Abstract
Outbreak investigations are essential to control and prevent the dissemination of pathogens. This study developed and validated a complete analysis protocol for faster and more accurate surveillance and outbreak investigations of antibiotic-resistant microbes based on Oxford Nanopore Technologies (ONT) DNA whole-genome sequencing. The protocol was developed using 42 methicillin-resistant Staphylococcus aureus (MRSA) isolates identified from former well-characterized outbreaks. The validation of the protocol was performed using Illumina technology (MiSeq, Illumina). Additionally, a real-time outbreak investigation of six clinical S. aureus isolates was conducted to test the ONT-based protocol. The suggested protocol includes: (1) a 20 h sequencing run; (2) identification of the sequence type (ST); (3) de novo genome assembly; (4) polishing of the draft genomes; and (5) phylogenetic analysis based on SNPs. After the sequencing run, it was possible to identify the ST in 2 h (20 min per isolate). Assemblies were achieved after 4 h (40 min per isolate) while the polishing was carried out in 7 min per isolate (42 min in total). The phylogenetic analysis took 0.6 h to confirm an outbreak. Overall, the developed protocol was able to at least discard an outbreak in 27 h (mean) after the bacterial identification and less than 33 h to confirm it. All these estimated times were calculated considering the average time for six MRSA isolates per sequencing run. During the real-time S. aureus outbreak investigation, the protocol was able to identify two outbreaks in less than 31 h. The suggested protocol enables identification of outbreaks in early stages using a portable and low-cost device along with a streamlined downstream analysis, therefore having the potential to be incorporated in routine surveillance analysis workflows. In addition, further analysis may include identification of virulence and antibiotic resistance genes for improved pathogen characterization.
Keywords: outbreak, nanopore-sequencing, multidrug-resistant bacteria, methicillin-resistant Staphylococcus aureus
Data Summary
S. aureus sequences were deposited in the NCBI Sequence Read Archive under the BioProjects PRJNA658251 and PRJNA658260. The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement.
Surveillance and outbreak investigations are critical to limit the transmission of pathogens. In the healthcare setting, rapid outbreak investigations make it possible to implement adequate limitation strategies, saving patients from serious infections and saving healthcare costs. Next-generation sequencing (NGS) could provide unprecedented resolution in discriminating highly related bacterial strains. However, the implementation of NGS is usually far from ideal for routine diagnostics due to time-consuming procedures, expensive equipment and the requirement of specialized personnel. To get the high resolution of NGS and overcome some of the limitations, this study developed and validated a rapid and straightforward protocol using Oxford Nanopore Technology (ONT)-based DNA sequencing to investigate outbreaks related to antibiotic-resistant bacteria. Using methicillin-resistant Staphylococcus aureus (MRSA) as the model, the suggested protocol was time-efficient and presented results as expected by identifying outbreaks in early stages. Additionally, the approach has a low capital cost and it uses a portable sequencing device, which facilitates its implementation in space- and resource-limited settings without high bioinformatic competence.
Introduction
Performing surveillance and identifying outbreaks at early stages are of vital importance to limit the spread of antimicrobial-resistant bacteria in hospitals, in long-term care institutions and in the community in general. By identifying the source(s) of the pathogen and possible transmission pathways it is possible to implement preventive measures [1]. When accumulation of antibiotic-resistant bacteria occurs, molecular genotyping of the bacterial isolates will help to determine the extent and source of the outbreak. Conventional genotyping methods rely on selected genes or small DNA sequences as biomarkers for the whole genome and will therefore in many situations be inadequate [2–4].
The advances of next-generation sequencing (NGS) technology has led to decreasing cost, growing speed and high-throughput of whole-genome sequencing (WGS), allowing the discrimination required to distinguish among isolates of the same lineage, overcoming the limitation in previous typing methods [1, 5]. However, the suggested protocols are so far usually suitable only for larger reference labs/core facilities as they require expensive equipment, trained laboratory personnel with specialized bioinformatics skills and a high number of bacterial isolates for cost-efficiency. Those characteristics usually place the use of WGS far from ideal for routine diagnostics [6, 7].
Nanopore-based sequencing from Oxford Nanopore Technologies (ONT) has brought miniaturized DNA sequencing devices with high-throughput. Consequently, many previous limitations in NGS technology have been overcome since the devices are portable and less expensive; they have shorter turnaround times and streamlined downstream analysis; and their ultra-long real-time available reads are well suited for rapidly distinguishing low-diversity bacterial strains [1, 7]. Some studies have been reporting the ONT-based sequencing potential as a pathogen surveillance tool [8–11]. However, none of them have described a complete protocol for outbreak investigations related to antimicrobial-resistant bacteria.
Staphylococcus aureus is one of the bacteria for which new antibiotics are urgently needed [12]. Recent surveillance data from European countries show a general trend towards increasing methicillin-resistant S. aureus (MRSA) prevalence from the north to the south of the continent [13, 14]. In Norway, a country with low prevalence of MRSA, recent publications have reported that global circulation and import of cases from abroad are related to the increasing rate of MRSA infections [15, 16]. Due to the low diversity among some MRSA isolates belonging to the same clone, methods such as multilocus sequencing typing (MLST), multiple locus variable-number tandem repeat analysis (MLVA), pulsed-field gel electrophoresis (PFGE and S. aureus Protein A (spa) typing cannot discriminate all epidemiologically linked cases in certain settings. Therefore, those cases require higher-resolution methods to identify the exact demarcation of the outbreaks [3, 4].
Our hypothesis was that using ONT-based sequencing analysis on antimicrobial-resistant bacteria will lead to significantly faster and more accurate surveillance and outbreak investigations than other NGS technologies and conventional genotyping methods. Therefore, this study developed a complete analysis protocol from bacterial isolate to NGS genotype for use in real-time surveillance, using MRSA as the bacterial model. We also tested the ONT-based protocol on a S. aureus outbreak in real-time. This type of protocol using nanopore technology is not readily available on today’s market.
Methods
Bacterial isolates and DNA extraction
The study was conducted at Akershus University Hospital (Ahus) in Lørenskog, Norway. Ahus is the largest acute-care hospital in Norway serving about 625 000 people (>10 % of the Norwegian population). The hospital has since 2000 performed molecular outbreak investigations, including genotyping of S. aureus and MRSA from the hospital and collaborating healthcare institutions. From this capacity, several thousand of S. aureus isolates have been collected from carriers, infections and outbreaks. In this study, a total of 42 MRSA isolates were included after careful selection from known, well-characterized outbreaks in the hospital and nearby long-term care institutions, as well as sporadic cases from Ahus’ catchment area. They represent both conserved clones in Norway and internationally (ST8-SCCmecIV-t304-PVL negative, ST772- SCCmecV-t657) [3, 4] as well as common clones in Norway (spa types t002 and t223) (Table 1). The study included isolates ranging a time span of 11 years (2004–2014). An outbreak was defined as two or more cases epidemiologically linked through individual, time and space, and genotypically linked based on genotyping methods at hand at the time of the original outbreak investigation, such as spa typing, MLST, MLVA and PFGE. A sporadic case was defined as having no genotypical and/or epidemiological link to any known outbreak. All bacterial isolates came from diagnostic or screening samples and they were originally cultured as part of routine diagnostics at the hospital.
Table 1.
Epidemiological and genotyping data on 42 MRSA isolates from known, well-characterized outbreaks as well as sporadic cases. The collection date is represented by the month and year of the bacterial isolation from the clinical sample. The outbreak ID describes isolates selected among outbreaks (O) and sporadic cases (S) with the following number (1, 2, 3 or 4) representing the lineage (according to the previous genotyping methods; fourth column) and the letters (A, B, C, D or E) representing different outbreaks
|
Isolate |
Collection date |
Outbreak ID |
Lineage |
|---|---|---|---|
|
MRSA_01 |
Feb-2004 |
O-1A |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_02 |
Aug-2004 |
O-1B |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_03 |
Oct-2004 |
O-1B |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_04 |
Apr-2005 |
O-1E |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_05 |
Jun-2007 |
O-1C |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_06 |
Jun-2007 |
O-1C |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_07 |
Jun-2007 |
O-1C |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_08 |
Jul-2007 |
O-1C |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_09 |
Jul-2007 |
O-1C |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_10 |
Aug-2010 |
O-1D |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_11 |
Aug-2010 |
O-1D |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_12 |
Nov-2012 |
S-1 |
ST8-t304-SCCmecIV-PVLneg (lineage 1) |
|
MRSA_13 |
Aug-2010 |
O-2D |
t223-PVLneg (lineage 2) |
|
MRSA_14 |
Dec-2010 |
O-2E |
t223-PVLneg (lineage 2) |
|
MRSA_15 |
Mar-2011 |
O-2D |
t223-PVLneg (lineage 2) |
|
MRSA_16 |
Jul-2011 |
O-2C |
t223-PVLneg (lineage 2) |
|
MRSA_17 |
Feb-2012 |
O-2B |
t223-PVLneg (lineage 2) |
|
MRSA_18 |
Mar-2012 |
O-2B |
t223-PVLneg (lineage 2) |
|
MRSA_19 |
Jun-2012 |
O-2C |
t223-PVLneg (lineage 2) |
|
MRSA_20 |
Aug-2012 |
O-2A |
t223-PVLneg (lineage 2) |
|
MRSA_21 |
Aug-2012 |
O-2A |
t223-PVLneg (lineage 2) |
|
MRSA_22 |
Oct-2006 |
O-3B |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_23 |
Nov-2006 |
O-3B |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_24 |
Dec-2006 |
O-3G |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_25 |
Jan-2007 |
O-3G |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_26 |
Jan-2007 |
O-3G |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_27 |
Sep-2007 |
O-3F |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_28 |
Sep-2007 |
O-3F |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_29 |
Nov-2007 |
O-3E |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_30 |
Nov-2007 |
O-3E |
ST5-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_31 |
Dec-2009 |
O-3D |
ST1637-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_32 |
Dec-2009 |
O-3D |
ST1637-t002-SCCmecIV-PVLneg (lineage 3) |
|
MRSA_33 |
Jan-2011 |
O-3C |
t002-PVLneg (lineage 3) |
|
MRSA_34 |
Jan-2011 |
O-3C |
t002-PVLneg (lineage 3) |
|
MRSA_35 |
Oct-2011 |
O-3A |
t002-PVLneg (lineage 3) |
|
MRSA_36 |
Oct-2011 |
O-3A |
t002-PVLneg (lineage 3) |
|
MRSA_37 |
Oct-2011 |
O-3A |
t002-PVLneg (lineage 3) |
|
MRSA_38 |
Oct-2011 |
O-3A |
t002-PVLneg (lineage 3) |
|
MRSA_39 |
Oct-04 |
S-4 |
ST772-t657-SCCmecV-PVLpos (lineage 4) |
|
MRSA_40 |
Aug-13 |
S-4 |
ST772-t657-SCCmecV-PVLpos (lineage 4) |
|
MRSA_41 |
Dec-13 |
O-4A |
ST772-t657-SCCmecV-PVLpos (lineage 4) |
|
MRSA_42 |
Jan-14 |
O-4B |
ST772-t657-SCCmecV-PVLpos (lineage 4) |
ST, sequence type; t, spa-typing; PVLneg/pos, isolate negative or positive for the genes encoding the Panton-Valentine leukocidin.
Genomic DNA isolation was performed using the PureLink genomic DNA kit (Thermo Fisher Scientific, MA, USA) according to the manufacturer’s recommendations with the following modifications: for bacterial cell lysis, 5 mg ml−1 of lysostaphin (Sigma-Aldrich, St Louis; MO, USA) in PBS was used and the cell-pellet incubation at 37 °C for 45 min. DNA yield was quantified by NanoDrop spectrophotometer or Qubit fluorometer (Thermo Fisher Scientific).
ONT library preparation and sequencing
ONT sequencing libraries were prepared by multiplexing the DNA from six MRSA isolates per flow cell using the Rapid Barcoding Sequencing kit (SQK-RBK004; ONT, Oxford, UK). The protocol (version RBK_9054_v2_revJ_14Aug2019) from the manufacturer was followed, apart from using the double of input DNA (800 ng) and therefore double of the kit components during the barcoding step. Sequencing libraries were loaded onto a FLO-MIN106 R9.4.1 SpotON flow cell and sequenced in the GridION X5 Mk1 sequencing device (ONT). Primary acquisition of data and real-time basecalling was carried out using the graphical user interface MinKNOW v2.0 and Guppy basecaller v3.0.6 (both from ONT). The demultiplexing of barcodes and quality control of the reads were also accomplished in real-time using EPI2ME platform (ONT). All quality reads (quality score above 7) were extracted after 20 h of the sequencing run for downstream analysis.
Protocol for real-time surveillance and outbreak investigations
Unless stated otherwise, the bioinformatics tools were used with default parameter settings. Multilocus sequence-type (ST) prediction from uncorrected long reads was performed with Krocus v 0.2.3 [17]. Assembly was performed using Flye v.2.7.1 [18] to a minimum coverage of 30X and Bandage v.0.8.1 [19] was used to visualize the assemblies. The draft genomes were then submitted to polishing using two iterations of Medaka v.1.0.1 [20]. Exact commands for Flye and Medaka are given in the Table S1 (available in the online version of this article). The polished fasta files were analysed with realphy v. 1.12 [21]. The following reference genomes were used individually on realphy according to the corresponding lineage from the isolates: USA300_TCH1516 (for isolates ST8) (GenBank accession number: CP000730), HO-5096–0412 (isolates ST22) (GenBank accession number: HE681097), CHU15-056 (isolates spa type t002) (GenBank: CP021171) and DAR4145 (isolates ST772) (GenBank: NZ_CP010526). The output from realphy generated a file corresponding to the phylogenetic tree, which was visualized using FigTree v1.4.4 [22]. To compute pairwise distances (number of SNPs between the genomes), the realphy output file ‘polymorphisms_move.fas’ was analysed on Geneious Prime software v2019.2.3 [23]. All cited tools, except for Geneious Prime, are freeware. All cited tools, except from realphy, were run on Ubuntu 16.04, which contained 28 vCPU and 64 Gb of RAM.
Method validation
For validation purposes, all MRSA isolates were also sequenced by Illumina technology (Illumina, San Diego, USA). The sequencing libraries were accomplished using Nextera XT DNA Library Preparation Kit (Illumina), according to the instructions of the manufacturer except from the following modifications: half of the recommended quantity of the working volumes and library amplification and clean up were used [24]. KK4835 KAPA Library Quantification Kit qPCR (Roche, Rotkreuz, Switzerland) was performed to quantification and normalization of libraries. The whole-genome sequencing was carried out on the Illumina’s MiSeq platform and MiSeq v3 Reagent Kit (Illumina) for 56 h. The generated sequencing data were subjected to quality control using FastQC [25]. The trimming and de novo assembly were carried out on Geneious Prime software v2019.2.3 using BBDuk trimmer v1.0 and SPAdes v3.13.0, respectively, to a minimum coverage of 30X. Multilocus sequence-type identification was performed on the assemblies with MLST 2.0 v 2.0.4 from Center for Genomic Epidemiology; CGE [26]. Further analyses were conducted following the same protocol described for nanopore-based data, excluding the polishing step.
To observe and control for a potential impact of recombination on the phylogenetic analysis performed with realphy, the isolate assemblies were analysed with Snippy [27]. Snippy is a command-line tool for identification of core genome polymorphic sites. It uses Gubbins [28] to predict genomic locations affected by recombination and removes the polymorphic sites present in the recombination regions from the further analysis. The phylogenetic trees have been visualized with ggtree [29].
Real-time outbreak investigation
During the study, the hospital had an ongoing outbreak investigation on methicillin sensitive S. aureus (MSSA) in the newborn intensive care unit. Six MSSA isolates were included to test our nanopore-based DNA sequencing protocol in real-time investigation. The DNA extraction, ONT-based DNA sequencing and further analysis were performed using the same protocol described for MRSA isolates. For the phylogenetic analysis, we used as reference genome the EDCC5464 strain (ST22) (GenBank accession number: NZ_CP022291.1). For comparison purposes, the six MSSA isolates were also analysed by the conventional molecular methods spa-typing [30] and MLVA [31], as previously described.
Results
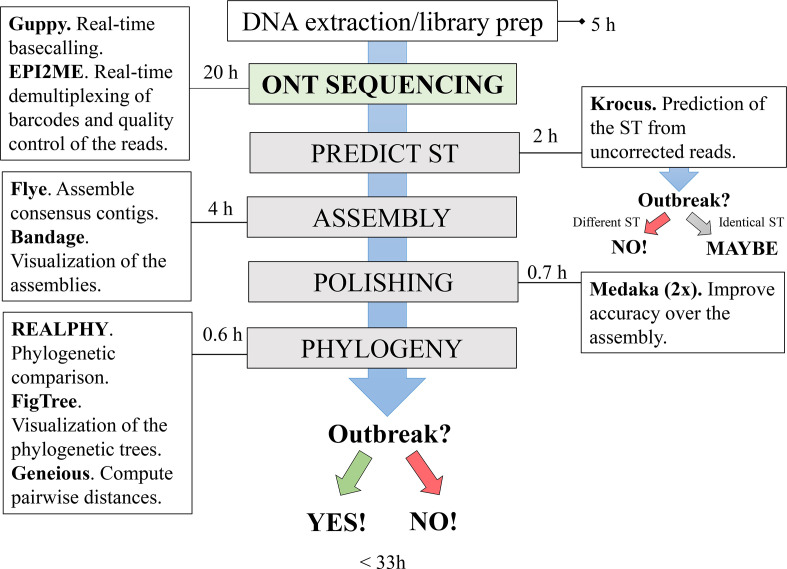
A summary of the developed protocol with all required software and the average time per analysis is presented in Fig. 1.
Fig. 1.
Summary of the rapid nanopore-based DNA sequencing protocol for real-time surveillance and outbreak investigations of antimicrobial-resistant bacteria. Adapted from Salazar and co-workers [37]. The estimated time per analysis was based on the average time among all sequencing runs considering six MRSA isolates per run. The DNA extraction, the preparation of the library and the load of the flow cell take approximately 5 h. After 20 h of the sequencing run on ONT device (which includes real-time basecalling and demultiplexing of the barcodes), Krocus [17] is used to predict the ST (sequence type) from the long reads. Obtaining different STs among the isolates may indicate that they are not part of the same outbreak. However, acquiring the same ST for two or more isolates indicates that the outbreak has to be confirmed or discarded following the next steps of the protocol. The genomes are assembled in 4 h using Flye [18] and visualized using Bandage [19]. Then the drafts genomes are submitted to polishing using two iterations of Medaka [20]. After approximately 0.7 h, the polished genomes are uploaded in realphy [21] to run the phylogenetic analysis, which takes 0.6 h to finish, including both visualization of the phylogenetic tree (FigTree; [22] and the computation of the pairwise distances (Geneious Prime software [23]). An outbreak can be confirmed or discarded in less than 33 h.
Based on the developed protocol and taking the previous epidemiological and genomic information from the isolates into account, the new definition of an outbreak is: two or more genomes that are linked through the epidemiological data (time and space), presenting the same ST and ≤22 SNPs' difference. Genomes that present between 23–30 SNPs' difference are classified as ‘closely related’ and, depending on epidemiological data, they can also be classified as part of the outbreak. A 31–60 SNPs' difference is classified as ‘possibly closely related’, which should not be classified as part of the same outbreak, unless the outbreak stretches over a long time period or includes a large number of individuals. Finally, the classification ‘sporadic isolates’ was applied when two isolates have more than 61 SNPs' difference. This classification was based on the analysis by Cunningham and co-workers [32] with threshold modifications according to our data.
Overview of the sequenced data
Considering six S. aureus isolates per analysis/run, the estimated time for DNA extraction was 3 h and for ONT library preparation was 2 h. For nanopore-based DNA sequencing, 20 h of sequencing run was sufficient to achieve enough genome coverage for further analysis. Considering all the 42 isolates, 1.0 Gb of yield (on average; ranging from 352 Mb to 2.7 Gb) and 3.9 Kb of reads length (on average; ranging from 1.3 to 8.4kb) were obtained. The quality score was 12.9 (on average; ranging from 11.7 to 13.6) (Table S2).
ONT-based protocol
All the following estimated times described here were calculated considering the average time for six MRSA isolates. Krocus was able to identify the ST directly from uncorrected long reads for all the 42 MRSA isolates (100 %; Table S2) in accordance to the previous molecular characterization (Table 1). After the sequencing run, it was possible to identify the ST in 2 h (20 min per isolate). Assemblies were achieved after 4 h (ranging from 13 min to 1 h 30 min; 40 min per isolate on average) and the assembler managed to create circular contigs for all the isolates. The average of coverage was 344.3X (ranging from 115 to 946X). All information regarding the assemblies is presented in Table S2. The polishing was carried out in 7 min per isolate (approximately 42 min for the six isolates, varying from 4 to 11 min for the two iterations). Although not part of our protocol, we additionally tested spa-Typer [33] using WGS data to identify the spa-type. All the MRSA genomes perfectly match the spa type previously genotyped (100 % of the isolates; Tables 1 and S2).
Phylogenetic comparison
Phylogenetic analysis was performed using realphy pipeline comparing the genomes belonging to the same lineage (defined according to Table 1). Phylogenetic trees and pairwise SNP distances are depicted in Fig. 2. The results from realphy were compared to the epidemiological data to define whether an outbreak was identified or not. realphy took from 19 min (for three isolates, which is the minimum number of input files for the pipeline) to 1 h (17 isolates) to finish the analysis. For six MRSA isolates, it took an average time of 0.6 h to finish the run and to perform the SNP analysis. Altogether, all the steps of the protocol from DNA extraction to the outbreak confirmation took <33 h (six MRSA isolates).
Fig. 2.
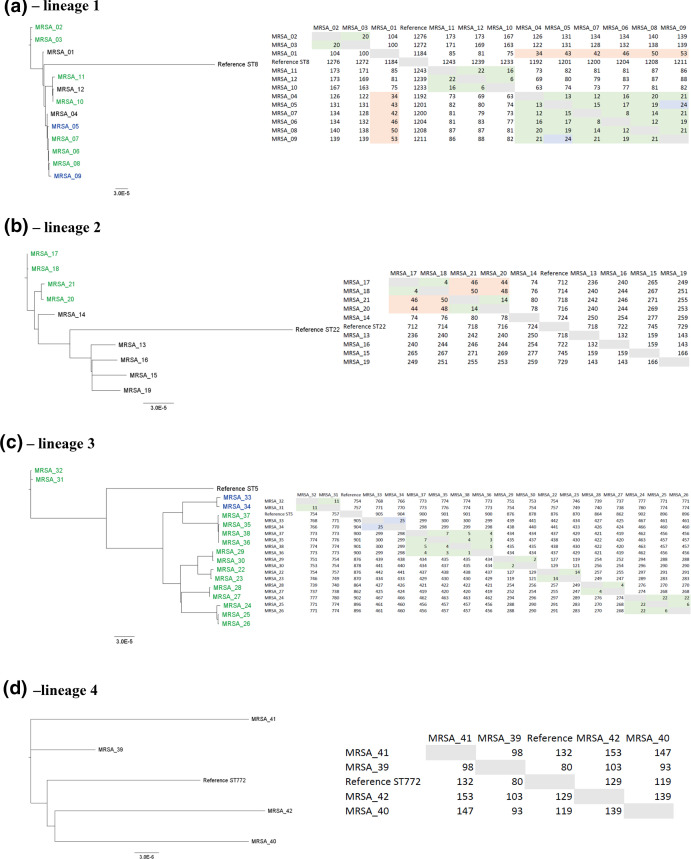
Phylogenetic comparison of the MRSA isolates using the nanopore-based protocol. The isolates were grouped by lineage (defined according to Table 1). Phylogenetic trees are shown on the left, while the table containing the pairwise distances (number of SNPs) between the isolates are shown on the right. The name of the isolates highlighted in green or blue were designated as part of outbreaks (left) defined after nanopore analysis. The numbers highlighted in green display≤22 SNPs, representing the isolates that were genotypically indistinguishable. The numbers highlighted in blue display 23–30 SNPs, representing samples defined as genotypically closely related. The numbers highlighted in orange (31–60 SNPs) represent isolates defined as possibly closely related. Finally, names and numbers not highlighted represent sporadic isolates. All the phylogenetic trees and SNP analysis were carried out using realphy online pipeline [21].
The analysis of the isolates belonging to lineage 1 (12 isolates) revealed three different outbreaks (Fig. 2a): the first one in 2004 involving two isolates (MRSA_02 and MRSA_03), the second one in 2007 (five isolates: MRSA_05 to MRSA_09) and a third one in 2010 (two isolates; MRSA_10 and MRSA_11). These are in concordance with previous outbreak demarcation (Table 1) apart from the isolates MRSA_05 and MRSA_09 from the second outbreak (Fig. 2a; highlighted in blue), which here were classified as closely related. However, due to the epidemiological link, and because they were both very similar to the other isolates included in the outbreak, these isolates were also classified as belonging to the same outbreak. Although the isolate MRSA_12 is genotypically indistinguishable from the other isolates from the third outbreak, there is no known epidemiological link among them (outbreak ID S-1; Table 1). A similar situation was observed for the isolate MRSA_04, which was classified as genotypically indistinguishable from the others isolates from the second outbreak (Fig. 2a), without any known epidemiological link (Table 1; outbreak ID O-1E). Additionally, although the isolate MRSA_01 was classified as sporadic in both nanopore-based protocol and previous genotyping methods, it was considered as possible closely related to the isolate MRSA_04 and to the isolates from the second outbreak (Fig. 2a; highlighted in orange on the right).
Regarding lineage 2 (nine isolates), two different outbreaks that occurred in 2012 were identified (Fig. 2b): one outbreak related to the isolates MRSA_17 and MRSA_18 and the other related to the isolates MRSA_20 and MRSA_21. These results are in concordance to what was previously observed from conventional genotyping (Table 1; outbreak ID O-2A and O-2B). Additionally, the four isolates related to these two outbreaks were classified as possibly closely related (Fig. 2b; highlighted in orange). Furthermore, the previous investigation using more limited methods had classified the isolates MRSA_16 and MRSA_19 as well as the isolates MRSA_13 and MRSA_15 as part of two different outbreaks (outbreak ID O-2C and O-2D in Table 1, respectively), which was not confirmed by our protocol (Fig. 2b).
Considering all isolates belonging to lineage 3, our protocol detected seven different outbreaks (Fig. 2c; highlighted in green and blue) in 2006, 2007, 2009 and 2011, which agrees with the previous genotyping (Table 1; outbreak ID O-3A to O-3G). Although the isolates MRSA_33 and MRSA_34 exhibited a 25 SNP difference (Fig. 2c; highlighted in blue), they could be linked to the same outbreak through the epidemiological data (Table 1; outbreak ID O-3C). Interestingly, the isolates MRSA_25 and MRSA_26, which were isolates with different phenotypical resistance patterns collected from one patient sample, exhibited only 6 SNP difference. Finally, among the four isolates belonging to lineage 4, an outbreak was not identified (Fig. 2d), corroborating with the previous outbreak investigation using the conventional methods [Table 1; two isolates sporadic (outbreak ID S-4) and two belonging to two different outbreaks (outbreak ID O-4A and O-4B)].
Illumina validation of the protocol
For Illumina sequencing data an average of 106.8X of coverage (ranging from 50.3X to 143.3X) was obtained when considering all the isolates. MLST 2.0 was able to correctly identify the ST from MiSeq data for all the 42 isolates (100%). Additional information can be found in Table S2 (second sheet).
Illumina MiSeq data agreed with the classification used to confirm or discard an outbreak in the nanopore-based protocol (Fig. S1). However, when comparing the results from ONT and MiSeq, minor differences among the numbers of SNPs were observed in some occasions. Although the isolates MRSA_05 and MRSA_09 (both from lineage 1) and MRSA_33 and MRSA_34 (both from lineage 3) were classified as part of two distinct outbreaks in both sequencing methods, ONT classified them as closely related (between 23–30 SNPs; highlighted in blue in Fig. 2) while MiSeq classified them as indistinguishable (≤22 SNPs; highlighted in green in Fig. S1). Similarly, the isolate MRSA_01 (lineage 1) was classified by ONT as sporadic (≥61 SNPs; not highlighted in Fig. 2a) and by MiSeq as only possibly closely related to MRSA_02–09 (between 31–60 SNPs; highlighted in orange in Fig. S1a), concluding that in both sequencing technologies, this isolate is not part of any outbreak.
Validation of the phylogenetic analysis
realphy phylogenetic analyses were compared with analyses performed with Snippy. One of the main differences between the two approaches is that Snippy explicitly predicts recombination-affected locations. However, the results of the two approaches seem almost identical and likely suggest one of the following: (1) the majority of the recombination-affected SNPs were excluded by realphy prior to the final phylogenetic analysis (possibly for not fulfilling some other filtering criteria) and thus did not have an impact on the results, or (2) even when a certain proportion of recombination-affected SNPs have been present in realphy analysis, these SNPs did not affect the observed signal in the data.
The numbers of SNPs identified with both data analysis approaches for each lineage and both sequencing platforms are presented in Table 2. From the numbers for lineage 4, realphy identifies ~20 % more SNPs than Snippy even when (almost) no recombination-affected SNPs are identified. Taking also into account that the phylogenetic trees constructed with the Snippy pipeline are almost identical to the realphy phylogenetic trees (compare Figs S2 and S3 with Figs 2 and S1, respectively), it is likely that the recombination-affected SNPs do not affect the realphy analysis heavily.
Table 2.
Overview of SNP counts for all analysed lineages, both sequencing platforms, and both phylogenetic analyses (realphy and Snippy). The last column of this table contains additional SNPs identified by Snippy as SNPs located within recombination-affected regions
|
Lineage |
Sequencing platform |
realphy SNP counts |
Snippy SNP counts |
Snippy SNPs in recombination regions |
|---|---|---|---|---|
|
Lineage 1 |
Illumina |
1412 |
601 |
1011 |
|
Lineage 1 |
ONT |
1411 |
651 |
926 |
|
Lineage 2 |
Illumina |
1173 |
858 |
2909 |
|
Lineage 2 |
ONT |
1144 |
891 |
2957 |
|
Lineage 3 |
Illumina |
1978 |
1504 |
3484 |
|
Lineage 3 |
ONT |
1939 |
1538 |
3460 |
|
Lineage 4 |
Illumina |
283 |
217 |
0 |
|
Lineage 4 |
ONT |
351 |
308 |
6 |
Real-time MSSA outbreak investigation
All reads from the six MSSA isolates were extracted after 20 h of the sequencing run for downstream analysis (details in Table S2; third sheet). Krocus was able to identify the STs in approximately 1.5 h (Table 3). The assembly of the genomes was then performed in 3 h and the polishing in 37 min. realphy took 0.6 h to run and the phylogenetic analysis matched the MLVA results (Fig. 3) implying two outbreaks and two singletons. spa-type using the conventional molecular method identified two isolates as t4565, two isolates as t712, one isolate as t015 and one isolate presenting a new spa-type. By comparison, spa-Typer using WGS data identified the same spa-types as the conventional method (Table 3). The confirmation of two different outbreaks at the hospital ward was confirmed in <31 h using the suggested protocol.
Table 3.
Comparison of conventional genotyping methods and the nanopore-based protocol on a real-time MSSA outbreak investigation. The uppercase letters (A–D) represent the different branches of the phylogenetic trees from Fig. 3a, b represent two separate outbreaks while C and D represent sporadic isolates
|
Conventional genotyping methods |
Nanopore-based protocol |
||||
|---|---|---|---|---|---|
|
Isolate |
spa type |
Outbreak (MLVA) |
ST (Krocus) |
spa type |
Outbreak (realphy) |
|
MSSA_01 |
t4565 |
A |
ST22 |
t4565 |
A |
|
MSSA_02 |
t4565 |
A |
ST22 |
t4565 |
A |
|
MSSA_03 |
t712 |
B |
ST22 |
t712 |
B |
|
MSSA_04 |
t712 |
B |
ST22 |
t712 |
B |
|
MSSA_05 |
t015 |
C |
ST45 |
t015 |
C |
|
MSSA_06 |
nd |
D |
ST101 |
nd |
D |
nd, not determined.
Fig. 3.
Phylogenetic comparison of six MSSA isolates from a suspected outbreak. The MSSA isolates were analysed by both conventional genotyping methods [(a) MLVA results] and by the suggested nanopore-based protocol [(b) realphy results]. Reference strain EDCC5464 (ST22) was used for realphy analysis. Both in (a) and (b), the uppercase letters A and B represent two separated outbreaks while C and D represent sporadic isolates. (c) Represents the pairwise distance analysis showing the number of SNPs between the isolates. Numbers and names highlighted in green represent the threshold to characterize an outbreak (≤22 SNPs).
Discussion
Our proposed protocol can predict ST after 27 h of receiving the bacterial isolates and confirm/refute an outbreak in <33 h. The resolution level was similar to Illumina sequencing, the current gold standard in NGS analysis, and equal or higher than conventional genotyping methods. The protocol can be used in resource-limited settings without a high bioinformatic knowledge. The applicability was shown in a real-time outbreak investigation revealing two different outbreaks in the same hospital ward.
The proposed protocol shows to be very time-efficient and to render results as expected. Using Krocus directly on nanopore raw reads allowed for ST identification within minutes (on average of 20 min per isolate). Obtaining different STs among the isolates may indicate that the isolates are not related to an outbreak. However, acquiring the same ST for two or more isolates indicates that the outbreak has to be confirmed or discarded following the next steps of the protocol. This fast identification may help discarding an outbreak only 2 h after the genome sequencing or during the first 27 h after the bacteria identification when considering for six MRSA isolates (3 h for DNA extraction, 2 h for library preparation, 20 h for DNA sequencing and 2 h for ST identification). These times will vary depending on the number of isolates per sequencing run. The assembly can be done on an average of 40 min per isolate using Flye de novo assembler, which is reliable and faster when compared to other long-read assemblers [34]. Since ONT has still higher sequencing error rates (4–20 %) compared to other NGS platforms [7], the polishing of the assemblies is a required step to increase accuracy. For MRSA isolates, two iterations of Medaka (7 min per isolate in average) was enough to improve the assemblies and to perform the phylogenetic analysis. The computational environment (performance) may impact the estimated times reported here for the assemblies and polishing since we used 20 cores (-t 20) to perform both steps (Table S1).
In the phylogenetic analysis using realphy, it was possible to confirm previous outbreaks in less than 33 h. realphy was chosen since the pipeline infers phylogenetic trees from WGS data using a simple and online approach. It also runs in Windows operational system. Therefore, this pipeline is easier to non-bioinformaticians to use, thus facilitating the outbreak analysis in more facilities. Compared to other online pipelines (e.g. CSI phylogeny from CGE; 35) realphy showed better discrimination of the strains in terms of SNP distances in both Illumina MiSeq and Oxford Nanopore platforms´ data (data not showed).
The suggested protocol could confirm most of the outbreaks and sporadic isolates identified by the conventional genotyping. However, these conventional methods identified two outbreaks related to lineage 2 (outbreaks ID O-2C and O-2D) that were, in fact, sporadic cases (Fig. 2b). Since isolates from the same clone can be very conserved, this inconsistency is probably related to the low discriminatory power of conventional genotyping methods compared to ONT sequencing, once again corroborating to the higher resolution of NGS. Interestingly, according to the suggested protocol, two isolates belonging to lineage 1 (MRSA_04 and MRSA_12) were genetically indistinguishable from other isolates of the same lineage without a known epidemiological link. This could be due to low diversity of the isolates belonging to the same lineage, or because there were possible transmission links between the infected patients, which the infection control staff had failed to discover during the initial investigations. Therefore, it is especially important to always consider all the epidemiological information to draw conclusions regarding outbreaks, and to perform quick genotyping to spur the detection of possible transmission links that are not always evident.
The result of ONT vs Illumina regarding the phylogenetic analysis were not completely overlapping, but the overall results showed excellent discriminatory power. The incompatibilities observed when comparing the results from the two sequencing platforms could be related to discrepancies originated during ONT basecalling, which may be improved using more recent versions of Guppy basecaller (as Guppy 3.6 or above) and flow cells (as R10.3). Recently, Greig and co-workers [36] compare Illumina (HiSeq) and ONT sequencing data from two isolates of Shiga toxin-producing Escherichia coli (STEC) O157:H7 to determine whether inference of relatedness based on single-nucleotide variants was consistent with the two technologies. Using ONT, the two isolates were unambiguously identified in <15 h and it was possible to distinguish the genetic relatedness between the isolates in approximately 6 h, whereas using the Illumina workflow the time from DNA extraction to sequencing and analysis was 3–6 days. After optimization for the ONT variant filtering, they observed few discrepant variants (six and seven difference for the two isolates) identified by the two technologies, however both technologies conclude that the isolates originated from different sources without an epidemiological link, similarly to the present study. Comparing to our approach, the authors were able to give faster answers by using only two isolates and running Krocus and serotyping during the sequencing run. Those analysis in real-time can also be implemented in the suggested protocol.
Although the Illumina platform is still the gold standard for NGS, this technology requires expensive equipment, numerous bacterial isolates per run for high-performance and it takes between 1 to 2 days for the sequencing to be available for analysis [7]. As mentioned earlier, the advantages of nanopore long-read sequencing are numerous, including time and the ease of execution and analysis. However, although the cost for setting up ONT sequencing facilities are expected to be low, the costs related to consumables are considerable and hence a factor that may limit the use of this method [7].
In the MSSA outbreak investigation, the results from the protocol perfectly matched the results from the conventional genotyping. Although MLVA and spa type are cheaper and take around the same time as our protocol to finish (1.5 working days), their resolution level may be suboptimal [2–4]. By confirming an outbreak, the potential reservoirs and the transmission dynamics may be identified and, more importantly, targeted preventive measures to contain the spread of the bacteria can be implemented.
Despite the advantages of the suggested protocol, the main limitation of the study included testing only S. aureus isolates from Norway. Although the protocol could be suitable for outbreak investigations related to other antibiotic-resistant bacteria from other world settings, the estimated times to run each step/pipeline and the thresholds established to define an outbreak may vary. Additionally, phylogenetic analyses were done on the different lineages separately, which would not mimic a true outbreak investigation. However, when testing only epidemiologically linked isolates without considering the ST, the same classification to define the outbreak was maintained, although in some cases a small variation of one to two additional SNPs was observed when analysing samples of different STs together (results not showed). Therefore, in order to better discriminate genetically conserved isolates, it is suggested analysing only isolates from the same ST at time using the accordant reference.
In conclusion, the suggested protocol will be especially valuable to identify outbreaks and their dynamics/source(s) in early stages allowing for implementation of immediate actions to contain the spread of the antibiotic-resistant bacteria. Moreover, since the portable devices from ONT have a low capital cost and a relatively user-friendly bioinformatics, it can also be implemented in small laboratories without sequencing facilities and high bioinformatic competence, thus enabling the investigation of outbreaks locally. Therefore, this protocol has the potential to be incorporated in routine surveillance analysis workflows. Beyond the outbreak investigations, further analysis of the generated genome sequences may include identification of antimicrobial-resistance and virulence-related genes, which can be used to improve the characterization of human pathogens.
Supplementary Data
Funding information
This study was funded by the the Norwegian Surveillance Programme for Antimicrobial Resistance (NORM) and strategic funding from Akershus University Hospital (No. 268902).
Acknowledgements
We thank the staff of the Department of Clinical Microbiology and Infection Control at Akershus University Hospital for their cooperation in sample collection and outbreak surveillance.
Author contributions
This study was conceptualized by H.V.A. and S.B.J.; H.V.A. acquired the financial support for the project leading to this publication; S.B.J. headed the outbreak surveillance at the hospital; K.H. performed laboratory culture, DNA extractions and Illumina sequencing with help from F.A.F. Formal analyses were performed by F.A.F. and H.V.A. Investigation and methodology was designed by F.A.F. and H.V.A. with T.V. supporting the bioinformatic analysis. F.A.F. wrote the original draft of the manuscript with input from H.V.A., S.B.J. and T.V. All authors reviewed and approved the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The study was approved by the local Data Protection Official at Akershus University Hospital (2017_147).
Footnotes
Abbreviations: MLST, multilocus sequence typing; MLVA, multiple locus variable-number tandem repeat analysis; MRSA, methicillin-resistant Staphylococcus aureus; NGS, next-generation sequencing; ONT, Oxford Nanopore Technologies; PFGE, pulsed-field gel electrophoresis; SNP, single nucleotide polymorphism; spa typing, S. aureus protein A typing; ST, sequence type; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary tables and three supplementary figures are available with the online version of this article.
References
- 1.Quainoo S, Coolen JPM, van Hijum SAFT, Huynen MA, Melchers WJG, et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev. 2017;30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fossum Moen AE, Holberg-Petersen M, Andresen LL, Blomfeldt A. Spa typing alone is not sufficient to demonstrate endemic establishment of meticillin-resistant Staphylococcus aureus in a low-prevalence country. J Hosp Infect. 2014;88:72–77. doi: 10.1016/j.jhin.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Blomfeldt A, Hasan AA, Aamot HV. Can MLVA differentiate among endemic-like MRSA isolates with identical Spa-Type in a low-prevalence region? PLoS One. 2016;11:e0148772. doi: 10.1371/journal.pone.0148772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomfeldt A, Larssen KW, Moghen A, Haugum K, Steen TW, et al. Bengal Bay clone ST772-MRSA-V outbreak: conserved clone causes investigation challenges. J Hosp Infect. 2017;95:253–258. doi: 10.1016/j.jhin.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Ahrenfeldt J, Skaarup C, Hasman H, Pedersen AG, Aarestrup FM, et al. Bacterial whole genome-based phylogeny: construction of a new benchmarking dataset and assessment of some existing methods. BMC Genomics. 2017;18:1–13. doi: 10.1186/s12864-016-3407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossen JWA, Friedrich AW, Moran-Gilad J, ESCMID Study Group for Genomic and Molecular Diagnostics (ESGMD Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect. 2018;24:355–360. doi: 10.1016/j.cmi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Berry M, I, Melendrez MC, Bishop-Lilly KA, Rutvisuttinunt W, Pollett S, et al. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: approaches, applications, and considerations for development of laboratory capacity. J Infect Dis. 2019;221:S292–307. doi: 10.1093/infdis/jiz286. [DOI] [PubMed] [Google Scholar]
- 8.Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, et al. Real-time, portable genome sequencing for Ebola surveillance . Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017;72:104–114. doi: 10.1093/jac/dkw397. [DOI] [PubMed] [Google Scholar]
- 10.Golparian D, Donà V, Sánchez-Busó L, Foerster S, Harris S, et al. Antimicrobial resistance prediction and phylogenetic analysis of Neisseria gonorrhoeae isolates using the Oxford nanopore MinION sequencer. Sci Rep. 2018;8:17596. doi: 10.1038/s41598-018-35750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Payne M, Octavia S, Luu LDW, Sotomayor-Castillo C, Wang Q, et al. Enhancing genomics-based outbreak detection of endemic Salmonellaenterica serovar Typhimurium using dynamic thresholds. Microb Genom. 2019 doi: 10.1099/mgen.0.000310. 04 Nov 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO Who publishes list of bacteria for which new antibiotics are urgently needed. Available from. [Dec 2;2020 ]; https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
- 13.Lee AS, de Lencastre H, Garau J, Kluytmans J, Malhotra-Kumar S, et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers. 2018;4:1–23. doi: 10.1038/nrdp.2018.33. [DOI] [PubMed] [Google Scholar]
- 14.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blomfeldt A, Larssen KW, Moghen A, Gabrielsen C, Elstrøm P, et al. Emerging multidrug-resistant Bengal Bay clone ST772-MRSA-V in Norway: molecular epidemiology 2004-2014. Eur J Clin Microbiol Infect Dis. 2017;36:1911–1921. doi: 10.1007/s10096-017-3014-8. [DOI] [PubMed] [Google Scholar]
- 16.Di Ruscio F, Guzzetta G, Bjørnholt JV, Leegaard TM, Moen AEF, et al. Quantifying the transmission dynamics of MRSA in the community and healthcare settings in a low-prevalence country. Proc Natl Acad Sci U S A. 2019;116:14599–14605. doi: 10.1073/pnas.1900959116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page AJ, Keane JA. Rapid multi-locus sequence typing direct from uncorrected long reads using Krocus . PeerJ. 2018;6:e5233. doi: 10.7717/peerj.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 19.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies: Fig. 1. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oxford Nanopore Technologies Medaka. Available from. [Dec 2;2020 ]; https://nanoporetech.github.io/medaka/
- 21.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol. 2014;31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rambaut, A FigTree. Available from. [Dec 2;2020 ]; https://github.com/rambaut/figtree
- 23.Geneious Geneious prime. Available from. [Dec 2;2020 ]; https://www.geneious.com/prime
- 24.Noone JC, Stegger M, Lilje B, Stavem K, Helmersen K, et al. Molecular characteristics of Staphylococcus aureus associated prosthetic joint infections after hip fractures treated with hemiarthroplasty: a retrospective genome-wide association study. Sci Rep. 2020;10:16553. doi: 10.1038/s41598-020-73736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babraham Bioinformatics FastQC. Available from. [Dec 2;2020 ]; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 26.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seemann T Snippy. Available from. [Dec 2;2020 ]; https://github.com/tseemann/snippy
- 28.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G. Using ggtree to visualize data on Tree-Like structures. Curr Protoc Bioinformatics. 2020;69:e96. doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
- 30.Fossum AE, Bukholm G. Increased incidence of methicillin-resistant Staphylococcus aureus ST80, novel ST125 and SCCmecIV in the south-eastern part of Norway during a 12-year period. Clin Microbiol Infect. 2006;12:627–633. doi: 10.1111/j.1469-0691.2006.01467.x. [DOI] [PubMed] [Google Scholar]
- 31.Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, et al. Multiple-Locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One. 2009;4:e5082. doi: 10.1371/journal.pone.0005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham SA, Chia N, Jeraldo PR, Quest DJ, Johnson JA, et al. Comparison of whole-genome sequencing methods for analysis of three methicillin-resistant Staphylococcus aureus outbreaks. J Clin Microbiol. 2017;55:1946–1953. doi: 10.1128/JCM.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartels MD, Petersen A, Worning P, Nielsen JB, Larner-Svensson H, et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2014;52:4305–4308. doi: 10.1128/JCM.01979-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wick RR, Holt KE. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Res. 2019;8:2138. doi: 10.12688/f1000research.21782.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greig DR, Jenkins C, Gharbia S, Dallman TJ. Comparison of single-nucleotide variants identified by illumina and Oxford nanopore technologies in the context of a potential outbreak of Shiga toxin–producing Escherichia coli. Gigascience. 2019;8:1–12. doi: 10.1093/gigascience/giz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salazar AN, Nobrega FL, Anyansi C, Aparicio-Maldonado C, Costa AR, et al. An educational guide for nanopore sequencing in the classroom. PLoS Comput Biol. 2020;16:e1007314. doi: 10.1371/journal.pcbi.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.