Abstract
The classical factors for predicting prognosis currently cannot meet the developing requirements of individualized and accurate prognostic evaluation in lung adenocarcinoma (LUAD). With the rapid development of high-throughput DNA sequencing technologies, genomic changes have been discovered. These sequencing data provide unprecedented opportunities for identifying cancer molecular subtypes. In this article, we classified LUAD into two distinct molecular subtypes (Cluster 1 and Cluster 2) based on Copy Number Variations (CNVs) and mRNA expression data from the Cancer Genome Atlas (TCGA) based on non-negative matrix factorization. Patients in Cluster 1 had worse outcomes than that in Cluster 2. Molecular features in subtypes were assessed to explain this phenomenon by analyzing differential expression genes expression pattern, which involved in cellular processes and environmental information processing. Analysis of immune cell populations suggested different distributions of CD4+ T cells, CD8+ T cells, and dendritic cells in the two subtypes. Subsequently, two novel genes, TROAP and RASGRF1, were discovered to be prognostic biomarkers in TCGA, which were confirmed in GSE31210 and Tianjin Medical University Cancer Institute and Hospital LUAD cohorts. We further proved their crucial roles in cancers by vitro experiments. TROAP mediates tumor cell proliferation, cycle, invasion, and migration, not apoptosis. RASGRF1 has a significant effect on tumor microenvironment. In conclusion, our study provides a novel insight into molecular classification based on CNVs related genes in LUAD, which may contribute to identify new molecular subtypes and target genes.
Keywords: Molecular subtypes, CNVs, Prognostic biomarkers, LUAD, TROAP, RASGRF1
Introduction
Lung cancer remains the most prevalent cancer type with the highest mortality throughout the world (1). Lung adenocarcinoma (LUAD) is the main histologic subtype and accounts for over 40% among lung cancer with enormous morphological and genomic heterogeneity (2). The classical factors for predicting prognosis in non-small-cell lung cancer (NSCLC), such as TNM stage and histological differentiation, currently cannot meet the developing requirements of individualized and accurate prognostic evaluation. Transcriptional, proteomic, genomic, and epigenomic profiles provided more strategies to classify molecular subtypes for precision management of tumors patients (3, 4, 5, 6, 7). With the increasing molecular targeted and immune therapies, the treatment options of lung adenocarcinoma have changed drastically over the past decade (8). However, improvements in clinical outcomes were limited to small subgroups because of phenotypic and molecular heterogeneity (9). More attention has been paid to the integration of molecular and genomic analysis in LUAD for the diagnosis and treatment (10), owing to high rates of somatic mutations and genome rearrangements occurs in LUAD (11).
Copy Number Variations (CNVs) is a kind of DNA structural variation that leads to the abnormal copy number of DNA fragments in genome during tumorigenesis (12). CNVs has been shown to be involved in the development of various cancers by regulating mRNA level and affecting transcription regulation (13). Whole genome duplication and CNVs are early evolution events in NSCLC, the number or the complexity of which were high risk indicator for recurrence or death (14). In recent years, CNVs has be used to distinguish malignant and non-malignant tissues by inferCNV algorithm (15).
Initially, the classification of cancer subtypes was mostly based on clinical pathology (16). With the rapid development of high-throughput DNA sequencing technologies, genomic changes have been discovered. These sequencing data provide unprecedented opportunities for identifying cancer molecular subtype (17). A recent research discovered that CNVs characteristics distinguished malignant ductal cells subtypes in pancreatic ductal adenocarcinoma using single-cell RNA-seq (18). Not only that, it has been found that CNVs can affect antitumor immunity by affecting immune cell subsets and tumoricidal activity, which is expected to become a new immunotherapy prediction biomarker (19). This indicates that there has been a growing recognition of the role of CNVs in tumorigenesis. At present, there are several researches on molecular genotyping identified using immune related transcriptome data in LUAD (20,21). Based on these reports, we attempted to use the DNA copy-number-correlated (CNVcor) genes to conducte molecular subtypes in LUAD.
In this research, we have identified two molecular subtypes (Cluster 1 and Cluster 2) with distinct molecular characteristics in LUAD based on CNVcor genes from the Cancer Genome Atlas (TCGA). Furthermore, expression features, functional characteristics, and the prognostic values of patients can be evaluated by the two subtypes. Then, we discovered two key differentially expressed genes (DEGs) between the two subtypes: Trophinin-associated protein (TROAP) and Ras protein specific guanine nucleotide releasing factor 1 (RASGRF1), which served as prognostic predictors. Finally, we explored their prognostic values by immunohistochemistry (IHC) in Tianjin Medical University Cancer Institute and Hospital (TMUCIH) LUAD tissue microarray (TMA) cohorts, and functional roles in vitro experiments. TROAP and RASGRF1 have been extensively studied as critical genes that play a major role in cancer cell survival or immune regulation. In conclusion, our study provides a novel insight into molecular subtypes by CNVcor genes in LUAD, which might be helpful in revealing more mechanism behind cancers, identifying new prognostic indicators, and finding new therapeutic targets in cancers.
Materials and methods
Download and preprocessing of DNA copy numbers and mRNA expression
DNA copy numbers, mRNA expression FPKM values and the corresponding clinical information of patients in the LUAD project were obtained from the TCGA data portal (https://portal.gdc.cancer.gov/). A total of 443 samples with CNVs, RNA-Seq and clinical data were included in the research. DNA copy numbers from TCGA were preprocessed as follows. Regions with probes < 5 of copy number segment data in LUAD were removed. Two regions with 50% overlaps were considered as the same region. We quantified the CNVs region with its corresponding gene using the GENCODE GRCh38 release 22 annotations. Multiple CNVs regions annotated to the same gene were merged into one region, then we took the average of the multiple CNVs regions as the final CNVs region value. Genes with FPKM = 0 had less than 0.5% of all the samples were filtered out. SNVs with a mutation allelic fraction (MAF) were downloaded for copy number alterations, tumor mutational burden (TMB), and microsatellite instability (MSI) calculation, by “Maftools” package.
Determination of molecular subgroups by CNVcor genes
The correlation coefficients between CNVs data and FPKM value were calculated by Pearson's correlations and then converted into Z-values by using formula ln ([1+r]/[1-r]). Genes with P values < 0.05 were considered as the CNVcor gene sets, which were provided in Table S1. Non-negative matrix factorization (NMF) clustering was performed to cluster CNVcor datasets using NMF bioconductor package (R version 3.3.5). With standard “brunet” method, 50 iterations were employed and cluster number K was set at 2 to 5, which were sufficient to achieve the optimal cluster K = 2 on the basis of cophenetic, dispersion and silhouette. Two molecular subtypes (CNVcorC1 and CNVcorC2 cluster) were identified as Cluster 1 and Cluster 2.
Identification of DEGs and fuctional enrichment analysis
Differential expression genes between Cluster 1 and Cluster 2 cluster were calculated using the Limma package with False Discovery Rate (FDR) < 0.05 and cut-off values of |log2FC| > 1. The defined DEGs were subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathyway (http://www.kegg.jp) and Gene Ontology (GO) (http://www.geneontology.org) functional enrichment analysis using “ggplot2” and “clusterProfiler” packages in R version 3.3.5. Two-side wilcoxon test was used to compare RASGRF1 or TROAP between two subgroups. Kruskal-Wallis test was used to test for multiple groups. Gene Set Enrichment Analysis (GSEA) was conducted using GSEA v3.0 (http://www.broadinstitute.org/gsea), the results of which were provided in Table S2.
Tumor Tissue Samples and Immunohistochemistry
All samples were obtained from TMUCIH, which was approved by the Ethical Committee of TMUCIH and each participator with informed consent. A total of 216 NSCLC patients receiving pulmonary resection and systemic lymph node dissection had no neoadjuvant therapy before the sample collection between January 2013 and June 2014. All patients were followed up until April 2020. The 2 mm TMA were generated from formalin-fixed paraffin sections of NSCLC. TMA were deparaffinized and rehydrate. Then, antigen retrieval was performed by EDTA pH 9.0 in a microwave. After blocking with 3% hydrogen peroxide and 5% goat serum, slides were incubated with primary antibodies overnight at 4°C, followed by EIVISON plus (kit-9903, MXB, China) and DAB kit (ZLI-9019, ZSGB-BIO, China). Immunostaining was evaluated under light microscopy at 400X magnification. RASGRF1 and TROAP IHC staining were assessed by two pathologists using histologic score (H score) by multiplying fraction score and intensity score (range, 0-300). The absolute number of immune cells were manually counted. Primary antibodies and specific concentrations are listed in Table S3.
Immune cells infiltration analysis
The tumor immunological infiltration features between Cluster 1 and Cluster 2 were systematically and comprehensively analyzed by ESTIMATE in the Tumor immune estimation resource (TIMER) (https://cistrome.shinyapps.io/timer/), and GSVA algorithm in TISIDB (http://cis.hku.hk/TISIDB/), which contains samples from TCGA. The immune cells included CD4+ T cell, CD8+ T cell, CD20+ B cell, CD68+ macrophage, and CD11c+ dendritic cells (DCs) were calculated by IHC. Correlations of RASGRF1 and immune cells were tested by Spearman rank test.
Kaplan-Meier survival analysis
Overall survival (OS) was the period between surgical resection and death or the last follow-up. Progression-free survival (PFS) was the time between surgical resection and progression. Disease-free survival (DFS) was the time to relapse or death from any cause. Survival estimations were employed to assess the survival of the patients from TCGA project or TMUCIH cohort using Kaplan-Meier curves with a log-rank test. HR calculated by cox regression analysis. Maxstat package was used to determine the optimal cutoff point for the continuous variables in R 3.3.5. GSE31210 dataset was used to validated the prognostic values of RASGRF1 and TROAP from Kaplan-Meier plotter (http://kmplot.com/analysis/).
Cell culture and transfection
BEAS-2B, A549, and NCI-H1299 cell lines were purchased from ATCC. GLC-82 and LTEP-A2 cell lines were purchased from the Tumor Cell Bank of Chinese Academy of Medical Science (Shanghai, China). All the cell lines were cultured in the indicated media as instructed. Cells were transfected by small interfering RNA (siRNA) specifically targeted TROAP using Lipofectamine RNAiMAX, or RASGRF1 plasmid using Lipofectamine 3000 transfection reagent (Invitrogen, USA). The sequences of TROAP siRNA were provided in Table S2.
RT-qPCR and Western blot
Total RNA was extracted by Trizol (Invitrogen) according to the manufacturer's instruction. cDNA was synthesized by PrimeScript RT Master Mix (TaKaRa). Quantitative RT-PCR (RT-qPCR) was performed with RASGRF1 or TROAP primer sequences and analyzed by the comparative Ct value (2−ΔΔCt). The sequences of primers were provided in Table S2. Protein extraction and concentration measurement were conducted and isolated by SDS-PAGE. And then the protein was transferred onto a PVDF membrane and immunoblotted with anti-RASGRF1 or anti-TROAP at 4°C overnight. HRP-conjugated secondary antibodies were applied and incubated for 1 hour at room temperature.
Cell Functional Experiments
A549 and H1299 cells transfected with TROAP siRNA/control or RASGRF1 plasmid/vector were applied in the following experiments. Each experiment was conducted in triplicate. Three thousand A549 cells or 2000 H1299 cells were seeded in each well in 96-well plates for CCK-8 assay. CCK-8 reagent (US Everbright Inc., China) was added into each well and incubated for 2 hours, measured at 450 nm. Cells were seeded into 6-well plate (1000/well) and incubated for 7 days in clonogenic assay. Transwell chamber filters (BD, 8-um) were coated with Matrigel (40ul/filter), followed by cells placed in the upper chamber (2 × 104 cells/well) in serum-free PRMI-1640 medium. The chamber was then placed in a well containing 500 ul 1640 medium with 20% fetal bovine serum. After 48 hours of incubation, cells on the upper chambers removed to the lower chambers, stained with 0.2% crystal violet. For scratch assay, cells in 6-well plates were allowed to reach 80% confluence and then changed to serum-free medium for 48 hours. Cell cycle analysis was detected by flow cytometric analysis of propidium iodide (PI) staining (US Everbright Inc., China). Annexin V staining was performed using Apoptosis Kit (US Everbright Inc., China).
Results
Molecular subtypes defined by CNVcor genes
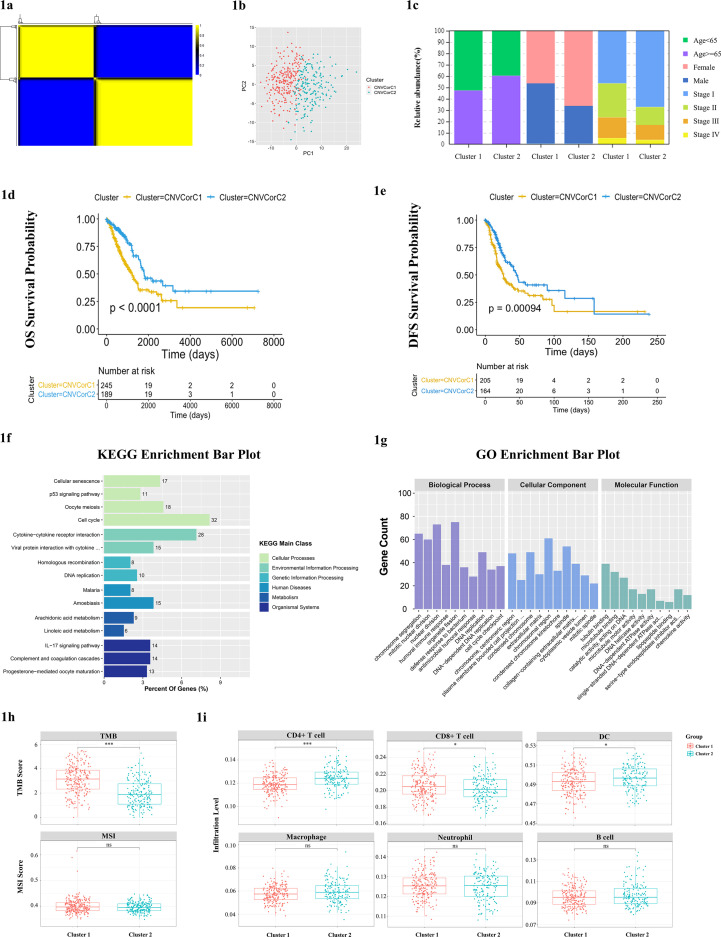
Firstly, genomic profiles of DNA CNVs, gene expression and clinical information were obtained from 443 samples in TCGA LUAD subject. We then filtered out differential gene mRNA expression analysis to identify 1802 differential genes (|log2 fold change (log2FC) | > 1 and FDR < 0.05), 343 of which had effect on patient outcomes (P < 0.05). Pearson correlations were performed between gene CNVs and mRNA expression and identified 160 genes which mRNA expression were correlated with CNVs as CNVcor genes in the 343 genes (P < 0.05). Subsequently, two molecular subtypes (Cluster 1 and Cluster 2) were identified by the CNVcor gene sets using NMF. With standard “brunet” method, 50 iterations were employed and cluster number K was set at 2 to 5, which were sufficient to achieve the optimal cluster K = 2 (Fig. 1A). The principal component analysis showed that samples were separated into two distinct clusters with little samples overlaps (Fig. 1B), in which Cluster 1 accounted for 56.7% and Cluster 2 accounted for 43.3% (Table S1). The two subtypes were significantly associated with age, gender, and stage using Chi-square test. Cluster 1 had a higher proportion of younger patients, male patients, and patients with II-IV stages compared with Cluster 2 (Fig. 1C). In addition, there were significant differences between the two clusters with regard to OS (P < 0.0001) and DFS (P = 0.00094) and Cluster 1 showed poorer outcomes than Cluster 2 (Fig. 1D, E). The copy number alterations of these 160 genes between the two clusters were shown in Fig. S1A.
Fig. 1.
Molecular subtypes defined by CNVcor genes in LUAD. (A) CNVcor genes expressions were correlated with DNA CNV based on Pearson correlations (p<0.05). Two distinct molecular subtypes were identified by the CNVcor gene sets by NMF. (B) The PCA of gene expression depicted little overlaps of samples between the two subgroups: patients were clearly classified into two clusters. (C) The distribution of samples in the two clusters revealed the distribution of clinical features in subtypes by Chi-square test. (D) Survival analysis with Kaplan-Meier plot showed the OS rate in Cluster 2 subgroup was significantly higher than that in Cluster 1 (p<0.0001); (E) the DFS rate in Cluster 2 subgroup was significantly higher than that in Cluster 1 (p=0.00094). (F) KEGG enrichment of 944 DEGs between Cluster 1 and 2, and six main categories were enriched. (G) GO functional annotation analysis of DEGs between Cluster 1 and 2 in terms of three categories including BP, CC, and MF. (H) TMB and MSI levels in the two clusters. (I) Immune cell infiltration feature between the two clusters using TIMER, including CD8+ T cells, CD4+ T cells, B cells, macrophage, neutrophil, and DCs. DCs (p=0.011) and CD4 +T cells (p=6e-09) were significantly higher in Cluster 2 than that in Cluster 1, while CD 8+T cells scores were lower (p=0.038). No significant difference was observed between Cluster1 and Cluster2 cluster in B cells, macrophage, and neutrophil. P-values from wilcoxon test. *P value ≤ 0.05; **P value ≤ 0.01; ***P value ≤ 0.001; ****P value ≤ 0.0001.
Molecular features and immune cells infiltrations characteristics in subtypes
In order to compare molecular features between Cluster 1 and Cluster 2, we analyzed the DEGs using Limma package. Heatmap represented 495 genes overexpressed and 449 genes underexpressed in Cluster 1 (|log2-FC|> 1 and FDR < 0.05) (Fig. S1B). To clarify the potential functions of the DEGs, we performed the KEGG enrichment analysis, which suggested that the dysregulated genes were predominantly implicated in cellular processes and environmental information processing pathways significantly, such as “p53 signaling pathway”, “cell cycle” and “cytokine-cytokine receptor interaction” (Fig. 1F). Then, GO annotation and enrichment analysis of DEGs were applied to further assay the functional characteristics from three aspects covering biological process (BP), cellular component (CC) and molecular function (MF), respectively (Fig. 1G). The significant results revealed that the DEGs were enriched in “DNA-dependent DNA replication”, “cell cycle checkpoint”, “humoral immune response”, “antibacterial humoral response”, and “defense response to bacterium” in terms of BP. Regarding CC, the DEGs were mainly located in “chromosome, centromeric region”, “chromosomal region”, “collagen-containing extracellular matrix”, “extracellular matrix”, and “cytoplasmic vesicle lumen”. Under MF the DEGs were enriched in “microtubule binding”, “catalytic activity, acting on DNA”, and “chemokine activity”.
Above results prompted us to evaluate the immune related features between the two clusters by wilcoxon test. Higher TMB level was found in Cluster 1 than Cluster 2 (Fig. 1H). Tumor-infiltrating immune cells were estimated by TIMER, including CD8+ T cells, CD4+ T cells, B cells, macrophage, neutrophil, and DCs. The immune proportions of six immune cells were displayed in Fig. 1H. We observed that the immune proportions of DCs (P = 0.011) and CD4+ T cells (P = 6e-09) were significantly higher in Cluster 2 than that in Cluster 1, while CD8+ T cells scores were lower (P = 0.038) (Fig. 1I). No significant difference was observed between Cluster 1 and Cluster 2 in B cells, macrophage, and neutrophil.
RASGRF1 and TROAP identified as the potential key genes in LUAD
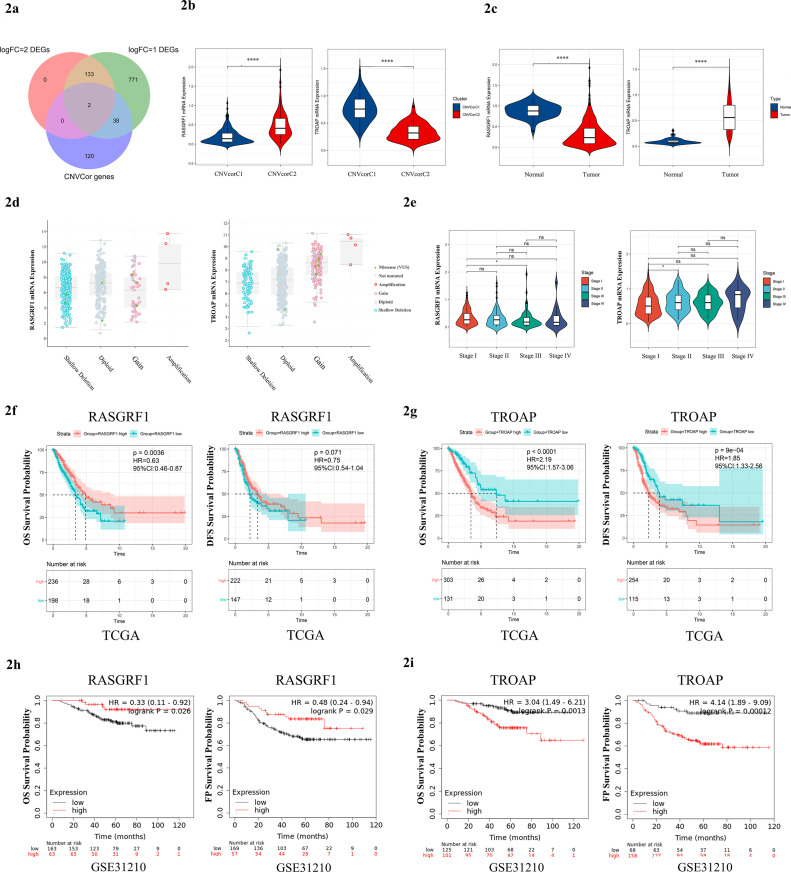
Prognostic value and immune cell infiltration proportions differences between the two subgroups suggested that specific molecular biomarkers may contribute to patient's prognostication. To find these key genes, we took the intersection of the DEGs and CNVcor genes. We found that RASGRF1 and TROAP were both DEGs and CNVcor genes (Fig. 2A), which implied that RASGRF1 and TROAP may play essential roles in the two different clusters. RASGRF1 was lowly expressed in the Cluster 1 than Cluster 2 (P = 5.4e-24) and tumor tissues than normal tissues (P = 1.641e-26), and TROAP was highly expressed in the Cluster 1 than Cluster 2 (P = 8.687e-52) and tumor tissues than normal tissues (P = 2.92e-31) by wilcoxon test (Fig. 2B, C). The different relationship between gene copy number alterations and transcript levels for RASGRF1 and TROAP is shown in Fig. 2D. RASGRF1 and TROAP were differentially expressed among different tumor stage (Fig. 2E). Significant association was found between RASGRF1 expression and age (P = 0.16), or gender (P = 9.6e-05) (Fig. S2A), meanwhile the expression level of TROAP was associated with age (P = 0.00019), or gender (P = 0.000069) (Fig. S2B). The patients with high RASGRF1 expression had an obviously longer OS (HR = 0.63, P = 0.0036) than those with low RASGRF1 expression, and longer DFS (HR = 0.75, P = 0.071) without any statistical significance (Fig. 2F). However, the patients with high TROAP expression had a significantly shorter OS (HR = 2.19, P < 0.0001) and DFS (HR = 1.85, P = 9e-04) than those with low TROAP expression (Fig. 2G). Subsequently, GSE31210 dataset was chosen to further verified the prognostic values of RASGRF1 (OS HR = 0.33 P = 0.026, FP HR = 0.48 P = 0.029) (Fig. 2H) and TROAP (OS HR = 3.04 P = 0.0013, FP HR = 4.14 P = 0.00012) (Fig. 2I). Based on the results above, we identified RASGRF1 and TROAP as significant prognostic factors in LUAD.
Fig. 2.
Identification of RASGRF1 and TROAP as the potential key genes. (A) Intersection of DEGs and CNVCor genes contains two key genes: RASGRF1 and TROAP. (B) RASGRF1 expression was lowly expressed in Cluster1 (p=5.4e-24), and TROAP was highly expressed in Cluster1 (p=8.687e-52). (C) RASGRF1 expression was lowly expressed in tumor tissue (p=1.641e-26), and TROAP was highly expressed in tumor tissues (p=2.92e-31). (D) The association between gene copy-number alterations and transcript levels using cBioportal. (E) Relative expression level of RASGRF1 and TROAP in different clinical stages. Statistical test: Kruskal-Wallis test. (F) Survival analysis with Kaplan-Meier plot shows that patients with high RASGRF1 expression had an obviously longer OS (HR=0.63, p=0.0036) than those with low RASGRF1 expression, and longer DFS (HR=0.75, p=0.071) without any statistical significance; (G) Patients with high TROAP expression had a significantly shorter OS (HR=2.19, p<0.0001) and DFS (HR=1.85, p=9e-04) than those with low TROAP expression. (H) GSE31210 dataset was chosen to further verified the prognostic values of RASGRF1 and (I) TROAP. P-values were calculated using the log-rank test.
RASGRF1 and TROAP protein expression characteristics and prognostic value
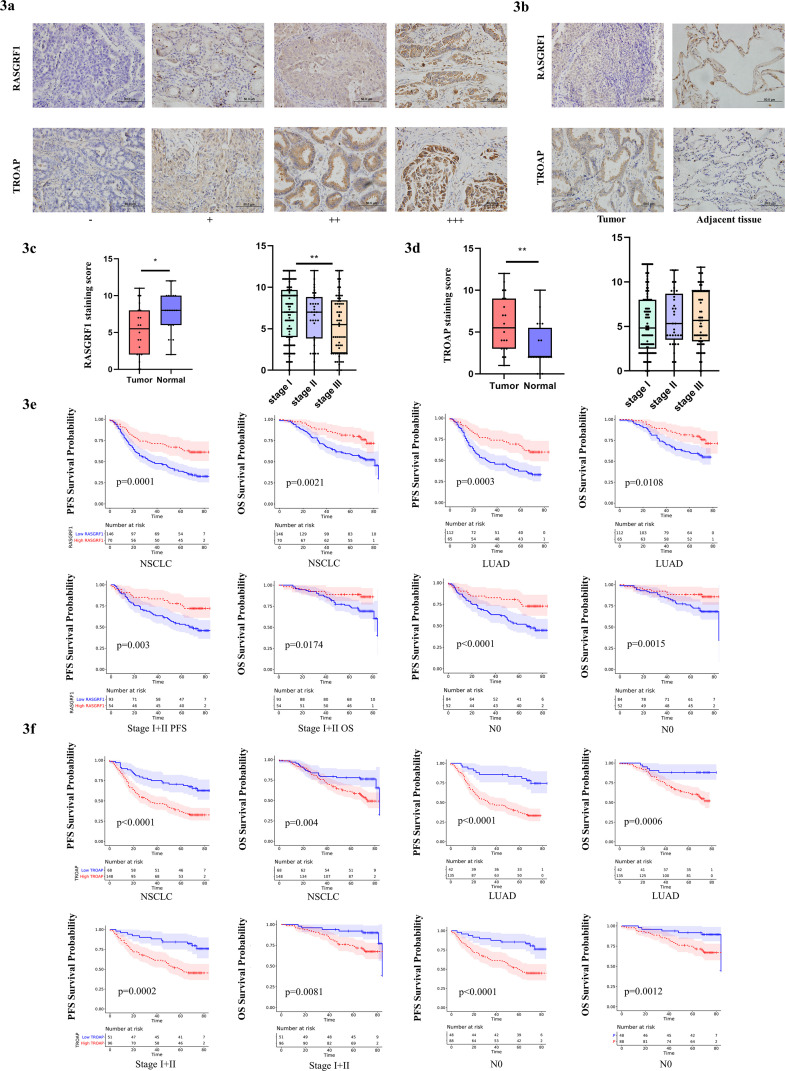
Subsequently, we investigate the expression of RASGRF1 and TROAP in 216 NSCLC samples and 20 adjacent tissues from TMUCIH by IHC. And we further evaluated the relationship between clinical information and RASGRF1 or TROAP expression by Chi-square test (Table 1). Decreased RASGRF1 expression was associated with gender (P = 0.016), histologic type of lung cancer (P = 0.004), clinical stage (P = 0.024), lymph node classification (P = 0.017), and smoking status (P = 0.025). Increased TROAP expression was significantly associated with gender (P = 0.044), histologic type of lung cancer (P < 0.0001), and smoking status (P = 0.007). Representative staining of RASGRF1 and TROAP expression was predominantly visible in the cytoplasm at 400x magnification (Fig. 3A). Consistent with the preceding analysis in TCGA, RASGRF1 was lowly expressed in tumor tissue (P = 0.0181) (Fig. 3C), and TROAP was highly expressed in tumor tissue than that in adjacent normal tissue (P = 0.0061) (Fig. 3D). We then evaluated the expression in different clinical stage and found that RASGRF1 showed significant differences in expression levels between stage I and III (P = 0.0084, Fig. 3C).
Table 1.
Patient clinical parameters and their association with RASGRF1 or TROAP expression.
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| Hazard Ratio | Hazard Ratio | |||||
| Clinical parameters | n | (95%CI) | P Value | (95%CI) | P Value | |
| Age(years) | ||||||
| < 65 | 152 | 1.098 | 0.6314 | 1.128 | 0.599 | |
| ≥ 65 | 64 | (0.742∼1.624) | (0.710∼1.794) | |||
| Gender | ` | |||||
| Males | 118 | 0.806 | 0.2403 | 0.635 | 0.0408* | |
| Females | 98 | (0.563∼1.154) | (0.414∼0.975) | |||
| Type | ||||||
| LUAD | 177 | 1.059 | 0.8067 | 1.559 | 0.0651 | |
| LUSC | 39 | (0.661∼1.696) | (0.889∼2.733) | |||
| Clinical stage | ||||||
| I+II | 147 | 3.331 | <0.0001*** | 4.361 | <0.0001*** | |
| III | 69 | (2.161∼5.133) | (2.614∼7.274) | |||
| T classification | ||||||
| T1 | 97 | 1.988 | 0.0002*** | 1.962 | 0.0028** | |
| T2+T3+T4 | 119 | (1.390∼2.844) | (1.279∼3.010) | |||
| N classification | ||||||
| N0 | 136 | 2.924 | <0.0001*** | 3.787 | <0.0001*** | |
| N1+N2+N3 | 80 | (1.964∼4.353) | (2.364∼6.067) | |||
| Smoke | ||||||
| No smoking | 112 | 1.109 | 0.5698 | 1.551 | 0.0426* | |
| Smoking | 104 | (0.774∼1.588) | (1.009∼2.384) | |||
| RASGRF1 | ||||||
| Low | 146 | 0.438 | 0.0001*** | 0.445 | 0.0021** | |
| High | 70 | (0.304∼0.633) | (0.286∼0.695) | |||
| TROAP | ||||||
| Low | 68 | 2.612 | <0.0001*** | 2.12 | 0.004** | |
| High | 148 | (1.807∼3.774) | (1.354∼3.320) | |||
Note. Bold font indicates P < 0.05. *P value ≤ 0.05; **P value ≤ 0.01; ***P value ≤ 0.001; ****P value ≤ 0.0001.
Fig. 3.
RASGRF1 and TROAP expression characteristics and prognostic value. (A) Representative IHC images were taken at 400x magnification: negative (-), weak positive (+), moderate positive (++), and strong positive (+++). (B) Representative staining in tumor and tumor-adjacent normal tissue; RASGRF1 staining score was statistically higher in normal tissue compared to tumor tissue (n=20, p=0.0181), and TROAP staining score was statistically higher in tumor tissue compared to normal tissue (n=20, p=0.0061). (C) RASGRF1 expression in LUAD and different clinical stages. (D) TROAP expression in LUAD and different clinical stages. (E) RASGRF1 expression was protective factors for PFS (HR=0.438, p=0.0001) and OS (HR=0.445, p=0.0021) in NSCLC. The prognostic value of RASGRF1 was related to histologic type, clinical stage, and N classification in both PFS and OS. (F) TROAP expression was risk factors for PFS (HR=2.612, p<0.0001) and OS (HR=2.12, p=0.004) in NSCLC. The prognostic value of TROAP was associated with histologic type, clinical stage, and N classification in both PFS and OS. HR calculated by cox regression analysis based on TMUCIH data. P-Values were calculated using the log-rank test.
Simultaneously, we employed univariate analysis of variable factors correlated to PFS and OS (Table 2). The results exhibited that clinical stage, tumor size classification, and lymph node classification were risk factors for PFS and OS, as well as gender and smoking status for OS. To assess potential impact of clinical parameters on the prognostic value of RASGRF1 or TROAP, we investigated the prognostic value of RASGRF1 (Table 3) and TROAP (Table 4) in selective clinical subgroups. The prognostic value of RASGRF1 was related to age, histologic type, clinical stage, and N classification in both PFS and OS (Fig. 3E). The survival curves demonstrated that RASGRF1 expression was protective factors for PFS (HR = 0.438, P = 0.0001) and OS (HR = 0.445, P = 0.0021) in NSCLC. RASGRF1 predicts a better prognosis for PFS (HR = 0.445, P = 0.0003) and OS (HR = 0.488, P = 0.0108) in patients with LUAD; PFS (HR = 0.42, P = 0.003) and OS (HR = 0.385, P = 0.0174) in patients in stage I and II; PFS (HR = 0.404, P = 0.0028) and OS (HR = 0.402, P = 0.0264) in patients without lymph node invasion; PFS (HR = 0.339, P < 0.0001) and OS (HR = 0.361, P = 0.0015) in younger patients. The prognostic value of TROAP was associated with histologic type, clinical stage, N classification, and smoking status in both PFS and OS ((Fig. 3F). TROAP expression was risk factors for PFS (HR = 2.612, P < 0.0001) and OS (HR = 2.12, P = 0.004) in NSCLC. TROAP predicts a poorer prognosis for PFS (HR = 3.971, P < 0.0001) and OS (HR = 4.317, P = 0.0006) in patients with LUAD; PFS (HR = 3.246, P = 0.0002) and OS (HR = 2.832, P = 0.0081) in patients in stage I and II; PFS (HR = 3.338, P = 0.0002) and OS (HR = 2.899, P = 0.0128) in patients without lymph node invasion.
Table 2.
Univariate survival analysis of clinical parameters and RASGRF1 or TROAP expression with PFS and OS in patients with NSCLC.
| RASGRF1 | TROAP | |||||||
|---|---|---|---|---|---|---|---|---|
| Clinical parameters | n (%) | Low | High | P Value | Low | High | P Value | |
| Age(years) | ||||||||
| <65 | 152(70.4) | 98 | 54 | 0.131 | 48 | 104 | 0.962 | |
| ≥65 | 64(29.6) | 48 | 16 | 20 | 44 | |||
| Gender | ||||||||
| Males | 118(54.6) | 88 | 30 | 0.016* | 44 | 74 | 0.044* | |
| Females | 98(45.4) | 58 | 40 | 24 | 74 | |||
| Type | ||||||||
| LUAD | 177(81.9) | 112 | 65 | 0.004** | 42 | 135 | <0.0001*** | |
| LUSC | 39(18.1) | 34 | 5 | 26 | 13 | |||
| Clinical stage | ||||||||
| I | 113(52.3) | 67 | 46 | 0.024* | 42 | 71 | 0.166 | |
| II | 34(15.7) | 26 | 8 | 9 | 25 | |||
| III | 69(31.9) | 53 | 16 | 17 | 52 | |||
| T classification | ||||||||
| T1 | 97(44.9) | 59 | 38 | 0.152 | 35 | 62 | 0.095 | |
| T2 | 83(38.4) | 60 | 23 | 19 | 64 | |||
| T3+T4 | 36(16.7) | 27 | 9 | 14 | 22 | |||
| N classification | ||||||||
| N0 | 136(63.0) | 84 | 52 | 0.017* | 48 | 88 | 0.116 | |
| N1+N2+N3 | 80(37.0) | 62 | 18 | 20 | 60 | |||
| Smoke | ||||||||
| No smoking | 112(51.9) | 68 | 44 | 0.025* | 26 | 86 | 0.007** | |
| Smoking | 104(48.1) | 78 | 26 | 42 | 62 | |||
Note. Bold font indicates P < 0.05.
Table 3.
Univariate survival analysis of RASGRF1 expression in subgroups with different clinical parameters.
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| Hazard Ratio | Hazard Ratio | |||||
| Clinical parameters | n | (95%CI) | P Value | (95%CI) | P Value | |
| Age(years) | ||||||
| <65 | 152 | 0.339(0.219∼0.524) | <0.0001*** | 0.361(0.212∼0.616) | 0.0015** | |
| ≥65 | 64 | 0.856(0.417∼1.759) | 0.6825 | 0.809(0.345∼1.897) | 0.6372 | |
| Gender | ||||||
| Males | 118 | 0.475(0.286∼0.791) | 0.0152* | 0.419(0.233∼0.755) | 0.0185* | |
| Females | 98 | 0.430(0.247∼0.749) | 0.0053** | 0.550(0.270∼1.120) | 0.1189 | |
| Type | ||||||
| LUAD | 177 | 0.445(0.299∼0.664) | 0.0003*** | 0.488(0.295∼0.806) | 0.0108* | |
| LUSC | 39 | 0.257(0.081∼0.813) | 0.1468 | 0.331(0.093∼1.174) | 0.2486 | |
| Clinical stage | ||||||
| I+II | 147 | 0.420(0.253∼0.696) | 0.003** | 0.385(0.198∼0.748) | 0.0174* | |
| III | 69 | 0.572(0.329∼0.997) | 0.0743 | 0.571(0.310∼1.054) | 0.1053 | |
| T classification | ||||||
| T1 | 97 | 0.436(0.237∼0.799) | 0.0141* | 0.388(0.181∼0.832) | 0.033* | |
| T2+T3+T4 | 119 | 0.503(0.313∼0.809) | 0.0133* | 0.549(0.312∼0.966) | 0.0679 | |
| N classification | ||||||
| N0 | 136 | 0.404(0.238∼0.685) | 0.0028** | 0.402(0.201∼0.867) | 0.0264* | |
| N1+N2+N3 | 80 | 0.637(0.370∼1.096) | 0.1365 | 0.617(0.336∼1.135) | 0.1609 | |
| Smoke | ||||||
| No smoking | 112 | 0.427(0.260∼0.704) | 0.0019** | 0.565(0.294∼1.083) | 0.1054 | |
| Smoking | 104 | 0.450(0.255∼0.793) | 0.022* | 0.367(0.195∼0.691) | 0.0156* | |
Note. Bold font indicates P < 0.05.
Table 4.
Univariate survival analysis of RASGRF1 expression in subgroups with different clinical parameters.
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| Hazard Ratio | Hazard Ratio | |||||
| Clinical parameters | n | (95%CI) | P Value | (95%CI) | P Value | |
| Age(years) | ||||||
| <65 | 152 | 2.233(1.426∼3.497) | 0.0022** | 1.801(1.031∼3.144) | 0.0654 | |
| ≥65 | 64 | 3.646(1.908∼6.965) | 0.0015** | 3.255(1.535∼6.905) | 0.0064** | |
| Gender | ||||||
| Males | 118 | 2.118(1.322∼3.396) | 0.0038** | 1.761(1.019∼3.044) | 0.0576 | |
| Females | 98 | 5.242(2.884∼9.530) | 0.0003*** | 6.066(2.803∼13.13) | 0.0032** | |
| Type | ||||||
| LUAD | 177 | 3.971(2.589∼6.091) | <0.0001*** | 4.317(2.477∼7.524) | 0.0006*** | |
| LUSC | 39 | 1.758(0.695∼4.448) | 0.1813 | 1.764(0.693∼4.492) | 0.1859 | |
| Clinical stage | ||||||
| I+II | 147 | 3.246(1.957∼5.383) | 0.0002*** | 2.832(1.466∼5.471) | 0.0081** | |
| III | 69 | 1.461(0.821∼2.598) | 0.2313 | 1.086(0.549∼2.149) | 0.8144 | |
| T classification | ||||||
| T1 | 97 | 3.452(1.866∼6.387) | 0.0013** | 2.641(1.212∼5.757) | 0.0407* | |
| T2+T3+T4 | 119 | 2.153(1.355∼3.422) | 0.0043** | 1.781(1.022∼3.104) | 0.0632 | |
| N classification | ||||||
| N0 | 136 | 3.338(1.963∼5.674) | 0.0002*** | 2.899(1.441∼5.831) | 0.0128** | |
| N1+N2+N3 | 80 | 1.539(0.893∼2.653) | 0.1549 | 1.288(0.690∼2.405) | 0.4303 | |
| Smoke | ||||||
| No smoking | 112 | 5.202(3.042∼8.897) | <0.0001*** | 6.987(3.423∼14.26) | 0.0012** | |
| Smoking | 104 | 1.949(1.164∼3.265) | 0.0152* | 1.602(0.899∼2.855) | 0.1255 | |
Note. Bold font indicates P < 0.05.
TROAP promotes tumor cells proliferation, invasion, and migration
All the above results implied that RASGRF1 or TROAP might be considered as a valuable prognostic factor in LUAD. As a consequence, we hypothesize that RASGRF1 or TROAP might have a modest effect on tumor biological behavior in cancer cells. To elucidate the hypothesis, we carried out the subsequent experiments.
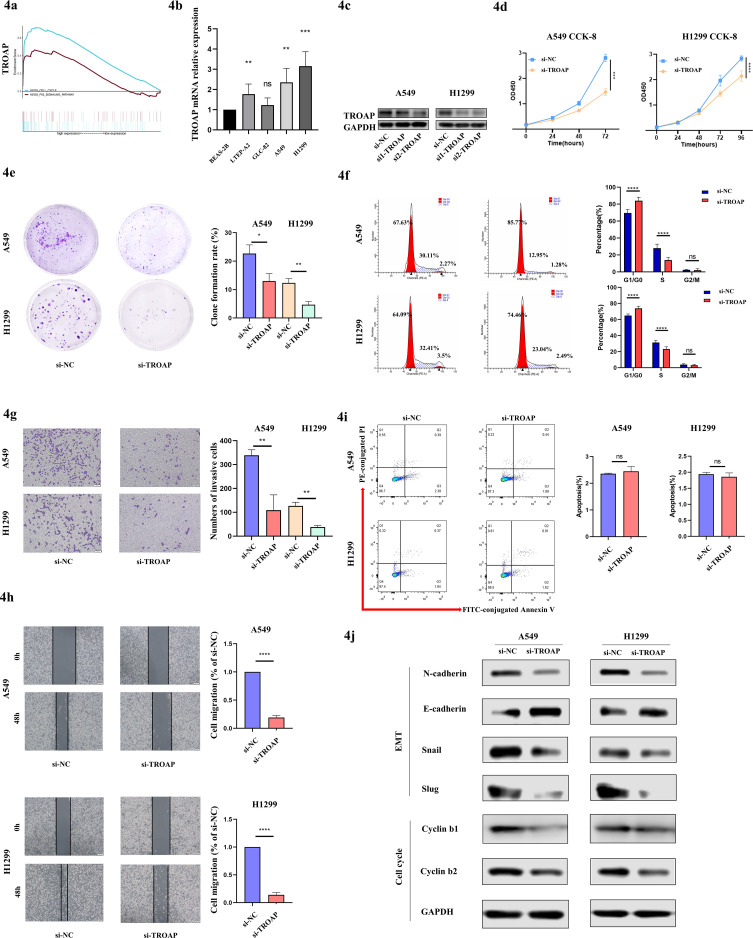
In the following research, we firstly performed GSEA in LUAD, and found overexpressed TROAP was associated with cell cycle and P53 signaling pathway (Fig. 4A), which was consistent with our conjecture. And then, we explored gene mRNA expression in NSCLC cell lines by RT-qPCR and found that TROAP was upregulated in LTEP-A2, A549, and H1299 compared with BEAS-2B (Fig. 4B). TROAP expression was specifically knocked down by transfecting siRNA into A549 and H1299 cells, followed by the confirmation of knockdown efficiency by western blot (Fig. 4C). Next, the cell proliferation ability was measured by CCK-8, 2D clone formation, and cell cycle assays. As indicated in Figs. 4D, E, F, A549 and H1299 cells proliferated slowly and the colony numbers were reduced obviously along with decreased TROAP. Meanwhile, the TROAP deficiency induced a partial cell cycle arrest at the G1-S transition with elevated G0/G1 populations and reduced S population in A549 or H1299 transfected TROAP siRNA. However, TROAP had on effect on the apoptosis (Fig. 4I).
Fig. 4.
TROAP promotes tumor cells proliferation, invasion, and migration. (A) GSEA revealed that TROAP was most predominantly enriched “cell cycle” and “P53 signaling pathway”. (B) TROAP mRNA expression in human NSCLC cell lines. Significance tested by Kruskal-Wallis. (C) Western blot confirmation of TROAP knockdown efficiency by siRNA in A549 and H1299 cells. (D) CCK-8 assay of A549 and H1299 cells with TROAP knockdown at 24, 48, and 72 hours after transfection. Significance tested by two-way ANOVA. (E) Representative images for clonogenic assay after transfection for 7 days. Significance tested by wilcoxon rank-sum test. (F) Cell cycle analysis indicated a strong cell-cycle bias. (G) Transwell invasion assays of cancer cells with TROAP siRNA transfection. (H) Scratch monolayer assays of cancer cells with TROAP siRNA transfection. (I) TROAP deficiency did not affect cell apoptosis. (J) Western blot showed the expression levels of EMT and cell cycle related regulated proteins.
Furthermore, the metastatic capacity was suppressed when deregulated TROAP in A549 and H1299 cells via transwell (Fig. 4G) and wound healing assays (Fig. 4H). Our data revealed that both the amount of tumor cells that migrated onto the lower surface of the membrane, and the wound closure speed were slackened remarkably when transfected with specific TROAP siRNA. It is known that epithelial-to-mesenchymal transition (EMT) is crucial for tumor metastasis, and cyclins contributed to cell cycle progression. We then detected the metastasis related proteins and cell cycle associated proteins. The results demonstrated that TROAP was correlated with the increase of N-cadherin, Snail, Slug, Cyclin b1, and Cyclin b2, as well as reduction of E-cadherin (Fig. 4J). The current data demonstrated that overexpressed TROAP could promote cell proliferation and metastatic potential via regulation EMT and specific cell cycle regulatory proteins.
RASGRF1 have potential function of regulating TME
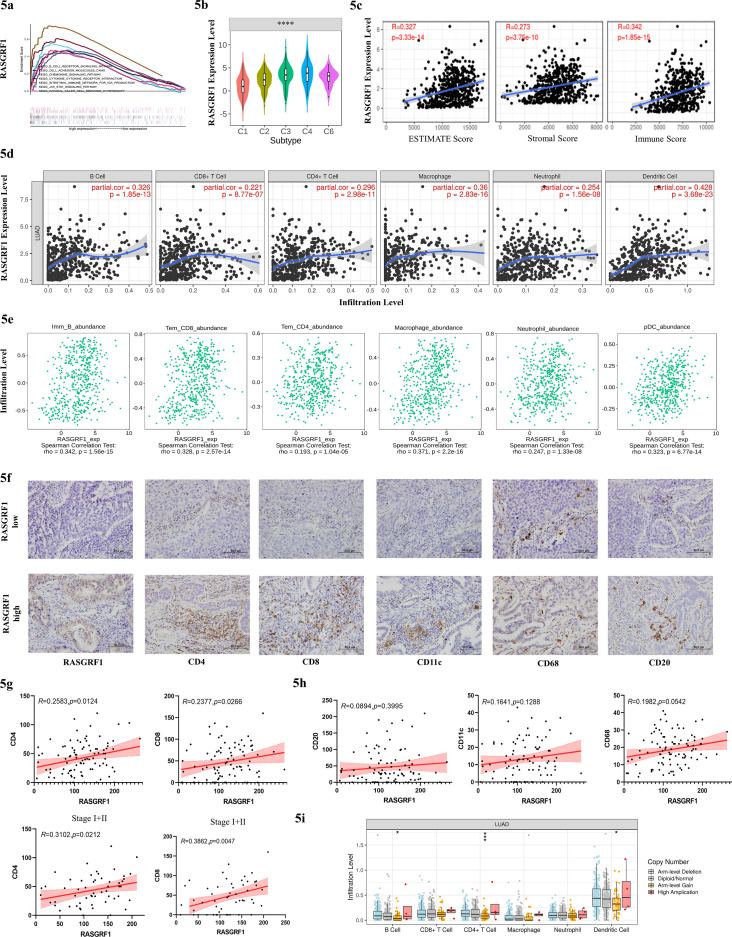
We also explored the potential function of RASGRF1 in NSCLC cell lines and found that RASGRF1 had no influence on tumor initiation and progression (Fig. S3). GSEA revealed that RASGRF1 was primarily enriched in immune related pathways like “B cell receptor signaling pathway”, “chemokine signaling pathway”, “Jak stat signaling pathway” (Fig. 5A). In the next step, we sought to explore the effect of RASGRF1 on tumor immune microenvironment. Hence, we queried RASGRF1 expression in different immune subtypes from TISIDB database, which showed that RASGRF1 was different among immune subtypes (Fig. 5B). Then, ESTIMATE algorithm was conducted to calculate the stromal and immune score, which showed positive correlations between RASGRF1 and ESTIMATE, stromal, and immune score (Fig. 5C). In order to further explore the potential immunomodulatory function of RASGRF1, TIMER database was used to identify the correlation between RASGRF1 and six immune cells. Consistent with the results above, RASGRF1 was significantly positively correlated with the infiltration levels of B+ cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, DCs (partial.cor > 0.2, P < 0.0001) (Fig. 5D). For a more accurate evaluation of association between RASGRF1 and more specific immune cell types, GSVA algorithm was used for analysis via TISIDB (Fig. 5E). Here as well, the strongest correlations were presented, and others can be found in Figure S4a. We have then further confirmed these by IHC in 116 LUAD patients via IHC. Patients with high expression of RASGRF1 were accompanied by increasing infiltration levels of CD4+ and CD8+ T cells, especially in patients with stage I and II (R = 0.3102, P = 0.0212 and R = 0.3862, P = 0.0047, respectively) (Figs. 5F, G). No clearly correlation was found between RASGRF1 and CD20+ B cell, CD11c+ DCs, or CD68+ macrophage (Fig. 5H). We identified a significantly negative association with RASGRF1 and TMB (R = -0.31, P = 6.23e-13) (Fig. S4B). Finally, we distinguished immune cell infiltration levels across each type of RASGRF1 CNVs (Fig. 5I). We can find the highest immune cells infiltration level in the group of RASGRF1 with high amplication, but the trend was statistically insignificant, which may be partly due to the small sample size. Totally, our results demonstrated that RASGRF1 may work as an immune modulator in TME.
Fig. 5.
RASGRF1 have potential function of regulation in TME. (A) GSEA revealed that RASGRF1 was most significantly enriched in immune related pathways. (B) Different RASGRF1 expression among immune subtypes from TISIDB database (C1 (wound healing); C2 (IFN-gamma dominant); C3 (inflammatory); C4 (lymphocyte depleted); C5 (immunologically quiet); C6 (TGF-b dominant)). (C) Positive correlation between RASGRF1 and ESTIMATE, stromal, and immune score. (D) RASGRF1 was significantly and positively correlated with the infiltration levels of B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, DCs from TIMER database (Spearman's partial.cor>0.2, p<0.0001). (E) Positive association between RASGRF1 and specific immune cell types by GSVA from TISIDB database tested by Spearman rank test. (F-H) The relationship between RASGRF1 and immune cell was confirmed by IHC on TMA of 115 LUAD samples. (I) Immune cell infiltration level across each type of RASGRF1 CNV. The infiltration level for each SCNA category is compared with the normal using a two-sided wilcoxon rank-sum test.
Discussion
The rapid advancement of high-throughput sequencing which boots the deep and efficient exploration of genomes and transcriptomes, has revolutionized the understanding of cancers in molecular alterations and biological mechanisms. With further studies, it is now recognized that individual tumors in the same type are highly heterogeneous and even have diverse genomic alterations. The classical factors for predicting prognosis in NSCLC, such as TNM stage and histological differentiation, currently cannot meet the developing requirements of individualized and accurate prognostic evaluation. At the same time, the constantly updated public databases provided a large number of genome and transcriptome sequencing results with corresponding clinical characteristics (22). Without question, cancer therapy has entered a personalized medicine era based on molecular subtyping and pathology guidance. However, it is a bumpy road to achieve truly molecular typing clinical treatment. Thus, sustained efforts have been dedicated toward searching for effective typing system using various approaches. NMF is an unsupervised clustering approach for molecular typing, which were currently widely used to identify special clusters by features in cancers (23, 24, 25, 26). DNA CNVs is a hallmark of genome abnormality in cancers. Transcriptional deregulation by the aberrations is playing a pivotal role in cancer heterogeneous progression. More attention has been paid to the integration of multiple omics data analysis for cancer diagnosis and treatment.
In 2018, diffuse large B-cell lymphoma can be divided into different genetic subtypes according to the characteristics of gene mutation, translocation, and copy number abnormality using technologies such as whole exon sequencing, whole transcript sequencing, DNA copy number analysis and gene targeted amplicon sequencing, respectively (27,28). Recent studies suggested that CNVs characteristics can distinguish malignant ductal cells in pancreatic ductal adenocarcinoma using single-cell RNA-seq (18). Inspired by these unexpected results, we seek to identify CNVcor genes and use the CNVcor genes to differentiate entirely distinct subtypes in LUAD. We hope this theory can explore more unknown mechanisms underlying for tumor heterogeneity, which will help to predict outcomes, determine appropriate treatment options, and even define candidate treatment targets in the future.
In our study, the CNVcor genes were identified by the DNA CNVs and transcriptional profiles from the 443 LUAD patients in TCGA database. These samples were separated into two distinct subtypes (Cluster 1 and Cluster 2). Gene expression pattern, survival status, gene copy aberration, and immune cell infiltration levels were markedly different between Cluster 1 and Cluster 2. Nevertheless, our current study was limited by the lack of available RNA sequencing data in the current study to validate our genotyping findings. To further test this investigation, RNA tissues extracted from LUAD samples in our cohort will be sequenced, and then clustered with defined features in our next work. A deeper understanding is needed to verify the potential clinical value of molecular subtypes in other malignancies.
Next, we chosen to explore the intersection of CNVcor genes and DEGs, and discovered two candidate gene, RASGRF1 and TROAP, which were related to OS and DFS. These results therefore confirmed that the method adopting CNVcor genes partially discriminated patients, which may provide us more clues to underly tumor heterogeneity and develop novel therapeutic targets. These results were validated by the independent GEO dataset and specimens from TMUCIH. RASGRF1 is a guanine-nucleotide exchange factor which catalyzes the GDP/GTP exchange and involves in a variety of neural functions, such as neurite outgrowth and neuron soma changes (29). RASGRF1 has been reported to facilitate tumor initiation and progression by acting as an oncogene in several cancer, such as alveolar rhabdomyosarcoma (30), astrocytoma (31), chronic (32) or acute lymphocytic leukemia (33), and sever as a possible risk factor in gastric (34) and colorectal cancer (35). In melanoma, RASGRF1 upregulates the expression of MMP-9 to promote cell invasion and metastasis (36). However, according to Fernando et al, RasGRF1/2 could suppress tumor cell movement via binding to Cdc42 in in A375M2 cells (37). The role of RASGRF1 in lung tumorigenesis and the association between RASGRF1 and tumor microenvironment (TME) have not been explored. TROAP is a cytoplasmic protein firstly identified to be involved in the embryo implantation, which participates in microtubule regulation (38). Until now, TROAP overexpression was found to be correlated with bad outcomes in ovarian cancer (39), gastric cancer (40), colorectal cancer (41) and liver cancer (42,43). However, Lian et al considered TROAP has a protective role in hepatocellular carcinoma (44). TROAP was reported to enhance cancer progression via activating Wnt3/survivin signaling pathways in prostate cancer as an adverse prognostic factor (45). Recently, TROAP has been regarded as an independent prognostic marker in LUAD (46). However, the mechanism by which TROAP exerts as an oncogene in cancer is still barely known. Little available studies about the roles of these two genes in past were even inconsistent with each other. These may be due to small sample size, or tumor interpatient/intratumor heterogeneity. In our studies, we identified RASGRF1 as a tumor suppressor gene and TROAP as an oncogene through database analysis, and confirmed the accuracy by in vitro experimentation and independent validation cohort.
RASGRF1 expression patterns observed across different immune subtypes showed that RASGRF1 exhibits highest expression in C4, the second highest expression in C3. As for the C4 subtypes (n = 20) predicted worst outcome which was the opposite of RASGRF1 as a protective factor, it remains too early to make a more definitive conclusion due to the small size. To avoid the limitation of one single algorithm and produce a more comprehensive assessment, we analyzed tumor infiltrated immune cells by different algorithm (47). Based on our results, patients with high RASGRF1 expression had an improved prognosis. RASGRF1 may be more involved in immunomodulatory pathways and its expression was significantly associated with immune cells infiltrations. However, we conducted the correlation between RASGRF1 and immune cells in TMUCIH cohort by IHC to examine the immunoregulatory function of RASGRF1. Despite our limitations of our single-center retrospective study, we preliminary revealed the relevance between RASGRF1 and CD4+ or CD8+ T cell. Meanwhile, the correlation between TMB and clusters or RASGRF1 were assessed, which revealed a negative correlation. The high TMB was a bad prognostic factor for OS or DFS in NSCLC (48), which was consistent with the role of RASGRF1 as a good prognostic marker.
In conclusion, we investigated the possible pathogenic mechanisms from the perspective of genomic alterations in LUAD via the data analysis of genomics and transcriptomics. In the current study, we highlighted that DNA CNVs plays a critical role in LUAD and samples could be subdivided into two distinct clusters by CNVcor genes. We compared the molecular characteristics and immune microenvironment of samples according to the two clusters. In addition, we identified two candidate genes, RASGRF1 or TROAP, both present in CNVcor genes and DEGs as the key genes in the two subtypes. We further demonstrated the tumor suppressor role of RASGRF1 and the oncogenic ability of TROAP in vitro experiments. In summary, we thought this novel classification may be useful in risk stratification and determining a more precise survival prediction, possibly as well as beneficial in the development of therapies in cancers.
Author contributions
Conceived and designed the study: Lili Yang and Baihui Li. Performed the experiments: Ziqi Huang, Baihui Li, and Wenwen Yu. Analyzed the data: Baihui Li carried out the bioinformatics analysis, and Ziqi Huang analyzed the experiments data. Clinical sample collection: Jian Zhang, Qingqing Wang, Lei Wu, and Fan Kou. Wrote the paper: Baihui Li. Reviewed the manuscript: Shaochuan Liu and Lili Yang. Revised the manuscript: Lili Yang. All authors read and approved the final manuscript.
Acknowledgments
We would like to acknowledge tumor tissue banking facility of TMUCIH for providing us tissues from patients with NSCLC.
Footnotes
✩ Funding: This work was supported by a grant from the National Key Technology R&D Program (No.2018YFC1313400), National Natural Science Foundation of China (No.81974246), Tianjin Research Innovation Project (Grant No. 2020YJSB164) for Postgraduate Students, and Scientific Research Program of Tianjin Education Commission (No.2019KJ185).
✩✩ Conflict of Interest: The authors declare no potential conflicts of interest.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.05.006.
Appendix. Supplementary materials
Reference
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A, Watanabe S, Tsuta K, Kamatani N, Yoshida T, Nakamura Y, Yokota J, Kubo M, Kohno T. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44(8):900–903. doi: 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]
- 3.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 4.Huang KL, Li SQ, Mertins P, Cao S, Gunawardena HP, Ruggles KV, Mani DR, Clauser KR, Tanioka M, Usary J, Kavuri SM, Xie L, Yoon C, Qiao JW, Wrobel J, Wyczalkowski MA, EG P, Snider JE, Hoog J, Singh P, Niu B, Guo ZF, Sun S QC, Sanati S, Kawaler E, Wang X, Scott A, Ye K, McLellan MD, Wendl MC, Malovannaya A, Held JM, Gillette MA, Fenyo D, Kinsinger CR, Mesri M, Rodriguez H, Davies SR, Perou CM, Ma C, Townsend RR, Chen X, Carr SA, Ellis MJ, Ding L. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat Commun. 2017;8:14864. doi: 10.1038/ncomms14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, Kreuz M, Felsberg J, Beyer U, Löffler‑Wirth H, Kaulich K, Steinbach JP, Hartmann C, Gramatzki D, Schramm J, Westphal M, Schackert G, Simon M, Martens T, Boström J, Hagel C, Sabel M, Krex D, Tonn JC, Wick W, Noell S, Schlegel U, Radlwimmer B, Pietsch T, Loeffler M, Deimling AV, Binder H, Reifenberger G. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015;129(5):679–693. doi: 10.1007/s00401-015-1409-0. [DOI] [PubMed] [Google Scholar]
- 6.Woo HG, Choi JH, Yoon S, Jee BA, Cho EJ, Lee JH, Yu SJ, Yoon JH, Yi NJ, Lee KW, Suh KS, Kim YJun. Integrative analysis of genomic and epigenomic regulation of the transcriptome in liver cancer. Nat Commun. 2017;8(1):839. doi: 10.1038/s41467-017-00991-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu W, Xu M, Wang L, Zhou W, Xiang R, Shi Y, Zhang YS, Piao YJ. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduct Target Ther. 2019;4:55. doi: 10.1038/s41392-019-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y, Zhang Y, Ye T, Zhao Y, Gao Z, Yuan H, Zheng D, Zheng SB, Li H, Li Y, Jin Y, Sun YH, Chen HQ. Detection of Novel NRG1, EGFR, and MET Fusions in Lung Adenocarcinomas in the Chinese Population. J Thorac Oncol. 2019;14(11):2003–2008. doi: 10.1016/j.jtho.2019.07.022. [DOI] [PubMed] [Google Scholar]
- 9.Dietz S, Lifshitz A, Kazdal D, Harms A, Endris V, Winter H, Stenzinger A, Warth A, Sill M, Tanay A, Sültmann H. Global DNA methylation reflects spatial heterogeneity and molecular evolution of lung adenocarcinomas. Int J Cancer. 2019;144(5):1061–1072. doi: 10.1002/ijc.31939. [DOI] [PubMed] [Google Scholar]
- 10.Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization classification of lung tumors. Semin Roentgenol. 2005;40(2):90–97. doi: 10.1053/j.ro.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, Beer DG, Cope L, Creighton CJ, Danilova L. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. The Lancet Oncology. 2015;16(7):e342. doi: 10.1016/S1470-2045(15)00077-7. –ee51. [DOI] [PubMed] [Google Scholar]
- 13.Waddell N, Pajic M, Patch A-M, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren Y, Huang S, Dai C, Xie D, Zheng L, Xie H, Zheng H, She YL, Zhou FY, Wang Y. Germline Predisposition and Copy Number Alteration in Pre-stage Lung Adenocarcinomas Presenting as Ground-Glass Nodules. Front Oncol. 2019;9:288. doi: 10.3389/fonc.2019.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, Rodman C, Luo CL, Mroz EA, Emerick KS. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171(7):1611–1624. doi: 10.1016/j.cell.2017.10.044. , e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calvayrac O, Pradines A, Pons E, Mazieres J, Guibert N. Molecular biomarkers for lung adenocarcinoma. Eur Respir J. 2017;49(4) doi: 10.1183/13993003.01734-2016. O Calvayrac, A Pradines, E Pons, J Mazieres and N. Guibert, Molecular biomarkers for lung adenocarcinoma, Eur Respir J 49 (4), 2017. [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, Goldenberg A. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods. 2014;11(3):333–337. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- 18.Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, Liu LL, Huang D, Jiang JL, Cui GS. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Bai X, Wang J, Tang XR, Wu DH, Du SS, Du XJ, Zhang YW, Zhu HBo, Fang Y. Combination of TMB and CNA stratifies prognostic and predictive responses to immunotherapy across metastatic cancer. Clin Cancer Res. 2019;25(24):7413–7423. doi: 10.1158/1078-0432.CCR-19-0558. [DOI] [PubMed] [Google Scholar]
- 20.Jang HJ, Lee HS, Ramos D, Park IK, Kang CH, Burt BM, Kim YT. Transcriptome-based molecular subtyping of non-small cell lung cancer may predict response to immune checkpoint inhibitors. J Thorac Cardiovasc Surg. 2020;159(4):1598–1610. doi: 10.1016/j.jtcvs.2019.10.123. , e3. [DOI] [PubMed] [Google Scholar]
- 21.Song Y, Yan S, Fan W, Zhang M, Liu W, Lu H, Cao MR, Hao CG, Chen L, Tian FL. Identification and validation of the immune subtypes of lung adenocarcinoma: implications for immunotherapy. Front Cell Dev Biol. 2020;8:550. doi: 10.3389/fcell.2020.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network Electronic address aadhe, cancer genome atlas research n. integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203. doi: 10.1016/j.ccell.2017.07.007. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Yang H, Teo ASM, Amer LB, Sherbaf FG, Tan CQ, Alvarez JJS, Lu B, Lim JQ, Takano A. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet. 2020;52(2):177–186. doi: 10.1038/s41588-019-0569-6. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, Xing BC, Sun W, Ren LL, Hu B, Li CY, Zhang L. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567(7747):257–261. doi: 10.1038/s41586-019-0987-8. [DOI] [PubMed] [Google Scholar]
- 25.Gobin M, Nazarov PV, Warta R, Timmer M, Reifenberger G, Felsberg J, Vallar L, Chalmers AJ, Herold-Mende CC, Goldbrunner R. A DNA Repair and Cell-Cycle gene expression signature in primary and recurrent glioblastoma: prognostic value and clinical implications. Cancer Res. 2019;79(6):1226–1238. doi: 10.1158/0008-5472.CAN-18-2076. [DOI] [PubMed] [Google Scholar]
- 26.Papaioannou MD, Djuric U, Kao J, Karimi S, Zadeh G, Aldape K, Diamandis P. Proteomic analysis of meningiomas reveals clinically distinct molecular patterns. Neuro Oncol. 2019;21(8):1028–1038. doi: 10.1093/neuonc/noz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, Roulland S, Kasbekar M, Young RM, Shaffer AL. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med. 2018;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, Lawrence MS, Roemer MGM, Li AJ, Ziepert M. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Medarde A, Santos E. The RasGrf family of mammalian guanine nucleotide exchange factors. Biochim Biophys Acta. 2011;1815(2):170–188. doi: 10.1016/j.bbcan.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Tarnowski M, Schneider G, Amann G, Clark G, Houghton P, Barr FG, Kenner L, Ratajczak MZ, Kucia M. RasGRF1 regulates proliferation and metastatic behavior of human alveolar rhabdomyosarcomas. Int J Oncol. 2012;41(3):995–1004. doi: 10.3892/ijo.2012.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng D, Xue L, Shao N, Qu H, Wang Q, Wang S, Xia XW, Yang YL, Zhi F. miR-137 acts as a tumor suppressor in astrocytoma by targeting RASGRF1. Tumour Biol. 2016;37(3):3331–3340. doi: 10.1007/s13277-015-4110-y. [DOI] [PubMed] [Google Scholar]
- 32.Liao W, Jordaan G, Coriaty N, Sharma S. Amplification of B cell receptor-Erk signaling by Rasgrf-1 overexpression in chronic lymphocytic leukemia. Leuk Lymphoma. 2014;55(12):2907–2916. doi: 10.3109/10428194.2014.898759. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Sanda T, Look AT, Novina CD, von Boehmer H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J Exp Med. 2011;208(4):663–675. doi: 10.1084/jem.20102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takamaru H, Yamamoto E, Suzuki H, Nojima M, Maruyama R, Yamano HO, Yoshikawa K, Kimura T, Harada T, Ashida M. Aberrant methylation of RASGRF1 is associated with an epigenetic field defect and increased risk of gastric cancer. Cancer Prev Res (Phila) 2012;5(10):1203–1212. doi: 10.1158/1940-6207.CAPR-12-0056. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Xu Z, Yang B, Zhou X, Kong H. RASGRF1 Hypermethylation, a Putative Biomarker of Colorectal Cancer. Ann Clin Lab Sci. 2018;48(1):3–10. [PubMed] [Google Scholar]
- 36.Zhu TN, He HJ, Kole S, D'Souza T, Agarwal R, Morin PJ, Bernier M. Filamin A-mediated down-regulation of the exchange factor Ras-GRF1 correlates with decreased matrix metalloproteinase-9 expression in human melanoma cells. J Biol Chem. 2007;282(20):14816–14826. doi: 10.1074/jbc.M611430200. [DOI] [PubMed] [Google Scholar]
- 37.Calvo F, Sanz-Moreno V, Agudo-Ibanez L, Wallberg F, Sahai E, Marshall CJ, Crespo P. RasGRF suppresses Cdc42-mediated tumour cell movement, cytoskeletal dynamics and transformation. Nat Cell Biol. 2011;13(7):819–826. doi: 10.1038/ncb2271. [DOI] [PubMed] [Google Scholar]
- 38.Yang S, Liu X, Yin Y, Fukuda MN, Zhou J. Tastin is required for bipolar spindle assembly and centrosome integrity during mitosis. FASEB J. 2008;22(6):1960–1972. doi: 10.1096/fj.07-081463. [DOI] [PubMed] [Google Scholar]
- 39.Partheen K, Levan K, Osterberg L, Claesson I, Fallenius G, Sundfeldt K, Horvath G. Four potential biomarkers as prognostic factors in stage III serous ovarian adenocarcinomas. Int J Cancer. 2008;123(9):2130–2137. doi: 10.1002/ijc.23758. [DOI] [PubMed] [Google Scholar]
- 40.Jing K, Mao Q, Ma P. Decreased expression of TROAP suppresses cellular proliferation, migration and invasion in gastric cancer. Mol Med Rep. 2018;18(3):3020–3026. doi: 10.3892/mmr.2018.9230. [DOI] [PubMed] [Google Scholar]
- 41.Ye X, Lv H. MicroRNA-519d-3p inhibits cell proliferation and migration by targeting TROAP in colorectal cancer. Biomed Pharmacother. 2018;105:879–886. doi: 10.1016/j.biopha.2018.04.114. [DOI] [PubMed] [Google Scholar]
- 42.Jiao Y, Li Y, Lu Z, Liu Y. High Trophinin-Associated Protein Expression Is an Independent Predictor of Poor Survival in Liver Cancer. Dig Dis Sci. 2019;64(1):137–143. doi: 10.1007/s10620-018-5315-x. [DOI] [PubMed] [Google Scholar]
- 43.Hu H, Xu L, Chen Y, Luo SJ, Wu YZ, Xu SH, Liu MT, Lin F, Mei Y, Yang Q. The Upregulation of Trophinin-Associated Protein (TROAP) Predicts a Poor Prognosis in Hepatocellular Carcinoma. J Cancer. 2019;10(4):957–967. doi: 10.7150/jca.26666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lian Y, Fan W, Huang Y, Wang H, Wang J, Zhou L, Wu XJ, Deng MH, Huang YH. Downregulated trophinin-associated protein plays a critical role in human hepatocellular carcinoma through upregulation of tumor cell growth and migration. Oncol Res. 2018;26(5):691–701. doi: 10.3727/096504017X15101398724809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J, Chu C, Chen M, Shi Z, Gan S, Qu F, Pan XW, Yang QW, Tian YJ, Wang L. TROAP regulates prostate cancer progression via the WNT3/survivin signalling pathways. Oncol Rep. 2019;41(2):1169–1179. doi: 10.3892/or.2018.6854. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Zhou Y, Luo R, Liu K, Chen Z. Trophinin-associated protein expression is an independent prognostic biomarker in lung adenocarcinoma. Journal of Thoracic Disease. 2019;11(5):2043–2050. doi: 10.21037/jtd.2019.04.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, List M, Aneichyk T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436. doi: 10.1093/bioinformatics/btz363. –ii45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Owada-Ozaki Y, Muto S, Takagi H, Inoue T, Watanabe Y, Fukuhara M, Yamaura T, Okabe N, Matsumura Y, Hasegawa T. Prognostic impact of tumor mutation burden in patients with completely resected nonsmall cell lung cancer: brief report. J Thorac Oncol. 2018;13(8):1217–1221. doi: 10.1016/j.jtho.2018.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.