate. Here, we present our view that it would be better to understand the functional bacterial and ecological structure of the reactor microbiome before we discuss the effect of operating conditions (i.e., lactate supplement) on n-caproate production. We noticed that the sufficient lactate supplementation does not necessarily lead to the synthesis of high concentrations of n-caproate (more than 20 g·liter−1), and we also found little connection between propionate accumulation and excessive lactate addition in this paper. In the following sections, we will uncover the mechanism for high concentrations of n-caproate production.
Ecological network of reactor microbiome.
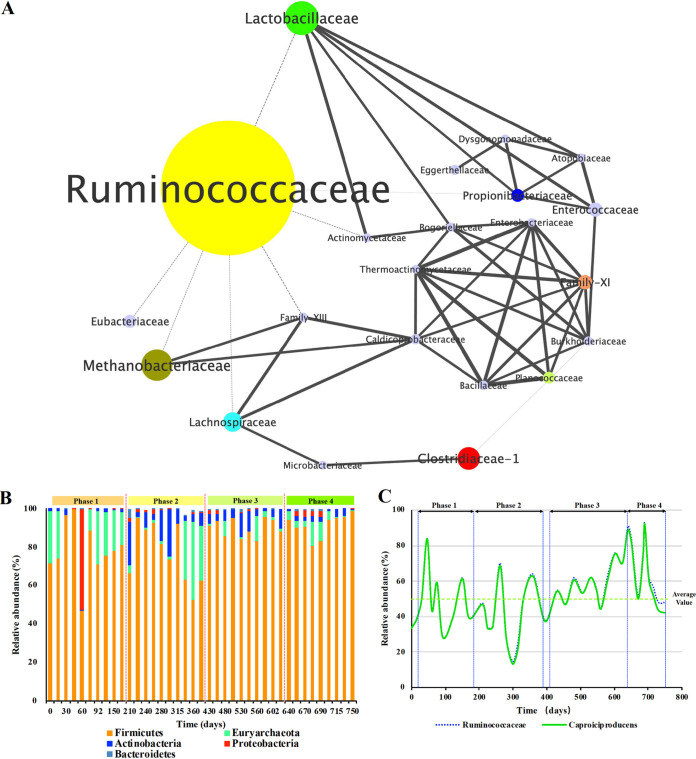
We performed a long-term study to gain overall insight into the microbial compositional structure. During the 735-day n-caproate-producing period, 39 samples were sequenced, and the structural biodiversity, including Chao1, Shannon index results, observed operational taxonomic unit (OTU) numbers, coverage values, and sequence read numbers, are presented in Table S1 in the supplemental material. In total, 22 bacterial families with an average relative abundance of greater than 0.1% in the samples were identified (Fig. S1). The 22 families made up 98.7% ± 1.40% of the total microbial community.
The phylum-level analysis revealed that Firmicutes predominated in the community during the entire fermentation period (Fig. 2B). Euryarchaeota were detected as a minor group, with an average relative abundance of 8.49% ± 11.0%. Other minor phyla in the reactor microbiome were Actinobacteria (4.88% ± 5.56%), Proteobacteria (2.01% ± 8.18%), and Bacteroidetes (0.370% ± 0.815%).
FIG 2.
Ecological network of the reactor microbiome. (A) Microbial cooccurrence network in the reactor microbiome, constructed on the basis of the relative abundance profiles of the reactor microbiome at the family level over the 780 consecutive days. Each node represents a family. The sizes of the nodes are proportional to the average relative abundances of the families. Solid edges represent a positive relationship (+), and dashed edges represent a negative relationship (−). Edges are also weighted by the strength of the correlation. (B) Variations in relative abundance at the phylum level. (C) Dynamics of the relative abundance of the core taxon Ruminococcaceae (mean value, 53.9%) and its predominant genus, Caproiciproducens.
As shown in Fig. 2A, there were three dominant families in the network, Ruminococcaceae (Firmicutes), Lactobacillaceae (Firmicutes), and Methanobacteriaceae (Euryarchaeota). Obviously, Ruminococcaceae were the most dominant, with an average relative abundance of 53.8% ± 17.9%. The Caproiciproducens genus (also named Clostridium IV in the RDP database and Ruminococcus in the Greengenes database) accounted for greater than 98% of Ruminococcaceae (Fig. 2C). By using the 16S rRNA sequence information of strain CPB6, we found that the strain made up 35.3% ± 23.4% to 45.4% ± 17.3% of the total taxa (Table 4). Strain CPB6, affiliated with Caproiciproducens, is a model species for n-caproate production from lactate in pure-culture studies (11, 25, 26). Therefore, these results indicate that the dominant taxon, Ruminococcaceae, is the functional group that is responsible for n-caproate production from lactate. The second most abundant family was Lactobacillaceae, whose relative abundance fluctuated between 26.1% and 1.22%, with a mean value of 10.2% ± 8.03% (Fig. S1). Lactobacillaceae were mainly from real wastewater (Table S2). The Methanobacteriaceae, the sole methane-producing archaea, with a relative abundance greater than 0.1%, made up the third most abundant (8.70% ± 11.0% mean value) family (Fig. S1) in the reactor microbiome. Additionally, Methanobacterium, which is a genus known to produce methane from hydrogen and carbon dioxide, accounted for 99.9% of the family Methanobacteriaceae. The growth source of this population was derived from fermentation of lactate (carbon dioxide and hydrogen), which was detected in the gas produced from the chain elongation process (Fig. S3).
TABLE 4.
Variations in n-caproate production-associated speciesa during the long-term operation period
| Species | Relative abundance (%) in phase: |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Caproiciproducens_unclassified | 10.3 ± 3.78 | 5.50 ± 5.58 | 10.3 ± 6.30 | 15.6 ± 6.58 |
| Caproiciproducens_uncultured_bacterium | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.048 ± 0.122 |
| Caproiciproducens_uncultured_clostridia_bacterium | 0.001 ± 0.001 | 0.009 ± 0.015 | 0.026 ± 0.076 | 0.467 ± 0.711 |
| Caproiciproducens_uncultured_clostridium_sp. | 0.447 ± 0.393 | 0.332 ± 0.405 | 0.790 ± 1.272 | 0.945 ± 0.867 |
| Caproiciproducens_uncultured_organism | 0.154 ± 0.143 | 0.029 ± 0.043 | 0.175 ± 0.268 | 0.819 ± 2.013 |
| Caproiciproducens_uncultured_ruminococcaceae_bacterium | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.007 ± 0.015 |
| Caproiciproducens_uncultured_ruminococcus_sp. | 2.55 ± 1.25 | 0.729 ± 0.688 | 2.71 ± 1.51 | 3.35 ± 3.50 |
| Ruminococcaceae_uncultured_clostridium_sp. | 0.000 ± 0.000 | 0.003 ± 0.008 | 0.001 ± 0.002 | 0.193 ± 0.248 |
| Ruminococcaceae_bacterium_CPB6 | 35.3 ± 23.4 | 30.9 ± 19.9 | 41.1 ± 13.5 | 45.4 ± 17.3 |
| Ruminococcaceae_bacterium_CPC-11 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.001 ± 0.001 | 0.043 ± 0.070 |
| Ruminococcaceae_bacterium_HAWD2 | 0.000 ± 0.000 | 0.009 ± 0.017 | 0.005 ± 0.013 | 0.036 ± 0.079 |
| Ruminococcaceae_NK4A214_group_unclassified | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.015 ± 0.022 |
| Ruminococcaceae_UCG-005_unclassified | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.007 ± 0.014 |
| Ruminococcaceae_UCG-013_uncultured_clostridiaceae_bacterium | 0.000 ± 0.000 | 0.021 ± 0.025 | 0.018 ± 0.028 | 0.032 ± 0.072 |
| Ruminococcaceae_UCG-014_unclassified | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.001 ± 0.002 | 0.015 ± 0.025 |
| Ruminococcaceae_unclassified | 0.024 ± 0.038 | 0.068 ± 0.073 | 0.082 ± 0.097 | 0.543 ± 0.565 |
| Ruminiclostridium_5_unclassified | 0.009 ± 0.016 | 0.000 ± 0.000 | 0.003 ± 0.005 | 0.017 ± 0.020 |
| Lachnospiraceae_uncultured_coprococcus_sp. | 2.98 ± 3.11 | 0.133 ± 0.132 | 0.290 ± 0.30 | 2.45 ± 2.58 |
| n-Caproate production-associated species, in total no. of OTUs | 55.8 ± 16.9 | 38.1 ± 18.7 | 55.9 ± 11.7 | 71.9 ± 14.7 |
| n-Caproate production-associated species except CPB6, in total no. of OTUs | 20.41 ± 8.18 | 7.19 ± 6.39 | 14.9 ± 7.68 | 24.8 ± 8.17 |
Species affiliated with genera in which relative abundance variations were positively correlated with n-caproate production (P < 0.05) (Fig. 5A and Tables S5 and S6).
The network demonstrates that the core microbe Ruminococcaceae is independent of the microbial community. The population only presents negative correlations (shown as dashed lines) with other nodes (six cooccurrences), including Lactobacillaceae, Methanobacteriaceae, Family XIII, Enterococcaceae, Eubacteriaceae, and Propionibacteriaceae. In addition, there is another negative interaction between Clostridiaceae 1 and Burkholderiaceae. However, the network also shows intense and complex relationships within the other microbial cooccurrences, forming a couple of subnetworks in the topological structure. Among them, eight families showed greater than 1% relative abundance, including Clostridiaceae 1 (6.32% ± 6.57%), Lachnospiraceae (4.64% ± 3.64%), Family XI (2.44% ± 3.29%), Enterococcaceae (2.04% ± 4.05%), Eubacteriaceae (1.85% ± 8.83%), Propionibacteriaceae (1.84% ± 2.78%), and Planococcaceae (1.41% ± 8.04%). Another 11 organisms were identified with a relative abundance between 0.1 and 1.0% at the family level (Fig. S1).
Linking n-caproate production concentration to dynamic changes in the microbial community.
Despite the steady long-term n-caproate production, the efficiency showed significant swings at different HRTs over time, e.g., short-term setbacks or enhancement. In this section, we examine the subtle details of these swings and their associated microbiome ecology to explore the inner connection between network structure and the concentration of n-caproate production. Therefore, this section consists of three parts: the variations in performance of the bioprocess and their connections to dynamic changes in the microbial community (inferred from 16S rRNA gene amplicon sequencing), including network structure and taxonomic composition.
(i) The variations in performance of the bioprocess.
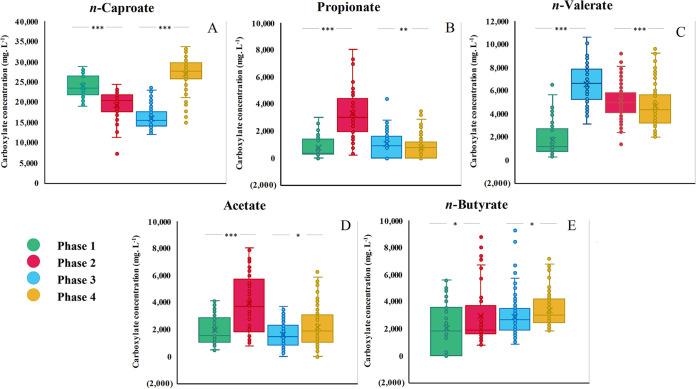
The HRT of first two phases was 15 days. In phase one (16 to 180 days), the reactor microbiome steadily produced n-caproate at an average concentration of 23.9 ± 2.74 g·liter−1. The average specificity of n-caproate reached 83.2 ± 9.55 mmol C%, and the main by-product was n-butyrate (Table S3). However, after 180 days of the trial, acetate, propionate, and n-valerate simultaneously started to increase (P < 0.001), significantly decreasing the n-caproate concentration (P < 0.001) (Fig. 3). Meanwhile, the methane in the biogas significantly increased from 12.7% ± 5.53% to 16.9% ± 7.10% (P < 0.001), and the hydrogen content dropped from 12.6% ± 3.09% to 9.67% ± 3.64% (P < 0.001) (Table S4). By shortening the HRT to 8 days, the concentrations of acetate, propionate, and n-valerate decreased notably (P < 0.001), while the n-butyrate concentration increased slightly (P < 0.01) in the subsequent 200 days (phase 3). The specificity of n-caproate slightly increased from 58.2% ± 9.75% (at phase 2) to 63.7% ± 10.0%, although the n-caproate concentration decreased (Table S3). The content of methane in biogas significantly dropped by 36.7% ± 1.55% to 10.7% ± 1.55% (P < 0.001), while hydrogen increased by 36.5% ± 1.63% to 13.2% ± 1.63% (P < 0.001) (Table S4). After 200 days of acclimation at an HRT of 8 days, it was surprising to find that the n-caproate concentration rapidly increased from 13.4 g·liter−1 (day 604) to 28.9 g·liter−1 (day 653), with a maximum rate of 11.5 g·liter−1·day−1 (26.2 g chemical oxygen demand [COD]·liter−1·day−1). On subsequent days, the concentration of n-caproate continued to increase to the highest level of 33.7 g·liter−1 and stabilized at an average concentration of 28.4 ± 2.40 g·liter−1 for 96 days (Fig. 1). At the same time, we observed a slow increase in even-numbered SCCs (acetate and n-butyrate, P < 0.02) and a further decline in odd-numbered SCCs (propionate, P < 0.028; n-valerate, P < 0.002). The hydrogen further increased by 38.6% ± 1.1.16% to 18.3% ± 1.1.16% (P < 0.001), whereas methane declined to the lowest level of 7.00% ± 1.21% (P < 0.001) (Table S4). During phase 4 (calculated from Table S3), the calculated average n-caproate production rate was 3.54 g·liter−1·day−1 (8.07 g COD·liter−1·day−1), which was 2.25 times higher than that in phase 1 (1.57 g·liter−1·day−1), suggesting a higher n-caproate production efficiency at short HRTs than at long HRTs.
FIG 3.
Comparison of carboxylate (acetate, propionate, n-butyrate, n-valerate, and n-caproate) production from lactate in four phases. Asterisks indicate P values of <0.05 (*), 0.01 (**), and 0.001 (***). Phase 1, 16 to 180 days (HRT, 15 days); phase 2, 181 to 385 days (HRT, 15 days); phase 3, 408 to 635 days (HRT, 8 days); phase 4, 636 to 752 days (HRT, 8 days).
(ii) Connection between network structure and the variations in performance of the bioprocess.
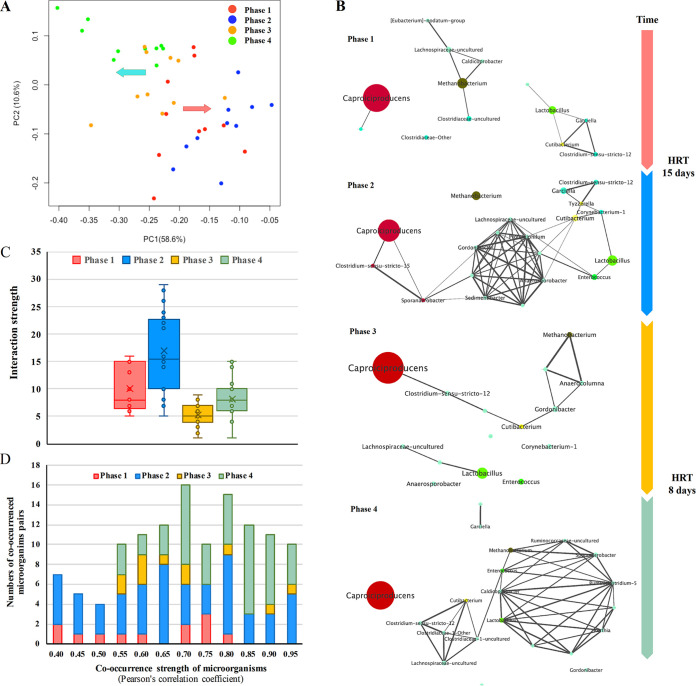
The principal component analysis (PCA) (Fig. 4A) suggests a progressive separation of the microbial community under two different HRT conditions over time. In the same HRT, microbial communities were more similar to each other. Phase 2 demonstrates the period when the microbial ecosystem became degraded (propionate and n-valerate significantly increased, accompanied by n-caproate decline), whereas phase 4 represents an improvement period when the concentration of n-caproate continuously increased and reached the highest level of 33.7 g liter−1. Additionally, the microbial community composition in phase 2 was significantly different from that in phase 4.
FIG 4.
Dynamic changes in the microbial community in the four stages. (A) Principal-component analysis (PCA) of 39 samples in four phases. (B) Microbial cooccurrence in the n-caproate-producing mixed microbiome in four phases at the genus level. Each node represents a genus whose size is proportional to the average relative abundance. Edges are also weighted by the strength of the correlation. Each edge represents a positive correlation between the two genera with a Pearson’s correlation coefficient above 0.40. (C) Cooccurrence node interaction strength plot showing the number of interacting genera per node in each sample (including false-negative and -positive degrees). (D) The strength distribution of the microbial cooccurrence at four phases.
To further explore dynamic changes in the microbial community in detail, we constructed a cooccurrence network of microorganisms in the four phases. We found that although the dominant microbiome was always Caproiciproducens, the topological structure of the cooccurrence network was significantly different under different HRT conditions, and it also varied over time (Fig. 4B). In the nonsterile open environment, the network was simple at the beginning, but after 180 days of operation, the observed number of OTUs increased (Table S1), forming a subnetwork with fully connected topology. Our analysis shows that the number of genus-genus interactions (i.e., interaction strength) and the strength of the microbial cooccurrence were both significantly increased (Fig. 4C and D). The change in taxonomic community negatively affected the production of n-caproate from lactate.
When we shortened the HRT, the structure of the cooccurrence network became loose again (Fig. 4B), and the average interactions per genus declined from 17 to 5 (Fig. 4C), implying that some species that potentially interact with each other were seriously affected or even diluted from the reactor. Interestingly, approximately 200 days later, the topology of the cooccurrence network changed again (Fig. 4B), and the strength of the microbial cooccurrence became greater in phase 4 than in phase 3 (Fig. 4D). However, the interaction strength in phase 4 was still significantly lower than those in phases 1 and 2 (Fig. 4C). Combined with the reactor performance, we found that this change significantly enhanced the n-caproate production efficiency (concentration, production rate, and specificity) from lactate.
In summary, under nonsterile open conditions, the reactor microbiome was always occupied by the n-caproate-producing bacterium Caproiciproducens, suggesting that the controlled environment (mild acid, lactate rich) was selected for the specific metabolic pathway of lactate-driven chain elongation. However, the microbial community changed dynamically with changes in operating conditions (HRTs) and time, gradually corrupting or improving the performance of the lactate-driven chain elongation bioprocess. This result raises a question regarding the roles of different genera in bioprocess efficiency.
(iii) Connection between taxonomic composition and the variations in performance of the bioprocess.
We performed correlation analysis between the variation in n-caproate concentration and the dynamic change in Caproiciproducens abundance, and we observed no significant correlation (Spearman's ρ = 0.02). Hence, we infer that the decrease or increase in n-caproate concentration was mainly caused by taxonomic variability and biotic interactions in the reactor microbiome.
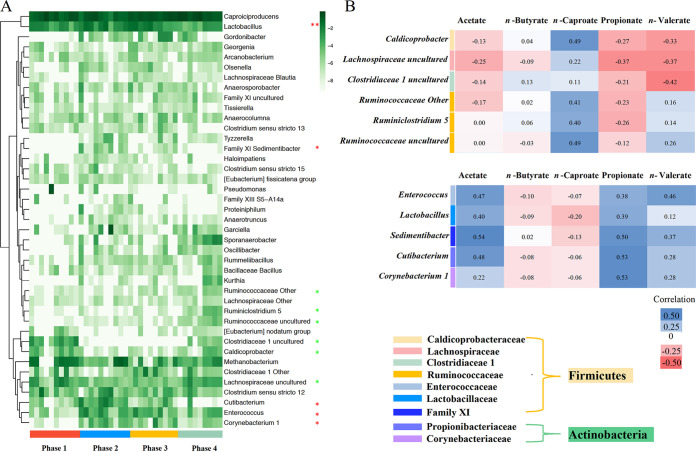
To investigate how variation in taxonomic composition was related to n-caproate production, we split the community variation into 41 distinct genus (with average relative abundance of more than 0.1%) variations (Fig. 5A). We noticed an approximately 10-fold decrease in the relative abundance of three genera from phase 1 to phase 2 but a recovery at phases 3 and 4 (P < 0.05) (Tables S5 and S6). These genera are Lachnospiraceae uncultured, Clostridiaceae 1 uncultured, and Caldicoprobacter. Furthermore, we observed another three genera, Ruminiclostridium 5, uncultured Ruminococcaceae, and Ruminococcaceae other, that increased significantly at phase 4 (P < 0.05) (Tables S5 and S6).
FIG 5.
Variations in taxonomic community and taxon function prediction. (A) Heatmap showing variations of relative abundances in microbial communities at the genus level (41 bacterial genera that had an average relative abundance of greater than 0.1%; Fig. S2). To highlight taxa with low relative abundances, values of relative abundances were converted to logarithm and clustered as n-caproate production-positive and -negative associated organisms, respectively (Table S5 and Table S6; P < 0.05 [*] and P < 0.01 [**]). (B) Spearman’s rank correlation between carboxylate production and related genera. Blue and red colors indicate positive and negative correlations, respectively. Color saturation is proportional to the absolute correlation.
At the same time, several genera showed opposing trends in changes in relative abundances. Cutibacterium (formerly Propionibacterium), which is known as a propionate-producing genus (42), was the most typical genus among them. Its relative abundance increased by more than 17 times (0.351% ± 0.418% versus 6.08% ± 6.08%, P < 0.02) from phase 1 to phase 2, but it finally declined back to 0.480% ± 1.36% (phase 4) as the HRT shortened over time. Another four taxa observed were Lactobacillus, Enterococcus, Family XI Sedimentibacter, and Corynebacterium 1.
According to correlation analysis, Lachnospiraceae uncultured, Clostridiaceae 1 uncultured, Ruminiclostridium 5, Ruminococcaceae uncultured, and Ruminococcaceae other are positively associated with n-caproate production. The other five taxa, i.e., Cutibacterium, Lactobacillus, Enterococcus, Family XI Sedimentibacter, and Corynebacterium 1, showed a negative correlation with n-caproate production but a positive correlation with acetate, propionate, and n-valerate production (Fig. 5B).
Prediction of the metabolic network profile by metagenome analysis.
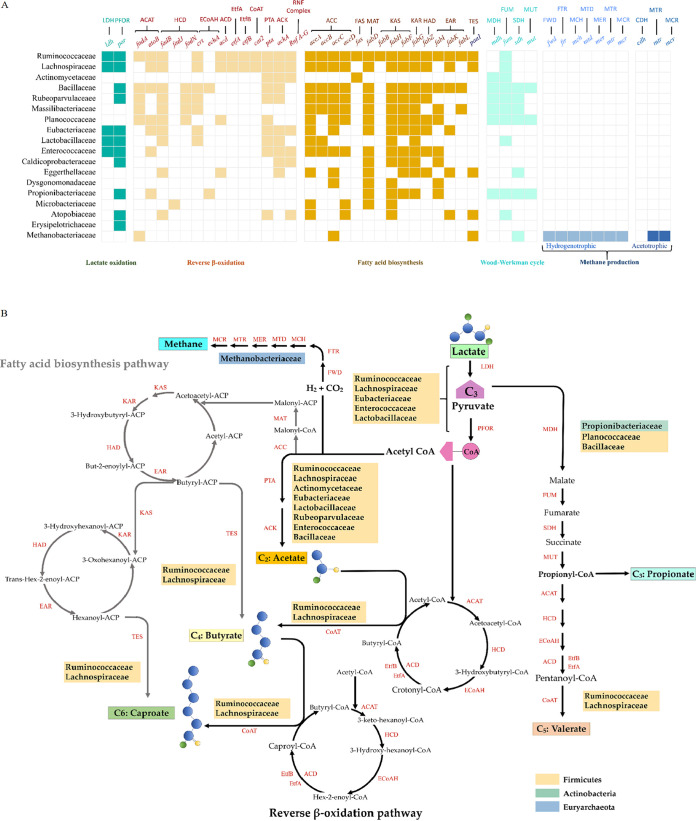
In the nonsterile open environment with lactate as the sole carbon source, the metabolites were mainly SCCs (acetate, propionate, n-butyrate, and n-valerate) and MCC n-caproate (Fig. 1). This section provides insights into the predictive microbial metabolic network of the reactor microbiome on the basis of an analysis of the metagenome. Detailed information about the metagenome results, including sequencing quality, metagenome assembly information, and categories of functional genes at different levels, is presented in the supplemental material (Tables S7 to S11 and Fig. S4 and S5). Based on the sequencing data, we successfully reconstructed 43 metagenome-assembled genomes (>70% complete, <5% contamination; Table S12), which almost coincided with the families (except Burkholderiaceae) in the network based on 16S rRNA (Fig. 2). In addition, we found two other genomes outside the network, affiliating with Rubeoparvulaceae and Massilibacteriaceae. No bacteria carry both coding genes for ethanol oxidization, i.e., adh (K19955) and ada (K00132), indicating that ethanol is not the source for n-caproate production in our biosystem. All of the functional genes of the metagenome were annotated against the KO database to carry out enzyme assays to further investigate lactate metabolic pathways, including lactate oxidation, chain elongation, propionate production, and methane production (Table S13).
In this analysis, two cyclic pathways for chain elongation were identified: fatty acid biosynthesis (FAB) and reverse β oxidation (RBO). Additionally, instead of the acrylate pathway, we only found key genes associated with the Wood-Werkman cycle for the conversion of lactic acid to propionate via a succinate intermediate. We summarized the major microbiomes and their hosted key genes encoding related enzymes in Fig. 6A. In the sequencing gene bank, we found that the main family Ruminococcaceae has all the necessary genes in the FAB pathway as well as the RBO pathway, indicating that Ruminococcaceae contributes to n-caproate production at the gene level. In addition, we found that Lachnospiraceae also have all the necessary chain elongation genes, but we did not find all of the necessary genes in the reconstructed microbe. Scarborough et al. reported that a species, Shuttleworthia satelles, from the family Lachnospiraceae contains genes needed for the RBO pathway, and we found the species in our metagenome analysis, representing 0.002% of total microorganisms. Whether the family can produce n-caproate still requires further clarification, but these results implied that Lachnospiraceae is an n-caproate production-associated family.
FIG 6.
Metabolic pathway profile with lactate as the sole electron donor and taxon function summarization and prediction based on 16S rRNA and metagenome sequences.
The lactate fermentation-associated bacteria Enterococcaceae and Lactobacillaceae have the key genes encoding enzymes that catalyze the oxidation of lactate to acetyl-CoA. Eight families, Bacillaceae, Actinomycetaceae, Eubacteriaceae, Enterococcaceae, Rubeoparvulaceae, Lactobacillaceae, Runimococcaceae, and Lachnospiraceae, have the pta and ackA genes, encoding phosphate acetyltransferase and acetate kinase for acetate formation from acetyl-CoA (42). Three families, Bacillaceae, Planococcaceae, and Propionibacteriaceae, possess all of the genes of the Wood-Werkman cycle needed for propionate production. We did not find the necessary genes in the acetotrophic methane production pathway, but all of the genes in the hydrogenotrophic methane production pathway were found in Methanobacteriaceae.
Based on the gene information and the metabolic products from lactate, we predicted a metabolic network, which is illustrated in Fig. 6B. Eight bacteria were associated with lactate oxidation or acetate production. Three bacteria (Bacillaceae, Planococcaceae, and Propionibacteriaceae) may contribute to propionate production. The identified n-caproate producer belongs to the Ruminococcaceae family, while Lachnospiraceae may be positively associated with n-caproate production. In the LCE biosystem, no acetotrophic methanogen was present, but a hydrogenotrophic methanogen was present. In addition, we found no gene information about the function of six families, Bogoriellaceae, Thermoactinomycetaceae, Clostridiaceae 1, Burkholderiaceae, and Clostridiales Family XI and Family XIII, according to metagenome data.
DISCUSSION
Functional group and competitive group.
According to our present results, we divided the microbial ecology into two groups: a functional group that may directly or indirectly contribute to n-caproate production and a competitive group that may reduce n-caproate production efficiency.
The functional group is mainly Ruminococcaceae, which may include Caproiciproducens, Ruminiclostridium 5, Ruminococcaceae uncultured, and other Ruminococcaceae. There are 17 species based on 16S rRNA sequencing belonging to these four genera (Table 4). The variation trend in the relative abundance of all these species was consistent with that of the n-caproate concentration. Among them, one species, Ruminococcaceae bacterium CPC-11, was reported to produce n-caproate from lactate in a pure-culture system (43). Based on the metagenome and Spearman's rank correlation analysis, we classified Lachnospiraceae as a functional group. There was only one species, Lachnospiraceae uncultured Coprococcus sp., in this family. Scarborough et al. predicted that Lachnospiraceae-affiliated organisms play an important role in MCCs production, so our results were partly consistent with the conclusion of Scarborough et al. (41).
Although the relative abundance of Ruminococcaceae has no direct relationship with bioreactor efficiency (Spearman's correlation coefficient [ρ], 0.02), our results showed a connection: the relative abundance is positively related to performance. This may not be caused by functional bacteria; underneath the surface may be the expansion or shrinkage of competitive bacteria. Combined with the metagenome analysis results, Bacillaceae, Planococcaceae, and Propionibacteriaceae were classified as potential competitive groups. Among these families, Cutibacterium, affiliated with Propionibacteriaceae, was confirmed to be a strong competitor. The research of Candry et al. (34) also demonstrated that propionate producer is the dominant competitor for the chain elongator, Caproiciproducens. Candry et al. (34) suggested that the main propionate producers are Veillonella and Aminobacterium, and pH value (lower than 6.0) is the key regulating factor to inhibit the two bacteria. Therefore, the strategy to inhibit propionate production depends on the genus of propionate producers. For Cutibacterium in Propionibacteriaceae, we should give priority to optimizing HRT, and for Veillonella, we should further consider the pH value. Acetotrophic methanogens have been regarded as a typical competitor for acetate used in chain elongation. Here, we found no acetotrophic methanogens but hydrogentrophic methanogens, Methanobacteriaceae, which consume hydrogen for methane production. Our results indicated that the methane ratio in biogas was negatively related to production efficiency of n-caproate. The variation in hydrogen was consistent with the trend of n-caproate production (Fig. S3, Table S4). Given that hydrogen is very important in maintaining the thermodynamic conditions preventing anaerobic oxidation (1, 44), we infer that the existence of Methanobacteriaceae brings the risk of lowering n-caproate production efficacy.
Some taxa in the bioreactor may utilize lactate to produce acetyl-CoA, such as Ruminococcaceae, Lachnospiraceae, Eubacteriaceae, Lactobacillaceae, and Enterococcaceae. Acetyl-CoA can be further catalyzed into acetate by Ruminococcaceae, Lachnospiraceae, Actinomycetaceae, Rubeoparvulaceae, Lactobacillaceae, Enterococcaceae, Eubacteriaceae, and Bacillaceae (Fig. 6B). On the one hand, acetyl-CoA and acetate are both necessary metabolites for chain elongation (Fig. 6B); on the other hand, excessive acetate accumulation may consume too much lactate, leading to a low lactate/acetate ratio, which may be thermodynamically unfavorable for n-caproate production (1). Combined with Spearman's correlation analysis, Enterococcaceae, Lactobacillaceae, and Rubeoparvulaceae are primarily suggested to be conditioned competitive groups until we find additional evidence. In research by Scarborough et al. (41), the Eubacteriaceae-affiliated species Pseudoramibacter alactolyticus was recognized as a functional n-caproate-producing bacterium. Here, only one species was found in the family, Eubacteriaceae uncultured bacterium, which is different from Pseudoramibacter alactolyticus. Therefore, the function of Eubacteriaceae in the LCE biosystem may depend on the particular species. At present, there are still many taxa in the LCE biosystem whose function could not be determined.
Hypothetical microbial ecological model for the LCE reactor microbiome that produces a high concentration of n-caproate and implications.
Based on our research, we propose a hypothetical model to reveal the mechanism by which an LCE reactor microbiome produces a high concentration of n-caproate. The model is comprised of four parts. (i) The core microbiome involves efficient, n-caproic acid toxicity-tolerant functional groups. Here, the core microbiome for the LCE biosystem is Ruminococcaceae, which fundamentally underlies the observed network structure. Without this microbiome, the biosystem would perform much differently. (ii) The core bacteria can maintain stability for long-term operation regardless of the variations in the microbial network structure, which might be caused by changes in operation conditions or disrupted feedback. (iii) Microbial network analysis revealed that the microbial community yielding higher n-caproate concentrations showed relatively lower microbe-microbe interaction strength. The core bacteria have all of the genes needed for lactate oxidization and chain elongation, and they can also utilize glucose or sucrose for cell growth (26). The functional microbes were relatively independent of the community and showed a negative relationship with other bacteria. A relatively short HRT could reduce the interaction strength. (iv) Low relative abundances and taxonomic diversity of competitors in the community were also observed.
To date, many bacteria for n-caproate production from lactate have been identified, such as Megasphaera hexanoica, Megasphaera indica, Pseudoramibacter alactolyticus, clostridial species strains BL-3, BL-4, and BL-6, and so on (41, 45–47), but none of these species produced n-caproate at the same high concentration as Ruminococcaceae strain CPB6. Therefore, we suggest that the functional species are very important for the LCE process. Fortunately, strain CPB6, or Ruminococcaceae-affiliated Caproiciproducens, has been found in many LCE bioreactors (13, 27, 29, 34, 48). The colonization and accumulation of functional bacteria is usually related to the environment, e.g., the operation conditions of the process and feedback, which can select for specific microbiomes. For example, long-term operation at a long HRT led to the growth of a competitor, Propionibacteriaceae, which significantly reduced the concentration of n-caproate in the effluent. In some works (31, 33), researchers prefer quite short HRTs (e.g., 4 h) to increase the production rate, but we are worried that the functional anaerobic n-caproate producer may be washed out from the biosystem for their relatively longer growth cycle. pH is also critical for n-caproate producers, and Candry et al. (34) found that a lower pH (5.5) was a favorable condition for Caproiciproducens.
Previous studies usually found two dominant by-products, i.e., n-butyrate (12, 28, 41, 49) and propionate (31) accumulation, decreasing n-caproate production efficacy. Here, we discussed propionate accumulation, which was mainly caused by competitors, e.g., Propionibacteriaceae. Kucek et al. (31) indicated that excessive lactate is the main reason for propionate accumulation, but we found no significant correlation in our system in which lactate was always oversupplied. The difference may be caused by different microbial communities, especially the dominant bacteria.
In our research, a significant increase in the n-caproate concentration was always accompanied by a slight increase in n-butyrate accumulation, which was soon minimized with steady n-caproate production (Fig. 1). The enzyme analysis in pure culture of strain CPB6 indicated that the production rate of n-caproate was 2.34 times higher than that of n-butyrate (25). In our system, one key functional bacterium is strain CPB6, which provides a reason for the low level of n-butyrate accumulation. In addition, because n-butyrate is intermediate in chain elongation, maintaining n-butyrate at some critical level could permit favorability for additional n-caproate production (26). In some studies, n-butyrate was observed as the dominant product of chain elongation (12, 41). We suggest that the phenomenon is related to the structure of reactor microbiome and the types of functional n-caproate bacteria. Therefore, we speculate that the selection (or domestication) of the chain elongation reactor microbiome will take a long time (usually years) before a balance has been reached. During long-term selection in the proper environment (e.g., mild acid conditions (pH 5.5 to 6.0), moderate temperature (30 to 40°C), and continuous lactate supplementation), severe n-butyrate accumulation could be eliminated by gradual utilization for chain elongation. At present, we have no better understanding of any other operation conditions that can minimize n-butyrate accumulation.
Challenge and opportunity. (i) Challenge: acid inhibition and recovery.
Under mild acid conditions, a high concentration of n-caproate can easily induce acid inhibition according to reported works (1, 50). In the ethanol-driven chain elongation (ECE) process, the reported toxic limit of undissociated n-caproate was 6.8 to 7.5 mM (18, 51), and in the LCE process, the inhibitory concentration was 17.2 mM based on the work of Duber et al. (32). In this paper, at a pH of 6.0, the calculated undissociated n-caproate was 10.3 ± 3.20 mM (average value), and it could reach 15.4 ± 2.40 mM when a high concentration of n-caproate was produced (phase 4). During two periods of severe inhibition (days 377 to 407 and days 753 to 777), the calculated undissociated n-caproate was 10.7 and 12.4 mM, respectively, much lower than the reported toxic limit of 17.2 mM as well as the highest concentration of 15.4 ± 2.40 mM in phase 4. However, we noticed excessive lactate accumulation (17.7 and 22.4 g liter−1, respectively) just before both inhibition cases. Generally, lactate is relatively nontoxic; therefore, lactate accumulation may be a signal that inhibition is about to occur. At present, we do not understand the specific reason for the inhibition, but we suggest that pH control (involved in acid, e.g., HCl, addition) would induce a drastic increase in the concentrations of undissociated n-caproic acid and other short-chain fatty acids in a short time, resulting in synergistic toxic effects on the reactor microbiome. However, lactate-driven chain elongation requires pH control to maintain mild acid conditions; therefore, approaches to resolve this dilemma need additional research.
To recover from inhibition, we have no better strategy than the following: discharge the fermentation liquid and replenish fresh feedback to reduce n-caproate and/or lactate concentration, sharing the same basis as in-line extraction to circumnavigate the toxicity. Therefore, the LCE process is facing other dilemmas: (i) it is uneconomical to extract n-caproate at low concentrations, and (ii) high concentrations may cause serious inhibition at one point, leading to stagnation and restarting.
(ii) Opportunity.
Our research illustrates that functional strains are the keystone for the LCE process, implying that it is possible to overcome n-caproate inhibition and improve synthetic efficiency when additional efforts could be made in strain domestication, gene modification, and novel bacterial discovery. Currently, many new biotechnologies, e.g., omics technology and gene editing technology, have great potential to improve n-caproic acid tolerance, enhancing the efficacy of functional bacteria. Additionally, we believe that various chain elongation bacteria may inhabit lactate-rich environments, such as a factory containing lactate fermentation (51), and carbohydrate-rich wet waste treatment plants (29, 32), which may provide us with a natural strain bank. Emerging artificial intelligence, single-cell sorting, and new-generation microbial culture technologies will bring us additional opportunities for the discovery, isolation, and cultivation of novel species.
Furthermore, reported works usually utilized similar bioreactors for anaerobic digestion (such as CSTR, UASB, and EGSB) (33, 35, 52). Indeed, the fermentation characteristics of LCE are significantly different from those of anaerobic digestion, for instance, continuous pH control, product inhibition, and intense competition (SCC producers) for environmental niches. Thus, we need a more targeted design for the LCE process, including reactor design, microbial fixation material, operation principle, and so on.
We anticipate that the understanding of microbial ecology for high concentrations of n-caproate production will provide guidance or inspiration for researchers to promote the development of the LCE process, which will become one of the important drivers for the sustainable development of human society.
MATERIALS AND METHODS
Long-term n-caproate production from lactate.
The artificial wastewater contained (per liter) 5 ml of solution A (containing 10 g liter−1 MgSO4·7H2O, 4.5 g liter−1 CaCl2·H2O, 93.6 g liter−1 NH4Cl), 2 ml of solution B (containing 29 g liter−1 KH2PO4 and 33 g liter−1 K2HPO4), a certain amount of the carbon source (dl-lactate), 5%, vol/vol, real wastewater (containing dl-lactate, 70 to 100 g liter−1; acetate, 1.5 ± 0.08 g liter−1; n-butyrate, 1.0 ± 0.02 g liter−1; ethanol, 20 to 40 g liter−1; pH 3.80 ± 0.20), 1 ml of vitamin solution [containing biotin, 2.0 mg liter−1; folic acid, 2.0 mg liter−1; pyridoxine hydrochloride, 10.0 mg liter−1; thiamine HCl, 5.0 mg liter−1; riboflavin, 5.0 mg liter−1; nicotinic acid, 5.0 mg liter−1; calcium d-(+)-pantothenate, 5.0 mg liter−1; vitamin B12, 0.1 mg liter−1; p-aminobenzoic acid, 5.0 mg liter−1; thioctic acid, 5.0 mg liter−1] and 1 ml of trace element solution [containing nitrilotriacetic acid, 2.0 g liter−1; MnSO4·H2O, 1.0 g liter−1; Fe(SO4)2(NH4)2·6H2O, 0.8 g liter−1; CoCl2.6H2O, 0.2 g liter−1; ZnSO4·7H2O, 0.2 mg liter−1; CuCl2·2H2O, 20.0 mg liter−1; NiCl2·6H2O, 20.0 mg liter−1; Na2MoO4·2H2O, 20.0 mg liter−1; Na2SeO4, 20.0 mg liter−1; Na2WO4, 20.0 mg liter−1].
In the setup stage, 100 ml of preculture of the mixed microbiome (13) was inoculated into the bioreactor. After 15 days of culture, the reactor was operated for a period of 24 h with a sequence of four steps: step 1, external circulation (30 min); step 2, drainage (5 min); step 3, feeding (25 min); and step 4, treatment (23 h). The initial lactate concentration of every cycle was controlled at approximately 10 g·liter−1. The pH was typically adjusted to 6.0 by 5 M NaOH and 5 M HCl at the beginning of each cycle. During the entire reaction period of each cycle, the pH was monitored by a pH transmitter (H100; Hamilton, Switzerland) equipped with a pH electrode (120 mm; Hamilton, Switzerland). The experiment was performed in reactors with a working volume of 3 liters, and the HRT was controlled at 15 days for 400 days. After 400 days, the HRT was decreased to 8 days. Reactors were placed in a thermostatic room at a temperature of 30 ± 1°C. The experiment was performed in duplicate. The long-term operation was divided into three stages: stage 1, setup (0 to 15 days); stage 2, continuous n-caproate production; and stage 3, inhibition period. Stage 2 was separated into four phases: (i) phase 1 (16 to 180 days), stable n-caproate production at an HRT of 15 days; (ii) phase 2 (181 to 385 days), decreased efficiency of n-caproate production at an HRT of 15 days; (iii) phase 3 (408 to 635 days), the domestication period for n-caproate production at an HRT of 8 days; and (iv) phase 4 (636 to 752 days), high n-caproate production efficiency at an HRT of 8 days. Stage 3 contained two inhibition periods (days 386 to 407 and days 753 to 777).
Analysis of the microbial community.
Samples (50 ml) were withdrawn from the reactors at different phases. All of these samples were centrifuged at 12,857 × g for 10 min, and then pellets were used for genomic DNA extractions. The Power Soil DNA isolation kit (MoBio Laboratories, USA) was used. DNA density and quality were checked using a NanoDrop spectrophotometer. Extracted DNA was diluted to a concentration of 10 ng μl−1 and stored at –40°C for downstream use. The universal primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with 10-nucleotide (nt) barcodes were used to amplify the V4 hypervariable region of 16S rRNA genes for next-generation sequencing using a MiSeq sequencer (53, 54). The sequence data were processed using QIIME Pipeline, version 1.8.0. All sequence reads were trimmed and assigned to each sample based on their barcodes. Multiple steps were required to trim the sequences, such as the removal of sequences of <150 bp and those with an average base quality score (Q) of <30. We aligned cleaned sequence reads against the SILVA rRNA database (version 132). The relative abundances of genera in each sample were calculated on the basis of the 16S rRNA sequences with unique assignment of the classified genera. Alpha diversity (Shannon index) and beta diversity (weighted UniFrac) were calculated using QIME. Principal component analysis (PCA) was calculated using R based on the OTU table. Nonparametric multivariate analysis of Tukey’s test was employed to analyze community structure (analysis of similarity [ANOSIM]) in SPSS and R.
Correlation and cooccurrence analysis.
Hierarchical clustering was performed on the relative abundance profiles of the families and genera found in samples with Cluster 3.0. The distance measurement was based on the Spearman rank correlation between samples. The heatmap was generated using R package. To construct the cooccurrence network, we first filtered the low-abundance taxa that had average coverage by the Illumina reads of ≤0.1%, and then we calculated the pairwise interspecies Pearson correlations between sequencing depths (abundance) throughout the 39 samples (or samples in each phase). From the resulting numerous interspecies correlations, correlations above 0.4 were visualized using Cytoscape, displaying the average relative abundance of each taxon as the node size in the graph. Finally, to exclude coabsence, cooccurrences were not counted for taxa for which the sum of relative abundance in the four phases was less than 1%.
Metagenomic DNA extraction and sequencing.
Two samples (50 ml) were withdrawn from the reactors at the end of the experiment. Two samples were centrifuged at 12,857 × g for 10 min, and then pellets were used for DNA extraction. Total DNA was extracted from samples using a phenol-chloroform extraction method (55). The DNA concentration was measured using a Qubit dsDNA assay kit in a Qubit 2.0 fluorometer (Life Technologies, CA, USA).
The optical density at 600 nm (OD600) was between 1.8 and ∼2.0, and DNA samples with contents above 1.0 μg were used to construct a library. A total of 1.0 μg of DNA per sample was used as the input material for the DNA sample preparations. Sequencing libraries were generated using the NEBNext Ultra DNA library preparation kit for Illumina (NEB, USA) by following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Briefly, the DNA sample was fragmented by sonication to a size of 350 bp, and then DNA fragments were end-polished, A-tailed, and ligated with the full-length adaptor for Illumina sequencing with further PCR amplification. Finally, PCR products were purified (AMPure XP system), and libraries were analyzed for size distribution by an Agilent 2100 Bioanalyzer and quantified using real-time PCR. Clustering of the index-coded samples was performed on a cBot Cluster Generation System according to the manufacturer’s instructions. After cluster generation, paired-end sequencing (2× 150 bp) was performed on an Illumina NovaSeq 6000 platform (Illumina, San Diego, CA), and paired-end reads were generated.
Metagenomic analysis.
The preprocessing of the raw data obtained from the Illumina HiSeq sequencing platform using Readfq (V8; https://github.com/cjfields/readfq) was conducted to acquire clean data for subsequent analysis. All the reads of all samples were combined, and MEGAHIT software (v1.0.4-beta) was used for mixed assembly. The mixed assembled scaffolds were broken from N connections, and scaftigs were obtained. Filtered fragments shorter than 500 bp in all scaftigs were used for statistical analysis generated from mixed assembly. The scaftigs (≥500 bp) assembled from mixed assembly were predicted to the open reading frame level by MetaGeneMark (V2.10, http://topaz.gatech.edu/GeneMark/) software. DIAMOND (56) software (V0.9.9, https://github.com/bbuchfink/diamond/) was used for BLAST analysis of the unigenes to the sequences of bacteria and archaea, which were all extracted from the NR database (version 2018-01-02; https://www.ncbi.nlm.nih.gov/) of NCBI. For the final aligned results of each sequence, the result with an e value equal to or smaller than the smallest e value times 10 (57) was chosen for the LCA algorithm, which was applied to the system classification of MEGAN (58) software to verify the species annotation information of sequences. DIAMOND software (V0.9.9) was used to perform BLAST analysis of unigenes in the functional database (59, 60). The functional databases used included the KEGG database (version 2018-01-01; http://www.kegg.jp/kegg/) (61), eggNOG database (version 4.5; http://eggnogdb.embl.de/#/app/home) (62), and CAZy database (version 201801; http://www.cazy.org/) (63).
Metagenomic assembly and binning.
The clean reads and contigs were used for binning and refining genomes using MetaBAT2 (64). The genome bins were then checked for completeness and contamination using CheckM (65), and a total of 43 bins were selected after screening bins whose completeness was more than 70% and contamination was less than 5%. Genome taxonomy annotation was performed by GTDBtk (version 1.3.0) based on the GTDB database (66). Genome functional annotation was performed by Prokka (version 1.14.6) based on the UniProtKB database (67).
Calculations.
The average productivity of each product (n-butyrate, n-valerate, and n-caproate) was defined by its average concentration divided by the hydraulic retention time. The average specificity of each product was based on the product-to-carboxylate production ratio (in % mol C). The concentration of undissociated acid was calculated from the Henderson-Hasselbach equation: undissociated acid (mmol liter−1) = total acid (mmol liter−1)/(1 + 10pH − pKa). The pKa of acetate, propionate, n-butyrate, n-valerate, and n-caproate were 4.74, 4.86, 4.87, 4.82, 4.86, and 4.83, respectively.
Chemical analysis.
Liquid samples were collected daily, centrifuged for 5 min at 12,857 × g, diluted 20 times with distilled water, and subsequently sterilized using a 0.22-μm filter. Carboxylate (C1 to C6) and lactate concentrations were determined using an Agilent 1260 Infinity liquid chromatography system (Agilent, Santa Clara, CA, USA), which was equipped with a Hi-Plex H high-performance liquid chromatography column (300 by 6.5 mm) and a differential refraction detector (RID). The column temperature was controlled at 55°C. H2SO4 (0.005 M) at a flow rate of 0.6 ml/min was used as the mobile phase. The gas produced was collected every 3 days and analyzed with an Agilent 7890B gas chromatography system (Agilent, Santa Clara, CA, USA), which was equipped with a thermal conductivity detector and a 1.83-m stainless steel column packed with Porapak Q (80/100 mesh). The operating temperatures at the injection port, column oven, and detector were 150, 80, and 200°C, respectively. Argon, at a flow rate of 25 ml/min, was used as the carrier gas.
Data availability.
The raw sequence data (raw metagenomic sample data and raw 16S amplicon sample data) reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under accession number CRA003483, which is publicly accessible at https://bigd.big.ac.cn/gsa.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Natural Science Foundation of China (21777153), Youth Innovation Promotion Association CAS, Western Light Class A project CAS (2018XBZG_XBQNXZ_A_006), Special Talent Project of Organization Department of Sichuan Provincial Party Committee, Science and Technology Support Program of Sichuan Province (2016JZ0010), and Technology Program of Chengdu City (2016-HM01-00355-SF).
X.Z., C.L., and J.J. designed the experiments; X.Z., X.F., C.L., J.J., and L.F. performed the experiments; X.Z., X.F., J.L., and Y.T. analyzed and interpreted the data; X.Z., X.F., and Y.C. drafted the manuscript. All authors reviewed and critically revised the manuscript and approved the final version for publication.
We declare that we have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Angenent LT, Richter H, Buckel W, Spirito CM, Steinbusch KJJ, Plugge CM, Strik D, Grootscholten TIM, Buisman CJN, Hamelers HVM. 2016. Chain elongation with reactor microbiomes: open-culture biotechnology to produce biochemicals. Environ Sci Technol 50:2796–2810. 10.1021/acs.est.5b04847. [DOI] [PubMed] [Google Scholar]
- 2.Marshall CW, LaBelle EV, May HD. 2013. Production of fuels and chemicals from waste by microbiomes. Curr Opin Biotechnol 24:391–397. 10.1016/j.copbio.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Spirito CM, Richter H, Rabaey K, Stams AJM, Angenent LT. 2014. Chain elongation in anaerobic reactor microbiomes to recover resources from waste. Curr Opin Biotechnol 27:115–122. 10.1016/j.copbio.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Urban C, Xu J, Sträuber H, Dantas T, Mühlenberg J, Härtig C, Angenent L, Harnisch F. 2017. Production of drop-in fuel from biomass at high selectivity by combined microbial and electrochemical conversion. Energy Environ Sci 10:2231–2244. 10.1039/C7EE01303E. [DOI] [Google Scholar]
- 5.Harvey BG, Meylemans HA. 2014. 1-Hexene: a renewable C6 platform for full-performance jet and diesel fuels. Green Chemistry 16:770–776. 10.1039/C3GC41554F. [DOI] [Google Scholar]
- 6.Augustin K, Khabbush A, Williams S, Eaton S, Orford M, Cross JH, Heales SJR, Walker MC, Williams RSB. 2018. Mechanisms of action for the medium-chain triglyceride ketogenic diet in neurological and metabolic disorders. Lancet Neurol 17:84–93. 10.1016/S1474-4422(17)30408-8. [DOI] [PubMed] [Google Scholar]
- 7.Woolford M. 1975. Microbial screening of the straight chain fatty acids (C1-C12) as potential silage additives. J Sci Food Agric 26:219–228. 10.1002/jsfa.2740260213. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Hao J, Guzman J, Spirito C, Harroff L, Angenent L. 2018. Temperature-phased conversion of acid whey waste into medium-chain carboxylic acids via lactic acid: no external e-donor. Joule 2:280–295. 10.1016/j.joule.2017.11.008. [DOI] [Google Scholar]
- 9.Barker HA, Kamen MD, Bornstein BT. 1945. The synthesis of butyric and caproic acids from ethanol and acetic acid by Clostridium kluyveri. Proc Natl Acad Sci U S A 31:373–381. 10.1073/pnas.31.12.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seedorf H, Fricke WF, Veith B, Brüggemann H, Liesegang H, Strittmatter A, Miethke M, Buckel W, Hinderberger J, Li F, Hagemeier C, Thauer RK, Gottschalk G. 2008. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci U S A 105:2128–2133. 10.1073/pnas.0711093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao Y, Zhu X, Wang H, Wang Y, Li X, Jin H, Rui J. 2017. Complete genome sequence of Ruminococcaceae bacterium CPB6: a newly isolated culture for efficient n-caproic acid production from lactate. J Biotechnol 259:91–94. 10.1016/j.jbiotec.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 12.Han W, He P, Shao L, Lü F. 2018. Metabolic interactions of a chain elongation microbiome. Appl Environ Microbiol 84:e01614-18. 10.1128/AEM.01614-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Tao Y, Liang C, Li X, Wei N, Zhang W, Zhou Y, Yang Y, Bo T. 2015. The synthesis of n-caproate from lactate: a new efficient process for medium-chain carboxylates production. Sci Rep 5:14360. 10.1038/srep14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weimer PJ, Stevenson DM. 2012. Isolation, characterization, and quantification of Clostridium kluyveri from the bovine rumen. Appl Microbiol Biotechnol 94:461–466. 10.1007/s00253-011-3751-z. [DOI] [PubMed] [Google Scholar]
- 15.Roddick F, Britz M. 1997. Production of hexanoic acid by free and immobilised cells of Megasphaera elsdenii: influence of in‐situ product removal using ion exchange resin. J Chem Technol Biotechnol 69:383–391. . [DOI] [Google Scholar]
- 16.Agler M, Spirito C, Usack J, Werner J, Angenent L. 2012. Chain elongation with reactor microbiomes: upgrading dilute ethanol to medium-chain carboxylates. Energy Environ Sci 5:8189–8192. 10.1039/c2ee22101b. [DOI] [Google Scholar]
- 17.Grootscholten TIM, Strik D, Steinbusch KJJ, Buisman CJN, Hamelers HVM. 2014. Two-stage medium chain fatty acid (MCFA) production from municipal solid waste and ethanol. Appl Energy 116:223–229. 10.1016/j.apenergy.2013.11.061. [DOI] [Google Scholar]
- 18.Ge S, Usack JG, Spirito CM, Angenent LT. 2015. Long-term n-caproic acid production from yeast-fermentation beer in an anaerobic bioreactor with continuous product extraction. Environ Sci Technol 49:8012–8021. 10.1021/acs.est.5b00238. [DOI] [PubMed] [Google Scholar]
- 19.Steinbusch K, Hamelers HVM, Plugge C, Buisman C. 2011. Biological formation of caproate and caprylate from acetate: fuel and chemical production from low grade biomass. Energy Environ Sci 4:216–224. 10.1039/C0EE00282H. [DOI] [Google Scholar]
- 20.Roghair M, Liu Y, Strik D, Weusthuis RA, Bruins ME, Buisman CJN. 2018. Development of an effective chain elongation process from acidified food waste and ethanol into n-caproate. Front Bioeng Biotechnol 6:50. 10.3389/fbioe.2018.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin M, Dai X, Weimer P. 2019. Shifts in fermentation end products and bacterial community composition in long-term, sequentially transferred in vitro ruminal enrichment cultures fed switchgrass with and without ethanol as a co-substrate. Bioresour Technol 285:121324. 10.1016/j.biortech.2019.121324. [DOI] [PubMed] [Google Scholar]
- 22.Wu Q, Feng X, Chen Y, Liu M, Bao X. 2021. Continuous medium chain carboxylic acids production from excess sludge by granular chain-elongation process. J Hazard Mater 402:123471. 10.1016/j.jhazmat.2020.123471. [DOI] [PubMed] [Google Scholar]
- 23.Han W, He P, Shao L, Lü F. 2019. Road to full bioconversion of biowaste to biochemicals centering on chain elongation: a mini review. J Environ Sci 86:50–64. 10.1016/j.jes.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Chen WS, Strik D, Buisman C, Kroeze C. 2017. Production of caproic acid from mixed organic waste: an environmental life cycle perspective. Environ Sci Technol 51:7159–7168. 10.1021/acs.est.6b06220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X, Zhou Y, Wang Y, Wu T, Li X, Li D, Tao Y. 2017. Production of high-concentration n-caproic acid from lactate through fermentation using a newly isolated Ruminococcaceae bacterium CPB6. Biotechnol Biofuels 10:102. 10.1186/s13068-017-0788-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Li X, Wang Y, Tao Y, Lu S, Zhu X, Li D. 2018. Improvement of n-caproic acid production with Ruminococcaceae bacterium CPB6: selection of electron acceptors and carbon sources and optimization of the culture medium. Microb Cell Fact 17:99. 10.1186/s12934-018-0946-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Contreras-Dávila CA, Carrión VJ, Vonk VR, Buisman CNJ, Strik D. 2020. Consecutive lactate formation and chain elongation to reduce exogenous chemicals input in repeated-batch food waste fermentation. Water Res 169:115215. 10.1016/j.watres.2019.115215. [DOI] [PubMed] [Google Scholar]
- 28.Khor WC, Andersen S, Vervaeren H, Rabaey K. 2017. Electricity-assisted production of caproic acid from grass. Biotechnol Biofuels 10:180. 10.1186/s13068-017-0863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nzeteu CO, Trego AC, Abram F, O'Flaherty V. 2018. Reproducible, high-yielding, biological caproate production from food waste using a single-phase anaerobic reactor system. Biotechnol Biofuels 11:108. 10.1186/s13068-018-1101-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marounek M, Fliegrova K, Bartos S. 1989. Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii. Appl Environ Microbiol 55:1570–1573. 10.1128/AEM.55.6.1570-1573.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kucek LA, Nguyen M, Angenent LT. 2016. Conversion of L-lactate into n-caproate by a continuously fed reactor microbiome. Water Res 93:163–171. 10.1016/j.watres.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Duber A, Jaroszynski L, Zagrodnik R, Chwialkowska J, Juzwa W, Ciesielski S, Oleskowicz-Popiel P. 2018. Exploiting the real wastewater potential for resource recovery n-caproate production from acid whey. Green Chem 20:3790–3803. 10.1039/C8GC01759J. [DOI] [Google Scholar]
- 33.Wu Q, Feng X, Guo W, Bao X, Ren N. 2020. Long-term medium chain carboxylic acids production from liquor-making wastewater: parameters optimization and toxicity mitigation. Chem Eng J 388:124218. 10.1016/j.cej.2020.124218. [DOI] [Google Scholar]
- 34.Candry P, Radić L, Favere J, Carvajal-Arroyo JM, Rabaey K, Ganigué R. 2020. Mildly acidic pH selects for chain elongation to caproic acid over alternative pathways during lactic acid fermentation. Water Res 186:116396. 10.1016/j.watres.2020.116396. [DOI] [PubMed] [Google Scholar]
- 35.Oleskowicz-Popiel P. 2018. Designing reactor microbiomes for chemical production from organic waste. Trends Biotechnol 36:747–750. 10.1016/j.tibtech.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O'Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA, Doebeli M, Parfrey LW. 2018. Function and functional redundancy in microbial systems. Nat Ecol Evol 2:936–943. 10.1038/s41559-018-0519-1. [DOI] [PubMed] [Google Scholar]
- 37.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim S-T, Weigel D, Kemen EM. 2016. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol 14:e1002352. 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi B, Chang M, Martin J, Mitreva M, Lux R, Klokkevold P, Sodergren E, Weinstock GM, Haake SK, Li H. 2015. Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. mBio 6:e01926-14. 10.1128/mBio.01926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen SJ, Candry P, Basadre T, Khor WC, Roume H, Hernandez-Sanabria E, Coma M, Rabaey K. 2015. Electrolytic extraction drives volatile fatty acid chain elongation through lactic acid and replaces chemical pH control in thin stillage fermentation. Biotechnol Biofuels 8:221. 10.1186/s13068-015-0396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambrecht J, Cichocki N, Schattenberg F, Kleinsteuber S, Harms H, Müller S, Sträuber H. 2019. Key sub-community dynamics of medium-chain carboxylate production. Microb Cell Fact 18:92. 10.1186/s12934-019-1143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scarborough M, Lawson C, Hamilton J, Donohue T, Noguera D. 2018. Metatranscriptomic and thermodynamic insights into medium-chain fatty acid production using an anaerobic microbiome. mSystems 3:e00221-18. 10.1128/mSystems.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross DE, Marshall CW, Gulliver D, May HD, Norman RS. 2020. Defining genomic and predicted metabolic features of the Acetobacterium genus. mSystems 5:e00277-20. 10.1128/mSystems.00277-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu ZJ, Tao Y, Xu ZC, He XH, Tang LQ, Fan KQ. 2018. Lactate utilizing n-caproate producing bacteria Clostridium sp. and its application. China patent CN105420168B.
- 44.Wu Q, Guo W, You S, Bao X, Luo H, Wang H, Ren N. 2019. Concentrating lactate-carbon flow on medium chain carboxylic acids production by hydrogen supply. Bioresour Technol 291:121573. 10.1016/j.biortech.2019.121573. [DOI] [PubMed] [Google Scholar]
- 45.Jeon B, Kim S, Sang BI. 2017. Megasphaera hexanoica sp. nov., a medium-chain carboxylic acid-producing bacterium isolated from a cow rumen. Int J Syst Evol Microbiol 67:2114–2120. 10.1099/ijsem.0.001888. [DOI] [PubMed] [Google Scholar]
- 46.Lanjekar VB, Marathe NP, Ramana VV, Shouche YS, Ranade DR. 2014. Megasphaera indica sp. nov., an obligate anaerobic bacteria isolated from human faeces. Int J Syst Evol Microbiol 64:2250–2256. 10.1099/ijs.0.059816-0. [DOI] [PubMed] [Google Scholar]
- 47.Liu B, Popp D, Sträuber H, Harms H, Kleinsteuber S. 2 August 2020. Lactate-based microbial chain elongation for n-caproate and iso-butyrate production: genomic and metabolic features of three novel Clostridia isolates. Res Square 10.21203/rs.3.rs-50186/v1. [DOI] [Google Scholar]
- 48.Wu Q, Guo W, Bao X, Meng X, Yin R, Du J, Zheng H, Feng X, Luo H, Ren N. 2018. Upgrading liquor-making wastewater into medium chain fatty acid: insights into co-electron donors, key microflora, and energy harvest. Water Res 145:650–659. 10.1016/j.watres.2018.08.046. [DOI] [PubMed] [Google Scholar]
- 49.Liu B, Kleinsteuber S, Centler F, Harms H, Sträuber H. 2020. Competition between butyrate fermenters and chain-elongating bacteria limits the efficiency of medium-chain carboxylate production. Front Microbiol 11:336. 10.3389/fmicb.2020.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weimer PJ, Nerdahl M, Brandl DJ. 2015. Production of medium-chain volatile fatty acids by mixed ruminal microorganisms is enhanced by ethanol in co-culture with Clostridium kluyveri. Bioresour Technol 175:97–101. 10.1016/j.biortech.2014.10.054. [DOI] [PubMed] [Google Scholar]
- 51.Tao Y, Li J, Rui J, Xu Z, Zhou Y, Hu X, Wang X, Liu M, Li D, Li X. 2014. Prokaryotic communities in pit mud from different-aged cellars used for the production of Chinese strong-flavored liquor. Appl Environ Microbiol 80:2254–2260. 10.1128/AEM.04070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sträuber H, Lucas R, Kleinsteuber S. 2016. Metabolic and microbial community dynamics during the anaerobic digestion of maize silage in a two-phase process. Appl Microbiol Biotechnol 100:479–491. 10.1007/s00253-015-6996-0. [DOI] [PubMed] [Google Scholar]
- 53.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao M, Rui J, Li J, Dai Y, Bai Y, Heděnec P, Wang J, Zhang S, Pei K, Liu C, Wang Y, Zhi LH, Frouz J, Li X. 2014. Rate-specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus chinensis steppe. Soil Biol Biochem 79:81–90. 10.1016/j.soilbio.2014.09.009. [DOI] [Google Scholar]
- 55.Teramoto K, Okubo T, Yamada Y, Sekiya S, Iwamoto S, Tanaka K. 2019. Classification of Cutibacterium acnes at phylotype level by MALDI-MS proteotyping. Proc Jpn Acad Ser B Phys Biol Sci 95:612–623. 10.2183/pjab.95.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 57.Oh J, Byrd AL, Deming C, Conlan S, Barnabas B, Blakesley R, Bouffard G, Brooks S, Coleman H, Dekhtyar M, Gregory M, Guan X, Gupta J, Han J, Ho S-l, Legaspi R, Maduro Q, Masiello C, Maskeri B, McDowell J, Montemayor C, Mullikin J, Park M, Riebow N, Schandler K, Schmidt B, Sison C, Stantripop M, Thomas J, Thomas P, Vemulapalli M, Young A, Kong HH, Segre JA, NISC Comparative Sequencing Program. 2014. Biogeography and individuality shape function in the human skin metagenome. Nature 514:59–64. 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huson D, Mitra S, Ruscheweyh H-J, Weber N, Schuster S. 2011. Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560. 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, Juncker AS, Manichanh C, Chen B, Zhang W, Levenez F, Wang J, Xu X, Xiao L, Liang S, Zhang D, Zhang Z, Chen W, Zhao H, Al-Aama JY, Edris S, Yang H, Wang J, Hansen T, Nielsen HB, Brunak S, Kristiansen K, Guarner F, Pedersen O, Doré J, Ehrlich SD, Pons N, Le Chatelier E, Batto J-M, Kennedy S, Haimet F, Winogradski Y, Pelletier E, LePaslier D, Artiguenave F, Bruls T, Weissenbach J, Turner K, Parkhill J, Antolin M, Casellas F, MetaHIT Consortium. 2014. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32:834–841. 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 60.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, Su L, Li X, Li X, Li J, Xiao L, Huber-Schönauer U, Niederseer D, Xu X, Al-Aama JY, Yang H, Wang J, Kristiansen K, Arumugam M, Tilg H, Datz C, Wang J. 2015. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun 6:6528. 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 61.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita K, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:D354–D357. 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Powell S, Forslund S, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldón T, Rattei T, Creevey C, Kuhn M, Jensen L, von Mering C, Bork P. 2014. EggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 42:D231–D239. 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cantarel B, Coutinho P, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The carbohydrate-active ENZYMES database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang D, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. Peer J 7:e7359. 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parks D, Imelfort M, Skennerton C, Hugenholtz P, Tyson G. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 67.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data (raw metagenomic sample data and raw 16S amplicon sample data) reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under accession number CRA003483, which is publicly accessible at https://bigd.big.ac.cn/gsa.