Abstract
Meticillin-resistant Staphylococcus aureus (MRSA) sequence type (ST) 45 was reported in the literature to have been first identified in 2006 in Taiwan. The present study was carried out to explore and trace the emergence, transmission and evolutional dynamics of MRSA ST45 in Taiwan. We identified MRSA ST45 isolates retrospectively from two collections of MRSA isolates, namely TSAR (Taiwan Surveillance of Antimicrobial Resistance) surveys and the CGMH (Chang Gung Memorial Hospital)-based laboratory collection. Representative ST45 isolates were selected for whole-genome sequencing (WGS) analysis. A total of 9554 MRSA isolates was included in this study. Among the 3766 MRSA isolates biennially collected from TSAR surveys between 1998 and 2014, ST45 accounted for 133 (3.53 %) MRSA isolates, was first identified in 2004, and the prevalence rate peaked in 2010 (up to 10.77 %). Among the 5788 MRSA isolates collected between 1995 and 2017 by the CGMH-based laboratory, 257 isolates (4.44 %) were characterized as ST45, with most identified from nursing homes since 2012. Of the 75 isolates randomly selected for WGS, two clades were identified. The major clade, clade II, comprised 63 isolates and was phylogenetically relatively close to those isolates identified from Singapore. All but one of the isolates in clade I, the minor clade, were identified from non-Taiwanese people, mostly from newly recruited foreign workers in 2017, and were phylogenetically relatively close to one isolate from the USA (CA-347). Conclusively, the emergence of MRSA ST45 strain in Taiwan can be traced back to 2004 and the strain is connected to South-East Asian countries. Since its emergence, transmission and spread of MRSA ST45 has occurred in Taiwan.
Keywords: foreign workers, meticillin-resistant Staphylococcus aureus, nursing homes, ST45, whole-genome sequencing
Data Summary
This whole-genome shotgun project has been deposited at GenBank/ENA/DDBJ under accession number PRJNA649874.
Impact Statement.
Meticillin-resistant Staphylococcus aureus (MRSA) has prevailed in hospitals worldwide and in some countries (regions) also in communities. There have been five major clones of MRSA internationally, including clonal complex 45 (CC45). This clone was first identified in Germany in 1993, spread worldwide subsequently and successfully established itself in long-term-care facilities (LTCFs). In Taiwan, MRSA has prevailed in hospitals and communities since it was identified, but declined gradually in the 2010s. There have been three major clones in the past two decades in Taiwan, namely sequence type (ST) 239, ST5 and ST59. Recently, dissemination of ST45 in nursing homes/LTCFs was identified in Taiwan. In this work, we identified 390 (4.1 %) ST45 isolates from a pool of 9554 MRSA isolates collected between 1995 and 2017. This clone was first identified in 2004 and has been increasingly identified since 2010. Whole-genome phylogeny of 75 isolates revealed one major clade, which comprised 63 isolates (84%) and was phylogenetically relatively close to those isolates identified from Singapore. The isolates of this clade harboured a greater number of mobile genetic elements and antimicrobial-resistance genes conferring resistance to aminoglycosides (aac6/aph2/aadD), β-lactam (blaZ) and tetracycline (tetK). A global epidemiological analysis of ST45 isolates may help figure out the perspective of ST45 worldwide.
Introduction
Staphylococcus aureus is a common cause of skin and soft-tissue infections. It can also cause more serious infections such as myositis, bone/joint infection, pneumonia, endocarditis, bacteraemia, and even the life-threatening conditions of necrotizing fasciitis and toxic shock syndrome. It is a challenge to treat infections due to S. aureus , particularly isolates that are meticillin-resistant (meticillin-resistant S. aureus , MRSA) and even recently emerging vancomycin-resistant isolates (vancomycin-resistant S. aureus , VRSA) [1, 2]. Vancomycin is among the first-line drugs for the treatment of MRSA infections. S. aureus isolates with complete resistance to vancomycin (VRSA) have emerged in recent years, which is mediated by a vanA gene cluster, and 52 VRSA strains have been isolated worldwide up until 2019 [2].
MRSA strains are traditionally classified as community-associated MRSA (CA-MRSA) and health-care-associated MRSA (HA-MRSA) according to epidemiological and/or molecular characteristics [1, 3]. There have been several major clones of HA-MRSA and CA-MRSA that prevailed in different continents, countries and regions worldwide [1, 3]. The rise and fall of specific clones of MRSA in human populations seems to be a common process that has occurred multiple times and in multiple locations [3].
In Taiwan, MRSA was first documented in the early 1980s and rapidly increased in the 1990s. In 2000, meticillin resistance had been identified in 53–83 % of all S. aureus isolates in 12 major hospitals of Taiwan [4]. However, a declining trend of HA-MRSA incidence was observed after 2000 [5, 6] and continued throughout 2020. In contrast, CA-MRSA infections have been increasingly reported, particularly in paediatric patients, since being identified in the early 2000s. The proportion of MRSA among community-associated S. aureus infections in children without risk factors increased from 9.8–36 % between 1997 and 2003 to 64.6 % in 2012, even higher than that among the health-care-associated isolates [7]. In Taiwan, there are three major clones of MRSA prevailing in the past two decades, namely sequence type (ST) 239, ST5 and ST59 [5, 6]. The former two clones, also well-known epidemic clones and traditionally HA-MRSA clones worldwide, have prevailed in health-care settings, while ST59, which is an endemic CA-MRSA clone and is further stratified to two subclones, namely Taiwan clone (ST59/SCCmec VT/PVL-positive) and Asian clone (ST59/SCCmec IV/PVL-negative), initially prevailed in the community and later penetrated into health-care settings [5, 6].
The MRSA ST45 strain, also called ‘Berlin epidemic MRSA’, was first observed in Berlin in 1993 [8, 9]. In Taiwan, MRSA ST45 was first reported in the literature from a study conducted in 2006, and the strain was identified in an infected patient and a colonized health-care worker (HCW) in a respiratory care ward [10]. Since then, clinical isolates collected in several studies also identified this strain and its frequency of identification has increased gradually, mostly in the elderly population [11, 12]. In 2012, in a survey for MRSA nasal carriage among both residents and HCWs in nursing homes, we identified the dissemination of MRSA ST45, which unexpectedly accounted for half of all colonizing MRSA isolates identified in 14 nursing homes island-wide [13]. In the subsequent years, the trend of increasing identification of ST45 among MRSA isolates was found continuously either in the surveys for MRSA in nursing homes or clinical isolates collected from hospital settings. The emergence of the ST45 clone in Taiwan cannot be overlooked. Therefore, we conducted this study to explore and trace the potential source, the trend and transmission of MRSA ST45 in Taiwan.
Methods
MRSA collections
In this study, we identified MRSA ST45 isolates retrospectively from two collections, which included a total of 9554 MRSA isolates. The first collection included MRSA isolates from the TSAR (Taiwan Surveillance of Antimicrobial Resistance) surveys: TSAR is a surveillance project, which was conducted by the investigators from the National Health Research Institute of Taiwan [14]. The TSAR project involved the collection of clinical isolates of various bacteria for determination of antimicrobial resistance (AMR) from up to 44 major hospitals island-wide in Taiwan biennially since 1998. A total of 3766 clinical MRSA isolates was collected from the first nine surveys between 1998 and 2014, ranging from 243 isolates in the 2000 survey to 476 isolates in the 2012 survey [12] (Table 1). The second collection included MRSA isolates from the CGMH (Chang Gung Memorial Hospital)-based laboratory: for surveillance of molecular epidemiology of MRSA in Taiwan, we collected and molecularly characterized (at least by PFGE) a total of 5788 MRSA isolates island-wide in Taiwan between 1995 and 2017 in the laboratory at the CGMH (Table 2). In addition to the CGMH, isolates were also collected from at least 10 hospitals island-wide in Taiwan. The isolates included clinical isolates and colonizing isolates from neonates, the paediatric population, adults and the elderly. A total of 2613 MRSA isolates (45 %) was collected between 1995 and 2007, 2529 MRSA isolates (44 %) between 2008 and 2012, plus 646 MRSA isolates (11 %) between 2013 and 2017. Of the 5788 MRSA isolates, 3186 (55 %) were clinical (infecting) isolates, 2494 (43 %) were human colonizing isolates (collected for study purposes but not for routine surveillance) and 108 (1.9 %) were non-human colonizing isolates (93 from pigs, 12 from the environmental objects, 2 from a bus and 1 from pork; also collected for study purposes). A total of 2759 isolates was identified from paediatric patients, 1438 isolates from adult patients and 1483 isolates from all age groups for surveillance of molecular epidemiology of MRSA in Taiwan (we estimated that more than 90 % of the isolates were from adult patients). The clinical isolates were identified from any body site, and most isolates were estimated to be health-care associated. The human colonizing isolates were identified from infants hospitalized in neonatal intensive care units (NICUs), neonates in nurseries, children at well-baby clinic visits, school children, case patients, adult patients, parturient mothers and elderly people resident in nursing homes. Fifty-two per cent of the 2494 human-colonizing isolates were identified from subjects without risk factors for MRSA acquisition.
Table 1.
Distribution of MRSA isolates, number of ST45 and number of ST45 selected for WGS analysis from the TSAR programme collection
|
Year |
No. of MRSA collected and characterized |
No. of ST45 (%) |
No. of ST45 selected for WGS analysis |
|---|---|---|---|
|
1998 |
373 |
0 |
0 |
|
2000 |
243 |
0 |
0 |
|
2002 |
495 |
0 |
0 |
|
2004 |
466 |
1 (0.21) |
1 |
|
2006 |
407 |
3 (0.74) |
3 |
|
2008 |
475 |
18 (3.79) |
3 |
|
2010 |
390 |
42 (10.77) |
3 |
|
2012 |
476 |
38 (7.98) |
3 |
|
2014 |
441 |
31 (7.03) |
3 |
|
Total |
3766 |
133 (3.53) |
16 |
Table 2.
Distribution of MRSA isolates, number of ST45 and number of ST45 selected for WGS analysis from the CGMH-based collection
|
Time period/population |
No. of MRSA collected and characterized |
No. of ST45 (%) |
No. of ST45 selected for WGS analysis |
||
|---|---|---|---|---|---|
|
Subtotal |
Clinical |
Colonizing |
|||
|
1995–2001 |
814 |
814 |
0 |
0 |
0 |
|
2002–2007 |
1799 |
711 |
1088 |
0 |
0 |
|
2008–2009 |
701 |
181 |
520 |
8 (1.14) |
0 |
|
2010–2012 |
1828 |
1177 |
651 |
120 (6.56) |
43 |
|
2013–2015 |
328 |
223 |
105 |
25 (7.62) |
0 |
|
2016–2017 |
318 |
80 |
238 |
104 (32.7) |
16 |
|
Total |
5788 |
3186 |
2602 |
257 (4.44) |
59 |
|
Paediatric |
2759 |
1135 |
1624 |
10 (0.36) |
2 |
|
Adults |
1438 |
630 |
808 |
195 (13.6) |
45 |
|
Unclassified |
1483 |
1421 |
62 |
49 (3.30) |
12 |
|
Non-human |
108 |
0 |
108 |
3 (2.78) |
0 |
Identification of MRSA ST45 isolates
In both laboratories in Chang Gung Memorial Hospital and National Health Research Institutes, S. aureus was identified by morphology, Gram stain and coagulase tests of strains grown on agar plates. To identify MRSA, cefoxitin susceptibility was tested by the disc-diffusion method according to the recommendations of the Clinical and Laboratory Standards Institute [15]. We characterized all the MRSA isolates by PFGE [7] and staphylococcal chromosome cassette mec (SCCmec) types [16]. Isolates with representative pulsotypes were selected for multilocus sequence typing (MLST) [17] and spa typing [18]. The isolates confirmed by MLST were identified as ST45. The other ST45 suspects, compared with these confirmed isolates, were identified as ST45 based on the same pulsotype and/or SCCmec type and spa type combinations. ST45 isolates from both collections were selected for whole-genome sequencing (WGS) based on representative specimen and host sources, pulsotype subtypes and collection years (see Results for details).
WGS
WGS was performed on an MiSeq sequencer (Illumina). The raw data of each strain were cleaned by adapter trimming and exclusion of reads in which greater than 45 % of bases were of quality score <20 (<Q20). The estimated genome coverages for all isolates ranged from 85.2 to 137.1. De novo assembly was conducted using SPAdes 3.11.1 [19]. The k-mer lengths were set to 21, 33, 55, 77, 99 and 127. Scaffold outputs greater than 200 bp were used in the following analysis. The numbers of scaffolds for each isolate ranged from 34 to 221 except for one isolate (up to 829). The other data of the project were deposited at GenBank/ENA/DDBJ under accession number PRJNA649874.
To compare the phylogenetic relatedness of the ST45 MRSA isolates in Taiwan with those from other geographical areas, one ST45 isolate from the USA (CA-347, GenBank accession no. CP006044) [20] and the short-read raw data of 107 ST45 strains isolated in Singapore in 2014 were downloaded from the Sequence Read Archive (SRA) using the fast-dump tool kit [21]. The assembly of draft genome of each isolate was also conducted with the same procedure as described above using SPAdes 3.11.1 [19].
Phylogenetic analysis
The draft genomes were firstly aligned to each other with the Parsnp script provided in the Harvest suite and subsequently analysed with the program Gubbins [22, 23]. Parsnp is capable of aligning multiple bacterial genomes and outputs the variants call, core-genome phylogeny and multi-alignment. The methicillin-susceptible S. aureus strain MCRF184 was used as the reference genome during the procedure. Gubbins is capable of localizing potential recombination and the phylogeny was reconstructed based on the SNPs outside of the potential recombination regions. SNPs in the core genome were used to reconstruct the phylogenies of the ST45 MRSA strains. The final phylogenetic tree was input into Interactive Tree Of Life (iTOL) (http://itol.embl.de) or FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) for further manipulation and annotation.
Determination of the resistance genes, virulence genes and plasmids harboured
The detection of resistance genes, virulence genes and plasmids was conducted with the srst2 script using the databases of ARG-ANNOT (version 2; https://github.com/katholt/srst2/tree/master/data/ARGannot_r2.fasta, assessed on December 4 2017), Staphylococcus_VF (Virulence Factors of Pathogenic Bacteria website, http://www.mgc.ac.cn/VFs/main.htm, Staphylococcus_VF_clustered.fasta, assessed on August 8 2017) and PlasmidFinder (https://github.com/katholt/srst2/blob/master/data/PlasmidFinder.fasta, assessed on December 4 2017), respectively [24].
Results
Identification of ST45 isolates
Overall, 390 (4.1 %) ST45 MRSA isolates were identified from the two collections. Among the 3766 MRSA isolates collected from TSAR surveys between 1998 and 2014, ST45 isolates accounted for 133 (3.53 %) isolates, and the rate ranged from none before 2004, 0.21 % in 2004 to 10.77 % in 2010. ST45 was first identified in the 2004 survey, then persistently identified in each later survey (Table 1) and an increasing trend was noted.
Among the 5788 isolates collected by the CGMH-based laboratory, 257 isolates (4.44 %) were characterized as ST45 (Table 2). None of the 2613 isolates collected and characterized between 1995 and 2007 was ST45, whereas the proportion of ST45 among all the MRSA isolates characterized increased from 1.14 % in 2008–2009, 6–8 % in 2010–2015 to 32.7 % in 2016–2017. Most of the ST45 isolates were identified from adults, particularly elderly people living in nursing homes. The first three ST45 isolates were identified in 2008 from infants staying in NICUs (neonatal intensive care units) [25], up to a total of 12 isolates before 2010, and then numbers increased markedly. In 2010, ST45 accounted for 34 (5.9 %) of 577 MRSA isolates collected from six major hospitals in Taiwan [8]. In 2012, ST45 accounted for 52 (50 %) of 105 MRSA isolates collected from residents and HCWs in 14 nursing homes island-wide during a one-time-point survey for MRSA carriage [11]. From July 2016 to February 2017, ST45 accounted for 85 (49.4 %) of 172 MRSA isolates collected from residents and HCWs in four nursing homes in northern Taiwan during a longitudinal survey (bimonthly for 6 months with four surveys) for MRSA carriage (unpublished data). ST45 also accounted for 18 (34.0 %) of 53 MRSA isolates identified from 1935 foreign workers recruited to Taiwan in 2017 [26] (Table 2).
ST45 isolates selected for WGS
Overall, 75 (19 %) ST45 MRSA isolates were selected for WGS. Among the 133 MRSA ST45 isolates identified from TSAR surveys, 16 isolates were selected for WGS and included the first ST45 isolate identified in 2004 (strain Sau41) and three isolates in each later survey (Tables 1 and S1, available with the online version of this article). Among the 257 ST45 isolates identified from the CGMH-based laboratory, 59 isolates were selected for WGS and included 1 isolate (strain DR4) from a Vietnamese attendee at an international conference held in Taiwan in 2010 [27], 22 clinical (infecting) isolates from 6 hospitals during 2010 and 2011 [11], 20 colonizing isolates from 14 nursing homes in a 2012 survey (15 isolates from foreign HCWs, 4 isolates from Taiwanese HCWs and 1 from a resident) [13], 5 colonizing isolates from two nursing homes in 2016 (2 isolates from Indonesian HCWs, and 1 each from a Filipino HCW, a Vietnamese HCW and a resident, respectively) and 11 colonizing isolates from foreign workers recruited from south-eastern countries in the 2017 survey (7 isolates from Vietnamese workers, 3 isolates from Indonesian workers and 1 from a Filipino worker) [26] (Tables 2 and S1).
Phylogenetic analysis and genomic compositions
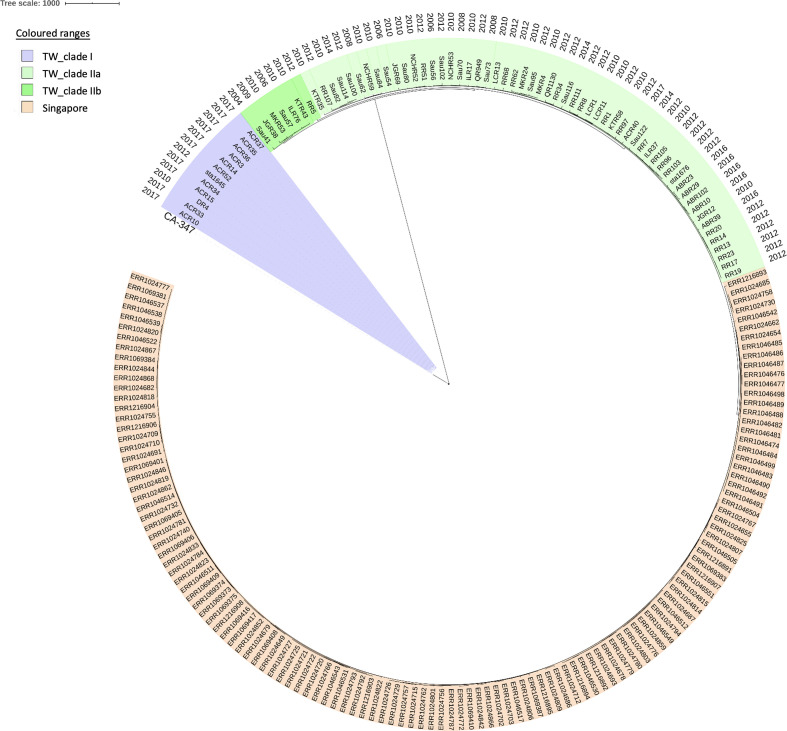
A total of 5616 nucleotide alterations could be identified in the core genomes of the ST45 isolates including the reference strain MCRF184 and were used for maximum-likelihood phylogeny reconstruction. Phylogenetic analysis disclosed that all of the ST45 isolates were classified into two distinct clades, clade I and II (Figs 1 and S1), with a range of 3601–3711 SNPs in pairwise comparisons of the isolates in the two different clades. There were 12 isolates clustered in clade I, the minor clade, and these included the isolate from a Vietnamese attendee of an international conference in 2010 (strain DR4), 1 clinical isolate from a 7-month-old infant in 2012 (strain sta1645) and 10 isolates from foreign workers (7 from Vietnam and 3 from Indonesia, named as 'ACR' strains) in the 2017 survey. The isolate from the USA (CA-347) was relatively phylogenetically close to clade I. The other 63 isolates, including 20 colonizing isolates from 19 foreign HCWs and 1 foreign worker, were clustered as clade II. The isolates in clade II were further split into two subclades, IIa (56 isolates) and IIb (7 isolates). The 107 ST45 isolates (with 1533 SNPs) identified in Singapore also belonged to clade II (Fig. 1), relatively phylogenetically close to those in subclade IIa. However, the isolates of clade II identified from Taiwan were separated by those identified from Singapore.
Fig. 1.
Phylogenetic analysis of 75 MRSA ST45 isolates from Taiwan based on SNPs in the core genomes. One ST45 isolate from the USA (CA-347) and 107 ST45 isolates from Singapore in 2014 were included for comparison. Twelve isolates were clustered as clade I and were relatively phylogenetically close to CA-347. The other 63 isolates were clustered as clade II (56 isolates as subclade IIa and 7 isolates as subclade IIb) and were relatively phylogenetically close to those identified in Singapore. The scale bar indicates the numbers of SNPs.
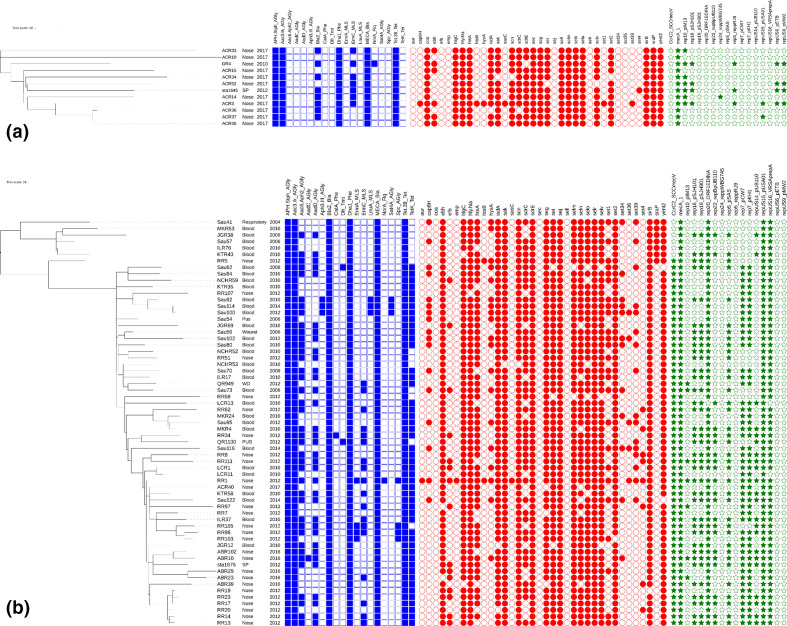
The AMR genes, mobile genetic elements (including plasmids) and virulence genes harboured by the ST45 MRSA isolates differed significantly between the isolates of both clades. Detailed information on the distributions of these genes and elements carried by the isolates of these two clades are shown in Table 3 and Fig. 2. None of the ST45 isolates carried PVL genes. Fifty-seven (90.5 %) of 63 isolates in clade II carried the mobile genetic elements for SCCmec V, while the other 6 isolates in clade II and all the 12 isolates in clade I carried SCCmec IV. When compared to the isolates of clade I, the isolates of clade II harboured a greater number of mobile genetic elements and AMR genes conferring resistance to aminoglycosides (aac6/aph2/aadD), β-lactam (blaZ) and tetracycline (tetK), and carried more virulence genes of ebh, sej, selr and set1.
Table 3.
Distribution of AMR genes, virulence genes and mobile genetic elements differentially carried by 75 ST45 MRSA isolates of two major clades in Taiwan
|
Element |
N (%) |
P |
|
|---|---|---|---|
|
Clade I (n=12) |
Clade II (n=63) |
||
|
Resistance genes (class of antibiotic) |
|||
|
aac6 aph2 (AGly) |
0 (0) |
39 (61.9) |
<0.001 |
|
aadD (AGly) |
0 (0) |
34 (54.0) |
<0.001 |
|
blaZ (Bla) |
8 (66.7) |
63 (100.0) |
<0.001 |
|
ermC (MLS) |
7 (58.3) |
18 (28.6) |
0.036 |
|
tetK (Tet) |
0 (0) |
49 (77.8) |
<0.001 |
|
Virulence genes (encoded protein) |
|||
|
coa (staphylocoagulase) |
12 (100.0) |
0 (0) |
<0.001 |
|
ebh (cell wall associated fibronectin binding protein) |
6 (50.0) |
61 (96.8) |
<0.001 |
|
sec (staphylococcal enterotoxin C) |
12 (100.0) |
0 (0) |
<0.001 |
|
sej (staphylococcal enterotoxin J) |
0 (0) |
63 (100) |
<0.001 |
|
sell (staphylococcal enterotoxin-like L) |
12 (100.0) |
0 (0) |
<0.001 |
|
selr (staphylococcal enterotoxin-like R) |
0 (0) |
63 (100) |
<0.001 |
|
set1 (exotoxin 1) |
1 (8.33) |
25 (39.7) |
0.048 |
|
sraP (LPXTG cell wall surface anchor family protein) |
12 (100.0) |
7 (11.1) |
<0.001 |
|
Mobile elements including plasmids |
|||
|
ccrC2 SCCmecV |
0 (0) |
57 (90.5) |
<0.001 |
|
rep10 pIM13 |
7 (58.3) |
7 (11.1) |
<0.001 |
|
rep19 pSJH901 |
0 (0) |
37 (58.7) |
<0.001 |
|
rep20 ORF1EDINA |
0 (0) |
61 (96.8) |
<0.001 |
|
rep22 repBpUB110 |
0 (0) |
22 (34.9) |
0.014 |
|
rep5 pSAS |
0 (0) |
37 (58.7) |
<0.001 |
|
rep5 reppRJ9 |
3 (25.0) |
0 (0) |
0.003 |
|
rep7 pKH1 |
0 (0) |
49 (77.8) |
<0.001 |
|
repUS14 pUB110 |
0 (0) |
22 (34.9) |
0.014 |
|
repUS15 pUSA01 |
2 (16.7) |
63 (100.0) |
<0.001 |
|
repUS16 VRSAprepA |
0 (0) |
61 (96.8) |
<0.001 |
|
repUS6 pETB |
4 (33.3) |
0 (0) |
<0.001 |
|
repUS9 pMW2 |
4 (33.3) |
0 (0) |
<0.001 |
AGly, Aminoglycoside; Bla, β-lactamase; MLS, macrolide–lincosamide–streptomycin; Tet, tetracycline.
Fig. 2.
Detailed distributions of AMR genes, virulence genes and plasmids in the ST45 MRSA isolates of clade I (a) and clade II (b). Blue colour indicates AMR genes, red colour indicates toxin genes and green colour indicates plasmids. The scale bar indicates the numbers of SNPs. Abbreviations: WD, wound; SP, sputum.
Discussion
Since its emergence, MRSA ST45 has spread to other European countries [17], including the Netherlands [28, 29], Belgium [30], the UK, etc., as well as to the USA (as USA600) [31, 32], and later to Asian countries such as Hong Kong, SAR, China [33–36], and Singapore [21, 37], and more recently to Australia [38], particularly in New South Wales. The strain emerging in the USA is associated with higher rates of AMR and more patient mortality [32], whereas the strain prevailing in Australia is a unique strain, which contains deletions in the spa gene [38]. So far, ST45 isolates seemed to have successfully established themselves in long-term care facilities.
In this systematic analysis of MRSA isolates from two large collections in Taiwan, we found that ST45 has also established itself in the general population in Taiwan. The detection (emergence) of MRSA ST45 strain in Taiwan could be traced back to the year 2004, earlier than previously reported (2006) [10]. Since its emergence, this strain increased gradually, and by 2010 it accounted for 34 (5.9 %) of 577 clinical MRSA bloodstream isolates from six major hospitals in Taiwan [11], and 8.5 % of 1307 clinical isolates collected between 2010 and 2014 from the TSAR programme. In a recent report, Huang et al. [39] found that ST45, though next to ST59, accounted for 39 % of 64 mecA-positive borderline MRSA isolates identified from 2001 to 2015 in a medical centre in northern Taiwan and has replaced ST59 as the predominant clone since 2012. The impact of ST45 in Taiwan should be continuously monitored.
In addition to clinical isolates, MRSA ST45 was identified from colonizing isolates. In a one-time-point survey conducted in 2012 [13], nasal MRSA carriage was identified among one-fifth of 523 participants (including 360 residents and 163 staff) in 14 nursing homes located throughout different regions of Taiwan, and ST45 accounted for half of the 105 MRSA isolates. More recently, in a longitudinal survey (bimonthly for 6 months with four surveys) for MRSA carriage amongst residents and HCWs in four nursing homes in northern Taiwan between August 2016 and February 2017 (unpublished data), we found that 40 % of the participants had MRSA carriage and again ST45 accounted for nearly half of the 172 MRSA isolates. In two other surveys for MRSA colonization among residents and environments conducted in six nursing homes in 2015 and 2016, respectively, MRSA ST45 accounted for nearly 30 and 20 % of the MRSA isolates, respectively [40, 41]. These findings suggest that the MRSA ST45 strain has prevailed in nursing homes in Taiwan since 2012 or probably earlier. In addition, Wu et al. [42] reported that ST45 accounted for 30 % of 36 MRSA isolates identified from a survey for MRSA carriage amongst emergency room HCWs and patients of two regional hospitals in central Taiwan in 2015, suggesting increasing identification of this strain in hospital settings too. Asymptomatic carriage of MRSA ST45 may accelerate the spread in the community as well as in the health-care settings. Continuing surveillance is needed.
In our previous study [13], we also found that foreign nursing staff were a significant risk factor for MRSA carriage and, thus, the issue of whether the MRSA ST45 strain was imported by the foreign (care) workers was raised. In this study, WGS classified the ST45 isolates into two clades. All but one isolate in clade I, the minor clade, were identified from non-Taiwanese people, mostly from newly recruited foreign workers on arrival. The only one isolate from a Taiwanese person in clade I (strain sta1645) was identified from a 7-month-old male infant in 2012. Since this study was retrospective, detailed information about whether there were any connections to south-eastern countries (such as being immigrants, travelling) among his parents, as well as his household members, could not be traced. In contrast, clade II, the major clade, accounted for 84 % of the 75 ST45 isolates analysed, and included clinical and colonizing isolates from patients and HCWs (Taiwanese and non-Taiwanese) collected between 2004 and 2016 in Taiwan. All the colonizing isolates from foreign HCWs were identified while they were working in nursing homes but not on their arrival, so we cannot delineate whether the colonizing isolates they carried were acquired before or after their arrival in Taiwan. Therefore, we cannot definitely identify the origin of ST45 in Taiwan. However, we found that since its emergence, transmission and spread of MRSA ST45 has occurred in Taiwan.
Phylogenetically, while ST45 isolates of clade I in this study were relatively close to strain CA-347 identified from the USA, the isolates in clade II were relatively close to those identified from Singapore (Fig. 1). In addition, Gruteke et al. [29] recently also sequenced five t1081 isolates (clonal complex 45/SCCmec V), three from the Netherlands and two from Hong Kong SAR, China, and they found the isolates from the Netherlands were more similar to those from Hong Kong (nearly 97 % identity) than CA-347 (91 % identity). According to the characteristics they described, the five isolates they sequenced were similar to the isolates in clade II in this study, though no sequence data can be compared directly. These findings suggest that the strain ST45 circulating in Taiwan is likely connected to those circulating in South-East Asian countries.
Recently, Effelsberg et al. [43] conducted a study on the global epidemiology of S. aureus ST45, and collected and sequenced a total of 451 ST45 S. aureus isolates from 26 countries, including 10 isolates from Asia. The tree topology showed a split into two distinct sublineages separated by long branches with strong geographical signatures. The larger sublineage (European/North American sublineage, n=373) primarily consisted of isolates from Europe (n=276) and North America (n=70), while the second sublineage (African/Australian sublineage, n=78) could be further divided into two groups, namely an African group (n=31) and an Australian group (n=47). All five isolates from Singapore and the only one isolate from Hong Kong SAR, China, were close to those in the Australian group. We further downloaded and analysed the sequence data of 290 isolates from their study. Phylogenetic analysis revealed that all the 12 isolates of clade I in this study were interspersed in a phylogenetic tree with European isolates and the two Vietnamese isolates in the larger sublineage. In contrast, all the 63 isolates of clade II in this study were embedded in the Australian group isolates of the smaller sublineage; the 56 isolates of clade IIa and 7 isolates of clade IIb, though clustered together in each subclade, were separated by a small group of Australian isolates.
There are several limitations of this study. Firstly, since all the MRSA ST45 isolates identified and characterized in this study were only from two collections, further studies are needed to obtain a more comprehensive picture of ST45 in Taiwan. However, both collections were big collections and may provide adequate information on this issue. Secondly, not every MRSA ST45 suspect was confirmed by MLST. However, all the 75 isolates selected for WGS were ultimately confirmed to be ST45 by WGS, suggesting our screening methods were acceptable. Thirdly, the nature of the present study is retrospective, so detailed travel abroad history could not be obtained from the case patients and HCWs, nor their household members. Thus, whether the isolates of ST45 from the study subjects were imported or not cannot be traced clearly.
Conclusions
The MRSA ST45 strain was detected in Taiwan as early as 2004 and could be connected to those identified in South-East Asian countries. After its emergence in Taiwan, this strain has gradually been identified in increasing frequency in the community, as well as in health-care settings, particular since 2010. Local transmission and spread of this strain have occurred, particularly in nursing homes. Further surveillance of local MRSA molecular epidemiology is needed.
Supplementary Data
Funding information
The study was supported by a grant from the Ministry of Science and Technology, Executive Yuan, Taiwan (MOST 105–2314-B-182A-139), and a grant from the CGMH (CMRPG3F1852, CMRPG3J1601).
Author contributions
Y.-C. H. designed the study, applied for the grants, provided the laboratory work, analysed and interpreted the data, and wrote the manuscript. C.-J. C. and T.-L. Y. L. analysed and interpreted the data. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was approved by the Institutional Review Board of the CGMH (201 509 728B0).
Footnotes
Abbreviations: AMR, antimicrobial resistance; CA-MRSA, community-associated MRSA; CGMH, Chang Gung Memorial Hospital; HA-MRSA, health-care-associated MRSA; HCW, health-care worker; MLST, multilocus sequence typing; MRSA, meticillin-resistant Staphylococcus aureus; ST, sequence type; TSAR, Taiwan Surveillance of Antimicrobial Resistance; VRSA, vancomycin-resistant Staphylococcus aureus; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary table and one supplementary figure are available with the online version of this article.
References
- 1.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus . Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong Y, Yang S, Rao X. Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J Adv Res. 2020;21:169–176. doi: 10.1016/j.jare.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31:e00020-18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsueh P-R, Liu C-Y, Luh K-T. Current status of antimicrobial resistance in Taiwan. Emerg Infect Dis. 2002;8:132–137. doi: 10.3201/eid0802.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chuang YY, Huang YC. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis. 2013;13:698–708. doi: 10.1016/S1473-3099(13)70136-1. [DOI] [PubMed] [Google Scholar]
- 6.Chen CJ, Huang YC. New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect. 2014;20:605–623. doi: 10.1111/1469-0691.12705. [DOI] [PubMed] [Google Scholar]
- 7.Wang HK, Huang CY, Huang YC. Clinical features and molecular characteristics of childhood community-associated methicillin-resistant Staphylococcus aureus infection in a medical center in northern Taiwan, 2012. BMC Infect Dis. 2017;17:470. doi: 10.1186/s12879-017-2560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witte W. Antibiotic resistance in Gram-positive bacteria: epidemiological aspects. J Antimicrob Chemother. 1999;44:1–9. doi: 10.1093/jac/44.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- 9.Ghebremedhin B, Konig W, Konig B. Heterogeneity of methicillin-resistant Staphylococcus aureus strains at a German university hospital during a 1-year period. Eur J Clin Microbiol Infect Dis. 2005;24:388–398. doi: 10.1007/s10096-005-1339-1. [DOI] [PubMed] [Google Scholar]
- 10.Lee YT, Lin DB, Wang WY, Tsao SM, Yu S-F, et al. First identification of methicillin-resistant Staphylococcus aureus MLST types ST5 and ST45 and SCCmec types IV and Vt by multiplex PCR during an outbreak in a respiratory care ward in central Taiwan. Diagn Microbiol Infect Dis. 2011;70:175–182. doi: 10.1016/j.diagmicrobio.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 11.Chen CJ, Huang YC, Su L-H, Wu T-L, Huang S-H, et al. Molecular epidemiology and antimicrobial resistance of methicillin-resistant Staphylococcus aureus bloodstream isolates in Taiwan, 2010. PLoS One. 2014;9:e101184. doi: 10.1371/journal.pone.0101184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauderdale T-LY. The Third Symposium of Methicillin-Resistant Staphylococcus aureus. Taipei, Taiwan: 2015. Pulsed-field gel electrophoresis typing of MRSA isolates from Taiwan: establishing a national database. [Google Scholar]
- 13.Tsao FY, Kou HW, Huang YC. Dissemination of methicillin-resistant Staphylococcus aureus sequence type 45 among nursing home residents and staff in Taiwan. Clin Microbiol Infect. 2015;21:451–458. doi: 10.1016/j.cmi.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Wang JT, Huang IW, Chang SC, Tan M-C, Lai J-F, et al. Increasing resistance to fusidic acid among clinical isolates of MRSA. J Antimicrob Chemother. 2017;72:616–618. doi: 10.1093/jac/dkw430. [DOI] [PubMed] [Google Scholar]
- 15.CLSI Performance Standards for Antimicrobial Susceptibility Testing, 20th Informational Supplement, M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. [Google Scholar]
- 16.Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci USA. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegger M, Driebe EM, Roe C, Lemmer D, Bowers JR, et al. Genome sequence of Staphylococcus aureus strain CA-347, a USA600 methicillin-resistant isolate. Genome Announc. 2013;1:e00517-13. doi: 10.1128/genomeA.00517-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chow A, Lim VW, Khan A, Pettigrew K, Lye DCB, et al. MRSA transmission dynamics among interconnected acute, intermediate-term, and long-term healthcare facilities in Singapore. Clin Infect Dis. 2017;64:S76–S81. doi: 10.1093/cid/cix072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y-C, Lien R-I, Lin T-Y. Effect of mupirocin decolonization on subsequent methicillin-resistant Staphylococcus aureus infection in infants in neonatal intensive care units. Pediatr Infect Dis J. 2015;34:241–245. doi: 10.1097/INF.0000000000000540. [DOI] [PubMed] [Google Scholar]
- 26.Chen KH, Chuang WC, Wong WK, Chuang CH, Chen CJ, et al. Nasal methicillin-resistant Staphylococcus aureus carriage among foreign workers recruited to Taiwan from Southeastern Asian countries. Open Forum Infect Dis. 2021;8:ofaa586. doi: 10.1093/ofid/ofaa586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang YC, Su L-H, Wu T-L, Lin T-Y. Methicillin-resistant Staphylococcus aureus nasal carriage in international medical conference attendees. J Microbiol Immunol Infect. 2019;52:242–247. doi: 10.1016/j.jmii.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Wannet WJ, Spalburg E, Heck ME, Pluister GN, Willems RJ, et al. Widespread dissemination in the Netherlands of the epidemic Berlin methicillin-resistant Staphylococcus aureus clone with low-level resistance to oxacillin. J Clin Microbiol. 2004;42:3077–3082. doi: 10.1128/JCM.42.7.3077-3082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruteke P, Ho P-L, Haenen A, Lo W-U, Lin C-H, et al. MRSA spa t1081, a highly transmissible strain endemic to Hong Kong, China, in the Netherlands. Emerg Infect Dis. 2015;21:1074–1076. doi: 10.3201/eid2106.141597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denis O, Jans B, Deplano A, Nonhoff C, De Ryck R, et al. Epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) among residents of nursing homes in Belgium. J Antimicrob Chemother. 2009;64:1299–1306. doi: 10.1093/jac/dkp345. [DOI] [PubMed] [Google Scholar]
- 31.Moore CL, Osaki-Kiyan P, Perri M, Donabedian S, Haque NZ, et al. USA600 (ST45) methicillin-resistant Staphylococcus aureus bloodstream infections in urban Detroit. J Clin Microbiol. 2010;48:2307–2310. doi: 10.1128/JCM.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakoulas G, Guram K, Reyes K, Nizet V, Zervos M. Human cathelicidin LL-37 resistance and increased daptomycin MIC in methicillin-resistant Staphylococcus aureus strain USA600 (ST45) are associated with increased mortality in a hospital setting. J Clin Microbiol. 2014;52:2172–2174. doi: 10.1128/JCM.00189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho P-L, Chow K-H, Lo P-Y, Lee K-F, Lai EL. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus associated with spread of the ST45 lineage in Hong Kong. Diagn Microbiol Infect Dis. 2009;64:131–137. doi: 10.1016/j.diagmicrobio.2009.01.030. [DOI] [PubMed] [Google Scholar]
- 34.Cheng VC, Chan JF, Lau EH, Yam WC, Ho SKY, et al. Studying the transmission dynamics of meticillin-resistant Staphylococcus aureus in Hong Kong using spa typing. J Hosp Infect. 2011;79:206–210. doi: 10.1016/j.jhin.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 35.Cheng VC, Tai JW, Wong ZS, Chen JH, Pan KB, et al. Transmission of methicillin-resistant Staphylococcus aureus in the long term care facilities in Hong Kong. BMC Infect Dis. 2013;13:205. doi: 10.1186/1471-2334-13-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luk S, Ho AYM, Ng TK, Tsang IH, Chan EH, et al. Prevalence, prediction, and clonality of methicillin-resistant Staphylococcus aureus carriage at admission to medical units in Hong Kong, China. Infect Control Hosp Epidemiol. 2014;35:42–48. doi: 10.1086/674393. [DOI] [PubMed] [Google Scholar]
- 37.Teo J, Tan TY, Hon PY, Lee W, Koh TH, et al. ST22 and ST239 MRSA duopoly in Singaporean hospitals: 2006-2010. Epidemiol Infect. 2013;141:153–157. doi: 10.1017/S0950268812000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beukers AG, Newton P, Hudson B, Ross K, Gottlieb T, et al. A multicentre outbreak of ST45 MRSA containing deletions in the spa gene in New South Wales, Australia. J Antimicrob Chemother. 2020;75:1112–1116. doi: 10.1093/jac/dkz560. [DOI] [PubMed] [Google Scholar]
- 39.Huang YT, Liao CH, Chen SY, Hsu HS, Teng LJ, et al. Emergence of multidrug-resistant sequence type 45 strains among mecA -positive borderline oxacillin-resistant Staphylococcus aureus causing bacteraemia in a medical centre in Taiwan. Int J Antimicrob Agents. 2018;52:70–75. doi: 10.1016/j.ijantimicag.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Lai CC, Lee CM, Chiang HT, Lu M-C, Wang L-F, et al. Methicillin-resistant Staphylococcus aureus sequence type 45 with high rates of ciprofloxacin and tetracycline resistance in the residents and environments of long-term care facilities in Taiwan. J Infect. 2018;76:305–307. doi: 10.1016/j.jinf.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Liu CY, Lai CC, Chiang HT, Lu M-C, Wang L-F, et al. Predominance of methicillin-resistant Staphylococcus aureus in the residents and environments of long-term care facilities in Taiwan. J Microbiol Immunol Infect. 2019;52:62–74. doi: 10.1016/j.jmii.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Wu T-H, Lee C-Y, Yang H-J, Fang Y-P, Chang Y-F, et al. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus among nasal carriage strains isolated from emergency department patients and healthcare workers in central Taiwan. J Microbiol Immunol Infect. 2019;52:248–254. doi: 10.1016/j.jmii.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 43.Effelsberg N, Stegger M, Peitzmann L, Altinok O, Coombs GW, et al. Global epidemiology and evolutionary history of Staphylococcus aureus ST45. J Clin Microbiol. 2020;59:e02198-20. doi: 10.1128/JCM.02198-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.