Abstract
The mechanisms underlying the therapeutic effect of Salvia miltiorrhiza (SM) on diabetic nephropathy (DN) were examined using a systematic network pharmacology approach and molecular docking. The Traditional Chinese Medicine Systems Pharmacology (TCMSP) database was used to screen active ingredients of SM. Targets were obtained using the SwissTargetPrediction and TCMSP databases. Proteins related to DN were retrieved from the GeneCards and DisGeNET databases. A protein–protein interaction (PPI) network was constructed using common SM/DN targets in the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database. The Metascape platform was used for Gene Ontology (GO) function analysis, and the Cytoscape plug-in ClueGO was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Molecular docking was performed using iGEMDOCK and AutoDock Vina software. Pymol and LigPlos were used for network mapping. Sixty-six active ingredients and 189 targets of SM were found. Sixty-four targets overlapped with DN-related proteins. The PPI network revealed that AKT serine/threonine kinase 1 (AKT1), VEGFA, interleukin 6 (IL6), TNF, mitogen-activated protein kinase 1 (MAPK1), tumor protein p53 (TP53), epidermal growth factor receptor (EGFR), signal transducer and activator of transcription 3 (STAT3), mitogen-activated protein kinase 14 (MAPK14), and JUN were the ten most relevant targets. GO and KEGG analyses revealed that the common targets of DN and SM were mainly involved in advanced glycation end-products, oxidative stress, inflammatory response, and immune regulation. Molecular docking revealed that potential DN-related targets, including tumor necrosis factor (TNF), NOS2, and AKT1, more stably bound with salvianolic acid B than with tanshinone IIA. In conclusion, the present study revealed the active components and potential molecular therapeutic mechanisms of SM in DN and provides a reference for the wide application of SM in clinically managing DN.
Keywords: diabetic nephropathy, molecular docking, molecular mechanism, network pharmacology, Salvia miltiorrhiza
Introduction
Diabetic nephropathy (DN) is a serious complication that is common in diabetic patients. As the incidence and mortality of diabetes increase yearly, the prevalence of DN rises sharply [1] and DN has become one of the leading causes of chronic renal failure [2]. DN accounts for approximately 40% of end-stage kidney disease (ESRD) cases [3]. DN starts with microalbuminuria, which progresses to macroalbuminuria and a decreased glomerular filtration rate, eventually terminating in ESRD. Histologically, glomerular basement membrane thickening and mesangial expansion are the earliest lesions observed in patients with DN [4]. These alterations are followed by nodular glomerulosclerosis and tubulointerstitial changes, including inflammatory cell infiltration, accumulation of activated myofibroblasts, and loss of capillary architecture [5]. Although multiple factors have been shown to be involved in the pathogenesis of DN, its specific molecular mechanism is complex and unclear, leading to a lack of effective therapies.
In recent years, the effects of therapeutics used in traditional Chinese medicine (TCM) in regulating blood sugar and lipid metabolism, reducing kidney damage, delaying kidney disease, and preventing glomerular sclerosis and fibrosis have gradually been uncovered. Salvia miltiorrhiza (SM) has a longstanding history of use for promoting blood circulation in TCM. Its main actions are reducing blood viscosity, improving hemorheological characteristics, accelerating fibrin degradation [6], antioxidant activity [7], anti-infection activity [8], and improving glucose metabolism disorders [9]. It is often used for microvascular-related diseases, such as DN and diabetic retinopathy.
Herbal remedies used in TCM act via a multitarget and multipathway intervention strategy that exerts overall regulatory and synergistic effects, which has certain advantages for DN prevention and individualized treatment. However, the mechanism of action of SM against DN is unclear.
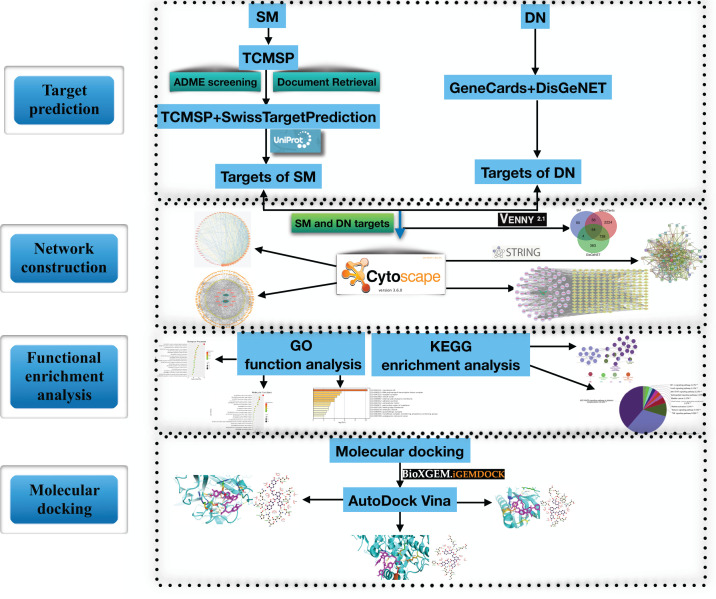
Network pharmacology methods are effective for studying and clarifying the mechanisms underlying drug actions and include chemoinformatics, bioinformatics, network biology, and pharmacology [10,11]. The research strategy of network pharmacology is in line with the integral view on disease in TCM [12,13] and provides new ideas and methods for research on TCM [14]. In this study, we utilized a network pharmacology approach to explore the main bioactive components of SM and predict their effective molecular targets and potential mechanisms in the treatment of DN. A flowchart of the study approach is shown in Figure 1.
Figure 1. Flowchart of the network pharmacology and molecular docking study.
Materials and methods
Screening of active components of SM
SM active components were retrieved from the TCM Systems Pharmacology Database [TCMSP, http://tcmspw.com/tcmsp.php]. We used pharmacokinetic information retrieval filters for absorption, distribution, metabolism, and excretion (ADME) screening based on oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18. Active compounds without potential target information were excluded. Some active compounds that were not predicted but reported in the literature were also included.
Construction of an active component–target network
Targets were obtained from the TCMSP and SwissTargetPrediction databases (http://www.Swisstargetprediction.ch). Then, the targets were standardized in the UniProt (https://www.uniprot.org) database with status set as ‘reviewed’ and species set as ‘human’ [15]. After removing duplicates, a database of SM compounds and their targets was constructed. Finally, a visual network was established using Cytoscape v.3.6.0 software.
Determination of potential DN-related targets
Potential DN-related targets were retrieved from the Human Gene Database (GeneCards, https://www.genecards.org/) and the DisGeNET Database (https://www.disgenet.org/home/) using the search term ‘diabetic nephropathy.’
Determination of DN-related targets of the active components
Screened targets of the active components and DN-related proteins were imported into a Venn diagram webtool (http://bioinformatics.psb.ugent.be/webtools/Venn/) for analysis, and common targets were identified as DN-related targets of the active components for further analysis.
Construction of a protein–protein interaction network of DN-related targets of the active components
To study the interactions between the active components of SM and their target proteins, drug–disease intersection target genes were searched using the interaction database platform STRING v.11.0 (https://string-db.org/), and a protein–protein interaction (PPI) network was constructed. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a comprehensive multifunctional data platform [16,17] that aims to provide PPI evaluation and integration [18]. In our database search, the species was set to ‘Homo sapiens,’ the confidence score cutoff was set at 0.4, and other settings were set to default.
Network construction and analysis
The targets of SM among DN-related proteins identified using STRING were further analyzed using the Cytoscape software v.3.6.0 to visualize and analyze the interaction network. We used the network analysis plug-in in the software to count the nodes in the network and analyze their roles in the network.
Gene Ontology functional analysis
The Metascape platform (http://metascape.prg/gp/index.html) has a comprehensive annotation function, and gene annotation data are updated on a monthly basis [19]. SM targets regulating DN abnormalities were entered into the Metascape platform, and we analyzed their main biological processes (BPs) and performed enrichment analysis. The results were visualized using biological online tools.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was conducted using the Cytoscape plug-in ClueGO. The candidate DN-related genes targeted by SM were entered into the ClueGO plug-in, with P set to <0.01 and the κ score set to ≥ .53.
Molecular docking
Using KEGG pathway enrichment analysis, we identified the potential DN-related genes targeted by SM active components. These targets were confirmed by molecular docking with the experimentally verified SM active components. The verified components were tanshinone IIA, which can improve kidney hypertrophy and 24-h urine protein excretion [20], and salvianolic acid B, which can inhibit the proliferation of mesangial cells and the production of extracellular matrix induced by high glucose in a dose-dependent manner [21]. Crystal structures of the verified components were obtained from the RCSB Protein Data Bank (PDB, https://www.rcsb.org/) [22]. The compound structure was saved as a docking ligand in MOL2 format. The iGEMDOCK software was used for molecular docking. The software automatically uses default parameters during standard docking.
From the molecular docking results, we selected the top five receptor proteins with the lowest energy value and the ligand that bound to these receptor proteins most stably and ran AutoDock Vina for docking. Pymol and LigPlos software was used for network visualization and construction, respectively.
Results
Screening of active components of SM
From the TCMSP database, we obtained 202 known active compounds of SM, which we screened based on OB ≥ 30% and DL ≥ 0.18, yielding 65 active ingredients that met the conditions. In addition, we also included the known active compound salvianolic acid B, which was screened out by ADME. Finally, 66 active components were selected for further analysis (Table 1).
Table 1. Basic information on the main active ingredients of SM.
| Mol ID | Molecule name | OB% | DL |
|---|---|---|---|
| MOL001601 | 1,2,5,6-tetrahydrotanshinone | 38.75 | 0.36 |
| MOL001659 | poriferasterol | 43.83 | 0.76 |
| MOL001771 | poriferast-5-en-3β-ol | 36.91 | 0.75 |
| MOL001942 | isoimperatorin | 45.46 | 0.23 |
| MOL002222 | sugiol | 36.11 | 0.28 |
| MOL002651 | dehydrotanshinone II A | 43.76 | 0.4 |
| MOL002776 | baicalin | 40.12 | 0.75 |
| MOL000569 | digallate | 61.85 | 0.26 |
| MOL000006 | luteolin | 36.16 | 0.25 |
| MOL007036 | 5,6-dihydroxy-7-isopropyl-1,1-dimethyl-2,3-dihydrophenanthren-4-one | 33.77 | 0.29 |
| MOL007041 | 2-isopropyl-8-methylphenanthrene-3,4-dione | 40.86 | 0.23 |
| MOL007045 | 3α-hydroxytanshinoneIIa | 44.93 | 0.44 |
| MOL007048 | (E)-3-[2-(3,4-dihydroxyphenyl)-7-hydroxy-benzofuran-4-yl]acrylic acid | 48.24 | 0.31 |
| MOL007049 | 4-methylenemiltirone | 34.35 | 0.23 |
| MOL007050 | 2-(4-hydroxy-3-methoxyphenyl)-5-(3-hydroxypropyl)-7-methoxy- 3-benzofurancarboxaldehyde |
62.78 | 0.4 |
| MOL007058 | formyltanshinone | 73.44 | 0.42 |
| MOL007059 | 3-β-hydroxymethyllenetanshiquinone | 32.16 | 0.41 |
| MOL007061 | methylenetanshinquinone | 37.07 | 0.36 |
| MOL007063 | przewalskin a | 37.11 | 0.65 |
| MOL007064 | przewalskin b | 110.32 | 0.44 |
| MOL007068 | przewaquinone B | 62.24 | 0.41 |
| MOL007069 | przewaquinone c | 55.74 | 0.4 |
| MOL007070 | (6S,7R)-6,7-dihydroxy-1,6-dimethyl-8,9-dihydro-7H-naphtho[8,7-g]benzofuran-10,11-dione | 41.31 | 0.45 |
| MOL007071 | przewaquinone f | 40.31 | 0.46 |
| MOL007077 | sclareol | 43.67 | 0.21 |
| MOL007079 | tanshinaldehyde | 52.47 | 0.45 |
| MOL007081 | danshenol B | 57.95 | 0.56 |
| MOL007082 | danshenol A | 56.97 | 0.52 |
| MOL007085 | salvilenone | 30.38 | 0.38 |
| MOL007088 | cryptotanshinone | 52.34 | 0.4 |
| MOL007093 | dan-shexinkum d | 38.88 | 0.55 |
| MOL007094 | danshenspiroketallactone | 50.43 | 0.31 |
| MOL007098 | deoxyneocryptotanshinone | 49.4 | 0.29 |
| MOL007100 | dihydrotanshinlactone | 38.68 | 0.32 |
| MOL007101 | dihydrotanshinone I | 45.04 | 0.36 |
| MOL007105 | epidanshenspiroketallactone | 68.27 | 0.31 |
| MOL007107 | C09092 | 36.07 | 0.25 |
| MOL007108 | isocryptotanshi-none | 54.98 | 0.39 |
| MOL007111 | isotanshinone II | 49.92 | 0.4 |
| MOL007115 | manool | 45.04 | 0.2 |
| MOL007119 | miltionone I | 49.68 | 0.32 |
| MOL007120 | miltionone II | 71.03 | 0.44 |
| MOL007121 | miltipolone | 36.56 | 0.37 |
| MOL007122 | miltirone | 38.76 | 0.25 |
| MOL007124 | neocryptotanshinone ii | 39.46 | 0.23 |
| MOL007125 | neocryptotanshinone | 52.49 | 0.32 |
| MOL007127 | 1-methyl-8,9-dihydro-7H-naphtho[5,6-g]benzofuran-6,10,11-trione | 34.72 | 0.37 |
| MOL007130 | prolithospermic acid | 64.37 | 0.31 |
| MOL007132 | (2R)-3-(3,4-dihydroxyphenyl)-2-[(Z)-3-(3,4-dihydroxyphenyl)acryloyl]oxy-propionic acid | 109.38 | 0.35 |
| MOL007141 | salvianolic acid g | 45.56 | 0.61 |
| MOL007142 | salvianolic acid j | 43.38 | 0.72 |
| MOL007143 | salvilenone I | 32.43 | 0.23 |
| MOL007145 | salviolone | 31.72 | 0.24 |
| MOL007150 | (6S)-6-hydroxy-1-methyl-6-methylol-8,9-dihydro-7H-naphtho[8,7-g] benzofuran-10,11-quinone |
75.39 | 0.46 |
| MOL007151 | tanshindiol B | 42.67 | 0.45 |
| MOL007152 | przewaquinone E | 42.85 | 0.45 |
| MOL007154 | tanshinone iia | 49.89 | 0.4 |
| MOL007155 | (6S)-6-(hydroxymethyl)-1,6-dimethyl-8,9-dihydro-7H-naphtho[8,7-g]benzofuran-10,11-dione | 65.26 | 0.45 |
| MOL007156 | tanshinone VI | 45.64 | 0.3 |
| MOL006824 | α-amyrin | 39.51 | 0.76 |
| MOL007118 | microstegiol | 39.61 | 0.28 |
| MOL007123 | miltirone II | 44.95 | 0.24 |
| MOL007149 | NSC 122421 | 34.49 | 0.28 |
| MOL007140 | (Z)-3-[2-[(E)-2-(3,4-dihydroxyphenyl)vinyl]-3,4-dihydroxy-phenyl]acrylic acid | 88.54 | 0.26 |
| MOL007051 | 6-o-syringyl-8-o-acetyl shanzhiside methyl ester | 46.69 | 0.71 |
| MOL007074 | salvianolic acid b | 3.01 | 0.41 |
Determination of targets of SM active components and active component–target network construction
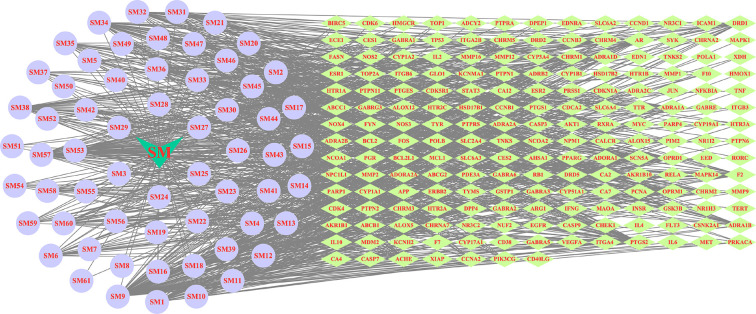
Candidate targets of active components in SM were searched from the TCMSP and SwissTargetPrediction databases. After UniProt standardization, deduplication occurred. After the removal of duplicate data, 189 targets were identified (Supplementary Table S1). Next, we used CytoScape 3.6.0 software to build a ‘active ingredients–targets’ interaction network, as shown in Figure 2.
Figure 2. ‘Ingredients–targets’ network construction.
The light cyan prism nodes represent the targets, the light purple round nodes represent SM ingredients.
Determination of DN-related targets
We retrieved 1189 and 3084 DN-related disease targets obtained from the GeneCards and DisGeNET databases, respectively.
Drug–disease intersection targets
Venn analysis was performed using the 189 targets of SM active components and 1189 and 3084 DN-related target genes, and 64 drug–disease intersection gene targets were obtained for further analysis, as shown in Figure 3 and Table 2. Information on these targets is provided in Supplementary Table S2.
Figure 3. SM/DN common target genes.
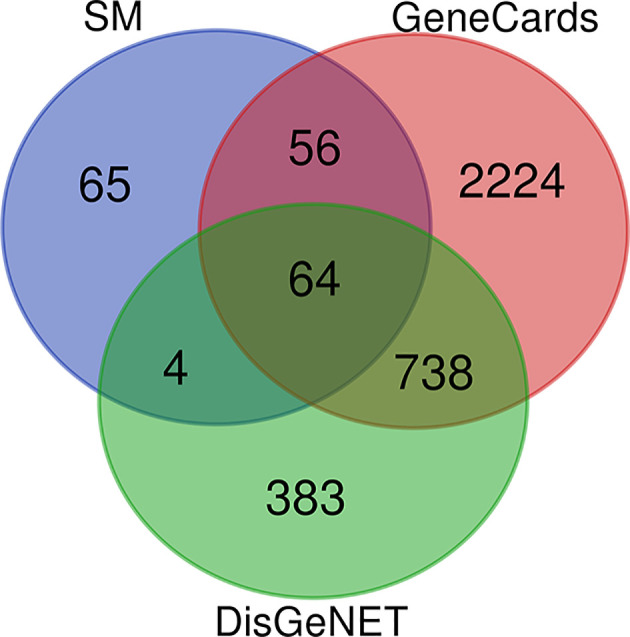
Table 2. Potential targets of SM against DN.
| Serial number | Protein name | Gene name | UniProt ID |
|---|---|---|---|
| 1 | peroxisome proliferator activated receptor γ | PPARG | P37231 |
| 2 | vascular endothelial growth factor A | VEGFA | P15692 |
| 3 | insulin receptor | INSR | P06213 |
| 4 | interleukin 6 | IL6 | P05231 |
| 5 | nitric oxide synthase 3 | NOS3 | P29474 |
| 6 | tumor necrosis factor | TNF | P01375 |
| 7 | solute carrier family 2 member 4 | SLC2A4 | P14672 |
| 8 | AKT serine/threonine kinase 1 | AKT1 | P31749 |
| 9 | signal transducer and activator of transcription 3 | STAT3 | P40763 |
| 10 | dipeptidyl peptidase 4 | DPP4 | P27487 |
| 11 | intercellular adhesion molecule 1 | ICAM1 | P05362 |
| 12 | tumor protein p53 | TP53 | P04637 |
| 13 | interleukin 10 | IL10 | P22301 |
| 14 | endothelin 1 | EDN1 | P05305 |
| 15 | CD40 ligand | CD40LG | P29965 |
| 16 | matrix metallopeptidase 9 | MMP9 | P14780 |
| 17 | interleukin 4 | IL4 | P05112 |
| 18 | nitric oxide synthase 2 | NOS2 | P35228 |
| 19 | interferon gamma | IFNG | P01579 |
| 20 | interleukin 2 | IL2 | P60568 |
| 21 | heme oxygenase 1 | HMOX1 | P09601 |
| 22 | matrix metallopeptidase 2 | MMP2 | P08253 |
| 23 | mitogen-activated protein kinase 1 | MAPK1 | P28482 |
| 24 | prostaglandin-endoperoxide synthase 2 | PTGS2 | P35354 |
| 25 | mitogen-activated protein kinase 14 | MAPK14 | Q16539 |
| 26 | nuclear receptor subfamily 3 group C member 2 | NR3C2 | P08235 |
| 27 | caspase 3 | CASP3 | P42574 |
| 28 | Jun proto-oncogene, AP-1 transcription factor subunit | JUN | P05412 |
| 29 | xanthine dehydrogenase | XDH | P47989 |
| 30 | estrogen receptor 1 | ESR1 | P03372 |
| 31 | matrix metallopeptidase 1 | MMP1 | P03956 |
| 32 | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma | PIK3CG | P48736 |
| 33 | endothelin receptor type A | EDNRA | P25101 |
| 34 | epidermal growth factor receptor | EGFR | P00533 |
| 35 | serine protease 1 | PRSS1 | P07477 |
| 36 | RELA proto-oncogene, NF-kB subunit | RELA | Q04206 |
| 37 | caspase 9 | CASP9 | P55211 |
| 38 | prostaglandin-endoperoxide synthase 1 | PTGS1 | P23219 |
| 39 | 5-hydroxytryptamine receptor 2A | HTR2A | P28223 |
| 40 | fatty acid synthase | FASN | P49327 |
| 41 | cyclin dependent kinase inhibitor 1A | CDKN1A | P38936 |
| 42 | coagulation factor X | F10 | P00742 |
| 43 | glutathione S-transferase pi 1 | GSTP1 | P09211 |
| 44 | BCL2 apoptosis regulator | BCL2 | P10415 |
| 45 | integrin subunit beta 3 | ITGB3 | P05106 |
| 46 | protein tyrosine phosphatase non-receptor type 2 | PTPN2 | P17706 |
| 47 | MDM2 proto-oncogene | MDM2 | Q00987 |
| 48 | integrin subunit alpha 2b | ITGA2B | P08514 |
| 49 | matrix metallopeptidase 12 | MMP12 | P39900 |
| 50 | nuclear receptor subfamily 1 group I member 2 | NR1I2 | O75469 |
| 51 | caspase 7 | CASP7 | P55210 |
| 52 | lymphocyte differentiation antigen CD38 | CD38 | P28907 |
| 53 | Glyoxalase I | GLO1 | Q04760 |
| 54 | arachidonate 12-lipoxygenase | ALOX12 | P18054 |
| 55 | aldose reductase (by homology) | AKR1B1 | P15121 |
| 56 | protein-tyrosine phosphatase 1C | PTPN6 | P29350 |
| 57 | LXR-α | NR1H3 | Q13133 |
| 58 | protein-tyrosine phosphatase 2C | PTPN11 | Q06124 |
| 59 | arginase-1(by homology) | ARG1 | P05089 |
| 60 | poly[ADP-ribose] polymerase-1 | PARP1 | P09874 |
| 61 | adenosine A1 receptor (by homology) | ADORA1 | P30542 |
| 62 | NADPH oxidase 4 | NOX4 | Q9NPH5 |
| 63 | tyrosine-protein kinase SYK | SYK | P43405 |
| 64 | cytochrome P450 19A1 | CYP19A1 | P11511 |
PPI network analysis
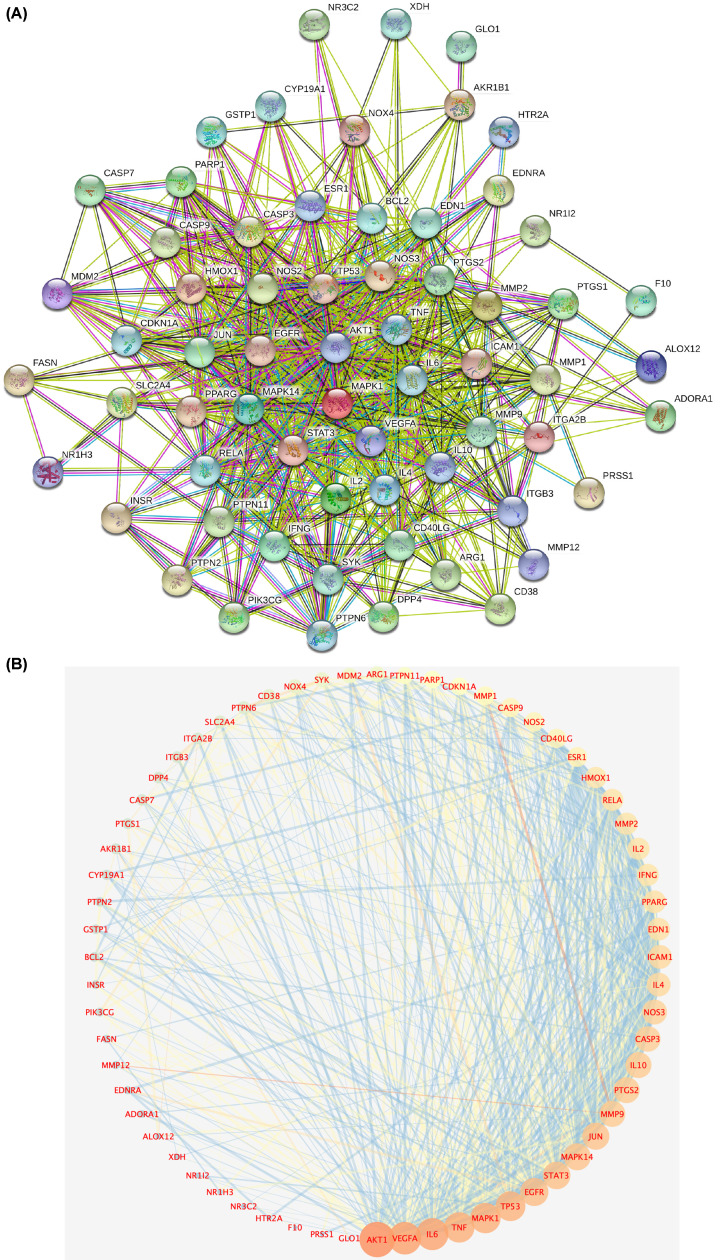
The 64 drug–disease intersection gene targets were analyzed using a PPI network constructed using the STRING database, as shown in Figure 4A. The network 64 nodes and 704 edges, and the average node degree was 21.3, with a PPI enrichment P-value of <1.0e-16 (Figure 4A). The results of STRING analysis were imported into Cytoscape software. The network analysis plug-in was used to count the nodes in the network graph and analyze their connectivity according to the node degree; the greater the node degree, the more biological functions the node has in the network. The network was constructed as shown in Figure 4B. The ten most-connected targets were AKT serine/threonine kinase 1 (AKT1), vascular endothelial growth factor A (VEGFA), interleukin 6 (IL6), tumor necrosis factor (TNF), mitogen-activated protein kinase 1 (MAPK1), tumor protein p53 (TP53), epidermal growth factor receptor (EGFR), signal transducer and activator of transcription 3 (STAT3), mitogen-activated protein kinase 14 (MAPK14), and transcription factor AP-1 (JUN), indicating their significance in the network (Figure 4B).
Figure 4. PPI network analysis.
(A) PPI network of targets generated using STRING 11.0. Nodes represent proteins. Edges represent PPIs. (B) Potential targets are arranged counterclockwise according to the degree value from large to small.
Gene Ontology functional analysis
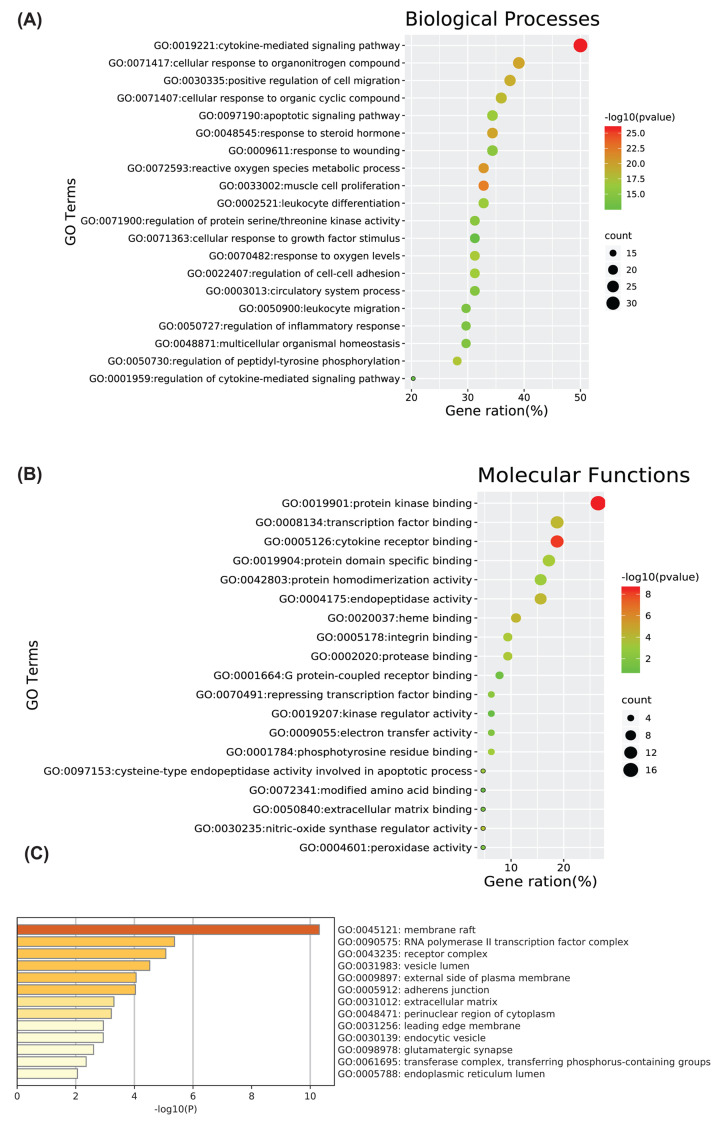
The Metascape data platform was used for enrichment analysis of the 64 relevant DN-related targets of SM, and the results were visualized using biological online tools. A total of 1557 BP Gene Ontology (GO) terms were enriched, and the 20 most significantly enriched BP terms (P<0.01) were selected for analysis. The results showed that BPs enriched in DN-related SM targets mainly included cytokine-mediated signaling pathway, apoptotic signaling pathway, positive regulation of cell migration, reactive oxygen species metabolic process, regulation of inflammatory response, regulation of cell–cell adhesion, response to oxygen levels, cellular response to growth factor stimulus, and regulation of protein serine/threonine kinase activity (Figure 5A).
Figure 5. GO enrichment analysis.
Included are (A) BP terms, (B) molecular function (MF) terms, and (C) cellular component (CC) terms. (A,B) Node color is displayed in a gradient from red to green in descending order of the P-value. The size of the nodes is arranged in ascending order of the number of genes. (C) Sorted by the importance of –log10(P) of each lane.
A total of 90 molecular function (MF) GO terms were enriched, and the 19 most significantly enriched MF terms based on P<0.01 were selected for analysis. The results showed that the intersection genes were mainly enriched in protein kinase binding, transcription factor binding, cytokine receptor binding, integrin binding, cysteine-type endopeptidase activity involved in apoptotic process, kinase regulator activity, heme binding, protein domain-specific binding, endopeptidase activity, nitric-oxide synthase regulator activity, and many other MFs related to the above genes (Figure 5B).
A total of 38 cellular component (CC) GO terms were enriched, and the 13 most significantly enriched CC terms based on P<0.01 were selected for analysis. The results showed that the intersection genes were mainly enriched in membrane rafts, RNA polymerase II transcription factor complex, external side of plasma membrane, extracellular matrix, adherens junction, and glutamatergic synapse (Figure 5C). Detailed node attribute information of the GO analysis results is provided in Supplementary Table S3.
KEGG pathway enrichment analysis
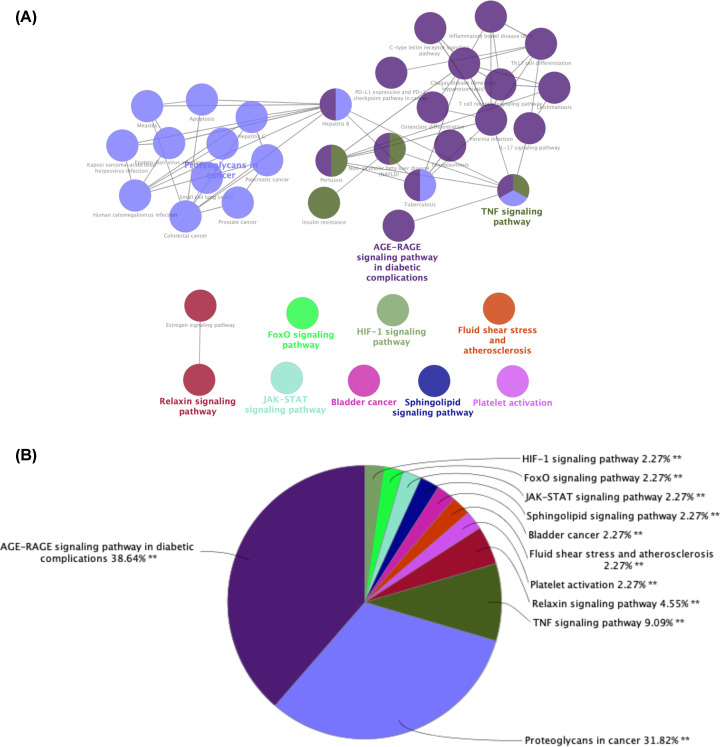
To reveal the potential mechanism underlying the therapeutic effect of SM in DN, we performed KEGG pathway enrichment analysis of the 64 intersection gene targets using the Cytoscape plug-in ClueGO. The screen was based on P<0.01 and a κ score ≥ 0.53 in order to visualize the results of KEGG enrichment (Figure 6A), and we used a pie chart to describe the percentages of genes involved in the different biological functions and signal pathways among the total number of intersection genes (Figure 6B). The results showed that 38 terms were enriched, including the AGE-RAGE signaling pathway in diabetic complications, TNF signaling pathway, JAK-STAT signaling pathway, FoxO signaling pathway, and HIF-1 signaling pathway. In addition, we found some other pathways, including fluid shear stress and atherosclerosis, platelet activation, and relaxin signaling pathway. These results revealed that SM alleviated DN by improving human immunity, anti-inflammatory action, reducing levels of advanced glycation end-products, antioxidant stress response, and regulating other pathways in response to harmful alien organisms. Detailed node attribute information of the KEGG analysis results is provided in Supplementary Table S4.
Figure 6. KEGG pathway analysis of potential targets of SM among DN-related proteins using the ClueGO plug-in.
(A) The KEGG term is indicated as a node, and the size of the node indicates its importance. Only the most significant terms in the group are labeled. (B) Pie chart presenting the percentage of genes involved in different biological functions and signaling pathways in the total number of genes that are intersected.
Molecular docking study
According to the results of KEGG pathway enrichment analysis, we selected the AGE-RAGE signaling pathway in diabetic complications, which had the largest percentage of genes involved in different biological functions and signaling pathways among the total number of intersection genes, for further analysis. Based on drug–target correspondence, the target proteins in this pathway were molecularly docked. The 16 target proteins enriched in the AGE-RAGE signaling pathway in diabetic complications were AKT1, BCL2, CASP3, EDN1, ICAM1, IL6, JUN, MAPK1, MAPK14, MMP2, NOS3, NOS2, RELA, STAT3, TNF, and VEGFA. We selected the experimentally verified SM active molecules tanshinone IIA and salvianolic acid B and molecularly docked the 16 target proteins with them. The most stable conformation is the one with the lowest binding energy. The 16 potential DN-related targets more stably bound to salvianolic acid B than to tanshinone IIA (Table 3).
Table 3. Docking scores of targets with tanshinone IIA and salvianolic acid B (kcal.mol–1).
| Target name | PDBID | Tanshinone IIA | Salvianolic acid B | Canagliflozin |
|---|---|---|---|---|
| MMP2 | 1EAK | −88.21 | −127.25 | −109.23 |
| EDN1 | 1EDP | −62 | −110.92 | −92.83 |
| RELA | 1NFI | −81.98 | −151.32 | −92.46 |
| NOS3 | 1NIW | −92.71 | −131.05 | −113.3 |
| JUN | 1S9K | −87.31 | −147.78 | −99.95 |
| AKT1 | 1UNQ | −79.22 | −148.03 | −102.74 |
| BCL2 | 1YSW | −78.34 | −129.71 | −113.85 |
| TNF | 2E7A | −90.62 | −146.4 | −113.05 |
| MAPK14 | 2NPQ | −84.34 | −139.24 | −102.14 |
| BCL2 | 2O2F | −82.16 | −133.24 | −99.02 |
| BCL2 | 2O21 | −89.69 | −136.58 | −105.02 |
| BCL2 | 2O22 | −97.25 | −135.9 | −104.91 |
| NOS2 | 3E7G | −89.58 | −162.49 | −109.28 |
| CASP3 | 3KJF | −81.44 | −130.26 | −91.52 |
| VEGFA | 3V2A | −76.85 | −121.29 | −91.19 |
| IL6 | 4CNI | −89.55 | −125.52 | −98.18 |
| MAPK1 | 4IZ5 | −78.5 | −144.95 | −97.71 |
| STAT3 | 4ZIA | −84.1 | −136.45 | −103.68 |
| ICAM1 | 5MZA | −80.39 | −125.56 | −98.36 |
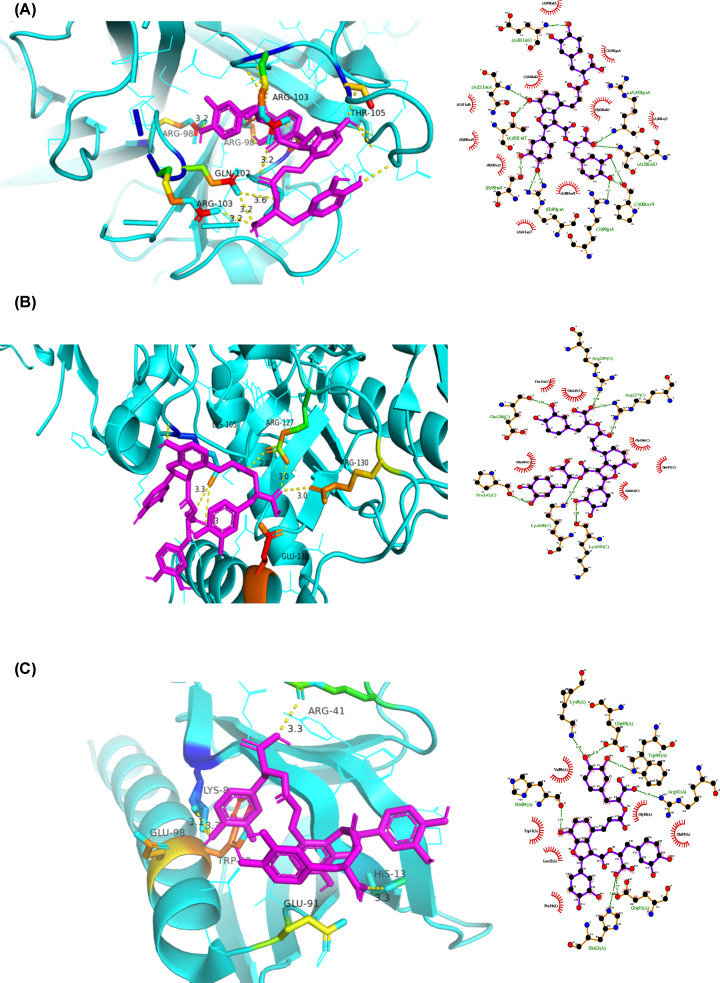
Using AutoDock Vina software, the five target proteins with the lowest energy value in the molecular docking (AKT1, NOS2, TNF, JUN, and RELA) were molecularly docked with the active component salvianolic acid B. Figure 7 shows the best docking combinations for the target proteins and salvianolic acid B, including TNF, NOS2, and AKT1, with binding energies of –9.3, –6.6, and –6.4 kcal/mol, respectively. This shows that salvianolic acid B has a good binding ability to these targets.
Figure 7. Molecular docking.
Molecular models of the binding of salvianolic acid B with (A) TNF, (B) NOS2, and (C) AKT1 shown as 3D and 2D diagrams.
Discussion
The mechanisms of action of TCM therapeutics are complex, with multiple components and targets. When the pathogenesis of disease has not been clarified, it becomes more difficult to analyze the mechanism of action of a TCM therapeutic. Network pharmacology combines system network analysis and pharmacology. It allows systematic study of the effective components, targets, and pathways of drugs at the molecular level, improving our understanding of the interactions between components, targets, and pathways.
In the present study, TCM active component–target network analysis revealed that luteolin, tanshinone IIA, salviolone, salvianolic acid B, dihydrotanshinlactone, and other active ingredients can act on multiple targets in the network. This finding suggests that these components may be important for the therapeutic effect of SM in DN and warrant further exploration. Luteolin has the most potential targets, followed by tanshinone IIA. According to previous reports, luteolin not only increases insulin-mediated glucose uptake and enhances insulin sensitivity [23] but also inhibits high glucose-induced vascular endothelial growth factor (VEGF) [24], reducing reactive oxygen species generation and lipid accumulation [25]. This shows that luteolin can improve insulin resistance and regulate glucose and lipid metabolism. Tanshinone IIA reduces vascular intimal hyperplasia, improves tissue blood perfusion, improves kidney microcirculation, removes intracellular oxygen free radicals, improves blood lipids, promotes anticoagulation, and exerts various other actions [26], such as anti-inflammatory and antioxidant actions [27,28], thereby reducing kidney damage. Salvianolic acid B is a water-soluble compound with the highest activity and content in SM [29]. A number of basic studies have shown that salvianolic acid B has potential therapeutic effects on renal microcirculation. Salvianolic acid B has anti-oxidation [30], anti-inflammatory [31], neuroprotection [32], and anti-fibrosis [33] effects. Salvianolic acid B can be delivered to the kidneys to reduce the progression of renal fibrosis [34], thereby protecting renal function and delaying DN progression.
In the PPI network of 64 targets of SM acting in DN, AKT1, VEGFA, IL6, TNF, MAPK1, TP53, EGFR, STAT3, MAPK14, and JUN were the top ten targets based on node degree. These proteins are regarded core proteins and may play important roles in the therapeutic effect of SM in DN. These proteins are involved in oxidative stress, inflammation, vascular permeability, and immune regulation. For example, the activation of AKT1 promotes cell proliferation and inhibits cell apoptosis. It is an important player in the immune inflammatory mechanism of DN [35]. It is closely related to mesangial matrix proliferation, basement membrane thickening, podocyte damage, and renal tubular epithelial cell transdifferentiation [36]. IL6 and TNF have immunomodulatory and pro-inflammatory effects [37]. VEGFA is related to vascular permeability in patients with DN [38]. Activated eGFR up-regulates reactive oxygen species production and endoplasmic reticulum stress, and this mechanism plays an important role in DN onset [39]. Therefore, it can be inferred that luteolin, tanshinone IIA, and salvianolic acid B, the main active components of SM, reduce oxidative stress and inhibit the expression of inflammatory mediators, such as IL-10, IL-6, and TNF, thus delaying DN progression.
To predict the mechanism underlying the therapeutic effect of SM in DN, we performed GO enrichment analysis of the 64 potential targets. As shown in Figure 5A, the 20 most significantly enriched BP terms were mainly related to cytokines, apoptosis, reactive oxygen species, and inflammation regulation. Relevant studies have shown that DN onset and development are related to cell dysfunction and damage [40,41], chronic inflammatory infiltration [42], cell apoptosis, and oxidative stress [43]. This indicates that the main targets are important for multiple BPs. MFs enriched in targets mainly included cytokine receptor binding, integrin binding, endopeptidase activity, transcription factor binding, protein kinase binding, and heme binding (Figure 5B). The targets involved mainly included VEGFA, PTGS2, DDP4, TNF, and NOS2, which are mainly involved in oxidative stress, the inflammatory response, and immune regulation. DDP4 inhibitors are of great significance for reducing blood sugar levels in diabetic patients and delaying the onset and development of DN [44,45]. In addition, as shown in Figure 5C, CCs enriched in targets mainly included membrane raft, RNA polymerase II transcription factor complex, external side of plasma membrane, extracellular matrix, and adherens junction. These enriched functions involved top targets, such as TNF and JUN. Together, these findings illustrate the complexity of the pathological mechanism of DN.
To further explore the potential mechanism of SM in treating DN, we conducted KEGG analysis of the 64 potential targets of SM acting in DN. As shown in Figure 6, the pathways related to DN, including the AGE-RAGE signaling pathway in diabetic complications, TNF signaling pathway, JAK-STAT signaling pathway, and FoxO signaling pathway, mainly involve three aspects: (1) accumulation of advanced glycation end-products: normally, the glycation reaction proceeds very slowly. However, the response is obviously accelerated in the hyperglycemic state, and the aggregation of AGEs in tissues and their binding with RAGE, a specific receptor, produces cytotoxic effects and damages the kidneys, which may be the key factors contributing to DN. Studies have shown that the AGE/RAGE signaling pathway can promote the expression of NF-κB [46], up-regulate TGF-β1, VEGF [47], activate NADPH oxidases, induce the expression and release of inflammatory factors and adhesion factors, increase vascular permeability, increase the expression of connective tissue growth factor, and enhance oxidative stress, thus increasing proteinuria, promoting renal fibrosis, leading to DN onset and development. Studies have shown that the interaction between AGEs and RAGE leads to vasoconstriction and a procoagulant state [48], accelerates renal vascular aging and injury [49], and further promotes DN progression. (2) Immune inflammation regulation: TNF has immunomodulatory and pro-inflammatory effects [37]. TNF-α stimulates the aggregation and adhesion of inflammatory cells, increases the permeability of microvessels, and impairs glomeruli through an inflammatory response [50]. Some studies have confirmed that TNF-α levels are significantly increased in DN patients and positively correlate with the course of disease [51,52]. In addition, studies have shown that JAK/STAT signaling activation can cause immune inflammation in the kidneys [53,54] and mediates mesangial proliferation and renal tissue fibrosis associated with DN [55]. (3) Oxidative stress: FoxO mainly regulates oxidative stress, apoptosis, and immune responses through the transcription and transmission of various growth factors and cytokine signals, among which FoxO1 plays an important role in the pathogenesis of kidney disease [56]. FoxO1 activation can inhibit podocyte epithelial–mesenchymal cell transformation induced by high glucose and improve proteinuria and renal damage in diabetic mice [57]. In addition, we found other pathways, such as proteoglycans in cancer, fluid shear stress, and atherosclerosis, indicating that SM has potential applications in tumors, atherosclerosis, and other diseases. Based on the aforementioned multiple pathways, it is speculated that SM delays the progression of DN and protects renal function by participating in advanced glycation end products, oxidative stress, inflammatory response, immune regulation, and other processes.
To further explore the potential molecular mechanism of SM in the treatment of DN, we conducted molecular docking studies of 16 targets closely related to DN according to a KEGG-based screening, using the experimentally validated key components tanshinone IIA and salvianolic acid B as ligands. Results showed that the 16 potential targets had a good binding ability to salvianolic acid B, and the interactions were more stable than those with tanshinone IIA.
The present study had some limitations. We only explored the effect of SM in DN at the network pharmacology level. However, current network information technology is not comprehensive, and the accuracy of database data and real-time updates needs to be improved. Therefore, the results obtained in the present study require verification in terms of pharmacodynamics, and mechanistic experiments are needed to explain the complex multitarget, multipathway, and synergistic interactions involved in the therapeutic effects of SM.
Conclusions
The present study analyzed the mechanisms underlying the therapeutic effect of SM in DN using network pharmacology, by constructing an SM ingredient–target–DN-related pathway network (Figure 8). Our findings revealed that SM exerts pharmacological effects in DN in a multicomponent–multitarget–multipathway manner, including advanced glycation end-products, oxidative stress, inflammatory response, and immune regulation. Our findings offer a reference for further investigation of the mechanism underlying the therapeutic effect of SM in DN.
Figure 8. Network structure of ‘SM–component–target pathway–DN’.
Ovals represent SM and DN, diamonds represent components, hexagons represent targets, and rectangles represent pathways.
Supplementary Material
Abbreviations
- ADME
absorption, distribution, metabolism, and excretion
- AKT1
AKT serine/threonine kinase 1
- BP
biological process
- CC
cellular component
- DL
drug-likeness
- DN
diabetic nephropathy
- EGFR
epidermal growth factor receptor
- ESRD
end-stage kidney disease
- GO
Gene Ontology
- IL6
interleukin 6
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
mitogen-activated protein kinase
- MF
molecular function
- OB
oral bioavailability
- PPI
protein–protein interaction
- SM
Salvia miltiorrhiza
- STAT3
signal transducer and activator of transcription 3
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins database
- TCM
traditional Chinese medicine
- TCMSP
TCM Systems Pharmacology
- TNF
tumor necrosis factor
- TP53
tumor protein p53
Contributor Information
Linhua Zhao, Email: melonzhao@163.com.
Xiaolin Tong, Email: tongxiaolin@vip.163.com.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81430097]; and the Major Achievement Guidance Project of Traditional Chinese Medicine Science and Technology [grant number 44223].
Author Contribution
X.‐l.T., L.‐h.Z., and L.-l.Z. conceived and designed the research methods. L.-l.Z., L.H., X.‐m.W., Y.W., and J.-h.Z. collected the data. L.-l.Z. and L.H. analyzed the data. L.-l.Z. and L.H. wrote the original draft. X.‐l.T., L.‐h.Z., and L.-l.Z. reviewed and edited the manuscript. All authors read and approved the final manuscript.
References
- 1.Reutens A.T.et al. (2011) Epidemiology of diabetic nephropathy. Diabetes Kidney 170, 1–7 10.1159/000324934 [DOI] [PubMed] [Google Scholar]
- 2.Gembillo G.et al. (2019) Role of Vitamin D status in diabetic patients with renal disease. Medicina (Kaunas). 55, 273–294 10.3390/medicina55060273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keri K.et al. (2018) Diabetic nephropathy: newer therapeutic perspectives. J. Commun. Hosp. Int. Med. Pers. 8, 200–207 10.1080/20009666.2018.1500423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponchiardi C.et al. (2013) Temporal profile of diabetic nephropathy pathologic changes. Curr. Diabetes Rep. 13, 592–599 10.1007/s11892-013-0395-7 [DOI] [PubMed] [Google Scholar]
- 5.Zhang L.et al. (2020) Research progress on the pathological mechanisms of podocytes in diabetic nephropathy. J. Diabetes Res. 2020, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L.et al. (2012) Tanshinone IIA protects rabbits against LPS-induced disseminated intravascular coagulation (DIC). Acta Pharmacol. Sin. 33, 1254–1259 10.1038/aps.2012.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nickavar B.et al. (2016) Effect-directed analysis for the antioxidant compound in Salvia verticillata. Iranian J. Pharm. Res. 15, 241–246 [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X.et al. (2018) Tanshinone IIA ameliorates lipopolysaccharide-induced inflammatory response in bronchial epithelium cell line BEAS-2B by down-regulating miR-27a. Biomed. Pharmacother. 104, 158–164 10.1016/j.biopha.2018.05.021 [DOI] [PubMed] [Google Scholar]
- 9.Kim E.J.et al. (2007) Antidiabetes and antiobesity effect of cryptotanshinone via activation of AMP-activated protein kinase. Mol. Pharmacol. 72, 62–72 10.1124/mol.107.034447 [DOI] [PubMed] [Google Scholar]
- 10.Zhang J.et al. (2019) Effects and mechanisms of Danshen-Shanzha herb-pair for atherosclerosis treatment using network pharmacology and experimental pharmacology. J. Ethnopharmacol. 229, 104–114 10.1016/j.jep.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 11.Chen L.et al. (2018) Network pharmacology-based strategy for predicting active ingredients and potential targets of Yangxinshi tablet for treating heart failure. J. Ethnopharmacol. 219, 359–368 10.1016/j.jep.2017.12.011 [DOI] [PubMed] [Google Scholar]
- 12.Xu J.et al. (2020) Pharmacological mechanisms underlying the neuroprotective effects of Miq. on Alzheimer’s disease. Int. J. Mol. Sci. 21, 2071–2085 10.3390/ijms21062071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding Z.et al. (2020) Systems pharmacology reveals the mechanism of activity of Ge-Gen-Qin-Lian decoction against LPS-induced acute lung injury: a novel strategy for exploring active components and effective mechanism of TCM formulae. Pharmacol. Res. 156, 1047–1059 10.1016/j.phrs.2020.104759 [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y.et al. (2016) Network pharmacology-based approaches capture essence of Chinese herbal medicines. Chinese Herbal Med. 8, 107–116 10.1016/S1674-6384(16)60018-7 [DOI] [Google Scholar]
- 15.Consortium T.U. (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, 71–75 10.1093/nar/gkr981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D.et al. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C.et al. (2019) Discovery of the anti-tumor mechanism of calycosin against colorectal cancer by using system pharmacology approach. Med. Sci. Monit. 25, 5589–5593 10.12659/MSM.918250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D.et al. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y.et al. (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523–1533 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S.K.et al. (2009) Protective effect of Tanshinone IIA on the early stage of experimental diabetic nephropathy. Biol. Pharm. Bull. 32, 220–224 10.1248/bpb.32.220 [DOI] [PubMed] [Google Scholar]
- 21.Luo P.et al. (2008) Inhibitory effects of salvianolic acid B on the high glucose-induced mesangial proliferation via NF-kappaB-dependent pathway. Biol. Pharm. Bull. 31, 1381–1386 10.1248/bpb.31.1381 [DOI] [PubMed] [Google Scholar]
- 22.Berman H.et al. (2003) Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10, 980 10.1038/nsb1203-980 [DOI] [PubMed] [Google Scholar]
- 23.Ding L.et al. (2010) Luteolin enhances insulin sensitivity via activation of PPARγ transcriptional activity in adipocytes. J. Nutr. Biochem. 21, 941–947 10.1016/j.jnutbio.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 24.Lu H.et al. (2015) Effects of luteolin on retinal oxidative stress and inflammation in diabetes. RSC Adv. 5, 4898–4904 10.1039/C4RA10756J [DOI] [Google Scholar]
- 25.Liu J.et al. (2011) Reduction of lipid accumulation in HepG2 cells by luteolin is associated with activation of AMPK and mitigation of oxidative stress. Phytother. Res. 25, 588–596 10.1002/ptr.3305 [DOI] [PubMed] [Google Scholar]
- 26.Ruano A.et al. (2016) Residential radon, EGFR mutations and ALK alterations in never-smoking lung cancer cases. Eur. Respir. J. 48, 1462–1470 10.1183/13993003.00407-2016 [DOI] [PubMed] [Google Scholar]
- 27.Gao H.et al. (2017) Total tanshinones exhibits anti-inflammatory effects through blocking TLR4 dimerization via the MyD88 pathway. Cell Death Dis. 8, e3004 10.1038/cddis.2017.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X.et al. (2017) Tanshinone IIA attenuates renal damage in STZ-induced diabetic rats via inhibiting oxidative stress and inflammation. Oncotarget 8, 31915–31922 10.18632/oncotarget.16651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhai J.et al. (2019) Salvianolic acid B attenuates apoptosis of HUVEC cells treated with high glucose or high fat via Sirt1 activation. Evid. Based Complement. Alternat. Med. 2019, 1–11 10.1155/2019/9846325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y.et al. (2019) Protective effect of salvianolic acid B against oxidative injury associated with cystine stone formation. Urolithiasis 47, 503–510 10.1007/s00240-019-01114-4 [DOI] [PubMed] [Google Scholar]
- 31.Liu Q.et al. (2018) Salvianolic acid B attenuates experimental pulmonary inflammation by protecting endothelial cells against oxidative stress injury. Eur. J. Pharmacol. 840, 9–19 10.1016/j.ejphar.2018.09.030 [DOI] [PubMed] [Google Scholar]
- 32.Wang Q.et al. (2019) Salvianolic acid B inhibits the development of diabetic peripheral neuropathy by suppressing autophagy and apoptosis. J. Pharm. Pharmacol. 71, 417–428 10.1111/jphp.13044 [DOI] [PubMed] [Google Scholar]
- 33.Liu Q.et al. (2019) Salvianolic acid B attenuates experimental skin fibrosis of systemic sclerosis. Biomed. Pharmacother. 110, 546–553 10.1016/j.biopha.2018.12.016 [DOI] [PubMed] [Google Scholar]
- 34.Li J.et al. (2017) Coordination-driven assembly of catechol-modified chitosan for the kidney-specific delivery of salvianolic acid B to treat renal fibrosis. Biomater. Sci. 6, 179–188 10.1039/C7BM00811B [DOI] [PubMed] [Google Scholar]
- 35.Patel R.K.et al. (2005) PI3K/AKT signaling and systemic autoimmunity. Immunol. Res. 31, 47–55 10.1385/IR:31:1:47 [DOI] [PubMed] [Google Scholar]
- 36.Ribback S.et al. (2015) PI3K/AKT/mTOR pathway plays a major pathogenetic role in glycogen accumulation and tumor development in renal distal tubules of rats and men. Oncotarget 6, 13036–13048 10.18632/oncotarget.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi H.et al. (2020) Hepatitis B core antigen impairs the polarization while promoting the production of inflammatory cytokines of M2 macrophages via the TLR2 pathway. Front. Immunol. 11, 535–549 10.3389/fimmu.2020.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onions K.L.et al. (2019) VEGFC reduces glomerular albumin permeability and protects against alterations in VEGF receptor expression in diabetic nephropathy. Diabetes 68, 172–187 10.2337/db18-0045 [DOI] [PubMed] [Google Scholar]
- 39.Xu Z.et al. (2017) EGFR inhibition attenuates diabetic nephropathy through decreasing ROS and endoplasmic reticulum stress. Oncotarget 8, 32655–32667 10.18632/oncotarget.15948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu P.et al. (2018) Activity of Group 2 innate lymphoid cells is associated with chronic inflammation and dysregulated metabolic homoeostasis in type 2 diabetic nephropathy. Scand. J. Immunol. 87, 99–107 10.1111/sji.12637 [DOI] [PubMed] [Google Scholar]
- 41.Tesch G.H. (2017) Diabetic nephropathy - is this an immune disorder? Clin. Sci. (Lond.) 131, 2183–2199 10.1042/CS20160636 [DOI] [PubMed] [Google Scholar]
- 42.Fathy S.A.et al. (2019) Influence of IL-6, IL-10, IFN-γ and TNF-α genetic variants on susceptibility to diabetic kidney disease in type 2 diabetes mellitus patients. Biomarkers 24, 43–55 10.1080/1354750X.2018.1501761 [DOI] [PubMed] [Google Scholar]
- 43.Wu N.et al. (2016) Acute blood glucose fluctuation enhances rat aorta endothelial cell apoptosis, oxidative stress and pro-inflammatory cytokine expression in vivo. Cardiovasc. Diabetol. 15, 109–121 10.1186/s12933-016-0427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamont B.J.et al. (2008) Differential antidiabetic efficacy of incretin agonists versus DPP-4 inhibition in high fat fed mice. Diabetes 57, 190–198 10.2337/db07-1202 [DOI] [PubMed] [Google Scholar]
- 45.Moritoh Y.et al. (2008) Chronic administration of alogliptin, a novel, potent, and highly selective dipeptidyl peptidase-4 inhibitor, improves glycemic control and beta-cell function in obese diabetic ob/ob mice. Eur. J. Pharmacol. 588, 325–332 10.1016/j.ejphar.2008.04.018 [DOI] [PubMed] [Google Scholar]
- 46.Tóbon J.C.et al. (2014) Receptor for AGEs (RAGE) as mediator of NF-kB pathway activation in neuroinflammation and oxidative stress. CNS Neurol. Disord. Drug Targets 13, 1615–1626 10.2174/1871527313666140806144831 [DOI] [PubMed] [Google Scholar]
- 47.Pang R.et al. (2015) Effects of metformin on apoptosis induced by advanced glycation end-products and expressions of caspase-3, Bax and Bcl-2 in human dermal fibroblasts in vitro. J. Southern Med. Univ. 35, 898–902 [PubMed] [Google Scholar]
- 48.Liu F.et al. (2014) The expression of GPR109A, NF-kB and IL-1β in peripheral blood leukocytes from patients with type 2 diabetes. Ann. Clin. Lab. Sci. 44, 443–448 [PubMed] [Google Scholar]
- 49.Yamagishi S.et al. (2018) Role of ligands of receptor for advanced glycation end products (RAGE) in peripheral artery disease. Rejuv. Res. 21, 456–463 10.1089/rej.2017.2025 [DOI] [PubMed] [Google Scholar]
- 50.Rakitianskaia I.A.et al. (2013) Role of intrarenal product TNF-alpha in the development of glomerular and tubulointerstitial tissues changes in elderly patients with diabetic nephropathy. Adv. Gerontol. 26, 658–665 [PubMed] [Google Scholar]
- 51.Moresco R.N.et al. (2013) Diabetic nephropathy: traditional to proteomic markers. Clin. Chim. Acta 421, 17–30 10.1016/j.cca.2013.02.019 [DOI] [PubMed] [Google Scholar]
- 52.Wu C.et al. (2010) Aberrant cytokines/chemokines production correlate with proteinuria in patients with overt diabetic nephropathy. Clin. Chim. Acta 411, 700–704 10.1016/j.cca.2010.01.036 [DOI] [PubMed] [Google Scholar]
- 53.Liu Q.et al. (2014) Therapeutic effects of suppressors of cytokine signaling in diabetic nephropathy. J. Histochem. Cytochem. 62, 119–128 10.1369/0022155413512493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiezel D.et al. (2014) Impaired renal growth hormone JAK/STAT5 signaling in chronic kidney disease. Nephrol. Dial. Transplant. 29, 791–799 10.1093/ndt/gfu003 [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y.et al. (2014) Suppressor of cytokine signaling (SOCS) 2 attenuates renal lesions in rats with diabetic nephropathy. Acta Histochem. 116, 981–988 10.1016/j.acthis.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 56.Shi W.et al. (2018) Research progress of Foxo in kidney diseases. J. Chinese Pract. Diagn. Ther. 32, 205–208 [Google Scholar]
- 57.Du M.et al. (2016) Overexpression of FOXO1 ameliorates the podocyte epithelial-mesenchymal transition induced by high glucose in vitro and in vivo. Biochem. Biophys. Res. Commun. 471, 416–422 10.1016/j.bbrc.2016.02.066 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.