Abstract
Nonulosonic acids (NulOs) are a group of nine-carbon monosaccharides with different functions in nature. N-acetylneuraminic acid (Neu5Ac) is the most common NulO. It covers the membrane surface of all human cells and is a central molecule in the process of self-recognition via SIGLECS receptors. Some pathogenic bacteria escape the immune system by copying the sialylation of the host cell membrane. Neu5Ac production in these bacteria is catalysed by the enzyme NeuB. Some bacteria can also produce other NulOs named pseudaminic and legionaminic acids, through the NeuB homologues PseI and LegI, respectively. In Opisthokonta eukaryotes, the biosynthesis of Neu5Ac is catalysed by the enzyme NanS. In this study, we used publicly available data of sequences of NulOs synthases to investigate its distribution within the three domains of life and its relationship with pathogenic bacteria. We mined the KEGG database and found 425 NeuB sequences. Most NeuB sequences (58.74 %) from the KEGG orthology database were classified as from environmental bacteria; however, sequences from pathogenic bacteria showed higher conservation and prevalence of a specific domain named SAF. Using the HMM profile we identified 13 941 NulO synthase sequences in UniProt. Phylogenetic analysis of these sequences showed that the synthases were divided into three main groups that can be related to the lifestyle of these bacteria: (I) predominantly environmental, (II) intermediate and (III) predominantly pathogenic. NeuB was widely distributed in the groups. However, LegI and PseI were more concentrated in groups II and III, respectively. We also found that PseI appeared later in the evolutionary process, derived from NeuB. We use this same methodology to retrieve sialic acid synthase sequences from Archaea and Eukarya. A large-scale phylogenetic analysis showed that while the Archaea sequences are spread across the tree, the eukaryotic NanS sequences were grouped in a specific branch in group II. None of the bacterial NanS sequences grouped with the eukaryotic branch. The analysis of conserved residues showed that the synthases of Archaea and Eukarya present a mutation in one of the three catalytic residues, an E134D change, related to a Neisseria meningitidis reference sequence. We also found that the conservation profile is higher between NeuB of pathogenic bacteria and NanS of eukaryotes than between NeuB of environmental bacteria and NanS of eukaryotes. Our large-scale analysis brings new perspectives on the evolution of NulOs synthases, suggesting their presence in the last common universal ancestor.
Keywords: Bacteria, evolution, NeuB/NanS, nonulosonic acid synthase, pathogenicity
Data Summary
The data underlying this article are available at https://github.com/alezanatta/neub. The deposited files include the sequences used, the IDs, the associated metadata and the alignments used in the reconstruction of the phylogenetic trees. The datasets were retrieved from public databases: Nonulosonic acid synthase sequences:
KEEG database (NeuB): https://www.genome.jp/dbget-bin/www_bget?K01654
KEEG database (NanS): https://www.genome.jp/dbget-bin/www_bget?K05304
KEEG database (PseI): https://www.genome.jp/dbget-bin/www_bget?K15898
KEEG database (LegI): https://www.genome.jp/dbget-bin/www_bget?K18430
Uniprot: https://www.uniprot.org/
Organisms’ metadata:
PATRIC: https://www.patricbrc.org/
Impact Statement.
Evolutionary studies have shown that the hominid lineage changed the pattern of sialylation of their cells after a mutation in the gene encoding CMP-Neu5Ac hydroxylase (CMAH). In this way, N-glycolylneuraminic acid (Neu5Gc) was replaced by N-acetylneuraminic acid (Neu5Ac). This change also led to adaptations in sialic-acid-binding immunoglobulin-like lectin (SIGLEC) receptors that started to recognize cells covered with Neu5Ac as their own. Currently, it is believed that pathogenic bacteria exploit this characteristic of our immune system by adding Neu5Ac molecules in terminal regions of their lipopolysaccharides. In bacteria, the biosynthesis of Neu5Ac is catalysed by the enzyme NeuB. Here we performed a massive phylogenetic analysis of nonulosonic acid (NulOs) synthase sequences. We showed that NeuB is widely distributed among hundreds of bacterial genera and are more abundant in environmental species, contrary to previous belief. On the other hand, pseudaminic acid synthase had a strong association with pathogenic bacteria. The comparison between conserved NeuB residues of environmental and pathogenic bacteria showed that sequences of pathogenic organisms are more conserved than those from environmental organisms, possibly due to the selective pressure determined by host–pathogen interactions. The inclusion of the eukaryotic sialic acid synthase NanS sequences in the study revealed that this set of sequences formed a specific group, separate from the sequences identified as bacterial NanS. Finally, we present a new view on the evolution of sialic acid synthases suggesting their origin in the last universal common ancestor.
Introduction
Sialic acids are nine-carbon monosaccharides belonging to a group known as nonulosonic acids (NulOs) that constitute a family of approximately 50 alpha-keto acids [1, 2]. Human cells use a specific type of sialic acid, called N-acetylneuraminic acid (Neu5Ac), in the terminal portions of their membrane glycoconjugates. This sialyation pattern allows the cells of the immune system to recognize other cells as their own via sialic acid binding Ig-like lectin (SIGLEC) receptors [3]. Evolutionary studies on the sialylation of Hominidae lineage cells have shown that a mutation in the gene encoding the CMP-Neu5Ac hydroxylase (CMAH), responsible for the production of the sialic acid N-glycolylneuraminic acid (Neu5Gc), caused accumulation of its precursor Neu5Ac [4]. This mutation occurred between 3 and 2 million years ago and changed the sialylation pattern of ancestral hominid cells, making them resistant to an ancestral parasite of the genus Plasmodium. The change in the sialylation pattern of the cells was also accompanied by changes in the innate immune system, more precisely, in the SIGLEC receptors. Khan and collaborators showed that mutations in the SIGLEC gene cluster occurred approximately 600 000 years ago, after the separation of primates of the chimpanzee/bonobo lineage, but are earlier than the common ancestor of Neanderthals and Denisovans [5].
Host cell self-identification mimicry, through incorporation of Neu5Ac in the terminal portion of the lipopolysaccharides (LPS) [6], is a known mechanism used by pathogenic bacteria to evade the immune system [7, 8]. Bacteria obtain Neu5Ac by two processes: acquisition from the environment and de novo endogenous biosynthesis [9]. Acquisition from the environment usually occurs through the uptake of Neu5Ac available in the environment or cleavage of Neu5Ac from host cells via neuraminidases [10, 11]. The biosynthetic process, in general, is dependent on the enzyme N-acetylneuraminate synthase (NeuB). This enzyme catalyses the condensation reaction between N-acetylmannosamine (ManNAc) and phosphoenolpyruvate (PEP) to produce Neu5Ac [12]. In vitro analyses have demonstrated that NeuB alone has the ability to catalyse the reaction in the presence of reagents, regardless of the action of other proteins [13]. Previous studies have shown that mutations in the neuB gene of Streptococcus suis considerably alter the sialic acid biosynthetic pathway. Mutations in the neuB gene have been shown to modify the thickness of the capsule structure, making it thinner, and these organisms more susceptible to phagocytosis by host macrophages. In some cases, hosts infected with neuB mutants did not even present symptoms of the disease [14]. Bacteria also have other types of NulOs that share structural similarities with Neu5Ac, of which legionaminic acid (N,N'-diacetyl legionaminic acid) and pseudaminic acid (5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-manno-non-2-ulopyronosonic acid) stand out [15, 16]. The biosynthetic pathways of legionamic acid and pseudaminic acid involve more reactions than those in the biosynthetic pathway of Neu5Ac; however, in both pathways, the condensation reaction is catalysed by the NeuB homologues LegI, for legionaminic acid, and PseI, for pseudaminic acid. Both LegI and PseI are considered virulence factors, given their ability to interact with the host immunological system [15–17]. In Opisthokonta, the biosynthesis of Neu5Ac is catalysed by the enzyme NanS. The main difference is in the phosphorylation of the substrate and the product. NanS uses ManNAc-6-phosphate instead of ManNAc and produces Neu5Ac-9-phosphate instead of Neu5Ac, a reaction favoured by the interaction with the α2β2 loop present in the human enzyme [18]. The phosphate of Neu5Ac-9-phosphate is subsequently removed by the NanP phosphatase [12].
The Neisseria meningitidis NeuB crystal structure revealed that the enzyme is a homodimer and has two main domains: an N-terminal domain, known as the NeuB domain, and a C-terminal domain, called the antifreeze-like domain (SAF) due to its similarity to some type III antifreeze proteins [19, 20]. The NeuB domain has approximately 250 residues and binds the substrates ManNAc and PEP for the subsequent condensation reaction. It has approximately 15 conserved residues, which are directly involved in its catalytic function and have a fold similar to a TIM barrel. The SAF domain has approximately 75 residues, 10 of which are in a linker region between the two domains. The SAF domain has been suggested to be important for dimer stability, as three conserved residues are positioned within the cavity adjacent to the TIM barrel and play a direct role in substrate binding [19, 20].
Accepted theories regarding the evolution of the biosynthesis of sialic acids suggest that this process was developed after the evolution of deuterostomes (higher invertebrates and vertebrates) and transferred to bacteria [21]. This could explain why sialic acids are less present in Archaea and remarkably absent from plants [22]. Other theories state that there has been convergent evolution from the microbial biosynthetic pathway [23], but these theories still remain in debate. To further understand the evolution and role of NeuB and its homologues in bacterial pathogenicity, we investigated its distribution in the domains Bacteria, Archaea and Eukarya using data mining and a large-scale phylogenetic analysis.
Methods
Identification of NeuB reference sequences and inference of species lifestyle
All entries available in the KEGG orthology database [24] identified as N-acetylneuraminate synthase (KEGG entry K01654) were recovered. Sequences with less than 240 amino acids or not of bacterial origin were filtered out. All sequences were affiliated with a particular species, and this feature was used to retrieve information about species lifestyle from publicly available data in the JGI GOLD [25], KEGG [24] and PATRIC [26] databases. We classified the microorganisms according to three factors: (1) if the microorganism was a reported pathogen; (2) the isolation site of the microorganism; and (3) the host of the microorganism, if applicable. An organism identified as pathogenic in at least one of the three databases mentioned above was considered a pathogen. If there was no pathology described for an organism, it was classified as environmental. If there were no available data in any database for the organism, it was manually classified based on the literature. The presence of the SAF domain was evaluated using the NCBI Batch CD-Search tool against NCBI’s Conserved Domain Database (CDD) [27] and through an SAF domain PFAM family sequence entry (Pfam PF08666) [28]. Organisms with sequences containing the SAF domain were identified in the phylogenetic tree.
Phylogenetic analysis
The NeuB amino acid sequences retrieved from KEGG were aligned using MAFFT v.7.402 [29]. The resulting alignment was trimmed to remove positions with more than 95 % gaps and less than 0.05 similarity scores using TrimAl v.1.2.59 [30]. Prior to phylogenetic analysis, we performed a model test using IQ-Tree [31]. The best model (LG+I+G4) was used to reconstruct a phylogenetic tree through maximum likelihood (ML), with 456 bootstrap replicates and MRE-based bootstrapping stop criterion, using RaxML v.8.2.10 software [32]. The generated tree was visualized in iTOL [33].
Analysis of conserved amino acids in NeuB
The sequences of NeuB retrieved from KEGG were divided into three groups: pathogenic, environmental with an SAF domain and environmental without an SAF domain. The sequences in each group were aligned independently using MAFFT v.7.402. We used the Emboss Prophecy tool [34] to create a frequency-based profile of the groups and a Python script to identify the frequency of each amino acid. The profiles were compared against the N. meningitidis NeuB ultrastructure [19] to determine the conserved catalytic residues.
Mining of NeuB sequences and massive phylogenetic analysis
A Markov profile was created based on the alignment of 412 amino acid sequences using Hmmer v.3.2.1 [35] and used to mine data from UniProtKB [36] through EBI Hmmer Web. Taxa were restricted to only Bacteria. All retrieved sequences were scanned for the presence of the NeuB domain. Sequences that did not have the NeuB N-terminal domain according to the InterPro database (InterPro IPR013132) were discarded [37]. We also excluded duplicate entries or marked as obsolete in UniProtKB. The remaining sequences were analysed using the NCBI Batch CD-Search tool, and the sequences that lacked complete N-terminal and C-terminal sequences were removed, resulting in 13 941 amino acid sequences. A neighbour-joining phylogenetic tree was inferred using the alignment-free SWeeP tool [38]. The tree was visualized in IToL [33].
Mining of prokaryotic and eukaryotic sialic acid synthases and phylogenetic analysis
To clarify the evolutionary relationship between prokaryotic and eukaryotic nonulosonic acid synthases, two Markov profiles were created: one based on the alignment of 412 NeuB amino acid sequences using Hmmer v.3.2.1 [35] and the other based on all available eukaryotic sequences in the KEGG orthology database identified as sialic acid synthase (NanS, K05304). Both were used to extract data from UniProtKB [36] using the EBI Hmmer Web. Taxa were restricted to Bacteria and Archaea only when using the NeuB profile and Eukarya when using the NanS profile. Sequences that did not have the NeuB N terminus according to the InterPro database (InterPro IPR013132) were discarded [37]. Duplicate entries or marked as obsolete in UniProtKB have also been deleted. A chi-square distribution was performed using IQTree [31], resulting in 7714 amino acid sequences. The resulting sequences were aligned using MAFFT 7.402 [29] and the resulting alignment was trimmed using TrimAl 1.2.59 [30] to remove positions with more than 95 % gaps and less than 0.05 similarity scores. A phylogenetic tree was inferred using the ML estimate and 1000 replicates of fast bootstrap using RaxML 8.2.10 [32]. The tree was visualized in ItoL [33].
Results
Identification and phylogenetic analysis of NeuB sequences from the KEGG database
The NeuB enzyme is part of the amino sugar and nucleotide sugar metabolism pathway according to the KEGG pathway database. Identified NeuB sequences, specifically, are grouped in the KEGG pathway database under ID K01654, allowing us to retrieve all NeuB sequences available on KEGG. In total, 412 sequences, with an average of 365 amino acids and from 377 species, were identified as NeuB sequences. The presence of the NeuB N-terminal domain in these sequences was confirmed through a search against the CDD looking for the NeuB domain ID in the Pfam database (PF03102). The retrieved sequences were from 18 bacterial phyla, mostly Proteobacteria , Firmicutes , Bacteroidetes and Cyanobacteria (Fig. S1).
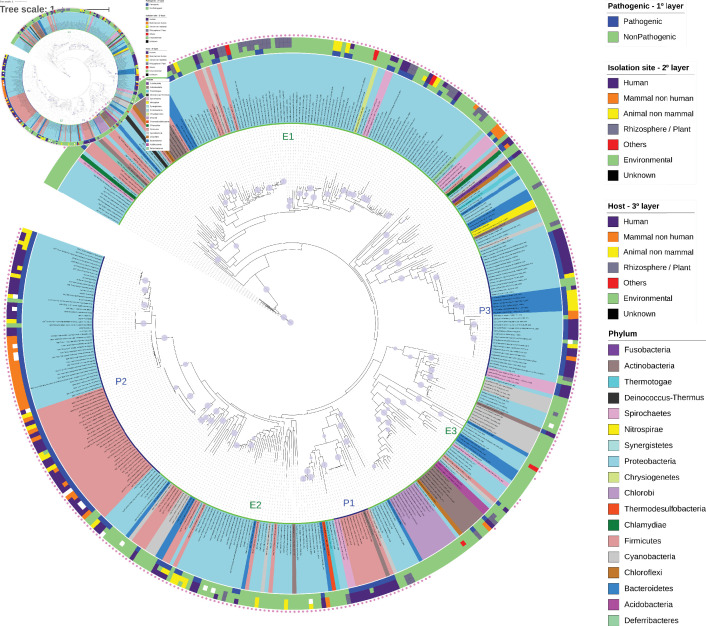
To further understand the relationship between organisms capable of producing Neu5Ac and their lifestyles and the relationship between the presence of the Neu5Ac biosynthetic enzyme and pathogenicity, we classified all species from which NeuB were obtained according to pathogenicity, isolation site and host based on data available in the GOLD, PATRIC and KEGG databases. In addition, we searched the literature for specific data relating to these organisms. Using this methodology, we determined that 58.74 % of the bacterial species that have NeuB were classified as environmental organisms, suggesting that NeuB may have a broader distribution than previously thought and that the presence of NeuB is not an exclusive feature of pathogenic bacteria. To investigate the relationships between these sequences, we reconstructed a NeuB phylogenetic tree (Fig. 1). Most of the sequences from organisms that were identified as pathogens clustered into three main groups identified as P1, P2 and P3. Group P1 contained sequences of the genus Clostridium ( Firmicutes ), a sequence of Eggerthella ( Actinobacteria ) and two sequences of Leptospira (Spirochaetes). Group P2, the largest group, contained sequences derived from different species of the class Gammaproteobacteria and the genus Streptococcus from the phylum Firmicutes . Group P3 contained sequences of the genera Neisseria ( Gammaproteobacteria ), Campylobacter ( Epsilonproteobacteria ), and Flavobacterium ( Bacteroidetes ), and Leptospira ( Spirochaetes ). Approximately 71.93 % of the pathogenic organisms in these three groups were identified as isolated from humans or non-human mammals, such as cattle and pigs. In addition, it is important to note that the Gammaproteobacteria in P1 were different from the Gammaproteobacteria in P2, just as the Firmicutes in P1 were different from those in P3. Additionally, despite the large amount of Proteobacteria present in the tree, it was possible to observe that they did not group, so clusters do not reflect the phylogeny of the species.
Fig. 1.
Phylogenetic tree of NeuB sequences retrieved from the KEGG database. The NeuB sequences were retrieved from the KEGG database using ID K01654. The taxon ID associated with each sequence was used to identify the species from which the sequence originated and to extract metadata related to lifestyle, isolation site and host. NeuB sequences were trimmed to the NeuB domain and then aligned using MAFFT in the default configuration. The alignment was used to infer the phylogenetic relationships through the ML method on the IQTree server. The automatic bootstrap, suggested by IQTree, ended with 456 replicates. The completed tree was viewed and edited in the iTOL program. The species names are displayed at the end of each branch. The colours of the species names represent the phylum to which they belong. The first layer after the species name represents the lifestyle. The second layer represents the isolation site, and the third layer represents the identified host. The outermost pink dot indicates the presence of the SAF domain in the respective sequence. Clusters of pathogenic (P1, P2 and P3, inner blue line) and environmental (E1, E2 and E3, inner green line) bacterial species are indicated. The purple dots on the tree branches represent bootstrap values above 75 %. Bar, 10 amino acid substitutions per site per million years.
Three large groups of environmental bacteria could also be observed; these bacteria grouped remarkably according to the isolation site. The first group (E1) contained 24.75 % of the organisms isolated from the rhizosphere or in plants or with plant hosts. Several of the bacteria found in this group were nitrogen fixers, including members of the genera Azospirillum , Paraburkholderia , Sinorhizobium and Pseudomonas , and saprophytic species of the genus Leptospira (identified in the GOLD database as pathogens). This group contained some opportunistic pathogens, such as Acinetobacter baumannii , Bacillus cereus and Aeromonas salmonicida , as well as classic pathogens, such as Clostridium tetani . The second group (E2) appeared between P3 and P1 groups and contained mainly bacteria isolated from water or fish. A smaller number of pathogenic representatives from the genera Vibrio , Bacteroides and Clostridium appeared in this group. The last group (E3) appeared before P3 and contained bacteria isolated from oceans and extreme habitats. Only two species of pathogens appeared in this group: Rothia mucilaginosa and Thermobifda fusca.
Study of the NeuB ultrastructure of N. meningitidis revealed that the SAF domain may stabilize the homodimer and substrates [19]. We found that only 40 sequences did not have the SAF domain. Of those, 95 % belonged to environmental organisms. When we specifically considered the remaining 5 % of strains that were not environmental organisms, we identified Clostridium botulinum B strain Eklund 17B (NRP) and Streptomyces scabiei strain 87.22. The former, although capable of producing the botulism neurotoxin, was isolated from marine sediments and has not been reported in a clinical case [39]. The latter is the causative agent of potato scab [40], and there is no known relationship between bacterial sialic acid biosynthesis and plants [22]. Taking this into account, we can say that 100 % of the sequences that did not have the SAF domain originated from environmental organisms and tended to cluster in the phylogenetic tree. On the other hand, 98.82 % of the sequences classified as originating from pathogenic bacteria had an SAF domain. Curiously, the E3 group was the only one formed by sequences with and without the SAF domain. This result suggests that the SAF domain could have a role in pathogenicity or in changing lifestyle from environmental to opportunistic. Also, the SAF domain may have been lost in this group of environmental bacteria
Conservation of catalytic and non-catalytic residues of NeuB between different lifestyles
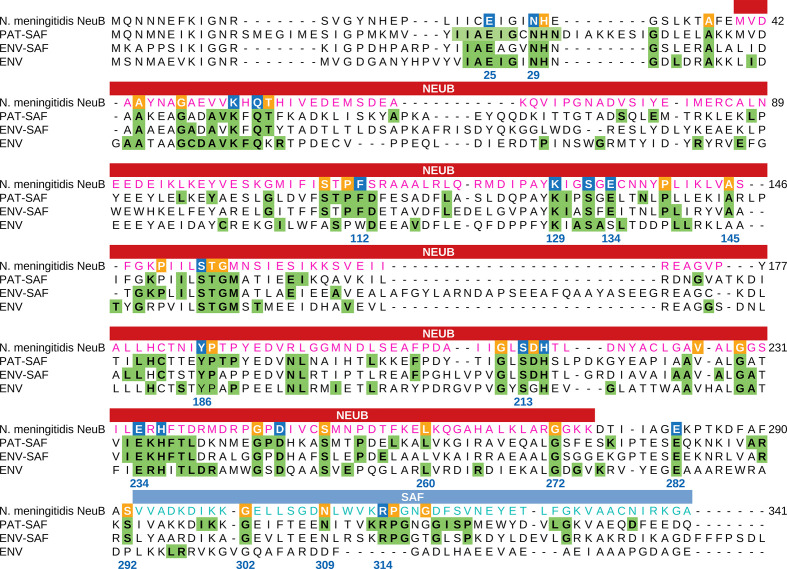
The crystal structure of N. meningitidis NeuB obtained in the presence of its two substrates, ManNAc and PEP, revealed the residues important for stabilization and catalysis. N. meningitidis is a pathogenic bacterium capable of causing meningococcal meningitis. Thus, to investigate the conservation of catalytic residues in NeuB sequences of environmental and pathogenic origin, we divided the sequences into three groups: PAT-SAF, comprising 169 sequences with an SAF domain from pathogenic organisms; ENV-SAF, representing 202 sequences from environmental organisms with an SAF domain; and ENV, 40 environmental organisms without an SAF domain. For each group, we performed separated sequence alignment. Based on the frequency of each residue in each position, we created a sequence logo (Figs S2–S4) and a sequence consensus profile for each group. These profiles were compared with the N. meningitidis NeuB sequence as a reference (Fig. 2). This comparison allowed us to identify the conserved catalytic residues in each group. As a result, we found 17 catalytic amino acid residues and 30 non-catalytic potential conserved residues. As determined for N. meningitidis NeuB, the amino acid residues E25, E134 and E234 are potentially involved in catalysis. In the PAT-SAF group, these three residues were extremely conserved. However, in the ENV-SAF and ENV groups, residue E134 was replaced by aspartic acid in 29.2 % of the sequences. Other residues potentially involved in the catalysis or stabilization of the substrates were also conserved with few exceptions, such as phenylalanine 112 (F112), which, in the ENV group, was eventually replaced by tryptophan. In the C-terminal domain, the only conserved catalytic residue found in both groups with an SAF domain was arginine 314 (R314). Phenylalanine 288 (F288), which was previously cited by Gunawan et al. [19] as being potentially involved in catalysis, was found only in 19.3 % of the PAT-SAF group. In the ENV-SAF group, F288 was replaced by valine in 23.76 % of the sequences. A replacement also occurred in the ENV group, but in this case, the residue was replaced by tryptophan. Of the 30 non-catalytic residues, we found two not conserved in the ENV-SAF group: leucine 260 (L260) in the NeuB domain and asparagine 309 (N309) in the SAF domain.
Fig. 2.
Comparison between the NeuB sequence of N. meningitidis and the NeuB sequence profiles of pathogenic and environmental bacteria. The sequences identified as NeuB in the KEGG database were divided into three groups: PAT-SAF, NeuB from pathogenic bacteria with an SAF domain (n=169); ENV-SAF, NeuB from environmental bacteria with an SAF domain (n=202); and ENV, NeuB from environmental bacteria without an SAF domain (n=40). Each group of sequences were aligned separately using MAFFT. The alignments were used to build a conservation profile for each group considering each amino acid residue in each position. The tree profiles were compared with the NeuB sequence of N. meningitidis . The residues important for catalysis were identified by Gunawan et al. [19] based on the NeuB structure crystallized with its substrates ManNAc and PEP. The positions marked in blue in the N. meningitidis sequence represent catalytic residues, while the positions marked in orange represent non-catalytic residues. Green tagged residues in the profiles represent positions with at least 70 % conservation in the alignment. The amino acid residue positions listed in the alignment are related to NeuB of N. meningitidis . The NeuB domain and the SAF domain are indicated in the box above the alignment.
Our analysis allowed the identification of a new highly conserved residue in the NeuB structure, a glycine at position 229 (G229), according to the N. meningitidis amino acid sequence. This residue was not identified as important in the structure but was conserved in 100 % of the sequences in the three groups. The four preceding amino acids, A225, V226, A227 and L228, also exhibited high conservation in all groups. Finally, we identified 77 conserved positions in the ENV-SAF group and 99 in the PAT-SAF group in at least 70 % of the sequences (green residues in Fig. 2). These amino acids are primarily located near catalytic residues. We also found 80 conserved amino acid residues among the profiles and the reference sequence. This result suggests that PAT-SAF sequences are more conserved than ENV-SAF and non-SAF sequences.
NeuB distribution in the domain Bacteria
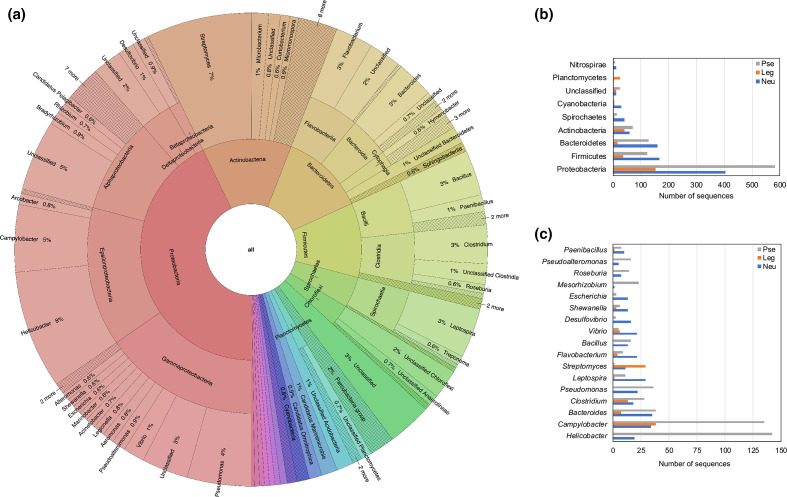
The results presented so far show evidence of the wide distribution of NeuB within the domain Bacteria. Despite being found in many pathogenic bacteria, NeuB was predominantly found in environmental bacteria. However, the number of sequences analysed was small compared to the number of described bacterial species. To obtain a complete overview of the evolution of NeuB and the biosynthesis of nonulosonic acids in the domain Bacteria, we used the sequences previously found in KEGG to create a hidden Markov model (HMM) profile. This HMM-based methodology allowed us to perform a broader search for NeuB sequences in the UniProtKB database. The sequence mining of NeuB and its homologues returned 13 941 sequences. The taxonomic distribution of the NeuB and homologous sequences among the 100 most well-represented genera was concentrated in the phylum Proteobacteria (Fig. 3a) with a prevalence of PseI sequences in relation to NeuB and LegI (Fig. 3b, c). The other identified phyla showed a more proportional distribution between NeuB and PseI with a slight dominance of NeuB. In general, with the exception of the phylum Planctomycetes , LegI was the homologue with the fewest representatives. Looking specifically at the best represented genera, it was possible to identify that the majority of the PseI sequences came from Helicobacter and Campylobacter . Interestingly, no LegI sequences were identified in the genus Helicobacter . On the other hand, in the genus Campylobacter , sequences of the three homologues, PseI, LegI and NeuB, were identified, respectively in that order of quantity. Unlike the other genera, Streptomyces was the only one to present a higher number of LegI sequences than PseI and NeuB.
Fig. 3.
Diversity of bacterial species that have NeuB and NeuB homologues. (a) Taxonomic distribution of 13 941 NeuB sequences and their homologues identified through data mining using HMM profile. The recovered sequences were classified according to unambiguous E.C. number in synthases of N-acetylneuraminic acid (NeuB), pseudaminic acid (PseI) and legionaminic acid (LegI) and distributed among the most represented bacterial phyla (b) and genera (c).
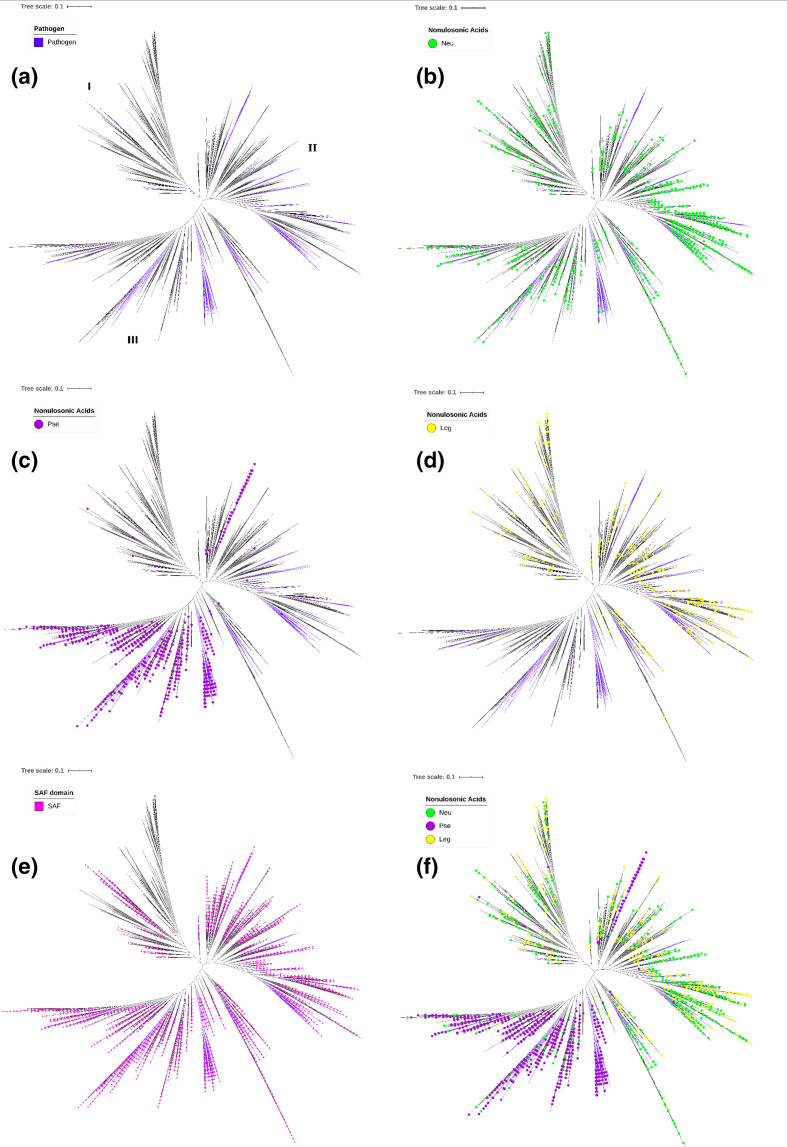
The length of the retrieved NeuB homologue sequences showed a wide variation, between 250 and 1500 aa. To eliminate the bias that could be introduced in the analysis due to the disparity of the sequences, we decided to do the phylogenetic analysis only with the protein domains. The identification of the domains was carried out by comparison with the CDD. Due to the large number of sequences, we used SWeeP, an alignment-free vectoral alignment tool, to cluster sequences. Based on the clusters (Fig. S5), we created a neighbour-joining tree of all sequences recovered (Fig. 4). The topology of the final tree presented a division of three large groups (I, II and II). As done for the KEGG tree, we looked for information about pathogenicity in the GOLD and KEGG databases. Thus, we were able to classify 604 additional sequences as originating from pathogenic bacteria. Combining these sequences with those identified as originating from pathogenic organisms in the KEGG tree, we obtained a total of 773 sequences (5.5 %) with evidence of belonging to pathogenic bacteria (Fig. 4a). Most pathogens were located mainly in groups II and III, and they were not grouped taxonomically, as in the previous phylogenetic analysis (Fig. 1). In addition, as in the previous analysis, sequences from pathogenic organisms tended to form concise groups within larger groups of sequences.
Fig. 4.
Phylogenetic analysis of bacterial NeuB and NeuB homologues. The NeuB HMM profile, reconstructed with the sequences from the KEGG database, was used to mine NeuB sequences and their homologues in the UniprotKB database. A total of 13 941 sequences were recovered and the presence of the NeuB domain was confirmed by comparison with the CDD database. The NeuB domains (~250 aa) were separated from the rest of the sequences and used in phylogenetic analysis. The total neighbour-joining tree was calculated using SWeeP [38], an alignment-free clustering tool. (a) The coloured clades represent the pathogenic organisms identified through the GOLD and KEGG databases. NeuB sequences and homologues were identified based on an unambiguous E.C. number. The coloured dots represent sequences identified as: (b) neuraminic acid synthase (Neu), (c) pseudaminic acid synthase (Pse) and (d) legionaminic acid synthase (Leg). (e) SAF domain identified by Interpro. (f) All previous E.C. numbers identified. Bars, 0.1 amino acid substitutions per site per million years.
As mentioned above, NeuB has homologues that participate in the biosynthesis of other nonulosonic acids, such as PseI and LegI. To identify these counterparts in the phylogenetic tree, we retrieved the Enzyme Commission (E.C.) number for all sequences in which it was available unambiguously, that is, one sequence and one E.C. number. We found 2196 unambiguous E.C. numbers. Of these, 923 were N-acetylneuraminic acid synthase (Neu, E.C. 2.5.1.56), 971 were pseudaminic acid synthase (Pse, E.C. 2.5.1.97) and 302 were N,N’-diacetyllegionaminate synthase (Leg, E.C. 2.5.1.101). These data were plotted on the phylogenetic tree (Fig. 4b–d). Through this analysis, it was possible to observe that Neu sequences appeared in the three groups (Fig. 4b), while Pse sequences appeared mainly in group III (Fig. 4c), and Leg sequences were distributed mainly in groups I and II (Fig. 4d). This analysis was also performed for the NeuB, LegI and PseI sequences retrieved only from the KEGG database and we found the same distribution (Fig. S6). We analysed organisms of the genera Acinetobacter , Legionella , Neisseria , Pseudomonas , Campylobacter , Streptococcus, Clostridium and Leptospira and observed that Pse sequences from these organisms were clustered in group III, while Neu and Leg sequences were present in groups I and II. The exceptions were organisms identified as pathogens of the genus Campylobacter , whose Pse sequences were also present in group II. Considering the clear division that we observed in the tree between Pse and Neu/Leg, it is possible that the Campylobacter in group II were erroneously annotated and were assigned the wrong E.C. number as well (Fig. 4f). The SAF domain was identified in 70 % of the sequences and was widely distributed in the tree but had a higher concentration in groups II and III than in group I. In contrast, in group I, it was possible to identify whole branches with sequences without the SAF domain (Fig. 4e). Additionally, organisms predicted to be environmental, such as Thermoanaerobacterium thermosaccharolyticum , Moorea producens , Crocosphaera watsonii, Nodularia spumigena, Brevibacterium aurantiacum and representatives of the genus Streptomyces were found only in group I, reinforcing the potential role of the SAF domain in lifestyle adaptation. Some opportunistic pathogens present in the environment or in animals appeared in the three groups. The high concentration of pathogenic bacteria and PseI in group III suggests that pseudaminic acid and not N-acetylneuraminic acid is truly associated with pathogenicity. PseI was isolated in group III with a small group of NeuB sequences at its basal branch suggesting that PseI may have diverged from NeuB.
Evolutionary relationship between prokaryotic and eukaryotic sialic acid synthases
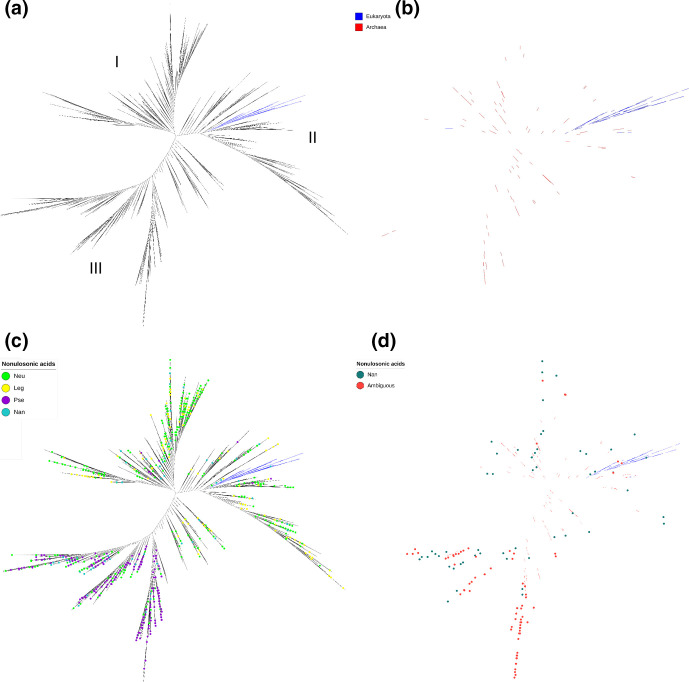
Theories regarding Neu5Ac biosynthesis postulate that bacterial NeuB synthase may have been transferred horizontally from eukaryotes to prokaryotes. To investigate this hypothesis, we performed a phylogenetic analysis with sequences of synthase from prokaryotes, NeuB, and eukaryotes, NanS. Again, the HMM profile created for NeuB from the sequences from KEGG was used to mine the UniProtKB database, but now including sequences from the domain Archaea. The NanS sequences were obtained in a similar way. First, we retrieved the NanS sequences from the KEGG database (KO K05304). These sequences were used to create an HMM profile and mine the UniProt database. All sequences were filtered to remove duplicates, obsolete sequences, sequences that did not have the catalytic domain and more than 95 % of informative regions in the alignment. At the end, we obtained a dataset with 7714 sequences: 6905 from Bacteria, 160 from Archaea, 267 from Eukarya and 382 that were not identified. These NeuB and NanS sequences were used to build a phylogenetic tree (Fig. 5a). Similar to the analysis presented only for bacterial NeuB, this new tree presented a division into three large groups, identified as I, II and III. From our analysis it was possible to see that, with the exception of five sequences, all other eukaryotic synthase sequences grouped into a single branch within group II (Fig. 5b). On the other hand, sequences of archaea synthases were spread over the three groups (Fig. 5b). The sequences that received a classification referring to the E.C. number (996 out of 7714) were marked on the tree (Fig. 5c). Again, the sequences with E.C. number referring to NeuB and LegI were distributed between groups I and II, while PseI was concentrated in group III (Fig. 5c). Analysing specifically the sequences with an E.C. number referring to NanS, we found a large concentration in group III followed by group I. We did not find an overlap between the eukaryotic NanS sequences and the E.C. number for NanS, suggesting that there may be problems with the identification of these enzymes. This hypothesis was more evident when we marked the ambiguous sequences, which received more than one E.C. number. We saw a higher concentration in group III referring to PseI (Fig. 5d). The only sequences of eukaryotic sialic acid synthases that grouped in the group III branch were: Emiliania husleyi, Aureococcus anophagefferens, Alexandrium fundyense, Micromonas commoda and Chromera velia, the first four representatives of the Algae group and the last one representative of the Alveolate group. All other NanS sequences identified in the group III branch were of bacterial origin.
Fig. 5.
Phylogenetic analysis of NeuB, NanS and NeuB homologues. We used the NeuB and NanS sequences retrieved from the KEGG database to create two individual HMM profiles, one for NeuB and the other for NanS. These profiles were used to mine the UniProt database and retrieve more NeuB and NanS sequences. A chi-square distribution was performed in the retrieved data using IQ-Tree, resulting in 7714 sequences. The presence of the NeuB domain was confirmed by comparison with the CDD. The sequences were aligned using MAFFT and an ML tree was inferred using the RaXML tool with 1000 fast bootstrap replicates. (a) ML tree of prokaryotic NeuB and eukaryotic NanS inferred using RaXML. The groups (i) predominantly environmental, (II) intermediate and (III) predominantly pathogenic are marked on the tree. (b) Branches representing NeuB from Archaea and NanS from Eukarya. The blue branches represent Eukarya sequences, the red branches represent Archaea sequences. (c) Phylogenetic tree of NeuB and NanS highlighting sequences with unambiguous E.C. number. (d) Phylogenetic tree of NeuB and NanS highlighting sequences with ambiguous E.C. numbers (multiple identifications for one sequence).
Similar to what was done previously, the sequences of sialic acid synthases of pathogenic bacteria, environmental bacteria, archaea and eukaryotes were aligned separately to build a conservation profile. The alignment of these profiles showed that there is a closer relationship between the sialic acid synthases of archaea and eukaryotes, followed by pathogenic bacteria and environmental bacteria. It is important to highlight two striking differences among Archaea and Eukarya and Bacteria. First is the mutation that occurred in one of the three catalytic residues in the profiles of archaea and eukaryotes in relation to bacteria: the glutamic acid residue 134 (E134 in the N. meningitidis sequence) was replaced by an aspartic acid residue. Second is the substitution of the conserved residue in the SAF domain of the eukaryotic sequence: the arginine residue was replaced by a valine residue (R314 in the N. meningitidis sequence) (Fig. 6). The R314 residue, in particular, was shown to be essential for the enzyme activity in bacteria [41].
Fig. 6.
Conservation analysis of sialic acid synthase sequences and profiles. The sequences identified as NeuB in the KEGG database were divided into four groups: NeuB from pathogenic bacteria (n=169); NeuB from environmental bacteria with an SAF domain (n=242); and NeuB from Archaea (n=10). NanS sequences from eukaryotes were also retrieved from KEGG (n=160). Each group of sequences were aligned separately using MAFFT. The alignments were used to build a conservation profile for each group considering each amino acid residue in each position. The tree profiles were compared with the NeuB sequence of N. meningitidis and the NanS from Homo sapiens. The residues important for catalysis were identified by Gunawan et al. [19] based on the NeuB structure crystallized with its substrates ManNAc and PEP. The positions marked in blue in the N. meningitidis sequence represent the catalytic residues, while the positions marked in orange represent the non-catalytic residues. Green tagged residues in the profiles represent positions with at least 70 % conservation in the alignment. The amino acid residue positions listed in the alignment are related to NeuB of N. meningitidis . The NeuB domain and the SAF domain are indicated in the box above the alignment.
Discussion
The enzyme NeuB catalyses the formation of Neu5Ac from ManNAc and PEP. The consensus is that some pathogenic gram-negative bacteria transfer Neu5Ac to their LPS to mimic host cell glycosylation and evade the immune system [3, 7, 9]. Due to features such as this, NeuB is often classified as a virulence factor. However, here we have presented a massive phylogenetic analysis showing that NeuB is widely distributed among the domain Bacteria, including pathogens and environmental representatives, and, for the first time, we identified that the SAF domain could be possibly related to lifestyle adaptation. Considering the evolution of sialic acid synthases, we present evidence that these genes were present in the last universal common ancestor (LUCA).
Our KEGG phylogenetic tree (Fig. 1) showed that NeuB does not follow the phylogeny, but instead showed a strong tendency to build clusters according to the lifestyle of the bacteria from which it originates. NeuB sequences from facultative or opportunistic pathogens such as Acinetobacter baumannii and Ralstonia pickettii (strain 12D) tended to cluster with known environmental organisms. In the case of the three A. baumannii strains present in the analysis, A.r baumannii 1656-2 and A. baumannii ACICU grouped with environmental organisms isolated from plants or the rhizosphere, whereas A. baumannii SDF grouped with marine environmental organisms. A. baumannii SDF, unlike the first two strains, which were isolated from clinical cases [42], was isolated in body lice and is susceptible to antibiotics [43]. A similar finding occurred with Bacteroides fragilis , which causes opportunistic infections and had sequences grouped with both environmental organisms and pathogens.
Gárcia and collaborators compared the kinetics of NeuB activity among the pathogenic bacteria N. meningitidis , E. coli and an environmental bacterium of the species Idiomarina loihiensis [44]. The calculated Km for substrates ManNAc and PEP for I. loihiensis NeuB was greater than that calculated for E. coli and N. meningitidis NeuB. Similarly, the catalytic efficiency (Kcat/Km) of I. loihiensis NeuB was half that of E. coli and N. meningitidis NeuB. The three enzymes tested have the SAF domain and high conservation in catalytic residues. However, the enzyme derived from the environmental bacteria showed worse kinetics. Our profile-based amino acid residue conservation analysis revealed that there are, in fact, differences in residue conservation between NeuB sequences of pathogenic and environmental organisms (Fig. 2). Some positions even presented the same residue but with different degrees of conservation. However, the sequences from pathogenic organisms showed higher conservation, which was mainly concentrated in the vicinity of amino acids designated as catalytic [19]. A higher accumulation of amino acid substitutions was observed in the NeuB proteins of environmental organisms without an SAF domain compared to the NeuB proteins with an SAF domain. This observation suggests a higher selective pressure on genes and proteins with SAF domains, even those of environmental origin. In the KEGG tree, all sequences that did not have the SAF domain were considered to be from environmental bacteria. Similar results were obtained for the largest tree. Sequences without an SAF domain clustered in group I, where the majority of the sequences originating from environmental bacteria were concentrated. On the other hand, the presence of an SAF domain in environmental bacteria could facilitate changes in lifestyle, from environmental to opportunistic, for example, probably due to better reaction kinetics. Previous studies suggest that the SAF domain is related to the stability of the NeuB dimer [19]. It is possible that this higher stability results in an enzyme that is more efficient and capable of responding quickly to changing environments. Iyer et al. [45] carried out an extensive search for proteins that contained the SAF domain. They found, in addition to sialic acid synthase, other proteins with the SAF domain. Among them were proteins related to flagellum synthesis and assembly, dehydrogenases and dehydratases. In all cases, the function predicted for this domain was carbohydrate bond [45]. Whether the lack of an SAF domain makes the enzyme less efficient or less important within the cellular metabolic context, allowing accumulation of mutations, remains to be investigated. One possibility is convergent evolution between pathogenic organisms and their respective hosts with the objective of making the biosynthesis process more efficient and allowing survival in this environment [46].
The smaller size of NeuB sequences from pathogenic bacteria could be related to the translation efficiency of the protein, as the gene length is inversely proportional to the rate of protein synthesis [47], giving these organisms an advantage of quickly adapting and surviving host immune system clearance [48]. Additionally, because the gene loss process has been proven to be advantageous to organism evolution [49] and the neuB gene was not lost throughout the evolutionary process, NeuB could have more functions in bacteria in addition to protecting them from the host immune system, although with lesser selective pressure for environmental than for pathogenic bacteria. Also, considering the distribution of sialic acid synthases across the three domains of life, it is possible that these carbohydrates may have had some influence on cell evolution [50].
Lewis et al. [23] performed an analysis of the biosynthetic pathway of nonulosonic acids in 1000 genomes. They obtained the first evidence of the wide distribution of this pathway within the domain Bacteria. Nevertheless, phylogenetic analysis based on their dataset showed that there is a separation between the biosynthesis of Neu5Ac, Leg and Pse. In addition, the use of blast in this case influences the result, since blast identifies sequences based on similarity that would tend to group together in phylogenetic analysis. On the other hand, in our analysis based on 13 941 sequences using HMM, it was not possible to separate NeuB and LegI. Only PseI formed a defined and separate group from NeuB and LegI. Therefore, it is also not possible to say which nonulosonic acid appeared first, Neu5Ac or Leg; however, we can infer that Pse appeared last on the evolutionary scale, given that a branch formed by NeuB and Leg is at the base of the Pse branch (Fig. 4f). This suggests that PseI diverged from others over the course of evolution and, at some point, began to accept 6-deoxy-Altdi-NAc instead of N-acetyl-d-mannosamine as a substrate giving rise to pseudaminic acid. The differentiation between NeuB and LegI seems to have been more recent.
The evolution of sialic acid synthases, mainly Neu5Ac synthases, is still an issue of debate. Recent theories point to convergent evolution or horizontal transfer of genes that encode synthases [23, 51]. Considering the analyses carried out in this work, based on thousands of sequences, we offer a different view on the evolution of sialic acid synthase. Petit et al. [51] in their study on sialidases suggest, through phylogenetic analyses, the presence of biosynthesis of sialic acids in the last eukaryote common ancestor (LECA), the presence of NanS in Bacteria/Archaea in addition to the presence of NeuB/LegI in eukaryotes. To help answer these questions and deepen the discussion about the origin of synthases, we performed a phylogenetic analysis including NeuB, LegI and PseI from the domain Bacteria, NeuB from Archaea and NanS from Eukarya. In fact, sialic acid biosynthesis is not only present in the LECA, but also before it. Our analysis showed that synthases are found in Archaea, Bacteria and Eukarya. The high conservation of sequences between the three domains of life suggests that this gene may be present in the LUCA.
Our results showed that sialic acid synthases suffer from a deep lack of annotation accuracy, which makes interpretation of the results challenging. When observing the sequences identified as NanS in prokaryotes (Fig. S7), we realize that these are grouped with other bacterial synthases: NeuB, LegI and PseI. This may be due to the sequence similarity between NeuB and NanS, of approximately 33 % [6], which can lead to errors in the annotation of both. In the phylogenetic analysis made including Archaea and Eukarya (Fig. 5a, b) we noticed that, while Archaea appear distributed along with Bacteria, eukaryotic proteins form a separate clade. In this exclusive clade of eukaryotes, bacterial sialic acid synthases have not been identified. Some of the sequences reported as NanS in Bacteria [51], as in Legionella pneumophila (RefSeq entry WP_027268045), despite having the N-terminal NeuB were identified as being PseI under analysis against TIGRFAM (TIGR03586) using CD-Search. Annotation errors regarding the differentiation between NeuB and NanS also appeared in our analysis: Homo sapiens, for example, is ambiguously identified with E.C. numbers 2.5.1.56 and 2.5.1.57, NeuB and NanS respectively.
Another point that caught our attention was the mutation in one of the three conserved catalytic residues. In bacteria, pathogenic and environmental, there is a residue of aspartic acid, while in archaea and eukaryotes there is a residue of glutamic acid. Analysing the NeuB structure of N. meningitidis (PDB accession number 1XUZ), it is possible to verify that this residue is close to the region where the ManNAc-6P phosphate group would be found. It is possible that the mutation in this residue is related to the use of the phosphorylated and non-phosphorylated versions of ManNAc. This implies that archaea also use ManNAc-6P instead of ManNAC and that the mutation actually occurred in the NeuB sequence of bacteria. Unfortunately, no archaeal Neu5Ac synthase has been characterized to date to verify this hypothesis. The wide distribution of Neu5Ac synthases and the higher conservation of the NeuB sequence in pathogenic bacteria in relation to environmental bacteria suggests that, in fact, there may have been a convergent evolution of this gene only in pathogenic organisms that brought it closer to its host sequence. Considering the analysis presented in the phylogenetic tree of bacterial NeuB and its homologues (Fig. 4a), we see that the branch of the eukaryotic NanS (Fig. 5a) fits in group II, characterized by presenting NeuB and LegI of pathogenic bacteria. Considering that group III, which includes the majority of pathogenic bacteria, is formed mainly by PseI, this result reinforces our hypothesis that the pathogenic bacteria NeuB showed convergent evolution with the eukaryotic NanS.
Conclusion
The presence and function of glycoconjugates containing Neu5Ac in pathogenic bacteria is well known, given their importance in evading the immune system. On the other hand, for environmental bacteria, the function of glycoconjugates containing Neu5Ac remains an intriguing issue. In contrast to previous studies, we have shown here that NeuB is widely distributed within the domain Bacteria and occurs predominantly in environmental bacteria. It is possible that the SAF domain, identified in all sequences originating from pathogenic bacteria, has some relationship with virulence, but this needs further clarification. We conclude, based on our analyses and previous studies, that (1) NeuB has always been present in the domain Bacteria and possibly in the LUCA; (2) infection by pathogenic bacteria capable of producing Neu5Ac was facilitated by a mutation in the human CMAH and SIGLEC; and (3) the change in the pattern of human cell sialylation triggered a conservation process via natural selection of the NeuB enzyme in pathogenic bacteria.
Supplementary Data
Funding information
InovaFiocruz/Fundação Oswaldo Cruz (Grant number VPPCB-07-FIO-18-2-38); CNPq (Grant number 424410/2018-4).
Acknowledgements
We thank the Coordination of Superior Level Staff Improvement (CAPES), Carlos Chagas Institute (ICC) and Graduation Program in Bioinformatics for their support.
Author contributions
A.Z.V. carried out the analyses and wrote the article; R.T.R. helped in the reconstruction of the phylogenetic tree and interpretation of the results; H.F. conducted the analysis and interpretation of the results and assisted in the writing process.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CDD, conserved domain database; E.C., enzyme commission; HMM, hidden Markov model; LPS, lipopolysaccharide; ManNAc, N-acetylmannosamine; ML, maximum likelihood; Neu5Ac, N-acetylneuraminic acid; NeuB, N-acetylneuraminate synthase; Neu5Gc, N-glycolylneuraminic acid; NulOs, nonulosonic acids; PEP, phosphoenolpyruvate; SIGLEC, sialic acid binding Ig-like lectin.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Seven supplementary figures are available with the online version of this article.
References
- 1.Schauer R. Sialic acids as regulators of molecular and cellular interactions. Curr Opin Struct Biol. 2009;19:507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Chen X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl Microbiol Biotechnol. 2012;94:887–905. doi: 10.1007/s00253-012-4040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacAuley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci U S A. 1998;95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan N, de Manuel M, Peyregne S, Do R, Prufer K, et al. Multiple genomic events altering hominin siglec biology and innate immunity predated the common ancestor of humans and archaic hominins. Genome Biol Evol. 2020;12:1040–1050. doi: 10.1093/gbe/evaa125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakata D, Close BE, Colley KJ, Matsuda T, Kitajima K. Molecular cloning and expression of the mouse N-acetylneuraminic acid 9-phosphate synthase which does not have deaminoneuraminic acid (KDN) 9-phosphate synthase activity. Biochem Biophys Res Commun. 2000;273:642–648. doi: 10.1006/bbrc.2000.2983. [DOI] [PubMed] [Google Scholar]
- 7.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 8.Varki A, Angata T. Siglecs—the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- 9.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 10.Uchiyama S, Carlin AF, Khosravi A, Weiman S, Banerjee A, et al. The surface-anchored nana protein promotes pneumococcal brain endothelial cell invasion. J Exp Med. 2009;206:1845–1852. doi: 10.1084/jem.20090386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ercoli G, Fernandes VE, Chung WY, Wanford JJ, Thomson S, et al. Intracellular replication of Streptococcus pneumoniae inside splenic macrophages serves as a reservoir for septicaemia. Nat Microbiol. 2018;3:600–610. doi: 10.1038/s41564-018-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanner ME. The enzymes of sialic acid biosynthesis. Bioorg Chem. 2005;33:216–228. doi: 10.1016/j.bioorg.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Chou WK, Dick S, Wakarchuk WW, Tanner ME. Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J Biol Chem. 2005;280:35922–35928. doi: 10.1074/jbc.M507483200. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, Cao M, Shi J, Zhang H, Hu D, et al. Attenuation of Streptococcus suis virulence by the alteration of bacterial surface architecture. Sci Rep. 2012;2:1–9. doi: 10.1038/srep00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan MI, Lundgren BR, Chaumun M, Whitfield DM, Clark B, et al. Total biosynthesis of legionaminic acid, a bacterial sialic acid analogue. Angew Chem Int Ed Engl. 2016;55:12018–12021. doi: 10.1002/anie.201606006. [DOI] [PubMed] [Google Scholar]
- 16.Schoenhofen IC, McNally DJ, Brisson JR, Logan SM. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology. 2006;16:8–14. doi: 10.1093/glycob/cwl010. [DOI] [PubMed] [Google Scholar]
- 17.Stephenson HN, Mills DC, Jones H, Milioris E, Copland A, et al. Pseudaminic acid on Campylobacter jejuni flagella modulates dendritic cell IL-10 expression via Siglec-10 receptor: a novel flagellin-host interaction. J Infect Dis. 2014;210:1487–1498. doi: 10.1093/infdis/jiu287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotton TR, Joseph DDA, Jiao W, Parker EJ. Probing the determinants of phosphorylated sugar-substrate binding for human sialic acid synthase. Biochim Biophys Acta. 2014;1844:2257–2264. doi: 10.1016/j.bbapap.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Gunawan J, Simard D, Gilbert M, Lovering AL, Wakarchuk WW, et al. Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J Biol Chem. 2005;280:3555–3563. doi: 10.1074/jbc.M411942200. [DOI] [PubMed] [Google Scholar]
- 20.Davies PL, Baardsnes J, Kuiper MJ, Walker VK, et al. Structure and function of antifreeze proteins. Philosophical Transactions of the Royal Society B: Biological Sciences. 2002;357:927–935. doi: 10.1098/rstb.2002.1081. pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varki A. Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc Natl Acad Sci U S A. 2010;107:8939–8946. doi: 10.1073/pnas.0914634107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeleny R, Kolarich D, Strasser R, Altmann F. Sialic acid concentrations in plants are in the range of inadvertent contamination. Planta. 2006;224:222–227. doi: 10.1007/s00425-005-0206-8. [DOI] [PubMed] [Google Scholar]
- 23.Lewis AL, Desa N, Hansen EE, Knirel YA, Gordon JI, et al. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci U S A. 2009;106:13552–13557. doi: 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Verezemska O, et al. Genomes online database (gold) v.6: data updates and feature enhancements. Nucleic Acids Res. 2017;45:D446–D456. doi: 10.1093/nar/gkw992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 33.Letunic I, Bork P. Interactive tree of life (iTOL) V3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 35.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bateman A, Martin MJ, O’Donovan C, Magrane M, Alpi E, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Pierri CR, Voyceik R, Santos de Mattos LGC, Kulik MG, Camargo JO, et al. Sweep: representing large biological sequences datasets in compact vectors. Sci Rep. 2020;10:1–10. doi: 10.1038/s41598-019-55627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stringer SC, Carter AT, Webb MD, Wachnicka E, Crossman LC, et al. Genomic and physiological variability within group II (non-proteolytic) Clostridium botulinum . BMC Genomics. 2013;14:333–18. doi: 10.1186/1471-2164-14-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bignell DRD, Selpke RF, Huguet-Tapla JC, Chambers AH, Parry RJ, et al. Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant-microbe interactions. Mol Plant Microbe Interact. 2010;23:161–175. doi: 10.1094/MPMI-23-2-0161. [DOI] [PubMed] [Google Scholar]
- 41.Joseph DDA, Jiao W, Parker EJ. Arg314 is essential for catalysis by N-acetyl neuraminic acid synthase from Neisseria meningitidis. Biochemistry. 2013;52:2609–2619. doi: 10.1021/bi400062c. [DOI] [PubMed] [Google Scholar]
- 42.Wallace L, Daugherty SC, Nagaraj S, Johnson JK, Harris AD, et al. Use of comparative genomics to characterize the diversity of Acinetobacter baumannii surveillance isolates in a health care institution. Antimicrob Agents Chemother. 2016;60:5933–5941. doi: 10.1128/AAC.00477-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, et al. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One. 2008;3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.García García MI, Lau K, Von Itzstein M, García Carmona F, Sánchez Ferrer Álvaro. Molecular characterization of a new N-acetylneuraminate synthase (NeuB1) from Idiomarina loihiensis. Glycobiology. 2015;25:115–123. doi: 10.1093/glycob/cwu096. [DOI] [PubMed] [Google Scholar]
- 45.Iyer LM, Aravind L. The emergence of catalytic and structural diversity within the beta-clip fold. Proteins. 2004;55:977–991. doi: 10.1002/prot.20076. [DOI] [PubMed] [Google Scholar]
- 46.Nee S. Antagonistic co-evolution and the evolution of genotypic randomization. J Theor Biol. 1989;140:499–518. doi: 10.1016/S0022-5193(89)80111-0. [DOI] [PubMed] [Google Scholar]
- 47.Li G-W. How do bacteria tune translation efficiency? Curr Opin Microbiol. 2015;24:66–71. doi: 10.1016/j.mib.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor RC, Webb Robertson BJM, Markillie LM, Serres MH, Linggi BE, et al. Changes in translational efficiency is a dominant regulatory mechanism in the environmental response of bacteria. Integr Biol. 2013;5:1393–1406. doi: 10.1039/c3ib40120k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. 2016;17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 50.Boyd EF. Structural and biosynthetic diversity of Nonulosonic acids (NulOs) that Decorate surface structures in bacteria. Trends Microbiol. 1867;2020:1–16. doi: 10.1016/j.tim.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petit D, Teppa E, Cenci U, Ball S, Harduin-Lepers A. Reconstruction of the sialylation pathway in the ancestor of eukaryotes. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-20920-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.