Abstract
Pancreatic diseases such as pancreatitis, type 1 diabetes and pancreatic cancer impose substantial health-care costs and contribute to marked morbidity and mortality. Recent studies have suggested a link between gut microbiota dysbiosis and pancreatic diseases; however, the potential roles and mechanisms of action of gut microbiota in pancreatic diseases remain to be fully elucidated. In this review, we summarize the evidence that supports relationship between alterations of gut microbiota and development of pancreatic diseases, and discuss the potential molecular mechanisms of gut microbiota dysbiosis in the pathogenesis of pancreatic diseases. We also propose current strategies toward gut microbiota to advance a developing research field that has clinical potential to reduce the cost of pancreatic diseases.
Keywords: gut microbiota, pancreatic diseases, type 1 diabetes, acute pancreatitis, chronic pancreatitis, pancreatic cancer
Introduction
The gut microbiota has recently emerged as an essential mediator of human health, acting by interfering with host functions, including metabolism, digestion, and gut mucosal immune responses and integrity. The interplay among the gut microbiota, intestinal barrier and immune system maintains gut homeostasis and protects the host from pathogenic flora [1], and disruption of this homeostasis leads to an imbalance of the gut microbiota called “dysbiosis” [2]. To date, gut dysbiosis has been reported to participate in the pathogenesis of several gastrointestinal diseases, including inflammatory bowel disease [3], irritable bowel syndrome [3], and other diseases, such as obesity [4], metabolic syndrome [5, 6], Parkinson’s disease [7], and pancreatic diseases [8].
The human pancreas is crucial for maintenance of metabolism and health due to its endocrine (the vital metabolic hormone insulin) and exocrine (digestive enzymes) functions [9]. Recent studies support the existence of interactions between the gut microbiota and pancreas. On the one hand, the pancreas is connected to the gastrointestinal tract via the pancreatic duct, exocrine pancreatic function is an important host factor affecting the composition and diversity of gut microbiota [10], and pancreas-derived antimicrobial peptides can influence the gastrointestinal microbiota [11]. On the other hand, gut microbes and their derivatives can migrate into the pancreas to influence the pancreatic microenvironment. Alterations in the gut microbiota are associated with several pancreatic diseases, such as type 1 diabetes (T1D), acute pancreatitis (AP), chronic pancreatitis (CP), and pancreatic cancer (PC) [8, 12–15]. However, no direct causal relationship has been established between gut dysbiosis and pancreatic diseases. Thus, here, were view relevant animal and human studies to explore the crosstalk between the gut microbiota and pancreas and discuss the roles and potential mechanisms of dysbiosis in the pathogenesis of pancreatic diseases.
Role of the microbiota in the development of type 1 diabetes
T1D is an autoimmune disorder characterized by T-cell-mediated destruction of insulin-producing β-cells in pancreatic islets [16]. Genetic predisposition (low-risk human leukocyte antigen, balance of protective and susceptibility alleles for T1D) and environmental factors (antibiotics, hygiene, diet, and seasonality) are important elements in the development of T1D [17]. Environmental factors related to T1D development, such as diet, antibiotics and pH of drinking water, also have an impact on the gut microbiota. Changes in the gut microbiota and its metabolites (such as short-chain fatty acids (SCFAs)) may subsequently increase intestinal permeability, instigate abnormal immune cell functioning, and promote the development of proinflammatory niches, thus stimulating β-cell autoimmunity and T1D development [18–20]. Here, we summarize the current knowledge about the relationship between the microbiota and T1D and explore possible mechanisms of gut dysbiosis in T1D development based on both animal and human studies.
Animal studies
Evidence supporting the role of the gut microbiota in T1D development is largely derived from two rodent models of autoimmune diabetes, including nonobese diabetic (NOD) mice and Bio-Breeding diabetes-prone (BB-DP) rats. Both NOD mice and BB-DP rats carry the risk genes of T1D and develop spontaneous T1D, similar to humans. In both models, gut microbes are necessary for host health, and perturbations to the normal composition of commensal communities disrupt homeostasis and increase susceptibility to T1D [21, 22]. Specifically, Roesch et al. found that the abundances of bacteria of the genera Bacteroides, Eubacterium, and Ruminococcus increased, whereas those of Lactobacillus, Bryantella, Bifidobacterium, and Turicibacter decreased, in BB-DP rats compared with Bio-Breeding diabetes-resistant rats [23] (Table 1). Similarly, different microbiota compositions were also found between NOD and nonobese diabetes-resistant (NOR) mice, and NOD mice had a lower Firmicutes/Bacteroidetes ratio as well as a lower abundance of Prevotella than the NOR mice [24]. Meanwhile, a reduced abundance of SCFA-producing bacteria was found in individuals diagnosed with T1D, indicating that certain commensal bacterial populations and their metabolites contribute to protection against T1D [25–27].
Table 1.
Summaries of major gut microbiota involved in pancreatic diseases.
| Reference | Sample number | Species | Sample | Disease states vs control | Bacterial population evaluation | Increased bacterial population | Decreased bacterial population |
|---|---|---|---|---|---|---|---|
| Roesch et al. [23] | 10/10 | Rats | Feces | T1D vs normal | 16S rRNA sequencing | Bacteroides, Eubacterium, Ruminococcus | Lactobacillus, Bryantella, Bifidobacterium, Turicibacter |
| Hu et al. [134] | 39T1D/24 normal | Mouse | Feces | Pre-T1D vs normal | Gram-negative, Gram-positive | Bacteroidetes Erysipelotrichaceae | |
| Leiva-Gea et al. [37] | 15T1D/13 normal | Human | Feces | T1D vs normal | 16S rRNA sequencing | Bacteroides, Ruminococcus, Veillonella, Blautia, Streptococcus | Bifidobacterium, Roseburia, Faecalibacterium, Lachnospira |
| Murri et al. [36] | 16T1D/16 normal | Human | Feces | T1D vs normal | PCR-DGGE and RT-qPCR | Clostridium, Bacteroides, Veillonella | Lactobacillus, Bifidobacterium, Blautia coccoides/Eubacterium rectale, Prevotella |
| de Goffau et al. [39] | 18T1D/18 normal | Human | Feces | T1D vs normal | 16S rRNA sequencing | Bacteroides | Bifidobacterium adolescentis, Bifidobacterium pseudocatenulatum |
| Giongo et al. [38] | 4T1D/4 normal | Human | Feces | T1D vs normal | 16S rRNA sequencing | Bacteroides ovatus | Firmicutes |
| Davis- Richardson et al. [135] | 29T1D/47 normal | Human | Feces | T1D vs normal | 16S rRNA sequencing | Bacteroides dorei, Bacteroides vulgatus | NA |
| Huang et al. [42] | 12T1D/10 normal | Human | Feces | T1D vs normal | 16S rRNA sequencing | Bacteroidetes | Firmicutes |
| Zhu et al. [14] | 41MAP/59MSAP/30SAP/35 normal | Human | Feces | SAP vs MAP and MSAP | 16S rRNA sequencing | Escherichia-Shigella | Blautia |
| Zhang et al. [136] | 45AP/44 normal | Human | Feces | AP vs normal | 16S rRNA sequencing | Bacteroidetes, Proteobacteria | Firmicutes, Actinobacteria |
| Tan et al. [78] | 32MAP//44SAP/32 normal | Human | Feces | AP vs normal | PCR-DGGE and RT-qPCR | Enterobacteriaceae, Enterococcus | Bifidobacterium |
| Chen et al. [65] | 20 AP/20 normal | Rats | Feces | AP vs normal | 16S rRNA sequencing | Escherichia-Shigella, Phascolarctobacterium | Saccharibacteria, Tenericutes |
| Han et al. [92] | 8CP/8 normal | Mouse | Colon feces | CP vs normal | 16S rRNA sequencing | Bacteroides, Alloprevotella | Lachnospiraceae_NK4A136, Ruminiclostridium, Roseburia |
| Zhou et al. [100] | 71CP/69 normal | Human | Feces | CP vs normal | 16S rRNA sequencing | Proteobacteria | Firmicutes, Actinobacteria |
| Jandhyala et al. [104] | 30CP/10 normal | Human | Feces | CP vs normal | 16S rRNA sequencing | Firmicutes/Bacteroidetes | Faecalibacterium prausnitzii, Ruminococcus bromii |
| Gorovits et al. [137] | 96 CP | Human | Feces | CP vs normal | gas-liquid chromatography analysis | Enterobacter, Proteus, Klebsiella, Morganella | Bifidobacterium, Lactobacillus |
| Farrell et al. [115] | 27CP/28 normal | Human | Oral | CP vs normal | qPCR | Streptococcus mitis | Granulicatella adiacens |
| Michaud et al. [138] | 405PC/416 normal | Human | Plasma | PC vs normal | Antibiotics to oral bacteria | Porphyromonas gingivalis | Streptococcus mitis |
| Fan et al. [139] | 361/371 normal | Human | Oral | PC vs normal | 16S rRNA sequencing | Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans | Fusobacteria, Leptotrichia |
| Torres et al. [140] | 8PC/22 normal | Human | Oral | PC vs normal | 16S rRNA sequencing | Leptotrichia, Bacteroides, | Porphyromonas gingivalis, Neisseria elongate, Aggregatibacter actinomycetemcomitans |
| Half et al. [141] | 15PC/15 normal | Human | Feces | PC vs normal | 16S rRNA sequencing | Bacteroides Verrucomicrobia, Sutterella, Veillonella, Bacteroides, Odoribacter and Akkermansia | Firmicutes, Actinobacteria |
| Lin et al. [142] | 13PC/12 normal | Human | Oral | PC vs normal | 16S rRNA sequencing | Bacteroides | Corynebacterium |
| Farrell et al. [115] | 38PC /38 normal | Human | Oral | PC vs normal | 16S microarray | Granulicatella adiacens | Neisseria elongate, Streptococcus mitis |
| Ren et al. [129] | 85PC/57 normal | Human | Feces | PC vs normal | 16S rRNA sequencing | Bacteroidetes | Firmicutes, Proteobacteria |
T1D type 1 diabetes, AP acute pancreatitis, MAP mild acute pancreatitis, MSAP moderately severe AP, SAP severe acute pancreatitis, CP chronic pancreatitis, PC pancreatic cancer, qPCR real-time quantitative PCR, SAP severe acute pancreatitis.
Indeed, interventions such as antibiotic treatment, probiotic supplementation, dietary supplementation, or FMT to perturb the gut microbiota composition were proven to delay or accelerate T1D progression [28–33]. It was reported that alterations in the gut bacterial profile caused by vancomycin markedly increased the incidence of T1D [28, 29]. Dietary factors such as gluten and fiber may change the incidence of T1D by altering the composition of the gut microbiota, and low-ester pectin, a novel dietary fiber, could decrease the diabetes incidence in NOD mice by selectively enriching specific microbial species that produce SCFAs [30]. In addition, early oral administration of the probiotic Clostridium butyricum (CB0313.1) in NOD mice induced the onset of diabetes by selectively modulating the structure of the intestinal microbiota, including increasing the Firmicutes/Bacteroidetes ratio and changing the abundances of Clostridium and butyrate-producing bacteria [31]. Transfer of Akkermansia muciniphila was also effective in delaying the onset of diabetes in NOD mice, which is mediated by increased expression of the antimicrobial peptide Reg3γ, lowered serum endotoxin levels, reduced islet toll-like receptor (TLR) expression, increased forkhead box P3 positive (Foxp3+) regulatory T-cell (Treg) abundance in islets, and increased interleukin-10 and transforming growth factor beta expression in pancreatic lymph nodes [32]. Similarly, a protective role against T1D was found for segmented filamentous bacteria in NOD mice [33], and segmented filamentous bacteria-positive females could more strongly induce a robust T-helper cell type 17 (Th17) population in the small-intestinal lamina propria than males. Therefore, it is clear that the gut microbiota and its metabolites exert profound effects on T1D development and can be used as biomarkers for T1D prediction, and therapeutic strategies targeting the gut microbiota might be effective in controlling diabetes [34, 35].
Human studies
The role of the intestinal microbiota as an important regulator of autoimmune diabetes in animal models is well established, and consistent alterations in the gut microbiota have also been observed in humans with T1D [36, 37] (Table 1). A study by Giongo et al. analyzed bacteria in fecal samples of infants and young children and discovered that children who developed T1D had lower proportions of bacteria from the Firmicutes phylum and higher proportions of bacteria from the Bacteroidetes phylum than age-matched healthy controls at 4–8 months of age [38]. De Goffau et al. compared the intestinal microbiota composition between children with at least two diabetes-associated autoantibodies and autoantibody-negative children matched for age, sex, and early feeding history. They found that a decreased abundance of Bifidobacterium and an increased abundance of the Bacteroides genus in children with β-cell autoimmunity and a low abundance of lactate-producing and butyrate-producing species were associated with β-cell autoimmunity [39]. In another study, De Goffau et al. analyzed the gut microbiota of children aged 1–5 years with new-onset T1D vs age-matched healthy controls and demonstrated that the healthy children had a more balanced microbiota than the diabetic children and that butyrate-producing species appear to hold a pivotal position in T1D prevention [25]. Consistent with this, a case-control study by Murri et al. also demonstrated that gut microbiota dysbiosis was a stimulant of T1D, and the Firmicutes/Bacteroidetes ratio was negatively correlated with the plasma glucose level, while the quantity of Clostridium was positively correlated with the plasma glucose level. In addition, diabetic children had decreased abundances of Bifidobacterium, Lactobacillus, the Blautia coccoides/Eubacterium rectale group and Prevotella compared with healthy children, and the quantity of bacteria that was essential for maintenance of gut integrity was significantly lower in children with diabetes than in healthy children [36]. Moreover, Mejía-León et al. compared the structure of the fecal microbiota in mestizo children with T1D at onset, T1D after 2 years of treatment, and healthy controls [40]. They found that the newly diagnosed T1D cases had high levels of the genus Bacteroides, whereas the gut microbiota in healthy controls was dominated by Prevotella, and children with T1D treated for ≥2 years had similar levels of Bacteroides and Prevotella compared to those in the control group [40]. Based on the above findings from human studies, we can conclude that gut microbiota dysbiosis is strongly associated with β cell autoimmunity or T1D development; however, whether microbial alteration is involved in disease causation or is a consequence of selection by the host remains unclear.
To reveal the causal relationship, several cohort studies have been conducted and identified that changes in gut microbiota composition occurred before T1D development [37, 38]. For instance, Endesfelder et al. investigated the fecal microbiomes of 22 children who developed anti-islet cell autoantibodies and 22 matched control children who remained islet autoantibody negative from ages 3 to 36 months. During the first year of life, the microbiome changed markedly and was affected by breastfeeding, food introduction, and birth delivery mode, and there were no differences between anti-islet cell autoantibody positive and negative children in terms of bacterial diversity, microbial composition, or single-genus abundances. However, at ages 0.5 and 2 years in the children who developed anti-islet cell autoantibodies, substantial alterations in microbial interaction networks were observed [41]. Similarly, Huang et al. investigated relationships between the composition of the gut microbiome and T1D progression in Han Chinese subjects between the ages of 12 and 33, and they observed that Han Chinese T1D patients had an increased Bacteroidetes/Firmicutes ratio and a positive correlation of Bacteroides abundance with the presence of anti-islet cell autoantibodies compared to healthy subjects [42]. In addition to cohort studies, intervention studies have also been performed to certify a causal contribution of the microbiota to the T1D pathology. Recently, a single-center, randomized, double-blind, placebo-controlled pilot study conducted in children with T1D for at least 1 year showed that consumption of prebiotics could alter the gut microbiota composition and decrease intestinal permeability, leading to improved beta cell function [43]. In addition, Uusitalo et al. reported that early supplementation with probiotics during the first 4 post natal weeks reduced the risk of beta cell autoimmunity in infants genetically susceptible to T1D compared to those with no supplementation [44]. Nevertheless, such randomized controlled clinical trials are still rare, and individual bacterial species that are associated with T1D have not been identified. Overall, these findings in human studies have strongly demonstrated that the gut microbiota plays a pivotal role in the pathogenesis of anti-islet cell autoimmunity and T1D development. Altered microbial interactions in early life result in aberrant microbial development in later life and thus influence T1D development [13, 45].
Mechanisms by which the gut microbiota affects T1D development
The above animal and human studies suggested that compositional changes in the gut microbiota are involved in the pathophysiology of T1D, and gut dysbiosis-mediated immunological deregulation and gut leakage are possible pathogenic mechanisms (Fig. 1). The gut microbiota is considered the largest organ of the immune system owing to its ability to constantly interact with immunological cells [46]. On the one hand, gut dysbiosis causes the disassembly of tight junctions, thereby disrupting the integrity of the intestinal barrier and allowing unregulated passage of environmental antigens such as the microbiota and their products [47]. The translocated microbial antigens could be taken up by antigen-presenting cells, including macrophages and dendritic cells (DCs), and then processed and presented to autoreactive T cells, resulting in the conversion of Th0 to Th1 and Th17 cells and in the destruction of islet beta cells [20]. In addition, the maturation of the Tregs expressing the transcription factor Foxp3 is crucial for immune homeostasis, and the reduced frequency or function of Foxp3+ Tregs in NOD mice stimulates the onset of T1D [41, 48]. Moreover, several studies have pointed out that not only the gut microbiota but also its metabolites, especially SCFAs play pivotal roles in promoting mucosal immune homeostasis and host health via their direct immune-modulatory effects on immune cells. Among metabolites, butyrate provides energy for colonic epithelial cells and enhances the abundance and function of splenic and colonic Foxp3+ Tregs via histone modification [49]. More interestingly, we previously showed that gut microbiota-derived SCFAs could promote cathelicidin-related antimicrobial peptide expression in pancreatic cells, which protects against autoimmune diabetes in NOD mice [11]. Similarly, Miani et al. demonstrated that gut microbiota-derived molecules, namely, aryl hydrocarbon receptor ligands and butyrate, promote interleukin-22 secretion by pancreatic innate lymphoid cells, which induce the expression of mouse β-defensin 14 in endocrine cells and subsequently prevent autoimmune diabetes in NOD mice [50].
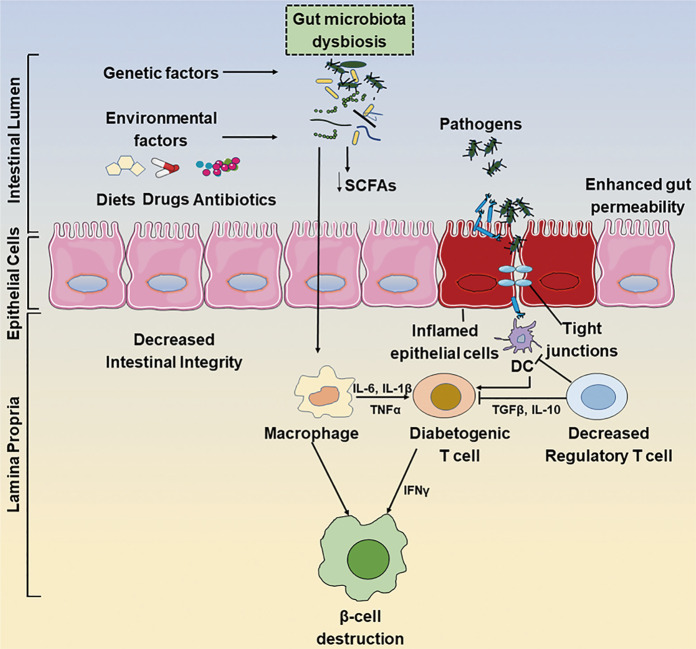
Fig. 1. Interactions between gut microbiota dysbiosis and the development of type 1 diabetes.
Gut microbiota dysbiosis causes the disassembly of tight junctions, thereby disrupting the integrity of the intestinal barrier and allowing unregulated passage of environmental pathogens such as microbes and their products. The translocated pathogens could be taken up by antigen-presenting cells, including macrophages and dendritic cells, which can process and present antigens to autoreactive T cells and subsequently promote the destruction of pancreatic beta cells in genetically predisposed individuals. DC dendritic cells, SCFAs short-chain fatty acids.
On the other hand, gut microbe-derived pathogen-associated molecular patterns can be recognized by pathogen recognition receptors such as TLRs and subsequently initiate the innate immune response to address T1D. The first attempt to investigate the innate immune pathway associated with microbial exposure in T1D was conducted in myeloid differentiation primary response protein (MyD88)-deficient NOD mice. MyD88 is an adaptor protein of multiple TLR family receptors that can recognize microbial stimuli and contribute to downstream signaling pathways of TLRs [51]. SPF NOD mice lacking MyD88 were completely protected from T1D, and the protective effect was derived from the beneficial microbial composition [52]. Conversely, MyD88-deficient mice had an increased risk of developing T1D under germ-free conditions, while the incidence of diabetes was attenuated in these mice under exposure to a defined microbial mixture, which further supports the notion that the microbial community interacts closely with the host innate immune system [52]. In addition, Peng et al. found that the transfer of the gut microbiota from MyD88-deficient NOD mice to wild-type NOD mice delayed the onset of diabetes, while the destruction of the gut microbiota by broad-spectrum antibiotics increased the incidence of T1D [53]. Moreover, TLR2 and TLR4 are used to modulate T1D development through the “balanced signal” hypothesis, in which microbes provide prediabetic or tolerizing signaling to promote or inhibit autoimmune T1D through TLR2 and TLR4, respectively [54, 55]. Collectively, the above findings indicate that the commensal microbiota is important for the prevention of T1D, and the different compositions of the intestinal microbiota could affect the mucosal innate immune system and modify the T1D pathology.
Role of the microbiota in the development of acute pancreatitis
In addition to T1D, AP is another common pancreatic disease and is caused by premature intra-acinar activation of trypsinogen and other proteolytic enzymes, resulting in pancreatic acinar injury and an inflammatory response. AP is characterized by the following two criteria: acute abdominal pain and increased circulation of pancreatic enzymes (amylase, lipase) due to pancreatic acinar cell death [56]. The common causes of AP are biliary obstruction by gallstones (40%), alcohol misuse (30%), and hypertriglyceridemia (2%–5%) [57]. Systemic injury manifested in the form of organ failure is the result of severe AP, and severe AP has high morbidity and mortality in up to 20% of cases, which contributes to substantial hospitalization and health-care costs for many people worldwide [58–61]. Disruptions of the epithelial barrier and increased gut permeability have been frequently observed in AP pathology, including alterations in tight junction proteins [62, 63]. For instance, Sonika et al. reported reduced expression of claudin-4 in duodenal biopsies of AP patients [63]. The gut microbiota and antimicrobial peptides have also been recognized as key components of the intestinal barrier, and they interact with each other to maintain gut homeostasis and barrier function [64–67]. Once the enteric microenvironment is altered, some intestinal bacteria (Staphylococcus, Enterococcus, Escherichia coli and Klebsiella) migrate into the pancreas and worsen the local inflammatory condition, resulting in bacterial translocation and overgrowth and mucosal immune dysfunction [68–70]. Identification of the underlying role of the altered gut microbiota in AP could lead to novel therapeutic strategies that would improve the clinical outcome. Therefore, several animal and human studies have been used to interpret the correlation between the gut microbiota and AP development.
Animal studies
Bacterial infections, especially those caused by pathogenic bacteria, are commonly associated with AP. However, whether intestinal microbiota dysbiosis is involved in the progression of AP remains largely unknown. To reveal the role of the gut microbiota in AP, Zhu et al. compared the microbial structure between AP mice and healthy controls. AP mice had a higher level of Escherichia-Shigella, Enterococcus and an unclassified member of Enterococcaceae and a lower level of Blautia than healthy mice. In addition, pancreatic injury was alleviated in antibiotic-treated mice and germ-free mice after AP induction. While fecal microbiota transplantation (FMT) into gut microbiota-depleted mice could exacerbate the disease, these mice exhibit more serious morphological damage, such as necrosis, inflammatory infiltrate and pancreatic edema, compared to mice without FMT. Thus, it was concluded that gut dysbiosis could worsen the severity of AP in mouse models [14]. In addition, Chen et al. found increased numbers of Escherichia-Shigella and Phascolarctobacterium and decreased numbers of Candidatus_Saccharimonas, Prevotellaceae_UCG-001, Lachnospiraceae_UCG-001, Ruminiclostridium_5, and Ruminococcaceae_UCG-008 in AP rats. Simultaneously, they found that lysozyme and α-defensin5 mRNA expression levels (Paneth cell antimicrobial peptide) decreased significantly in the AP group, and the abundance of Escherichia-Shigella was correlated inversely with the decrease in lysozyme levels [65]. Concomitant with gut dysbiosis, gut barrier dysfunction occurred, as indicated by higher plasma diamine oxidase and D-lactate levels in AP mice [65]. Based on the fact that the intestinal microbiota and antimicrobial peptides participate in the protection of the intestinal barrier, it was concluded that gut dysbiosis and decreased antimicrobial peptide levels destroyed the intestinal barrier during acute necrotizing pancreatitis [65]. In another study, Zheng et al. investigated the effect of another Escherichia commensal strain, E. coli MG1655, on AP in a rat model, and E. coli MG1655-monocolonized rats presented more severe injury in the pancreas and intestinal barrier dysfunction than gut microbiota-depleted rats [71]. The above two studies indicated that nonpathogenic commensals could also exhibit adverse effects in the context of AP. More interestingly, enteric viruses, as a part of the gut microbiota, also play a crucial role in experimental AP. We previously showed that the depletion of enteric viruses by an oral antiviral cocktail (AVC) could alleviate experimental AP. Meanwhile, AVC treatment suppressed innate immune cell infiltration and TLR9 expression and signaling and modulated inflammatory responses, thereby protecting mice from experimental AP by inhibiting TLR9 signaling [72]. From the above findings, altered gut microbiota composition is closely associated with AP development, and gut dysbiosis and subsequently impaired antimicrobial peptide production in Paneth cells worsen AP.
To date, the exact predominant bacteria in AP pathogenesis unknown, but previous studies (listed in Table 1) have suggested that animals with AP have greater abundances of the phyla Proteobacteria and Bacteroidetes and lower abundances of the phylum Firmicutes than healthy controls. Considering the role of gut dysbiosis in AP development, some potential therapeutic strategies (such as enteral nutrition, probiotics, and symbiotics) targeting the gut microbiota have emerged for the treatment of AP in animals. In the study of van Minnen et al., multispecies probiotics (Lactobacillus acidophilus W70, Lactobacillus casei W56, Lactobacillus salivarius W24, and Lactococcus lactis W58, Bifidobacterium bifidum W23 and Bifidobacterium infantis W52; each probiotic at 5 × 109 CFU/mL) were used to explore whether the modulation of the intestinal flora could reduce bacterial translocation or improve outcome in a rat model of AP. The results showed that modification of the gut microbiota with multispecies probiotics contributed to reduced bacterial translocation and mortality in the course of AP [73]. The beneficial effects of probiotics on AP can be attributed to their capability to enhance pancreatic glutathione biosynthesis and reduce oxidative stress [74]. In addition, Akyol et al. evaluated the effects of probiotics alone (Saccharomyces boulardii, 25 mg/d) or combined with two antibiotics (ciprofloxacin and meropenem) on experimental AP, and they found that probiotic treatment alone could reduce bacterial translocation, and probiotic–antibiotic combination therapy was shown to improve histopathologic scores and oxidative parameters in experimental AP [75]. Interestingly, a study found that pretreatment but not treatment with multispecies probiotics alleviated intestinal barrier dysfunction in a murine model of AP [76]. Furthermore, enteral nutrition was proven to be an effective way to attenuate AP, and the protection is attributed to specific immunomodulatory nutrients, such as glutamine, arginine, and n-3 fatty acids, which help to maintain mucosal health and gut function as well as stabilize the gut microbiota [77].
Human studies
Based on animal studies of AP, a large number of clinical studies have been used to explore the relationship between gut dysbiosis and AP development in humans. Zhu et al. collected fecal samples from 165 adult participants, including 41 with mild AP, 59 with moderately severe AP, 30 with severe AP, and 35 healthy controls. They found that the composition of the gut microbiota between AP patients and healthy subjects was significantly different, and gut dysbiosis was closely correlated with systematic inflammation and gut barrier dysfunction. In addition, the microbial composition changed further with the worsening of AP. In severe AP cases, the abundances of beneficial bacteria such as Blautia decreased compared with those in other groups. It was also observed that bacterial invasion of epithelial cells in AP is highly correlated with the abundances of Escherichia-Shigella and Enterococcus, potential pathogenic bacteria that are enriched in AP. This study indicated that some members of the gut microbiota could form niche-specific relationships and that gut microbiota dysbiosis is associated with gut barrier impairment and subsequent AP severity, suggesting its role as a potential therapeutic target [14]. Similarly, Tan et al. conducted a clinical study of 108 participants, including 44 with severe AP, 32 with mild AP, and 32 healthy volunteers, with ages ranging between 25 and 65 years. Fecal sample sequencing suggested that Enterobacteriaceae and Enterococcus populations were more abundant in all patients with AP than in healthy subjects, while the abundance of the beneficial bacterium Bifidobacterium was lower in all AP patients than in healthy participants, which further indicates that gut dysbiosis worsens the severity of AP in humans [78].
Recently, modulation of the gut microbiota was considered as a potential therapeutic approach for AP control. In a study by Roberts et al., early enteral nutrition was beneficial in severe AP patients, and its use was linked to improved glycemic control, reduced infectious complications, and reduced multiorgan failure and mortality [79]. These results can be attributed to the role of nutrient levels in the maintenance of the gut flora. In addition, a few studies have examined probiotic prophylaxis in patients with AP, and conflicting results have been observed. Oláh et al. first conducted a human study of probiotics (Lactobacillus plantarum 299) for the treatment of pancreatitis, and they found that L. plantarum 299 had no adverse effects and was effective in reducing pancreatic sepsis and the number of surgical interventions [80]. In a subsequent study by Oláh et al., four species of lactic acid bacteria (LAB)—Lactobacillus paracasei subsp. paracasei 19, L. plantarum 2362, Pediococcus pentosaceus 5–33:3, and Leuconostoc mesenteroides 32–77:1—together with four prebiotic fibers were administered to 31 severe AP patients, and the above symbiotic therapy significantly decreased the cumulative incidence of systemic inflammatory response and multiorgan failure and increased the recovery rate of patients [81]. However, in a study by Besselink et al., the “Probiotics in Pancreatitis Trial” indicated that a multispecies probiotic mixture (L. acidophilus, L. casei, L. salivarius, L. lactis, B. bifidum, and Bifidobacterium lactis) did not reduce the risk of infectious complications and was associated with an increased risk of mortality [82], which prevented many researchers from further exploring the therapeutic use of probiotics in patients with pancreatitis. In another retrospective study, no positive or negative impact of probiotic treatment was demonstrated in patients with predicted severe AP without initial organ failure [83]. This controversial effect of probiotics on clinical AP across studies is mainly attributed to the different compositions, dosages, and treatments of probiotic therapy, since only a few of the thousands of bacteria that have been investigated as potential probiotics have probiotic and anti-inflammatory properties. Therefore, new specific probiotics or new cocktails of probiotics are needed, and extensive preclinical studies are essential for application in clinical practice.
Mechanisms by which the gut microbiota affects AP development
The development of animal model studies and human studies helps provide a full understanding of the immune mechanisms by which the gut microbiota affects AP development. As shown in Fig. 2, gut dysbiosis and the resultant suppression of antimicrobial peptide production lead to gut barrier impairment and pathogenic microbiota translocation through the intestinal epithelium. Subsequently, microbial components such as lipopolysaccharide can activate the host innate immune system via pattern recognition receptors such as TLRs. Some studies reported that the lack of TLR4 could ameliorate the severity of AP in mice and protect against damage, while TLR2 does not participate in the pathogenesis of AP in mice [84, 85]. In addition, we previously suggested that the depletion of enteral viruses could modulate experimental AP by suppressing TLR9 signaling. It was observed that AP mice treated with a TLR9 agonist had aggravated AP-related symptoms and decreased chemokine production. Conversely, TLR9 antagonists exert protective effects against AP [72]. Furthermore, damage-associated molecular pattern molecules (DAMPs) released from injured tissue can serve as TLR ligands and are important mediators of the pathogenesis of AP. Extracellular high mobility group box 1 (HMGB1), a kind of DAMP molecule, can activate TLR9 and help immune activation and sensing of necrotic cells [86, 87], and increased HMGB1 production is positively associated with severe AP [88, 89].
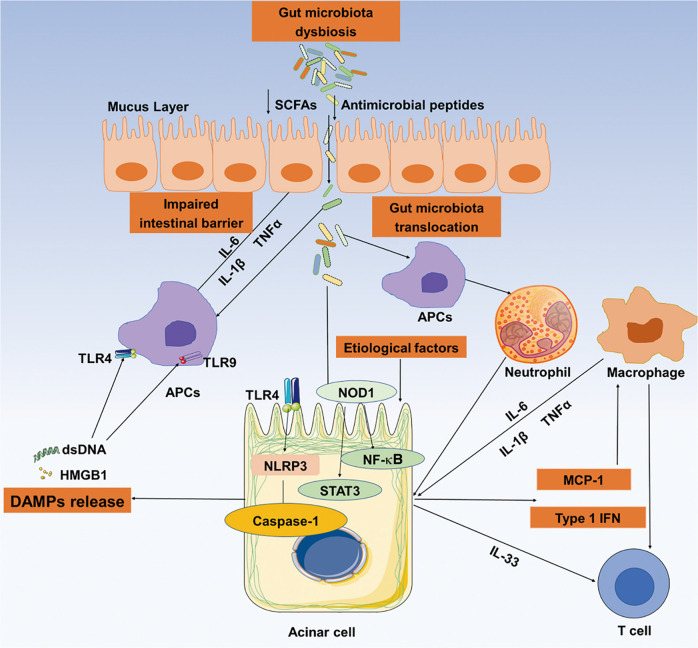
Fig. 2. Interactions between intestinal microbiota dysbiosis and the development of acute pancreatitis.
Gut microbiota dysbiosis and the resultant suppressed antimicrobial peptide production lead to gut barrier impairment and pathogenic microbiota translocation through the intestinal epithelium. The translocated gut microbiota can activate the host innate immune system via TLR-mediated signaling. Following gut dysbiosis, neutrophil infiltration, and macrophages are recruited into the pancreas to promote the progression of acute pancreatitis (AP). Furthermore, damage-associated molecular pattern molecules (DAMPs) released from damaged acinar cells can serve as TLR ligands and are important mediators of the pathogenesis of AP. AP acute pancreatitis, APCs antigen-presenting cells, DC dendritic cells, DAMPs damage-associated molecular pattern molecules, SCFAs short-chain fatty acids, TLRs Toll-like receptors.
According to Watanabe et al., the activation of innate immune responses by translocated commensal organisms is necessary to trigger inflammation during AP, and the activation of nucleotide-binding oligomerization domain 1 in acinar cells is an essential component of the innate immune response, resulting in the activation/production of nuclear factor-κB (NF-κB) and type I interferon [90]. Following gut dysbiosis-mediated activation of innate immune signaling pathways and associated tissue injury, neutrophil infiltration and macrophage recruitment into the pancreas accelerate the progression of AP. In addition, a member of the IL-1 superfamily of cytokines, IL-33, released during cell injury, was shown to be protective against AP by well-defined wound healing and reparative roles of activated macrophages. However, there is limited evidence supporting the role of the gut microbiota in modulating these immune cells and cytokines. Therefore, exploring the role of the gut microbiota in the innate immune system and adaptive immune system will help us to better understand the potential effects of the gut microbiota in AP pathology.
Role of the microbiota in the development of chronic pancreatitis
Long-term and recurrent AP could progress to CP. CP is characterized by a long-term or recurrent process of inflammation with concurrent sequelae of an acute episode, which is often accompanied by pancreatic exocrine insufficiency, eventually affecting the gut microenvironment and microbiota [91]. Recent evidence indicates that the intestinal microbiota impacts the course of CP, and these findings deserve further exploration in humans.
Animal studies
The microbial influence on CP is being increasingly recognized, supported by emerging evidence in experimental models. The study by Han et al. demonstrated that there were significant alterations in the gut microbiota in CP mice compared to control groups, as revealed by decreased abundances of Lachnospiraceae_NK4A136, Ruminiclostridium, and Roseburia and increased abundances of Bacteroides and Alloprevotella, indicating that gut dysbiosis is closely associated with CP development [92]. Likewise, Leppkes et al. found that when specific pathogen-free (SPF) mice were pretreated with the feces of CP mice by oral gavage before IL-17A expression was induced, they developed severe CP, whereas untreated SPF mice did not. It has been demonstrated that the gut microbiota is a necessary factor in IL-17A-induced CP [93]. In addition, CP could affect the exocrine function of the pancreas, and pancreatic exocrine insufficiency was proven to be the most important host factor involved in shaping the human intestinal microbiome. For example, Nishiyama et al. found that supplementation with pancreatic digestive enzymes induced the colonization of beneficial bacteria, including A. muciniphila and Lactobacillus reuteri and inhibited proinflammatory bacteria, thereby contributing to gut barrier enhancement and attenuation of CP [94]. These findings suggest the potential of FMT and probiotics to treat CP.
Similar to AP, enteral nutrition via a jejunal tube is replacing parenteral nutrition but requires repeated trials [79, 95]. Moreover, antibiotic use in CP mice is also under debate [96, 97]. However, prebiotics such as natural polysaccharides are efficient in the attenuation of CP symptoms and modulation of the gut microbiota. For instance, Hu et al. observed that Inonotus obliquus polysaccharide could regulate gut microbiota composition and diversity in mice with CP. The microbial diversity of I. obliquus polysaccharide-treated groups was lower than that of the controls. In addition, I. obliquus polysaccharide treatment increased the proportion of Bacteroidetes and decreased that of Firmicutes [98]. Similarly, Ganoderma lucidum polysaccharide treatment could also alter the composition and diversity of the intestinal microbiota by decreasing the relative abundance of Bacteroidetes and increasing that of Firmicutes at the phylum level. In addition, supplementation with G. lucidum polysaccharides increased the relative abundances of beneficial bacteria such as Lactobacillus, Roseburia and Lachnospiraceae at the genus level [99]. These results revealed that the potential mechanism of action of prebiotics on CP might be intestinal microbiota dependent. However, whether probiotics are capable of decreasing the risk of CP in mice has not been thoroughly studied.
Human studies
Given that gut dysbiosis is closely associated with CP, the investigation of potential microbes involved in CP has proven to be beneficial for the identification of high-risk patients. A study conducted by Zhou et al. showed that the composition and diversity of the gut microbiota were changed in the CP group compared to the healthy group, and the CP group showed higher Proteobacteria abundances and lower Firmicutes and Actinobacteria abundances than the healthy group (Table 1). This alteration was closely associated with fecal elastase 1 activity, an index of pancreatic exocrine insufficiency [100]. In addition, exocrine pancreatic function is the most important host factor involved in shaping the human intestinal microbiome, as suggested by Frost et al. They found a significant correlation between pancreatic elastase levels and changes in microbial diversity. Meanwhile, an increase in Prevotella and a decrease in Bacteroides were found in CP patients in a population-based study [10]. In addition, small-intestinal bacterial overgrowth in patients with CP is supportive evidence for the involvement of gut microbial dysbiosis in CP, which often causes chronic intestinal symptoms, including abdominal pain, bloating, and malabsorption [101, 102]. In turn, gut dysbiosis leads to reduced pancreatic synthesis of antimicrobial peptides, exacerbating small-intestinal bacterial overgrowth and pancreatic exocrine insufficiency, thus playing a critical role in the CP pathology. In a meta-analysis conducted by Capurso et al., the combined prevalence of bacterial overgrowth in patients with CP was 36%, accounting for one-third of CP patients [101]. The above findings were also proven by Kurdi et al. [102]. In a subsequent study, the proportions of Bacteroides, Streptococcus, and Clostridium species were higher in patients with CP. The authors speculated that gut dysbiosis may further reflect malabsorption and/or decreased levels of pancreatic enzymes, thus affecting CP development [103]. Jandhayala et al. examined the taxonomic and functional alterations in the intestinal microbiota in 16 patients with CP, 14 patients with CP combined with diabetes and 10 healthy controls. They observed an increase in the Firmicutes/Bacteroidetes ratio in all CP patients and a reduction in the abundances of Faecalibacterium prausnitzii and Ruminococcus bromii in patients with CP with and without diabetes compared to the controls. The abundance of F. prausnitzii was negatively correlated with the plasma endotoxin level and glycemic status, and plasma endotoxin levels were positively correlated with blood glucose levels and negatively correlated with plasma insulin levels, indicating that gut microbial dysbiosis was associated with the metabolic alterations of CP [104]. These data indicate that particular changes in intestinal flora composition are associated with CP, gut dysbiosis may act as a diagnostic biomarker for CP, and future approaches involving therapeutic modulation of the intestinal microflora may be effective for CP treatment. However, the literature related to probiotic application in CP patients is still limited, and this aspect needs to be further explored.
Mechanisms by which the gut microbiota affects CP development
As illustrated in Fig. 3, long-term inflammation, acinar cell atrophy, and invariable pancreatic stellate cell activation associated with pancreatic fibrosis are notable features in the pathological process of CP [105, 106]. In CP development, T cells and macrophages are the two predominant types of immune cells [107, 108]. Acinar cells are injured or apoptotic in chronic inflammation due to the release of TNFα in macrophages, resulting in IL-33 production, which could act via the suppression of the tumorigenicity 2 (ST2) receptor on T cells to induce fibrogenic mediators such as IL-13, the profibrogenic mediator acting on M2 macrophages to induce the production of TGF-β1 [90]. Furthermore, gut dysbiosis also affects mucosal immune cell phenotypes, but the molecular mechanisms underlying the microbiota–immune interactions in the pathogenesis of CP are still unclear [109]. Therefore, elucidation of the crosstalk between the gut microbiota and mucosal immune system will provide us with a mechanistic understanding of CP development. With a deeper understanding of their interaction, precise manipulations of the intestinal flora may be possible and will lead to innovative new approaches for CP treatment.
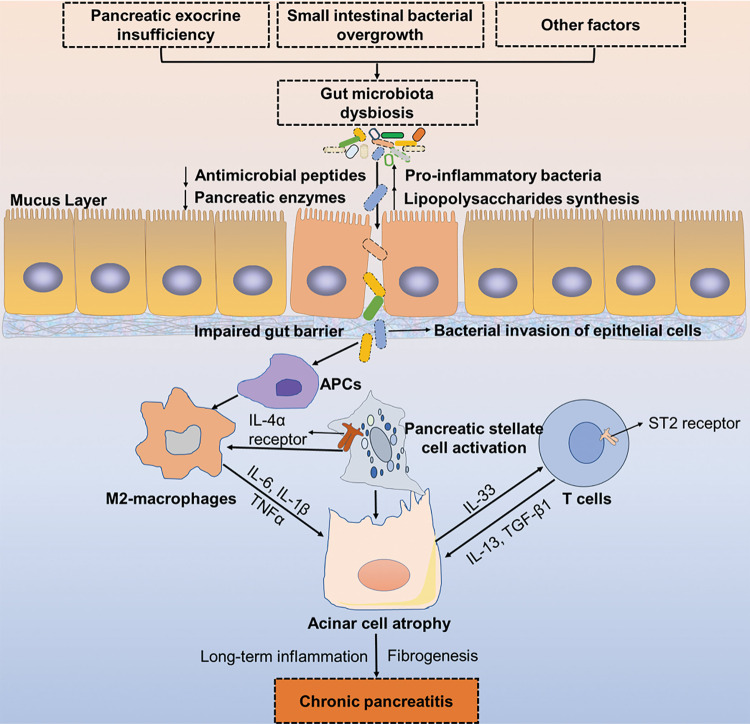
Fig. 3. Proposed relationship between gut microbiota dysbiosis and the development of chronic pancreatitis.
In the context of CP, pancreatic exocrine insufficiency, gut bacterial overgrowth, and other environmental factors account for gut dysbiosis and suppressed antimicrobial peptide production, resulting in an impaired gut barrier and leaky gut. Thereafter, proinflammatory bacteria and toxins translocate through the leaky gut to the pancreas and recognize antigen-presenting cells (APCs) to activate M2 macrophages. Acinar cells are injured or apoptotic due to the release of TNFα, IL-6, and IL-1β in macrophages, resulting in IL-33 production. The proinflammatory cytokine IL-33 acts on T cells via the ST2 receptor to induce fibrogenic mediators such as IL-13 and TGF-β1, which further aggravates acinar cell atrophy and pancreatic inflammation, resulting in chronic pancreatitis. APCs antigen-presenting cells, CP chronic pancreatitis, TNFα tumor necrosis factor α, TGF-β1 transforming growth factor-β1, ST2 suppression of tumorigenicity 2.
Gut microbiota and pancreatic cancer development
CP is a high-risk factor for PC development, and PC is a highly lethal disease, leading to the death of 93% of patients within 5 years of diagnosis [110]. Genetic factors, pancreatitis, smoking, and excess body weight are risk factors for PC [111–114]. In addition, H. pylori infection is another considerable risk factor for PC. Fungi and the oral microbiota may also play a role in pancreatic carcinogenesis [115–117]. Early detection of PC would provide the optimal opportunity to improve the survival rate of patients, but to date, there are no well-recognized screening tools or biomarkers for PC at the population level. Recently, an increasing number studies have demonstrated that the gut microbiota might influence PC susceptibility and tumor progression and can eventually influence therapeutic efficacy by promoting inflammation, activating the immune response, and perpetuating cancer-associated inflammation [116, 118–120]. Therefore, we discuss the role of the gut microbiota in PC in studies conducted in animals and humans in the following section, hoping to reveal microbiota-targeted interventions with therapeutic potential for PC.
Animal studies
The gut microbiota might play an important role in PC development. A study conducted by Pushalkar et al. suggested that ablation of the microbiome with antibiotics protected against PC, whereas transfer of the fecal samples from PC-bearing hosts to wild-type mice reversed the tumor protection [121]. Bifidobacterium pseudolongum (B. pseudolongum) occupies a large portion of the gut and tumor and finally accelerates oncogenesis in a TLR-dependent manner, and cell-free extracts from B. pseudolongum can upregulate tolerogenic cytokines such as IL-10 [121]. Moreover, antibiotic treatment in KrasG12D/+/PTENlox/+/Pdx1-Cre mice (a genetically engineered mouse model of pancreatic adenocarcinoma) showed a decreased proportion of poorly differentiated tumors compared with nontreated mice, as reported by Thomas et al. [122]. A study conducted by Sethi et al. showed that gut microbiota depletion significantly reduced tumor burden in models of PC, colon cancer and melanoma, and the protective effects of the gut microbiota on PC were related to a significant increase in interferon gamma-producing T cells and decrease in IL-17A and IL-10-producing T cells [120]. Collectively, these data strongly suggest that gut dysbiosis promotes pancreatic oncogenesis in mouse models, and certain commensal microbes may play a protective role in the tumor microenvironment by modulating immune systems. Furthermore, gut dysbiosis has been proven to be correlated with inflammation, which could further aggravate gut dysbiosis and increase the vulnerability to pathogens [123]. Many studies have demonstrated that the gut microbiota and its metabolic components lead to the susceptibility of PC via multiple pathways. Mendez et al. tested the role of the gut microbiome and its metabolites in the early detection of PC. Their results showed that Proteobacteria and Firmicutes were dominant in the early stages of PC development, and microbial metabolites such as polyamine were significantly elevated in the serum of KPC mice (a genetically engineered PC murine model). Moreover, there was a strong correlation between microbial alterations and the release of metabolites. Therefore, they concluded that microbial dysbiosis and polyamine metabolism could be predictive markers for the early detection of PC [124]. Thus, combining metabolites and microbiome analyses could help to elucidate interactions between the gut microbiota, metabolism, and the host, and clarifying how gut dysbiosis impacts host response and inflammation will be critical to obtaining an accurate picture of the role of the microbiome in PC development.
Human studies
Accumulating evidence from animal models shows that certain microbes and gut dysbiosis can potentiate PC tumor development by releasing tumor-related metabolites and activating immune responses. Thus, the gut microbiota may also participate in the pathogenesis of PC patients (see Table 1) [115, 125]. Riquelme et al. analyzed the tumor microbiome composition in PC patients with short-term survival and long-term survival. They found that the higher tumor microbial diversity in patients exhibiting long-term survival and an intra-tumoral microbiome signature (Pseudoxanthomonas-Streptomyces-Saccharopolyspora-Bacillus clausii) can be used to predict long-term survival in patients with PC [126]. Meanwhile, the authors found that the tumor microbiome can be modulated and tumor growth as well as tumor immune infiltration can be affected by humans-to-mice FMT experiments [126]. In addition, an altered oral microbiota has also been reported in relation to PC (Table 1). In study by Farrell et al., the oral microbial compositions of 28 patients with PC and 28 healthy controls were investigated, and they found that the taxa Firmicutes, Proteobacteria, Actinobacteria, Cytophaga, Fusobacterium, and Bacteroides were the predominant microbes in PC patients, and Neisseria elongata and Streptococcus mitis, as valid biomarkers for managing PC progression, were significantly decreased in PC patients compared with healthy controls [115]. Similarly, Pseudomonas aeruginosa, a unique ligand for taste receptor 2 member 38 (T2R38, expressed in both oral cells and PC cells), has been proven to be involved in cancer invasion and metastasis [127]. In addition, Fusobacterium species are regarded as independent negative prognostic biomarkers of PC due to their low abundance in PC tissues [128]. Furthermore, another study showed that PC patients had decreased gut microbial diversity compared with healthy controls, characterized by an increase in certain pathogens and LPS-producing bacteria and a decrease in probiotics and SCFA-producing bacteria, indicating that the unique gut microbial profile can be regarded as a biomarker for PC diagnosis [129]. These data provide evidence for the potential role of the gut microbiota and its metabolites in influencing PC susceptibility, while the related mechanisms of gut dysbiosis in the carcinogenesis of PC have still not been deciphered and require more attention.
Mechanisms by which the gut microbiota affects pancreatic cancer development
During PC, the composition of the gut microbiota changes, and the gut barrier is impaired. As a result, certain gut microbes could translocate to the pancreas and colonize the pancreas to induce a suppressive microenvironment that facilitates PC progression (Fig. 4). Specifically, microbiome ablation improves tumor immune surveillance and responses to programmed cell death protein 1 blockade, which contributes to a reduction in myeloid-derived suppressor cells (MDSCs), induces macrophage polarization to M2-like tumor-associated macrophages, and increases CD4+ T-helper 1 cell and CD8+ T-cell activation [55, 121, 130]. Gut microbes interact with the innate immune system via pattern recognition receptors in PC, since TLR-deficient mice (including TLR4, TLR7, TLR9) exhibit slower progression of PC [131–133], and then activate pro-tumorigenic signaling pathways such as NF-κB, signal transducer and activator of transcription 3 (STAT3) and mitogen-activated protein kinase (MAPK) signaling pathways, finally promoting the progression of PC [131, 132]. In addition, Sethi et al. found that depletion of the gut microbiota by oral antibiotics significantly reduced tumor burden in wild-type C57BL/6J mice but did not reduce tumor growth in Rag1 knockout mice (lacking mature T and B lymphocytes), suggesting that the regulatory effect of the gut microbiota on PC requires active participation of adaptive immunity [120]. Meanwhile, gut microbiome depletion by oral antibiotics significantly increased the numbers of Th1 (IFNγ+CD4+CD3+), Tc1 (IFNγ+CD8+CD3+), and IFNγ-secreting T cells (IFNγ+CD3+) and decreased the numbers of protumor IL-17a (IL17a+CD3+) and IL-10 (IL10+CD4+CD3+) secreting T cells [120]. Moreover, the bitter receptor T2R38 can be activated by N-acetyl-dodecanoyl homoserine, a quorum sensing molecule of P. aeruginosa, and then increase the expression of multidrug resistance protein 1, thus linking the gut microbiota and PC [127]. Above all, the gut microbiota plays a pivotal role in PC pathogenesis by modulating innate and adaptive immune systems. With a better understanding of the role of the gut microbiota in PC progression, manipulation of the gut bacteria could emerge as a novel immunotherapeutic strategy.
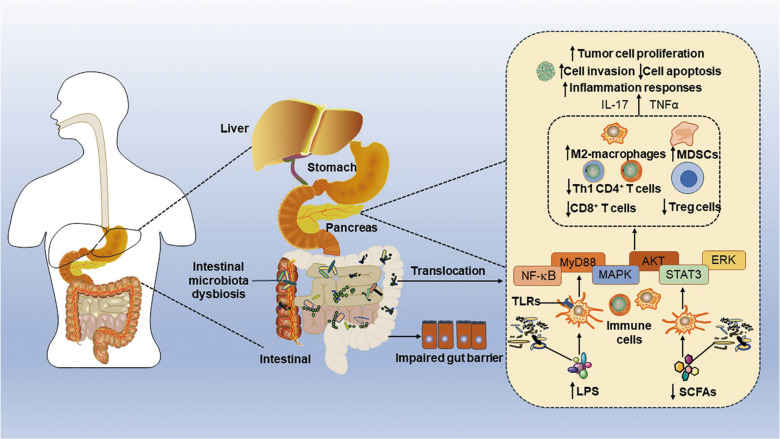
Fig. 4. Proposed relationship between gut microbiota dysbiosis and pancreatic cancer development.
Gut dysbiosis during pancreatic cancer (PC) is responsible for impairment of the gut barrier and translocation of the microbiota and microorganism-associated molecular patterns, such as lipopolysaccharide (LPS), to the pancreas. The leakage of LPS, which activates Toll-like receptors (TLRs) that are expressed on various immune cells, activates the myeloid differentiation primary response protein (MyD88), nuclear factor-κB (NF-κB), mitogen-activated protein kinase (MAPK), and signal transducer and activator of transcription 3 (STAT3) signaling pathways, ultimately inhibiting Treg polarization of the T cell response and decreasing Th1 CD4+ T cell and CD8+ T cell recruitment, and supporting M2 macrophage polarization and myeloid-derived suppressor cell (MDSC) production. Consequently, these processes initiate the inflammatory response and tumor cell invasion and proliferation, and finally promote the progression of pancreatic cancer. LPS lipopolysaccharide, MAPK mitogen-activated protein kinase, MDSCs myeloid-derived suppressor cells, MyD88 myeloid differentiation primary response protein, NF-κB nuclear factor-κB, PC pancreatic cancer, SCFAs short-chain fatty acids, STAT3 signal transducer and activator of transcription 3, TLRs Toll-like receptors.
Conclusions
In conclusion, the crosstalk between the gut microbiota and host response indicates the role of bacteria in pancreatic diseases. Although the mechanistic understanding of this relationship is still limited, it is clear that this field of research is moving forward and that novel therapeutic interventions based on bacteria-related functions could be generated in the near future.
Acknowledgements
The work was supported by funds from the National Natural Science Foundation of China (Grant Nos.: 81870439, 82070666, 31900644, and 81973322), Natural Science Foundation for Distinguished Young Scholars of Jiangsu Province, Jiangsu Province Recruitment Plan for High-level, Innovative and Entrepreneurial Talents (Innovative Research Team), Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province, the Fundamental Research Funds for the Central Universities (Grant Nos.: 81870439 and JUSRP22007), National First-Class Discipline Program of Food Science and Technology (Grant No.: JUFSTR20180103), Wuxi Social Development Funds for International Science & Technology Cooperation (Grant No.: WX0303B010518180007PB), Jiangsu Province Qing Lan Project, Jiangsu Province “Six Summit Talents” Program (Grant No.: 2019-YY-038) and Wuxi Taihu talent plan.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Li-long Pan, Bin-bin Li, Xiao-hua Pan
References
- 1.Baumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pagliari D, Piccirillo CA, Larbi A, Cianci R. The interactions between innate immunity and microbiota in gastrointestinal diseases. J Immunol Res. 2015;2015:898297. doi: 10.1155/2015/898297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vila AV, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10:eaap8914. doi: 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–9. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–79. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 7.Sun MF, Shen YQ. Dysbiosis of gut microbiota and microbial metabolites in Parkinson’s Disease. Ageing Res Rev. 2018;45:53–61. doi: 10.1016/j.arr.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Akshintala VS, Talukdar R, Singh VK, Goggins M. The gut microbiome in pancreatic disease. Clin Gastroenterol Hepatol. 2019;17:290–5. doi: 10.1016/j.cgh.2018.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Q, Melton DA. Pancreas regeneration. Nature. 2018;557:351–8. doi: 10.1038/s41586-018-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost F, Kacprowski T, Ruhlemann M, Bulow R, Kuhn JP, Franke A, et al. Impaired exocrine pancreatic function associates with changes in intestinal microbiota composition and diversity. Gastroenterology. 2019;156:1010–5. doi: 10.1053/j.gastro.2018.10.047. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Furio L, Mecheri R, vanderDoes AM, Lundeberg E, Saveanu L, et al. Pancreatic β-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity. 2015;43:304–17. doi: 10.1016/j.immuni.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–35. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 13.Tai N, Wong FS, Wen L. The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Rev Endocr Metab Disord. 2015;16:55–65. doi: 10.1007/s11154-015-9309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, He C, Li X, Cai Y, Hu J, Liao Y, et al. Gut microbiota dysbiosis worsens the severity of acute pancreatitis in patients and mice. J Gastroenterol. 2019;54:347–58. doi: 10.1007/s00535-018-1529-0. [DOI] [PubMed] [Google Scholar]
- 15.Wei MY, Shi S, Liang C, Meng QC, Hua J, Zhang YY, et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer. 2019;18:97. doi: 10.1186/s12943-019-1008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383:69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra SP, Wang S, Nagpal R, Miller B, Singh R, Taraphder S, et al. Probiotics and prebiotics for the amelioration of type 1 diabetes: present and future perspectives. Microorganisms. 2019;7:67. [DOI] [PMC free article] [PubMed]
- 18.Bibbo S, Dore MP, Pes GM, Delitala G, Delitala AP. Is there a role for gut microbiota in type 1 diabetes pathogenesis? Ann Med. 2017;49:11–22. doi: 10.1080/07853890.2016.1222449. [DOI] [PubMed] [Google Scholar]
- 19.Sane F, Scuotto A, Pierrat V, Kacet N, Hober D, Romond MB. Diabetes progression and alterations in gut bacterial translocation: prevention by diet supplementation with human milk in NOD mice. J Nutr Biochem. 2018;62:108–22. doi: 10.1016/j.jnutbio.2018.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Sorini C, Cosorich I, Conte ML, Giorgi LD, Facciotti F, Lucianò R, et al. Loss of gut barrier integrity triggers activation ofislet-reactive T cells and autoimmune diabetes. Proc Natl Acad Sci U S A. 2019;116:15140–9. doi: 10.1073/pnas.1814558116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullaney JA, Stephens JE, Costello ME, Fong C, Geeling BE, Gavin PG, et al. Type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome. 2018;6:35. doi: 10.1186/s40168-018-0417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alkanani AK, Hara N, Gottlieb PA, Ir D, Robertson CE, Wagner BD, et al. Alterations in intestinal microbiota correlate with susceptibility totype 1 diabetes. Diabetes. 2015;64:3510–20. doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM, et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. 2009;3:536–48. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daft JG, Ptacek T, Kumar R, Morrow C, Lorenz RG. Cross-fostering immediately after birth induces a permanent microbiota shift that is shaped by the nursing mother. Microbiome. 2015;3:17. doi: 10.1186/s40168-015-0080-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, et al. Aberrant gut microbiota composition at the onsetof type 1 diabetes in young children. Diabetologia. 2014;57:1569–77. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 26.Roop RM, Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18:552–62. doi: 10.1038/ni.3713. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Jin P, Peng J, Zhang X, Wong FS, Wen L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J Autoimmun. 2016;72:47–56. doi: 10.1016/j.jaut.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Candon S, Perez-Arroyo A, Marquet C, Valette F, Foray AP, Pelletier B, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One. 2015;10:e0125448. doi: 10.1371/journal.pone.0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu C, Pan LL, Niu W, Fang X, Liang W, Li J, et al. Modulation of gut microbiota by low methoxyl pectin attenuates type 1 diabetes in non-obese diabetic mice. Front Immunol. 2019;10:1733. doi: 10.3389/fimmu.2019.01733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia L, Shan K, Pan LL, Feng N, Lv Z, Sun Y, et al. Clostridium butyricum CGMCC0313.1 protects against autoimmune diabetes by modulating intestinal immune homeostasis and inducing pancreatic regulatory T cells. Front Immunol. 2017;8:1345. doi: 10.3389/fimmu.2017.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hänninen A, Toivonen R, Pöysti S, Belzer C, Plovier H, Ouwerkerk JP, et al. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67:1445–53. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- 33.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–53. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng P, Li Z, Zhou Z. Gut microbiome in type 1 diabetes: a comprehensive review. Diabetes Metab Res Rev. 2018;34:e3043. doi: 10.1002/dmrr.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehuen A, Diana J, Zaccone P, Cooke A. Immune cell crosstalk in type 1 diabetes. Nat Rev Immunol. 2010;10:501–13. doi: 10.1038/nri2787. [DOI] [PubMed] [Google Scholar]
- 36.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leiva-Gea I, Sanchez-Alcoholado L, Martin-Tejedor B, Castellano-Castillo D, Moreno-Indias I, Urda-Cardona A, et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case-control study. Diabetes Care. 2018;41:2385–95. doi: 10.2337/dc18-0253. [DOI] [PubMed] [Google Scholar]
- 38.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Goffau MC, Luopajärvi K, Knip M, Ilonen J, Ruohtula T, Härkönen T, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 2013;62:1238–44. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mejía-León ME, Petrosino JF, Ajami NJ, Domínguez-Bello MG, de la Barca AMC. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. 2014;4:3814. doi: 10.1038/srep03814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endesfelder D, zu Castell W, Ardissone A, Davis-Richardson AG, Achenbach P, Hagen M, et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes. 2014;63:2006–14. doi: 10.2337/db13-1676. [DOI] [PubMed] [Google Scholar]
- 42.Huang Y, Li SC, Hu J, Ruan HB, Guo HM, Zhang HH, et al. Gut microbiota profiling in Han Chinese with type 1 diabetes. Diabetes Res Clin Pract. 2018;141:256–63. doi: 10.1016/j.diabres.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 43.Ho J, Nicolucci AC, Virtanen H, Schick A, Meddings J, Reimer RA, et al. Effect of prebiotic on microbiota, intestinal permeability, and glycemic control in children with type 1 diabetes. J Clin Endocrinol Metab. 2019;104:4427–40. doi: 10.1210/jc.2019-00481. [DOI] [PubMed] [Google Scholar]
- 44.Uusitalo U, Liu X, Yang J, Aronsson CA, Hummel S, Butterworth M, et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr. 2016;170:20–8. doi: 10.1001/jamapediatrics.2015.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sofi MH, Gudi R, Karumuthil-Melethil S, Perez N, Johnson BM, Vasu C. pH of drinking water influences the composition of gut microbiome and type 1 diabetes incidence. Diabetes. 2014;63:632–44. doi: 10.2337/db13-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDermott AJ, Huffnagle GB. The microbiome and regulation of mucosal immunity. Immunology. 2014;142:24–31. doi: 10.1111/imm.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watts T, Berti I, Sapone A, Gerarduzzi T, Not T, Zielke R, et al. Role of the intestinal tight junction modulator zonulin in the pathogenesis of type I diabetes in BB diabetic-prone rats. Proc Natl Acad Sci U S A. 2005;102:2916–21. doi: 10.1073/pnas.0500178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoka A, Barna G, Somogyi A, Muzes G, Olah A, Al-Aissa Z, et al. Extension of the CD4(+)Foxp3(+)CD25(-/low) regulatory T-cell subpopulation in type 1 diabetes mellitus. Autoimmunity. 2015;48:289–97. doi: 10.3109/08916934.2014.992518. [DOI] [PubMed] [Google Scholar]
- 49.Tanca A, Palomba A, Fraumene C, Manghina V, Silverman M, Uzzau S. Clostridial butyrate biosynthesis enzymes are significantly depleted in the gut microbiota of nonobese diabetic mice. mSphere. 2018;3:e00492–18. [DOI] [PMC free article] [PubMed]
- 50.Miani M, Le Naour J, Waeckel-Enee E, Verma SC, Straube M, Emond P, et al. Gut microbiota-stimulated innate lymphoid cells support beta-defensin 14 expression in pancreatic endocrine cells, preventing autoimmune diabetes. Cell Metab. 2018;28:557–72.e6. doi: 10.1016/j.cmet.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 51.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng J, Narasimhan S, Marchesi JR, Benson A, Wong FS, Wen L. Long term effect of gut microbiota transfer on diabetes development. J Autoimmun. 2014;53:85–94. doi: 10.1016/j.jaut.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burrows MP, Volchkov P, Kobayashi KS, Chervonsky AV. Microbiota regulates type 1 diabetes through Toll-like receptors. Proc Natl Acad Sci U S A. 2015;112:9973–7. doi: 10.1073/pnas.1508740112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol. 2020;17:53–64. doi: 10.1038/s41575-019-0242-7. [DOI] [PubMed] [Google Scholar]
- 56.Frossard JL, Steer ML, Pastor CM. Acute pancreatitis. Lancet. 2008;371:143–52. doi: 10.1016/S0140-6736(08)60107-5. [DOI] [PubMed] [Google Scholar]
- 57.Adolph TE, Mayr L, Grabherr F, Schwarzler J, Tilg H. Pancreas-microbiota cross talk in health and disease. Annu Rev Nutr. 2019;39:249–66. doi: 10.1146/annurev-nutr-082018-124306. [DOI] [PubMed] [Google Scholar]
- 58.Garg PK, Singh VP. Organ failure due to systemic injury in acute pancreatitis. Gastroenterology. 2019;156:2008–23. doi: 10.1053/j.gastro.2018.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lund H, Tonnesen H, Tonnesen MH, Olsen O. Long-term recurrence and death rates after acute pancreatitis. Scand J Gastroenterol. 2006;41:234–8. doi: 10.1080/00365520510024133. [DOI] [PubMed] [Google Scholar]
- 60.Ahmed Ali U, Issa Y, Hagenaars JC, Bakker OJ, van Goor H, Nieuwenhuijs VB, et al. Risk of recurrent pancreatitis and progression to chronic pancreatitis after a first episode of acute pancreatitis. Clin Gastroenterol Hepatol. 2016;14:738–46. doi: 10.1016/j.cgh.2015.12.040. [DOI] [PubMed] [Google Scholar]
- 61.Tu J, Zhang J, Ke L, Yang Y, Yang Q, Lu G, et al. Endocrine and exocrine pancreatic insufficiency after acute pancreatitis: long-term follow-up study. BMC Gastroenterol. 2017;17:114. doi: 10.1186/s12876-017-0663-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu HY, Li WQ, Wang XY, Li JS, Yu WK. Early gut mucosal dysfunction in patients with acute pancreatitis. Pancreas. 2008;36:192–6. doi: 10.1097/MPA.0b013e31815a399f. [DOI] [PubMed] [Google Scholar]
- 63.Sonika U, Goswami P, Thakur B, Yadav R, Das P, Ahuja V, et al. Mechanism of increased intestinal permeability in acute pancreatitis: alteration in tight junction proteins. J Clin Gastroenterol. 2017;51:461–6. doi: 10.1097/MCG.0000000000000612. [DOI] [PubMed] [Google Scholar]
- 64.Ahuja M, Schwartz DM, Tandon M, Son A, Zeng M, Swaim W, et al. Orai1-mediated antimicrobial secretion from pancreatic acini shapes the gut microbiome and regulates gut innate immunity. Cell Metab. 2017;25:635–46. doi: 10.1016/j.cmet.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J, Huang C, Wang J, Zhou H, Lu Y, Lou L, et al. Dysbiosis of intestinal microbiota and decrease in paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS One. 2017;12:e0176583. doi: 10.1371/journal.pone.0176583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong W. Shaping the gut microbiome from the pancreas. Sci Signal. 2017;10:eaan3016. doi: 10.1126/scisignal.aan3016. [DOI] [PubMed] [Google Scholar]
- 68.Bu¨ chler MW GB, Mu¨ ller CA, Friess H, Seiler CA, Uh W. Acute necrotizing pancreatitis: treatment strategy according to the status of infection. Ann Surg. 2000;232:619–26. doi: 10.1097/00000658-200011000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beger HG, Bittner R, Block S, Büchler M. Bacterial contamination of pancreatic necrosis. Gastroenterology. 1986;91:433–8. doi: 10.1016/0016-5085(86)90579-2. [DOI] [PubMed] [Google Scholar]
- 70.Isenmann R, Runzi M, Kron M, Kahl S, Kraus D, Jung N, et al. Prophylactic antibiotic treatment in patients with predicted severe acute pancreatitis: a placebo-controlled, double-blind trial. Gastroenterology. 2004;126:997–1004. doi: 10.1053/j.gastro.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 71.Zheng JY, Lou LH, Fan JJ, Huang CL, Mei QX, Wu JH, et al. Commensal Escherichia coli aggravates acute necrotizing pancreatitis through targeting of intestinal epithelial cells. Appl Environ Microbiol. 2019;85:e00059–19. doi: 10.1128/AEM.00059-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J, Pan X, Yang J, Jia L, Wu C, Liu H, et al. Enteral virus depletion modulates experimental acute pancreatitis via toll-like receptor 9 signaling. Biochem Pharmacol. 2020;171:113710. doi: 10.1016/j.bcp.2019.113710. [DOI] [PubMed] [Google Scholar]
- 73.van Minnen LP, Timmerman HM, Lutgendorff F, Verheem A, Harmsen W, Konstantinov SR, et al. Modification of intestinal flora with multispecies probiotics reduces bacterial translocation and improves clinical course in a rat model of acute pancreatitis. Surgery. 2007;141:470–80. doi: 10.1016/j.surg.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 74.Lutgendorff F, Trulsson LM, van Minnen LP, Rijkers GT, Timmerman HM, Franzen LE, et al. Probiotics enhance pancreatic glutathione biosynthesis and reduce oxidative stress in experimental acute pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1111–21. doi: 10.1152/ajpgi.00603.2007. [DOI] [PubMed] [Google Scholar]
- 75.Akyol S, Mas MR, Comert B, Ateskan U, Yasar M, Aydogan H, et al. The effect of antibiotic and probiotic combination therapy on secondary pancreatic infections and oxidative stress parameters in experimental acute necrotizing pancreatitis. Pancreas. 2003;26:363–7. doi: 10.1097/00006676-200305000-00009. [DOI] [PubMed] [Google Scholar]
- 76.Rychter JW, Minnen Van, Verheem A, Timmerman HM, Rijkers GT, Schipper MEI, et al. Pretreatment but not treatment with probiotics abolishes mouse intestinal barrier dysfunction in acute pancreatitis. Surgery. 2009;145:157–67. doi: 10.1016/j.surg.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 77.Hegazi RAF, O’Keefe SJD. Nutritional immunomodulation of acute pancreatitis. Curr Gastroenterol Rep. 2007;9:99–106. doi: 10.1007/s11894-007-0904-4. [DOI] [PubMed] [Google Scholar]
- 78.Tan CC, Ling ZX, Huang Y, Cao YD, Liu Q, Cai T, et al. Dysbiosis of intestinal microbiota associated withinflammation involved in the progression of acute pancreatitis. Pancreas. 2015;44:868–75. doi: 10.1097/MPA.0000000000000355. [DOI] [PubMed] [Google Scholar]
- 79.Roberts KM, Nahikian-Nelms M, Ukleja A, Lara LF. Nutritional aspects of acute pancreatitis. Gastroenterol Clin N Am. 2018;47:77–94. doi: 10.1016/j.gtc.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 80.Oláh A, Belágyi T, Issekutz A, Gamal ME, Bengmark S. Randomized clinical trial of specific lactobacillus and fibre supplement to early enteral nutrition in patients with acute pancreatitis. Br J Surg. 2002;89:1103–7. doi: 10.1046/j.1365-2168.2002.02189.x. [DOI] [PubMed] [Google Scholar]
- 81.Oláh A, Belágyi T, Pótó L, Romics L, Jr., Bengmark S. Synbiotic control of inflammation and infection in severe acute pancreatitis: a prospective, randomized, double blind study. Hepatogastroenterology. 2007;54:590–4. [PubMed] [Google Scholar]
- 82.Besselink MGH, van Santvoort HC, Buskens E, Boermeester MA, van Goor H, Timmerman HM, et al. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet. 2008;371:651–9. doi: 10.1016/S0140-6736(08)60207-X. [DOI] [PubMed] [Google Scholar]
- 83.van Baal MC, Kohout P, Besselink MG, van Santvoort HC, Benes Z, Zazula R, et al. Probiotic treatment with Probioflora in patients with predicted severe acute pancreatitis without organ failure. Pancreatology. 2012;12:458–62. doi: 10.1016/j.pan.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 84.Awla D, Abdulla A, Regnér S, Thorlacius H. TLR4 but not TLR2 regulates inflammation and tissue damagein acute pancreatitis induced by retrograde infusionof taurocholate. Inflamm Res. 2011;60:1093–8. doi: 10.1007/s00011-011-0370-1. [DOI] [PubMed] [Google Scholar]
- 85.Sharif R, Dawra R, Wasiluk K, Phillips P, Dudeja V, Kurt-Jones E, et al. Impact of toll-like receptor 4 on the severity of acute pancreatitis and pancreatitis-associated lung injury in mice. Gut. 2009;58:813–9. doi: 10.1136/gut.2008.170423. [DOI] [PubMed] [Google Scholar]
- 86.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–96. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 87.Miyake Y, Yamasaki S. Sensing necrotic cells. Adv Exp Med Biol. 2012;738:144–52. [DOI] [PubMed]
- 88.Yasuda T, Ueda T, Takeyama Y, Shinzeki M, Sawa H, Nakajima T, et al. Significant increase of serum high-mobility group box chromosomal protein 1 levels in patients with severe acute pancreatitis. Pancreas. 2006;33:359–63. doi: 10.1097/01.mpa.0000236741.15477.8b. [DOI] [PubMed] [Google Scholar]
- 89.Luan ZG, Zhang H, Ma XC, Zhang C, Guo RX. Role of high-mobility group box 1 protein in the pathogenesis of intestinal barrier injury in rats with severe acute pancreatitis. Pancreas. 2010;39:216–23. doi: 10.1097/MPA.0b013e3181bab5c5. [DOI] [PubMed] [Google Scholar]
- 90.Watanabe T, Kudo M, Strober W. Immunopathogenesis of pancreatitis. Mucosal Immunol. 2017;10:283–98. doi: 10.1038/mi.2016.101. [DOI] [PubMed] [Google Scholar]
- 91.Ramsey ML, Conwell DL, Hart PA. Complications of chronic pancreatitis. Dig Dis Sci. 2017;62:1745–50. doi: 10.1007/s10620-017-4518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Han MM, Zhu XY, Peng YF, Lin H, Liu DC, Li L. The alterations of gut microbiota in mice with chronic pancreatitis. Ann Transl Med. 2019;7:464. doi: 10.21037/atm.2019.08.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leppkes M, Nowecki S, Wirtz SJ, Becker C, Neurath MF. 284 intestinal microbiota contributes to the pathogenesis of IL-17A induced chronic pancreatitis. Gastroenterology. 2014;146:S–68. [Google Scholar]
- 94.Nishiyama H, Nagai T, Kudo M, Okazaki Y, Azuma Y, Watanabe T, et al. Supplementation of pancreatic digestive enzymes alters the composition of intestinal microbiota in mice. Biochem Biophys Res Commun. 2018;495:273–9. doi: 10.1016/j.bbrc.2017.10.130. [DOI] [PubMed] [Google Scholar]
- 95.O’Brien SJ, Omer E. Chronic pancreatitis and nutrition therapy. Nutr Clin Pract. 2019;34:S13–26. doi: 10.1002/ncp.10379. [DOI] [PubMed] [Google Scholar]
- 96.Vlodov J, Tenner SM. Acute and chronic pancreatitis. Prim Care. 2001;28:607–28. doi: 10.1016/s0095-4543(05)70056-7. [DOI] [PubMed] [Google Scholar]
- 97.Ozturk K, Tasci I, Yasar M, Akay C, Alcigir M, Vural S, et al. Effects of rapamycin treatment on pancreatic fibrosis, cellular apoptosis and oxidative stress in experimental chronic pancreatitis model. Acta Gastroenterol Belg. 2015;78:3–7. [PubMed] [Google Scholar]
- 98.Hu Y, Teng C, Yu S, Wang X, Liang J, Bai X, et al. Inonotus obliquus polysaccharide regulates gut microbiota of chronic pancreatitis in mice. AMB Express. 2017;7:39. doi: 10.1186/s13568-017-0341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li K, Zhuo C, Teng C, Yu S, Wang X, Hu Y, et al. Effects of Ganoderma lucidum polysaccharides on chronic pancreatitis and intestinal microbiota in mice. Int J Biol Macromol. 2016;93:904–12. doi: 10.1016/j.ijbiomac.2016.09.029. [DOI] [PubMed] [Google Scholar]
- 100.Zhou CH, Meng YT, Xu JJ, Fang X, Zhao JL, Zhou W, et al. Altered diversity and composition of gut microbiota in Chinese patients with chronic pancreatitis. Pancreatology. 2020;20:20–4. doi: 10.1016/j.pan.2019.11.013. [DOI] [PubMed] [Google Scholar]
- 101.Capurso G, Signoretti M, Archibugi L, Stigliano S, Delle Fave G. Systematic review and meta-analysis: small intestinal bacterial overgrowth in chronic pancreatitis. United Eur Gastroenterol J. 2016;4:697–705. doi: 10.1177/2050640616630117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kurdi BE, Babar S, Iskandarani ME, Bataineh A, Lerch MM, Young M, et al. Factors that affect prevalence of small intestinal bacterial overgrowth in chronic pancreatitis: a systematic review, meta-analysis, and meta-regression. Clin Transl Gastroenterol. 2019;10:e00072. doi: 10.14309/ctg.0000000000000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hamada S, Masamune A, Nabeshima T, Shimosegawa T. Differences in gut microbiota profiles between autoimmune pancreatitis and chronic pancreatitis. Tohoku J Exp Med. 2018;244:113–7. doi: 10.1620/tjem.244.113. [DOI] [PubMed] [Google Scholar]
- 104.Jandhyala SM, Madhulika A, Deepika G, Rao GV, Reddy DN, Subramanyam C, et al. Altered intestinal microbiota in patients with chronic pancreatitis: implications in diabetes and metabolic abnormalities. Sci Rep. 2017;7:43640. doi: 10.1038/srep43640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Apte MV, Pirola RC, Wilson JS. Pancreatic stellate cells: a starring role in normal and diseased pancreas. Front Physiol. 2012;3:344. doi: 10.3389/fphys.2012.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Witt H, Apte MV, Keim V, Wilson JS. Chronic pancreatitis: challenges and advances in pathogenesis, genetics, diagnosis, and therapy. Gastroenterology. 2007;132:1557–73. doi: 10.1053/j.gastro.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 107.Goecke H, Forssmann U, Uguccioni M, Friess H, Conejo-Garcia JR, Zimmermann A, et al. Macrophages infiltrating the tissue in chronic pancreatitis express the chemokine receptor CCR5. Surgery. 2000;128:806–14. doi: 10.1067/msy.2000.108613. [DOI] [PubMed] [Google Scholar]
- 108.Deng XY, Wang L, Elm MS, Gabazadeh D, Diorio GJ, Eagon PK, Whitcomb DC. Chronic alcohol consumption accelerates fibrosisin response to cerulein-induced pancreatitis in rats. Am J Pathol. 2005;166:93–106. doi: 10.1016/S0002-9440(10)62235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang C, Li Q, Ren J. Microbiota-immune interaction in the pathogenesis of gut-derived infection. Front Immunol. 2019;10:1873. doi: 10.3389/fimmu.2019.01873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Howlader N, Noone A, Krapcho M, Miller D, Brest A, Yu M, et al. SEER cancer statistics review, 1975–2016. National Cancer Institute. Bethesda, MD, https://seer.cancer.gov/csr/1975_2016/, based on November 2018 SEER data submission, posted to the SEER web site, April 2019.
- 111.Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144:1252–61. doi: 10.1053/j.gastro.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Iodice S, Gandini S, Maisonneuve P, Lowenfels AB. Tobacco and the risk of pancreatic cancer: a review and meta-analysis. Langenbecks Arch Surg. 2008;393:535–45. doi: 10.1007/s00423-007-0266-2. [DOI] [PubMed] [Google Scholar]
- 113.Chari ST, LeibsonC, Rabe KG, Timmons LJ, Ransom J, de Andrade M, et al. Pancreatic cancer-associated diabetes mellitus prevalence andtemporal association with diagnosis of cancer. Gastroenterology. 2008;134:95–101. doi: 10.1053/j.gastro.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de Gonzalez AB, SweetlandS, Spencer E. A meta-analysis of obesity and the risk of pancreatic cancer. Br J Cancer. 2003;89:519–23. doi: 10.1038/sj.bjc.6601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582–8. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang C, Li J. Pathogenic microorganisms and pancreatic cancer. Gastrointest Tumors. 2015;2:41–7. doi: 10.1159/000380896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574:264–7. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Michaud DS, Izard J. Microbiota, oral microbiome, and pancreatic cancer. Cancer J. 2014;20:203–6. doi: 10.1097/PPO.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zambirinis CP, Pushalkar S, Saxena D, Miller G. Pancreatic cancer, inflammation, and microbiome. Cancer J. 2014;20:195–202. doi: 10.1097/PPO.0000000000000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sethi V, Kurtom S, Tarique M, Lavania S, Malchiodi Z, Hellmund L, et al. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology. 2018;155:33–7. doi: 10.1053/j.gastro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403–16. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thomas RM, Gharaibeh RZ, Gauthier J, Beveridge M, Pope JL, Guijarro MV, et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis. 2018;39:1068–78. doi: 10.1093/carcin/bgy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sheflin AM, Whitney AK, Weir TL. Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. 2014;16:406. doi: 10.1007/s11912-014-0406-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mendez R, Kesh K, Arora N, Di Martino L, McAllister F, Merchant N, et al. Microbial dysbiosis and polyamine metabolism as predictive markers for early detection of pancreatic cancer. Carcinogenesis. 2020;41:561–70. doi: 10.1093/carcin/bgz116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Half E, Keren N, Reshef L, Dorfman T, Lachter I, Kluger Y, et al. Fecal microbiome signatures of pancreatic cancer patients. Sci Rep. 2019;9:16801. doi: 10.1038/s41598-019-53041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178:795–806.e12. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gaida MM, Mayer C, Dapunt U, Stegmaier S, Schirmacher P, Wabnitz GH, et al. Expression of the bitter receptor T2R38 in pancreatic cancer localization in lipid droplets and activation by a bacteria-derived quorum-sensing molecule. Oncotarget. 2016;7:12623–32. doi: 10.18632/oncotarget.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mitsuhashi K, NoshoK, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–20. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ren ZG, Jiang JW, Xie HY, Li A, Lu HF, Xu SY. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget. 2017;8:95176–91. doi: 10.18632/oncotarget.18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Riquelme E, Maitra A, McAllister F. Immunotherapy for pancreatic cancer: more than just a gut feeling. Cancer Discov. 2018;8:386–8. doi: 10.1158/2159-8290.CD-18-0123. [DOI] [PubMed] [Google Scholar]
- 131.Ochi A, Nguyen AH, Bedrosian AS, Mushlin HM, Zarbakhsh S, Barilla R, et al. MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. J Exp Med. 2012;209:1671–87. doi: 10.1084/jem.20111706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ochi A, Graffeo CS, Zambirinis CP, Rehman A, Hackman M, Fallon N, et al. Toll-like receptor 7 regulates pancreatic carcinogenesis in mice and humans. J Clin Investig. 2012;122:4118–29. doi: 10.1172/JCI63606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zambirinis CP, Ochi A, Barilla R, Greco S, Deutsch M, Miller G. Induction of TRIF- or MYD88-dependent pathways perturbs cell cycle regulation in pancreatic cancer. Cell Cycle. 2013;12:1153–4. doi: 10.4161/cc.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hu Y, Peng J, Li F, Wong FS, Wen L. Evaluation of different mucosal microbiota leads to gut microbiota-based prediction of type 1 diabetes in NOD mice. Sci Rep. 2018;8:15451. doi: 10.1038/s41598-018-33571-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Davis-Richardson AG, Ardissone AN, Dias R, Simell V, Leonard MT, Kemppainen KM, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang XM, Zhang ZY, Zhang CH, Wu J, Wang YX, Zhang GX. Intestinal microbial community differs between acute pancreatitis patients and healthy volunteers. Biomed Environ Sci. 2018;31:81–6. doi: 10.3967/bes2018.010. [DOI] [PubMed] [Google Scholar]
- 137.Gorovits ES, Tokareva EV, Khlynova OV, Zhelobov VG, El’kin VD. Complex evaluation of intestine microbiocenosis condition in patients with chronic pancreatitis. Zh Mikrobiol Epidemiol Immunobiol. 2013;4:73–6. [PubMed]
- 138.Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjønneland A, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancerin a large European prospective cohort study. Gut. 2013;62:1764–70. doi: 10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67:120–7. doi: 10.1136/gutjnl-2016-312580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Torres PJ, Fletcher EM, Gibbons SM, Bouvet M, Doran KS, Kelley ST. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3:e1373. doi: 10.7717/peerj.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Half E, Keren N, Dorfman T, Reshef L, Lachter I, Kluger Y, et al. P-165 Specific changes in fecal microbiota may differentiate pancreatic cancer patients from healthy individuals. Ann Oncol. 2015;26:iv48. [Google Scholar]
- 142.Lin IH, Wu J, Cohen SM, Chen C, Bryk D, Marr M, et al. Abstract 101: Pilot study of oral microbiome and risk of pancreatic cancer. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research; 2013 Apr 6-10; Washington, DC. Philadelphia (PA): AACR; Cancer Res 2013;73(8 Suppl):Abstract nr 101.