Abstract
Heterotaxy (HTX), a condition characterized by internal organs not being arranged as expected relative to each other and to the left-right axis, is often accompanied with congenital heart disease (CHD). The purpose was to detect the pathogenic variants in a Chinese family with HTX and CHD. A non-consanguineous Han Chinese family with HTX and CHD, and 200 unrelated healthy subjects were enlisted. Exome sequencing and Sanger sequencing were applied to identify the genetic basis of the HTX family. Compound heterozygous variants, c.3426-1G>A and c.4306C>T (p.(Arg1436Trp)), in the dynein axonemal heavy chain 11 gene (DNAH11) were identified in the proband via exome sequencing and further confirmed by Sanger sequencing. Neither c.3426-1G>A nor c.4306C>T variant in the DNAH11 gene was detected in 200 healthy controls. The DNAH11 c.3426-1G>A variant was predicted as altering the acceptor splice site and most likely affecting splicing. The DNAH11 c.4306C>T variant was predicted to be damaging, which may reduce the phenotype severity. The compound heterozygous variants, c.3426-1G>A and c.4306C>T, in the DNAH11 gene might be the pathogenic alterations resulting in HTX and CHD in this family. These findings broaden the variant spectrum of the DNAH11 gene and increase knowledge used in genetic counseling for the HTX family.
Introduction
Human and other vertebrate visceral organs are asymmetric to the left-right (LR) axis. Normal organ placement relative to the LR axis is referred to as situs solitus (SS). Abnormal positioning is heterotaxy (HTX) or situs inversus (SI) [1]. A randomized placement of visceral organs relative to each other and LR axis is HTX. Visceral organ placement in a perfect mirror-image position along the LR axis is SI [2].
HTX is an extremely clinical heterogeneous disorder. HTX prevalence is about 1 in 10,000 newborns [3]. HTX, SI, and SS may present in different families or accidentally in the same family [1]. HTX is often connected with congenital heart disease (CHD) and is classified as either isolated or syndromic [4, 5]. At least 59 syndromes have been reported to associate either HTX or SI [5].
HTX is a genetic heterogeneous disorder with incomplete penetrance. It may be inherited in an autosomal recessive, autosomal dominant or X-linked fashion. It associates with genetic factors, environmental modifiers, and developmental randomness [4, 5]. As of the date of this writing (November 2020), nine loci (HTX1-9) and eight causative genes, including the Zic family member 3 gene (ZIC3), the cripto, FRL-1, cryptic family 1 gene (CFC1), the activin A receptor type 2B gene (ACVR2B), the nodal growth differentiation factor gene (NODAL), the cilia and flagella associated protein 53 gene (CFAP53), the matrix metallopeptidase 21 gene (MMP21), the polycystin 1 like 1, transient receptor potential channel interacting gene (PKD1L1) and the meiosis specific nuclear structural 1 gene (MNS1) have been reported as associating with isolated HTX [6–14].
The great clinical and genetic heterogeneity presents challenges to correctly identifying causative variants in individuals with HTX phenotype using Sanger sequencing. Exome sequencing, a cost-saving measure, has been used in the genetic investigation of HTX [12–14]. Our previous study has successfully identified pathogenic variants in two Chinese families with SI phenotype via exome sequencing [15, 16]. In this study, exome sequencing and Sanger sequencing detected the compound heterozygous variants, c.3426-1G>A and c.4306C>T (p.(Arg1436Trp)), in the dynein axonemal heavy chain 11 gene (DNAH11, NM_001277115.1) in a Han Chinese family with HTX and CHD.
Materials and methods
Subjects
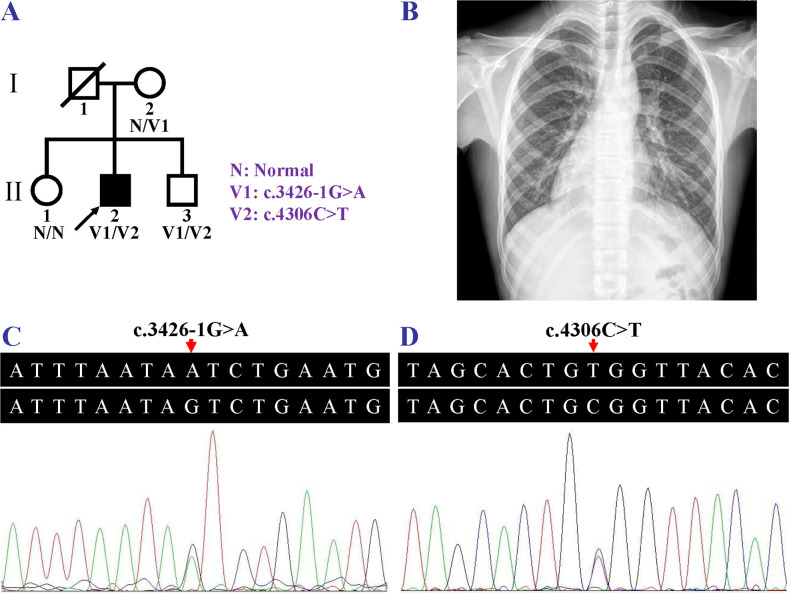
A two-generation, non-consanguineous Han family, from Yiyang, Hunan, China, with HTX and CHD was enrolled from May to July 2019. Four family members including the proband (II:2), his unaffected mother (I:2), and two unaffected siblings (II:1 and II:3) participated (Fig 1A). The proband (II:2) was diagnosed, at age five, as having cyanotic complex CHD due to dyspnea, cyanosis, poor activity endurance and cardiac murmurs. His first operation occurred at sixteen after he was admitted to hospital due to severe shortness of breath. He presented with cyanotic lips. The apical impulse was detected at the right fifth intercostal space. A systolic ejection murmur was heard in the right second intercostal space. Chest X-ray revealed dextrocardia (Fig 1B). Echocardiography showed complex CHD, including single ventricle, incomplete transposition of the great arteries, and pulmonary valve stenosis. No abdominal organ abnormalities were detected in the proband. Dyspnea, cyanosis, dextrocardia, cardiac murmurs, or cardiovascular malformations were not detected in his parents or other siblings. Two hundred unrelated subjects (100 males and 100 females, aged 16.5±5.5 years) who did not have dyspnea, cyanosis, dextrocardia, or cardiovascular anomalies were recruited as healthy controls from May to October 2019. The study was approved by the Institutional Review Board of the Third Xiangya Hospital, Central South University (Changsha, China) in accordance with the Declaration of Helsinki. The study was conducted from May to December 2019. Written informed consent forms were obtained, and venous blood samples were taken from all participants. The authors had no access to information that could identify individual participants during or after data collection.
Fig 1. Pedigree of a Chinese heterotaxy family, chest X-ray of the heterotaxy patient and the DNAH11 Sanger sequencing electropherograms.
(A) Pedigree of a Chinese heterotaxy family. N, normal; V1, the DNAH11 c.3426-1G>A variant; V2, the DNAH11 c.4306C>T variant. The slash indicates deceased individual, the fully shaded symbol indicates the affected individual, and the arrow indicates the proband. (B) Chest X-ray of the family member (II:2) revealed dextrocardia. (C, D) The compound heterozygosity for the DNAH11 variants, c.3426-1G>A and c.4306C>T, in the individual (II:2) with heterotaxy.
Exome sequencing
Genomic DNA was obtained from blood samples via a saturated phenol-chloroform extraction method [17]. A microgram of genomic DNA from the proband (II:2) was exome sequenced at BGI-Shenzhen, China. The library with targeted exome was captured by the Agilent SureSelect Human All Exon V6 and sequenced using a BGISEQ-500 platform. Average sequencing depth was 267.63×. Debased reads were filtered and the clean reads were mapped to the human reference genome (UCSC database version hg19, http://genome.ucsc.edu/) using a Burrows-Wheeler Aligner tool (BWA, version 0.7.15). Duplicate reads were marked by Picard-tools (version 2.5.0). Single nucleotide polymorphisms (SNPs) and insertions/deletions (Indels) were detected by a HaplotypeCaller of Genome Analysis Toolkit (GATK, version 3.3.0). Variant annotation and prediction were performed by SnpEff tool (https://pcingola.github.io/SnpEff/). Variants with minor allele frequency ≥1% in the following databases were eliminated: the SNP database version 141 (dbSNP141), the 1000 Genomes Project (1000 genomes release phase 3), and the NHLBI Exome Sequencing Project (ESP) 6500 [18]. The functional impacts of nonsynonymous SNPs or Indels were predicted by the Sorting Intolerant from Tolerant (SIFT) and the Protein Variation Effect Analyzer (PROVEAN, version 1.1.3, http://provean.jcvi.org/index.php). The effects of splice site variants on splicing were predicted by the Human Splicing Finder (HSF, version 3.1, http://www.umd.be/HSF3/).
Sanger sequencing, variant analysis and protein structure modeling
Candidate variants were validated by Sanger sequencing. A co-segregation analysis between possible causative variants and this family’s HTX phenotype was performed [19]. Paired primer sequences for the candidate variants (c.3426-1G>A and c.4306C>T) in the DNAH11 gene were synthesized respectively as follows: 5′-TGTTGCCAGTTTCATGATAGAGA-3′ and 5′-TACAGCCAGAAGATGCACCA-3′; 5′-TTCACCAGCCTTTAGGCAAA-3′ and 5′-TCTCAGTCCCCAGCTCTTTC-3′. The recorded frequencies of the candidate variants in the Genome Aggregation Database (gnomAD, version 3.0, http://gnomad.broadinstitute.org) were further checked, and pathogenic variant databases, including the Leiden Open Variation Database (LOVD, v.3.0, https://www.lovd.nl/3.0/home), the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php), and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) were referred to interpreting variants. The American College of Medical Genetics and Genomics (ACMG) sequence variant interpretation guidelines, and recommendations for interpreting the loss of function pathogenic criterion (PVS1) created by ClinGen Sequence Variant Interpretation Working Group, were used to categorize the identified variants [20, 21].
The protein structures of wild-type and variant-type were predicted via the online SWISS-MODEL tool (http://www.swissmodel.expasy.org), and the visualized structures were constructed using the PyMOL software (version 1.7, Schrödinger, LLC, Portland, U.S.A.).
Results
Exome sequencing generated 240,827,260 clean reads. Approximately 99.94% of the clean reads were mapped to the human reference genome. A total of 105,788 SNPs and 18,688 Indels were detected. After a series of optimization screening measures, two variants, c.3426-1G>A and c.4306C>T in the DNAH11 gene, were discovered. Other possible disease-causing alterations in the pathogenic genes known for HTX, SI or CHD were eliminated.
The compound heterozygous variants, a splicing variant, c.3426-1G>A, and a missense variant, c.4306C>T (p.(Arg1436Trp)), in the DNAH11 gene, were further confirmed in the HTX patient (II:2) via Sanger sequencing (Fig 1C and 1D). The heterozygous c.3426-1G>A variant in the DNAH11 gene was detected in his mother (I:2). Neither c.3426-1G>A nor c.4306C>T variant in the DNAH11 gene was discovered in his elder sister (II:1). The compound heterozygous variants, c.3426-1G>A and c.4306C>T, in the DNAH11 gene were also identified in his brother (II:3) who had no HTX or SI phenotype. This was in accord with prior reports that SI and HTX may occur in about half of PCD patients, approximately 59.1% of individuals or 42.9% of affected siblings having the DNAH11 biallelic variants [22, 23]. His father’s genotyping was indefinable as he (I:1) passed away. Due to the presence of the DNAH11 c.4306C>T variant in two brothers (II:2 and II:3) and the absence in the mother (I:2), the variant might be not de novo, but inherited from his father (I:1), further supporting that the disorder was inherited in an autosomal recessive manner in this family. Neither DNAH11 c.3426-1G>A nor c.4306C>T variant was detected in the 200 healthy controls.
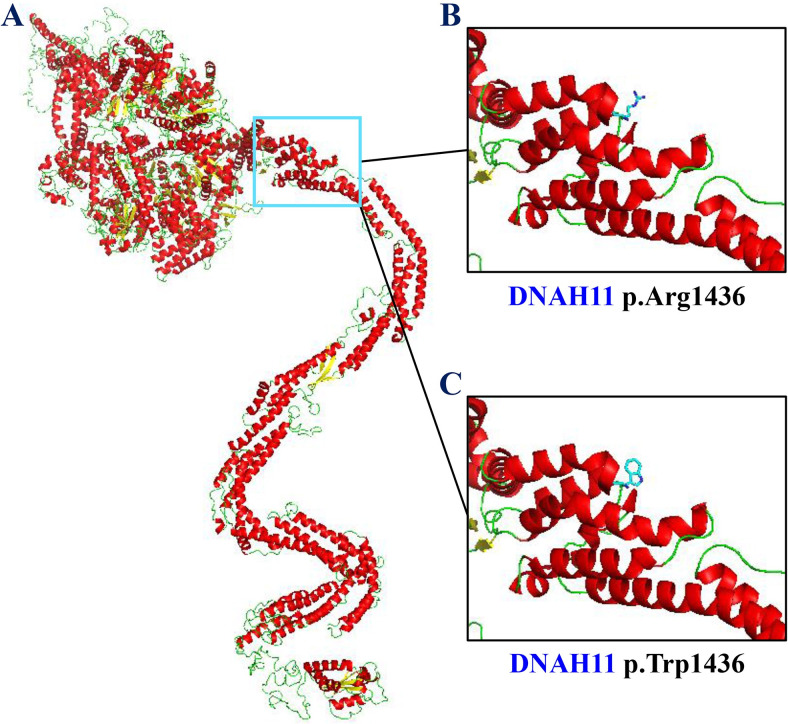
The HSF tool predicted that the DNAH11 c.3426-1G>A variant alters acceptor splice site and most probably affects splicing. The DNAH11 c.4306C>T variant was predicted to result in substituting tryptophan for arginine at codon 1436 (p.(Arg1436Trp)) and it would be damaging with a prediction score of 0.001 by SIFT, and be deleterious with a score of -3.10 by PROVEAN. The allele frequencies in the gnomAD were 1.31×10−5 for c.3426-1G>A and 3.09×10−4 for c.4306C>T. The two variants were not recorded in LOVD, but deposited in HGMD, responsible for ciliary dyskinesia and bronchiectasis, respectively. The DNAH11 c.3426-1G>A variant was interpreted as pathogenic in the ClinVar database, while the c.4306C>T variant had conflicting interpretations of pathogenicity (uncertain significance or benign). According to guidelines for the sequence variant interpretation, the variants, c.3426-1G>A and c.4306C>T, were classified as “pathogenic” (PVS1+PM2+PP5) and “uncertain significance” (PM2), respectively. Conformational changes after the arginine at residue 1436 (p.Arg1436) altered to tryptophan (p.Trp1436) were displayed by structural modeling (Fig 2).
Fig 2. Structural modeling displayed conformational changes of the DNAH11 p.(Arg1436Trp) variant.
(A) The cartoon representation of the DNAH11 protein. (B) The stick model of the arginine at residue 1436 (p.Arg1436). (C) The stick model of the mutant tryptophan at residue 1436 (p.Trp1436).
Discussion
HTX occurs in approximately 6.3%-12.1% of patients having primary ciliary dyskinesia (PCD) [24, 25]. In 2002, a homozygous c.8533C>T (p.(Arg2845*)) variant and a heterozygous c.8990G>A (p.(Arg2997Gln)) variant in the DNAH11 gene were firstly detected in two patients with SI and most likely PCD, respectively [26]. Subsequently compound heterozygous variants, c.12363C>G (p.(Tyr4121*)) and c.13531_*36del (p.(Ala4511_Ala4516delinsGln)), in the DNAH11 gene were identified in a family with PCD, including a patient with SI [27]. Biallelic variants in the DNAH11 gene may be responsible for about 22% of PCD patients with normal ciliary ultrastructure. It was reported that SI occurred in 54.5% of PCD patients, HTX in about 4.5% of PCD patients, and SS in about 41% of PCD patients with DNAH11 biallelic variants [22].
The DNAH11 gene locates in 7p15.3, contains 82 exons, and encodes a ciliary outer dynein arm with 4516 amino acids. The protein contains an N-terminal tail domain, six ATPase associated diverse cellular activities (AAA) domains, four P-loops, a helix-1-microtubule-binding-helix-2 domain, and a C-domain [26].
The embryonic expression of Dnah11, a mouse homolog of human DNAH11, also called “left-right dynein” (lrd), is detected primarily in non-ciliated cells and highly restricted in the node at mouse embryonic day 7.5. It also expresses in many ciliated cells in newborn and adult mice [28]. Dnah11-containing monocilia in the node generate a leftward nodal flow which may determine the normal left-right patterns in mouse embryos [29, 30].
The iv/iv mice, resulting from the homozygous missense p.Glu2271Lys variant in the Dnah11 gene, presented with an abnormal nodal flow, randomized expression of lateralized genes, a high incidence of cardiovascular anomaly, and randomized left-right development. Half of them exhibited either HTX or SI, and half exhibited SS [28, 30–34]. Phenotypes indicative of human PCD, including rhinitis, sinusitis, otitis media, static tracheal cilia, and sperm motility defects, were also discovered in the iv/iv mice [34].
Compound heterozygous DNAH11 variants, responsible for about 14.3% of patients with CHD and HTX, may be a frequent genetic basis of familial CHD and HTX syndrome [35]. The reported compound heterozygous variants in the DNAH11 gene and their various phenotypes were summarized in Table 1 [22, 23, 27, 34–43].
Table 1. Clinical phenotypes of individuals with DNAH11 compound heterozygous variants.
| Patients | Gender | Ethnicity | Sequence variants# | PCD phenotypes | Situs | CHD | Ciliary motion | References |
|---|---|---|---|---|---|---|---|---|
| - | F | Chinese | c.73G>A, p.(Ala25Thr) | BR, rhinosinusitis | SI | - | KS | [36] |
| c.5702A>C, p.(Glu1901Ala) | ||||||||
| OP41-II:1 | M | Caucasian | c.350A>T, p.(Glu117Val) | NRD, sinusitis, OM | SI | - | KS (H) | [22] |
| c.7148T>C, p.(Leu2383Pro) | ||||||||
| #5062 | F | Chinese | c.727A>G, p.(Ile243Val) | na | HTX | + | CD (I+R) | [35] |
| c.10829A>T, p.(Asp3610Val) | ||||||||
| #5031 | M | Chinese | c.846G>C, p.(Met282Ile) | na | HTX | + | CD (I+R+D) | [35] |
| P44 | M | c.2406G>A, p.(Trp802*) | Rhinosinusitis | HTX | + | KS (R) | [37] | |
| A | F | Italian | c.883-1G>A | A: BR, rhinitis, rhinosinusitis, otitis | SI | - | KS (I+H) | [38] |
| B | M | c.4130G>A, p.(Trp1377*) | SI | |||||
| B: Recurrent pneumonia, BR | ||||||||
| B212 | na | Chinese | c.1300T>C, p.(Phe434Leu) | BR | na | na | na | [39] |
| c.6983C>T, p.(Pro2328Leu) | ||||||||
| #5065 | M | Chinese | c.1339G>A, p.(Gly447Arg) | na | HTX | + | CD (I+R) | [35] |
| c. 3470T>G, p.(Leu1157Arg) | ||||||||
| Patient 1 | M | Polish | c.1648del, p.(Arg550Glyfs*16) | na | HTX | + | na | [40] |
| Patient 2 | na | c.2772G>A, p.(Met924Ile) | ||||||
| c.11662C>T, p.(Arg3888Cys) | ||||||||
| PCD761 | F | Caucasian | c.2275-1G>C, p.Tyr759_Glu889del | NRD, BR, sinusitis, OM | SI | - | KS (H) | [22] |
| c.13213del, p.(Arg4405Alafs*2) | ||||||||
| Family 1–1 | F | Finnish | c.2341G>A, p.(Glu781Lys) | Rhinosinusitis, OM | SI | - | KS (Static) PCD (D) | [41] |
| Family 1–2 | M | c.7645+5G>A | Rhinosinusitis | SS | ||||
| B082 | na | Chinese | c.2419G>C, p.(Asp807His) | BR | na | na | na | [39] |
| c.2542G>A, p.(Val848Met) | ||||||||
| B012 | na | Chinese | c.2419G>C, p.(Asp807His) | BR | na | na | na | [39] |
| c.12258C>A, p.(Tyr4086*) | ||||||||
| P28 | M | Chinese | c.2485C>T, p.(Arg829Cys) | Rhinosinusitis, OM | SS | - | PCD (R) | [37] |
| c.5608C>T, p.(Pro1870Ser) | ||||||||
| B036 | na | Chinese | c.2542G>A, p.(Val848Met) | BR | na | na | na | [39] |
| c.9260A>G, p.(Lys3087Arg) | ||||||||
| B170 | na | Chinese | c.2542G>A, p.(Val848Met) | BR | na | na | na | [39] |
| c.11624A>G, p.(Lys3875Arg) | ||||||||
| B156 | na | Chinese | c.2542G>A, p.(Val848Met) | BR | na | na | na | [39] |
| c.12071C>T, p.(Ser4024Phe) | ||||||||
| 11174 | M | Italian | c.2753G>T, p.(Gly918Val) | Bronchitis, rhinitis, sinusitis, otitis | SS | - | PCD (I) | [23] |
| c.12796_12801delinsATA, p.Phe4266_Asn4267delinsIle | ||||||||
| #4 | F | English | c.2832dup, p.(Gln945Serfs*10) | Recurrent chest infection, rhinitis | SS | - | PCD | [42] |
| c.13240dup, p.(Thr4414Asnfs*34) | ||||||||
| B142 | na | Chinese | c.2912A>G, p.(Asp971Gly) | BR | na | na | na | [39] |
| c.11396T>C, p.(Ile3799Thr) | ||||||||
| P25 | M | Chinese | c.3020T>G, p.(Leu1007*) | Rhinosinusitis, OM | SI | - | KS (N) | [37] |
| c.3470T>G, p.(Leu1157Arg) | ||||||||
| #6 | F | English | c.3220G>T, p.(Glu1074*) | Recurrent chest infection | SI | - | KS (Static) | [42] |
| c.13069C>T, p.(Arg4357*) | ||||||||
| II:2 | M | Chinese | c.3426-1G>A | - | HTX | + | na | This study |
| c.4306C>T, p.(Arg1436Trp) | ||||||||
| P45 | F | Chinese | c.3470T>G, p.(Leu1157Arg) | Rhinosinusitis, OM | SI | - | KS (R) | [37] |
| c.6727C>G, p.(Arg2243Gly) | ||||||||
| #7 | M | English | c.3544C>T, p.(Arg1182*) | Recurrent chest infection, rhinitis | SS | - | PCD | [42] |
| c.8798-5G>A | ||||||||
| #1 | M | English | c.3727G>T, p.(Glu1243*) | Recurrent chest infection, rhinitis | SS | - | PCD (Static) | [42] |
| c.13531_13532insTTCAGGCTGAAGA, p.(Ala4511Valfs*13) | ||||||||
| PCD1077 | F | Caucasian | c.3901G>T, p.(Glu1301*) | NRD, sinusitis, OM | SI | - | KS | [22] |
| c.11804C>T, p.(Pro3935Leu) | ||||||||
| OP406-II:1 | M | Caucasian | c.4254+5G>T | na | SI | KS (H) | [22] | |
| OP406-II:2 | F | c.4726-1G>A | NRD, sinusitis | SS | PCD (H) | |||
| B099 | na | Chinese | c.4306C>T, p.(Arg1436Trp) | BR | na | na | na | [39] |
| c.6118C>T, p.(Arg2040Cys) | ||||||||
| #2 | F | English | c.4395_4398del, p.(Ser1465Argfs*6) | Recurrent chest infection, rhinitis | SS | - | PCD (Static) | [42] |
| c.7642C>T, p.(Gln2548*) | ||||||||
| P32 | M | Chinese | c.4457T>A, p.(Leu1486Gln) | NRD, rhinosinusitis, OM | SS | - | PCD (N) | [37] |
| c.10006G>T, p.(Ala3336Ser) | ||||||||
| 9003 | F | American | c.4505A>C, p.(Gln1502Pro) | NRD, recurrent pneumonia, BR, sinusitis, OM | HTX | + | CD (H) | [43] |
| c.9376G>A, p.(Glu3126Lys) | ||||||||
| PCD106 | M | Caucasian | c.4516_4517del, p.(Leu1506Serfs*11) | Sinusitis, OM | SS | - | PCD | [22] |
| KS (H) | ||||||||
| PCD108 | M | NRD, sinusitis, OM | SI | |||||
| c.7266+1G>A | ||||||||
| B185 | na | Chinese | c.4898C>A, p.(Ser1633Tyr) | BR | na | na | na | [39] |
| c.9260A>G, p.(Lys3087Arg) | ||||||||
| 11228 | M | Italian | c.4922C>G, p.(Ser1641*) | Bronchitis, rhinitis | SI | - | KS | [23] |
| c.9304G>A, p.(Gly3102Ser) | ||||||||
| #5707 | M | Chinese | c.5473dup, p.(Gln1825Profs*23) | na | HTX | + | CD (I+R) | [35] |
| c.8275T>C, p.(Phe2759Leu) | ||||||||
| c.13183C>T, p.(Arg4395*) | ||||||||
| #3 | F | English | c.5506C>T, p.(Arg1836*) | Recurrent chest infection | SI | - | KS (Static) | [42] |
| c.5636T>A, p.(Leu1879*) | ||||||||
| PCD565 | M | Caucasian | c.5778+1G>A | NRD, BR, sinusitis, OM | SI | - | KS (H) | [22] |
| c.13061T>A, p.(Leu4354His) | ||||||||
| PCD812 | M | Caucasian | c.5815G>A, p.(Gly1939Arg) | NRD, sinusitis, OM | SI | - | KS | [22] |
| c.13373C>T, p.(Pro4458Leu) | ||||||||
| PCD157 | F | Caucasian | c.6244C>T, p.(Arg2082*) | NRD, BR, sinusitis, OM | SI | - | KS (H) | [22] |
| c.11929G>T, p.(Glu3977*) | ||||||||
| #5130 | F | Chinese | c.6785T>C, p.(Ile2262Thr) | na | HTX | + | CD (R+D) | [35] |
| c.11398G>C, p.(Asp3800His) | ||||||||
| B178 | na | Chinese | c.6905A>C, p.(His2302Pro) | BR | na | na | na | [39] |
| c.13112C>T, p.(Pro4371Leu) | ||||||||
| B073 | na | Chinese | c.7292G>T, p.(Ser2431Ile) | BR | na | na | na | [39] |
| c.9017C>T, p.(Thr3006Met) | ||||||||
| P17 | M | Chinese | c.7292G>T, p.(Ser2431Ile) | NRD, rhinosinusitis, OM | SI | - | KS (R) | [37] |
| P30 | M | SS | + | PCD (R) | ||||
| c.7364A>C, p.(Asp2455Ala) | NRD | |||||||
| c.9017C>T, p.(Thr3006Met) | ||||||||
| c.13373C>T, p.(Pro4458Leu) | ||||||||
| #730 | na | Caucasian | c.7772C>T, p.(Pro2591Leu) | NRD, rhinitis, sinusitis, OM | SS | - | PCD (erratic) | [34] |
| c.8698C>T, p.(Arg2900*) | ||||||||
| OP98-II:1 | M | Caucasian | c.7914G>C, p.Trp2604* | BR, sinusitis, OM | SI | - | KS (H) | [22] |
| PCD (H) | ||||||||
| OP98-II:2 | M | c.13330_13333dup, p.(Ile4445Asnfs*4) | SS | |||||
| C | M | Italian | c.8114A>G, p.(His2705Arg) | NRD, recurrent pneumonia, sinusitis, otitis | SI | + | KS (I+H) | [38] |
| c.10264G>A, p.(Gly3422Arg) | ||||||||
| B167 | na | Chinese | c.9260A>G, p.(Lys3087Arg) | BR | na | na | na | [39] |
| c.11647C>T, p.(Leu3883Phe) | ||||||||
| B163 | na | Chinese | c.9260A>G, p.(Lys3087Arg) | BR | na | na | na | [39] |
| c.13175C>T, p.(Thr4392Met) | ||||||||
| P23 | F | Chinese | c.9539T>A, p.(Leu3180*) | Rhinosinusitis, OM | SI | - | KS (R) | [37] |
| c.9706C>T, p.(Arg3236*) | ||||||||
| #5045 | M | Chinese | c.10379C>A, p.(Thr3460Lys) | na | HTX | + | CD (R+D) | [35] |
| c.13273G>A, p.(Gly4425Ser) | ||||||||
| PCD1126 | F | Asian | c.12064G>C, p.(Ala4022Pro) | BR, sinusitis | SS | - | PCD (H) | [22] |
| c.13500_13504dup, p.(Thr4502Argfs*15) | ||||||||
| II-2 | M | German | c.12363C>G, p.Tyr4121* | Recurrent pneumonia, bronchitis, sinusitis, BR (II-4, 6, 11), OM (II-3, 4, 9) | SI | - | KS | [27] |
| PCD | ||||||||
| PCD | ||||||||
| II-3, 4 | F | c.13531_*36del, p.Ala4511_Ala4516delinsGln | SS | |||||
| II-6, 9, 11 | M | SS | ||||||
| OP235-II:1 | F | Caucasian | c.12697C>T, p.(Gln4233*) | BR, sinusitis, OM | SS | - | PCD (H) | [22] |
| KS (H) | ||||||||
| OP235-II:2 | F | c.12980T>C, p.(Leu4327Ser) | NRD, BR, sinusitis, OM | SI | ||||
| PCD918 | F | Asian | c.13065_13067del, p.(Leu4356del) | BR, NRD, sinusitis, OM | SS | - | PCD | [22] |
| KS (H) | ||||||||
| PCD919 | M | HTX | ||||||
| c.13075C>T, p.(Arg4359*) |
# Description of the sequence variants is recalibrated in accordance with the Human Genome Variation Society nomenclature for variants (http://varnomen.hgvs.org/) using the reference sequence (NM_001277115.1).
BR, bronchiectasis; CD, ciliary dysfunction; CHD, congenital heart disease; D, discordance; F, female; H, hyperkinetic; HTX, heterotaxy; I, immotile; KS, Kartagener syndrome; M, male; N, nearly normal; na, not available; NRD, neonatal respiratory distress; OM, otitis media; PCD, primary ciliary dyskinesia; R, restricted; SI, situs inversus; SS, situs solitus; +, present; -, not present.
In this study, the compound heterozygous variants, c.3426-1G>A and c.4306C>T (p.(Arg1436Trp)), in the DNAH11 gene were identified in the HTX patient and his unaffected brother. Different clinical phenotypes (HTX, SI or SS) of the siblings with the DNAH11 gene biallelic variants have been reported in some families [22, 40]. HTX and SI occurring in about 59% of individuals with the DNAH11 biallelic variants may be connected with clinical heterogeneity of LR asymmetry disorders with incomplete penetration, or additional genetic variants in the family member II:3 serving a suppressor in the DNAH11 pathway. It might also be associated with environmental modifiers and randomized left-right development [1, 22].
The DNAH11 c.3426-1G>A variant in the homozygous state was previously identified in a fetus with structural abnormalities, including single atrium and ventricle, pulmonary stenosis, and right isomerism [44]. The compound heterozygous DNAH11 missense variants, c.4306C>T (p.(Arg1436Trp)) and c.6118C>T (p.(Arg2040Cys)), were reported in a patient with bronchiectasis, a clinical manifestation of PCD [39]. Neither variants were detected in the enrolled 200 healthy controls. Allele frequency of either variant in the total population was very low. The DNAH11 c.3426-1G>A variant was predicted to alter the acceptor splice site, and the c.4306C>T variant was predicted to be damaging. Together with the pathogenicity evidence, the c.3426-1G>A and c.4306C>T variants in the DNAH11 gene, as a novel combination, could be the pathogenic variants for HTX and CHD in this family, and the missense variant may reduce the phenotype severity.
DNAH11 biallelic variants were detected in patients with ciliary dysfunction, HTX, SI and CHD phenotypes [23, 40]. The patient refused to permit either the nasal curettage or the bronchial mucosal biopsy to perform an analysis on the ciliary ultrastructure and the waveform. No obvious chronic airway infection symptoms or signs, such as recurrent rhinitis, sinusitis, otitis media, bronchitis, frequent pneumonias, or bronchiectasis were observed in the two family members having the DNAH11 compound heterozygous variants. However, the very minor or preclinical phenotypes of PCD, such as rhinitis and bronchitis, might be overlooked and a diagnosis of mild PCD may be not excluded. In this study, there is a limitation that the father’s genotyping is unavailable due to his death. He died from an accidental injury, without complaining of PCD-related rhinitis, sinusitis, otitis media, bronchitis, bronchiectasis, or cardiovascular anomaly.
In conclusion, the compound heterozygous variants, c.3426-1G>A and c.4306C>T, in the DNAH11 gene might be the pathogenic alterations in this family with HTX and CHD. Our findings broaden the variant spectrum of the DNAH11 gene and provide additional information usable in genetic counseling for this family.
Acknowledgments
The authors are grateful to the participating individuals and investigators for their co-operation and efforts in collecting clinical and genetic information and DNA specimens.
Data Availability
The data involved in the study cannot be shared publicly because of ethical and legal concerns, regarding sensitive information. Reasonable requests to access the datasets by the qualified researchers can be sent to the Institutional Review Board of the Third Xiangya Hospital, Central South University (xy3irb@163.com).
Funding Statement
The study was supported by grants from National Natural Science Foundation of China (81670216, 81873686 and 81800219), Natural Science Foundation of Hunan Province, China (2019JJ50927, 2020JJ3057 and 2020JJ4830), Scientific Research Project of Health Commission of Hunan Province, China (B2019174), Special Emergency Project from Science and Technology Department of Hunan Province (2020SK3032) and the Hunan Provincial Innovation Foundation for Postgraduate, China (CX20190254). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Deng H, Xia H, Deng S. Genetic basis of human left-right asymmetry disorders. Expert Rev Mol Med. 2015; 16:e19. doi: 10.1017/erm.2014.22 . [DOI] [PubMed] [Google Scholar]
- 2.Jacobs JP, Anderson RH, Weinberg PM, Walters HL 3rd, Tchervenkov CI, Del Duca D, et al. The nomenclature, definition and classification of cardiac structures in the setting of heterotaxy. Cardiol Young. 2007; 17 Suppl 2:1–28. doi: 10.1017/S1047951107001138 . [DOI] [PubMed] [Google Scholar]
- 3.Lin AE, Ticho BS, Houde K, Westgate MN, Holmes LB. Heterotaxy: associated conditions and hospital-based prevalence in newborns. Genet Med. 2000; 2(3):157–72. doi: 10.1097/00125817-200005000-00002 . [DOI] [PubMed] [Google Scholar]
- 4.Sutherland MJ, Ware SM. Disorders of left-right asymmetry: heterotaxy and situs inversus. Am J Med Genet C Semin Med Genet. 2009; 151C(4):307–17. doi: 10.1002/ajmg.c.30228 . [DOI] [PubMed] [Google Scholar]
- 5.Peeters H, Devriendt K. Human laterality disorders. Eur J Med Genet. 2006; 49(5):349–62. doi: 10.1016/j.ejmg.2005.12.003 . [DOI] [PubMed] [Google Scholar]
- 6.Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth A, Penman-Splitt M, et al. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet. 1997; 17(3):305–8. doi: 10.1038/ng1197-305 . [DOI] [PubMed] [Google Scholar]
- 7.Bamford RN, Roessler E, Burdine RD, Saplakoğlu U, dela Cruz J, Splitt M, et al. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet. 2000; 26(3):365–9. doi: 10.1038/81695 . [DOI] [PubMed] [Google Scholar]
- 8.Kato R, Yamada Y, Niikawa N. De novo balanced translocation (6;18)(q21;q21.3) in a patient with heterotaxia. Am J Med Genet. 1996; 66(2):184–6. . [DOI] [PubMed] [Google Scholar]
- 9.Kosaki R, Gebbia M, Kosaki K, Lewin M, Bowers P, Towbin JA, et al. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am J Med Genet. 1999; 82(1):70–6. . [DOI] [PubMed] [Google Scholar]
- 10.Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach SD, et al. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009; 18(5):861–71. doi: 10.1093/hmg/ddn411 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perles Z, Cinnamon Y, Ta-Shma A, Shaag A, Einbinder T, Rein AJ, et al. A human laterality disorder associated with recessive CCDC11 mutation. J Med Genet. 2012; 49(6):386–90. doi: 10.1136/jmedgenet-2011-100457 . [DOI] [PubMed] [Google Scholar]
- 12.Perles Z, Moon S, Ta-Shma A, Yaacov B, Francescatto L, Edvardson S, et al. A human laterality disorder caused by a homozygous deleterious mutation in MMP21. J Med Genet. 2015; 52(12):840–7. doi: 10.1136/jmedgenet-2015-103336 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vetrini F D’Alessandro LC, Akdemir ZC, Braxton A, Azamian MS, Eldomery MK, et al. Bi-allelic mutations in PKD1L1 are associated with laterality defects in humans. Am J Hum Genet. 2016; 99(4):886–93. doi: 10.1016/j.ajhg.2016.07.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ta-Shma A, Hjeij R, Perles Z, Dougherty GW, Abu Zahira I, Letteboer SJF, et al. Homozygous loss-of-function mutations in MNS1 cause laterality defects and likely male infertility. PLoS Genet. 2018; 14(8):e1007602. doi: 10.1371/journal.pgen.1007602 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng S, Wu S, Xia H, Xiong W, Deng X, Liao J, et al. Identification of a frame shift mutation in the CCDC151 gene in a Han-Chinese family with Kartagener syndrome. Biosci Rep. 2020; 40(6):BSR20192510. doi: 10.1042/BSR20192510 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Deng S, Xia H, Yuan L, Xu H, Tang S, et al. Identification of a CCDC114 variant in a Han-Chinese patient with situs inversus. Exp Ther Med. 2020; 20(4):3336–42. doi: 10.3892/etm.2020.9059 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia H, Hu P, Yuan L, Xiong W, Xu H, Yi J, et al. A homozygous MYO7A mutation associated to Usher syndrome and unilateral auditory neuropathy spectrum disorder. Mol Med Rep. 2017; 16(4):4241–6. doi: 10.3892/mmr.2017.7053 . [DOI] [PubMed] [Google Scholar]
- 18.Hu P, Wu S, Yuan L, Lin Q, Zheng W, Xia H, et al. Compound heterozygous POMT1 mutations in a Chinese family with autosomal recessive muscular dystrophy-dystroglycanopathy C1. J Cell Mol Med. 2017; 21(7):1388–93. doi: 10.1111/jcmm.13068 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia H, Huang X, Xu H, Zhou YA, Gong L, Yang Z, et al. GJB2 c.235delC variant associated with autosomal recessive nonsyndromic hearing loss and auditory neuropathy spectrum disorder. Genet Mol Biol. 2019; 42(1):48–51. doi: 10.1590/1678-4685-gmb-2017-0318 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17(5):405–24. doi: 10.1038/gim.2015.30 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018; 39(11):1517–24. doi: 10.1002/humu.23626 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knowles MR, Leigh MW, Carson JL, Davis SD, Dell SD, Ferkol TW, et al. Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax. 2012; 67(5):433–41. doi: 10.1136/thoraxjnl-2011-200301 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boaretto F, Snijders D, Salvoro C, Spalletta A, Mostacciuolo ML, Collura M, et al. Diagnosis of primary ciliary dyskinesia by a targeted next-generation sequencing panel: molecular and clinical findings in Italian patients. J Mol Diagn. 2016; 18(6):912–922. doi: 10.1016/j.jmoldx.2016.07.002 . [DOI] [PubMed] [Google Scholar]
- 24.Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007; 115(22):2814–21. doi: 10.1161/CIRCULATIONAHA.106.649038 . [DOI] [PubMed] [Google Scholar]
- 25.Shapiro AJ, Davis SD, Ferkol T, Dell SD, Rosenfeld M, Olivier KN, et al. Laterality defects other than situs inversus totalis in primary ciliary dyskinesia: insights into situs ambiguus and heterotaxy. Chest. 2014; 146(5):1176–86. doi: 10.1378/chest.13-1704 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartoloni L, Blouin JL, Pan Y, Gehrig C, Maiti AK, Scamuffa N, et al. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc Natl Acad Sci U S A. 2002; 99(16):10282–6. doi: 10.1073/pnas.152337699 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwabe GC, Hoffmann K, Loges NT, Birker D, Rossier C, de Santi MM, et al. Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum Mutat. 2008; 29(2):289–98. doi: 10.1002/humu.20656 . [DOI] [PubMed] [Google Scholar]
- 28.Supp DM, Witte DP, Potter SS, Brueckner M. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature. 1997; 389(6654):963–6. doi: 10.1038/40140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature. 2002; 418(6893):96–9. doi: 10.1038/nature00849 . [DOI] [PubMed] [Google Scholar]
- 30.McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003; 114(1):61–73. 10.1016/s0092-8674(03)00511-7 . [DOI] [PubMed] [Google Scholar]
- 31.Brueckner M D ’Eustachio P, Horwich AL. Linkage mapping of a mouse gene, iv, that controls left-right asymmetry of the heart and viscera. Proc Natl Acad Sci U S A. 1989; 86(13):5035–8. doi: 10.1073/pnas.86.13.5035 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanzlik AJ, Binder M, Layton WM, Rowe L, Layton M, Taylor BA, et al. The murine situs inversus viscerum (iv) gene responsible for visceral asymmetry is linked tightly to the Igh-C cluster on chromosome 12. Genomics. 1990; 7(3):389–93. doi: 10.1016/0888-7543(90)90173-r . [DOI] [PubMed] [Google Scholar]
- 33.Okada Y, Nonaka S, Tanaka Y, Saijoh Y, Hamada H, Hirokawa N. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol Cell. 1999; 4(4):459–68. doi: 10.1016/s1097-2765(00)80197-5 . [DOI] [PubMed] [Google Scholar]
- 34.Lucas JS, Adam EC, Goggin PM, Jackson CL, Powles-Glover N, Patel SH, et al. Static respiratory cilia associated with mutations in Dnahc11/DNAH11: a mouse model of PCD. Hum Mutat. 2012; 33(3):495–503. doi: 10.1002/humu.22001 . [DOI] [PubMed] [Google Scholar]
- 35.Liu S, Chen W, Zhan Y, Li S, Ma X, Ma D, et al. DNAH11 variants and its association with congenital heart disease and heterotaxy syndrome. Sci Rep. 2019; 9(1):6683. doi: 10.1038/s41598-019-43109-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Feng X, Zhang J, Hao Y, Wang Y. Co-occurrence of Moyamoya syndrome and Kartagener syndrome caused by the mutation of DNAH5 and DNAH11: a case report. BMC Neurol. 2020; 20(1):314. doi: 10.1186/s12883-020-01895-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Z, Chen W, Wang L, Qian L. Clinical and genetic spectrum of children with primary ciliary dyskinesia in China. J Pediatr. 2020; 225:157–65.e5. doi: 10.1016/j.jpeds.2020.05.052 . [DOI] [PubMed] [Google Scholar]
- 38.Pifferi M, Michelucci A, Conidi ME, Cangiotti AM, Simi P, Macchia P, et al. New DNAH11 mutations in primary ciliary dyskinesia with normal axonemal ultrastructure. Eur Respir J. 2010; 35(6):1413–6. doi: 10.1183/09031936.00186209 . [DOI] [PubMed] [Google Scholar]
- 39.Guan WJ, Li JC, Liu F, Zhou J, Liu YP, Ling C, et al. Next-generation sequencing for identifying genetic mutations in adults with bronchiectasis. J Thorac Dis. 2018; 10(5):2618–30. doi: 10.21037/jtd.2018.04.134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Namavarian A, Eid A, Goh ES-Y, Thakur V. A novel DNAH11 variant segregating in a sibship with heterotaxy and implications for genetic counseling. Mol Genet Genomic Med. 2020; 8:e1358. doi: 10.1002/mgg3.1358 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz R, Elenius V, Lukkarinen H, Saarela T. Two novel mutations in the DNAH11 gene in primary ciliary dyskinesia (CILD7) with considerable variety in the clinical and beating cilia phenotype. BMC Med Genet. 2020; 21(1):237. doi: 10.1186/s12881-020-01171-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoemark A, Burgoyne T, Kwan R, Dixon M, Patel MP, Rogers AV, et al. Primary ciliary dyskinesia with normal ultrastructure: three-dimensional tomography detects absence of DNAH11. Eur Respir J. 2018; 51(2):1701809. doi: 10.1183/13993003.01809-2017 . [DOI] [PubMed] [Google Scholar]
- 43.Nakhleh N, Francis R, Giese RA, Tian X, Li Y, Zariwala MA, et al. High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation. 2012; 125(18):2232–42. doi: 10.1161/CIRCULATIONAHA.111.079780 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu F, Li R, Li Y, Nie ZQ, Lei T, Wang D, et al. Whole exome sequencing as a diagnostic adjunct to clinical testing in fetuses with structural abnormalities. Ultrasound Obstet Gynecol. 2018; 51(4):493–502. doi: 10.1002/uog.18915 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data involved in the study cannot be shared publicly because of ethical and legal concerns, regarding sensitive information. Reasonable requests to access the datasets by the qualified researchers can be sent to the Institutional Review Board of the Third Xiangya Hospital, Central South University (xy3irb@163.com).