Abstract
Background:
The Decipher test measures expression of 22 RNA biomarkers associated with aggressive prostate cancer used to improve risk stratification of patients to help guide management. To date, Decipher’s genomic classification has not been extensively correlated with specific histologic growth patterns in prostatic adenocarcinoma. With a growing understanding of the clinical aggressiveness associated with cribriform growth pattern (CF), intraductal carcinoma (IDC), and percent Gleason pattern 4 (G4%), we sought to determine if their presence was associated with an increased genomic risk as measured by the Decipher assay.
Design:
Clinical use of the Decipher assay was performed on the highest Gleason score (GS) tumor nodule of prostatectomy specimens from a prospective cohort of 48 patients, with GS varying from 7 through 9 to help guide clinical risk stratification. The tumors were reviewed for CF, IDC, and G4%, which were then compared to the Decipher score (0–1) and risk stratification (high vs not high).
Results:
The presence of CF/IDC was significantly associated with Decipher risk score (p=0.007), with a high risk Decipher score in 22% vs 56% of patients without or with CF/IDC. On binary logistic regression analysis, G4% (odds ratio (OR) 1.04 per percent increase [95%CI 1.02–1.06], p=0.0004) and CF predominant (OR 9.60 [95%CI 1.48–62.16], p=0.02) were significantly associated with a high risk GC score. IDC did not reach significance (OR 1.92 [95%CI 0.65–5.67, p=0.24).
Conclusions:
Our findings add to an expanding knowledge base that supports G4% and CF/IDC as molecularly unique and clinically relevant features in prostatic adenocarcinoma. These histologic features should be standardly reported as they are associated with more aggressive prostate cancer. Future work should determine the independent information these histologic findings have relative to genomic assessment on long-term outcomes.
Keywords: Prostate cancer, cribriform, intraductal, Gleason pattern 4, genomic, risk stratification, metastasis, mortality
1. INTRODUCTION:
The management of localized prostate cancer is grounded in risk stratification. This generally includes common prognostic variables, such as baseline serum PSA, tumor stage, Gleason score (GS), and percent positive biopsy cores. These are the key variables that comprise clinically used multivariable models, such as the National Comprehensive Cancer Network (NCCN)1 or the Cancer of the Prostate Risk Assessment (CAPRA)2 risk groups. However, there is additional histologic data readily available that has independent prognostic value, and yet is often not incorporated into these models.
In one of our previous studies, Cole et al.3 showed that the percent of Gleason pattern 4 (G4%) is predictive of both adverse pathology (at time of radical prostatectomy (RP)) and biochemical recurrence at more specific G4% thresholds (i.e. 1–10% G4 versus 20–30% G4) within Gleason 3+4 disease. Moreover, cribriform growth pattern (CF), classified as Gleason pattern 4, has been demonstrated to be an independent risk factor for metastasis and disease specific death, and the presence of CF combined with G4% outperforms G4% in predicting biochemical recurrence free-survival.4–7 Similarly, the presence of intraductal carcinoma (IDC-P) at the time of biopsy or prostatectomy has also been associated with poor outcomes, including biochemical recurrence, metastatic disease, and prostate cancer-specific mortality.4,8–11 Due to the morphologic overlap between invasive CF and IDC-P, some studies have grouped both patterns into a single group (presence of CF and/or IDC-P), which also carries prognostic value.4,9
In parallel to the identification of histologic features that are associated with prognostic groups, there are now several commercially available prognostic gene expression-based tests that assess prostate cancer tissue (e.g. Decipher12, Oncotype Dx13, Prolaris14). Of these, the Decipher test (GenomeDX Biosciences) is relatively commonly used after RP and measures the expression of 22 RNA biomarkers associated with aggressive prostate cancer. These biomarkers are comprised of coding and non-coding RNA in or near genes associated with aggressive prostate cancer. Attributed functions of these genes include cellular proliferation, differentiation, cell cycle progression, cell structure and adhesion, immune response, and yet-unknown functions; seven of these genes are androgen-regulated.12 This test issues a score and risk categorization validated to predict 5-year metastatic risk and 10-year prostate cancer specific mortality. Additionally, Decipher has been shown in a recent meta-analysis to independently predict metastatic development when adjusting for patient age, tumor stage, nodal stage, Gleason grade group, margin status, and adjuvant or salvage therapies.15 Furthermore, the addition of Decipher to NCCN or CAPRA significantly improves the ability to identify which patients will subsequently develop metastatic disease.16 For these reasons, Decipher testing has recently been included in NCCN guidelines for prostate cancer in specific clinical scenarios.1 However, to date, Decipher’s genomic classification has not been extensively correlated with histologic features beyond GS.17 Thus, we sought to determine if the presence of CF, IDC, or G4% are associated with an increased genomic risk score in prostate cancer patients.
2. MATERIALS AND METHODS:
2.1. Study cohort:
All patients were part of our prostate cancer registry maintained through the Michigan Prostate SPORE (Specialized Programs of Research Excellence), a collaborative and interdisciplinary translational research effort; through this translational research program, the National Cancer Institute (NCI) brings together the experience and expertise of research scientists from across the nation to collaborate in interdisciplinary translational prostate cancer research. All patients included in this study underwent RP and lymph node dissection for clinically localized prostate cancer followed by clinical use of the Decipher test between 2016–2018. Use of the Decipher test was considered in cases with pT3 disease and/or positive surgical margins and ultimately decided upon via clinician preference and patient consent at post-operative follow-up. Patients were included for the current analysis if their RP pathology had a component of Gleason pattern 4 along with available slides to review in order to quantify the presence of CF, IDC, and G4%. No patients underwent neoadjuvant therapy, including androgen deprivation, prior to RP. The study was reviewed and approved by the University of Michigan Institutional Review Board.
2.2. Histopathologic review:
Two reviewers, including one pathology resident (AST) and one genitourinary pathologist (RM), reviewed whole-mount prostatectomy slides from the cases on which Decipher testing was ordered. The tumors were assessed for CF pattern (including both dense and loose cribriform growth) on a 0–2 scale (0 or no CF = not present, 1 or CF minor = present in less than 50% of the Gleason 4 component, 2 or CF predominant= present in more than 50% of the Gleason 4 component). The presence of IDC was noted as not present, present, or a predominant feature. The percentage of Gleason pattern 4 (G4%) was estimated and CF and IDC were scored based on the index tumor nodule within the whole-mount slide representing the block sent for Decipher testing (consistent with the highest Gleason score); all slides from each case were reviewed to confirm that the block sent for testing was indeed representative of the index tumor/highest grade nodule. This cohort represents a real time assessment of prostate cancer patients for Decipher analyses; hence, FFPE tissue corresponding to the index tumor of area was submitted for Decipher testing; serial consecutive sections were not available to perform immunohistochemistry for basal cell markers to discern CF from IDC-P, which were distinguished upon morphologic basis only.
2.3. Specimen selection for Decipher testing:
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were selected from each case after review of whole-mount hematoxylin and eosin stained slides from the prostatectomy specimen. According to testing recommendations, the block felt to be best representative of the most prognostic (i.e. highest grade) tumor nodule was submitted for analysis for Decipher testing. For example, if one slide portrayed GS 7 carcinoma with 10% Gleason pattern 4 and another slide showed G4% of 40%, the latter block was submitted. Similarly, if one block demonstrated tertiary Gleason pattern 5, it was selected against other blocks that did not demonstrate a tertiary high-grade pattern.
2.4. Calculation of Decipher score and risk categorization (Decipher assay):
Described in greater depth in prior work, the Decipher assay (GenomeDX) measures the expression of 22 RNA biomarkers and delivers a continuous score (between 0 and 1) for which higher scores indicate an increased risk of clinical metastasis and cancer related mortality.12 The score is reported along with a validated categorization as “low risk” (<0.45), “intermediate risk” (0.45 to 0.60), and “high risk” (>0.60).18 In a recent patient-level meta-analysis, patients in the low, intermediate, and high risk categories had a 5-year incidence of metastasis of 2.4%, 5.8%, and 15.2%, respectively, and a 10-year incidence of metastasis of 5.5%, 15.0%, and 26.7%, respectively.15
2.5. Statistical analyses:
Statistical significance for risk category and scores were determined by Chi squared contingency analysis and Fisher’s t-test, respectively. Binary logistic regression analysis was performed to determine the association of clinical and pathologic features with the probability of having a high Decipher score (versus a low or intermediate score). Given the limited number of patients, a multivariable analysis was deemed to not be advised given the high probability of overfitting the model. Previous work4,9 has acknowledged the morphologic overlap between CF invasive carcinoma and IDC by combining them in a single category for analysis, so we also sought to establish if CF pattern and/or IDC were significantly associated with Decipher risk categorization. We also analyzed if CF pattern and IDC alone were associated with higher risk stratification by Decipher. Finally, the group of GS 3+4 adenocarcinoma was separated into groups of G4% at a potentially clinically relevant threshold of 15% (less than 15% and greater than or equal to 15%) based on outcome differences between 1–10% G4 versus 20–30% G4 in Cole et al.3; risk categorization and Decipher scores were analyzed for patients with G4% above and below this threshold.
3. RESULTS:
3.1. Clinical and pathological characteristics of the study cohort:
The cohort of 48 patients selected by clinicians for Decipher testing is characterized in Table 1. The average age of patients was 63 years (range 47–73 years). The median baseline PSA was 7.3 ng/mL. Gleason grade groups are represented as follows: group 2 (n=26, 54%), 3 (n=16, 33%), 4 (n=2, 4%), and 5 (n=4, 8%). Cribriform growth was observed in 38 (79%) cases and intraductal carcinoma was observed in 15 (31%). Pathologic staging demonstrated extraprostatic extension in 40 (83%) cases and seminal vesicle invasion in 10 (21%) cases. Positive surgical margins were demonstrated in 17 (35.4%) cases. 12 (25%) tumors were anterior dominant, 11 (23%) were transitional zone, and 25 (52%) were exclusively peripheral zone. All patients had clinical and pathologic node negative disease.
Table 1:
Patient and tumor Characteristics.
| N | % | ||
|---|---|---|---|
| Age (years) | |||
| Median (range) | 63 | 47–73 | |
| Baseline PSA (ng/mL) | |||
| Median (range) | 7.3 | 4.0–40.2 | |
| Grade Group | |||
| 2 | 26 | 54 | |
| 3 | 16 | 33 | |
| 4 | 2 | 4 | |
| 5 | 4 | 8 | |
| Pathologic T-stage | |||
| Extracapsular Extension | 40 | 83 | |
| Seminal Vesicle Invasion | 10 | 21 | |
| Surgical Margins | |||
| Positive | 17 | 35 | |
| Negative | 31 | 65 | |
| CAPRA-S Score | |||
| Median (range) | 4 | 2–8 | |
| Cribriform | |||
| Present | 38 | 79 | |
| Absent | 10 | 21 | |
| Intraductal Carcinoma | |||
| Present | 15 | 31 | |
| Absent | 33 | 69 | |
| Tumor Location* | |||
| Peripheral Zone | 25 | 52 | |
| Transitional Zone | 11 | 23 | |
| Anterior Zone | 12 | 25 | |
| Decipher Score | |||
| Median (range) | 0.6 | 0.05–0.92 | |
| Low | 17 | 35 | |
| Intermediate | 7 | 15 | |
| High | 24 | 50 | |
Note: all patients had clinical and pathologic node negative disease.
Location of the sample sent for Decipher testing.
The median duration of follow up after RP for the cohort was 291 days (range 39 to 571 days), at which point 7 patients (15.6%) had started adjuvant radiation therapy, 40 (83.3%) were on a surveillance protocol, and 1 (2.1%) patient had received salvage radiation therapy.
3.2. Correlation of Decipher score and categorization with routine clinicopathologic features
Baseline PSA was strongly correlated with the Decipher score (R2 = 0.97) (Fig 1A). Increases in Gleason grade group were correlated to a high Decipher score with an odds ratio of 4.78 (95%CI 1.58–14.45, p=0.01; Fig 1B). Pathologic stage also increased Decipher categorization as high-risk, with particular significance of pT3b disease when compared to pT2 disease (p=0.008; Fig 1C). These results are summarized in Table 2.
Figure 1:
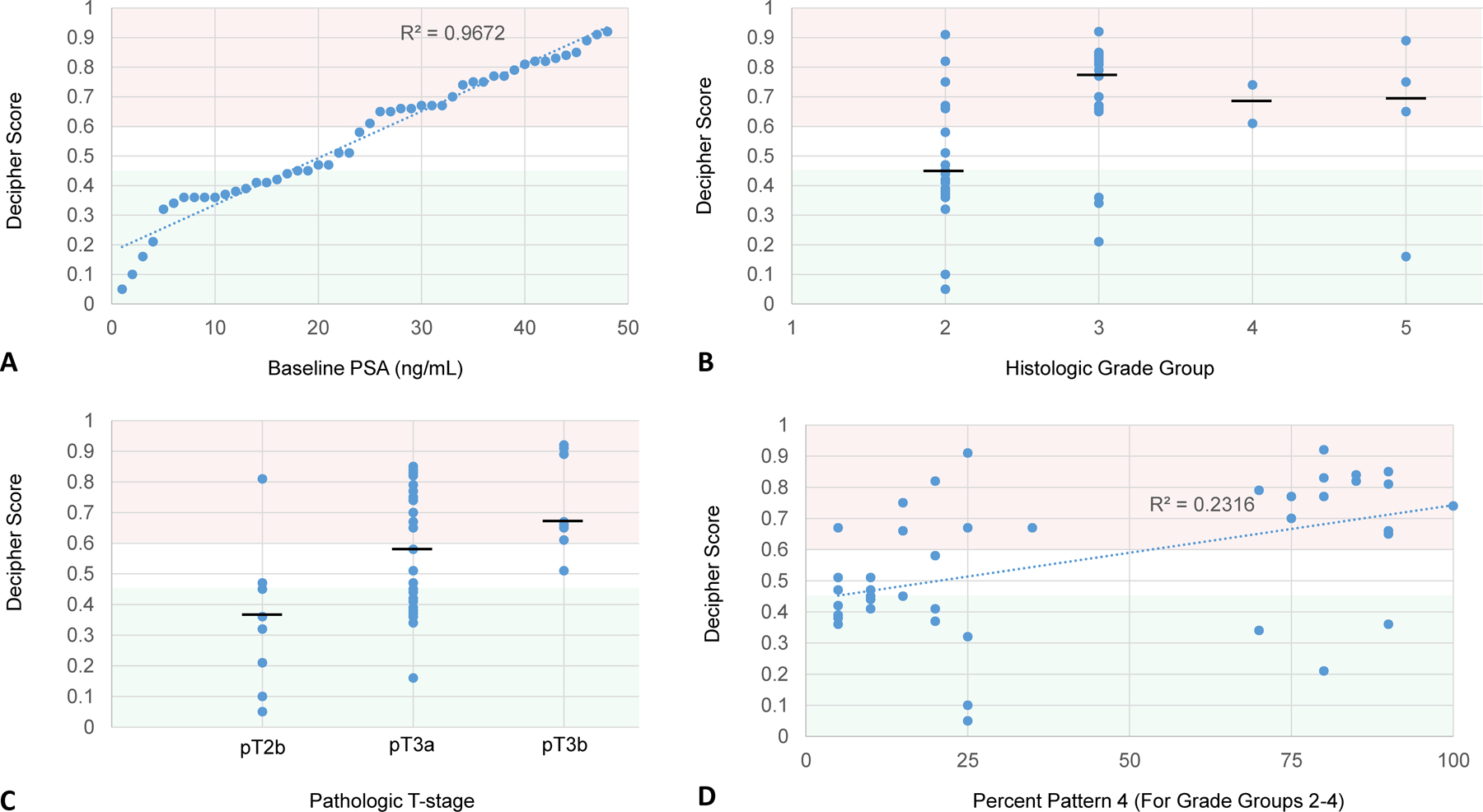
Decipher scores plotted against baseline PSA (A), histologic grade group (B), pathologic T-stage (C), and percent Gleason pattern 4 (D). Decipher scores from 0 to 0.45 (shaded green), 0.45 to 0.6 (shaded white), and 0.6 to 1.0 (shaded red), are respectively considered low, intermediate, and high risk. Black horizontal bar represents the median value.
Table 2:
Univariable Analysis of co-variables associated with Decipher High Risk
| 95%CI | |||||
|---|---|---|---|---|---|
| Variable | Odds Ratio | Lower | Upper | p-value | |
| Grade Group | (ordinal) | 4.78 | 1.58 | 14.45 | 0.01 |
| %GP4 | (continuous) | 1.04 | 1.02 | 1.06 | 0.0004 |
| Max Size of tumor | (continuous) | 1.07 | 0.51 | 2.24 | 0.87 |
| pT-stage | |||||
| pT2 | Reference | ||||
| pT3a | 6.562 | 0.72 | 59.85 | 0.095 | |
| pT3b | 56 | 2.93 | 1071.64 | 0.008 | |
| Cribriform Architecture (CF) | |||||
| Absent | Reference | ||||
| Present | 3.64 | 0.62 | 21.36 | 0.15 | |
| Predominant | 9.60 | 1.48 | 62.16 | 0.02 | |
| Intraductal Carcinoma (IDC) | (present vs absent) | 1.92 | 0.65 | 5.67 | 0.24 |
| CF and/or IDC | (present vs absent) | 4.53 | 0.83 | 24.65 | 0.08 |
3.2. Correlation of cribriform growth pattern and intraductal carcinoma to Decipher results:
The presence and predominance of cribriform growth increased high risk categorization by Decipher (Fig 2a); when compared to absent CF, present but not predominant CF non-significantly increased Decipher high-risk categorization (OR=3.64 [95%CI 0.62–21.36], p=0.15) and predominant CF significantly increased high-risk categorization (OR=9.60 [95%CI 1.48–62.16], p=0.02). The presence of IDC was non-significantly associated with increased high risk Decipher (OR 1.92 [95%CI 0.65–5.67], p=0.24) (Fig 2b). Specifically, within Gleason 3+4 (grade group 2) disease, risk categorization by Decipher significantly increased with the presence of cribriform growth (p=0.037). The presence of CF and/or IDC compared to neither pattern being present (Fig 2c) was non-significantly associated with increased high-risk categorization (OR=4.53 [95%CI 0.83–24.65], p=0.08) but significantly associated with increased median Decipher risk score (p=0.007).
Figure 2:
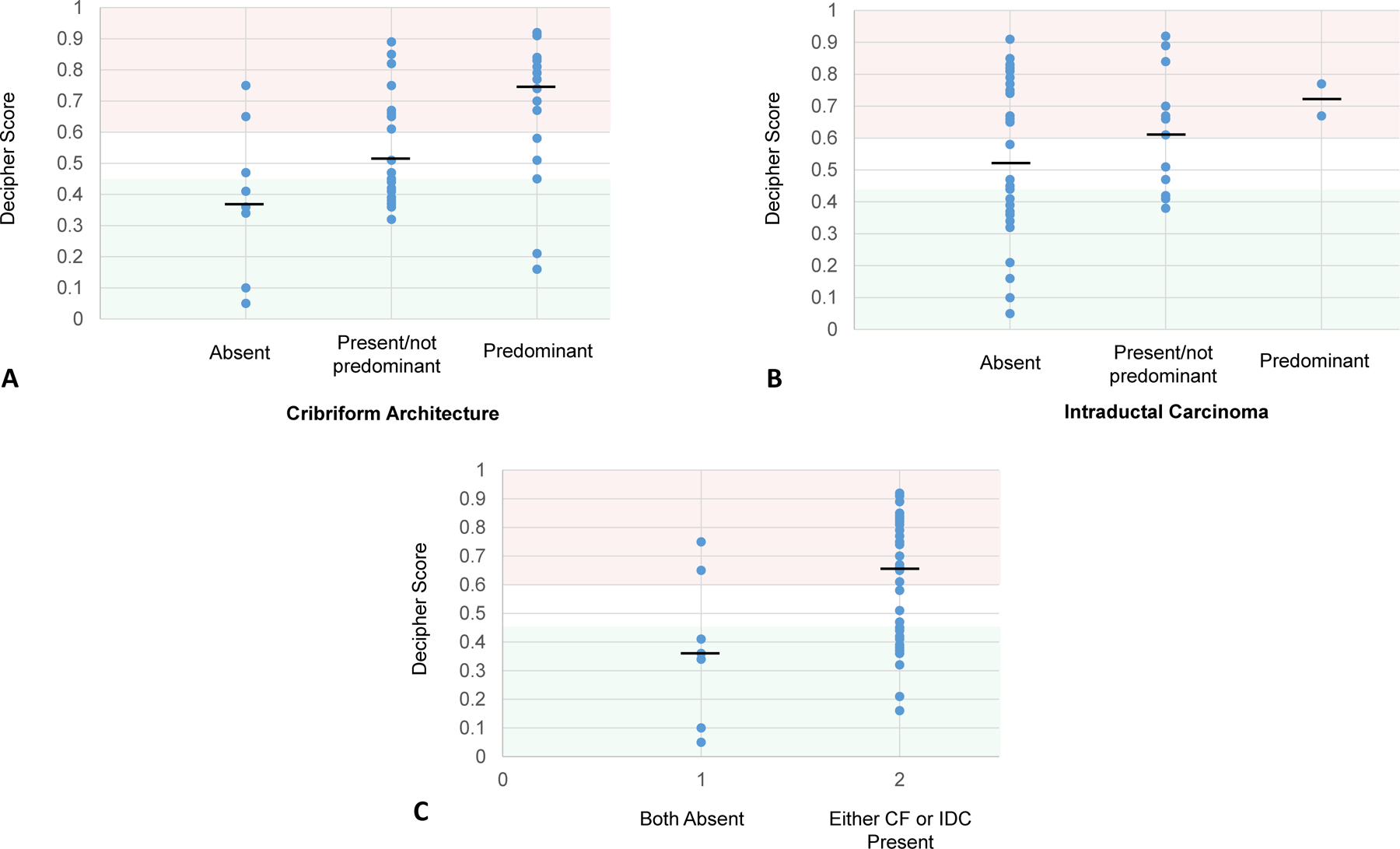
Decipher scores with cases grouped by A) the presence/predominance of cribriform growth, B), the presence/predominance of intraductal growth, and C) the presence of CF and/or IDC versus both patterns absent. Decipher scores from 0 to 0.45 (shaded green), 0.45 to 0.6 (shaded white), and 0.6 to 1.0 (shaded red), are respectively considered low, intermediate, and high risk. Black horizontal bar represents the median value.
3.3. Correlation of percent Gleason pattern 4 to genomic risk:
Among grade groups 2 through 4, the percent of Gleason pattern 4 had a modest correlation to Decipher score (R2=0.2316) (Fig 1D). However, for each percentage point increase in G4% there was a 4% increase in the odds of harboring a Decipher high risk score (OR 1.04 [95%CI 1.02–1.06], p=0.0004). GS 3+4 cancer (grade group 2) with greater than or equal to 15% G4 was associated with higher Decipher risk than GS 3+4 cancer with less than 15% G4 (p=0.019).
4. DISCUSSION:
In this study, the presence of CF and G4% were associated with increased Decipher genomic risk. The association with cribriform growth was significant when all cases were analyzed as well as when only Gleason score 3+4=7 cases were assessed. When cases were grouped by presence or absence of IDC, no statistically significant association was observed, which may be due to the limited number of cases with IDC present given the odds ratio estimate was over 1.0. In contrast, this finding suggests that cribriform growth pattern may carry more prognostic clinical relevance than IDC, and may be a stronger histologic correlate of an aggressive genomic signature in prostate cancer patients. In the presence of CF and/or IDC, there was a non-significant trend toward higher Decipher risk categorization, supporting, albeit inconclusively, the idea that these features have some prognostic connotation as a grouped category.
The prognostic importance of cribriform growth is further reinforced by our data demonstrating a difference in Decipher risk between patients with no cribriform growth, cribriform growth comprising less than 50% of Gleason pattern 4 disease, and cribriform growth comprising 50% or greater of Gleason pattern 4 disease. The volume of prostatic adenocarcinoma with cribriform growth pattern and its predominance within Gleason pattern 4 components may have clinically relevant importance that could encourage its use in routine pathology reporting after further investigation.
In addition, our study suggests that the specific percentage of Gleason pattern 4 in Gleason 3+4=7 cancer may have more prognostic impact than previously understood. Using a threshold of 15%, we found GS 3+4 cancer with greater than or equal to 15% Gleason pattern 4 to be associated with higher Decipher risk than GS 3+4 cancer with less than 15% Gleason pattern 4 (p=0.019). These results in addition to other work demonstrating the importance of similar thresholds of Gleason pattern 4 percentage in biopsy specimens3 raise the potential importance of G4% in explaining the molecular/clinical heterogeneity among GS 3+4 carcinoma. While there is no single accepted G4% threshold, GS 3+4 cancer with G4% of less than 15% may have clinical and biological behavior closer to Gleason score 6 than Gleason score 7 disease. Thus, the G4% in radical prostatectomy specimens of GS 3+4 may be a relevant component to include in pathology reports.
Our results do reflect a population from real-world, clinical use of the Decipher assay, indicating utility in a cohort of patients derived from day to day practice. A limitation of our study, on the other hand, includes ascertainment bias due to the method of accruing patients who were thought to benefit from genomic risk information for clinical decision making with respect to adjuvant therapy after radical prostatectomy. Our patient population is targeted to real world practice and does not include many patients with the extremes of low- and high-stage and grade disease for whom the decision for adjuvant therapy could be made without ancillary genomic information. While pathologic limitations such as inter-observer variability in the recognition of or in distinguishing CF from IDC may affect generalizability, cribriform growth is a relatively easily recognized feature in prostatic adenocarcinoma for genitourinary and general surgical pathologists alike. Finally, our inability to oversee the specific area of tumor processed during Decipher testing limits definitive conclusions regarding genomic risk within specific histologic areas; however, given that the predominance of CF was observed to affect a validated clinical genomic risk assessment tool, we add to the body of literature that CF-containing tumors do confer increased clinical risk. Based on our results, routine histopathologic assessment does not supplant the need for Decipher testing when recommended, as some tumors with no CF or IDC were classified as high genomic risk by Decipher. Rather, recognition and reporting of CF and IDC may add complementary prognostic value to molecular testing or act as a correlate to risk stratification in settings in which molecular testing is limited or not possible. Additional work with a larger cohort may further elucidate this potential benefit.
A larger scale study powered for multivariate analysis across Gleason scores would be beneficial to further assess the independent and grouped effects of cribriform growth pattern, intraductal carcinoma, and % Gleason pattern 4 on genomic risk while controlling for other clinicopathologic factors. Additional samples would also improve the ability to discern the association between Decipher risk and cribriform vs. intraductal carcinoma. Additional work should focus on long-term follow up of this or similar cohorts in order to assess if certain histopathologic variables are associated with mortality or metastasis specifically in cases with high (or low) genomic risk.
In summary, our findings add to an expanding knowledge base that supports cribriform growth pattern as a unique and clinically relevant pattern of prostatic adenocarcinoma. Based on Decipher assessment alone, cribriform growth pattern should be considered an indication of biologically aggressive disease, especially among clinically heterogeneous groups of GS 3+4 (Gleason grade group 2) cancers. Cribriform growth pattern and intraductal carcinoma should be considered for inclusion in templated histopathologic reporting, and may be considered in future revisions of grade group categorization. Moreover, reporting the percentage of Gleason pattern 4 in radical prostatectomy specimens can convey clinically meaningful information to urologists and radiation oncologists, and a threshold of 15% Gleason pattern 4 may carry prognostic importance. This and future work may aid in increasing the prognostic value of pathology reporting, improving selection of patients for genomic testing, and optimizing information available for shared decision making in clinical decisions about treatment options.
Acknowledgements:
Funding: DES: Prostate Cancer Foundation
Footnotes
Disclosures: DES: Advisory boards for Janssen and Blue Earth.
REFERENCES:
- 1.Network NCC. NCCN Clinical Practice Guidelines in Oncology: Prostate Cancer (Version 4.2018). 2018; https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf. Accessed November 18, 2018.
- 2.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: A straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117(22):5039–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole AI, Morgan TM, Spratt DE, et al. Prognostic Value of Percent Gleason Grade 4 at Prostate Biopsy in Predicting Prostatectomy Pathology and Recurrence. J Urol. 2016;196(2):405–411. [DOI] [PubMed] [Google Scholar]
- 4.Kweldam CF, Kummerlin IP, Nieboer D, et al. Presence of invasive cribriform or intraductal growth at biopsy outperforms percentage grade 4 in predicting outcome of Gleason score 3+4=7 prostate cancer. Mod Pathol. 2017;30(8):1126–1132. [DOI] [PubMed] [Google Scholar]
- 5.Kweldam CF, Wildhagen MF, Steyerberg EW, Bangma CH, van der Kwast TH, van Leenders GJ. Cribriform growth is highly predictive for postoperative metastasis and disease-specific death in Gleason score 7 prostate cancer. Mod Pathol. 2015;28(3):457–464. [DOI] [PubMed] [Google Scholar]
- 6.Choy B, Pearce SM, Anderson BB, et al. Prognostic Significance of Percentage and Architectural Types of Contemporary Gleason Pattern 4 Prostate Cancer in Radical Prostatectomy. Am J Surg Pathol. 2016;40(10):1400–1406. [DOI] [PubMed] [Google Scholar]
- 7.Dong F, Yang P, Wang C, et al. Architectural heterogeneity and cribriform pattern predict adverse clinical outcome for Gleason grade 4 prostatic adenocarcinoma. Am J Surg Pathol. 2013;37(12):1855–1861. [DOI] [PubMed] [Google Scholar]
- 8.Kimura K, Tsuzuki T, Kato M, et al. Prognostic value of intraductal carcinoma of the prostate in radical prostatectomy specimens. Prostate. 2014;74(6):680–687. [DOI] [PubMed] [Google Scholar]
- 9.Kweldam CF, Kummerlin IP, Nieboer D, et al. Disease-specific survival of patients with invasive cribriform and intraductal prostate cancer at diagnostic biopsy. Mod Pathol. 2016;29(6):630–636. [DOI] [PubMed] [Google Scholar]
- 10.Trudel D, Downes MR, Sykes J, Kron KJ, Trachtenberg J, van der Kwast TH. Prognostic impact of intraductal carcinoma and large cribriform carcinoma architecture after prostatectomy in a contemporary cohort. Eur J Cancer. 2014;50(9):1610–1616. [DOI] [PubMed] [Google Scholar]
- 11.Van der Kwast T, Al Daoud N, Collette L, et al. Biopsy diagnosis of intraductal carcinoma is prognostic in intermediate and high risk prostate cancer patients treated by radiotherapy. Eur J Cancer. 2012;48(9):1318–1325. [DOI] [PubMed] [Google Scholar]
- 12.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brand TC, Zhang N, Crager MR, et al. Patient-specific Meta-analysis of 2 Clinical Validation Studies to Predict Pathologic Outcomes in Prostate Cancer Using the 17-Gene Genomic Prostate Score. Urology. 2016;89:69–75. [DOI] [PubMed] [Google Scholar]
- 14.Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol. 2013;31(11):1428–1434. [DOI] [PubMed] [Google Scholar]
- 15.Spratt DE, Yousefi K, Deheshi S, et al. Individual Patient-Level Meta-Analysis of the Performance of the Decipher Genomic Classifier in High-Risk Men After Prostatectomy to Predict Development of Metastatic Disease. J Clin Oncol. 2017;35(18):1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spratt DE, Zhang J, Santiago-Jimenez M, et al. Development and Validation of a Novel Integrated Clinical-Genomic Risk Group Classification for Localized Prostate Cancer. J Clin Oncol. 2018;36(6):581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Den RB, Santiago-Jimenez M, Alter J, et al. Decipher correlation patterns post prostatectomy: initial experience from 2 342 prospective patients. Prostate Cancer Prostatic Dis. 2016;19(4):374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190(6):2047–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]


