Abstract
The vibrational, electronic and charge transfer studies on 2-bromo-6-methoxynaphthalene (2BMN) were done using DFT method with B3LYP/6-311++G(d,p) theory using GAUSSIAN 09W software. Theoretical and experimental investigations on FT-IR and FT Raman were executed on 2BMN. The calculated vibrational wavenumbers were scaled using suitable scaling factors and vibrational assignments were done to all modes of vibrations using Potential Energy Distribution (PED). Frontier Molecular Orbitals were calculated using TD-DFT method and the HOMO-LUMO energy gap was also obtained. Other electronic properties and global parameters for 2BMN were found using the HOMO-LUMO energy values. An energy gap of 4.208 eV shows the stability of the molecule. The reactive sites were predicted using Molecular Electrostatic Potential (MEP), Electron Localization Function (ELF) and Fukui calculations. Hence all electrophilic sites and nucleophilic areas of the molecule were determined. The delocalization of electron density was studied using NBO calculations. The intramolecular transitions and stability of structure were explained using in detail using the former. As the compound satisfies drug-like properties and has a softness value (indicating its less toxic nature), it may be used as a pharmaceutical product. Molecular docking studies were made and the protein-ligand binding properties were discussed. It was found out that title compound exhibits anti-cancer activities. The low binding energy predicts that the compound may be modified as a drug for treating Cancer.
Keywords: FT-IR, FT Raman, Molecular docking, NBO, ELF
FT-IR, FT Raman, Molecular docking, NBO, ELF
1. Introduction
Cancer is listed as one of the most life-threatening diseases of the present time. Several studies on prevention and cure of this are prevalent nowadays. Cancer is a disease in which abnormal growth of cells is observed that are malignant or benign [1, 2]. Advancement in technology has resulted in the detection of cancerous cells and their observation [3]. Several studies have shown that Cancer accounts for 16% of total death. In 2018, estimates showed that 18.1 million people are diagnosed with Cancer and deaths counting to 9.6 million globally [4, 5]. This calls for research in developing drugs for the treatment of this life-threatening disease. Studies on anti-inflammatory drugs have shown that taking Nonsteroidal Anti-Inflammatory Drugs (NSAIDs) has decreased cancer risk [6, 7, 8, 9]. NSAIDs can be very effective as they tend to work quickly. They generally have fewer side effects than other steroidal drugs that lower inflammation. A thorough study of literature shows that NSAIDs have a cancer-protective impact on various types of cancers such as breast, prostate, colourectal, ovarian, head and neck [10, 11, 12, 13, 14].
2-bromo-6-methoxynaphthalene is a compound that acts as an intermediate in the production of anti-inflammatory drugs like Naproxen and Nabumetone by Heck reaction [15, 16]. This compound also exhibits anti-inflammatory properties and therefore calls for further research in developing drugs. Literature shows that exhaustive spectroscopic evaluation and quantum computational observations using DFT were not carried out for 2BMN. In this study, meticulous calculations on vibrational spectroscopic data, DFT and molecular docking are performed. These results are interpreted to predict its anti-inflammatory properties and relevance in treatment of Cancer.
2. Materials and methods
2.1. Experimental details
2BMN was bought from Sigma Aldrich with a purity of 99%. Without further purifications, the study was carried out. The FT-Raman spectrum with a resolution 2cm−1 was obtained using Nd-YAG laser at wavelength 1064nm and power 100mW, in the region 4000-100cm−1 equipped in BRUCKER RFS 27 available at IIT SAIF, Chennai, India. The FT-IR spectrum of 2BMN was reported in the region 4000-400 cm−1, with a resolution of 1.0 cm−1. PERKIN ELMER FT-IR spectrophotometer at SAIF IIT-Chennai, India, which uses KBr pellet technique, was used to get the data. The UV-VIS analysis on 2BMN was done at SAS, VIT, Vellore in the range 200–800nm using DMSO as solvent.
2.2. Computational details
The quantum chemical calculations for 2BMN were done using DFT (B3LYP) with 6–311++G (d, p) as the theory level [17, 18]. Unlike other basis sets, this basis set has a more detailed description and lists the orbitals used for valance and core electrons as a separate entity. Further, the above basis set has been polarized (d, p) and diffused (++) for better approximations. The selected basis set is decent for calculating final accurate energies. Literature shows that 6-311++G (d, p) is quite effective in giving acceptable calculated geometries and energies, which can be achieved at a relatively low computational cost.
The structure was optimized using GAUSSIAN09W. The bond angles and bond length of the molecule were achieved from this optimized structure making use of CHEMCRAFT [19]. The harmonic vibrational frequencies were assigned with the help of VEDA software for this optimized structure [20]. A scaling factor of 0.967 was included to compare the experimental and theoretical vibrational wavenumber [21, 22]. TD-DFT/B3LYP theory was used in obtaining UV results. Frontier Molecular Orbital analysis was done and HOMO-LUMO energy levels were calculated. Electron density delocalization within donor and acceptor NBOs of the molecule was also estimated. This supported in evaluating the hyper-conjugation together with intramolecular interactions. Electron Localization Function (ELF) maps were obtained using MULTIWFN [23]. GAUSSSUM 2.2 was used to prepare the DOS and PDOS spectra [24]. Molecular docking simulations were done using AutoDock 4.2.6 [25] and to get the protein-ligand interaction, PyMOL 2.0 was used [26].
3. Results and discussions
3.1. Geometry
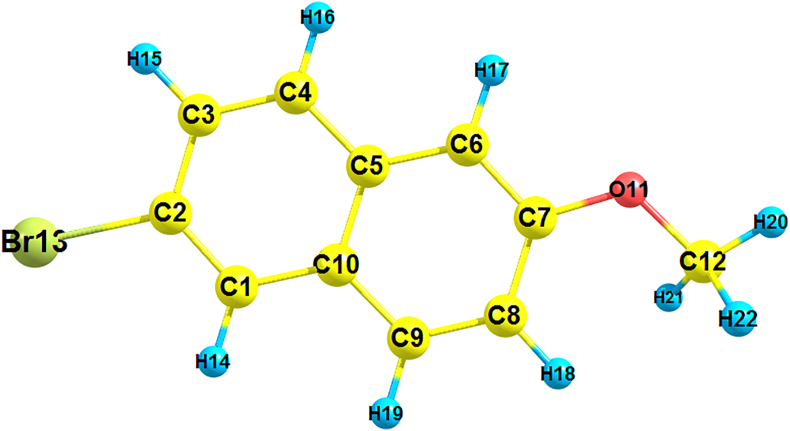
Table 1 gives the optimized structural parameters of 2BMN. The optimized structure of 2BMN following the scheme of atom numbering is shown in Figure 1. The basis set selected to do calculations yields final accurate energies and is quite effective in giving acceptable calculated geometries and energies. The geometry of molecule was optimized to minimum energy using DFT-B3LYP/6-311++G(d,p) basis set and was compared with the experimental determined by X-ray diffraction for 2-methoxynaphthalene determined by Bolte et al. [27]. Experimental and theoretical results were in concurrence.
Table 1.
Selected geometric parameters of the title compound.
| Bond Length(Å) |
Bond Angle (ᵒ) |
||||
|---|---|---|---|---|---|
| Atoms | Theoretical | Experimental∗ | Atoms | Theoretical | Experimental∗ |
| C1–C2 | 1.370 | 1.374 | C2–C1–C10 | 119.8 | 120.74 |
| C1–C10 | 1.421 | 1.424 | C1–C2–C3 | 121.6 | 120.06 |
| C2–C3 | 1.415 | 1.414 | C1–C10–C5 | 119.5 | 119.33 |
| C3–C4 | 1.372 | 1.375 | C1–C10–C9 | 122.1 | 121.97 |
| C4–C5 | 1.422 | 1.426 | C2–C3–C4 | 119.5 | 120.52 |
| C5–C6 | 1.411 | 1.419 | C3–C4–C5 | 121.3 | 120.72 |
| C5–C10 | 1.431 | 1.423 | C4–C5–C6 | 122.2 | 121.59 |
| C6–C7 | 1.381 | 1.378 | C4–C5–C10 | 118.4 | 118.62 |
| C7–C8 | 1.418 | 1.422 | C6–C5–C10 | 119.4 | 119.79 |
| C7–O11 | 1.365 | 1.427 | C5–C6–C7 | 120.8 | 119.53 |
| C8–C9 | 1.376 | 1.364 | C5–C10–C9 | 118.3 | 118.69 |
| C9–C10 | 1.415 | 1.426 | C6–C7–C8 | 120 | 120.8 |
| O11–C12 | 1.422 | 1.425 | C6–C7–O11 | 116.4 | 114.01 |
| C2–Br13 | 1.920 | - | C8–C7–O11 | 123.6 | 125.18 |
| C7–C8–C9 | 120 | 120.35 | |||
| C7–O11–C12 | 119.1 | 117.22 | |||
| C8–C9–C10 | 121.4 | 120.72 | |||
| C1–C2–Br13 | 119.9 | - | |||
| C3–C2–Br13 | 118.5 | - | |||
Taken from Ref [27].
Figure 1.
Optimized geometry of title compound using DFT B3LYP/6-311++G(d,p) basis set.
The C–O bonds C7–O11 and C12–O11 show bond distance 1.365 and 1.422Å respectively. C–Br bond has a value of 1.920Å. C–C bonds in the ring have bond distance values within the range of 1.370–1.431Å. The maximum bond length within the ring was observed at the junction of two benzene rings forming naphthalene (C5–C10) and shorter bond lengths were observed at C1–C2, C3–C4, C6–C7 and C8–C9. This can be explained by the fact that at the junction of a bicyclic compound, there is bond localization and change in electron density.
The angles between carbon atoms within the ring were found to be in the range 118.3–121.4ᵒ. Usually, in a benzene ring, the angles inside the ring are equal and 120ᵒ each, but in a naphthalene ring, due to conjugation, the bond angles inside the ring are different. Another significant angle was the angle C7–O11–C12 showing a value of 119.1ᵒ. At the same time, C6–C7–O11 and C8–C7–O11 have values 116.4ᵒ and 123.6ᵒ. Angles around Bromine atom were found to be 119.9ᵒ and 118.5ᵒ for C1–C2–Br13 and C3–C2–Br13, showing no much deviation from 120ᵒ.
3.2. Vibrational spectral analysis
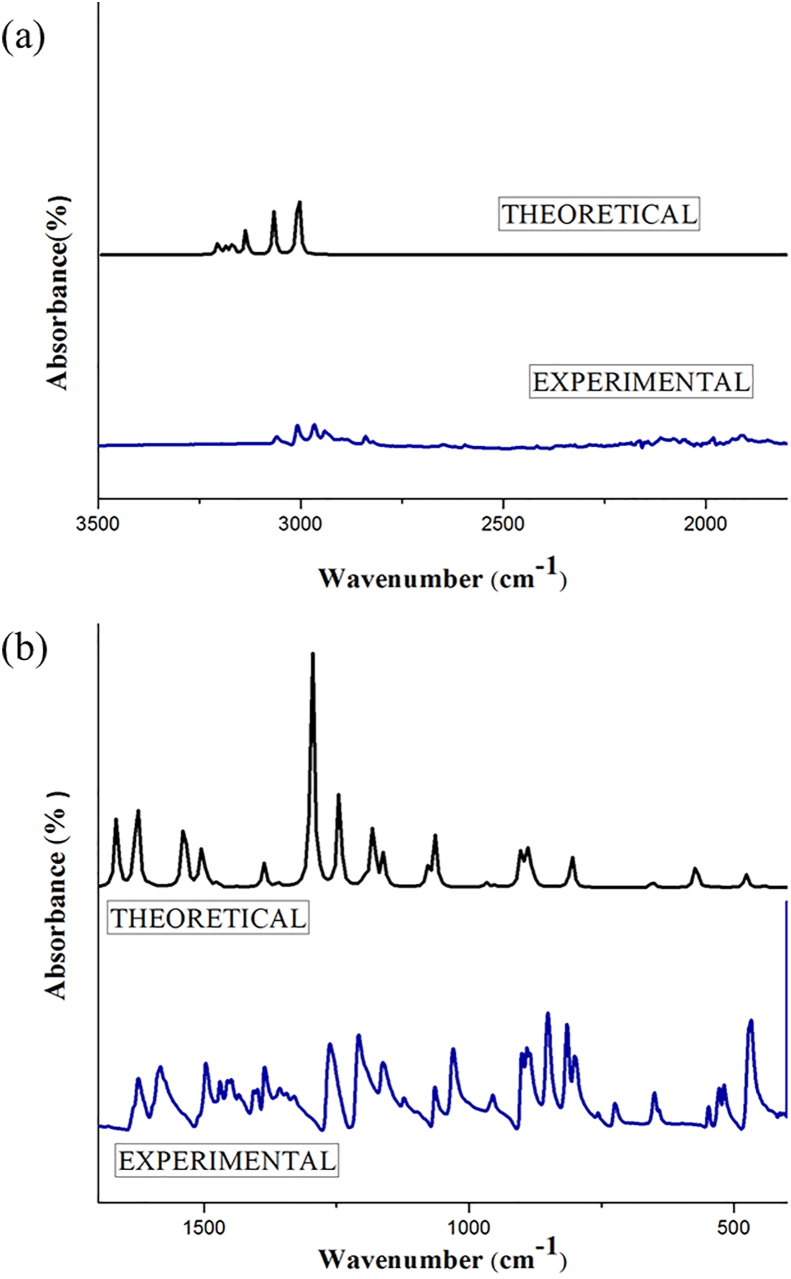
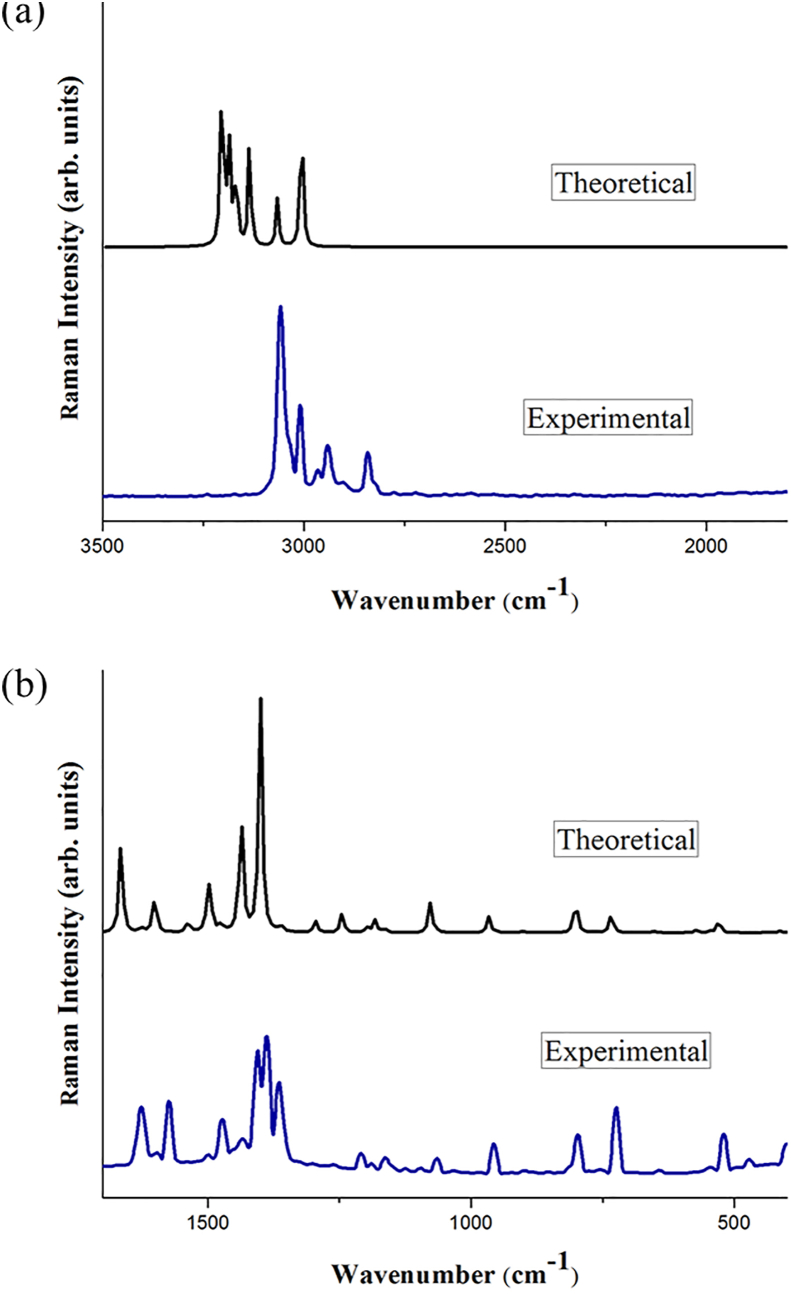
2BMN will show 60 modes of vibrations as it has 22 atoms. Wavenumbers (theoretical and experimental) of the molecule along with corresponding vibrational assignments and intensities are given in Table 2. Theoretical (scaled) and experimental vibrational spectra of 2BMN are shown in Figures 2 and 3.
Table 2.
Experimental and Theoretical (DFT B3LYP/6-311++G(d,p) basis set) vibrational spectroscopic data with vibrational assignments for title compound.
| Wavenumber(cm−1) |
IR intensity |
Raman activity |
Vibrational assignments(%PED) | |||||
|---|---|---|---|---|---|---|---|---|
| Experimental |
Theoretical |
|||||||
| IR | Raman | Unscaled | Scaled∗ | Absolute | Relative∗∗ | Absolute | Relative∗∗ | |
| 3205 | 3099 | 8 | 3 | 144 | 62 | as ʋ CH(99) | ||
| 3202 | 3096 | 4 | 1 | 131 | 56 | as ʋ CH(93) | ||
| 3188 | 3083 | 2 | 1 | 77 | 33 | as ʋ CH(92) | ||
| 3184 | 3079 | 6 | 2 | 113 | 48 | as ʋ CH(98) | ||
| 3061 (vw) | 3169 | 3064 | 6 | 2 | 61 | 26 | as ʋ CH(94) | |
| 3058 (vs) | 3167 | 3062 | 7 | 2 | 53 | 23 | as ʋ CH(94) | |
| 3009 (vw) | 3009 (ms) | 3135 | 3032 | 23 | 8 | 164 | 70 | as ʋ CH(92) |
| 2967 (vw) | 2941 (w) | 3066 | 2965 | 37 | 14 | 73 | 31 | as ʋ CH(100) |
| 2841 (w) | 3006 | 2907 | 71 | 26 | 207 | 89 | as ʋ CH(92) | |
| 1626 (m) | 1626 (w) | 1665 | 1610 | 81 | 30 | 95 | 41 | as ʋ CC(39) |
| 1581 (s) | 1574 (w) | 1627 | 1573 | 124 | 46 | 5 | 2 | as ʋ CC(47) |
| 1601 | 1548 | 2 | 1 | 42 | 18 | as ʋ CC(30) + δ CCC(17) | ||
| 1497 (s) | 1473 (w) | 1537 | 1486 | 98 | 36 | 13 | 6 | δ HCC(25) |
| 1506 | 1456 | 40 | 15 | 5 | 2 | δ HCH(58) + t HCOC(20) | ||
| 1448 (ms) | 1498 | 1449 | 8 | 3 | 38 | 16 | as ʋ CC(10) + δ HCC(16) | |
| 1433 (vw) | 1494 | 1445 | 9 | 3 | 18 | 8 | δ HCH(76) + t HCOC(16) | |
| 1405 (s) | 1475 | 1426 | 6 | 2 | 8 | 3 | δ HCH(68) | |
| 1388 (s) | 1388 (s) | 1437 | 1390 | 1 | 0 | 129 | 55 | as ʋ CC(37) |
| 1365 (ms) | 1400 | 1354 | 1 | 0 | 233 | 100 | as ʋ CC(41) + δ CCC(10) | |
| 1386 | 1340 | 27 | 10 | 0 | 0 | as ʋ CC(40) | ||
| 1361 | 1316 | 6 | 2 | 7 | 3 | as ʋ CC(15) + δ HCH(29) | ||
| 1262 (s) | 1296 | 1253 | 272 | 100 | 11 | 5 | as ʋ OC(28) + δ HCC(10) | |
| 1284 | 1242 | 8 | 3 | 0 | 0 | δ CCC(42) + δ HCC(14) | ||
| 1208 (s) | 1209 (vw) | 1245 | 1204 | 111 | 41 | 19 | 8 | as ʋ CC(17) + δ HCC(32) |
| 1162 (s) | 1163 (vw) | 1199 | 1159 | 10 | 3 | 6 | 2 | δ HCH(12) + t HCOC(42) |
| 1183 | 1144 | 55 | 20 | 12 | 5 | δ HCC(45) | ||
| 1178 | 1139 | 25 | 9 | 1 | 0 | s ʋ CC(14) + δ HCC(27) | ||
| 1166 | 1128 | 1 | 0 | 3 | 1 | δ HCH(28) + t HCOC(53) | ||
| 1161 | 1123 | 38 | 14 | 2 | 1 | as ʋ CC(13) + δ HCC(35) | ||
| 1072 (w) | 1065 (vw) | 1077 | 1041 | 22 | 8 | 29 | 12 | as ʋ CC(57) + δ HCC(14) |
| 1030 (s) | 1064 | 1029 | 59 | 22 | 1 | 0 | as ʋ OC(57) + δ HCC(12) | |
| 957 (w) | 974 | 942 | 1 | 0 | 0 | 0 | t HCCC(79) + t CCCC(11) | |
| 953 (w) | 966 | 934 | 5 | 2 | 15 | 6 | as ʋ CC(14) + δ CCC(39) | |
| 900 (vs) | 952 | 921 | 2 | 1 | 1 | 0 | t HCCC(77) + t CCCC(12) | |
| 901 | 871 | 51 | 19 | 2 | 1 | δ CCC(88) | ||
| 888 | 859 | 41 | 15 | 0 | 0 | t HCCC(60) | ||
| 851 (vs) | 880 | 851 | 13 | 5 | 0 | 0 | t HCCC(67) | |
| 814 (vs) | 797 (w) | 811 | 784 | 3 | 1 | 0 | 0 | t HCCC(66) |
| 800 (s) | 806 | 779 | 32 | 12 | 0 | 0 | t HCCC(61) | |
| 801 | 775 | 2 | 1 | 31 | 13 | as ʋ CC(17) + δ CCC(35) | ||
| 729 (vw) | 724 (w) | 746 | 721 | 0 | 0 | 0 | 0 | t CCCC(67) |
| 733 | 709 | 0 | 0 | 17 | 7 | s ʋ CC(17) + as ʋ OC(19) + δ HCC(24) | ||
| 655 (w) | 654 | 632 | 4 | 2 | 0 | 0 | t HCCC(11) + t CCCC(41) + t OCCC(16) | |
| 654 | 632 | 4 | 2 | 1 | 1 | s ʋ CC(13) + δ CCC(38) | ||
| 571 | 552 | 31 | 11 | 3 | 1 | δ CCC(20) + δ COC(23) + δ OCC(22) | ||
| 520 (w) | 546 | 528 | 0 | 0 | 1 | 0 | t HCCC(11) + γ BrCCC(13) + γ OCCC(22) | |
| 519 (w) | 529 | 512 | 1 | 0 | 11 | 5 | as ʋ CC(13) + δ HCC(32) | |
| 467 (vs) | 479 (vw) | 477 | 461 | 16 | 6 | 0 | 0 | t CCCC(81) |
| 401 (vw) | 443 | 428 | 2 | 1 | 1 | 0 | as ʋ BrC(12) + δ CCC(34) | |
| 413 | 399 | 0 | 0 | 2 | 1 | t CCCC(59) | ||
| 383 | 370 | 1 | 0 | 1 | 1 | as ʋ BrC(12) + δ CCC(26) + δ COC(26) + δ BrCC(11) | ||
| 350 | 338 | 0 | 0 | 0 | 0 | t CCCC(25) + γ BrCCC(25)+γ OCCC(14) | ||
| 282 (w) | 254 | 246 | 3 | 1 | 6 | 2 | δ OCC(32) + δ COC(20) + δ BrCC(20) | |
| 244 | 236 | 1 | 0 | 0 | 0 | t HOCC(72) | ||
| 208 (s) | 232 | 224 | 1 | 0 | 6 | 3 | s ʋ CC(10) + ʋ BrC(32) + δ CCC(10) + δ OCC(12) + δ BrCC(11) | |
| 187 | 181 | 0 | 0 | 1 | 1 | t CCCC(22) + γ BrCCC(17) +γ OCCC(14) | ||
| 175 | 169 | 1 | 1 | 1 | 0 | t CCCC(41) + γ BrCCC(12) +γ CCCC(14) | ||
| 104 (vs) | 139 | 134 | 0 | 0 | 1 | 0 | δ CCC(19) + δ OCC(17) + δ BrCC(44) | |
| 84 (vs) | 84 | 81 | 3 | 1 | 0 | 0 | t COCC(48) | |
| 49 | 47 | 1 | 0 | 0 | 0 | t CCCC(24) + t COCC(35) + γ CCCC(16) | ||
ʋ-stretching, δ-in-plane bending, γ - out-of-plane bending and t-torsion.
as-asymmetric stretching, s-symmetric stretching.
vs-very strong, s-strong, ms-medium strong, w-weak, vw-very weak.
scaling factor 0.967 for B3LYP/6-311++G(d,p) basis set.
Normalised to 100.
Figure 2.
Experimental and scaled theoretical FT-IR spectra for 2BMN for (a) higher wavenumbers (4000-2500 cm−1) and (b) lower wavenumbers (2500-50 cm−1).
Figure 3.
Experimental and scaled theoretical FT-Raman spectra for 2BMN for (a) higher wavenumbers (4000-2500 cm−1) and (b) lower wavenumbers (2500-50 cm−1).
3.2.1. CH vibrations
Asymmetric stretching of C–H bond for 2BMN was observed at higher wavenumbers. C–H stretching vibrations usually occur over 3000 cm−1 [28]. Theoretical wavenumbers 3064 cm−1 and 3062 cm−1 correspond to asymmetric stretching of C–H bond. FT IR peak at 3061 cm−1 and FT-Raman peak at 3058 cm−1 matches with theoretical C–H stretching.
For methyl group, vibrations occur at the region 3000-2925 cm−1 or 2940-2904 cm−1 [29]. FT-IR peaks at 3009 for experimental FT-Raman and 3009 cm−1 correspond to stretching of C–H in the methyl group. Peaks at 3058, 3009 and 2841 cm−1 for experimental FT-Raman spectrum falls in the methyl group stretching. Theoretical wavenumbers 3032, 2965, 2907 cm−1 falls in the desired region. These theoretical observations were found to agree with the results of experimental data.
Theoretical bending vibrations for methyl group can be assigned to wavenumbers 1486, 1449, 1445 and 1426 cm−1. As per literature, asymmetric bending modes involving CH3 vibrations were seen in the region 1485-1400 cm−1 [30]. Experimental peaks for the same were observed at 1497, 1448 and 1388 cm−1 for FT- IR spectrum. While in FT Raman spectrum, peaks at wavenumbers 1473, 1433 and 1405 cm−1 represent CH3 bending.
3.2.2. CC vibrations
For an aromatic ring, CC vibrations belong to the region 1300-1000 cm−1 while other CC vibrations are present in a range of 1650–1000 cm−1 [31, 32, 33]. For 2BMN, the theoretical peaks corresponding to CC stretching were at 1610, 1573, 1390, 1354, 1204 and 1041 cm−1. FT-IR peaks observed at 1626, 1581, 1388, 1208 and 1072 cm−1 were marked as CC bonds stretching. FT Raman peaks for compound were observed at 1626, 1574, 1388 and 1365 cm−1 respectively.
3.2.3. C–Br and C–O vibrations
C–Br stretching can be found between wavenumbers 650–395 cm−1 and for 2BMN, the same is assigned at 428, 370 and 224 cm−1 [34]. The matching experimental peaks were found at 401 cm−1 and 208 cm−1 in FT-Raman. C–O stretching peaks for 2BMN were at 1262 and 1030 cm−1 in the IR spectrum. Equivalent theoretical spectrum was at 1253 and 1029 cm−1. This is matching with an expected range of 1310–1095 cm−1 [35].
3.3. NBO
NBO studies are essential in examining the intra and intermolecular bonding within the molecule [36]. It gives a clear understanding of the interactions between the bonds. Donor and acceptor orbitals together with stabilization energy were computed using the same theory and are listed in Table 3. A considerable stabilization energy value denotes a significant interaction of donors and acceptors. This means that there is an extra donation propensity for the electron donors to acceptors and a more substantial degree of conjugation.
Table 3.
Second-order perturbation theory analysis of fock matrix in NBO basis.
| Donor | Type | ED/e | Acceptor | Type | ED/e | E(2) (a) kcal/mol |
E(j)-E(i) (b) a.u. |
F(i,j) (c) a.u. |
|---|---|---|---|---|---|---|---|---|
| C 1 - C 2 | π | 1.741 | C 3 - C 4 | π∗ | 0.2669 | 18.21 | 0.29 | 0.065 |
| C 1 - C 2 | π | C 5 - C 10 | π∗ | 0.4899 | 14.6 | 0.3 | 0.063 | |
| C 1 - C 10 | σ | 1.9664 | C 1 - C 2 | σ∗ | 0.0252 | 3.25 | 1.19 | 0.056 |
| C 1 - C 10 | σ | C 2 -Br 13 | σ∗ | 0.0299 | 4.83 | 0.79 | 0.055 | |
| C 1 - C 10 | σ | C 5 - C 10 | σ∗ | 0.0353 | 3.66 | 1.24 | 0.06 | |
| C 1 - H 14 | σ | 1.9804 | C 2 - C 3 | σ∗ | 0.2632 | 3.92 | 1.04 | 0.057 |
| C 1 - H 14 | σ | C 5 - C 10 | σ∗ | 0.0353 | 4.27 | 1.08 | 0.061 | |
| C 3 - C 4 | σ | 1.9726 | C 2 -Br 13 | σ∗ | 0.0299 | 4.92 | 0.8 | 0.056 |
| C 3 - C 4 | σ | C 5 - C 6 | σ∗ | 0.0192 | 3.1 | 1.24 | 0.055 | |
| C 3 - C 4 | π | 1.727 | C 1 - C 2 | π∗ | 0.3407 | 18.2 | 0.26 | 0.062 |
| C 3 - C 4 | π | C 5 - C 10 | π∗ | 0.4899 | 16.74 | 0.29 | 0.066 | |
| C 3 - H 15 | σ | 1.9806 | C 1 - C 2 | σ∗ | 0.0252 | 4.06 | 1.03 | 0.058 |
| C 3 - H 15 | σ | C 4 - C 5 | σ∗ | 0.0204 | 3.25 | 1.06 | 0.052 | |
| C 4 - C 5 | σ | 1.9746 | C 5 - C 6 | σ∗ | 0.0192 | 3.07 | 1.23 | 0.055 |
| C 4 - C 5 | σ | C 5 - C 10 | σ∗ | 0.0353 | 3.6 | 1.25 | 0.06 | |
| C 4 - H 16 | σ | 1.9809 | C 2 - C 3 | σ∗ | 0.0263 | 3.31 | 1.04 | 0.053 |
| C 4 - H 16 | σ | C 5 - C 10 | σ∗ | 0.0353 | 4.34 | 1.08 | 0.061 | |
| C 5 - C 6 | σ | 1.9724 | C 5 - C 10 | σ∗ | 0.0353 | 3.54 | 1.24 | 0.059 |
| C 5 - C 6 | σ | C 7 - O 11 | σ∗ | 0.0268 | 3.53 | 1.06 | 0.055 | |
| C 5 - C 10 | σ | 1.9625 | C 1 - C 10 | σ∗ | 0.0232 | 3.31 | 1.2 | 0.056 |
| C 5 - C 10 | σ | C 1 - H 14 | σ∗ | 0.0129 | 2.05 | 1.09 | 0.042 | |
| C 5 - C 10 | σ | C 4 - C 5 | σ∗ | 0.0240 | 3.41 | 1.22 | 0.058 | |
| C 5 - C 10 | σ | C 5 - C 6 | σ∗ | 0.0192 | 3.28 | 1.23 | 0.057 | |
| C 5 - C 10 | σ | C 6 - H 17 | σ∗ | 0.0117 | 2.02 | 1.1 | 0.042 | |
| C 5 - C 10 | σ | C 9 - C 10 | σ∗ | 0.0205 | 3.49 | 1.23 | 0.059 | |
| C 5 - C 10 | π | 1.5198 | C 1 - C 2 | π∗ | 0.3407 | 20 | 0.25 | 0.065 |
| C 5 - C 10 | π | C 3 - C 4 | π∗ | 0.2669 | 16.62 | 0.26 | 0.062 | |
| C 5 - C 10 | π | C 6 - C 7 | π∗ | 0.3121 | 16.25 | 0.26 | 0.061 | |
| C 5 - C 10 | π | C 8 - C 9 | π∗ | 0.2732 | 17.55 | 0.26 | 0.064 | |
| C 6 - C 7 | σ | 1.9776 | C 7 - C 8 | σ∗ | 0.0318 | 3.21 | 1.22 | 0.056 |
| C 6 - C 7 | π | 1.7067 | C 5 - C 10 | π∗ | 0.4899 | 18.03 | 0.29 | 0.068 |
| C 6 - C 7 | π | C 8 - C 9 | π∗ | 0.2732 | 17.59 | 0.27 | 0.062 | |
| C 6 - H 17 | σ | 1.9792 | C 5 - C 10 | σ∗ | 0.0353 | 4.38 | 1.08 | 0.062 |
| C 6 - H 17 | σ | C 7 - C 8 | σ∗ | 0.0318 | 4.04 | 1.04 | 0.058 | |
| C 7 - C 8 | σ | 1.9789 | C 6 - C 7 | σ∗ | 0.0257 | 3.18 | 1.24 | 0.056 |
| C 8 - C 9 | σ | 1.9781 | C 1 - C 10 | σ∗ | 0.0232 | 2.98 | 1.21 | 0.054 |
| C 8 - C 9 | σ | C 7 - C 8 | σ∗ | 0.0318 | 2.17 | 1.22 | 0.046 | |
| C 8 - C 9 | σ | C 7 - O 11 | σ∗ | 0.0268 | 3.63 | 1.06 | 0.055 | |
| C 8 - C 9 | π | 1.7395 | C 5 - C 10 | π∗ | 0.4899 | 15.71 | 0.29 | 0.064 |
| C 8 - C 9 | π | C 6 - C 7 | π∗ | 0.3121 | 19.8 | 0.28 | 0.067 | |
| C 8 - H 18 | σ | 1.9784 | C 6 - C 7 | σ∗ | 0.0257 | 4 | 1.06 | 0.058 |
| C 8 - H 18 | σ | C 9 - C 10 | σ∗ | 0.0205 | 3.27 | 1.07 | 0.053 | |
| C 9 - C 10 | σ | 1.9739 | C 1 - C 10 | σ∗ | 0.0232 | 3.35 | 1.2 | 0.057 |
| C 9 - C 10 | σ | C 5 - C 10 | σ∗ | 0.0353 | 3.66 | 1.25 | 0.06 | |
| C 9 - H 19 | σ | 1.9820 | C 5 - C 10 | σ∗ | 0.0353 | 4.37 | 1.08 | 0.062 |
| C 9 - H 19 | σ | C 7 - C 8 | σ∗ | 0.0318 | 3.3 | 1.05 | 0.053 | |
| O 11 - C 12 | σ | 1.9825 | C 12 - H 22 | σ∗ | 0.0181 | 25.23 | 4.26 | 0.293 |
| C 12 - H 20 | σ | 1.9911 | C 7 - O 11 | σ∗ | 0.0268 | 3.73 | 0.89 | 0.052 |
| C 12 - H 20 | σ | C 12 - H 22 | σ∗ | 0.0181 | 9.39 | 3.95 | 0.172 | |
| C 12 - H 21 | σ | 1.9937 | C 12 - H 22 | σ∗ | 0.0181 | 5.56 | 3.97 | 0.133 |
| C 12 - H 22 | σ | 1.9956 | C 12 - H 20 | σ∗ | 0.0112 | 4.78 | 0.97 | 0.061 |
| C 12 - H 22 | σ | C 12 - H 21 | σ∗ | 0.0180 | 5.91 | 1.02 | 0.069 | |
| C 12 - H 22 | σ | C 12 - H 22 | σ∗ | 0.0181 | 39.46 | 3.96 | 0.354 | |
| O 11 | LP (1) | 1.9521 | C 7 - C 8 | σ∗ | 0.0318 | 6.36 | 1.04 | 0.073 |
| O 11 | LP (2) | 1.8952 | C 6 - C 7 | σ∗ | 0.0257 | 4.62 | 0.89 | 0.058 |
| O 11 | LP (2) | C 6 - C 7 | π∗ | 0.3121 | 12.43 | 0.38 | 0.065 | |
| O 11 | LP (2) | C 12 - H 21 | σ∗ | 0.0180 | 4.91 | 0.85 | 0.059 | |
| Br 13 | LP (2) | 1.9725 | C 1 - C 2 | σ∗ | 0.0252 | 3.68 | 0.8 | 0.049 |
| Br 13 | LP (2) | C 2 - C 3 | σ∗ | 0.0263 | 3.87 | 0.82 | 0.05 | |
| Br 13 | LP (3) | 1.9300 | C 1 - C 2 | π∗ | 0.3407 | 11.48 | 0.3 | 0.056 |
E (2) represents the stabilization energy coupled with each donor (i) and acceptor (j) orbitals.
Ei and Ej are the diagonal elements expressed in a.u.
Fij is the second order Fock matrix.
Prominent σ to σ ∗ were observed at O11–C12 to C12–H22 and bonding orbital of C12–H22 to the anti-bonding orbital of C12–H22 of stabilization energies 25.23 kcal/mol and 39.46 kcal/mol respectively. This accounts for the maximum stabilization energy. At the same time, π to π∗ transitions with energies 20 kcal/mol and 19.8 kcal/mol were calculated for transitions from bonding orbital C5–C10 to anti-bonding orbital C1–C2 and π C8–C9 to π∗ C6–C7 respectively. Transfer of lone pair LP (1) of the O11 atom to anti-bonding orbital σ∗ (C7–C8) showed stabilization energy of 6.36 kcal/mol. While lone pair transition of LP (2) of O11 atom to π∗ (C6–C7) showed considerable stabilization energy of 12.43 kcal/mol. E(2) value for transition of lone pair from LP (3) of Br13 atom to π∗ (C1–C2) transition was 11.48 kcal/mol. This suggests an electron density transfer from the lone pair, which results in a substantial interaction within the molecule.
For the bond C5–C10 found at the junction of two benzene rings forming naphthalene ring, several transitions and electron density transfer were yielding to high stabilization energies. Transitions from σ (C5–C10) to σ∗ (C1–C10), σ∗ (C1–H14), σ∗ (C4–C5), σ∗ (C6–H17), σ∗ (C9–C10) showed stabilization energies 3.31, 2.05, 3.41, 3.28, 2.02 and 3.49 kcal/mol respectively. Very high stabilization energies (>16 kcal/mol) were observed for transitions of π (C5–C10) to π∗ (C1–C2), π∗ (C3–C4), π∗ (C6–C7), π∗ (C8–C9). This plays a major part in the stabilization of 2BMN molecule.
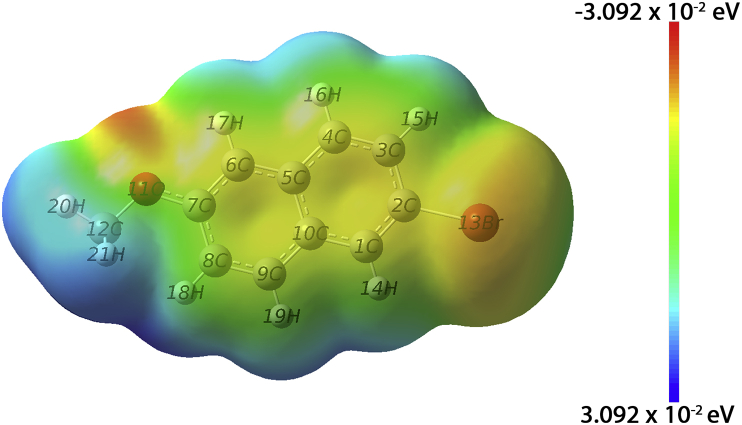
3.4. Molecular Electrostatic Potential
Molecular Electrostatic Potential maps give details of electron density. This helps in identifying sites of positive and negative electrostatic potential for electrophilic and nucleophilic attack or hydrogen bonding [37, 38, 39]. Different colours represent the difference in electrostatic potential. For high density regions, the colour codes used are red, orange or yellow. In case of low electron density, areas are marked with blue colour. Regions that are neutral within the molecule are denoted with green colour.
The optimized structure was used to map MEP which is given in Figure 4. Oxygen atoms are in red/orange regions denoting regions of electrophilic attack. Carbon atoms of ring are in yellow region while methyl group is in blue region.
Figure 4.
Molecular Electrostatic Potential Mapping for title compound.
3.5. Electron localization functions
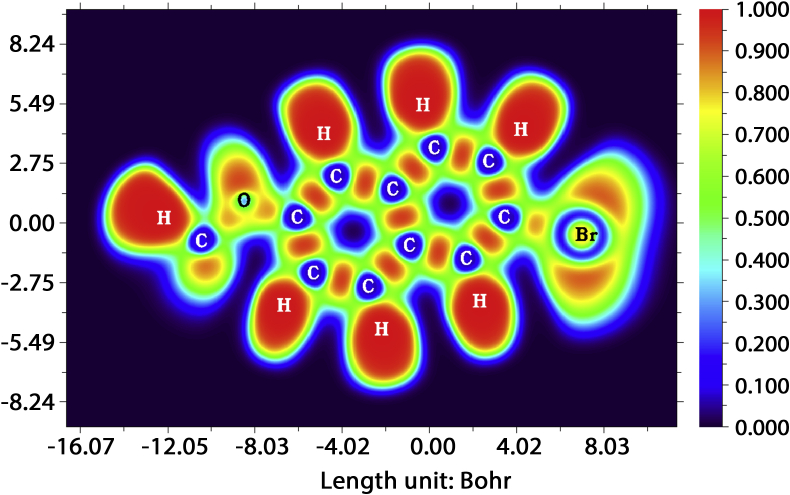
Electron Localization Function (ELF) uses the idea of finding an electron in the likely neighbourhood that has same spin like that of the reference electron. ELF analysis gives information on structure, bonding and reactivity [40]. The ELF values are colour coded. Red colour represents refers to high ELF value (0.8–1) and blue denotes the low ELF values while values that lie in the intermediate region are marked by green colour. ELF gives sensitive information on electron localization by determining additional local kinetic energy that results from Pauli repulsion [41]. The ELF representation for 2BMN is shown in Figure 5. Carbon atoms were found in blue area and Hydrogen atoms were in red area. This result compliment MEP result. Bonds between the carbon atoms of ring structure are in red areas. This was as the sp orbital of carbon overlapped with the s orbital of the hydrogen [41].
Figure 5.
Electron Localization Function mapping for 2-bromo-6-methoxynapthelene.
3.6. Fukui function calculations
To study the site of selectivity of the chemical system, local descriptors are to be introduced. Table 4 gives the values of condensed Fukui functions and dual descriptor Δf. The study is done to predict the changes in electron density on altering the number of electrons in the compound [42].
Table 4.
Fukui functions and dual descriptor values for all atoms of 2BMN.
| Atoms | Fukui functions |
|||
|---|---|---|---|---|
| fr + | fr - | fr 0 | Δf | |
| 1 C | 0.234 | -0.314 | -0.040 | 0.549 |
| 2 C | -0.108 | -0.133 | -0.121 | 0.025 |
| 3 C | -0.007 | 0.011 | 0.002 | -0.018 |
| 4 C | -0.071 | -0.049 | -0.060 | -0.022 |
| 5 C | 0.028 | 0.005 | 0.017 | 0.023 |
| 6 C | -0.122 | -0.094 | -0.108 | -0.028 |
| 7 C | 0.049 | -0.001 | 0.024 | 0.050 |
| 8 C | -0.046 | -0.020 | -0.033 | -0.026 |
| 9 C | -0.073 | -0.050 | -0.061 | -0.023 |
| 10 C | 0.051 | 0.011 | 0.031 | 0.040 |
| 11 O | -0.033 | -0.060 | -0.047 | 0.027 |
| 12 C | 0.075 | 0.018 | 0.047 | 0.057 |
| 13 Br | -0.138 | -0.184 | -0.161 | 0.045 |
| 14 H | -0.058 | -0.058 | -0.058 | 0.000 |
| 15 H | -0.063 | -0.052 | -0.057 | -0.011 |
| 16 H | -0.067 | -0.061 | -0.064 | -0.006 |
| 17 H | -0.063 | -0.067 | -0.065 | 0.004 |
| 18 H | -0.067 | -0.055 | -0.061 | -0.013 |
| 19 H | -0.068 | -0.061 | -0.065 | -0.007 |
| 20 H | -0.098 | -0.047 | -0.072 | -0.052 |
| 21 H | -0.020 | -0.020 | -0.020 | 0.000 |
| 22 H | -0.028 | -0.027 | -0.028 | 0.000 |
A clear understanding of electrophilic sites and nucleophilic sites of 2BMN was made from the dual descriptor investigation. It was seen that H14 and H17 atoms have positive dual descriptor values indicating locations of nucleophilic attack. While all other hydrogen atoms have negative dual descriptor values showing their affinity to electrophilic attack. Bromine atom has positive value implying that they favour nucleophilic attack. C2 atom, which was attached to Bromine atom and carbon atoms (C12 and C7) attached to Oxygen also has a positive value of Δf.
3.7. Frontier molecular orbital, UV ANALYSIS, DOS and PDOS spectrum
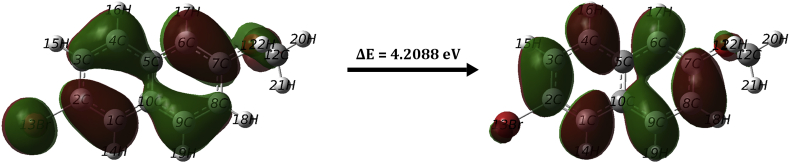
Electronic properties of a compound can be studied using Frontier Molecular Orbital (FMO) data. The energy gap (Eg) of a molecule is the difference in the energy values of HOMO and LUMO energies. HOMO and LUMO can be understood as the electron donor and electron acceptor respectively [43, 44]. This energy gap explains the reactivity as well as stability of molecule [45]. HOMO-LUMO values were used to determine the global reactivity parameters. These are listed in Table 5. Figure 6 shows the HOMO and LUMO energy levels and energy gap calculated for 2BMN. Using HOMO-LUMO, the energy gap was calculated to be 4.208eV. A high tendency to accept electrons can be attributed to low value (-1.881eV) of LUMO energy level. This low HOMO–LUMO energy gap value has a considerable impact on the IMCT and bioactivity. The high chemical hardness value (2.104eV) shows low stability of the molecule [46]. Electrophilicity index was 3.773eV. This displays that 2BMN is biologically active [47].
Table 5.
Calculated Energy values for 2BMN.
| Parameter | B3LYP/6-311++G(d,p) |
|---|---|
| EHOMO(eV) | -6.089 |
| ELUMO(eV) | -1.881 |
| Ionization potential | 6.089 |
| Electron affinity | 1.881 |
| Energy gap(eV) | 4.208 |
| Electronegativity | 3.985 |
| Chemical potential | -3.985 |
| Chemical hardness | 2.104 |
| Chemical softness | 0.237 |
| Electrophilicity index | 3.773 |
Figure 6.
Energy gap obtained from HOMO and LUMO energy levels of 2BMN.
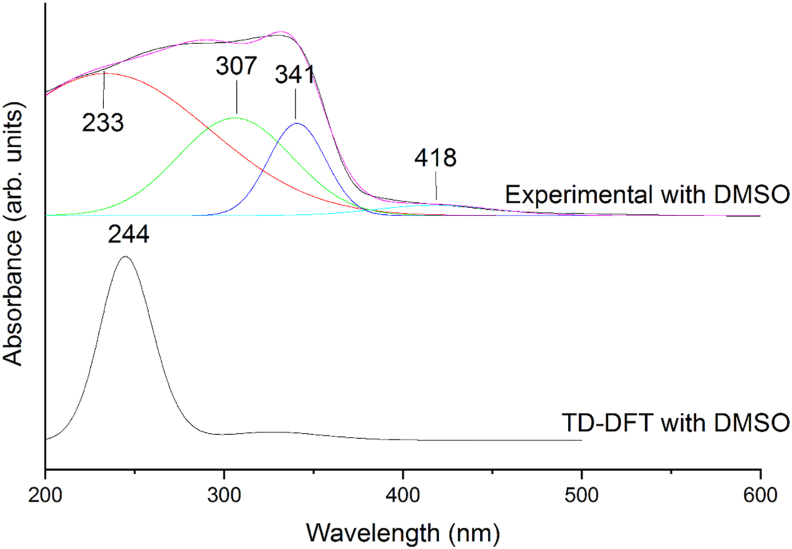
UV analysis was done by time-dependent DFT technique with basis set B3LYP/6-311++G(d,p) was used. The theoretical study was done, taking DMSO as the solvent and was calculated for ten excitation states. Theoretically obtained spectrum using TD-DFT B3LYP/6-311++G(d,p) and DMSO as solvent was compared with the UV-VIS spectrum (experimental) and is given in Figure 7. The theoretical electronic excitation wavelengths, oscillator strengths, energies, bandgap and assignments were obtained for first ten excitations using GAUSS SUM 2.2 software and are given in Table 6.
Figure 7.
Experimental deconvoluted UV spectrum compared with theoretical UV absorption spectra with DMSO as solvent.
Table 6.
UV-Vis. data obtained theoretically and experimentally for title compound using DMSO as solvent.
| Theoretical: TD-DFT B3LYP/6-311++G(d,p) |
Experimental |
||||||
|---|---|---|---|---|---|---|---|
| No. | Wavelength (nm) | Band Gap (eV) | Energy (cm-1) | Oscillatory Strength | Assignments (Major contributions) | Wavelength max (nm) | Band Gap (eV) |
| 1 | 330.38 | 3.75 | 30267.78 | 0.064 | HOMO- > LUMO (90%) | 418 | 2.97 |
| 2 | 298.82 | 4.15 | 33464.98 | 0.014 | H-1- > LUMO (51%), HOMO- > L+1 (41%) | 341 | 3.64 |
| 3 | 255.57 | 4.85 | 39128.65 | 0.003 | HOMO- > L+2 (96%) | 307 | 4.04 |
| 4 | 248.91 | 4.98 | 40174.75 | 0.432 | H-2- > LUMO (18%), H-1- > LUMO (14%) HOMO- > L+1 (18%), HOMO- > L+3 (46%) |
233 | 5.32 |
| 5 | 244.53 | 5.07 | 40894.21 | 1.036 | H-1- > LUMO (26%), HOMO- > L+1 (32%) HOMO- > L+3 (29%) | ||
| 6 | 239.13 | 5.18 | 41818.52 | 0.051 | H-2- > LUMO (70%), HOMO- > L+3 (14%) | ||
| 7 | 234.43 | 5.29 | 42656.54 | 0.018 | HOMO- > L+4 (93%) | ||
| 8 | 224.77 | 5.52 | 44489.85 | 0.164 | H-1- > L+1 (84%) | ||
| 9 | 222.33 | 5.58 | 44977.82 | 0.011 | HOMO- > L+5 (94%) | ||
| 10 | 221.51 | 5.6 | 45145.58 | 0.0004 | H-3- > LUMO (97%) | ||
The calculated UV absorption maxima of a compound are related to electron availability. The absorption maximum was observed at 244.53 nm. The maximum oscillator strength (1.0366), among the ten excited states, was observed for wavelength 244.53 nm with an energy of 40894.21 cm−1. HOMO→LUMO (90%) was observed at 330.38 nm. This has been compared to the experimentally deconvolved spectrum showing peaks at wavelengths 418, 341, 307 and 233 nm.
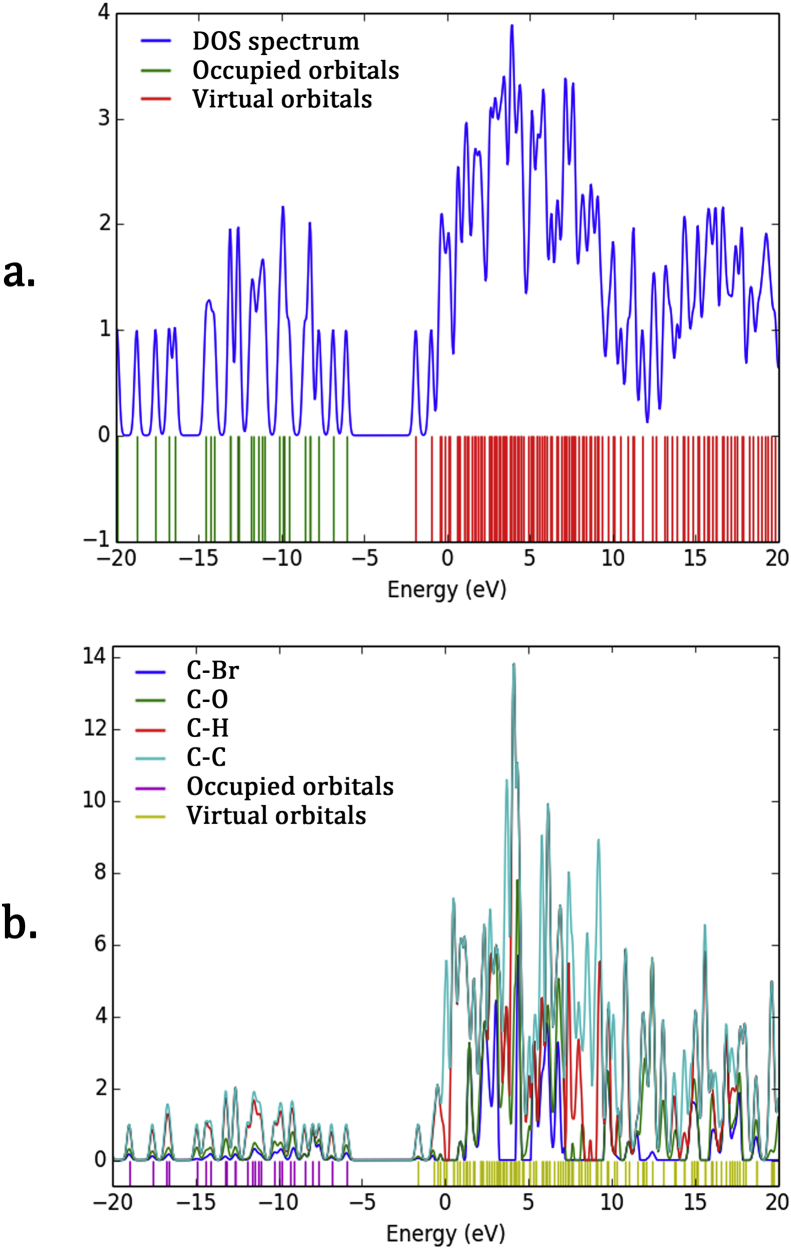
Molecular Orbital composition is shown by DOS spectrum with virtual orbital in red colour and occupied orbital in green colour. In the borderline region, the adjoining orbitals may depict quasi degenerate levels of energy. So the FMOs cannot be perfectly describing. Density of States allows in obtaining a graphical illustration of Molecular Orbital (MO) composition and their contribution to chemical bonding. The contribution of different orbitals MOs is given in PDOS [48]. The Gaussian curves containing information of MOs were convoluted to get DOS and PDOS spectrum. These spectra of unit height and Full Width at Half Maximum (FWHM) of 0.3 eV are shown in Figure 8 (a) and (b).
Figure 8.
(a) DOS and (b) PDOS spectrum of title compound.
3.8. Drug likeness
The drug-likeness properties of 2BMN have been studied using Veber's rule, Lipinski's rule of 5 and Ghose filter [49, 50, 51]. Lipinski's rule of 5 deals in predicting if a biologically active compound is orally active by studying its chemical and physical properties. The ADME (Absorption, Distribution, Metabolism and Excretion) of the compound on human body were analyzed using Veber's rule, which deals with polar surface area (3.52 Å2) and number of rotatable bonds (1). For 2BMN, the Rule of 5 was not violated. Drug likeness properties are listed in Table 7.
Table 7.
Drug Likeness parameters for title compound.
| Descriptor | Value |
|---|---|
| Hydrogen Bond Donor(HBD) | 0 |
| Hydrogen Bond Acceptor(HBA) | 1 |
| MlogP | 3.52 |
| Molar Refractivity | 58.14 g/mol |
| Number of Atoms | 22 |
| Number of Rotatable Bonds | 1 |
By checking molar refractivity (58.14 g/mol) and the number of atoms (22), which falls under the Ghose filter components, it was found that 2BMN satisfies the Ghose filter in addition to Lipinski's rule and Veber's rule. This illustrates the drug-liken nature of 2BMN.
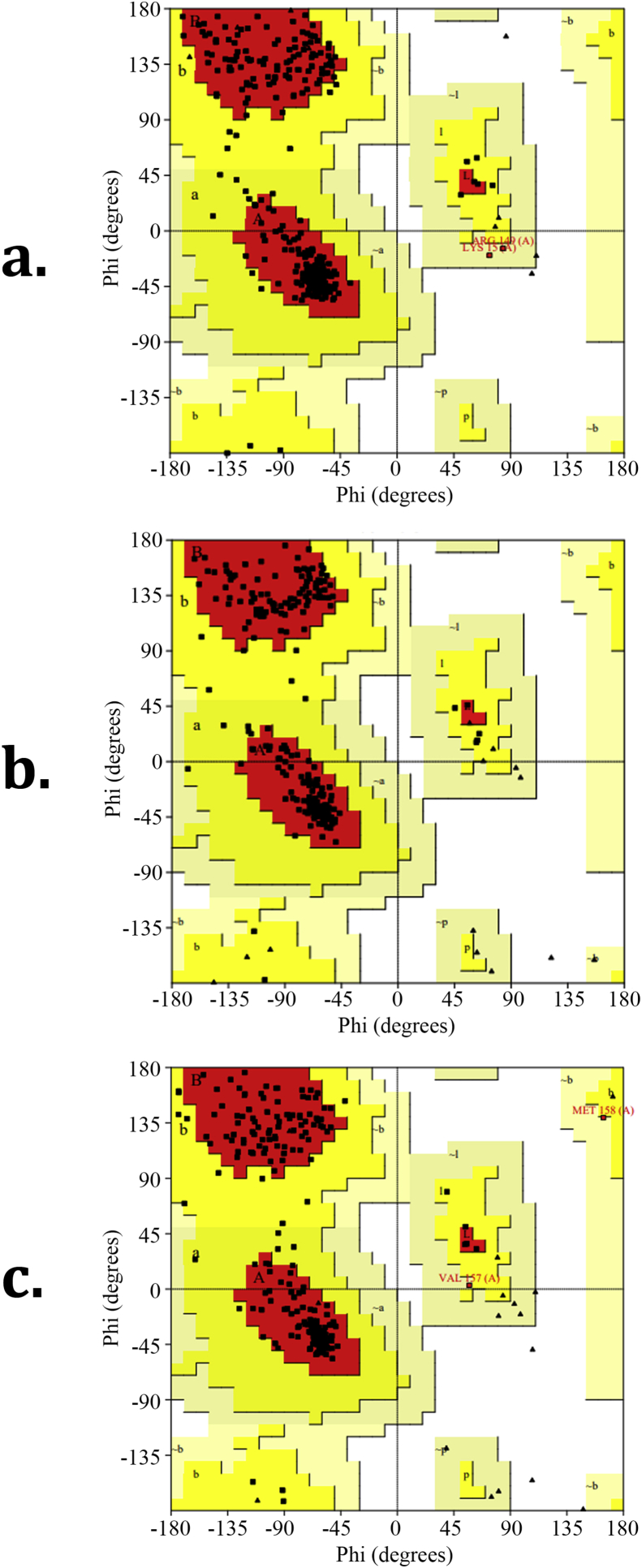
3.9. Ramachandran plot
The stability of protein molecules can be studied using the Ramachandran plot. It also provides data for protein structure determination, prediction and validation. The dihedral angles of protein's backbone are pictured on a plane, where various regions are known to be stable configurations [52, 53]. Ramachandran plot for proteins 6QDZ, 2Z7S and 6OAV are given in Figure 9 (a-c). This shows that the allowed regions are above 90% for both proteins. These proteins also have a large number of residues. In addition, there were no residues in the areas marked disallowed for both the proteins. The total number of residues for 6QDZ was 272 and for 2Z7S, it was 256. The residue counts for 6OAV, protein with anti-inflammatory property, were 327 respectively.
Figure 9.
Ramachandran plot for proteins (a) 6QDZ, (b) 2Z7S and (c) 6OAV.
3.10. Molecular docking
The structure-based drug design assay contains docking molecules into the binding site of proteins and through which binding affinity of the complex can be estimated. Molecular docking is vital as a visual inspection of predicted binding poses help in further development of a lead compound or to a drug with improved binding affinity.
Proteins 6QDZ, 2Z7S and 6OAV were taken from RCSB protein data bank [54, 55, 56]. 6QDZ and 2Z7S have properties related to Tyrosine kinase inhibition, while 6OAZ has anti-inflammatory properties. Figure 10 (a), (b) and (c) show protein-ligand interactions and Table 8 gives details on binding energy and residues. The molecular docking outcomes support that 2BMN has anti-inflammatory properties. In addition to this, 2BMN also exhibits Tyrosine-protein inhibitor properties. The compound shows low binding energy for protein-ligand binding for proteins used. From the low interaction energy and inhibition constant for 6QDZ and 2Z7S we can infer that 2BMN may be effective in cancer treatment.
Figure 10.
Molecular docking representation showing bonded residues and bond distances of 2BMN with proteins (a) 6QDZ, (b) 2Z7S and (c) 6OAV.
Table 8.
Docking parameters for title compound with selected proteins.
| Protein | Bonded Residues | Bond Distance (Å) |
Binding Energy (Kcal/mol) |
Inhibition Constant (micro molar) |
Reference RMSD (Å) |
|---|---|---|---|---|---|
| 6OAZ | His268(A) | 2.6 | -4.71 | 352.54 | 103.577 |
| Ser159(A) | 1.8 | ||||
| 6QDZ | Gly110(A) | 2.1 | -5.69 | 67.49 | 22.036 |
| Met109(A) | 2.6 | ||||
| 2Z7S | Leu144(A) | 2.4 | -6.03 | 37.9 | 27.076 |
4. Conclusion
Using DFT B3LYP/6-311++G(d,p) basis set, 2BMN was optimized and the geometrical values were determined. Theoretical FT-IR and FT Raman spectra (scaled) were compared to experimental data and significant peaks were discussed in detail. For all 60 modes of vibration of the molecule under study, vibrational assignments were done. Frontier Molecular Orbital examination helped to identify HOMO and LUMO energy levels of the molecule. Energy gap and global reactivity parameters were obtained from these results. The energy gap calculated indicates that the molecule is stable. The electrophilicity index was calculated and confirmed that 2BMN is biologically active. The calculated chemical softness value explains the low toxicity nature of 2BMN. All the active sites were studied using 2D colour-coded maps of MEP and ELF. Inter and intramolecular interactions were determined using NBO. Thus σ to σ∗, π to π∗ and lone pair transitions accounting to the stability of the molecule were calculated. Transitions containing C5–C10 bond of the naphthalene ring contribute to higher stabilization energy. This is mainly because of an electron density delocalization due to hyper-conjugation at the junction of naphthalene ring of 2BMN. The compound does not violate Lipinski's rule, Veber's rule and Ghose filter. Therefore, it can be concluded that 2BMN has drug-like properties and may be used as a pharmaceutical compound. The proteins selected for molecular docking studies showed a good quality model, with a considerable number of residues in the most allowed region. The proteins 6OAV, 6QDZ and 2Z7S were found to form hydrogen bonds with oxygen atom (O11) of 6BMN to form protein-ligand complexes. Molecular docking of 2BMN with 6OAV reveals that the compound has anti-inflammatory properties. A low value of binding energy and low inhibition constant of 2BMN with Tyrosine kinase inhibitor (6QDZ and 2Z7S) confirms the stability of protein-ligand complexes formed, which shows that the compound has anti-cancer properties. These results predict its ability to act as a possible drug in treating Cancer.
Declarations
Author contribution statement
Rinnu Sara Saji: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Johanan Christian Prasana: Conceived and designed the experiments; Analyzed and interpreted the data.
S. Muthu: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Jacob George: Performed the experiments.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Johanan Christian Prasana, Email: reachjcp@gmail.com.
S. Muthu, Email: mutgee@gmail.com.
References
- 1.“Cancer” . 12 September 2018. World Health Organization. [Google Scholar]
- 2.Anand P., Kunnumakkara A.B., Sundaram C., Harikumar K.B., Tharakan S.T., Lai O.S., Sung B., B Aggarwal B. Cancer is a preventable disease that requires major lifestyle changes. Pharm. Res. (N. Y.) 2008;25(9):2097–2116. doi: 10.1007/s11095-008-9661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.How is cancer diagonised? Am. Canc. Sco. 29 January 2013 [Google Scholar]
- 4.Latest Global Cancer Data: Cancer burden Rises to 18.1 Million New Cases and 9.6 Million Cancer Deaths in. 2018. [Google Scholar]
- 5.Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.August J.T., Anders M.W., Murad F., Coyle J. Vol. 39. Academic Press; San Diego: 1997. (Advances in Pharmacology). [Google Scholar]
- 7.Rostom A., Dubé C., Lewin G., Tsertsvadze A., Barrowman N., Code C., Sampson M., Moher D. Nonsteroidal anti-inflammatory drugs and cyclooxygenase-2 inhibitors for primary prevention of colorectal cancer: a systematic review prepared for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2007;146(5):376–389. doi: 10.7326/0003-4819-146-5-200703060-00010. [DOI] [PubMed] [Google Scholar]
- 8.Cooper K., Squires H., Carroll C., Papaioannou D., Booth A., Logan R.F., Maguire C., Hind D., Tappenden P. Chemoprevention of colorectal cancer: systematic review and economic evaluation. Health Technol. Assess. 2010;14(32):1–206. doi: 10.3310/hta14320. [DOI] [PubMed] [Google Scholar]
- 9.Rebecca S Y Wong. Adv Pharmacol Sci.; 2019. Role of nonsteroidal anti-inflammatory drugs (NSAIDs) in cancer prevention and cancer promotion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris R.E., Chlebowski R.T., D Jackson R., Frid D.J., Ascenseo J.L., Anderson G., Loar A., Rodabough R.J., White E. A. McTiernan “Breast cancer and nonsteroidal anti-inflammatory drugs: prospective results from the Women's Health Initiative. Canc. Res. 2003;63(18):6096–6101. [PubMed] [Google Scholar]
- 11.Vidal A.C., Howard L.E., Moreira D.M., Castro-Santamaria R., Andriole G.L., Freedland S.J. Aspirin, NSAIDs, and risk of prostate cancer: results from the REDUCE study. Clin. Canc. Res. 2015;21(4):756–762. doi: 10.1158/1078-0432.CCR-14-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friis S., Riis A.H., Erichsen R., Baron J.A., Sørensen H.T. Low-dose aspirin or nonsteroidal anti-inflammatory drug use and colorectal cancer risk. Ann. Intern. Med. 2015;163(5):347–355. doi: 10.7326/M15-0039. [DOI] [PubMed] [Google Scholar]
- 13.Trabert B., Ness R.B., Lo-Ciganic W.-H. Aspirin, nonaspirin nonsteroidal anti-inflammatory drug, and acetaminophen use and risk of invasive epithelial ovarian cancer: a pooled analysis in the ovarian cancer association consortium. J. Natl. Cancer Inst. 2014;106(2):431. doi: 10.1093/jnci/djt431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J., Leng W., Zhao L., Xu C., Wang J., Chen X., Wang Y., Peng X. “Nonsteroidal anti-inflammatory drugs using and risk of head and neck cancer: a dose–response meta-analysis of prospective cohort studies. Oncotarget. 2017;8(58):99066–99074. doi: 10.18632/oncotarget.21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai S.H. Preparation of Au, Ag, Pd trimetallic nanoparticles and their application as catalysts. J. Mater. Sci. 2003;13(5):978–980. [Google Scholar]
- 16.Owen Bussey R., III, Schuh Merlyn D. Quantitation of naproxen by quenching of phosphorescence from a ternary complex of 2-bromo-6-methoxynaphthalene and α-cyclodextrin. J. Inclusion Phenom. Macrocycl. Chem. 2007;57:163–167. [Google Scholar]
- 17.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., Li X., Caricato M., Marenich A.V., Bloino J., Janesko B.G., Gomperts R., Mennucci B., Hratchian H.P., Ortiz J.V., Izmaylov A.F., Sonnenberg J.L., Williams-Young D., Ding F., Lipparini F., Egidi F., Goings J., Peng B., Petrone A., Henderson T., Ranasinghe D., Zakrzewski V.G., Gao J., Rega N., Zheng G., Liang W., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Throssell K., Montgomery J.A., Jr., Peralta J.E., Ogliaro F., Bearpark M.J., Heyd J.J., Brothers E.N., Kudin K.N., Staroverov V.N., Keith T.A., Kobayashi R., Normand J., Raghavachari K., Rendell A.P., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Millam J.M., Klene M., Adamo C., Cammi R., Ochterski J.W., Martin R.L., Morokuma K., Farkas O., Foresman J.B., Fox D.J. Gaussian, Inc.; Wallingford CT: 2009. Gaussian 09W. [Google Scholar]
- 18.Becke A.D. Density-functional thermochemistry III, the role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 19.Zhurko, G.A. and Zhurko, D.A. Chemcraft. Version 1.7 (Build 132).
- 20.Jomroz M.H. VEDA4; Warsaw: 2004. Vibrational Energy Distribution Analysis. [Google Scholar]
- 21.Computational Chemistry comparison and benchmark database. NIST Stand. Ref. Datab. Aug 2020;101 [Google Scholar]
- 22.Osaki T., Eiko Soejima. 2010. Quadratic Scaling Functions for Obtaining normal Vibrational Wavenumbers from the B3LYP Calculation; p. ”. [Google Scholar]
- 23.Lu T., Chen F. Multiwfn: a multifunctional wavefunction analyzer. J. compt. Chem. 2012;33:580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- 24.O’Boyle N.M., Tenderholt A.L., Langner K.M., cclib A library for package-independent computational chemistry algorithms. J. Compt. Chem. 2008;29:839–845. doi: 10.1002/jcc.20823. [DOI] [PubMed] [Google Scholar]
- 25.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Compt. Chem. 1998;19:1639–1662. [Google Scholar]
- 26.The PyMOL Molecular Graphics System (Trial), Version 2.0. LLC; Schrodinger: 2017. [Google Scholar]
- 27.Bolte M., Bauch C. 2-Methoxynaphthalene at 173 K. Acta Crystallogr. 1998;C54:1862–1863. [Google Scholar]
- 28.Socrates G. third ed. John Wiley; New York: 2001. Infrared and Raman Characteristic Group Frequencies. Tables and Charts. [Google Scholar]
- 29.Colthup N.B., Daly L.H., Wiberley S.E. Academic Press; New York: 1990. Introduction to Infrared and Raman Spectroscopy. [Google Scholar]
- 30.Lin-Vien Daimay, Colthup Norman B., Fateley Willian G., Grasselli Jeanette G. Academic Press; Boston: 1991. The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules. [Google Scholar]
- 31.Muthu S., Prabhakaran A. Vibrational spectroscopic study and NBO analysis on tranexamic acid using DFT method. Spectrochim. Acta. 2014;129:184–192. doi: 10.1016/j.saa.2014.03.050. [DOI] [PubMed] [Google Scholar]
- 32.Sundaraganesan N., Illakiamani S., Meganathan C., Joshua B.D. Vibrational spectroscopy investigation using ab initio and density functional theory analysis on the structure of 3-aminobenzotrifluoride. Spectrochim. Acta, Part A. 2007;67:214–224. doi: 10.1016/j.saa.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Peesole R.L., Shield L.D., Mc Willam I.C. Wiley; New York: 1976. Modern Methods of Chemical Analysis. [Google Scholar]
- 34.Barnes A.J., Majid M.A., Stuckey M.A., Gregory P., Stead C.V. The resonance Raman spectra of Orange II and Para Red: molecular structure and vibrational assignment. Spectrochim. Acta, Part A. 1985;41:629–635. [Google Scholar]
- 35.Varsanyi G. Vol. 1. Adam Hilger; London: 1974. (Assignments for Vibrational Spectra of Seven Hundred Benzene Derivatives). [Google Scholar]
- 36.Socrates G. John Wiley and Sons; New York: 1981. Infrared Characteristic Frequencies. [Google Scholar]
- 37.Weinhold F., Landis C.R. 2001. Natural Bond Orbitals and Extensions of Localized. [Google Scholar]
- 38.Sheikhi M., Sheikh D. Quantum chemical investigations on phenyl-7,8-dihydro- [1,3] -dioxolo [4,5-G] quinolin-6(5h)-one. Rev. Roum. Chem. 2014;59:761. [Google Scholar]
- 39.Kuruvilla Tintu K., Prasana Johanan Christian, Muthu S. Jacob George, Vibrational spectroscopic (FT-IR, FT-Raman) and quantum mechanical study of 4-(2-chlorophenyl)-2-ethyl-9-methyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4] diazepine. J. Mol. Struct. 2018;1157:519–529. [Google Scholar]
- 40.Anuradha A., Saji Rinnu Sara, Varghese Mariam, Muthu S., Prasana Johanan Christian. Today Proc.; 2020. Vibrational spectroscopic, DFT studies and molecular docking on (2R)-2-acetamido-N-benzyl-3-methoxy propanamide as an antineuropathic pain drug, mater. [Google Scholar]
- 41.Saji Rinnu Sara, Prasana Johanan Christian, Muthu S., George Jacob, Kuruvilla Tintu K., Raajaraman B.R. Spectroscopic and quantum computational study on naproxen sodium. Spectrochim. Acta, Part A. 2019;222:117–185. doi: 10.1016/j.saa.2019.117614. [DOI] [PubMed] [Google Scholar]
- 42.Pandey Manju, Muthu S., Gowda N.M. Nanje. Quantum mechanical and spectroscopic (FT-IR, FT-Raman,1H,13C NMR, UV-Vis) studies, NBO, NLO, HOMO, LUMO and Fukui function analysis of 5-Methoxy-1H-benzo[d]imidazole-2(3H)-thione by DFT studies. J. Mol. Struct. 2017;1130:511–521. [Google Scholar]
- 43.Shahab S., Sheikhi M., Filippovich L., Dikusar Anatol’evich E., Yahyei H. Quantum-chemical modeling, spectroscopic (FT-IR, excited states, UV/Vis, polarization, and Dichroism) studies of two new benzo[d]oxazole derivatives. J. Mol. Struct. 2017;1137:335. [Google Scholar]
- 44.Rajesh P., Gunasekaran S., Manikandan A. Structural, Spectral analysis of Ambroxol using DFT methods. J. Mol. Struct. 2017;1144:379–388. [Google Scholar]
- 45.Ramazani A., Sheikhi M., Hanifehpour Y. Molecular structure, electronic properties, Homo–Lumo, MEP and NBO analysis of (N-Isocyanimino) triphenylphosphorane (Ph3PNNC): DFT calculations. J. Struct. Chem. 2018;59:529–540. [Google Scholar]
- 46.Poater J., Duran M., Sola M., Silvi B. Theoretical evaluation of electron delocalization in aromatic molecules by means of atoms in molecules (AIM) and electron Localization function (ELF) topological approaches. Chem. Rev. 2005;105:3911–3947. doi: 10.1021/cr030085x. [DOI] [PubMed] [Google Scholar]
- 47.Fathima Rizwana B., Prasana Johanan Christian, Muthu S., Abraham Christina Susan. Molecular docking studies, charge transfer excitation and wave function analyses (ESP, ELF, LOL) on valacyclovir: a potential antiviral drug. Comput. Biol. Chem. 2019;78:9–17. doi: 10.1016/j.compbiolchem.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 48.Chattaraj P.K., Maiti B., Sarkar U. Philicity: a unified treatment of chemical reactivity and selectivity. J. Phys. Chem. 2003;107:4973–4975. [Google Scholar]
- 49.Muthu S., Porchelvi E.E., Karabacak M., Asiri A.M., Swathi Sushmita S. Synthesis, structure, spectroscopic studies (FT-IR, FT-Raman and UV), normal coordinate, NBO and NLO analysis of salicylaldehyde p-chlorophenylthio semicarbazone. J. Mol. Struct. 2015;1081:400–412. [Google Scholar]
- 50.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 51.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 52.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1:55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 53.Georg R., Maack C., Gillmann C., Hagen H. IEEE digital library; 2019. Uncertainty-Aware Ramachandran Plots. [Google Scholar]
- 54.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Feng Z., Gilliland G.L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J.D., Zardecki C. The protein data bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 55.Arora R., V Nimonkar A., Baird D., Wang C., Chiu C.-H., Horton P.A., Hanrahan S., Cubbon R., Weldon S., Tschantz W.R., Mueller S., Brunner R., Lehr P., Meier P., Ottl J., Voznesensky A., Pandey P., Smith T.M., Stojanovic A., Flyer A., Benson T.E., Romanowski M.J., Trauger J.W. Structure of lipoprotein lipase in complex with GPIHBP1. Proc. Natl. Acad. Sci. 2019;116:10360–10365. doi: 10.1073/pnas.1820171116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikuta M., Kornienko M., Byrne N., Reid J.C., Mizuarai S., Kotani H., Munshi S.K. Crystal structures of the N-terminal kinase domain of human RSK1 bound to three different ligands: implications for the design of RSK1 specific inhibitors. Protein Sci. 2007;16(12):2626–2635. doi: 10.1110/ps.073123707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.