Abstract
Atrial fibrillation (AF) is a common cardiac arrhythmia leading to many adverse outcomes and increased mortality. Yet the molecular mechanisms underlying AF remain largely unknown. Recent advances in high-throughput technologies make large-scale molecular profiling possible. In the past decade, multiomics studies of AF have identified a number of potential biomarkers of AF. In this review, we focus on the studies of multiomics profiles with AF risk. We summarize recent advances in the discovery of novel biomarkers for AF through multiomics studies. We also discuss limitations and future directions in risk assessment and discovery of therapeutic targets for AF.
Keywords: Atrial fibrillation, Mechanism, Aetiology, Genomics, Multiomics
1. Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia with increasing prevalence and incidence globally.1–3 The lifetime risk of AF in individuals older than 55 years is ∼37%.4,5 It is estimated that ∼59.7 million people were affected by AF worldwide in 2019, about double the number in 1990. An additional five million individuals are diagnosed each year.1,6 The number of AF cases is estimated to reach 12.1 million by 2030 in USA alone.7 AF is associated with increased risk of many comorbidities, such as stroke,8 heart failure,9 myocardial infarction,10 dementia,11 as well as increased mortality.9,12,13 The adjusted annual incremental cost for individuals with AF is $18 601,14 resulting in an increase in the US health care costs of $28.4 billion (estimated for 2016).15 Therefore, it is important to identify new strategies to prevent AF.16
Many risk factors have been identified for AF,17–21 including advancing age, smoking, alcohol consumption, obesity, diabetes, elevated blood pressure, heart failure, and myocardial infarction. Risk scores, based on the epidemiologic cohorts’ data, have been developed to predict the 5- and 10-year risk of AF.22,23 However, the molecular mechanisms underlying AF pathogenesis are not yet fully understood, and therapies for AF are only partially effective with substantial morbidity.24,25
Recent advances in high-throughput technologies make large-scale molecular profiling possible. In the past decade, multiomics studies of AF have identified hundreds of potential biomarkers of AF. In this review, we focus on the studies of multiomics profiles with AF risk. In addition, we summarize recent advances in the discovery of novel biomarkers for AF through multiomics studies. We also discuss limitations and future directions in risk assessment and discovery of therapeutic targets for AF.
1.1 Search strategy
An electronic search for relevant publications was performed in the PubMed database. The major search terms included ‘atrial fibrillation OR AF’ AND ‘incident’ AND ‘biomarker’. Specific search terms included ‘genomic’ OR ‘transcriptomic’ OR ‘proteomic’ OR ‘metabolomic’. Two authors (J.K. and H.L) independently screened all retrieved records by titles and abstracts and then full-text articles. A detailed search strategy could be found in Supplementary material online, Table S1.
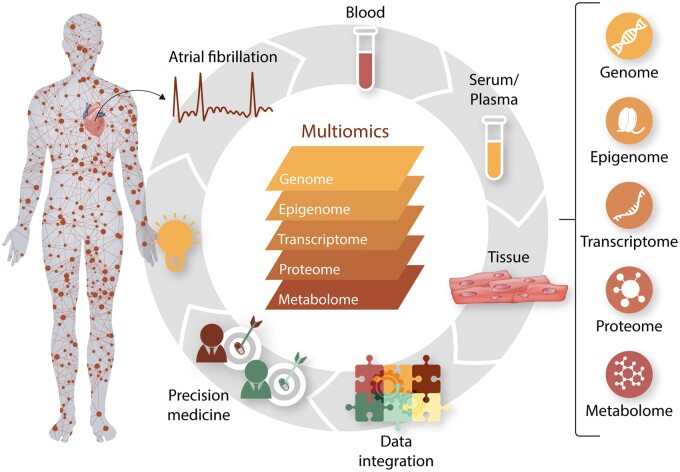
2. Multiomics study of AF
2.1 Genomics
2.1.1 Heritability
The familial nature of AF was first reported in 1936 with a case series noting in a footnote that three brothers were affected, all before the age of 30 years.26 Familial aggregation of AF was later validated in several other studies, suggesting that familial AF comprised up to 15% of lone AF cases.27,28 Fox et al.29 observed that approximately one-third of AF patients had at least one affected parent. Marcus et al.30 found that first-degree family history was associated with increased risk of lone AF. Christophersen et al.31 reported the importance of genetic heritability in same-sex twins and found that the co-twin of an AF-affected twin might be considered high risk for the development of AF. Lubitz et al.32 further validated the familial nature of AF and reported that having one first-degree family member was associated with a 40% increased risk. In addition, there was a 24% increased risk in AF per additional affected first-degree family member. A younger age of onset in familial members was also associated with increased risk of AF.32,33 Therefore, a better understanding of the genetic architecture of AF may pave the road for greater insight into AF pathogenesis.34
2.1.2 Candidate genes
The search for causal genes in AF started in the late 1990s.35,36 Since then, over 35 genes have been identified to associate with AF in familial studies.36–38 One example is SCN5A, which encodes a cardiac sodium channel essential to the cardiac action potential. SCN5A mutations were observed in up to 6% of AF patients, including lone AF and those with underlying cardiac co-morbidities.39 Makiyama et al.40 described a novel gain-of-function SCN5A mutation in familial AF.
Another example is GJA5, which encodes the connexin-40 responsible for atrial conduction. GJA5-knockout mice showed increased vulnerability to atrial arrhythmias.41,42 Loss-of-function variants in GJA5 inhibit cell–cell electrical coupling or gap-junction congregation at the surface.43 Chimeric mice with inconsistent expression of connexin-43 had conduction delays in their atrial myocardium, suggesting that mosaicism in the connexin tissue might be involved in arrhythmia initiation.44 However, most of the variants in candidate studies are rare, observed only in affected families, and replication efforts mostly failed.45 Hence, researchers have turned to comprehensive genome-wide screening of variants in large populations.
2.1.3 Genome-wide association studies
The most popular method to identify genetic variants for common diseases is the genome-wide association study (GWAS), which allows the comparison of allele frequency between disease cases and referents for each genetic variant [e.g. single-nucleotide polymorphisms (SNPs)]. The first GWAS of AF was performed in 2007 based on participants from Iceland, which identified the first common locus 4q25 for AF.46 Lubitz et al.47 later showed that at least four distinct AF susceptibility signals at the locus were associated with prevalent and incident AF. The closest gene at this locus is PITX2, which plays an important role defining right-left cardiac symmetry, developing pulmonary vein myocardium, and inhibiting sinus node formation in the left atrium.48,49 In animal models, PITX2c knockout-mice had shorter atrial refractory periods and were more predisposed to atrial arrhythmias.50 Using human left atrial samples, it had been shown that patients with AF have lower expression of the PITX2c isoform compared to non-AF individuals.51 In order to identify additional AF-related loci and since most genetic loci have small effects, the AFGen consortium was organized as an international effort for AF GWAS. The Consortium has led the field of AF genetics, describing the vast majority of genetic loci associated with AF in the past decade.47,52–56 The latest AF GWAS included over 65 000 AF cases from more than 50 cohorts, and identified 97 AF-related loci.57 Large biobanks, such as the UK BioBank, are also being utilized to identify additional genetic loci.58 Many of the genetic loci identified through GWAS implicated genes related to cardiopulmonary development, cardiac-expressed ion channels, channelopathies, and signalling molecules, emphasizing the polygenic nature of AF.59
Based on the GWAS results, a polygenic risk score (PRS) may be constructed that allows individualized projections of AF risk. PRSs represent the combined effect of multiple genetic variants on disease risk. Whereas individual SNPs carry relatively little effect (ranging from 0.07 to 0.3756), pooled SNPs register observable differences among individuals. By pooling SNPs to create the PRS, Weng et al.4 reported that individuals with the highest tertile PRS had 1.82-fold lifetime risk of those with lowest tertile PRS. Moreover, individuals with high PRS but low clinical risk were associated with a lifetime AF risk of 43.6%, which was comparable to those with high clinical risk.
Despite the remarkable success of GWAS, several challenges remain. Genetic variation accounted for 22.1% of variance in AF risk,60 but known AF-related loci may explain only a relatively small proportion of AF heritability, suggesting many more are yet to be identified. One possible reason for the missing heritability is unidentified rare variants with large effects compared to common genetic traits.61,62 Most of the GWAS analyses were conducted using microarray-based genotyping platforms, which have limited resolution to identify rare variants. In addition, the top variants from the GWAS are considered to tag, or serve as proxies, for the underlying functional variants. The functions of most GWAS loci are yet to be determined.63
2.1.4 Targeted sequencing and exome sequencing
The advance of next-generation sequencing allows the identification of rare variants in the general population. Lin et al.64 performed targeted sequencing on 77 GWAS loci and studied the association of both common and rare variants with AF. Rare damaging variants within PRRX1 were found to associate with AF. In addition, a common variant rs11265611 was found to associate with AF. This SNP is located in the first intron of IL6R, which codes the interleukin-6 receptor, a pro-inflammatory marker triggering acute-phase proteins.65,66 Inflammation is known as one of the causal pathways related to AF initiation and maintenance.67
Similar to targeted sequencing, exome sequencing aims to perform sequencing on the entire exome. Lubitz et al.68 performed exome sequencing on more than 1700 individuals with AF and 9000 controls from three cohorts. None of the rare variant regions were significant after adjusting for multiple testing, suggesting that rare coding variants may not be the predominant mechanism for AF in the community.
2.1.5 Whole-genome sequencing
As sequencing cost continues to decline, whole-genome sequencing is used with increasing frequency to identify genetic variants associated with complex disease. Several large-scale whole-genome sequencing projects have been initiated, including the Trans-Omics for Precision Medicine (TOPMed) project (https://www.nhlbiwgs.org/) that has generated variant calling from more than 200 K whole-genome sequencing samples. Whole-genome sequencing offers several advantages compared to target sequencing. First, whole-genome sequencing is able to cover the entire genome, which is particularly useful in identifying genetic loci that are located in non-coding regions. Second, since there is no requirement for the target region enrichment, whole-genome sequencing is capable of providing more consistent coverage. Moreover, whole-genome sequencing can also identify structural variants, which remain a challenge for targeted sequencing or exome sequencing.
Based on the TOPMed project, Choi et al.69 reported that loss-of-function variants in titin (TTN) were associated with early AF onset. The study included 2781 early-onset AF cases (<66 years) and 4959 referents. Titin is essential in myocardial function as a passive stabilizer of the sarcomere, suggesting that structural sarcomeric abnormalities might play a role in AF pathogenesis. Furthermore, the association with hypertrophic and dilated cardiomyopathies may explain the co-occurrence of AF in these patients.
2.2 Epigenomics
DNA methylation represents an important type of epigenetic modification without changing underlying DNA sequences. The methylation process adds a methyl group to the cytosine of cytosine-phosphate-guanine dinucleotides (CpG), and thus, modifies the conformational structure of chromatin and the binding of transcription factors. Different tissue types have different methylation states, and the methylation status may be modified by both genetic and environmental factors.70,71 Many AF risk factors such as obesity,72 smoking,73 inflammation,74 and alcohol consumption75 also alter methylation profiles. The first epigenome-wide association study of AF in the community included 183 prevalent AF cases, 220 incident AF cases, and 2236 referents from the Framingham Heart Study.76 Two CpG sites were significantly associated with prevalent AF, and five different CpG sites were associated with incident AF. One of the significant CpG sites was cg13639451, located upstream of WFIKKN2, which is known to be involved in the muscle fibre development and cardiac excitation-contraction coupling.77,78 Cg07191189 was another CpG site for prevalent AF. It is located near STRN that encodes striatin, which is able to bind with caveolin-134 and is known to be involved in cardiac development.79 Shen et al.80 studied the global DNA methylation in the right atrial myocardial tissue obtained from rheumatic valvular patients. From 10 AF patients and 10 referents, it was observed that AF patients tended to have higher global DNA methylation levels than referents (P < 0.05).
2.3 Transcriptomics
Gene expression is considered an intermediate phenotype between genetic variation and disease traits.81 Both animal models and human studies suggest that AF pathogenesis is accompanied by alterations in gene expression.82–84 Using whole blood transcriptomic profiles of individuals from the Framingham Heart Study, seven transcriptomic signatures were found to associate with prevalent AF.85 The most significant gene was PBX1 that has an important role in great-artery patterning, septation of outflow tract,86 and development of persistent truncus arteriosus.87 SLC7A1 was another significant gene, which has been related to endothelial dysfunction and hypertension.88 An AF-specific interaction network also was built and it was enriched with genes involved in multiple cardiovascular signalling pathways, such as the hypoxia signalling pathway responsible for oxygen deficit in cardiovascular organs, and the antiproliferative signalling pathway involved in the remodelling of cardiac myocardium.89,90 Of note, none of these genes were associated with incident AF, suggesting that different signalling pathways are responsible for AF onset and maintenance.
MicroRNAs (miRNAs) are a class of short non-coding RNAs that regulate mRNA expression. They increasingly have been recognized as potential biomarkers for cardiovascular disease, in part, due to their stability in peripheral circulation.91,92 Dawson et al.93 reported MiR-29 was a potential biomarker for AF. Liu et al.94 studied the association of plasma miRNAs with AF in a Chinese hospital-based cohort. Compared to referents, the expression of miRNA-150 was significantly down-regulated in paroxysmal AF and persistent AF patients. MiRNA-150 is involved in the regulation of cardiac fibrosis and cellular proliferation in myocardial infarction and heart failure.95 McManus et al.96 performed another study that included 153 prevalent AF cases, 107 incident AF cases, and 2185 referents from the Framingham Heart Study. The only miRNA associated with prevalent AF was miRNA-328. MiRNA-328 is known as an important gene regulator secreted by cardiomyocytes under stress.97 In an experimental model, the expression of miRNA-328 was up-regulated in the left atrial samples in animals with AF.98 As some circulating miRNAs are platelet-derived, anti-platelet medications might directly affect miRNA levels, and thus be important for AF patients with concurrent cardiovascular or cerebrovascular diseases with indication for anti-platelet treatment.99 The major part of miRNAs in plasma is localized in microparticles and originates in up to 45% from platelets.100 Furthermore, previous studies indicated an association between altered expression of miR-146b and the P38MAPK/COX-2 pathway, making a causal relationship between antiplatelet medication and Cox inhibition possible.101
In another study based on clinical samples, McManus et al.102 observed that circulating plasma expression of miRNA-21 and miRNA-150 were significantly associated with AF. Patients with persistent AF had lower miRNA-21 expression compared with paroxysmal AF, suggesting that higher AF burden could affect plasma miRNA expression. The miRNA-21 expression in the right atrium was down-regulated in patients undergoing cardiovascular surgery. In addition, a three-fold increase of miRNA-21 and miRNA-150 was observed after rhythm restoration in patients undergoing AF catheter ablation, suggesting significant dynamic changes after sinus rhythm restoration. Despite a relatively small study sample, this was the first study describing a strong association between circulating and tissue miRNAs linked to AF and significant improvement of miRNA levels after sinus rhythm restoration.102
2.4 Proteomics
Proteomics refers to simultaneous screening of large numbers of proteins as potential biomarkers for different diseases. Lind et al.103 used a custom-made proteomics chip to profile plasma proteins in participants from the Swedish study. Among the 92 screened proteins, 7 were significantly associated with incident AF after adjustment for age and sex. Two proteins, IL-6 and NT-proBNP, remained significant after adjusting for additional AF risk factors.103 Willeit et al.104 performed another proteomic study based on 880 participants from the Bruneck Study who were free of AF at the baseline. One hundred and seventeen participants developed AF during 20-year follow-up. Among the 13 inflammation markers measured at the baseline, sVCAM-1 was found to associate with incident AF after adjusting for age and sex. Ko et al.105 screened 1373 proteins by using the SOMAScan assay in the Framingham Heart Study and identified eight proteins associated with incident AF. Two of them, NT-proBNP and ADAMTS13 remained significant after adjusting for known risk factors for AF. NT-proBNP is a marker of myocardial stress and ventricular remodelling known to be associated with AF.106 ADAMTS13 is a protease of von Willebrand factor involved in left atrial remodelling,107 and it has been used as a biomarker of rhythm outcomes after cardioversion.108 In a recent study, Staerk et al.109 tested the association of 85 proteins with incident AF, which were measured using the Luminex xMAP platform. Higher levels of NT-proBNP and insulin-like growth factor-binding protein 1 as well as lower levels of insulin-like growth factor 1 were associated with increased risk of incident AF after adjusting for AF risk factors.
2.5 Metabolomics
Metabolomics are studies to perform systematic analyses of metabolites with diseases. Mayr et al.110 observed patients with persistent AF had elevated ketone body metabolism in the atria. De Souza et al.111 reported increased metabolic stress was associated with impaired energy utilization and increased ketoacid metabolism in a canine AF model. Alonso et al. performed the metabolomics profiling of blood samples from ∼1900 African American participants in the Atherosclerosis Risk in Communities (ARIC) study.112 The participants were followed over 20 years. Two bile acids (glycolithocholate sulfate and glycocholenate sulfate) were significantly associated with incident AF, suggesting a connection between liver function and AF, in agreement with previous studies.113,114 Potential arrhythmogenic effects of bile acids had been reported in humans,115 and elevated maternal bile acid levels during pregnancy were associated with cardiac arrhythmias in fetuses.116 In another study, Ko et al.117 analyzed liquid chromatography-tandem mass spectrometry metabolomics profiles in over 2000 individuals of European ancestry from the Framingham Heart Study; they did not reveal metabolites significantly associated with incident AF. In addition, neither bile acids identified in the ARIC study were replicated, possibly due to different ancestries, tissue types, or metabolic profiling platforms.118
3. Tissue consideration for multiomics
Unlike genetic profiles, other omics profiles, such as transcriptomic and epigenomic profiles, can change from one tissue to another. The most relevant tissue for AF is the left atrium. However, it is not feasible to perform invasive specimen collection on a large scale, especially in community-based cohorts. Most multiomics studies have measured molecular profiles from blood samples, which may be different from the heart samples. Furthermore, the left and right atria can have quite distinct expression patterns for some genes, resulting in different structure and function between these two chambers.119,120
One useful resource is the Genotype-Tissue Expression (GTEx) project, which quantified gene expression across 54 tissue types collected from 948 donors.121 Computational methods, such as PrediXcan,122 MetaXcan,123 RIVER,124 and ExPecto,125 are being developed to predict tissue-specific transcription profiles from genetic variants. For example, Roselli et al.57 performed a transcriptome-wide association study of AF using MetaXcan based on the GWAS summary association statistics. The pre-computed models were generated from the left and right atria from the GTEx project.121 The models predicted expression of 57 genes associated with AF, 42 of which were located in a single AF GWAS loci.
4. Data integration
As multiomics data become increasingly available, future studies might integrate different omics to obtain a more comprehensive picture of AF pathogenesis. Quantitative trait loci (QTL) analyses study the associations between genetic variants and other omics data.81 One study investigated the association between genetic variants and gene expression in the left and right atria.126 It identified 187 eQTLs from 53 left atrial samples and 259 eQTLs from 52 right atrial samples. Many of the eQTLs were shared between the left and right atrial samples, including rs3740293, which was one of the top AF-related variants. The SNP is located at the 3′-UTR of SYNPO2L, about 5 kb upstream of MYOZ1. It was significantly associated with the expression of MYOZ1 in both left and right atria, but not SYNPO2L, suggesting that the functional gene at this locus is likely to be MYOZ1 instead of SYNPO2L.126 The association was further validated using 70 additional samples collected from patients at Massachusetts General Hospital. In another study, the mQTL analyses were performed to investigate the association of AF-related variants with nearby DNA methylation.76 The most significant association was observed between rs6490029 at the CUX2 locus and cg10833066. The methylation of cg10833066 increased with increasing copies of the ‘A’ allele of rs6490029, which also was associated with higher AF risk as found in the previous GWAS.54 Further studies are needed to validate these results and apply QTL mapping to other AF-related electrophysiological traits like PR and RR intervals.127,128
Another approach to data integration is to perform multiomics modelling, which integrates the results from different omics association studies. Wang et al.129 recently developed a strategy to integrate the GWAS, the epigenome-wide association study, and the transcriptome-wide association study results for AF. The summary statistics from different omics data were meta-analyzed and weighted by the sample size of each omics data type. A tissue-specific network was then built130 and used to predict potential AF-related genes, which increased the proportion of heritability from 3.5% (by GWAS alone) to 10.4%. Importantly, a few potential drug targets were identified, including ADORA1, ATP1A3, ATP1B2, CACNA1D, KCNQ4, NR3C2, and THRA. Similarly, van Ouwerkerk et al.131 developed another approach to prioritize potentially functional variants by integrating transcriptomic and epigenomic data together with chromatin conformation information. A list of potentially target genes was identified from AF-associated variants, which could be further investigated for future studies.
5. Future directions
The summary of omics findings related to AF is presented in Tables 1 and 2 and Figure 1. Despite the success of omics study in AF, several aspects are important to consider for future studies.
Table 1.
Genomics association studies of AF
| Study | Population | Genes | Top variants |
|---|---|---|---|
| Gudbjartsson et al.46 |
Iceland AF n = 2.251 No AF n = 13.238 UK/US European ancestry AF n = 779 No AF n = 1.542 Chinese cohort AF n = 333 No AF n = 2.836 |
PITX2 |
rs2200733 rs10033464 |
| Benjamin et al.52 |
European ancestry Prevalent AF n = 896 Referents n = 15.768 Incident AF n = 2.517 Referents n = 21.337 Replication in German AFNET AF n = 2.145 Referents n = 4.073 |
MTHFR PITX2 ZFHX3 |
rs17375901 rs17042171 rs2106261 |
| Ellinor et al.54 |
European ancestry AF n = 6.707 Referents n = 52.426 Replication AF n = 5.381 Referents n = 10.030 Japanese cohort AF n = 843 Prevalent AF n = 3.350 |
KCNN3-PMVK PRRX1 PITX2 WNT8A CAV1 C9orf3 SYNPO2L SYNE2 HCN4 ZFHX3 |
rs6666258 rs3903239 rs6817105 rs2040862 rs3807989 rs10821415 rs10824026 rs1152591 rs7164883 rs2106261 |
| Lubitz et al.47 |
European ancestry n = 64 683 Prevalent AF n = 3.302 Incident AF n = 3.869 Japanese cohort n = 11 309 Prevalent AF n = 7916 |
KCNN3-PMVK PRRX1 PITX2 PITX2 PITX2 PITX2 CAV1 C9orf3 SYNPO2L SYNE2 HCN4 ZFHX3 |
rs6666258 rs3903239 rs1448818 rs6817105 rs4400058 rs6838973 rs3807989 rs10821415 rs10824026 rs1152591 rs7164883 rs2106261 |
| Sinner et al.55 |
European ancestry No AF n = 52.426 AF n = 6.707 Replication No AF n = 17.144 AF n = 6.691 Japanese cohort No AF n = 3.350 AF n = 843 Replication No AF n = 17.190 AF n = 7.530 |
NEURL TBX5 CAND2 GJA1 NEURL CUX2 |
rs12415501 rs10507248 rs4642101 rs13216675 rs6584555 rs6490029 |
| Christophersen et al.56 |
Multi-ancestry cohort GWAS AF n = 17 931 No AF n = 115 142 ExWAS and RVAS AF n = 22 346 No AF n = 132 086 |
rs72700118 rs3771537 rs2540949 rs2288327 rs337711 rs2967791 rs4946333 rs7508 rs35176054 rs75190942 rs6800541 rs89107 rs11047543 |
|
| Low et al.140 |
Japanese cohort AF n = 8180 Controls n = 28 612 Replication cohort: AF n = 3120 Controls n = 125 064 |
KCND3 PPFIA4 SLC1A4–CEP68 HAND2 HAND2 NEBL SH3PXD2A |
rs12044963 rs17461925 rs2540953 rs17059534 rs7698692 rs2296610 rs2047036 |
| Lee et al.141 |
Korean cohort AF n = 672 Controls n = 3700 Replication study AF n = 200 Controls n = 1812 |
PRRX1 PITX2 NEURL1 TBX5 ZFHX3 |
rs3903239 rs6817105 rs6584555 rs10507248 rs2106261 |
| Roselli et al.57 |
Multi-ancestry cohort AF n = 22 346 No AF n = 132 086 |
67 novel loci | |
| Nielsen et al.58 |
European ancestry AF n = 60 620 No AF n = 970 216 |
63 novel loci | |
| Choi et al.69 |
Lone AF n = 2781 No AF n = 4959 |
TTN |
OR (95% CI), P value 2.16 (1.34–3.48); 1.55×10−3 |
GWAS, genome-wide association study; ExWAS, exome-wide association study; RVAS, rare variant association study.
Table 2.
Multiomics association studies of AF
| Omics | Study | Population | Tissue type | Platform | Significant signatures |
|---|---|---|---|---|---|
| Epigenomics—Methylation | Lin et al.76 |
European ancestry FHS Offspring cohort n = 2639 Prevalent AF n = 183 Incident AF n = 220 Follow-up 9 years. |
Whole blood | lllumina Infinium Human Methylation 450 BeadChip |
Prevalent AF cg13639451 cg07191189 Incident AF cg26602477 cg15440392 cg04064828 cg27529934 cg06725760 |
| Transcriptomics—mRNA expression | Lin et al.85 |
European ancestry FHS Offspring cohort n = 2446 Prevalent AF n = 177 Incident AF n = 143 No AF n = 2126 Follow-up 7 years. |
Whole blood | Affymetrix Human Exon 1.0ST Array |
Prevalent AF PBX1 C17orf39 PNP C18orf10 SLC7A1 SPTB ANKH |
| Transcriptomics—MicroRNA expression | Liu et al.94 |
Chinese ancestry 5 healthy controls, 5 patients with paroxysmal atrial fibrillation (PAF) alone, and 5 patients with persistent atrial fibrillation (PersAF) |
Plasma | MirVana PARIS kit (Invitrogen) and massively parallel signature sequencing | 16 miRNAs for PAF and 11 miRNAs for PersAF |
| Transcriptomics—MicroRNA expression | McManus et al.96 |
European ancestry FHS offspring cohort n = 2445 Prevalent AF n = 153 Incident AF n = 107 Follow-up 5.4 years |
Whole blood | TaqMan (PAXgene, Applied Biosystems) |
Prevalent AF miR-328 |
| Transcriptomics—MicroRNA expression | McManus et al.102 |
AF n = 112 European ancestry n = 108 No AF n = 99 European ancestry n = 67 |
Right atrial tissue Plasma |
Quantitative reverse transcriptase–polymerase chain reaction Qiagen (Valencia, CA) miScriptAssays BioMark System (Fluidigm) |
miRs-21 miRs-150 |
| Proteomics | Lind et al.103 |
European ancestry PIVUS study No AF n = 830 AF n = 148 Follow-up 10.0 years European ancestry ULSAM study No AF N = 602 Incident AF = 123 Follow-up 7.9 years |
Plasma | A proximity extension assay (PEA) chip |
IL-6 NT-proBNP |
| Proteomics | Willeit et al.104 |
European ancestry No AF n = 763 AF n = 117 Follow-up 20 years |
Venous blood | A commercially available enzyme-linked immunosorbent assay kit (Bender MedSystems) | sVCAM-1 |
| Proteomics | Ko et al.105 |
European ancestry FHS Offspring cohort n = 1885 Incident AF n = 349 Follow-up 18.3 years. |
Citrate-plasma | SOMAscan proteomic profiling platform |
Analyzed plasma proteins (n = 1373) NCAM-120 WFIKKN2 Ntrk3 EGFR ADAMTS13 Angiopoietin-2 NT-proBNP BMPR1A |
| Proteomics | Staerk et al.109 |
European ancestry FHS Offspring and Third Generation cohorts (n = 3378) Incident AF n = 401 Follow-up 12.3 years. |
Plasma | Luminex xMAP platform |
Analyzed plasma proteins (n = 85) IGF1 IGFBP1 NT-proBNP |
| Metabolomics | Alonso et al.112 |
African ancestry ARIC study (n = 1919) Incident AF n = 183 Follow-up 22 years. |
Serum | Untargeted, gas chromatography/mass spectrometry and liquid chromatography/mass spectrometry–based metabolomic quantification (Metabolon, Inc) | Bile acids glycolithocholate sulfate and glycocholenate sulfate |
| Metabolomics | Ko et al.117 |
European ancestry FHS Offspring (n = 2458) Incident AF n = 156 Follow-up 10 years. |
Plasma | Targeted liquid chromatography-tandem mass spectrometry (AB SCIEX 4000 QTRAP triple quadrupole mass spectrometer for positively charged polar compounds and lipids) and an AB SCIEX 5500 QTRAP triple quadrupole mass spectrometer for negatively charged polar compounds |
Analyzed 217 metabolites (54 positively charged, 59 negatively charged, 104 lipids) None |
IGF1, insulin-like growth factor 1; IGFBP1, insulin-like growth factor-binding protein 1; NT-proBNP, N-terminal prohormone of brain natriuretic peptide.
Figure 1.
Multiomics studies in AF.
First, current omics studies have been based mostly on samples collected at a single examination, whereas longitudinal omics profiling is still very scarce. It is challenging to investigate the effects of longitudinal changes in omics profiles on AF pathogenesis since omics platforms and profiles change over time. In addition, the relationship between AF and omics profiles could be bidirectional and the causal effects remain unclear.132 Therefore, longitudinal omics methods should be added to the future investigation of AF pathogenesis.
Second, it would be valuable to explore the association of additional types of omics with AF, such as in samples collected from the gut microbiome,133 urine,134 saliva,135 and right and left atrial and left pulmonary vein tissues. These omics data might identify additional molecular signatures and biomarkers for AF. Given the limited availability of heart samples, the use of induced pluripotent stem (iPS) cells to derive heart tissue from fibroblasts or even peripheral blood cells may greatly expand research capabilities.136,137
Third, further development of data integration methodology is needed to combine different omics data to better understand the pathophysiologic pathways of AF. Such efforts would be important to the risk stratification and the identification of novel therapeutic targets for AF.138,139
Fourth, further clinical and observational studies are needed to define whether multiomics profiles are different in individuals with AF dependent on disease burden. As hundreds of potential biomarkers of AF have been reported in various studies, additional scrutiny will be necessary to understand any potential bias of omic biomarkers for risk profiling. Almost all studies were adjusted for age and sex; however, many studies did not take into account known clinical risk factors that might confound the results. In addition, some biomarkers reached only lenient significance; the associations will no longer be significant after adjusting for multiple testing.
Fifth, the mechanisms relating biomarkers are uncertain. Are they directly related, confounders, related via intermediate mechanisms, or merely spurious associations yet to be determined? In addition, even if the associations are replicated, it remains uncertain how the biomarkers are involved in the pathogenesis of AF.
Sixth, most of the existing omics studies were based on participants of European ancestry from Western Europe and North America; generalizability of these findings to other ancestries and regions is largely unknown. Therefore, it is imperative to include participants from diverse ancestries/ethnicities and regions for future omics studies, such as African American, Asian, Hispanic, and Indigenous individuals.94,112,140,141
Finally, from clinical perspective, the incorporation of omics profiling into clinical routine is yet to come. The risk prediction of AF using omics is limited mostly to genomics information, whereas omics information has not been used to improve prediction beyond clinical score. In addition, very few studies have analyzed C-statistics and reclassification metrics of predictive value after adjustment for biomarkers or genetic risk (Supplementary material online, Table S2). Most of them showed only modest improvement of predictive value.142–147 Future studies are needed to analyze risk prediction and to identify effect sizes, population attributable risks, and risk reclassification using different omics profiles.
In summary, the past decade has witnessed enormous progress in the multiomics study of AF, which has identified hundreds of potential biomarkers or targets for future investigations. Recent efforts paved the way towards more advanced analyses elucidating pathophysiological complexity of AF underlying processes. The next studies are needed to highlight not separate biomarkers based on genomic, proteomic, or metabolomics studies, but rather using a multiomics approach in diverse multiracial populations.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
J.K. received funding from the Marie Sklodowska-Curie Actions under the European Union’s Horizon 2020 research and innovation programme (Agreement No. 838259). L.T. was supported by the American Heart Association (18SFRN34150007). D.K. was supported by American College of Cardiology Foundation/Merck Research Fellowship in Cardiovascular Diseases and Cardiometabolic Disorders. S.R.P. was supported by NIH grant 5R01HL128914. E.J.B. was supported by NIH 2R01 HL092577, 1R01 HL141434 01A1, 2U54HL120163, and 1R01AG066010; American Heart Association, 18SFRN34110082. H.L. was supported by the European Commission Grant (Agreement No. 847770).
Supplementary Material
This article is part of the Spotlight Issue on Atrial Fibrillation.
References
- 1. Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ, Benziger CP, Bonny A, Brauer M, Brodmann M, Cahill TJ, Carapetis J, Catapano AL, Chugh SS, Cooper LT, Coresh J, Criqui M, DeCleene N, Eagle KA, Emmons-Bell S, Feigin VL, Fernández-Solà J, Fowkes G, Gakidou E, Grundy SM, He FJ, Howard G, Hu F, Inker L, Karthikeyan G, Kassebaum N, Koroshetz W, Lavie C, Lloyd-Jones D, Lu HS, Mirijello A, Temesgen AM, Mokdad A, Moran AE, Muntner P, Narula J, Neal B, Ntsekhe M, Moraes de Oliveira G, Otto C, Owolabi M, Pratt M, Rajagopalan S, Reitsma M, Ribeiro ALP, Rigotti N, Rodgers A, Sable C, Shakil S, Sliwa-Hahnle K, Stark B, Sundström J, Timpel P, Tleyjeh IM, Valgimigli M, Vos T, Whelton PK, Yacoub M, Zuhlke L, Murray C, Fuster V; GBD-NHLBI-JACC Global Burden of Cardiovascular Diseases Writing Group. Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J Am Coll Cardiol 2020;76:2982–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schnabel RB, Yin X, Gona P, Larson MG, Beiser AS, McManus DD, Newton-Cheh C, Lubitz SA, Magnani JW, Ellinor PT, Seshadri S, Wolf PA, Vasan RS, Benjamin EJ, Levy D. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the Framingham Heart Study: a cohort study. Lancet 2015;386:154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Magnussen C, Niiranen TJ, Ojeda FM, Gianfagna F, Blankenberg S, Njolstad I, Vartiainen E, Sans S, Pasterkamp G, Hughes M, Costanzo S, Donati MB, Jousilahti P, Linneberg A, Palosaari T, de Gaetano G, Bobak M, den Ruijter HM, Mathiesen E, Jorgensen T, Soderberg S, Kuulasmaa K, Zeller T, Iacoviello L, Salomaa V, Schnabel RB; BiomarCaRE Consortium. Sex differences and similarities in atrial fibrillation epidemiology, risk factors, and mortality in community cohorts: results from the BiomarCaRE consortium (Biomarker for Cardiovascular Risk Assessment in Europe). Circulation 2017;136:1588–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weng LC, Preis SR, Hulme OL, Larson MG, Choi SH, Wang B, Trinquart L, McManus DD, Staerk L, Lin H, Lunetta KL, Ellinor PT, Benjamin EJ, Lubitz SA. Genetic predisposition, clinical risk factor burden, and lifetime risk of atrial fibrillation. Circulation 2018;137:1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Staerk L, Wang B, Preis SR, Larson MG, Lubitz SA, Ellinor PT, McManus DD, Ko D, Weng LC, Lunetta KL, Frost L, Benjamin EJ, Trinquart L. Lifetime risk of atrial fibrillation according to optimal, borderline, or elevated levels of risk factors: cohort study based on longitudinal data from the Framingham Heart Study. BMJ 2018;361:k1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dai H, Zhang Q, Much AA, Maor E, Segev A, Beinart R, Adawi S, Lu Y, Bragazzi NL, Wu J. Global, regional, and national prevalence, incidence, mortality, and risk factors for atrial fibrillation, 1990-2017: results from the Global Burden of Disease Study 2017. Eur Heart J Qual Care Clin Outcomes 2020. qcaa061.doi: 10.1093/ehjqcco/qcaa061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Colilla S, Crow A, Petkun W, Singer DE, Simon T, Liu X. Estimates of current and future incidence and prevalence of atrial fibrillation in the U.S. adult population. Am J Cardiol 2013;112:1142–1147. [DOI] [PubMed] [Google Scholar]
- 8. Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: the Framingham Study. Stroke 1991;22:983–988. [DOI] [PubMed] [Google Scholar]
- 9. Odutayo A, Wong CX, Hsiao AJ, Hopewell S, Altman DG, Emdin CA. Atrial fibrillation and risks of cardiovascular disease, renal disease, and death: systematic review and meta-analysis. BMJ 2016;354:i4482. [DOI] [PubMed] [Google Scholar]
- 10. Soliman EZ, Safford MM, Muntner P, Khodneva Y, Dawood FZ, Zakai NA, Thacker EL, Judd S, Howard VJ, Howard G, Herrington DM, Cushman M. Atrial fibrillation and the risk of myocardial infarction. JAMA Intern Med 2014;174:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Santangeli P, Di Biase L, Bai R, Mohanty S, Pump A, Cereceda Brantes M, Horton R, Burkhardt JD, Lakkireddy D, Reddy YM, Casella M, Dello Russo A, Tondo C, Natale A. Atrial fibrillation and the risk of incident dementia: a meta-analysis. Heart Rhythm 2012;9:1761–1768. [DOI] [PubMed] [Google Scholar]
- 12. Benjamin EJ, Wolf PA, D’Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation 1998;98:946–952. [DOI] [PubMed] [Google Scholar]
- 13. Vinter N, Huang X, Fenger-Gron M, Frost L, Benjamin EJ, Trinquart L. 45-year trend in excess mortality associated with atrial fibrillation in the community-based Framingham Heart Study cohorts. BMJ 2020;370:m2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delaney JA, Yin X, Fontes JD, Wallace ER, Skinner A, Wang N, Hammill BG, Benjamin EJ, Curtis LH, Heckbert SR. Hospital and clinical care costs associated with atrial fibrillation for medicare beneficiaries in the Cardiovascular Health Study and the Framingham Heart Study. SAGE Open Med 2018;6:2050312118759444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dieleman JL, Cao J, Chapin A, Chen C, Li Z, Liu A, Horst C, Kaldjian A, Matyasz T, Scott KW, Bui AL, Campbell M, Duber HC, Dunn AC, Flaxman AD, Fitzmaurice C, Naghavi M, Sadat N, Shieh P, Squires E, Yeung K, Murray CJL. US health care spending by payer and health condition, 1996–2016. JAMA 2020;323:863–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benjamin EJ, Chen PS, Bild DE, Mascette AM, Albert CM, Alonso A, Calkins H, Connolly SJ, Curtis AB, Darbar D, Ellinor PT, Go AS, Goldschlager NF, Heckbert SR, Jalife J, Kerr CR, Levy D, Lloyd-Jones DM, Massie BM, Nattel S, Olgin JE, Packer DL, Po SS, Tsang TS, Van Wagoner DR, Waldo AL, Wyse DG. Prevention of atrial fibrillation: report from a national heart, lung, and blood institute workshop. Circulation 2009;119:606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benjamin EJ, Levy D, Vaziri SM, D’Agostino RB, Belanger AJ, Wolf PA. Independent risk factors for atrial fibrillation in a population-based cohort. The Framingham Heart Study. JAMA 1994;271:840–844. [PubMed] [Google Scholar]
- 18. Frost L, Hune LJ, Vestergaard P. Overweight and obesity as risk factors for atrial fibrillation or flutter: the Danish diet, cancer, and health study. Am J Med 2005;118:489–495. [DOI] [PubMed] [Google Scholar]
- 19. Wang TJ, Parise H, Levy D, D’Agostino RB Sr, Wolf PA, Vasan RS, Benjamin EJ. Obesity and the risk of new-onset atrial fibrillation. JAMA 2004;292:2471–2477. [DOI] [PubMed] [Google Scholar]
- 20. Psaty BM, Manolio TA, Kuller LH, Kronmal RA, Cushman M, Fried LP, White R, Furberg CD, Rautaharju PM. Incidence of and risk factors for atrial fibrillation in older adults. Circulation 1997;96:2455–2461. [DOI] [PubMed] [Google Scholar]
- 21. Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Shay CM, Spartano NL, Stokes A, Tirschwell DL, VanWagner LB, Tsao CW; American Heart Association Council on Epidemiology and Prevention Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation 2020;141(9):e139-e596.CIR0000000000000757. [DOI] [PubMed] [Google Scholar]
- 22. Alonso A, Krijthe BP, Aspelund T, Stepas KA, Pencina MJ, Moser CB, Sinner MF, Sotoodehnia N, Fontes JD, Janssens AC, Kronmal RA, Magnani JW, Witteman JC, Chamberlain AM, Lubitz SA, Schnabel RB, Agarwal SK, McManus DD, Ellinor PT, Larson MG, Burke GL, Launer LJ, Hofman A, Levy D, Gottdiener JS, Kaab S, Couper D, Harris TB, Soliman EZ, Stricker BH, Gudnason V, Heckbert SR, Benjamin EJ. Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the charge-af consortium. J Am Heart Assoc 2013;2:e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D’Agostino RB Sr, Newton-Cheh C, Yamamoto JF, Magnani JW, Tadros TM, Kannel WB, Wang TJ, Ellinor PT, Wolf PA, Vasan RS, Benjamin EJ. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet 2009;373:739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cappato R, Calkins H, Chen SA, Davies W, Iesaka Y, Kalman J, Kim YH, Klein G, Natale A, Packer D, Skanes A, Ambrogi F, Biganzoli E. Updated worldwide survey on the methods, efficacy, and safety of catheter ablation for human atrial fibrillation. Circ Arrhythm Electrophysiol 2010;3:32–38. [DOI] [PubMed] [Google Scholar]
- 25. January CT, Wann LS, Alpert JS, Calkins H, Cigarroa JE, Cleveland JC Jr, Conti JB, Ellinor PT, Ezekowitz MD, Field ME, Murray KT, Sacco RL, Stevenson WG, Tchou PJ, Tracy CM, Yancy CW; ACC/AHA Task Force Members. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on practice guidelines and the Heart Rhythm Society. Circulation 2014;130:2071–2104. [DOI] [PubMed] [Google Scholar]
- 26. Orgain ES, Wolff L, White PD. Uncomplicated auricular fibrillation and auricular flutter: frequent occurrence and good prognosis in patients without other evidence of cardiac disease. Arch Intern Med (Chic) 1936;57:493–513. [Google Scholar]
- 27. Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen WK, Hammill SC, Packer DL, Olson TM. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol 2003;41:2185–2192. [DOI] [PubMed] [Google Scholar]
- 28. Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet 2005;118:179–184. [DOI] [PubMed] [Google Scholar]
- 29. Fox CS, Parise H, D’Agostino RB Sr, Lloyd-Jones DM, Vasan RS, Wang TJ, Levy D, Wolf PA, Benjamin EJ. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA 2004;291:2851–2855. [DOI] [PubMed] [Google Scholar]
- 30. Marcus GM, Smith LM, Vittinghoff E, Tseng ZH, Badhwar N, Lee BK, Lee RJ, Scheinman MM, Olgin JE. A first-degree family history in lone atrial fibrillation patients. Heart Rhythm 2008;5:826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christophersen IE, Ravn LS, Budtz-Joergensen E, Skytthe A, Haunsoe S, Svendsen JH, Christensen K. Familial aggregation of atrial fibrillation: a study in Danish twins. Circ Arrhythm Electrophysiol 2009;2:378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, Villalon ML, Vasan RS, Pencina MJ, Levy D, Larson MG, Ellinor PT, Benjamin EJ. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA 2010;304:2263–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, Stefansson K. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J 2006;27:708–712. [DOI] [PubMed] [Google Scholar]
- 34. Mackay TF. The genetic architecture of quantitative traits. Annu Rev Genet 2001;35:303–339. [DOI] [PubMed] [Google Scholar]
- 35. Brugada R, Tapscott T, Czernuszewicz GZ, Marian AJ, Iglesias A, Mont L, Brugada J, Girona J, Domingo A, Bachinski LL, Roberts R. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med 1997;336:905–911. [DOI] [PubMed] [Google Scholar]
- 36. Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN, Yang YQ, Li YB, Liu Y, Xu HJ, Li XF, Ma N, Mou CP, Chen Z, Barhanin J, Huang W. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science 2003;299:251–254. [DOI] [PubMed] [Google Scholar]
- 37. Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, Sattiraju S, Ballew JD, Jahangir A, Terzic A. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet 2006;15:2185–2191. [DOI] [PubMed] [Google Scholar]
- 38. Tucker NR, Ellinor PT. Emerging directions in the genetics of atrial fibrillation. Circ Res 2014;114:1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, George AL Jr, Roden DM. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation 2008;117:1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Makiyama T, Akao M, Shizuta S, Doi T, Nishiyama K, Oka Y, Ohno S, Nishio Y, Tsuji K, Itoh H, Kimura T, Kita T, Horie M. A novel SCN5A gain-of-function mutation m1875t associated with familial atrial fibrillation. J Am Coll Cardiol 2008;52:1326–1334. [DOI] [PubMed] [Google Scholar]
- 41. Hagendorff A, Schumacher B, Kirchhoff S, LüDeritz B, Willecke K. Conduction disturbances and increased atrial vulnerability in connexin40-deficient mice analyzed by transesophageal stimulation. Circulation 1999;99:1508–1515. [DOI] [PubMed] [Google Scholar]
- 42. Kanno S, Saffitz JE. The role of myocardial gap junctions in electrical conduction and arrhythmogenesis. Cardiovasc Pathol 2001;10:169–177. [DOI] [PubMed] [Google Scholar]
- 43. Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, Liu X, Veinot JP, Tang AS, Stewart AF, Tesson F, Klein GJ, Yee R, Skanes AC, Guiraudon GM, Ebihara L, Bai D. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med 2006;354:2677–2688. [DOI] [PubMed] [Google Scholar]
- 44. Gutstein DE, Morley GE, Vaidya D, Liu F, Chen FL, Stuhlmann H, Fishman GI. Heterogeneous expression of gap junction channels in the heart leads to conduction defects and ventricular dysfunction. Circulation 2001;104:1194–1199. [DOI] [PubMed] [Google Scholar]
- 45. Sinner MF, Lubitz SA, Pfeufer A, Makino S, Beckmann BM, Lunetta KL, Steinbeck G, Perz S, Rahman R, Sonni A, Greenberg SM, Furie KL, Wichmann HE, Meitinger T, Peters A, Benjamin EJ, Rosand J, Ellinor PT, Kaab S. Lack of replication in polymorphisms reported to be associated with atrial fibrillation. Heart Rhythm 2011;8:403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007;448:353–357. [DOI] [PubMed] [Google Scholar]
- 47. Lubitz SA, Lunetta KL, Lin H, Arking DE, Trompet S, Li G, Krijthe BP, Chasman DI, Barnard J, Kleber ME, Dorr M, Ozaki K, Smith AV, Muller-Nurasyid M, Walter S, Agarwal SK, Bis JC, Brody JA, Chen LY, Everett BM, Ford I, Franco OH, Harris TB, Hofman A, Kaab S, Mahida S, Kathiresan S, Kubo M, Launer LJ, MacFarlane PW, Magnani JW, McKnight B, McManus DD, Peters A, Psaty BM, Rose LM, Rotter JI, Silbernagel G, Smith JD, Sotoodehnia N, Stott DJ, Taylor KD, Tomaschitz A, Tsunoda T, Uitterlinden AG, Van Wagoner DR, Volker U, Volzke H, Murabito JM, Sinner MF, Gudnason V, Felix SB, Marz W, Chung M, Albert CM, Stricker BH, Tanaka T, Heckbert SR, Jukema JW, Alonso A, Benjamin EJ, Ellinor PT. Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. J Am Coll Cardiol 2014;63:1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, Christoffels VM. PITX2c and NKX2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res 2007;101:902–909. [DOI] [PubMed] [Google Scholar]
- 49. Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. PITX2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A 2010;107:9753–9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, Rotering H, Fortmueller L, Laakmann S, Verheule S, Schotten U, Fabritz L, Brown NA. PITX2c is expressed in the adult left atrium, and reducing PITX2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet 2011;4:123–133. [DOI] [PubMed] [Google Scholar]
- 51. Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpon E, Tamargo J, Cinca J, Hove-Madsen L, Aranega AE, Franco D. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet 2011;4:269–279. [DOI] [PubMed] [Google Scholar]
- 52. Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D’Agostino RB, Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiríksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WHL, Agarwal SK, Stricker BHC, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Köttgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann H-E, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kääb S, Ellinor PT, Witteman JCM. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet 2009;41:879–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PIW, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel RB, Soliman EZ, Rice KM, Van Wagoner DR, Beckmann B-M, van Noord C, Wang K, Ehret GB, Rotter JI, Hazen SL, Steinbeck G, Smith AV, Launer LJ, Harris TB, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Köttgen A, Moebus S, Newton-Cheh C, Li M, Möhlenkamp S, Wang TJ, Kao WHL, Vasan RS, Nöthen MM, MacRae CA, Stricker BHC, Hofman A, Uitterlinden AG, Levy D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Gudnason V, Psaty BM, Roden DM, Meitinger T, Wichmann H-E, Witteman JCM, Barnard J, Arking DE, Benjamin EJ, Heckbert SR, Kääb S. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet 2010;42:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Müller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dörr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Völker U, Völzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann H-E, Witteman JCM, Kao WHL, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjögren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BHC, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kääb S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet 2012;44:670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sinner MF, Tucker NR, Lunetta KL, Ozaki K, Smith JG, Trompet S, Bis JC, Lin H, Chung MK, Nielsen JB, Lubitz SA, Krijthe BP, Magnani JW, Ye J, Gollob MH, Tsunoda T, Muller-Nurasyid M, Lichtner P, Peters A, Dolmatova E, Kubo M, Smith JD, Psaty BM, Smith NL, Jukema JW, Chasman DI, Albert CM, Ebana Y, Furukawa T, Macfarlane PW, Harris TB, Darbar D, Dorr M, Holst AG, Svendsen JH, Hofman A, Uitterlinden AG, Gudnason V, Isobe M, Malik R, Dichgans M, Rosand J, Van Wagoner DR, Consortium M, Consortium AF, Benjamin EJ, Milan DJ, Melander O, Heckbert SR, Ford I, Liu Y, Barnard J, Olesen MS, Stricker BH, Tanaka T, Kaab S, Ellinor PT; AFGen Consortium. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation 2014;130:1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Christophersen IE, Rienstra M, Roselli C, Yin X, Geelhoed B, Barnard J, Lin H, Arking DE, Smith AV, Albert CM, Chaffin M, Tucker NR, Li M, Klarin D, Bihlmeyer NA, Low S-K, Weeke PE, Müller-Nurasyid M, Smith JG, Brody JA, Niemeijer MN, Dörr M, Trompet S, Huffman J, Gustafsson S, Schurmann C, Kleber ME, Lyytikäinen L-P, Seppälä I, Malik R, Horimoto ARVR, Perez M, Sinisalo J, Aeschbacher S, Thériault S, Yao J, Radmanesh F, Weiss S, Teumer A, Choi SH, Weng L-C, Clauss S, Deo R, Rader DJ, Shah SH, Sun A, Hopewell JC, Debette S, Chauhan G, Yang Q, Worrall BB, Paré G, Kamatani Y, Hagemeijer YP, Verweij N, Siland JE, Kubo M, Smith JD, Van Wagoner DR, Bis JC, Perz S, Psaty BM, Ridker PM, Magnani JW, Harris TB, Launer LJ, Shoemaker MB, Padmanabhan S, Haessler J, Bartz TM, Waldenberger M, Lichtner P, Arendt M, Krieger JE, Kähönen M, Risch L, Mansur AJ, Peters A, Smith BH, Lind L, Scott SA, Lu Y, Bottinger EB, Hernesniemi J, Lindgren CM, Wong JA, Huang J, Eskola M, Morris AP, Ford I, Reiner AP, Delgado G, Chen LY, Chen Y-DI, Sandhu RK, Li M, Boerwinkle E, Eisele L, Lannfelt L, Rost N, Anderson CD, Taylor KD, Campbell A, Magnusson PK, Porteous D, Hocking LJ, Vlachopoulou E, Pedersen NL, Nikus K, Orho-Melander M, Hamsten A, Heeringa J, Denny JC, Kriebel J, Darbar D, Newton-Cheh C, Shaffer C, Macfarlane PW, Heilmann-Heimbach S, Almgren P, Huang PL, Sotoodehnia N, Soliman EZ, Uitterlinden AG, Hofman A, Franco OH, Völker U, Jöckel K-H, Sinner MF, Lin HJ, Guo X, Dichgans M, Ingelsson E, Kooperberg C, Melander O, Loos RJF, Laurikka J, Conen D, Rosand J, van der Harst P, Lokki M-L, Kathiresan S, Pereira A, Jukema JW, Hayward C, Rotter JI, März W, Lehtimäki T, Stricker BH, Chung MK, Felix SB, Gudnason V, Alonso A, Roden DM, Kääb S, Chasman DI, Heckbert SR, Benjamin EJ, Tanaka T, Lunetta KL, Lubitz SA, Ellinor PT; AFGen Consortium. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet 2017;49:946–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roselli C, Chaffin MD, Weng LC, Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson CD, Aragam KG, Arking DE, Barnard J, Bartz TM, Benjamin EJ, Bihlmeyer NA, Bis JC, Bloom HL, Boerwinkle E, Bottinger EB, Brody JA, Calkins H, Campbell A, Cappola TP, Carlquist J, Chasman DI, Chen LY, Chen YI, Choi EK, Choi SH, Christophersen IE, Chung MK, Cole JW, Conen D, Cook J, Crijns HJ, Cutler MJ, Damrauer SM, Daniels BR, Darbar D, Delgado G, Denny JC, Dichgans M, Dorr M, Dudink EA, Dudley SC, Esa N, Esko T, Eskola M, Fatkin D, Felix SB, Ford I, Franco OH, Geelhoed B, Grewal RP, Gudnason V, Guo X, Gupta N, Gustafsson S, Gutmann R, Hamsten A, Harris TB, Hayward C, Heckbert SR, Hernesniemi J, Hocking LJ, Hofman A, Horimoto A, Huang J, Huang PL, Huffman J, Ingelsson E, Ipek EG, Ito K, Jimenez-Conde J, Johnson R, Jukema JW, Kaab S, Kahonen M, Kamatani Y, Kane JP, Kastrati A, Kathiresan S, Katschnig-Winter P, Kavousi M, Kessler T, Kietselaer BL, Kirchhof P, Kleber ME, Knight S, Krieger JE, Kubo M, Launer LJ, Laurikka J, Lehtimaki T, Leineweber K, Lemaitre RN, Li M, Lim HE, Lin HJ, Lin H, Lind L, Lindgren CM, Lokki ML, London B, Loos RJF, Low SK, Lu Y, Lyytikainen LP, Macfarlane PW, Magnusson PK, Mahajan A, Malik R, Mansur AJ, Marcus GM, Margolin L, Margulies KB, Marz W, McManus DD, Melander O, Mohanty S, Montgomery JA, Morley MP, Morris AP, Muller-Nurasyid M, Natale A, Nazarian S, Neumann B, Newton-Cheh C, Niemeijer MN, Nikus K, Nilsson P, Noordam R, Oellers H, Olesen MS, Orho-Melander M, Padmanabhan S, Pak HN, Pare G, Pedersen NL, Pera J, Pereira A, Porteous D, Psaty BM, Pulit SL, Pullinger CR, Rader DJ, Refsgaard L, Ribases M, Ridker PM, Rienstra M, Risch L, Roden DM, Rosand J, Rosenberg MA, Rost N, Rotter JI, Saba S, Sandhu RK, Schnabel RB, Schramm K, Schunkert H, Schurman C, Scott SA, Seppala I, Shaffer C, Shah S, Shalaby AA, Shim J, Shoemaker MB, Siland JE, Sinisalo J, Sinner MF, Slowik A, Smith AV, Smith BH, Smith JG, Smith JD, Smith NL, Soliman EZ, Sotoodehnia N, Stricker BH, Sun A, Sun H, Svendsen JH, Tanaka T, Tanriverdi K, Taylor KD, Teder-Laving M, Teumer A, Theriault S, Trompet S, Tucker NR, Tveit A, Uitterlinden AG, Van Der Harst P, Van Gelder IC, Van Wagoner DR, Verweij N, Vlachopoulou E, Volker U, Wang B, Weeke PE, Weijs B, Weiss R, Weiss S, Wells QS, Wiggins KL, Wong JA, Woo D, Worrall BB, Yang PS, Yao J, Yoneda ZT, Zeller T, Zeng L, Lubitz SA, Lunetta KL, Ellinor PT. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 2018;50:1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, Surakka I, Mathis MR, Yamazaki M, Crawford RD, Gabrielsen ME, Skogholt AH, Holmen OL, Lin M, Wolford BN, Dey R, Dalen H, Sulem P, Chung JH, Backman JD, Arnar DO, Thorsteinsdottir U, Baras A, O’Dushlaine C, Holst AG, Wen X, Hornsby W, Dewey FE, Boehnke M, Kheterpal S, Mukherjee B, Lee S, Kang HM, Holm H, Kitzman J, Shavit JA, Jalife J, Brummett CM, Teslovich TM, Carey DJ, Gudbjartsson DF, Stefansson K, Abecasis GR, Hveem K, Willer CJ. BioBank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet 2018;50:1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roselli C, Rienstra M, Ellinor PT. Genetics of atrial fibrillation in 2020: GWAS, genome sequencing, polygenic risk, and beyond. Circ Res 2020;127:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weng LC, Choi SH, Klarin D, Smith JG, Loh PR, Chaffin M, Roselli C, Hulme OL, Lunetta KL, Dupuis J, Benjamin EJ, Newton-Cheh C, Kathiresan S, Ellinor PT, Lubitz SA. Heritability of atrial fibrillation. Circ Cardiovasc Genet 2017;10:e001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 2008;40:695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med 2010;363:166–176. [DOI] [PubMed] [Google Scholar]
- 63. Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009;106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin H, Sinner MF, Brody JA, Arking DE, Lunetta KL, Rienstra M, Lubitz SA, Magnani JW, Sotoodehnia N, McKnight B, McManus DD, Boerwinkle E, Psaty BM, Rotter JI, Bis JC, Gibbs RA, Muzny D, Kovar CL, Morrison AC, Gupta M, Folsom AR, Kaab S, Heckbert SR, Alonso A, Ellinor PT, Benjamin EJ; CHARGE Atrial Fibrillation Working Group. Targeted sequencing in candidate genes for atrial fibrillation: the cohorts for heart and aging research in genomic epidemiology (charge) targeted sequencing study. Heart Rhythm 2014;11:452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rafiq S, Frayling TM, Murray A, Hurst A, Stevens K, Weedon MN, Henley W, Ferrucci L, Bandinelli S, Corsi AM, Guralnik JM, Melzer D. A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes Immun 2007;8:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, Tandon A, Choy E, Hu D, Tamraz B, Pawlikowska L, Wassel-Fyr C, Huntsman S, Waliszewska A, Rossin E, Li R, Garcia M, Reiner A, Ferrell R, Cummings S, Kwok PY, Harris T, Zmuda JM, Ziv E. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am J Hum Genet 2007;80:716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Issac TT, Dokainish H, Lakkis NM. Role of inflammation in initiation and perpetuation of atrial fibrillation: a systematic review of the published data. J Am Coll Cardiol 2007;50:2021–2028. [DOI] [PubMed] [Google Scholar]
- 68. Lubitz SA, Brody JA, Bihlmeyer NA, Roselli C, Weng LC, Christophersen IE, Alonso A, Boerwinkle E, Gibbs RA, Bis JC, Cupples LA, Mohler PJ, Nickerson DA, Muzny D, Perez MV, Psaty BM, Soliman EZ, Sotoodehnia N, Lunetta KL, Benjamin EJ, Heckbert SR, Arking DE, Ellinor PT, Lin H, Project NGES. Whole exome sequencing in atrial fibrillation. PLoS Genet 2016;12:e1006284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Choi SH, Weng LC, Roselli C, Lin H, Haggerty CM, Shoemaker MB, Barnard J, Arking DE, Chasman DI, Albert CM, Chaffin M, Tucker NR, Smith JD, Gupta N, Gabriel S, Margolin L, Shea MA, Shaffer CM, Yoneda ZT, Boerwinkle E, Smith NL, Silverman EK, Redline S, Vasan RS, Burchard EG, Gogarten SM, Laurie C, Blackwell TW, Abecasis G, Carey DJ, Fornwalt BK, Smelser DT, Baras A, Dewey FE, Jaquish CE, Papanicolaou GJ, Sotoodehnia N, Van Wagoner DR, Psaty BM, Kathiresan S, Darbar D, Alonso A, Heckbert SR, Chung MK, Roden DM, Benjamin EJ, Murray MF, Lunetta KL, Lubitz SA, Ellinor PT; DiscovEHR Study and the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium. Association between titin loss-of-function variants and early-onset atrial fibrillation. JAMA 2018;320:2354–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597–610. [DOI] [PubMed] [Google Scholar]
- 71. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013;38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, Waldenberger M, Peters A, Erdmann J, Hengstenberg C, Cambien F, Goodall AH, Ouwehand WH, Schunkert H, Thompson JR, Spector TD, Gieger C, Tregouet DA, Deloukas P, Samani NJ. DNA methylation and body-mass index: a genome-wide analysis. Lancet 2014;383:1990–1998. [DOI] [PubMed] [Google Scholar]
- 73. Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, Guan W, Xu T, Elks CE, Aslibekyan S, Moreno-Macias H, Smith JA, Brody JA, Dhingra R, Yousefi P, Pankow JS, Kunze S, Shah SH, McRae AF, Lohman K, Sha J, Absher DM, Ferrucci L, Zhao W, Demerath EW, Bressler J, Grove ML, Huan T, Liu C, Mendelson MM, Yao C, Kiel DP, Peters A, Wang-Sattler R, Visscher PM, Wray NR, Starr JM, Ding J, Rodriguez CJ, Wareham NJ, Irvin MR, Zhi D, Barrdahl M, Vineis P, Ambatipudi S, Uitterlinden AG, Hofman A, Schwartz J, Colicino E, Hou L, Vokonas PS, Hernandez DG, Singleton AB, Bandinelli S, Turner ST, Ware EB, Smith AK, Klengel T, Binder EB, Psaty BM, Taylor KD, Gharib SA, Swenson BR, Liang L, DeMeo DL, O’Connor GT, Herceg Z, Ressler KJ, Conneely KN, Sotoodehnia N, Kardia SLR, Melzer D, Baccarelli AA, van Meurs JBJ, Romieu I, Arnett DK, Ong KK, Liu Y, Waldenberger M, Deary IJ, Fornage M, Levy D, London SJ. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet 2016;9:436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mendelson MM, Johannes R, Liu C, Huan T, Yao C, Miao X, Murabito JM, Dupuis J, Levy D, Benjamin EJ, Lin H. Epigenome-wide association study of soluble tumor necrosis factor receptor 2 levels in the Framingham Heart Study. Front Pharmacol 2018;9:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu C, Marioni RE, Hedman ÅK, Pfeiffer L, Tsai PC, Reynolds LM, Just AC, Duan Q, Boer CG, Tanaka T, Elks CE, Aslibekyan S, Brody JA, Kühnel B, Herder C, Almli LM, Zhi D, Wang Y, Huan T, Yao C, Mendelson MM, Joehanes R, Liang L, Love SA, Guan W, Shah S, McRae AF, Kretschmer A, Prokisch H, Strauch K, Peters A, Visscher PM, Wray NR, Guo X, Wiggins KL, Smith AK, Binder EB, Ressler KJ, Irvin MR, Absher DM, Hernandez D, Ferrucci L, Bandinelli S, Lohman K, Ding J, Trevisi L, Gustafsson S, Sandling JH, Stolk L, Uitterlinden AG, Yet I, Castillo-Fernandez JE, Spector TD, Schwartz JD, Vokonas P, Lind L, Li Y, Fornage M, Arnett DK, Wareham NJ, Sotoodehnia N, Ong KK, van Meurs JBJ, Conneely KN, Baccarelli AA, Deary IJ, Bell JT, North KE, Liu Y, Waldenberger M, London SJ, Ingelsson E, Levy D. A DNA methylation biomarker of alcohol consumption. Mol Psychiatry 2018;23:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lin H, Yin X, Xie Z, Lunetta KL, Lubitz SA, Larson MG, Ko D, Magnani JW, Mendelson MM, Liu C, McManus DD, Levy D, Ellinor PT, Benjamin EJ. Methylome-wide association study of atrial fibrillation in Framingham Heart Study. Sci Rep 2017;7:40377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hill JJ, Qiu Y, Hewick RM, Wolfman NM. Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: a novel protein with protease inhibitor and follistatin domains. Mol Endocrinol 2003;17:1144–1154. [DOI] [PubMed] [Google Scholar]
- 78. Maier LS, Bers DM. Role of Ca2+/calmodulin-dependent protein kinase (CAMK) in excitation-contraction coupling in the heart. Cardiovasc Res 2007;73:631–640. [DOI] [PubMed] [Google Scholar]
- 79. Mangoni ME, Couette B, Bourinet E, Platzer J, Reimer D, Striessnig J, Nargeot J. Functional role of l-type cav1.3 Ca2+ channels in cardiac pacemaker activity. Proc Natl Acad Sci U S A 2003;100:5543–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shen K, Tu T, Yuan Z, Yi J, Zhou Y, Liao X, Liu Q, Zhou X. DNA methylation dysregulations in valvular atrial fibrillation. Clin Cardiol 2017;40:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature 2008;452:423–428. [DOI] [PubMed] [Google Scholar]
- 82. Cervero J, Segura V, Macias A, Gavira JJ, Montes R, Hermida J. Atrial fibrillation in pigs induces left atrial endocardial transcriptional remodelling. Thromb Haemost 2012;108:742–749. [DOI] [PubMed] [Google Scholar]
- 83. Thijssen VL, van der Velden HM, van Ankeren EP, Ausma J, Allessie MA, Borgers M, van Eys GJ, Jongsma HJ. Analysis of altered gene expression during sustained atrial fibrillation in the goat. Cardiovasc Res 2002;54:427–437. [DOI] [PubMed] [Google Scholar]
- 84. Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kaab S, Hinterseer M, Kartmann H, Kreuzer E, Dugas M, Steinbeck G, Nabauer M. Reprogramming of the human atrial transcriptome in permanent atrial fibrillation: expression of a ventricular-like genomic signature. Circ Res 2005;96:1022–1029. [DOI] [PubMed] [Google Scholar]
- 85. Lin H, Yin X, Lunetta KL, Dupuis J, McManus DD, Lubitz SA, Magnani JW, Joehanes R, Munson PJ, Larson MG, Levy D, Ellinor PT, Benjamin EJ. Whole blood gene expression and atrial fibrillation: the Framingham Heart Study. PLoS One 2014;9:e96794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chang CP, Stankunas K, Shang C, Kao SC, Twu KY, Cleary ML. PBX1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development 2008;135:3577–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Stankunas K, Shang C, Twu KY, Kao SC, Jenkins NA, Copeland NG, Sanyal M, Selleri L, Cleary ML, Chang CP. PBX/MEIS deficiencies demonstrate multigenetic origins of congenital heart disease. Circ Res 2008;103:702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yang Z, Venardos K, Jones E, Morris BJ, Chin-Dusting J, Kaye DM. Identification of a novel polymorphism in the 3′-UTR of the l-arginine transporter gene SLC7A1: contribution to hypertension and endothelial dysfunction. Circulation 2007;115:1269–1274. [DOI] [PubMed] [Google Scholar]
- 89. Semenza GL. Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem Pharmacol 2000;59:47–53. [DOI] [PubMed] [Google Scholar]
- 90. Kido M, Du L, Sullivan CC, Li X, Deutsch R, Jamieson SW, Thistlethwaite PA. Hypoxia-inducible factor 1-alpha reduces infarction and attenuates progression of cardiac dysfunction after myocardial infarction in the mouse. J Am Coll Cardiol 2005;46:2116–2124. [DOI] [PubMed] [Google Scholar]
- 91. Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011;469:336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006;103:18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dawson K, Wakili R, Ordog B, Clauss S, Chen Y, Iwasaki Y, Voigt N, Qi XY, Sinner MF, Dobrev D, Kaab S, Nattel S. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 2013;127:1466–1475. [DOI] [PubMed] [Google Scholar]
- 94. Liu Z, Zhou C, Liu Y, Wang S, Ye P, Miao X, Xia J. The expression levels of plasma micornas in atrial fibrillation patients. PLoS One 2012;7:e44906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Goren Y, Meiri E, Hogan C, Mitchell H, Lebanony D, Salman N, Schliamser JE, Amir O. Relation of reduced expression of miR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am J Cardiol 2014;113:976–981. [DOI] [PubMed] [Google Scholar]
- 96. McManus DD, Lin H, Tanriverdi K, Quercio M, Yin X, Larson MG, Ellinor PT, Levy D, Freedman JE, Benjamin EJ. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham Offspring Study. Heart Rhythm 2014;11:663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang Q, Kandic I, Kutryk MJ. Dysregulation of angiogenesis-related microRNAs in endothelial progenitor cells from patients with coronary artery disease. Biochem Biophys Res Commun 2011;405:42–46. [DOI] [PubMed] [Google Scholar]
- 98. Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, Zhang Y, Shan H, Luo X, Bai Y, Sun L, Song W, Xu C, Wang Z, Yang B. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 2010;122:2378–2387. [DOI] [PubMed] [Google Scholar]
- 99. de Boer HC, van Solingen C, Prins J, Duijs JM, Huisman MV, Rabelink TJ, van Zonneveld AJ. Aspirin treatment hampers the use of plasma microRNA-126 as a biomarker for the progression of vascular disease. Eur Heart J 2013;34:3451–3457. [DOI] [PubMed] [Google Scholar]
- 100. Diehl P, Fricke A, Sander L, Stamm J, Bassler N, Htun N, Ziemann M, Helbing T, El-Osta A, Jowett JBM, Peter K. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res 2012;93:633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Su Z, Wu F. Inflammatory factors induce thrombosis through the miR-146b-3p/p38mapk/Cox-2 pathway. Biomed Res Int 2020;2020:8718321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McManus DD, Tanriverdi K, Lin H, Esa N, Kinno M, Mandapati D, Tam S, Okike ON, Ellinor PT, Keaney JF Jr, Donahue JK, Benjamin EJ, Freedman JE. Plasma microRNAs are associated with atrial fibrillation and change after catheter ablation (the MiRhythm Study). Heart Rhythm 2015;12:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lind L, Sundstrom J, Stenemo M, Hagstrom E, Arnlov J. Discovery of new biomarkers for atrial fibrillation using a custom-made proteomics chip. Heart 2017;103:377–382. [DOI] [PubMed] [Google Scholar]
- 104. Willeit K, Pechlaner R, Willeit P, Skroblin P, Paulweber B, Schernthaner C, Toell T, Egger G, Weger S, Oberhollenzer M, Kedenko L, Iglseder B, Bonora E, Schett G, Mayr M, Willeit J, Kiechl S. Association between vascular cell adhesion molecule 1 and atrial fibrillation. JAMA Cardiol 2017;2:516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ko D, Benson MD, Ngo D, Yang Q, Larson MG, Wang TJ, Trinquart L, McManus DD, Lubitz SA, Ellinor PT, Vasan RS, Gerszten RE, Benjamin EJ, Lin H. Proteomics profiling and risk of new-onset atrial fibrillation: Framingham Heart Study. Jaha 2019;8:e010976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Patton KK, Heckbert SR, Alonso A, Bahrami H, Lima JA, Burke G, Kronmal RA. N-terminal pro-b-type natriuretic peptide as a predictor of incident atrial fibrillation in the multi-ethnic study of atherosclerosis: the effects of age, sex and ethnicity. Heart 2013;99:1832–1836. [DOI] [PubMed] [Google Scholar]
- 107. Uemura T, Kaikita K, Yamabe H, Soejima K, Matsukawa M, Fuchigami S, Tanaka Y, Morihisa K, Enomoto K, Sumida H, Sugiyama S, Ogawa H. Changes in plasma von willebrand factor and ADAMTS13 levels associated with left atrial remodeling in atrial fibrillation. Thromb Res 2009;124:28–32. [DOI] [PubMed] [Google Scholar]
- 108. Freynhofer MK, Bruno V, Jarai R, Gruber S, Höchtl T, Brozovic I, Farhan S, Wojta J, Huber K. Levels of von Willebrand factor and ADAMTS13 determine clinical outcome after cardioversion for atrial fibrillation. Thromb Haemost 2011;105:435–443. [DOI] [PubMed] [Google Scholar]
- 109. Staerk L, Preis SR, Lin H, Lubitz SA, Ellinor PT, Levy D, Benjamin EJ, Trinquart L. Protein biomarkers and risk of atrial fibrillation: the FHS. Circ Arrhythm Electrophysiol 2020;13:e007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Mayr M, Yusuf S, Weir G, Chung YL, Mayr U, Yin X, Ladroue C, Madhu B, Roberts N, De Souza A, Fredericks S, Stubbs M, Griffiths JR, Jahangiri M, Xu Q, Camm AJ. Combined metabolomic and proteomic analysis of human atrial fibrillation. J Am Coll Cardiol 2008;51:585–594. [DOI] [PubMed] [Google Scholar]
- 111. De Souza AI, Cardin S, Wait R, Chung YL, Vijayakumar M, Maguy A, Camm AJ, Nattel S. Proteomic and metabolomic analysis of atrial profibrillatory remodelling in congestive heart failure. J Mol Cell Cardiol 2010;49:851–863. [DOI] [PubMed] [Google Scholar]
- 112. Alonso A, Yu B, Qureshi WT, Grams ME, Selvin E, Soliman EZ, Loehr LR, Chen LY, Agarwal SK, Alexander D, Boerwinkle E. Metabolomics and incidence of atrial fibrillation in African Americans: the Atherosclerosis Risk in Communities (ARIC) study. PLoS One 2015;10:e0142610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sinner MF, Wang N, Fox CS, Fontes JD, Rienstra M, Magnani JW, Vasan RS, Calderwood AH, Pencina M, Sullivan LM, Ellinor PT, Benjamin EJ. Relation of circulating liver transaminase concentrations to risk of new-onset atrial fibrillation. Am J Cardiol 2013;111:219–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Alonso A, Misialek JR, Amiin MA, Hoogeveen RC, Chen LY, Agarwal SK, Loehr LR, Soliman EZ, Selvin E. Circulating levels of liver enzymes and incidence of atrial fibrillation: the atherosclerosis risk in Communities cohort. Heart 2014;100:1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Rainer PP, Primessnig U, Harenkamp S, Doleschal B, Wallner M, Fauler G, Stojakovic T, Wachter R, Yates A, Groschner K, Trauner M, Pieske BM, von Lewinski D. Bile acids induce arrhythmias in human atrial myocardium – implications for altered serum bile acid composition in patients with atrial fibrillation. Heart 2013;99:1685–1692. [DOI] [PubMed] [Google Scholar]
- 116. Al Inizi S, Gupta R, Gale A. Fetal tachyarrhythmia with atrial flutter in obstetric cholestasis. Int J Gynaecol Obstet 2006;93:53–54. [DOI] [PubMed] [Google Scholar]
- 117. Ko D, Riles EM, Marcos EG, Magnani JW, Lubitz SA, Lin H, Long MT, Schnabel RB, McManus DD, Ellinor PT, Ramachandran SV, Wang TJ, Gerszten RE, Benjamin EJ, Yin X, Rienstra M. Metabolomic profiling in relation to new-onset atrial fibrillation (from the Framingham Heart Study). Am J Cardiol 2016;118:1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lewis GD, Asnani A, Gerszten RE. Application of metabolomics to cardiovascular biomarker and pathway discovery. J Am Coll Cardiol 2008;52:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kahr PC, Piccini I, Fabritz L, Greber B, Scholer H, Scheld HH, Hoffmeier A, Brown NA, Kirchhof P. Systematic analysis of gene expression differences between left and right atria in different mouse strains and in human atrial tissue. PLoS One 2011;6:e26389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hsu J, Hanna P, Van Wagoner DR, Barnard J, Serre D, Chung MK, Smith JD. Whole genome expression differences in human left and right atria ascertained by RNA sequencing. Circ Cardiovasc Genet 2012;5:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.GTEx Consortium. The genotype-tissue expression (GTEX) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Nicolae DL, Cox NJ, Im HK; GTEx Consortium. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 2015;47:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, Torstenson ES, Shah KP, Garcia T, Edwards TL, Stahl EA, Huckins LM, Nicolae DL, Cox NJ, Im HK; GTEx Consortium. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 2018;9:1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Li X, Kim Y, Tsang EK, Davis JR, Damani FN, Chiang C, Hess GT, Zappala Z, Strober BJ, Scott AJ, Li A, Ganna A, Bassik MC, Merker JD, Hall IM, Battle A, Montgomery SB; GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. The impact of rare variation on gene expression across tissues. Nature 2017;550:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhou J, Theesfeld CL, Yao K, Chen KM, Wong AK, Troyanskaya OG. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat Genet 2018;50:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lin H, Dolmatova EV, Morley MP, Lunetta KL, McManus DD, Magnani JW, Margulies KB, Hakonarson H, del Monte F, Benjamin EJ, Cappola TP, Ellinor PT. Gene expression and genetic variation in human atria. Heart Rhythm 2014;11:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Müller M, Sotoodehnia N, Sinner MF, Verwoert GC, Li M, Kao WHL, Köttgen A, Coresh J, Bis JC, Psaty BM, Rice K, Rotter JI, Rivadeneira F, Hofman A, Kors JA, Stricker BHC, Uitterlinden AG, van Duijn CM, Beckmann BM, Sauter W, Gieger C, Lubitz SA, Newton-Cheh C, Wang TJ, Magnani JW, Schnabel RB, Chung MK, Barnard J, Smith JD, Van Wagoner DR, Vasan RS, Aspelund T, Eiriksdottir G, Harris TB, Launer LJ, Najjar SS, Lakatta E, Schlessinger D, Uda M, Abecasis GR, Müller-Myhsok B, Ehret GB, Boerwinkle E, Chakravarti A, Soliman EZ, Lunetta KL, Perz S, Wichmann H-E, Meitinger T, Levy D, Gudnason V, Ellinor PT, Sanna S, Kääb S, Witteman JCM, Alonso A, Benjamin EJ, Heckbert SR. Genome-wide association study of PR interval. Nat Genet 2010;42:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Marroni F, Pfeufer A, Aulchenko YS, Franklin CS, Isaacs A, Pichler I, Wild SH, Oostra BA, Wright AF, Campbell H, Witteman JC, Kääb S, Hicks AA, Gyllensten U, Rudan I, Meitinger T, Pattaro C, van Duijn CM, Wilson JF, Pramstaller PP; EUROSPAN Consortium. A genome-wide association scan of RR and QT interval duration in 3 European genetically isolated populations: the Eurospan project. Circ Cardiovasc Genet 2009;2:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wang B, Lunetta KL, Dupuis J, Lubitz SA, Trinquart L, Yao L, Ellinor PT, Benjamin EJ, Lin H. Integrative omics approach to identifying genes associated with atrial fibrillation. Circ Res 2020;126:350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E, Sealfon SC, Chasman DI, FitzGerald GA, Dolinski K, Grosser T, Troyanskaya OG. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet 2015;47:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. van Ouwerkerk AF, Bosada FM, van Duijvenboden K, Hill MC, Montefiori LE, Scholman KT, Liu J, de Vries AAF, Boukens BJ, Ellinor PT, Goumans M, Efimov IR, Nobrega MA, Barnett P, Martin JF, Christoffels VM. Identification of atrial fibrillation associated genes and functional non-coding variants. Nat Commun 2019;10:4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tao H, Shi KH, Yang JJ, Li J. Epigenetic mechanisms in atrial fibrillation: new insights and future directions. Trends Cardiovasc Med 2016;26:306–318. [DOI] [PubMed] [Google Scholar]
- 133. Li J, Zuo K, Zhang J, Hu C, Wang P, Jiao J, Liu Z, Yin X, Liu X, Li K, Yang X. Shifts in gut microbiome and metabolome are associated with risk of recurrent atrial fibrillation. J Cell Mol Med 2020;24(22):13356–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zanetti D, Bergman H, Burgess S, Assimes TL, Bhalla V, Ingelsson E. Urinary albumin, sodium, and potassium and cardiovascular outcomes in the UK BioBank: observational and mendelian randomization analyses. Hypertension 2020;75:714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Abdul Rehman S, Khurshid Z, Hussain Niazi F, Naseem M, Al Waddani H, Sahibzada HA, Sannam Khan R. Role of salivary biomarkers in detection of cardiovascular diseases (CVD). Proteomes 2017;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 2007;448:313–317. [DOI] [PubMed] [Google Scholar]
- 137. Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell 2008;134:877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Dobrev D, Carlsson L, Nattel S. Novel molecular targets for atrial fibrillation therapy. Nat Rev Drug Discov 2012;11:275–291. [DOI] [PubMed] [Google Scholar]
- 139. Calvo D, Filgueiras-Rama D, Jalife J. Mechanisms and drug development in atrial fibrillation. Pharmacol Rev 2018;70:505–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Low SK, Takahashi A, Ebana Y, Ozaki K, Christophersen IE, Ellinor PT, Ogishima S, Yamamoto M, Satoh M, Sasaki M, Yamaji T, Iwasaki M, Tsugane S, Tanaka K, Naito M, Wakai K, Tanaka H, Furukawa T, Kubo M, Ito K, Kamatani Y, Tanaka T; AFGen Consortium. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet 2017;49:953–958. [DOI] [PubMed] [Google Scholar]
- 141. Lee JY, Kim TH, Yang PS, Lim HE, Choi EK, Shim J, Shin E, Uhm JS, Kim JS, Joung B, Oh S, Lee MH, Kim YH, Pak HN. Korean atrial fibrillation network genome-wide association study for early-onset atrial fibrillation identifies novel susceptibility loci. Eur Heart J 2017;38:2586–2594. [DOI] [PubMed] [Google Scholar]
- 142. Bundy JD, Heckbert SR, Chen LY, Lloyd-Jones DM, Greenland P. Evaluation of risk prediction models of atrial fibrillation (from the multi-ethnic study of atherosclerosis [mesa]). Am J Cardiol 2020;125:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Everett BM, Cook NR, Conen D, Chasman DI, Ridker PM, Albert CM. Novel genetic markers improve measures of atrial fibrillation risk prediction. Eur Heart J 2013;34:2243–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Lubitz SA, Yin X, Lin HJ, Kolek M, Smith JG, Trompet S, Rienstra M, Rost NS, Teixeira PL, Almgren P, Anderson CD, Chen LY, Engstrom G, Ford I, Furie KL, Guo X, Larson MG, Lunetta KL, Macfarlane PW, Psaty BM, Soliman EZ, Sotoodehnia N, Stott DJ, Taylor KD, Weng LC, Yao J, Geelhoed B, Verweij N, Siland JE, Kathiresan S, Roselli C, Roden DM, van der Harst P, Darbar D, Jukema JW, Melander O, Rosand J, Rotter JI, Heckbert SR, Ellinor PT, Alonso A, Benjamin EJ; AFGen Consortium. Genetic risk prediction of atrial fibrillation. Circulation 2017;135:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Sinner MF, Stepas KA, Moser CB, Krijthe BP, Aspelund T, Sotoodehnia N, Fontes JD, Janssens AC, Kronmal RA, Magnani JW, Witteman JC, Chamberlain AM, Lubitz SA, Schnabel RB, Vasan RS, Wang TJ, Agarwal SK, McManus DD, Franco OH, Yin X, Larson MG, Burke GL, Launer LJ, Hofman A, Levy D, Gottdiener JS, Kaab S, Couper D, Harris TB, Astor BC, Ballantyne CM, Hoogeveen RC, Arai AE, Soliman EZ, Ellinor PT, Stricker BH, Gudnason V, Heckbert SR, Pencina MJ, Benjamin EJ, Alonso A. B-type natriuretic peptide and c-reactive protein in the prediction of atrial fibrillation risk: the charge-af consortium of community-based cohort studies. Europace 2014;16:1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Smith JG, Newton-Cheh C, Almgren P, Melander O, Platonov PG. Genetic polymorphisms for estimating risk of atrial fibrillation in the general population: a prospective study. Arch Intern Med 2012;172:742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Tada H, Shiffman D, Smith JG, Sjogren M, Lubitz SA, Ellinor PT, Louie JZ, Catanese JJ, Engstrom G, Devlin JJ, Kathiresan S, Melander O. Twelve-single nucleotide polymorphism genetic risk score identifies individuals at increased risk for future atrial fibrillation and stroke. Stroke 2014;45:2856–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.