Abstract
The use of intracellular antibodies as templates to derive surrogate compounds is an important objective because intracellular antibodies can be employed initially for target validation in pre-clinical assays and subsequently employed in compound library screens. LMO2 is a T cell oncogenic protein activated in the majority of T cell acute leukaemias. We have used an inhibitory intracellular antibody fragment as a competitor in a small molecule library screen using competitive surface plasmon resonance (cSPR) to identify compounds that bind to LMO2. We selected four compounds that bind to LMO2 but not when the anti-LMO2 intracellular antibody fragment is bound to it. These findings further illustrate the value of intracellular antibodies in the initial stages of drug discovery campaigns and more generally antibodies, or antibody fragments, can be the starting point for chemical compound development as surrogates of the antibody combining site.
Keywords: LMO2, TAL1/SCL, GATA, Chromosomal translocations, Drug discovery, Intracellular antibodies, Leukaemia, SPR, Abd compounds
1. Introduction
Drug discovery relies on at least two stages, first target validation and second drug development. Intracellular antibody fragments can be developed as molecular tools for target validation and expressed in cells or mouse models as pre-clinical assays to show that affecting the function of a protein target has consequences for a clinical indication of interest. We have used this approach to validate the role of the T cell oncogenic protein LMO2 in T cell that is activated by chromosomal translocations. LMO2 was discovered as a gene activated by recurrent, specific (t11;14) chromosomal translocations in T cell neoplasias (Chambers and Rabbitts, 2015) and was subsequently found to be aberrantly expressed in almost all human T cell acute leukaemias (Ferrando et al., 2002). LMO2 forms a multiprotein, bipartite DNA binding complex in which LMO2 is a bridging protein, composed of two LIM domains between the DNA-binding portions comprising TAL1/SCL-E47 and GATA (Wadman et al., 1997).
By using either an anti-LMO2 intracellular single chain variable fragment (scFv) (Nam et al., 2008) or an anti-LMO2 intracellular domain antibody (an iDAb, also called VH576) (Tanaka et al., 2011), we confirmed that blocking LMO2 protein function prevents T cell tumour growth. The intracellular antibody prevents the formation of the LMO2-dependent protein complex, which was previously shown to be involved in leukaemia formation (Grutz et al., 1998), by bending and twisting the LMO2 protein out of its natural state (Sewell et al., 2014). The intracellular antibody was therefore an important potential target for guiding the selection of small molecules as surrogates of the antibody combining site. We have previously shown that intracellular antibodies could be used in small molecular library screening protocols to guide the selection of compounds that are surrogates of the antibody combining site. In this work, we used an anti-RAS intracellular antibody fragment to develop compounds that interfere with the protein-protein interaction of RAS with effectors by developing a competitive surface plasmon resonance (cSPR) screening strategy (Quevedo et al., 2018). In this selection method, we used a high affinity intracellular antibody to determine, by competition, which chemical fragments bind to the same RAS surface that the antibody binds. Thus RAS-binding chemical hit matter could be separated into those inhibited by the interaction of RAS with the high affinity antibody. This gave rise to the Antibody-derived (Abd) compounds that were developed by structure-based drug design medicinal chemistry into surrogates of the antibody combining site, indicating the power of using Intracellular antibody fragments to identify hit chemical matter.
We have characterized an anti-LMO2 iDAb that interferes with the formation of the LMO2 protein complex because its binding site spans the hinge region of LMO2 and causes it to bend and twist out the shape adopted when LMO2 binds to LDB1 (Sewell et al., 2014). This LMO2 fold is therefore different from the natural LMO2 structure (El Omari et al., 2013). We have implemented this Abd technology using the cSPR strategy to screen a small chemical library using LMO2 protein together with an anti-LMO2 intracellular antibody. Two related compounds were identified from this screen, showing that the Abd approach is a general strategy for finding chemical compound surrogates of antibody combining sites.
2. Results
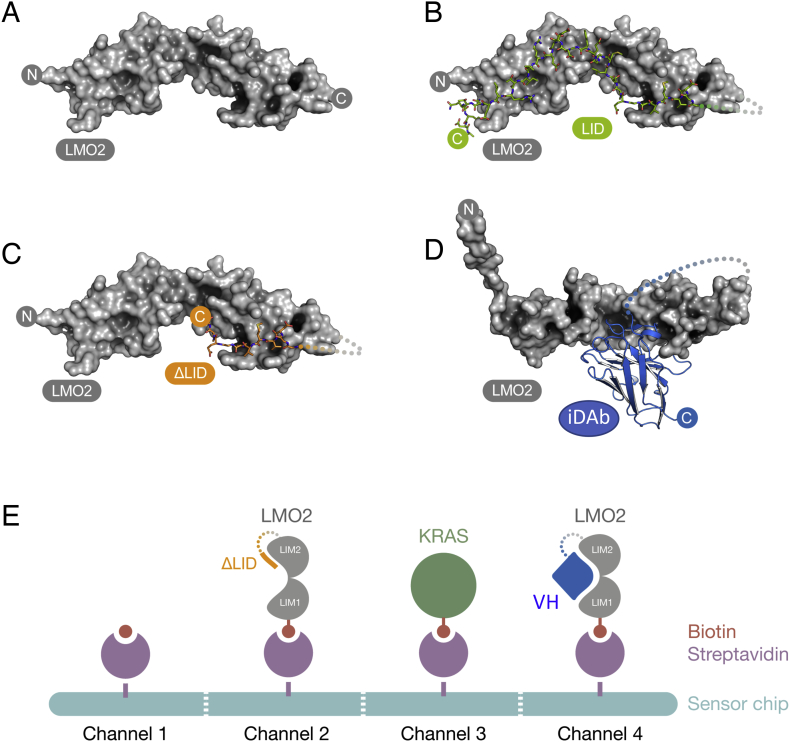
As a means to further evaluate the utility of the cSPR method using intracellular antibody fragments, we have studied further a compound library screen with the LMO2 T cell oncogenic protein. The different structural elements of LMO2 complexed with the LIM-binding domain of LDB1 (LID) are illustrated in Fig. 1A, B. In the screening, our objective was to obtain compounds that are surrogates of the two regions where the iDAb CDR bind to LMO2 (with a. longer term goal of fragment joining to generate a compound(s) that mimic the LMO2 contortion that makes the iDAb a powerful LMO2 inhibitor). For this reason, we did not employ the LMO2-LID protein on the chip, but rather a truncated version of LID that stops short of LMO2 hinge (LMO2ΔLID, illustrated Fig. 1C) and LMO2 bound to the anti-LMO2 iDAb (Fig. 1D).
Fig. 1.
LMO2 fusion proteins and organization of cSPR chip.
Molecular models of the recombinant proteins used for the cSPR library screen. (A) LMO2 only (B) LMO2-LDB1 LID domain fusion protein (Wilkinson-White et al., 2010) (C) LMO2-ΔLID (D) LMO2-VH576 (PDB ID: 4KFZ). LMO2 is shown as a surface representation, the LDB1 LID domain as sticks and VH576 in ribbon context. The LMO2 only, LMO2-LID and LMO2-ΔLID structures are based on the LMO2-LDB1 crystal structure (PDB ID: 2XJY). The connecting glycine-serine linker was disordered in the original crystal structure and not visible, this is shown as a dotted line. The glycine-serine linker in the LMO2-VH is drawn for completion but there are no structural data. The N- and C-termini of the proteins are marked for orientation purposes.
The SPR streptavidin chips used for the cSPR screen (depicted in panel E) were channel 1: blank; channel 2: LMO2-ΔLID; channel 3: KRAS; channel 4: LMO2-gly/ser-VH.
The cSPR compound library screen relies on blocking of the target molecule site by the binding of the intracellular antibody on the SPR chip and thus requires high affinity interaction (i.e. a very low Koff) since the antibody fragment is bound to the target molecule prior to passing the sample compound across the chip. Since the anti-RAS Intracellular antibody has a pM binding affinity, the stable interaction of RAS and antibody was achievable. The anti-LMO2 iDAb has a lower binding affinity (nM) and to expand the strength, we created a fusion protein between LMO2 and the iDAb with an extended flexible linker, thereby favoring the interaction between iDAb and LMO2 (Fig. 1D). The PPI-NET compound library was screened using the 4 channels of the SPR shown in Fig. 1E, comprising the target protein LMO2ΔLID in channel 2, an LMO2 competitor iDAb fusion in channel 4, a blank reference channel 1 and a negative control protein for this screen (KRAS in channel 3). This organization of the channels enabled us to sub-divide the chemical matter into those that might bind LMO2 in the iDAb binding region (Abd compounds as iDAb surrogates), those that bind LMO2 elsewhere and those that bind to RAS. The latter have been described previously (Cruz-Migoni et al., 2019; Quevedo et al., 2018).
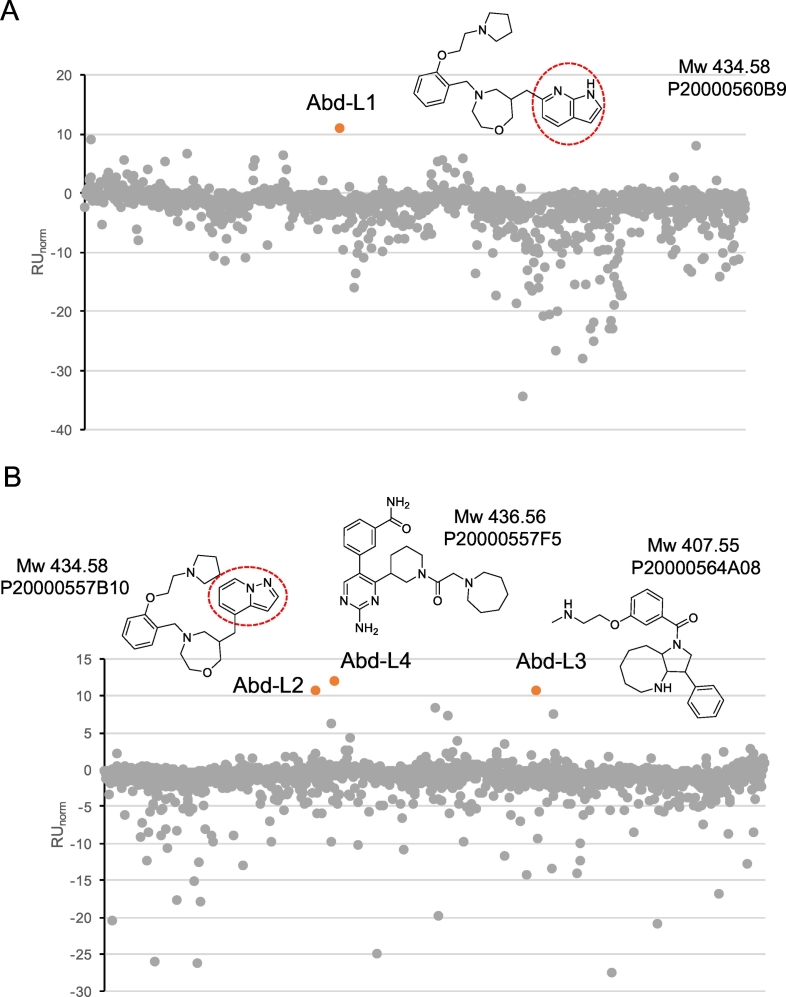
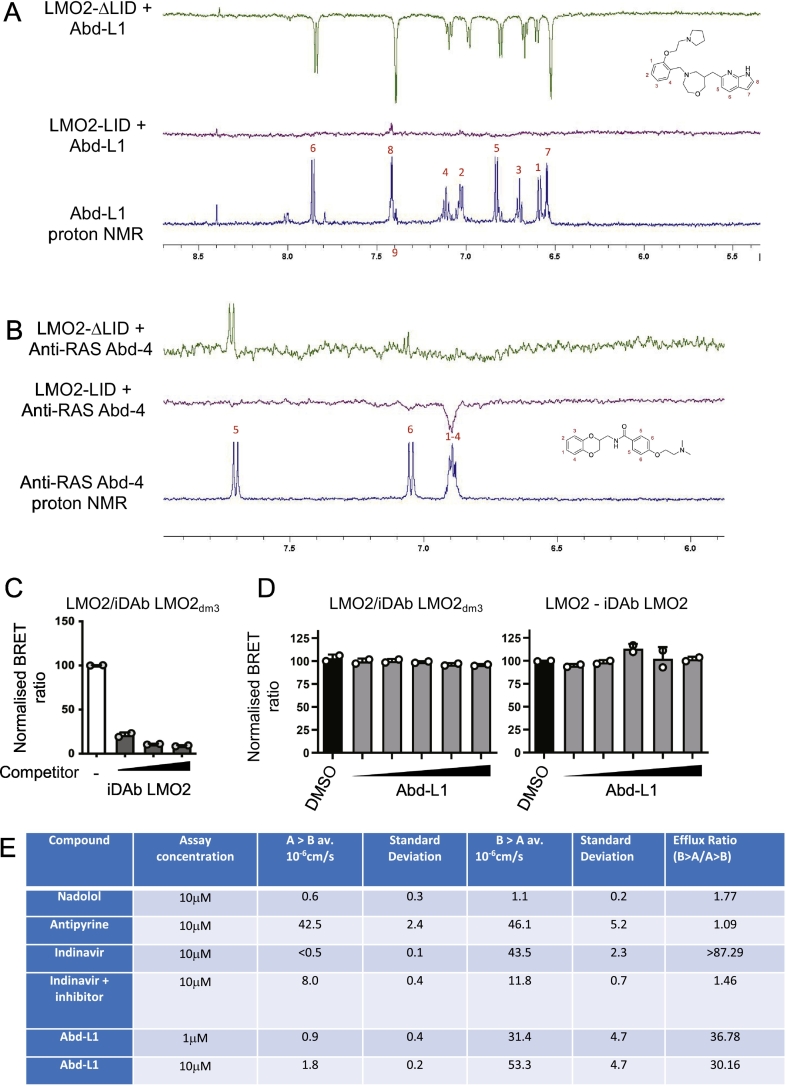
Four LMO2ΔLID Abd hits, that did not bind the LMO2-iDAb or KRAS, were identified as having response units (RU) above 10. The chemical structures and molecular weights of these are shown in Fig. 2 with the RU data from the screen. The compounds designated Abd-L1 and Abd-L4 were available commercially but Abd-L2 (an analogue of Abd-L1) and Abd-L3 were not. Further characterization of Abd-L1 was undertaken using two orthogonal assays,1D NMR waterLOGSY (Huang et al., 2017) and live cell Bioluminescence resonance energy transfer 2 (BRET2) (Bery et al., 2018). The binding of Abd-L1 with LMO2-LID and LMO2-ΔLID in waterLOGSY showed that Abd-L1 has extensive binding, in which most of the proton peaks showed the polarity shifts caused by interaction with the LMO2-ΔLID whereas limited change was observed with the LMO2-LID protein (Fig. 3A). As a negative control for the LMO2 proteins, the waterLOGSY spectra of a RAS-binding compound Abd-4 (Quevedo et al., 2018) showed no binding to either protein (Fig. 3B).
Fig. 2.
cSPR analysis of compounds.
Competitive SPR screening data are shown, utilizing the sensor chip configuration shown in Fig. 1E, for screening of the PPI-NET compound library. The cSPR screen was done at a single concentration of 150 μM for each compound. Response levels were measured and normalized by subtracting the response measured against one of the two control reference proteins: LMO2-VH576 fusion protein (panel A) or KRAS (panel B). The resulting normalized responses (RUnorm) are plotted against all of the compounds in the library. Hit compounds are indicated in orange. Both LMO2-VH and KRAS were used as reference proteins. Four hit compounds were identified from the cSPR screen in Fig. 2. The chemical structures and molecular weights (Mw) are shown, together with PPI-NET plate locations (P number) and the designated Antibody-derived (Abd) number. Abd-L1 and Abd-L2 are homologues that differ only by the presence of 1H-pyrrolo[2,3-b]pyridine in Abd-L1 or pyrazolo[1,5-a]pyridine in Abd-L2, both indicated by the red oval. Note: additional amounts of Abd-L2 and Abd-L3 were not available commercially. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Characterization of LMO2-binding compound Abd-1.
The LMO2-binding hit compound was assessed in vitro with NMR waterLOGSY and in cells using BRET2 assays. WaterLOGSY was carried out with compounds bound to LMO2-LID or to LMO2ΔLID comparing the spectra with the proton NMR of the compounds. Panel A shows waterLOGSY data with Abd-L1 and panel B waterLOGSY data with the KRAS-binding control compound Abd-4 (Quevedo et al., 2018). Cell-based interaction of the Abd-L1 compound with the target LMO2 protein was assessed by BRET2. Panels C and D show BRET2 assays comprising BRET signal from HEK293T cell interaction of LMO2/iDAb LMO2dm3 (in which the LMO2dm3 is a low affinity mutant of the anti-LMO2 iDAb VH576 (Sewell et al., 2014), characterized in (Bery et al., 2021) and competition of this interaction with an increased DNA quantity of iDAb LMO2 competitor (grey histograms, C) or with an increased dose of Abd-L1 at 1, 10, 20, 50 and 100 μM (grey histograms, panel D). In panel D, LMO2/iDAb LMO2 interaction was also tested in BRET competition assay with an increased dose of Abd-L1 (−) interaction with no competitor (white histogram in C) and in D, the negative control is DMSO treated cells (black histograms). Panel E shows CaCo-2 permeability assay with Abd-L1 compound.
The purpose of compound library screens is to identify chemical matter that can form the basis of drug development, which in turn requires efficacy of the compounds in cells by specifically interfering with the target protein. Accordingly, we assessed the anti-LMO2 binder Abd-L1 in live cell BRET2 analysis. An LMO2-iDAb BRET2 assay has been established (Bery et al., 2021) using the anti-LMO2 iDAb (Sewell et al., 2014) with mutations in its CDRs to reduce the affinity of the iDAb binding (dematured binding designated iDAb LMO2dm3) (Bery et al., 2021). While the BRET signal produced by interaction of LMO2 with iDAb LMO2dm2 was inhibited by expressing the high affinity iDAb LMO2 as competitor (Fig. 3C) an increasing amount of Abd-L1 failed to have any effect on the LMO2 - iDAb LMO2dm3 binding (Fig. 3D). The lack of interference in the protein-protein interaction (PPI) of LMO2 and iDAb could be due to the cellular properties of the Abd-L1 compound, as well as the low affinity of the compound in LMO2 binding. The cell permeability properties of Abd-L1 was assessed in a CaCo2 assay (Bavetsias et al., 2016) compared to low (nadolol) and high (antipyrine) permeability compounds and a compound with high export (indinavir). The CaCo2 data (Fig. 3E) show that Abd-L1 has low cellular permeability with high efflux in Caco-2 cells. Thus, Abd-L1 in its initial form has poor cell-based drug properties that will in part explain the lack of inhibition of the LMO2 - iDAb LMO2dm3 protein interaction.
3. Discussion
The use of intracellular antibodies (scFv or domain antibodies) for studies in cells has been widely employed as laboratory reagents (Visintin et al., 2004) but these macromolecules also have great potential as lead molecules for drug development per se, either in themselves as macrodrugs (Tanaka and Rabbitts, 2008) or templates for small molecule surrogates as recently described for RAS protein binding Abd compounds (Quevedo et al., 2018). The Abd technology can in principle be applied to any intracellular antibody interacting with a target and can identify surrogate compounds that can interfere with a function of the target protein defined by the intracellular antibody. The Abd technology relies on a high affinity interaction of the intracellular antibody with the target protein (Quevedo et al., 2018) because passing the analyte (compound) across the chip will be accompanied by melting of antibody-protein interaction if the Koff is too great. In the case of the RAS screen, the intracellular antibody bound with pM affinity to RAS (Tanaka et al., 2007). In the current study, our anti-LMO2 iDAb has affinity of 94 nM that could potentially allow loss of iDAb from the chip exposing LMO2 protein to interaction with the compounds. As a mechanism to limit this, we generated a fusion protein in which the iDAb was linked to LMO2 with a short flexible glycine-serine linker to achieve a de novo increase in strength of the iDAb-LMO2 interaction.
The cSPR screening for LMO2 binding compounds was designed to find for compounds that bind to LMO2 in the region where the anti-LMO2 iDAb binds. Our initial hits from the PPI-Net library were those that bind to the LMO2-ΔLID fusion, but not to LMO2-iDAb fusion protein that ensures the compounds do not interact with the IBD LID domain. Even though the compounds identified with LMO2-iDAb in the cSPR, we cannot be sure that these bind to the LMO2 surface where the iDAb binds because the LMO2 structural changes observed when LMO2 is bound by the iDAb, compared to LMO2-LID (Sewell et al., 2014), may affect their binding. Our four compounds that bound to LMO2-ΔLID include two were close analogues. The availability of the compounds was limited and we were only able to assess Abd-L1 in a cell-based assay examining the possible effect on the interaction of LMO2 with a low affinity version of the iDAb. This showed that Abd-L1 was not able to affect this PPI in cells and this will be exacerbated by the adverse properties of the compound in CaCo2 permeability assays. Similar compounds have recently been described from a screen with LMO2 protein (Milton-Harris et al., 2020).
Our results show that the competitive surface plasmon resonance screening strategy is a valuable tool to target proteins of interest with chemical matter, and that are active for in vitro assays. A screening campaign using this strategy with large chemical libraries of compounds triaged for cellular permeability properties, could identify compounds active in cells. Further, the use of antibodies as precursors for drug screening campaigns is a strategy that can be generally used to generate Abd compounds for drug hit to lead development. This can be applied to intracellular targets (Cruz-Migoni et al., 2019) but also to viruses (Xiao et al., 2020) and should be applicable to the range of antibody targets to replace antibodies (which have been used in target validation) with small molecules.
4. Materials and methods
4.1. Cloning
The DNA sequences encoding for LMO2-ΔLID (UniProt P25791; residues 26–156 and UniProt Q86U70; residues 334–344, joined by a GS linker, (Wilkinson-White et al., 2010)), and LMO2-iDAb (Sewell et al., 2014) were cloned into the expression vector pOPINS (Berrow et al., 2007) via the restriction sites KpnI and HindIII, incorporating an AviTag into the 5′ primer. The vector encodes an N-terminal hexahistidine tag and SUMO tag. The final protein expression constructs encoded for a single fusion protein consisting of His-SUMO-Avi-LMO2-GS-ΔLID and His-SUMO-Avi-LMO2-GS-VH. A construct encoding KRAS166G12V with N-terminal His tag and TEV protease cleavage site was modified via PCR to include an AviTag between the TEV site and protein-coding sequence (Fairhead and Howarth, 2015). For the SPR screening, all proteins were prepared with biotin tags.
4.2. Protein expression and purification
LMO2-ΔLID proteins were expressed in BL21(DE3)-Rosetta2 pLysS cells (Novagen) or in Lemo21(DE3). Bacterial cells were cultured at 37 °C in TB medium supplemented with 50 μg mL− 1 kanamycin and 34 μg mL− 1 chloramphenicol. At mid-log phase, expression was induced by addition of IPTG to a final concentration of 0.5 mM and the cultures incubated overnight at 18 °C with shaking at 220 rpm. Cells were harvested by centrifugation and resuspended in binding buffer (50 mM Hepes, pH 7.4, 500 mM NaCl, 5% glycerol, and 5 mM imidazole). Resuspended pellets were stored at −20 °C. Thawed cell pellets were supplemented with complete EDTA-free protease inhibitors at 1× concentration (Roche), 5 μL Benzonase per 1 L culture volume and MgSO4 to a final concentration of 1 mM. Cell suspensions were stirred on ice for 15 mins and lysed using a Constant Systems Cell Disruptor at 23kpsi, 5 °C. Cell extracts were clarified by centrifugation. His-tagged proteins were purified under gravity flow using nickel-Sepharose (GE Healthcare) columns. Bound proteins were washed in binding buffer containing 30 mM imidazole and eluted in a stepwise gradient with elution buffer containing 500 mM imidazole. SUMO tags were removed by incubating with SUMO protease overnight at 4 °C. For proteins to be used in SPR experiments, N-terminal AviTags were biotinylated by incubating with BirA enzyme overnight at 4 °C in the presence of 20 mM MgCl2, 500 μM biotin and 2 mM ATP. Proteins were further purified on a HiLoad 16/60 Superdex 200 Prep Grade column using an ÄKTA Avant system (GE Healthcare) buffered in 10 mM HEPES pH 7.4, 250 mM NaCl and 0.5 mM DTT. KRAS166G12V was purified as previously described (Cruz-Migoni et al., 2019), with the addition of the BirA incubation step for biotinylation of the purified protein.
4.3. SPR screening
Biotinylated LMO2-ΔLID, KRAS and LMO2-VH fusion were immobilized on a streptavidin-coated sensor chip SA (GE Healthcare) and the PPI-Net library compounds (comprising 1500 compounds) were injected over the surface at 150 μM concentration. SPR experiments were carried out, as described (Cruz-Migoni et al., 2019; Quevedo et al., 2018), using a Biacore T200 (GE Healthcare) at 10 °C to preserve the protein immobilized on the sensor surface. The surface was washed with 1 M NaCl, 50 mM NaOH before immobilizing ~4000 response units (RU) of LMO2-ΔLID. Control proteins KRAS166G12V and LMO2-VH were immobilized at ~4000RU and ~ 6000RU respectively to give approximately equimolar immobilization levels of all three proteins on the sensor surface. Flow cell 1 was blocked by injecting 10 mM biocytin over the surface for 5 min at 10 μL/min and used as the reference channel. Immobilization was carried out in HEPES running buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.005% P20, 5 mM MgCl2, 10 μM ZnCl2). Compound solutions were prepared by transferring 1.5 μL PPI-Net stock compounds at 10 mM in 100% DMSO into 96-well plates (Greiner) using a multichannel pipette. 98.5 μL running buffer with 3.5% DMSO was added to yield a solution of 150 μM compound in running buffer with 5% DMSO. Compound solutions were injected over all 4 flow channels for 30 s at 30 μL/min and dissociation monitored for 60 s. A negative control of running buffer with 5% DMSO was run after every 24 cycles. Between injections the flow system was washed with a solution of 50% DMSO. A solvent correction curve was used to correct for the effects of DMSO. Data were referenced, solvent corrected and processed using the T200 evaluation software. Single-point binding levels were exported to Excel. Data were baseline-corrected using the negative control binding levels as a reference and binding levels measured against LMO2-ΔLID plotted against binding levels measured against the control proteins.
Replacement Abd-L1 (PPI-NET identifier P20000560B9) and Abd-L4 (PPI-NET identifier P20000557F5) were purchased from Asinex. Their molecular weights were confirmed by mass spec: Abd-L1 LRMS m/z (ESI+) 435 [M + H]+; Abd-L2 LRMS m/z (ESI+) 437 [M + H]+. Low-resolution mass spectra were recorded on an Agilent 6120 spectrometer using solutions of MeOH.
4.4. waterLOGSY NMR
The waterLOGSY NMR method was used to measure LMO2 ligand interaction. WaterLOGSY experiments were conducted at a 1H frequency of 600 MHz using a Bruker Avance spectrometer equipped with a BBI probe. All experiments were conducted at 298 K. NMR tubes with a 3 mm diameter were used in all experiments. Solutions were buffered using an H2O 10 mM NaPO4, 50 mM NaCl buffer corrected to pH 7.4. In the sample preparation the compound (10 μL of a 10 mM solution in DMSO‑d6) was added to an Eppendorf tube before sequential addition of the buffer (130 μL), D2O (20 μL), and protein (40 μL, 50 μM), giving a final volume of 200 μL. The resulting solution was mixed by vortexing to ensure full mixing and transferred to a 3 mm NMR tube before running the NMR spectra.
4.5. BRET2 competition assays
650,000 HEK293T were seeded in each well of 6 well plates. After 24 h at 37 °C, cells were transfected with a total of 1.6 μg of DNA mix, containing 0.05 μg of donor (pEF-LMO2-RLuc8) + 0.1 μg of acceptor (pEF-GFP2-iDAb LMO2dm3) ± competitor plasmid, using Lipofectamine 2000 transfection reagent (Thermo-Fisher). In dose response competition experiments, competitor (iDAb LMO2) was transfected with the following amount of DNA: 0.1; 0.5 and 1 μg. Cells were detached 24 h later, washed with PBS and seeded in a white 96-well plate (clear bottom, PerkinElmer, Cat#6005181) in OptiMEM no phenol red medium complemented with 4% FBS and incubated for an additional 20–24 h at 37 °C before the BRET assay reading. For BRET competition assays with compounds, cells were incubated for an additional 4 h at 37 °C before the addition of the compounds on the cells. Cells were treated with Abd-L1 at concentrations of 1, 10, 20, 50 and 100 μM for 22 h. The compound was diluted in the BRET medium: OptiMEM no phenol red (Life Technologies) supplemented with 4% FBS and with a final concentration of 0.2% DMSO. A detailed BRET protocol is provided elsewhere (Bery and Rabbitts, 2019).
BRET2 measurements was determined immediately after injection of coelenterazine 400a substrate (10 μM final) to cells (Cayman Chemicals), using a CLARIOstar instrument (BMG Labtech) with a luminescence module. The BRET signal or BRET ratio corresponds to the light emitted by the GFP2 acceptor constructs (515 nm ± 30) upon addition of coelenterazine 400a divided by the light emitted by the RLuc8 donor constructs (410 nm ± 80). The background signal is subtracted from thw BRET ratio using the donor-only negative control where only the RLuc8 fusion plasmid is transfected into the cells. The normalized BRET ratio is the BRET ratio normalized to a negative control (iDAb control or DMSO control) during a competition assay.
Author's contributions
Originator of project: TR.
Participated in research design: PC, TR.
Conducted experiments: PC, CB, NB, SM, AH, AM.
Performed data analysis: All authors.
Wrote or contributed to the writing of the manuscript: All authors.
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Acknowledgements
We wish to thank Professor Andrew Wilson (University of Leeds) for providing the PPI-NET compound library screening collection and GlaxoSmithKline for making the PPI-net screening collection available. The work was supported by grants from Bloodwise (Blood Cancer UK #12051) and the Medical Research Council MR/J000612/1.
References
- Bavetsias V., Lanigan R.M., Ruda G.F., Atrash B., McLaughlin M.G., Tumber A., Mok N.Y., Le Bihan Y.V., Dempster S., Boxall K.J. 8-substituted Pyrido[3,4-d]pyrimidin-4(3H)-one derivatives as potent, cell permeable, KDM4 (JMJD2) and KDM5 (JARID1) histone lysine demethylase inhibitors. J. Med. Chem. 2016;59:1388–1409. doi: 10.1021/acs.jmedchem.5b01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrow N.S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D.I., Owens R.J. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bery N., Rabbitts T.H. Bioluminescence resonance energy transfer 2 (BRET2)-based RAS biosensors to characterize RAS inhibitors. Curr Protoc Cell Biol. 2019;83 doi: 10.1002/cpcb.83. [DOI] [PubMed] [Google Scholar]
- Bery N., Cruz-Migoni A., Bataille C.J., Quevedo C.E., Tulmin H., Miller A., Russell A., Phillips S.E., Carr S.B., Rabbitts T.H. BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions. Elife. 2018;7 doi: 10.7554/eLife.37122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bery N., Bataiile C.J.R., Russell A., Hayes A., Raynaud F., Milhas S., Anadd S., Tulimn H., Miller A., Rabbitts T.H. A cell-based screening method using an intracellular antibody for discovering small molecules targeting the translocation protein LMO2. Sci. Adv. 2021 doi: 10.1126/sciadv.abg1950. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J., Rabbitts T.H. LMO2 at 25 years: a paradigm of chromosomal translocation proteins. Open Biol. 2015;5:150062. doi: 10.1098/rsob.150062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Migoni A., Canning P., Quevedo C.E., Bataille C.J.R., Bery N., Miller A., Russell A.J., Phillips S.E.V., Carr S.B., Rabbitts T.H. Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2545–2550. doi: 10.1073/pnas.1811360116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Omari K., Hoosdally S.J., Tuladhar K., Karia D., Hall-Ponsele E., Platonova O., Vyas P., Patient R., Porcher C., Mancini E.J. Structural basis for LMO2-driven recruitment of the SCL:E47bHLH heterodimer to hematopoietic-specific transcriptional targets. Cell Rep. 2013;4:135–147. doi: 10.1016/j.celrep.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead M., Howarth M. Site-specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 2015;1266:171–184. doi: 10.1007/978-1-4939-2272-7_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A.A., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Grutz G.G., Bucher K., Lavenir I., Larson T., Larson R., Rabbitts T.H. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 1998;17:4594–4605. doi: 10.1093/emboj/17.16.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Bonnichon A., Claridge T.D., Leung I.K. Protein-ligand binding affinity determination by the waterLOGSY method: an optimised approach considering ligand rebinding. Sci. Rep. 2017;7:43727. doi: 10.1038/srep43727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton-Harris L., Jeeves M., Walker S.A., Ward S.E., Mancini E.J. Small molecule inhibits T-cell acute lymphoblastic leukaemia oncogenic interaction through conformational modulation of LMO2. Oncotarget. 2020;11:1737–1748. doi: 10.18632/oncotarget.27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam C.H., Lobato M.N., Appert A., Drynan L.F., Tanaka T., Rabbitts T.H. An antibody inhibitor of the LMO2-protein complex blocks its normal and tumorigenic functions. Oncogene. 2008;27:4962–4968. doi: 10.1038/onc.2008.130. [DOI] [PubMed] [Google Scholar]
- Quevedo C.E., Cruz-Migoni A., Bery N., Miller A., Tanaka T., Petch D., Bataille C.J.R., Lee L.Y.W., Fallon P.S., Tulmin H. Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment. Nat. Commun. 2018;9:3169. doi: 10.1038/s41467-018-05707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell H., Tanaka T., El Omari K., Mancini E.J., Cruz A., Fernandez-Fuentes N., Chambers J., Rabbitts T.H. Conformational flexibility of the oncogenic protein LMO2 primes the formation of the multi-protein transcription complex. Sci. Rep. 2014;4:3643. doi: 10.1038/srep03643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Rabbitts T.H. Interfering with protein-protein interactions: potential for cancer therapy. Cell Cycle. 2008;7:1569–1574. doi: 10.4161/cc.7.11.6061. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Williams R.L., Rabbitts T.H. Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J. 2007;26:3250–3259. doi: 10.1038/sj.emboj.7601744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Sewell H., Waters S., Phillips S.E., Rabbitts T.H. Single domain intracellular antibodies from diverse libraries: emphasizing dual functions of LMO2 protein interactions using a single VH domain. J. Biol. Chem. 2011;286:3707–3716. doi: 10.1074/jbc.M110.188193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin M., Meli G.A., Cannistraci I., Cattaneo A. Intracellular antibodies for proteomics. J. Immunol. Methods. 2004;290:135–153. doi: 10.1016/j.jim.2004.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I.A., Osada H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-White L.E., Dastmalchi S., Kwan A.H., Ryan D.P., Mackay J.P., Matthews J.M. 1H, 15N and 13C assignments of an intramolecular Lmo2-LIM2/Ldb1-LID complex. Biomol NMR Assign. 2010;4:203–206. doi: 10.1007/s12104-010-9240-y. [DOI] [PubMed] [Google Scholar]
- Xiao T., Frey G., Fu Q., Lavine C.L., Scott D.A., Seaman M.S., Chou J.J., Chen B. HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes. Nat. Chem. Biol. 2020;16:529–537. doi: 10.1038/s41589-020-0496-y. [DOI] [PMC free article] [PubMed] [Google Scholar]