Abstract
In the search for novel broad-spectrum therapeutics to fight chronic infections, inflammation, and cancer, host defense peptides (HDPs) have garnered increasing interest. Characterizing their biologically-active conformations and minimum motifs for function represents a requisite step to developing them into efficacious and safe therapeutics. Here, we demonstrate that metallating HDPs with Cu2+ is an effective chemical strategy to improve their cytotoxicity on cancer cells. Mechanistically, we find that prepared as Cu2+-complexes, the peptides not only physically but also chemically damage lipid membranes. Our testing ground features piscidins 1 and 3 (P1/3), two amphipathic, histidine-rich, membrane-interacting, and cell-penetrating HDPs that are α-helical bound to membranes. To investigate their membrane location, permeabilization effects, and lipid-oxidation capability, we employ neutron reflectometry, impedance spectroscopy, neutron diffraction, and UV spectroscopy. While P1-apo is more potent than P3-apo, metallation boosts their cytotoxicities by up to two- and seven-fold, respectively. Remarkably, P3-Cu2+ is particularly effective at inserting in bilayers, causing water crevices in the hydrocarbon region and placing Cu2+ near the double bonds of the acyl chains, as needed to oxidize them. This study points at a new paradigm where complexing HDPs with Cu2+ to expand their mechanistic reach could be explored to design more potent peptide-based anticancer therapeutics.
Subject terms: Membrane structure and assembly, Chemical modification, Biochemistry, Biophysics, Structural biology, Chemistry
Introduction
In recent years, global concerns have arisen regarding multi-drug resistant (MDR) pathogens and cancers1–7. Importantly, chronic bacterial infections represent prevalent comorbidity in cancer patients8–13. They also lead to chronic inflammation that can result in cancer14. Given the intersection between life-threatening infections and cancers, there is a strong motivation to develop therapeutics with broad-spectrum activity to treat both conditions simultaneously. With this regard, several membrane-interacting host-defense peptides (HDPs) with cytotoxic properties against cancer cells have emerged as promising templates7,8,15–22. However, a major challenge to developing them into efficacious, safe, and affordable therapeutics is the lack of structure-action relationship (SAR) studies on beneficial peptides, as needed to characterize their biologically-active conformations and minimum motifs required for function. Overall, there is also a strong need to identify chemical modifications that boost peptide potencies, and thereby reduce dosage, side effects, and cost7,19,22–30.
In this study, we investigate piscidins, which are cationic histidine-rich membrane-interacting HDPs widely expressed in teleost fish31–33. Through a combination of SAR studies, we demonstrate that coordinating these amphipathic peptides with Cu2+ is an effective chemical strategy to enhance their cytotoxicity on cancer cells. On a mechanistic level, we show that these biological effects correlate with the ability of the peptides to not only physically but also chemically impair lipid membranes.
As evolutionary-optimized molecules from the innate immune system, cationic HDPs favorably interact with anionic danger signals on the surface of pathogenic bacterial and cancer cells3,7,8,15,19,24,34–36. While most membrane-interacting HDPs target pathogens, several of them are cytotoxic to cancer cells, resulting in their classification as anticancer peptides (ACPs)7,8,15,19–21. Significant advantages of ACPs include their high specificity and low incidence of resistance due to the short timescale of their interactions with vital cell membranes, good biocompatibility due to low toxicity and side effects, and dual ability to kill neoplastic (abnormal) cells and exert anti-inflammatory function7,8,15,19,21,24,25,27,29,30,37. Anionic molecules more represented on cancer than healthy cells include phosphatidylserine (PS), and anionic carbohydrates on glycosylated lipids and proteins7,8,15,19,24,38. Other determinants that may impart specificity to ACPs include the altered cholesterol content, low pH, and increased membrane potential and surface area associated with neoplastic cells7,15.
Following their initial interactions with cell surfaces, ACPs induce cell death via necrotic and/or apoptotic pathways depending on their concentration and the specific cells that are targeted7,8,15,17,19,24,36,39. In the case of necrosis, cells display blebbing of their plasma membranes, suggesting that cell death is due to membrane lysis. Specific mechanisms of action for membrane disruption have been proposed, including membrane permeabilization or rupture via the formation of toroidal pores or carpets7,15,19,36. In terms of apoptosis, some ACPs get internalized inside the cells and interact with intracellular targets, ultimately triggering programmed cell death. Intracellular effects may include the disruption of mitochondrial membranes, which have a large membrane potential and anionic lipid content (e.g. cardiolipin, CL), and thus attract cationic ACPs15,17,19,40,41. In particular, high mitochondrial reactive oxygen species (ROS) levels have been associated with agents that induce apoptotic cell death17,40–43.
Marine animals produce HDPs that are highly potent against a wide range of harmful cells and diseases, including cancer21,44–46. Particularly promising peptides include the piscidins from hybrid striped bass31–33,47 and tilapia48, and their homologs in other fish species: pleurocidin from flounder49, epinecidin-1 from grouper50, and chrysophsin from red sea bream51. Piscidins are characterized by their abundance of histidine residues31. The isoforms piscidin 1 (P1) and piscidin 3 (P3) were co-discovered in the mast cells of hybrid striped bass47. Both peptides are highly potent against both Gram-positive and Gram-negative bacteria, with minimum inhibitory concentrations (MIC) in the range of 1 to 20 μmol/L33,47,52,53.
While P1 and P3 are 68% homologous and have almost identical α-helical structures in a lipid bilayer environment53–58, P1 is a more potent antimicrobial agent47,53,59. It is active against viruses such as HIV-160, coronaviruses61, and pseudorabies62. Its cytotoxic properties against several cancer cell lines were reported by Lin et al. (2012)39. The authors found that P1 induced apoptosis in HT1080 cells at low concentrations while it had necrotic effects at high concentrations.
In previous biophysical studies of P1/3, we demonstrated that the stronger membrane permeabilizing effects and pH-resiliency of P1 correlate with its higher histidine content and a membrane-inserted state that places its C-terminus in the bilayer core56. In contrast, P3 uses its N-terminus to direct membrane insertion. Recently, we showed that both P1 and P3 bind Cu2+/Ni2+ ions using an amino-terminal copper/nickel (ATCUN) binding motif59,63–65. The ATCUN motif (XXH), which is highly conserved in the piscidin family, exists in other proteins such as albumins66–69. The metal is coordinated by four nitrogens, including the nitrogen from the N-terminus, the two subsequent backbone amide nitrogens, and the δ-nitrogen of His-3 arranged in a distorted square planar coordination geometry68,70,71. The ATCUN motif requires the deprotonation of the three backbone nitrogen sites, and thus metallation is more favorable at basic pH59,63,64. Once bound to Cu2+, ATCUN-containing peptides become distinctively pink, a useful property to ascertain their metallation state under changing sample conditions63,64. Given the ability of Cu2+ to form ROS, synthetic peptides containing the ATCUN motif have been designed to generate radicals and damage harmful cells and pathogens, including in vivo67,69,71–77. Of particular interest here is the pioneering work of Pauling and colleagues who demonstrated the ability of the GGH-Cu2+ peptide to kill cancer cells through a mechanism involving the release of the metal ion when the complex reaches the acidic microenvironment of tumor cells76. Free Cu2+ then reacts with metabolites in the cancer cells to form ROS and damage vital cellular components (e.g., DNA).
As cell penetrating peptides with built-in host defense functions, P1 and P3 can specifically carry Cu2+ to and across bacterial cell membranes, enabling the metal ion to form ROS that oxidize unsaturated fatty acids in lipid membranes and nick intracellular DNA59,73,75. Recent studies of Cu2+-bound P1 showed that the presence of oxidized lipids in model membranes improves its membrane permeabilization effects, probably because the oxidized lipids weaken the integrity of the membrane barrier and the loss of a positive charge by the peptide upon metal-ion binding facilitates membrane insertion63.
As demonstrated here, Cu2+-coordination to P1 and P3 before they are added to cancer cells improves their cytotoxicities on cancer cells, with P3 responding to metallation much more dramatically than P1. In investigating this remarkable result on a mechanistic level, we establish a correlation between the enhanced cytotoxicity, deeper membrane insertion, and lipid peroxidation capability of P3-Cu2+ compared to P3. Thus, this study identifies the metallation of HDPs with Cu2+ as an effective strategy to improve their anticancer properties via the expansion of their mechanistic reach, so that both physical and chemical disruptive effects of membranes are elicited.
Results
Effect of Cu2+-metallation on the cytotoxicity of P1 and P3 against several cancer cell lines
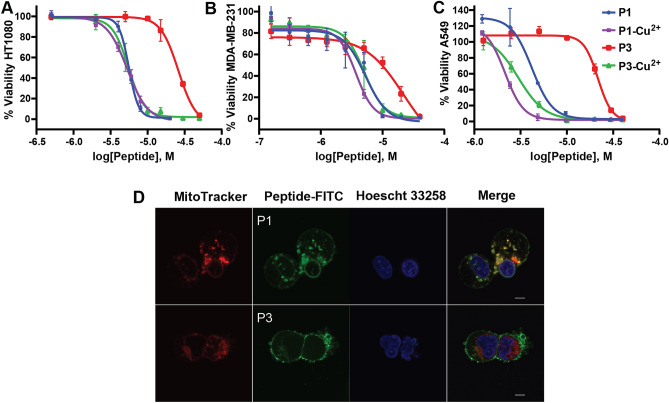
Prior studies by Lin et al. (2012) demonstrated that P1 induces apoptosis and necrosis in HT1080 (fibrosarcoma) cancer cells39. The authors also characterized the activity of P1 on A549 (lung epithelial) and HeLa (cervical) cancer cells. However, P3 was not tested. Here, we investigated the activities of P1 and P3 in the apo-state and holo (Cu2+-bound) form on these three cell lines, as well as the MDA-MB-231 (breast) cancer cells. As shown in Figs. 1 and S1A, and Table 1, and in agreement with the work of Lin et al., P1 has considerable cytotoxic effects against all cancer cell lines tested. Based solely on the cell viability profiles for P1 and P3 in the investigated concentration range (0–50 µmol/L), apo-P3 is much less cytotoxic than apo-P1. Remarkably, the Cu2+-bound form of P3 exhibits a significant boost in killing ability. For instance, based on Fig. 1 and the average IC50 values listed in Table 1, metallation with Cu2+ increases the efficacy of P3 by a factor of ≈7 on the A549 and ≈5 on the HT1080 cells. In the case of P1, Cu2+-binding improves activity by up to a factor of ≈2 (e.g., A549, MDA-MB-231). Free Cu2+ tested at up to 30 µmol/L was not cytotoxic under the same conditions (Fig. S1B). Thus, while it is not known whether the peptides remain bound to Cu2+ throughout the process of killing cancer cells, it is essential for Cu2+ and the peptides to be bound to each other in order to reach low IC50 values. The Cu2+-bound peptides did not exhibit this strong level of cytotoxicity on normal MRC-5 (lung) fibroblast cells (Fig. S1C), in agreement with the data previously obtained for apo-P1 on MRC-5, HGF-1 (gingival) and OMF (mucosal) fibroblast cells39,78. Higher selectivity of the cationic peptides for cancer cells is very likely related to different chemical conditions of their membrane environment, as explained in the introduction.
Figure 1.
Cytotoxicity assays for the apo- and holo-states of P1 and P3 acting on cancer cells. Fibrosarcoma (HT1080), breast (MDA-MB-231), and lung (A549) cancer cell lines (panels A, B, and C, respectively) were used to measure cell viability after 24 h of treatment with different concentrations (M = mol/L) of P1 and P3 in the apo- and Cu2+-bound states. Error bars are standard deviations based on triplicates. The IC50 values are summarized in Table 1. Similar results were obtained after 48 h (Fig. S1). The peptides were metallated using a 1:1 stoichiometric amount of CuCl2. Panel C: MD-MBA-231 cells exposed to 100 nM MitoTracker (red), 5 µg/mL Hoescht 33,258, and 4 µmol/L FITC-labeled P1 or P3 (green) for 20 min. Top row: FITC-P1 stains the cellular membrane and nuclear envelope and co-localizes with MitoTracker. Bottom row: FITC-P3 stains the cellular membrane and there is co-localization with MitoTracker (left cell). Scale bar represents 5 µm.
Table 1.
IC50 for P1 and P3 acting on cancer cells in the apo- and holo-forms.
| IC50 (µmol/L) | HT1080 | MDA-MB-231 | A549 | HeLa |
|---|---|---|---|---|
| P1 | 5.58a (5.41–5.77) | 5.83b (4.82–7.04) | 4.20a (3.71–4.65) | 2.22a (1.37–3.61) |
| P1-Cu2+ | 5.31 (4.91–5.75) | 3.92b (3.54–4.33) | 2.15 (1.94–2.30) | 2.17 (1.32–3.60) |
| P3 | 26.01 (23.7–28.6) | 17.5b (11.8–26.0) | 21.6 (20.4–23.7) | 18.1b (10.8–30.4) |
| P3-Cu2+ | 5.52 (5.03–6.05) | 5.25b (4.32–6.37) | 2.99 (2.60–3.34) | 1.97 (1.24–3.12) |
The range of concentrations corresponding to the 95% confidence interval are provided in parentheses.
aFrom this work. Comparison to data in Lin et al.39 is provided in the text.
bFirst data point at 0.16 µmol/L was not used to extract IC50 values due to large deviation from the trend line but all data points are displayed in Fig. 1.
We note that Lin et al.39 obtained slightly higher IC50 values on the HT1080, A549, and HeLa cells. In comparing their experimental protocol to ours, the most significant difference appears to be the use of different approaches to assess the concentrations of the peptide solutions. Piscidins 1 and 3 lack strong chromophores, preventing the common determination of molar concentration from the measurement of UV absorbance at 280 nm. As a result, we relied on amino acid analysis to derive molarity. In contrast, Lin et al.39 used peptide mass. Since highly cationic peptides have an undetermined number of counterions, the total sample weight will tend to overestimate both the net amount of peptide and IC50 values.
Microscopy images using P1 and P3 labeled with a green fluorescein isothiocyanate (FITC) label attached to their N-terminus show that they have a preference for membranous organelles (see Fig. 1 for MD-MBA-231 cells and Fig. S2 for HeLa cells). After 20 min of incubation with 4 μmol/L of the FITC-labeled peptides, the cells undergo morphological changes. In both MD-MBA-231 and HeLa cells, FITC-P1 co-localizes with the plasma membrane, the nuclear envelope, and red MitoTracker. FITC-P3 interacts with the plasma membrane of the MDMBA-231 as well as with the MitoTracker. Added to the HeLa cells, it also co-localizes with the nuclear membranes. We previously showed that P1 and P3 have strong affinities for membranes and P1 is more membrane active than P353,56–58,79. It is thus plausible that the stronger cytotoxicity of P1 compared to P3 relates to its stronger membrane activity observed on model and bacterial cell membranes.
Since the FITC group is attached to the amino terminal group of P1 and P3, FITC-P1/3 could not be metallated. As a result, we were not able to use these fluorescently-labeled peptides to determine how metallation affects cellular localization. However, P1 is highly membrane active and metallation does not affect its IC50 by more than a factor of 2 (Table 1), thus binding to Cu2+ very likely does not affect its mechanism of action and ability to strongly interact with membranous organelles. By comparison, P3 shows a much larger increase in IC50 upon Cu2+-binding (Table 1). To gain insights into the structural aspects of the peptide-bilayer interactions that could explain how metallation increases the cytotoxicity of P1 and P3 on cancer cells, we employed several complementary biophysical techniques, as described below (Fig. 2).
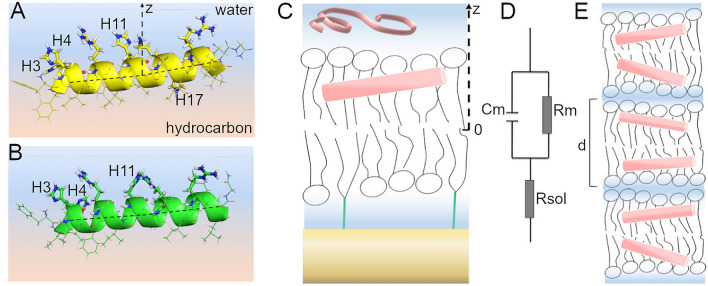
Figure 2.
Techniques for investigating peptide-bilayer structural interactions. (A) High-resolution structure of piscidin P1: FFHHIFRGIVHVGKTIHRLVTG (MW 2571) and (B) Piscidin P3: FIHHIFRGIVHAGRSIGRFLTG (MW 2492). The shown structures were determined by solid state NMR in 4:1 POPC/cholesterol bilayers at P/L = 1:4058. The corresponding Protein Data Bank IDs are 6PF0 (P1) and 6PEZ (P3). The α-helices of the peptides lay at the bilayer-water interface, adopting orientations that are almost parallel to the bilayer surface. (C) Cartoon describing the tethered bilayer membrane used for Surface Plasmon Resonance (SPR), Electrical Impedance Spectroscopy (EIS) and neutron reflectometry (NR). The tethered molecules (green) create a 10 Å thick sub-membrane aqueous space. (D) The simplest electric circuit model to describe the surface-supported bilayer, characterized by the membrane capacitance (Cm), resistance (Rm), and solvent resistance (Rsol). (E) Oriented lipid multilayers with peptide incorporated as described in the Methods for the neutron diffraction (ND) experiments. The repeat spacing (d) denotes the dimension of the repeat unit (thickness of the bilayer with its hydration layer).
Kinetics of lipid binding and membrane permeation for P1, P3, P1-Cu2+, and P3-Cu2+
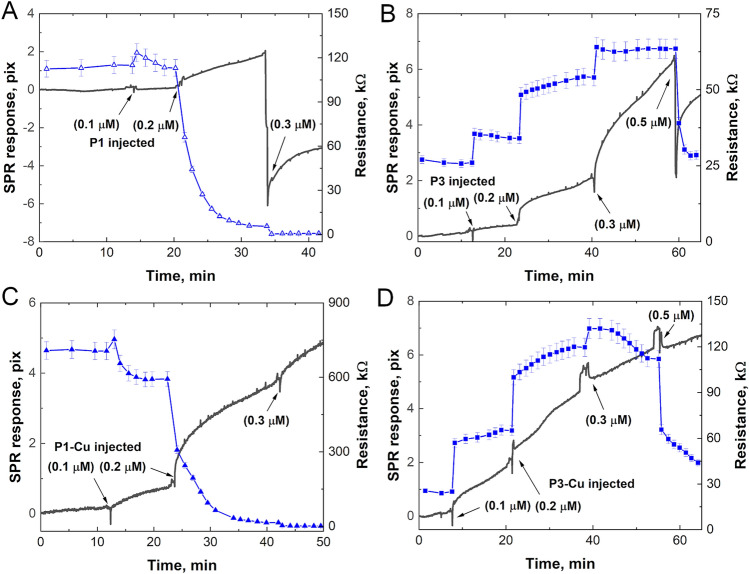
Planar, tethered bilayer membranes (tBLM) supported on gold-coated surfaces (Fig. 2C) were made using 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) to mimic cancer cell membranes80,81. Combined, surface plasmon resonance (SPR)/electrical impedance spectroscopy (EIS) measurements (Fig. 3) were conducted by injecting the peptide in small increments while simultaneously observing (1) the kinetics associated with the peptides binding to the bilayer (SPR) and (2) the formation of peptide-lined defects that increase membrane permeation and conductance (EIS). To capture the threshold needed for the peptides to bind to the bilayer and induce permeation, we started in the very low concentration range (0.1 to 0.5 μmol/L). While this concentration range is well below the IC50 values reported for P1 and P3 in Fig. 1, the SPR/EIS technique is sensitive enough to detect bilayer-peptide interactions in the sub-lethal regime. In this low peptide concentration regime, the SPR signal increases slowly as the peptides attack the membrane surface (Fig. 3A, B, black line). Fluctuations (drops) in the SPR signal, seen here at 0.3 μmol/L P1 and 0.5 μmol/L P3, are likely due to a dislocation of ions, lipid, or loosely bound material peptides as peptides accumulate on the surface. The aforementioned fluctuations are not as apparent in the SPR signals when using holo-P1/3 (Fig. 3C, D) compared to apo-P1/3 (Fig. 3A, B), suggesting that the Cu2+-bound peptides are more efficient at integrating themselves within the bilayer interior, precluding a significant mass dislocation at the tBLM surface.
Figure 3.
SPR/EIS of P1 and P3 with or without Cu2+. The black curves are the SPR responses, and the blue curves are the calculated bilayer resistance values from EIS measurements. Peptides were added in small increments on top of a stable POPC tBLM. SPR/EIS signals were collected simultaneously as a function of time: (A) P1, (B) P3, (C) P1-Cu2+, and (D) P3-Cu2+. The peptides were metallated using a 1:1 stoichiometric amount of CuCl2. See also Fig. S3. (µM = µmol/L).
As concentration increases further, peptides induce transbilayer defects that create pathways for ion conduction through the otherwise electrically sealing bilayer. This is best captured in the EIS signal (Fig. 3, blue curves). Such membrane defects are seen as a sharp drop in bilayer resistance from 125 to 0 kΩ, at a concentration as low as 0.2 μmol/L for P1 (Fig. 3A). In contrast to P1, P3 shows a step-wise accumulation on the surface of the bilayer as the concentration increases. It is only at 0.5 μmol/L that P3 induces a drop in resistance (Fig. 3B, blue line). Based on the EIS measurements, P1 exhibits a more aggressive behavior, requiring a dose five times lower than for P3 to open the bilayer. Interestingly, the IC50 of P1 is five-fold lower than that of P3 in the cytotoxicity assays presented above (Table 1), suggesting that when we compare the two peptides the more potent one is commensurately more membrane active. Overall, the SPR/EIS measurements reveal the differing bilayer interactions and efficacies of these two membrane-active peptide homologs. They also show that the holo-forms of P1/3 are generally more aggressive (larger drop in resistance) than the apo-forms. Tested at higher concentrations (e.g., 3 μmol/L) to match the IC50 concentrations listed in Table 1, the holo-forms remain more membrane disruptive than the apo-forms (Fig. S3). Clearly, the presence of the metal ion, carried through the ATCUN motif, modifies the interactions of P1 and P3 with the bilayer, resulting in a more efficient integration and permeabilization of the bilayer core. Next, to correlate these observations with direct structural information on the peptide-bilayer complexes, we employed surface scattering techniques to study planar bilayer systems exposed to the peptides.
Distributions of P1, P3, P1-Cu2+, and P3-Cu2+ in tethered lipid bilayers
NR was used here to determine the ensemble and time averaged spatial distribution of the peptides in the lipid bilayer. The NR experiments were set up on a POPC tBLM (Fig. 2C) for a direct comparison with the SPR/EIS studies presented above. Before the addition of a given peptide to the tBLM, NR measurements and density calculations of the tBLM reference system were performed without peptide to ensure the formation of a complete bilayer. Fresh tBLMs were independently prepared for P1, P3, P1-Cu2+, and P3-Cu2+. These peptides were injected at a fixed concentration of 3 µmol/L in the aqueous compartment above the tBLM and allowed to interact with the bilayer before taking measurements (see Methods and Fig. S4).
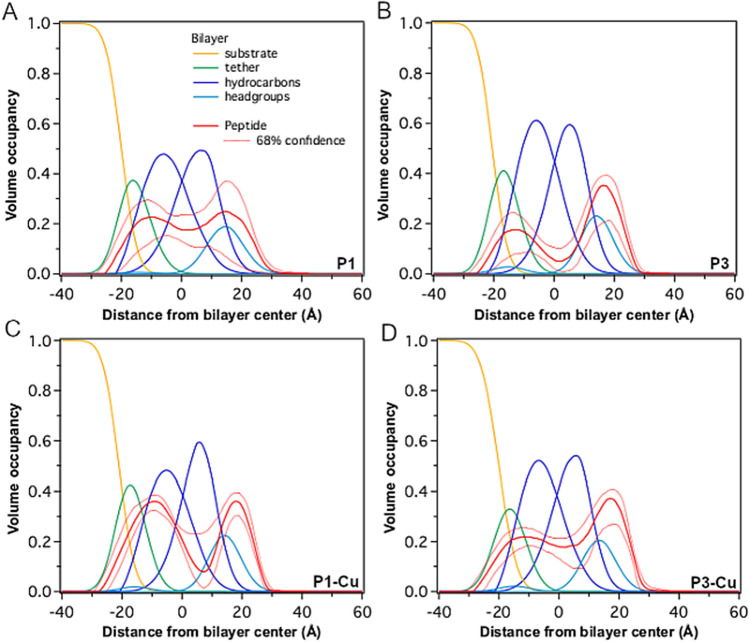
As many other helical HDPs, P1 and P3 adopt α-helical structures in the membrane-bound state56,57. The high resolution peptide structures derived from solid state NMR58, together with structural parameters of the lipid bilayer82,83 and scattering length densities (SLD) of the membrane components84, were integrated into a model to fit the depth-dependent distribution of each peptide in the bilayer. The best model fits were used to generate one-dimensional component volume occupancy (CVO) profiles85 (Fig. 4); these provide a way to quantify the fraction of the volume that is occupied by a particular molecular component as a function of the distance along the bilayer normal. For both P1 and P3 run at 3 µmol/L, peptide-to-lipid ratios (P/Ls) determined using the CVO profiles are close to 1/8 (see Table S1). This shows that the cationic P1/3 peptides freely bind to and robustly incorporate into the POPC bilayers, even in the absence of anionic lipids. Such a high P/L, as determined here, was previously associated with a regime of transition from surface-bound to tilted states and the subsequent formation of peptide-lined transmembrane defects56.
Figure 4.
Neutron reflectometry of P1 and P3 in the apo- and holo-states. The time and ensemble average spatial profiles of the various components of the tBLM (Fig. 2A) are shown as projections on the normal to the bilayer surface for (A) P1, (B) P3, (C) P1-Cu2+, and (D) P3-Cu2+. The peptides were metallated using a 1:1 stoichiometric amount of CuCl2. The inner bilayer leaflet is attached to the gold coated substrate (yellow) via molecular tethers (green). The outer leaflet of the bilayer is exposed to the aqueous compartment from which each peptide solution is injected.
It is important to note that the CVO profiles represent a time and ensemble average of the bilayer-peptide assemblies (Methods). In light of this, the profiles show that after initial partitioning into the outer (distal) bilayer leaflet from the aqueous injection compartment above it (Fig. 2C), P1 and P3 distribute into the two tBLM leaflets to different extents. While apo-P1 spreads out evenly between the two lipid leaflets and across the hydrocarbon region, apo-P3 remains predominantly confined to the headgroup region of the outer leaflet, unable to transition as efficiently as P1 into the hydrocarbon region and the inner (substrate-proximal) leaflet (Fig. 4A, B).
Next, we observed the effect of metalation on the distribution of P1/3 in the tBLM. While P1-Cu2+ shows a slightly higher accumulation in the inner leaflet compared to its apo-form (Fig. 4A, C), P3-Cu2+ remains preponderantly confined in the outer leaflet (Fig. 4B, D). However, P3-Cu2+ shows a deeper insertion, by 2 Å, into the hydrocarbon region compared to apo-P3, based on the calculated center-of-mass (COM) positions. The COM positions for each peptide, with respect to the bilayer center, are: P1: 2 ± 2 Å; P1-Cu2+: 1 ± 1 Å; P3: 5 ± 2 Å; and P3-Cu2+: 3 ± 1 Å. We note that these COM positions correspond to the average over the ensemble of all peptide molecules in the bilayer. Since the ensembles are characterized by broad distributions extending over both leaflets, it is important to consider the profile distributions of the peptides in the bilayer (Fig. 4A–D) and not just the positions of their COMs. The calculated relative amounts of each peptide in the inner bilayer leaflet, including the sub-membrane aqueous space (Fig. 2C1), are: P1:(45 ± 7) %, P3:(37 ± 6) %, P1-Cu2+ (54 ± 6) %, and P3-Cu2+ (39 ± 5) % (Table S1). For these calculations, we note that we used boundaries for the lipid bilayer regions based on the convention established by White and Nagle82,83.
Overall, the NR results indicate that P1 has an inherently higher ability to populate the hydrocarbon region of the bilayer and to cross the membrane barrier in both the apo- and holo- states. The presence of Cu2+ has a more consequential impact on P3, boosting its propensity to insert in the hydrocarbon region. In combination with the SPR/EIS studies, these structural results reveal that there is a direct correlation between the increased potency of Cu2+-bound P1 and P3 against cancer cells (Table 1) and their ability to insert and overcome the bilayer barrier.
Characterization of P1, P3, P1-Cu2+, and P3-Cu2+ in terms of bilayer conformations and disruptive effects to the bilayer structure
Following the NR experiments, we used ND to compare the effects of P1, P3, P1-Cu2+, and P3-Cu2+ on the bilayer structure and water distribution (Fig. 5). We worked at a P/L of 1:25 since we previously established that this value yields samples where the peptides are fully absorbed into the bilayer56. Because the multilayer samples are prepared by incubating liposomes with the peptides and the liposomes fuse once they are deposited on the glass substrate, the peptide molecules homogeneously populate both sides of the bilayer. The strong diffraction obtained from the multilayer systems allows us to analyze in greater detail the preferred position and conformation of each peptide in the bilayer57, together with perturbative effects of the peptides on the membrane structure.
Figure 5.
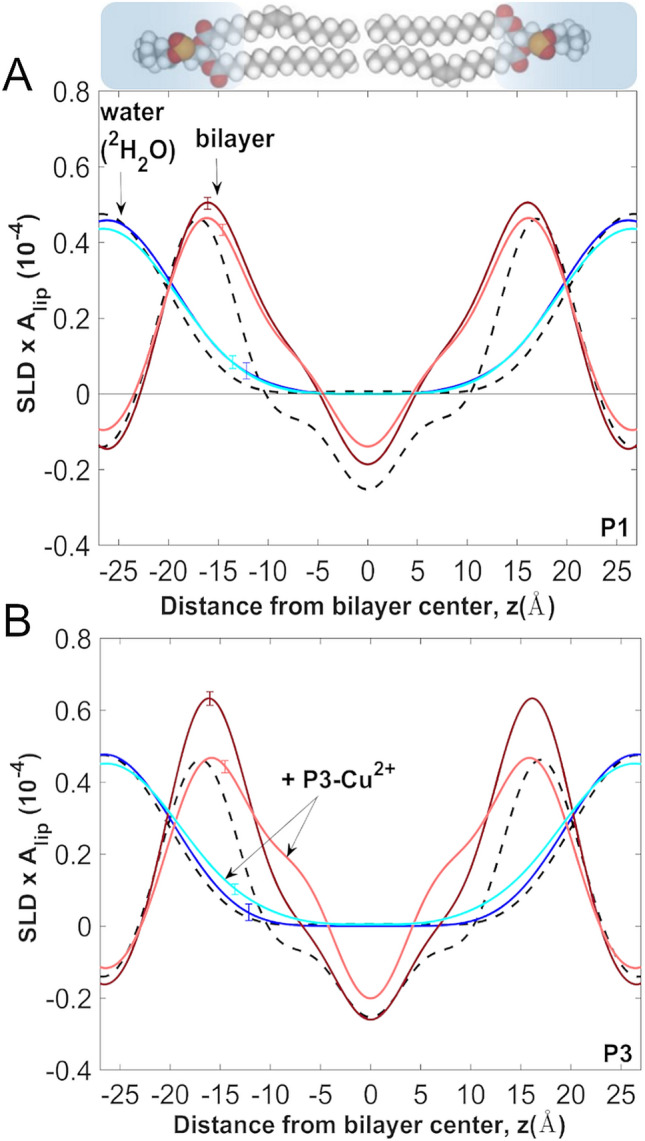
Bilayer scattering length density profiles from neutron diffraction on oriented lipid multilayers. (A) The scattering length density (SLD) profiles of the bilayer are shown as projections on the bilayer normal (z-axis) for POPC (black), P1/POPC (dark red), and P1-Cu2+/POPC (pink). The peptides were metallated using a 1:1 stoichiometric amount of CuCl2. The corresponding water profiles, determined from H2O/2H2O contrast are overlaid for POPC (black), P1/POPC (dark blue), and P1-Cu2+/POPC (light blue). Peptides distribute equally on both sides of the bilayer during their incubation with liposomes and deposition on the substrate (see Methods). (B) Same as in (A) but for P3. All oriented samples were prepared in POPC at P/L = 1:25, and measured at 23 °C and 93% relative humidity achieved by using the vapor phase of saturated salt solutions. The small error bars on the curves represent the uncertainty in the profiles, which were calculated using a 95% confidence interval in the Monte-Carlo sampling of the structure factors87. Structure factors and standard deviations are given in Table S2. All profiles were determined on a per-lipid scale using structure factors calibrated to reflect the composition of the unit cell and without explicitly determining the area per lipid87.
Figure 5 shows the centro-symmetric bilayer SLD profiles of POPC bilayers (with and without peptides) projected on the normal to the bilayer (z-axis). The most prominent features are the phospholipid headgroups of the two opposing bilayer leaflets (high SLD) and the hydrocarbon region in the middle (low SLD). Incorporation of the holo-form of each peptide leads to significant density redistributions, seen here as changes to the bilayer profile (Fig. 5A, B) relative to the undisturbed POPC bilayer. These changes are indicative of pronounced structural perturbations when the peptides become integrated in the bilayer, including a measurable thinning of the bilayer, often associated with the action of membrane–active peptides. The thinning is seen here as a reduction in the repeat distance (d) in the multilayered samples (Fig. 2E) with P1/3, compared to the unperturbed POPC. The values for d and their uncertainties, calculated from the positions of the Bragg peaks (Methods), were found to be: POPC: 53.5 ± 0.04 Å; + P1: 52.2 ± 0.1 Å; + P1-Cu2+: 52.6 ± 0.1 Å; + P3: 52.9 ± 0.1 Å; and + P3-Cu2+: 52.7 ± 0.2 Å.
The contrast in the behaviors of P1 and P3 upon metallation is clearly discernable in the density profiles (Fig. 5). We observe that while metallating P1 has little consequence on the density profile of the peptide/bilayer ensemble (Fig. 5A), it has a quite dramatic effect on P3, since P3-Cu2+ produces a clear density shift compared to apo-P3 (Fig. 5B). As indicated by the raised SLD in the center of the bilayer and the broader profile of the lipid headgroup region, this density shift is due to the dual effect of P3-Cu2+ migrating closer to the bilayer center and the lipid headgroups being pulled in by the peptide through hydrogen bonding56. To show how the water distribution relates to these structural changes, we specifically determined the water profiles in the POPC bilayer (Fig. 5A, B, blue shades) by using H2O/2H2O exchange (deuterium contrast) and ND86–88 (Methods). As the charged peptide helices insert in the bilayer, not only do they deform the bilayer surface by engaging lipid headgroups, but they also pull in water molecules, seen here as an extension of the water profile into the hydrocarbon region (Fig. 5A, B). The water penetration is most pronounced for the Cu2+-bound P1/3, indicating that the metal ion increases the water penetration, an effect that correlates with the increased conductance seen in the EIS measurements presented above.
Similar bilayer density redistribution effects are observed when the peptides are metallated with Ni2+ (Fig. S6) instead of Cu2+ (Fig. 5). This shows that, broadly speaking, the two metal ions similarly modulate the interactions of the peptides with the bilayer. Importantly, in further control experiments, we found that the Cu2+ ions by themselves (without peptides) have a completely different effect on the bilayer structure (Fig. S5). In contrast to the peptide- Cu2+, the free metal ions cause an increase in the repeat spacing (d), which is most likely due to the binding of the hydrated ions to the lipid headgroups89,90, causing further water accumulation at the membrane surfaces and a broadening of the inter-bilayer space.
Based on the results mentioned above, the bilayer structural perturbations that we observe in the presence of holo-P1 and -P3 are exclusively due to the complexation of the metal ions with the peptides and the complexation affects the interactions of P1 and P3 with the lipid bilayer. In particular, the resulting net loss of one positive charge at the amino end of the peptide upon metallation very likely facilitates the bilayer insertion of the metallated peptides63,64. Given that metallation more dramatically impacts the IC50 and membrane conformation of P3 compared to P1, we further examined P3-Cu2+ within the bilayer and used available 3D structures to characterize its orientation.
Position and orientation of P3-Cu2+ within the lipid bilayer
In earlier investigations, we specifically deuterated P1 and P3 near their N-terminus (positions 5–6) and C-terminus (positions 19–20) and determined their deuterium profiles by ND56. Using the 3D structures of the membrane-bound peptides determined by solid-state NMR and the specific deuterium profiles we solved the orientations of each peptide in the bilayer in terms of tilt angle and insertion depths of the deuterated N- and C-terminal regions. Here, we employed a similar approach on P3-Cu2+ bound to POPC (Fig. 6).
Figure 6.
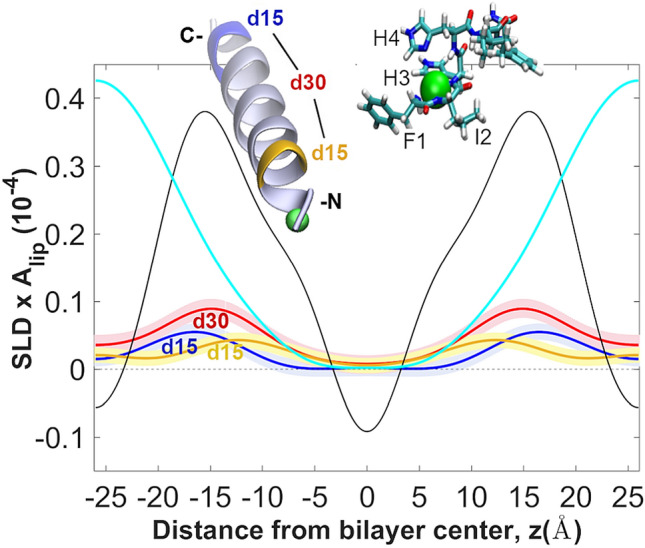
Positioning of P3-Cu2+ in the bilayer with ND and deuterium contrast. The SLD profiles of two groups of specifically deuterated amino acids near the N-terminus (I5d10F6d5; yellow) and C-terminus (F19d5L20d10; blue) of P3 were obtained, as well as their sum (red) (Methods). Each profile represents time and ensemble averages of each of deuterated groups in the thermally disordered bilayer. The overall profiles for the neat POPC bilayer with P3-Cu2+ (black) and the water distribution (blue) are overlaid on those for the deuterated groups. Measurements were done at P/L = 1:25, 23 °C, and 93% relative humidity. Uncertainty bands in the deuterium profiles (colored bands) were determined using a 68% confidence interval in the Monte-Carlo sampling of the measured structure factors87. The inset show representative average conformation of the P3 α-helix in the bilayer, using a 3D structure derived from the NMR structure (PDB ID # 6PEZ)58 and the predicted structure of the Cu2+-bound ATCUN motif based on density functional theory calculations64. The orientation of the α-helix in the bilayer is derived from the diffraction SLD profiles. The two deuterated sites are shown with yellow and blue; the Cu2+ ion (green sphere) is shown at a larger than true scale for better visibility.
As shown in Fig. 6, each of the two deuterated groups on the peptide define a broad, Gaussian-shaped distribution, when projected on the z-axis. The N-terminal group falls closer to the bilayer center than the C-terminal group. The distance between the maxima in the deuterium profiles of the two groups, which are both in the α-helical region of the peptide is 4.0 ± 0.4 Å. The distance between the COMs of these two groups along the α-helical axis, is 19.1 ± 1.2 Å. This was calculated by using the ensemble of structures previously solved by solid-state NMR for the apo-form (PDB ID # 6PEZ) and averaging their positions58. Based on these combined values, the estimated tilt of P3-Cu2+ is 78° relative to the normal to the bilayer (z-axis).
While we find the tilt of P3-Cu2+ in POPC to not be particularly pronounced, an important observation of this analysis is that the peptide is oriented so that the Cu2+-bound N-terminus is more inserted into the hydrocarbon region than the C-terminus (Fig. 6). This may appear surprising given the polar nature of the ion bound to the ATCUN motif. However, we also note that this peptide conformation is accompanied by a broader water distribution that extends into the hydrocarbon region (Figs. 5B and 6), trailing the Cu2+-bound ATCUN motif. Thus, we can infer the metal ion remains some degree of hydration in the bilayer interior, increasing water accessibility to the membrane core. The water penetration in the hydrocarbon is less pronounced for the undisturbed POPC bilayer, as well as for apo-P3/POPC system (Fig. 5B). We observe that P3-Cu2+ not only opens water accessibility into the hydrocarbon core, but the metal ion is located near the double bond of the oleic chain of POPC. Given the ability of Cu2+ to act as a catalyst for redox reactions, we decided to investigate whether the membrane-disruptive effects of the peptide go beyond the physical perturbations shown here and include the chemical oxidation of unsaturated fatty acids.
Oxidation of linoleic acid double bonds in lipid vesicles in the presence of Cu2+, P1, P3, P1-Cu2+, and P3-Cu2+
It has long been known that certain divalent metal ions, especially iron and copper, promote lipid peroxidation (oxidative deterioration of polyunsaturated fatty acids), a mechanism that has been strongly associated with cellular injury and death91. Divalent transition-metal ions participate in Fenton-like chemical reactions produces ROS (e.g., hydroxyl radicals, superoxide) that can oxidize double bonds within the membrane89. Here, to learn about the possible piscidin/Cu2+-mediated oxidation of double bonds in lipid bilayers, we used absorbance and mass spectrometry to measure the formation of the oxidation products in small unilamellar vesicles (SUV) constituted of di-linoleoyl-sn-glycero-3-phosphocholine (DLPC) and POPG mixed in a 3:1 molar ratio and prepared at a concentration of 0.50 mg total lipid/mL. We first tested the ability of free Cu2+ to oxidize the double bonds within the liposomes. Increasing amounts of Cu2+ for a fixed amount of lipids (expressed as Cu/L, the molar ratio of Cu2+ to total lipid) were mixed with the SUVs and the absorbance was measured at λ = 234 nm (Fig. 7A) (corresponding to λmax for the conjugated dienes that appear in lipid oxidation products)92,93. Because molecular oxygen (O2) found in the atmosphere readily dissolves in the bilayer, no exogenous oxidant is needed to initiate the oxidation of the double bonds94. As shown in Fig. 7A, a dose–response in oxidation yields is observed after 24 h with increasing Cu/L ratio. Similar results have been reported before on the oxidation of DLPC upon pre-incubation with Cu2+ for 24 h95. The Cu2+-catalyzed reactions were also measured in the presence of butylated hydroxytoluene (BHT), a free radical scavenger that interrupts the chain oxidation process96 (Fig. 7A). We see that BHT added in a proportion of 1 to 100 lipids stops lipid oxidation since the signal from the lipids/CuCl2/BHT sample is negligible at 234 nm.
Figure 7.
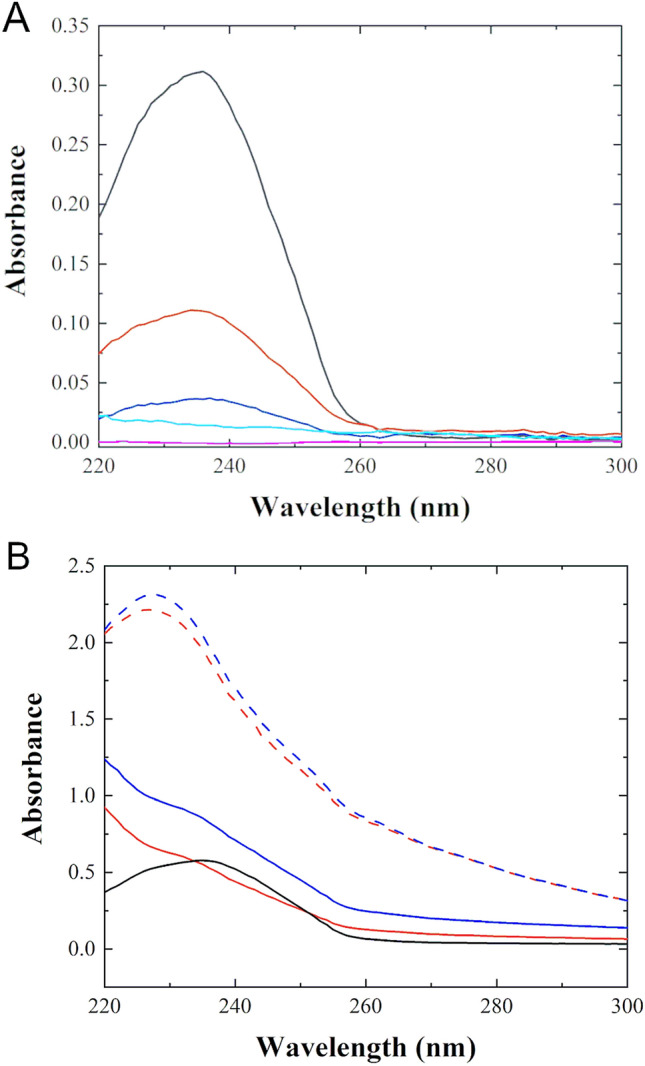
Absorbance measurements from vesicles containing a polyunsaturated lipid exposed to Cu2+ and piscidin bound to Cu2+. SUVs made of 3:1 DLPC/POPG were prepared and exposed to different forms and amounts of Cu2+ and measured by UV spectrophotometry after 24 h of exposure. (A) Samples containing SUVs at a fixed lipid concentration of 0.50 mg/mL (640 µmol/L) were mixed with various amounts of CuCl2, as follows: Cu2+/L = 1:2 molar ratio (black), Cu2+/L = 1:8 (red), Cu2+/L = 1:32 (blue), BHT/L = 1:100, and Cu2+/L = 1:8 (cyan) and lipid alone (magenta). (B) SUVs at a concentration of 0.20 mg/mL (260 µmol/L) were studied in the presence of the peptides at P/L = 1:10, corresponding to a peptide-Cu2+ complex concentration of 26 µmol/L. P1-Cu2+ (blue, dashed); P3- Cu2+ (red, dashed); P1 (blue); P3 (red); lipid alone (black). The source of free Cu2+ (CuCl2) was the same as that used for metallating P1 and P3. Measurements were taken in triplicates at 24 h after exposure. The spectra were corrected for the background signals from CuCl2 and the lipids at time zero as explained in the Methods. Uncertainties are smaller than the line thicknesses. Strong absorbance at 234 nm after 24 h of exposure points at the formation of lipid oxidation products, a result confirmed by mass spectrometry (Fig. S7).
It is important to note that divalent transition-metal ions can bind to both negatively-charged and zwitterionic lipid headgroups with high affinity, especially when amine moieties are present89,90. While it is difficult from the present studies to identify whether the free Cu2+ ions experience differential affinities for the PG and PC headgroups, our diffraction data (Fig. S5B) indicate that free Cu2+ binds to the headgroup region and causes lipid separation, presumably through a preferential segregation of the ions with the negatively-charged PG groups. This is consistent with our recent solid-state NMR data showing that Ni2+ preferentially interact with PG headgroups in PC/PG binary mixtures63. Due the sequestration of the Cu2+ on the membrane surface, it is conceivable that oxidizing agents that are generated near the membrane surface diffuse through the membrane, thus gaining access to the double bonds.
We compared the effect of free Cu2+ ions to that for P1 and P3, in both the apo- and holo- forms. As expected, apo-P1 and apo-P3 show little lipid oxidation (Fig. 7B), with the peak at 234 nm being swamped by a significant scattering contribution. This was consistent with our observation that the vesicle solutions show a faint turbidity upon peptide incorporation, even in the very dilute lipid solutions used to collect these data (0.20 mg/mL). However, samples containing the Cu2+-bound peptides display a prominent peak at 234 nm. Given that piscidins can serve as a Cu2+ carrier in the bilayer interior, according to our ND and NR results, it is concluded that the peptides facilitate the access of Cu2+ to the oxidation-prone double bonds, accelerating membrane damage. As revealed by mass spectrometry (Fig. S7), the lipid oxidation leads to fragmentation of the acyl chains and such a chemical damage could ultimately cause loss of bilayer structural integrity97–99.
Discussion
The most compelling result from our SAR studies on the host defense metallopeptides P1 and P3 is the boost in both function and membrane insertion that is elicited by Cu2+-coordination. This is significant because it identifies metallation as a chemical modification that can improve the biological activity of this family of HDPs, several of which have already proven to be promising templates for the development of novel strategies to treat infections, inflammation, and cancer7,8,15–22. Specific valuable features of P1 and P3 include their broad spectrum activity in the micromolar and sub-micromolar (for P1) range, anti-inflammatory properties, and pH-resiliency in the physiological range33,47,56,100–102. P1, which has been more tested in vivo than P3, has very good pharmacodynamics and pharmacokinetics characteristics61,62,102. The versatility of these HDPs derive partly from the abundance of histidine residues that endows them with buffering capacity as well as the ability to bind and carry metal ions (Cu2+/Ni2+) to their cellular site of action29,59,63,64. Bound to each other, the Cu2+ ion and piscidin become co-localized in cellular compartments. In the natural setting of the fish, this corresponds to the phagosomes of immune cells and the extracellular environment upon degranulation103.
To the best of our knowledge, this is the first study to report the cytotoxicity of P1-Cu2+, P3, and P3-Cu2+ on cancer cells. Apo-P1 was previously known to have anti-cancer activities39 but the effects of complexation with Cu2+ on this function had not been reported. We formerly showed that such metallation increases the direct antimicrobial potencies of P1 and P3 through effects that include the physical and chemical disruption of membranes and DNA59,63. This led us to assume that binding to Cu2+ could also enhance their cytotoxicity against cancer cells. The improved ability of piscidin to kill cancer cells upon Cu2+-coordination, as shown here, can be rationalized in part by examining peptide-bilayer interactions at the molecular level. In our earlier studies of apo-P1 and -P3, we demonstrated the abilities of the peptides to change their conformation from an unstructured form in solution to an amphipathic α-helix upon contacting the phosphate-containing groups of detergents and phospholipids53–55,57,104. This conformational change is induced by not only anionic but also zwitterionic membranes because the hydrophobic effect plays an important role in driving the partitioning of these peptides into amphipathic lipid membranes105–107. Using oriented CD, oriented-sample solid-state NMR57,58,79, ND56, and extensive all-atom MD simulations56,57,108,109, we found that P1 and P3 orient their amphipathic α-helices at an angle that depends on their concentration and the composition of the membrane. At low concentration, the peptides lie almost parallel to the bilayer plane. At higher concentrations, they become tilted and elicit membrane permeabilization. Lower P/L thresholds for tilting were observed for P1 than P3, and for zwitterionic than anionic bilayers, most likely because the anionic lipids retain the peptide on the bilayer surface56,58,79.
Here, we show that metallation with Cu2+ dramatically boosts the ability of P1 and P3 to kill cancer cells and produce conductive defects through otherwise electrically sealing model membranes. Indeed, our SPR/EIS measurements performed on a POPC-tBLM bilayer platform detected an immediate increase in conductance (drop in resistance) starting with apo-P1 at a concentration of 0.2 µmol/L. In the case of apo-P3, this response had a threshold of 0.5 µmol/L and was delayed by several minutes. Likewise, cancer cells were much more responsive to apo-P1 than apo-P3. Bound to Cu2+ before being added to cancer cells, P1 and P3 experienced IC50 values enhanced by a factor of up to ≈2 and ≈7 based on the data in Fig. 1, respectively, and this correlated well with increased bilayer conductances, as measured by SPR/EIS.
Our NR measurements done using the same supported bilayer as for the SPR/EIS experiments revealed the localization preferences of P1 and P3 in the bilayer milieu. Upon partitioning from the water phase into the bilayer outer leaflet (distal to the substrate), apo-P3 adopts a more superficial bilayer position than apo-P1. However, both peptides migrate to deeper locations in the hydrocarbon core when bound to Cu2+. Changes in the P1 distribution between the apo- and holo-forms have been difficult to assess because both states exhibit a high propensity for insertion in the hydrocarbon region. Nevertheless, while P1 remains broadly confined to the hydrocarbon core, metallation appears to induce a change in its bilayer position so that its COM moves closer to the inner leaflet. This suggests that complexation with the metal ion confers P1 with an increased ability to transfer from one side of the bilayer to the other.
In comparison to P1, P3 responds to Cu2+-complexation much more dramatically since its IC50 value improves by a factor of ≈7 versus ≈2 for P1 according to the data shown in Fig. 1. As shown by the NR and ND profiles, this effect correlates with Cu2+-binding having a more pronounced effect on P3 than P1 in terms of how the peptides get redistributed toward the bilayer center. ND offers additional information when used in conjunction with site-specific deuteration of P3: the Cu2+-carrying N-terminus of P3 is found to be strikingly more inserted in the hydrocarbon region than its C-terminus. For comparison, a similar orientation was reported for apo-P3 in POPC/POPG bilayers, albeit at a more pronounced tilt angle56, suggesting that both the presence of the metal ion and lipid composition can subtly influence the bilayer conformation of the peptide in terms of tilt and depth of insertion. The deeper insertion achieved by piscidin upon metallation can be explained by the loss of one positive charge at the amino end of the peptide when the metal ion binds59,63,64. Our earlier combined NMR and DFT structural investigations of P3-Ni2+ bound to 3:1 PC/PG suggested that the bulky and hydrophobic I2 and H4 side chains surround the metal ion to facilitate its insertion into the hydrocarbon region64. As revealed by our ND data and our lipid oxidation assays in model membranes, the inserted ATCUN-Cu2+ not only opens the bilayer for water penetration, but also appears to promote oxidation in the surrounding lipid. This dual ability to physically and chemically alter bilayers adds to the multifaceted actions that piscidins may be able to exert within living cells. As we previously showed, metallation of both P1 and P3 results in enhanced MICs and lipid peroxidation effects in bacterial cells59. In model membranes, the permeabilization effects of P1 are improved by metallating the peptide as well as adding oxidized lipids to the membrane63.
With regard to cancer cells, Lin et al. showed that apo-P1 exerts apoptotic effects on HT1080 cells at low concentrations while its mechanism of action is necrotic at higher doses39. Cheng et al. more recently observed that apo-P1 also has apoptotic effects on osteosarcoma (bone) cancer cells78. More specifically, the peptide enhanced the formation of mitochondrial ROS, resulting in oxidative damage. Apoptosis as a mechanism of cell death has been observed for other ACPs, including the synthetic peptide A9K and a derivative of magainin-219. This apoptotic cell death pathway is consistent with our microscopy studies presented here. Indeed, we found that P1 and P3 are cell penetrating at low concentrations and reach the mitochondria. Once in this location, it is plausible that they disrupt the mitochondrial membrane and initiate apoptosis. From these results, we would infer that at low concentrations, P1 and P3 are more disruptive to mitochondrial than plasma membranes. This is expected because of the differing lipid compositions of these organelles, with the former containing the anionic lipid cardiolipin (CL) that is well known to attract cationic HDPs7,15,17,110. Interestingly, we previously showed that P1 and P3 leverage membrane heterogeneity to disrupt membranes containing phosphatidylethanolamine (PE), another lipid abundant in mitochondrial membranes58. Further supporting the case that P1 and P3 interact favorably with mitochondrial membranes is the research by Tin et al. on tilapia P4, TP443. The authors showed that this piscidin, which was successfully used in vivo against triple-negative breast cancer, exhibits strong affinity for the mitochondrial membrane of cancer cells. Interestingly, they found evidence for necrotic cell death, with damaged mitochondria leaking calcium and experiencing disrupted calcium homeostasis and signaling.
Several peptides, including pleurocidin that is related to piscidin, use ROS formation to induce cell death via apoptosis or inhibition of nucleic acid synthesis111. Another cationic HDP with anti-cancer property, lactoferricin B interacts with the mitochondrial membrane, leading to the formation of ROS and inducing apoptosis112,113. Tilapia hepcidins also feature strong anti-cancer activity and the reduction of multi-drug resistance via a mechanism that involves ROS formation and the activation of a mitochondrial apoptosis pathway114–117.
Our study focused on showing that metallation enhances the cytotoxic effects of piscidins 1 and 3 on cancer cells and explaining this effect through a detailed characterization of peptide-lipid interactions. As a next step, it will be important to determine whether the mechanism of cell death by P1 and P3 bound to Cu2+ features apoptosis, as previously shown for apo-P139,78. For instance, it would be insightful to use cell-based assays to determine whether the Cu2+-bound peptides release Cu2+ intracellularly (e.g., acidic endosomes) resulting in enhanced ROS formation and increased apoptotic effects compared to apo-P1 used at low concentration. Combined with biophysical studies using model membranes that incorporate lipids present in the outer leaflets of cancer cell plasma (e.g., PS, PE, cholesterol) and mitochondrial membranes (e.g., CL, PE), this could help shed light on the propensity of the peptides to bind to and disrupt one type of membranes over the other. In the case of P3-Cu2+, which is known to be more damaging to bacterial DNA than membranes59,79, there is also the possibility that part of its mechanism involves using Cu2+ to form radicals that damage mitochondrial DNA.
Mitochondrial targeting has attracted a lot of attention in cancer therapy6,17,40,41,118–120. One of the major reasons is that cancer cells are more sensitized than healthy cells to mitochondrial perturbations due to their metabolic reprogramming. Cationic molecules that can accumulate preferentially in mitochondria based on charge and membrane potential have been particularly sought after since they can help deliver cargoes with anticancer properties and induce cell death17,18,40,118–120. The potential to use cationic host defense metallopeptides to cross cell membranes and reach mitochondria to deliver a cargo, such as a redox center is worth further investigating based on our results with piscidin. In particular, the spontaneous ability of metallated piscidin to release Cu2+ within acidic compartments (e.g., late endosomes) could contribute to their considerable efficacies. From this perspective, peptides with different potencies could be designed by varying their cell penetrating properties and pH-dependent affinities for Cu2+.
We note that before anticancer piscidin peptides bound to Cu2+ can be safely used in vivo, their stability in blood plasma has to be assessed since toxic effects could occur if Cu2+ were released and/or transferred to plasma proteins before reaching cancer cells121. Miyamoto et al. recently made constructs of the ATCUN motif and octreotide, a tumor homing sequence122. In terms of blood plasma stability, the authors found that bulky residues within the ATCUN motif stabilized the Cu2+-peptide complexes. YYH-octreotide, the most stable construct in blood plasma was successfully used in vivo to image tumors by 64Cu positron emission tomography. In the case of piscidin, we recently showed using NMR and quantum calculations that favorable cation-π interactions are possible between the metal ion and the bulky aromatic residues in the N-terminal region of the peptide, resulting in a stabilizing effect for the metal ion65. Given that piscidin and YYH-octreotide have metal-binding regions similarly rich in aromatic residues, we anticipate comparable stability of their Cu2+-complexes in vivo. Additional studies with piscidin peptides are needed to verify this prediction.
Conclusion
This work reveals that metallating HDPs is an effective strategy to enhance their anticancer properties through the expansion of their mechanism of action, so that both physical and chemical damage of membranes is achieved. In particular, our results from cell-based assays, electrochemistry and structural measurements clearly establish that coordinating Cu2+ to P3 tremendously enhances its cytotoxic effects on cancer cells and this effect correlates with a stronger ability of the holo- than apo-form to interact with model membranes. Since both P1 and P3 have cell penetrating properties at sub-lethal concentrations, they can not only use Cu2+-binding to enhance their bilayer penetrating abilities but also carry and release Cu2+ intracellularly, resulting in other disruptive effects such as ROS formation, lipid peroxidation, and the destabilization of mitochondrial membranes and/or DNA. As membrane-interacting Cu2+-binding peptides that have favorable biocompatibility properties and can reach mitochondria, piscidins could represent new opportunities in the development of novel mitochondria-targeting therapeutics to treat life-threatening diseases such as cancer.
Methods
Materials
Carboxyamidated P1 and P3, including the 2H-labeled forms, were chemically synthesized at the University of Texas Southwestern Medical Center. Once received, they were purified and lyophilized, as previously reported57. The peptides were dissolved in dilute HCl to substitute chloride for trifluoroacetate ions and dialyzed to remove excess salt, leading to 98% pure peptides57,100. Following reconstitution of the peptides in nanopure water, their molar concentrations were determined by amino acid analysis performed at the Protein Chemistry Center at Texas A&M. Metallation was done in a 1:1 molar ratio between the peptide and metal using aqueous CuCl2 (Hampton Research, Aliso Viejo, CA). The pH was carefully adjusted to 7.4 since protons are released by the peptide upon metal binding64. MTT reagent (Thiazolyl Blue Tetrazolium Bromide) was purchased from Sigma-Aldrich. Lipids, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), di-linoleoyl-sn-glycero-3-phosphocholine (DLPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidyl-rac-glycerol (POPG), were obtained from Avanti Polar Lipids (Alabaster, AL).
Cell cultures
A549 human lung cancer cells were obtained from ATCC (ATCC CCL-185) and were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM-F12, Lonza, catalog no. 12-719F) supplemented with 10% heat inactivated FBS and 1% Penn-Step. The HT1080 and MRC-5 cells were purchased from ATCC (catalog no. CCL-121 and CCL-171, respectively) and grown in Eagle’s Minimum Essential Medium (ATCC, catalog no. 30-2003TM) that was supplemented with FBS (ATCC, catalog no. 30-2021TM) such that the medium contained 10% FBS. The MDA-MB-231 and HeLa cells were obtained from ATCC (ATCC, catalog no. HTB-26 and CCL-2, respectively) and maintained in DMEM supplemented with 10% FBS. All cell cultures were grown at 37.5 °C in a humidified 5% CO2 atmosphere.
MTT/MTS assays on cancer cells to evaluate the cytotoxicity of P1 and P3 in the apo- and holo-states
The cytotoxicity assays followed well established procedure, with the details given in the Supplementary Information. The assays were performed in triplicates using 96 well plates Cell viability was calculated using the equation: % viability = [(AT-AB)/(AC-AB)] × 100, where AT is the absorbance of the treatment well, AB is the average background absorbance measured from three wells containing only media, and AC is the absorbance of the positive control well, which received only 100 µL of media and 40 µL of MTT solution (i.e. no peptide). Averages and error bars were calculated as the mean + / − standard deviation based on triplicates. The IC50 values were obtained by fitting the percentage viability (% viability) vs Log (M) concentration of peptides to generate inhibitor dose response curve using the GraphPad Prism software. The curves were fitted using a least-squares minimization method. Individual data points on the graphs are provided as the mean (± standard deviation) from the triplicate % viability values. The GraphPad Prism algorithm converted the logIC50 into IC50 and used the standard error of the mean on the logIC50 to yield the 95% confidence interval for the logIC50 and IC50.
Laser confocal microscopy
HeLa and MD-MBA-231 cells were propagated in DMEM supplemented with 10% FBS and containing 200 U/mL each of penicillin and streptomycin. Incubation was done at 37 °C with 5% CO2. Then 20,000 cells were seeded into each well of a chambered cover glass and allowed to adhere overnight. The cells were exposed to 100 nmol/L MitoTracker, 5 µg/mL Hoescht 33,258 (for DNA staining), and 4 µmol/L FITC-labeled peptides for 20 min. The medium was then aspirated, the cells were washed with PBS before adding fresh medium. The cells were imaged on a Nikon A1R spectral confocal microscope. Images are shown as acquired without any post-imaging processing.
Surface plasmon resonance (SPR)/electrochemical impedance spectroscopy (EIS)
SPR/EIS measurements were carried out on tethered bilayer membrane (tBLM) using a custom-built instrument previously described (Fig. 2B)123,124. Polished sapphire substrates were coated with a 5 Å layer of chromium followed by a 440 Å gold film by magnetron sputtering (Denton Vacuum LLC, Moorestown, NJ) at the nanoFab facility of the National Institute of Standards and Technology (NIST). Several POPC tBLMs were assembled on the gold surfaces using thiolated polyethyleneglycol linker molecules (1,2-di-O-myristyl-3-[ω-mercaptohexa(ethylene oxide) glycerol] (WC14) and β-mercaptoethanol (βME) at a ratio of 70:30 βME:WC14. The tBLMs were equilibrated in Tris buffer (20 mmol/L, 50 mmol/L NaCl, pH 7.4) and allowed to rest until both SPR and EIS signals were stable, before peptide injection. Two types of measurements were done. In the first type (Fig. 3), the peptide concentration was increased gradually, in steps of 0.1 μmol/L, and the SPR/EIS was monitored for 10 min after each addition. The peptide addition was repeated until the bilayer resistance dropped significantly. In the second type (Fig. S3), each peptide was introduced onto the tBLM-containing SPR/EIS cell at a fixed concentration of 3 μmol/L. After 10 min of incubation, the cell was rinsed with the buffer, and the SPR/EIS measurements were continued for another 10 min. In each case, intensity distribution versus pixel position on the camera chip was measured by SPR. The pixel positions of the SPR signal minima were plotted as a function of time. EIS data were recorded from 1 to 10 kHz at a rate of 1 spectrum per minute, resulting in a total of 22 spectra for each experiment. Each spectrum contained 100 data points. The Z-plot and Z-view software (Scribner Associates, Inc., NC) was employed for the impedance spectral collection and analysis. The simplest electrical circuit model (Fig. 2D) was used to fit the impedance data using a complex nonlinear least-squares minimization procedure based on a Levenberg − Marquardt algorithm implemented in the Zview software. The standard deviation in the fit parameters (e.g., capacitance and resistance) were calculated from the residuals of the fit (mean square error).
Neutron reflectometry (NR)
NR measurements were conducted on POPC tBLMs supported on 3 inch diameter, 5 mm thick n-type Si:P[100] wafers (El-Cat Inc., Ridgefield Park, NJ). The wafers were cleaned and coated with Cr (≈20 Å) and Au (≈150 Å) at the NIST nanoFab facility. Coated substrates were immersed in a 7:3 (mol/mol) ethanol solution of HC18125 and β-mercaptoethanol at a total concentration of 0.2 mmol/L to form a self-assembled monolayer (SAM). Substrates were assembled in fluids cells126 and 5 mg/mL solutions of POPC vesicles were added to the dry SAM for 2 h. Afterwards, the system was flushed with pure water to complete tBLM formation. Before the addition of peptide, NR measurements of the tBLM by itself was carried out to verify the formation of a complete bilayer. Peptide in buffer (20 mmol/L Tris, pH 7.5), at a solution concentration of 3 µmol/L, was then injected in the aqueous compartment adjacent to the tBLM, allowed to incubate with the bilayer for 30 min, and then rinsed with buffer to remove any loosely bound peptide. For each sample, data were sequentially collected with D2O and H2O-based buffers. Buffer exchange was achieved by flushing ≈10 mL of buffer through the cell (volume, 1.3 mL) using a syringe. NR measurements were performed at the CGD-Magik reflectometer at the NIST Center for Neutron Research (NCNR)127. Reflectivity curves were recorded at room temperature, for momentum transfer values 0.01 ≤ qz ≤ 0.25 Å−1. For data analysis, 1D-structural profiles of the substrate and the lipid bilayer along the lipid bilayer normal were parameterized using a continuous distribution model as described elsewhere85. Within such a model, spatial distributions of molecular groups along the bilayer normal are described with continuous component volume occupancy (CVO) profiles. Optimization of model parameters was performed using the ga_refl and Refl1D software packages developed at the NCNR126. All reflectivity curves of one data set were fitted simultaneously to the same model, sharing common fit parameters, for example for the solid substrate. A Monte-Carlo Markov chain-based global optimizer was used to determine fit parameter confidence limits126.
Neutron diffraction (ND)
Oriented lipid multilayers samples of POPC and piscidin at P/L = 1:25 (Fig. 2E) were prepared as previously described58. The Cu2+-bound peptides were solubilized initially in pure water and equilibrated in sodium phosphate buffer (4 mmol/L phosphate buffer, pH 7.8). Then, they were mixed with POPC liposomes in the same buffer and allowed to incubate for 1 h before fusing on glass substrates. ND measurements on oriented lipid multilayers (Fig. 2E), probing the direction orthogonal to the bilayer plane, were acquired with the instrument CGD-MAGIk at the NCNR. The data were processed and analyzed as described before86,128. In short, bilayer structure factors were obtained as the square root of the integrated Bragg peaks, corrected for background, absorption and extinction, and their phases determined by deuterium contrast, using H2O/2H2O exchange86–88. Tables with structure factors can be found in Supplementary Information (Tables S2 and S3). The bilayer one-dimensional SLD profile was calculated by Fourier synthesis of the structure factors. The contrasts between deuterium-containing and natural abundance samples arising from the higher neutron scattering length of deuterium (b2H = 6.67.10–5 Å) with respect to hydrogen (bH = − 3.74·10−5 Å) were used to parse out the specific profiles of the deuterated regions. The amplitudes of the profiles were determined to the best approximation using 2H2O for calibration87 and considering that there are 9.4 waters per lipid headgroup at 93% relative humidity and that the peptides bring in an additional 1.2 waters/lipid on average when the P/L is 1:2556. Repeat distances, d, (Fig. 2E) and their uncertainties were determined by a linear fit of the Bragg peak position versus diffraction order.
Lipid oxidation measurements
Fresh vials of DLPC and POPG in chloroform were open and mixtures of the two lipids were prepared in an oxygen-free chamber. After the evaporation of chloroform under a stream of nitrogen gas, SUVs were produced by suspending dry lipids in pure deionized water at a concentration of 0.50 mg/mL, followed by sonication to obtain a highly clear solution. Oxidation of polyunsaturated lipid chains of DLPC in the presence of Cu2+, P1-Cu2+, or P3-Cu2+ were monitored based on the appearance of an absorbance peak at 234 nm. This peak was previously reported to represent the formation of lipid oxidation products (e.g. conjugated dienes)92. For samples containing SUVs and CuCl2, oxidation was initiated by adding CuCl2 to the freshly prepared SUVs (0.50 mg/mL), at final copper/lipid (Cu/L) molar ratios of 1/2, 1/8, and 1/32. Control samples (without copper) or in the presence of the antioxidant butylated hydroxytoluene (BHT) were also measured. Samples containing P1 or P3 in the apo or holo states were prepared in water at a final P/L = 1/10 and stabilized in a sodium phosphate buffer (4 mmol/L, pH 7.8) to preserve the complexation of the two peptides with Cu2+. The lipid concentration in the peptide-containing samples was 0.20 mg/mL. The buffer concentration was kept minimal to avoid turbidity effects of the lipid dispersions. Since it takes several hours to achieve significant oxidation, the degree of lipid oxidation was assessed for all samples after exposure to air for 24 h, by measuring absorbance spectra in the range of 200 to 300 nm with a Molecular Devices Spectra Max M5 plate reader. The absorbance values reported and their standard deviations were obtained by averaging values from three simultaneous, independent measurements. The spectrophotometer was blanked using CuCl2 diluted at concentrations equivalent to those used in the samples measured in Fig. 7. To correct for the background from unoxidized lipids, we subtracted from the averaged spectra the signal collected for the SUVs alone at time zero.
Mass spectrometry
Aliquots of fresh DLPC were taken after opening the lipid vials, coordinating with the timeline of the oxidation measurements above. POPG was not used in order to avoid complicating the spectrum, unnecessarily. The lipid in chloroform was dried under a stream of nitrogen gas. Solubilized in water, CuCl2 was added to a Cu/L ratio of 1:2 and allowed to incubate for 20 h. Just before mass spectrometry measurements, the DLPC/Cu2+ samples were diluted in a 1:1 water/methanol solution to a final concentration of 100 µmol/L. Electrospray Mass Spectrometry (ESI–MS) was performed on an Orbitrap Elite (Thermo Fisher, San Jose, CA) in positive mode. Samples were directly infused into the mass spectrometer at a flow rate of 10 µL/min.
Supplementary Information
Acknowledgements
This research received support in part from the National Science Foundation (MCB-1714164 to M.M and V.S.; MCB-1716608 to M.L.C., and MCB-1715494 to A.M.A.B.). We thank Dr. William Stetler-Stevenson (NIH/NCI) for help with lung cancer assays. The study utilized neutron diffraction facilities at the US National Institute of Standards and Technology (NIST), Gaithersburg, MD. The identification of any commercial product or trade name does not imply endorsement or recommendation by NIST.
Abbreviations
- ACPs
Anticancer peptides
- AMPs
Antimicrobial peptides
- ATCUN
Amino terminal copper and nickel
- BHT
Butylated hydroxytoluene
- CL
Cardiolipin
- CD
Circular dichroism
- COM
Center of mass
- CVO
Component volume occupancy
- DLPC
Di-linoleoyl-sn-glycero-3-phosphocholine
- EIS
Electrical impedance spectroscopy
- FITC
Fluorescein isothiocyanate
- HDPs
Host defense peptides
- MDR
Multi-drug resistance
- MIC
Minimum inhibitory concentration
- MD
Molecular dynamics
- ND
Neutron diffraction
- NR
Neutron reflectometry
- P1
Piscidin 1
- P3
Piscidin 3
- PDB
Protein data bank
- PE
Phosphatidylethanolamine
- P/L
Peptide-to-lipid ratio
- POPC
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphatidyl-rac-glycerol
- ROS
Reactive oxygen species
- SAM
Self-assembled monolayer
- SLD
Scattering length density
- SARs
Structure-action relationships
- SPR
Surface plasmon resonance
- tBLM
Tethered bilayer membranes
- TP4
Tilapia piscidin 4
Author contributions
The peptides P1, P3, FITC-P1, and FITC-P3 were provided by M.L.C. P1 and P3 were metallated by M.L.C. The cytotoxicity cell assays were initiated by M.L.C and M.W.S., and performed and analyzed by A.C., M.L.C., M.D.J.L, and M.W.S. The microscopy experiments were designed by M.L.C., the samples prepared by M.D.J.L. and M.W.S., the microscopy performed by M.D.J.L., and the data analyzed by A.A.B., M.L.C., and M.D.J.L. The NR and ND experiments were designed, performed, and analyzed by F.C., F.H., and M.M. SPR/EIS experiments were designed, performed, and analyzed by F.C. and V.S. The oxidation and mass spectrometry measurements were performed by C.D., F.C. and K.A. The lipid peroxidation assays were designed, performed, and analyzed by F.C. and M.M. The manuscript was initiated by M.M. and written by M.M. and M.L.C. with significant insights and method section materials from all co-authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Myriam L. Cotten, Email: mcotten@wm.edu
Mihaela Mihailescu, Email: emihailescu@ibbr.umd.edu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91670-w.
References
- 1.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 2.Zavascki AP, Goldani LZ, Li J, Nation RL. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 2007;60:1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 3.Yount NY, Yeaman MR. Emerging themes and therapeutic prospects for anti-infective peptides. Annu. Rev. Pharmacol. Toxicol. 2012;52:337–360. doi: 10.1146/annurev-pharmtox-010611-134535. [DOI] [PubMed] [Google Scholar]
- 4.Baym M, Stone LK, Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;353:1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karam G, Chastre J, Wilcox MH, Vincent JL. Antibiotic strategies in the era of multidrug resistance. Crit. Care. 2016;20:136. doi: 10.1186/s13054-016-1320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui Q, et al. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist. Updat. 2018;41:1–25. doi: 10.1016/j.drup.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Gabernet G, Müller AT, Hiss JA, Schneider G. Membranolytic anticancer peptides. MedChemComm. 2016;7:2232–2245. doi: 10.1039/C6MD00376A. [DOI] [Google Scholar]
- 8.Felício MR, Silva ON, Gonçalves S, Santos NC, Franco OL. Peptides with dual antimicrobial and anticancer activities. Front Chem. 2017;5:5. doi: 10.3389/fchem.2017.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 10.Attiê R, Chinen LT, Yoshioka EM, Silva MC, de Lima VC. Acute bacterial infection negatively impacts cancer specific survival of colorectal cancer patients. World J. Gastroenterol. 2014;20:13930–13935. doi: 10.3748/wjg.v20.i38.13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vedham V, Divi RL, Starks VL, Verma M. Multiple infections and cancer: implications in epidemiology. Technol. Cancer Res. Treat. 2014;13:177–194. doi: 10.7785/tcrt.2012.500366. [DOI] [PubMed] [Google Scholar]
- 12.Fishman JA. Infections in immunocompromised hosts and organ transplant recipients: essentials. Liver Transpl. 2011;17(Suppl 3):S34–S37. doi: 10.1002/lt.22378. [DOI] [PubMed] [Google Scholar]
- 13.Xiao YF, et al. Peptide-based treatment: a promising cancer therapy. J. Immunol. Res. 2015;2015:761820. doi: 10.1155/2015/761820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rolston KV. The spectrum of pulmonary infections in cancer patients. Curr. Opin. Oncol. 2001;13:218–223. doi: 10.1097/00001622-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweizer F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009;625:190–194. doi: 10.1016/j.ejphar.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 17.Constance JE, Lim CS. Targeting malignant mitochondria with therapeutic peptides. Ther. Deliv. 2012;3:961–979. doi: 10.4155/tde.12.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thundimadathil J. Cancer treatment using peptides: current therapies and future prospects. J. Amino Acids. 2012;2012:967347. doi: 10.1155/2012/967347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaspar D, Veiga AS, Castanho MA. From antimicrobial to anticancer peptides. A review. Front Microbiol. 2013;4:294. doi: 10.3389/fmicb.2013.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi A, et al. CancerPPD: a database of anticancer peptides and proteins. Nucl. Acids Res. 2015;43:D837–843. doi: 10.1093/nar/gku892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Dong C, Li X, Han W, Su X. Anticancer potential of bioactive peptides from animal sources (review) Oncol. Rep. 2017;38:637–651. doi: 10.3892/or.2017.5778. [DOI] [PubMed] [Google Scholar]
- 22.Shoombuatong W, Schaduangrat N, Nantasenamat C. Unraveling the bioactivity of anticancer peptides as deduced from machine learning. EXCLI J. 2018;17:734–752. doi: 10.17179/excli2018-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabernet G, et al. In silico design and optimization of selective membranolytic anticancer peptides. Sci. Rep. 2019;9:11282. doi: 10.1038/s41598-019-47568-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papo N, Shai Y. Host defense peptides as new weapons in cancer treatment. Cell Mol. Life Sci. 2005;62:784–790. doi: 10.1007/s00018-005-4560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zambrowicz A, Timmer M, Polanowski A, Lubec G, Trziszka T. Manufacturing of peptides exhibiting biological activity. Amino Acids. 2013;44:315–320. doi: 10.1007/s00726-012-1379-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haney EF, Hancock REW. Peptide design for antimicrobial and immunomodulatory applications. Biopolymers. 2013;6:572–583. doi: 10.1002/bip.22250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov. Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Pearson CS, et al. Combined bioinformatic and rational design approach to develop antimicrobial peptides against mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2016;60:2757–2764. doi: 10.1128/AAC.00940-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Natale C, De Benedictis I, De Benedictis A, Marasco D. Metal-peptide complexes as promising antibiotics to fight emerging drug resistance: new perspectives in tuberculosis. Antibiotics (Basel) 2020;9:337. doi: 10.3390/antibiotics9060337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marqus S, Pirogova E, Piva TJ. Evaluation of the use of therapeutic peptides for cancer treatment. J. Biomed. Sci. 2017;24:21. doi: 10.1186/s12929-017-0328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noga EJ, Silphaduang U. Piscidins: a novel family of peptide antibiotics from fish. Drug News Perspect. 2003;16:87–92. doi: 10.1358/dnp.2003.16.2.829325. [DOI] [PubMed] [Google Scholar]
- 32.Silphaduang U, Colorni A, Noga EJ. Evidence for widespread distribution of piscidin antimicrobial peptides in teleost fish. Dis. Aquat. Organ. 2006;72:241–252. doi: 10.3354/dao072241. [DOI] [PubMed] [Google Scholar]
- 33.Lauth X, et al. Discovery and characterization of two isoforms of moronecidin, a novel antimicrobial peptide from hybrid striped bass. J. Biol. Chem. 2002;277:5030–5039. doi: 10.1074/jbc.M109173200. [DOI] [PubMed] [Google Scholar]
- 34.Mansour SC, Pena OM, Hancock RE. Host defense peptides: front-line immunomodulators. Trends Immunol. 2014;35:443–450. doi: 10.1016/j.it.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Yeung AT, Gellatly SL, Hancock RE. Multifunctional cationic host defence peptides and their clinical applications. Cell Mol. Life Sci. 2011;68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang YB, Zhai NC, Gao G, Chen YX. Effects of net charge on biological activities of alpha-helical anticancer peptides. Chem. J. Chin. U. 2012;33:1252–1258. [Google Scholar]
- 37.Uhlig T, et al. The emergence of peptides in the pharmaceutical business: from exploration to exploitation. EuPA Open Proteom. 2014;4:58–69. doi: 10.1016/j.euprot.2014.05.003. [DOI] [Google Scholar]
- 38.Le W, Chen B, Cui Z, Liu Z, Shi D. Detection of cancer cells based on glycolytic-regulated surface electrical charges. Biophys. Rep. 2019;5:10–18. doi: 10.1007/s41048-018-0080-0. [DOI] [Google Scholar]
- 39.Lin H-J, et al. Piscidin-1, an antimicrobial peptide from fish (Hybrid Striped Bass Morone saxatilis x M. chrysops), induces apoptotic and necrotic activity in HT1080 cells. Zool. Sci. 2012;29:327–332. doi: 10.2108/zsj.29.327. [DOI] [PubMed] [Google Scholar]
- 40.Li X, et al. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013;6:19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, et al. Rapid cytotoxicity of antimicrobial peptide tempoprin-1CEa in breast cancer cells through membrane destruction and intracellular calcium mechanism. PLoS ONE. 2013;8:e60462. doi: 10.1371/journal.pone.0060462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ting CH, Chen YC, Wu CJ, Chen JY. Targeting FOSB with a cationic antimicrobial peptide, TP4, for treatment of triple-negative breast cancer. Oncotarget. 2016;7:40329–40347. doi: 10.18632/oncotarget.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patrzykat A, Douglas SE. Gone gene fishing: how to catch novel marine antimicrobials. Trends Biotechnol. 2003;21:362–369. doi: 10.1016/S0167-7799(03)00145-8. [DOI] [PubMed] [Google Scholar]
- 45.Smith VJ, Desbois AP, Dyrynda EA. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar. Drugs. 2010;8:1213–1262. doi: 10.3390/md8041213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Destoumieux-Garzon D, et al. Antimicrobial peptides in marine invertebrate health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150300. doi: 10.1098/rstb.2015.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silphaduang U, Noga EJ. Peptide antibiotics in mast cells of fish. Nature. 2001;414:268–269. doi: 10.1038/35104690. [DOI] [PubMed] [Google Scholar]
- 48.Peng KC, et al. Five Different piscidins from Nile Tilapia, Oreochromis niloticus: analysis of their expressions and biological functions. PLoS ONE. 2012;7:e50263. doi: 10.1371/journal.pone.0050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole AM, Weis P, Diamond G. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 1997;272:12008–12013. doi: 10.1074/jbc.272.18.12008. [DOI] [PubMed] [Google Scholar]
- 50.Yin Z-X, et al. Cloning, expression and antimicrobial activity of an antimicrobial peptide, epinecidin-1, from the orange-spotted grouper, Epinephelus coioides. Aquaculture. 2006;253:204–211. doi: 10.1016/j.aquaculture.2005.10.002. [DOI] [Google Scholar]
- 51.Iijima N, et al. Purification and characterization of three isoforms of chrysophsin, a novel antimicrobial peptide in the gills of the red sea bream, chrysophrys major. Eur. J. Biochem. 2003;270:675–686. doi: 10.1046/j.1432-1033.2003.03419.x. [DOI] [PubMed] [Google Scholar]
- 52.Lee SA, et al. Solution structure and cell selectivity of piscidin 1 and its analogues. Biochemistry. 2007;46:3653–3663. doi: 10.1021/bi062233u. [DOI] [PubMed] [Google Scholar]
- 53.Chekmenev EY, et al. Investigating molecular recognition and biological function at interfaces using piscidins, antimicrobial peptides from fish. Biochim. Biophys. Acta. 2006;1758:1359–1372. doi: 10.1016/j.bbamem.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 54.Chekmenev EY, et al. High-field NMR studies of molecular recognition and structure-function relationships in antimicrobial piscidins at the water-lipid bilayer interface. J. Am. Chem. Soc. 2006;128:5308–5309. doi: 10.1021/ja058385e. [DOI] [PubMed] [Google Scholar]
- 55.De Angelis AA, et al. Amphipathic antimicrobial piscidin in magnetically aligned lipid bilayers. Biophys. J. 2011;101:1086–1094. doi: 10.1016/j.bpj.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mihailescu M, et al. Structure and function in antimicrobial piscidins: histidine position, directionality of membrane insertion, and pH-dependent permeabilization. J. Am. Chem. Soc. 2019;141:9837–9853. doi: 10.1021/jacs.9b00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perrin BS, Jr, et al. High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion. J. Am. Chem. Soc. 2014;136:3491–3504. doi: 10.1021/ja411119m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Comert F, et al. The host-defense peptide piscidin P1 reorganizes lipid domains in membranes and decreases activation energies in mechanosensitive ion channels. J. Biol. Chem. 2019;294:18557–18570. doi: 10.1074/jbc.RA119.010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Libardo MDJ, et al. Nuclease activity gives an edge to host-defense peptide piscidin 3 over piscidin 1, rendering it more effective against persisters and biofilms. FEBS J. 2017;284:3662–3683. doi: 10.1111/febs.14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G. Database-guided discovery of potent peptides to combat HIV-1 or superbugs. Pharmaceuticals. 2013;6:728–758. doi: 10.3390/ph6060728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lei Z, et al. Comparative pharmacokinetics and preliminary pharmacodynamics evaluation of piscidin 1 against PRV and PEDV in rats. Front Chem. 2018;6:244. doi: 10.3389/fchem.2018.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu H, et al. Antiviral activity of piscidin 1 against pseudorabies virus both in vitro and in vivo. Virol. J. 2019;16:95. doi: 10.1186/s12985-019-1199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paredes SD, et al. Enhancing the membrane activity of piscidin 1 through peptide metallation and the presence of oxidized lipid species: implications for the unification of host defense mechanisms at lipid membranes. Biochim. Biophys. Acta Biomembr. 2020;1862:183236. doi: 10.1016/j.bbamem.2020.183236. [DOI] [PubMed] [Google Scholar]
- 64.Rai RK, De Angelis A, Greenwood A, Opella SJ, Cotten ML. Metal-ion binding to host defense peptide piscidin 3 observed in phospholipid bilayers by magic angle spinning solid-state NMR. ChemPhysChem. 2018;20:295. doi: 10.1002/cphc.201800855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu R, Rooney MT, Zhang R, Cotten ML. Coordination of redox ions within a membrane-binding peptide: a tale of aromatic rings. J. Phys. Chem. Lett. 2021;12:4392–4399. doi: 10.1021/acs.jpclett.1c00636. [DOI] [PubMed] [Google Scholar]
- 66.Harford C, Sarkar B. Amino terminal Cu(II)- and Ni(II)-binding (ATCUN) motif of proteins and peptides: metal binding, DNA cleavage, and other properties. Acc. Chem. Res. 1997;30:123–130. doi: 10.1021/ar9501535. [DOI] [Google Scholar]
- 67.Joyner JC, Cowan JA. Target-directed catalytic metallodrugs. Braz. J. Med. Biol. Res. 2013;46:465–485. doi: 10.1590/1414-431X20133086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sankararamakrishnan R, Verma S, Kumar S. ATCUN-like metal-binding motifs in proteins: identification and characterization by crystal structure and sequence analysis. Proteins. 2005;58:211–221. doi: 10.1002/prot.20265. [DOI] [PubMed] [Google Scholar]
- 69.Alexander JL, Thompson Z, Cowan JA. Antimicrobial metallopeptides. ACS Chem. Biol. 2018;13:844–853. doi: 10.1021/acschembio.7b00989. [DOI] [PubMed] [Google Scholar]
- 70.Anast JM, Margerum DW. Electron-transfer reactions of copper(III)-peptide complexes with hexacyanoferrate(II) Inorg. Chem. 1982;21:3494–3501. doi: 10.1021/ic00139a045. [DOI] [Google Scholar]
- 71.Jin Y, Lewis MA, Gokhale NH, Long EC, Cowan JA. Influence of stereochemistry and redox potentials on the single- and double-strand DNA cleavage efficiency of Cu(II) and Ni(II) Lys-Gly-His-derived ATCUN metallopeptides. J. Am. Chem. Soc. 2007;129:8353–8361. doi: 10.1021/ja0705083. [DOI] [PubMed] [Google Scholar]
- 72.Joyner JC, Cowan JA. Targeted cleavage of HIV RRE RNA by rev-coupled transition metal chelates. J. Am. Chem. Soc. 2011;133:9912–9922. doi: 10.1021/ja203057z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Libardo MD, Cervantes JL, Salazar JC, Angeles-Boza AM. Improved bioactivity of antimicrobial peptides by addition of amino-terminal copper and nickel (ATCUN) binding motifs. ChemMedChem. 2014;9:1892–1901. doi: 10.1002/cmdc.201402033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Libardo MD, Nagella S, Lugo A, Pierce S, Angeles-Boza AM. Copper-binding tripeptide motif increases potency of the antimicrobial peptide anoplin via reactive oxygen species generation. Biochem. Biophys. Res. Commun. 2015;456:446–451. doi: 10.1016/j.bbrc.2014.11.104. [DOI] [PubMed] [Google Scholar]
- 75.Libardo MD, Paul TJ, Prabhakar R, Angeles-Boza AM. Hybrid peptide ATCUN-sh-buforin: influence of the ATCUN charge and stereochemistry on antimicrobial activity. Biochimie. 2015;113:143–155. doi: 10.1016/j.biochi.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 76.Kimoto E, Tanaka H, Gyotoku J, Morishige F, Pauling L. Enhancement of antitumor activity of ascorbate against Ehrlich ascites tumor cells by the copper:glycylglycylhistidine complex. Cancer Res. 1983;43:824–828. [PubMed] [Google Scholar]
- 77.Agbale CM, et al. Antimicrobial and antibiofilm activities of helical antimicrobial peptide sequences incorporating metal-binding motifs. Biochemistry. 2019;58:3802–3812. doi: 10.1021/acs.biochem.9b00440. [DOI] [PubMed] [Google Scholar]
- 78.Cheng MH, et al. Piscidin-1 induces apoptosis via mitochondrial reactive oxygen species-regulated mitochondrial dysfunction in human osteosarcoma cells. Sci. Rep. 2020;10:5045. doi: 10.1038/s41598-020-61876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hayden RM, et al. Complementary effects of host defense peptides piscidin 1 and piscidin 3 on DNA and lipid membranes: biophysical insights into contrasting biological activities. J. Phys. Chem. B. 2015;119:15235–15246. doi: 10.1021/acs.jpcb.5b09685. [DOI] [PubMed] [Google Scholar]
- 80.Nakagami K, et al. Increased choline kinase activity and elevated phosphocholine levels in human colon cancer. Jpn. J. Cancer Res. 1999;90:419–424. doi: 10.1111/j.1349-7006.1999.tb00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int. J. Cancer. 2007;120:1721–1730. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 82.Wiener MC, White SH. Fluid bilayer structure determination by the combined use of X-ray and neutron diffraction. II. "Composition-space" refinement method. Biophys. J. 1991;59:174–185. doi: 10.1016/S0006-3495(91)82209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta Biomembr. 2000;1469:159–195. doi: 10.1016/S0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sears VF. Neutron scattering lengths and cross sections. Neutron News. 1992;3:26–37. doi: 10.1080/10448639208218770. [DOI] [Google Scholar]
- 85.Heinrich F, Losche M. Zooming in on disordered systems: neutron reflection studies of proteins associated with fluid membranes. Biochim. Biophys. Acta Biomembr. 2014;1838:2341–2349. doi: 10.1016/j.bbamem.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Worcester DL, Franks NP. Structural analysis of hydrated egg lecithin and cholesterol bilayers. II. Neutrol diffraction. J. Mol. Biol. 1976;100:359–378. doi: 10.1016/S0022-2836(76)80068-X. [DOI] [PubMed] [Google Scholar]
- 87.Wiener MC, King GI, White SH. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. I. Scaling of neutron data and the distributions of double bonds and water. Biophys. J. 1991;60:568–576. doi: 10.1016/S0006-3495(91)82086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buldt G, Gally HU, Seelig A, Seelig J, Zaccai G. Neutron diffraction studies on selectively deuterated phospholipid bilayers. Nature. 1978;271:182–184. doi: 10.1038/271182a0. [DOI] [PubMed] [Google Scholar]
- 89.Poyton MF, Sendecki AM, Cong X, Cremer PS. Cu2+ binds to phosphatidylethanolamine and increases oxidation in lipid membranes. J. Am. Chem. Soc. 2016;138:1584–1590. doi: 10.1021/jacs.5b11561. [DOI] [PubMed] [Google Scholar]
- 90.Monson CF, et al. Phosphatidylserine reversibly binds Cu2+ with extremely high affinity. J. Am. Chem. Soc. 2012;134:7773–7779. doi: 10.1021/ja212138e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Halliwell, B. Transition-metal ions and oxidative damage—a citation-classic commentary on superoxide-dependent formation of hydroxyl radicals in the presence of iron chelates by Halliwell, B. and oxygen-toxicity, oxygen radicals, transition-metals and disease by Halliwell, B. and Gutteridge, J.M.C. Curr. Contents/Life Sci. 9–9 (1992).
- 92.Esterbauer H, Striegl G, Puhl H, Rotheneder M. Continuous monitoring of in vitro oxidation of human low density lipoprotein. Free Radic. Res. Commun. 1989;6:67–75. doi: 10.3109/10715768909073429. [DOI] [PubMed] [Google Scholar]
- 93.Patel RP, Svistunenko D, Wilson MT, DarleyUsmar VM. Reduction of Cu(II) by lipid hydroperoxides: implications for the copper-dependent oxidation of low-density lipoprotein. Biochem. J. 1997;322:425–433. doi: 10.1042/bj3220425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Battino R, Rettich TR, Tominaga T. The solubility of oxygen and ozone in liquids. J. Phys. Chem. Ref. Data. 1983;12:163–178. doi: 10.1063/1.555680. [DOI] [Google Scholar]
- 95.Knight JA, Voorhees RP. Peroxidation of linolenic acid–catalysis by transition metal ions. Ann. Clin. Lab. Sci. 1990;20:347–352. [PubMed] [Google Scholar]
- 96.Choe E, Min DB. Mechanisms of antioxidants in the oxidation of foods. Compr. Rev. Food Sci. F. 2009;8:345–358. doi: 10.1111/j.1541-4337.2009.00085.x. [DOI] [Google Scholar]
- 97.Khandelia H, Mouritsen OG. Lipid gymnastics: evidence of complete acyl chain reversal in oxidized phospholipids from molecular simulations. Biophys. J. 2009;96:2734–2743. doi: 10.1016/j.bpj.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wallgren, M. et al. Impact of oxidized phospholipids on the structural and dynamic organization of phospholipid membranes: a combined DSC and solid state NMR study. Faraday Discuss.161, 499–513; discussion 563–489 (2013). [DOI] [PubMed]
- 99.Mattila JP, Sabatini K, Kinnunen PK. Oxidized phospholipids as potential molecular targets for antimicrobial peptides. Biochim. Biophys. Acta. 2008;1778:2041–2050. doi: 10.1016/j.bbamem.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 100.Oludiran A, et al. How oxygen availability affects the antimicrobial efficacy of host defense peptides: lessons learned from studying the copper-binding peptides piscidins 1 and 3. Int. J. Mol. Sci. 2019;20:5289. doi: 10.3390/ijms20215289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kumar A, et al. Piscidin-1-analogs with double L- and D-lysine residues exhibited different conformations in lipopolysaccharide but comparable anti-endotoxin activities. Sci. Rep. 2017;7:39925. doi: 10.1038/srep39925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen WF, et al. The use of the antimicrobial peptide piscidin (PCD)-1 as a novel anti-nociceptive agent. Biomaterials. 2015;53:1–11. doi: 10.1016/j.biomaterials.2015.02.069. [DOI] [PubMed] [Google Scholar]
- 103.Mulero I, Noga EJ, Meseguer J, Garcia-Ayala A, Mulero V. The antimicrobial peptides piscidins are stored in the granules of professional phagocytic granulocytes of fish and are delivered to the bacteria-containing phagosome upon phagocytosis. Dev. Comp. Immunol. 2008;32:1531–1538. doi: 10.1016/j.dci.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 104.Chen W, Cotten ML. Expression, purification, and micelle reconstitution of antimicrobial piscidin 1 and piscidin 3 for NMR studies. Protein Expr. Purif. 2014;102:63–68. doi: 10.1016/j.pep.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 105.White SH, Wimley WC. Peptides in lipid bilayers: structural and thermodynamic basis of partitioning and folding. Curr. Opin. Struct. Biol. 1994;4:79–86. doi: 10.1016/S0959-440X(94)90063-9. [DOI] [Google Scholar]
- 106.Jacobs R, White SH. The nature of the hydrophobic binding of small peptides at the bilayer interface; implications for the insertion of transbilayer helices. Biochemistry. 1989;28:3421–3437. doi: 10.1021/bi00434a042. [DOI] [PubMed] [Google Scholar]
- 107.Hristova K, et al. An amphipathic alpha-helix at a membrane interface: a structural study using a novel X-ray diffraction method. J. Mol. Biol. 1999;290:99–117. doi: 10.1006/jmbi.1999.2840. [DOI] [PubMed] [Google Scholar]
- 108.Perrin BS, Fu R, Cotten ML, Pastor RW. Simulations of membrane-disrupting peptides II: AMP piscidin 1 favors surface defects over pores. Biophys. J. 2016;111:1258–1266. doi: 10.1016/j.bpj.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Perrin BS, Jr, Sodt AJ, Cotten ML, Pastor RW. The curvature induction of surface-bound antimicrobial peptides piscidin 1 and piscidin 3 varies with lipid chain length. J. Membr. Biol. 2015;248:455–467. doi: 10.1007/s00232-014-9733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hilchie AL, et al. Pleurocidin-family cationic antimicrobial peptides are cytolytic for breast carcinoma cells and prevent growth of tumor xenografts. Breast Cancer Res. 2011;13:R102. doi: 10.1186/bcr3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mader JS, Salsman J, Conrad DM, Hoskin DW. Bovine lactoferricin selectively induces apoptosis in human leukemia and carcinoma cell lines. Mol. Cancer Ther. 2005;4:612–624. doi: 10.1158/1535-7163.MCT-04-0077. [DOI] [PubMed] [Google Scholar]
- 113.Yoo YC, et al. Bovine lactoferrin and lactoferricin, a peptide derived from bovine lactoferrin, inhibit tumor metastasis in mice. Jpn. J. Cancer Res. 1997;88:184–190. doi: 10.1111/j.1349-7006.1997.tb00364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen JY, Lin WJ, Lin TL. A fish antimicrobial peptide, tilapia hepcidin TH2-3, shows potent antitumor activity against human fibrosarcoma cells. Peptides. 2009;30:1636–1642. doi: 10.1016/j.peptides.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 115.Juang V, Lee HP, Lin AM, Lo YL. Cationic PEGylated liposomes incorporating an antimicrobial peptide tilapia hepcidin 2–3: an adjuvant of epirubicin to overcome multidrug resistance in cervical cancer cells. Int. J. Nanomed. 2016;11:6047–6064. doi: 10.2147/IJN.S117618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lo Y-L, Lee H-P, Tu W-C. The use of a liposomal formulation incorporating an antimicrobial peptide from tilapia as a new adjuvant to epirubicin in human squamous cell carcinoma and pluripotent testicular embryonic carcinoma cells. Int. J. Mol. Sci. 2015;16:22711–22734. doi: 10.3390/ijms160922711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sarode GS, et al. Use of tilapia hepcidin in oral cancer therapeutics: a proposal. J. Contemp. Dent. Pract. 2019;20(4):403–404. doi: 10.5005/jp-journals-10024-2529. [DOI] [PubMed] [Google Scholar]
- 118.Battogtokh G, Cho YY, Lee JY, Lee HS, Kang HC. Mitochondrial-targeting anticancer agent conjugates and nanocarrier systems for cancer treatment. Front Pharmacol. 2018;9:922. doi: 10.3389/fphar.2018.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Redza-Dutordoir M, Averill-Bates DA. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta. 2016;1863:2977–2992. doi: 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 120.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat. Rev. Drug Discov. 2010;9:447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 121.Gaetke LM, Chow CK. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology. 2003;189:147–163. doi: 10.1016/S0300-483X(03)00159-8. [DOI] [PubMed] [Google Scholar]
- 122.Miyamoto T, Fukino Y, Kamino S, Ueda M, Enomoto S. Enhanced stability of Cu2+–ATCUN complexes under physiologically relevant conditions by insertion of structurally bulky and hydrophobic amino acid residues into the ATCUN motif. Dalton Trans. 2016;45:9436–9445. doi: 10.1039/C6DT01387B. [DOI] [PubMed] [Google Scholar]
- 123.McGillivray DJ, et al. Molecular-scale structural and functional characterization of sparsely tethered bilayer lipid membranes. Biointerphases. 2007;2:21–33. doi: 10.1116/1.2709308. [DOI] [PubMed] [Google Scholar]
- 124.Vanderah DJ, et al. Isostructural self-assembled monolayers. 2. Methyl 1-(3-mercaptopropyl)-oligo(ethylene oxide)s. Langmuir. 2004;20:1311–1316. doi: 10.1021/la035829g. [DOI] [PubMed] [Google Scholar]
- 125.Budvytyte R, et al. Structure and properties of tethered bilayer lipid membranes with unsaturated anchor molecules. Langmuir. 2013;29:8645–8656. doi: 10.1021/la401132c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kirby BJ, et al. Phase-sensitive specular neutron reflectometry for imaging the nanometer scale composition depth profile of thin-film materials. Curr. Opin. Colloid Interface Sci. 2012;17:44–53. doi: 10.1016/j.cocis.2011.11.001. [DOI] [Google Scholar]
- 127.Dura JA, et al. AND/R: a neutron diffractometer/reflectometer for investigation of thin films and multilayers for the life sciences. Rev. Sci. Instrum. 2006;77:074301. doi: 10.1063/1.2219744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mihailescu M, et al. Acyl-chain methyl distributions of liquid-ordered and -disordered membranes. Biophys. J. 2011;100:1455–1462. doi: 10.1016/j.bpj.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.