Highlights
-
•
Widely applicable UV mutagenesis method to improve transgene expression in the microalga Chlamydomonas.
-
•
New CC-1690*** chassis strain with shear stress tolerance, mating-competence and high transgene expression capacity.
-
•
Proof-of-concept cadaverine production from CO2 with CC-1690***.
Keywords: Chlamydomonas reinhardtii, Algal biotechnology, Transgene expression, Industrial strains, Robustness traits
Abstract
In the future, algae biotechnology could play an important role in sustainable development, especially with regard to the production of valuable chemicals. Among the established laboratory strains with efficient transgene expression, there are none that have demonstrated the required robustness for industrial applications, which generally require growth at larger scale. Here, we created a robust and mating-competent cell line of the green microalga Chlamydomonas reinhardtii, which also possesses a high transgene expression capacity. This strain shows a comparably high resistance to shear stress by accumulating increased amounts of biomass under these conditions. As a proof-of-concept, a high phototrophic productivity of cadaverine from CO2 and nitrate was demonstrated by efficiently expressing a bacterial l-lysine decarboxylase. In contrast to other established strains, this novel chassis strain for phototrophic production schemes is equipped with the traits required for industrial applications, by combining mating-competence, cell wall-mediated robustness and high level transgene expression.
1. Introduction
Green biotechnology could play an essential role in the reduction of CO2 emissions. Within these concepts, metabolically-engineered microalgae could become renewable and environment-friendly sources of diverse carbon-based compounds [1] and fuels [2]. Among biotechnologically suitable microorganisms, microalgae are especially promising, since they are photoautotrophic and readily cultivateable in cheap water-based media, using sunlight and carbon dioxide [3]. Metabolic engineering and synthetic biology approaches can substantially expand the range of products obtainable from microalgae [4] and increase their photoautotrophic productivity [5].
Chlamydomonas reinhardtii represents a well-established model organism for basic research in the field of photosynthesis research [6] and also microalgal biotechnology, including metabolic engineering [[7], [8], [9]], since for many fast-growing microlagal species suitable for large-scale cultivation, a molecular toolbox is not available [10].
In 2009 Neupert et al. created Chlamydomonas cell lines capable of efficiently expressing transgenes, by applying UV mutagenesis and subsequent selection of high transgene expressors based on their improved resitance towards the antibiotic emetine [11]. Since then, the UVM4/11 expression strains have successfully been used for the high-level expression of various nuclear transgenes [7,9,12,13].
Although, these strains efficiently express transgenes, UVM4/11 cell lines have two main disadvantages complicating their application in biotechnology. Firstly, the lack of a cell wall renders these strains susceptible to shear stress and thus unsuitable for cultivation at a larger scale, which is normally accompanied by intense mixing based on stirring or gassing with high flow rates [14]. Secondly, their mating inability prevents crossing experiments as a fast and efficient way of removing antibiotic selection markers or adding further genetic traits.
Recently, phototrophic production of the diamine cadaverine (1,5-pentanediamine) via decarboxylation of l-lysine, catalyzed by the enzyme l-Lysine decarboxylase (CadA, EC 4.1.1.18), was reported for C. reinhardtii, using UVM4-derivative strains [9]. 1,5-pentanediamine is used as a building block for the synthesis of (bio-) polyamides, which have excellent material properties and find application as medicinal plastics, fibers for textiles, or films and coatings [15,16].
In order to transfer sustainable cadaverine production into a robust C. reinhardtii phototrophic production chassis, we applied UV-mutagenesis to the cell wall-containing and nitrate-assimilating wildtype CC-1690 [17]. Here, we demonstrate that an iteration of UV mutagenesis and selection of improved transgene expressors, based on their enhanced resistance towards the antibiotic Zeocin, can be used to equip robust C. reinhardtii wildtype strains with the ability to express transgenes at a high level, which is a prerequisite for their application in sustainable production schemes.
2. Materials and methods
2.1. Algal strains and culture conditions
The Chlamydomonas reinhardtii cell wall-containing strain CC-1690 [17] was used for UV mutagenesis experiments and UVM4 was used as a reference strain for transgene expression at a high level [11]. Throughout the manuscript wildtype (WT) refers to strain CC-1690. These strains and derivative cell lines were cultivated at 20 °C under constant illumination (250−350 μmol photons m−2 s−1) by using either TAP medium [18] for mixotrophic growth or HS medium [19] and high cell density medium [9] for phototrophic growth.
2.2. Vector design and cloning
Cloning of vector DNA was performed using the C. reinhardtii MoClo Toolkit [20]. A Shble selection marker (PCM0-077) was assembled with the PSAD promoter (pCM0-016) and FDX1 terminator (Einhaus et al., 2021) as described previously [20] and the corresponding vector was used for initial transformation of CC-1690. UV-treated transformants were further transformed with pOpt2_Clover_Paro [7] to quantify gene expression based on fluorescence measurements. The E. coli lysine decarboxylase cadA (Uniprot: P0A9H3) was optimized for C. reinhardtii nuclear expression (as described in [21]) and fused to mRuby2 (NCBI: AFR60232) as previously described [9] and the resulting plasmid was used in combination with the aphVII hygromycin resistance gene (pCM0-073), [22] to probe heterologous cadaverine production.
2.3. Transformation of C. reinhardtii CC-1690
Nuclear transformation of C. reinhardtii was performed using the electroporation method according to Yamano et al. with some modification [23]. First, CC-1690 was cultivated in TAP medium to the early logarithmic phase (OD680 = 0.2 – 0.4 or 1–2 × 106 cells mL−1). Then, cells were harvested by centrifugation at 1,000 × g, the resulting pellet resuspended in electroporation buffer (100 mM sucrose, 1.5 mM phosphate buffer, pH 7.4) to a final concentration of 5–10 × 108 cells mL−1. For each transformation 2 μg of linearized plasmid DNA were mixed with 50 μl of cell suspension (2.5–5 × 107 cells) and the mixture was transferred to a 2 mm electroporation cuvette (Biorad). For electroporation, the square wave protocol was used with 400 V pulses and a duration of 5 ms. Then, the mixture was transfered to regeneration medium (40 mM glucose in TAP medium) and incubated with gentle shaking (55−65 rpm) for 40 h. The selection of transformants was conducted for 7–10 days on TAP agar plates with different concentrations of the antibiotics: Zeocin (5 μg/mL), Paromomycin (10 μg/mL) and Hygromycin b (20 μg/mL) under 150−200 μmol m−2 s−1 continuous light.
2.4. Zeocin resistance assay
Nuclear transformation of parental strain CC-1690 was performed using a ShBle selection marker [20] and individual transformants obtained were isolated on fresh Zeocin containing agar plates (5 μg mL−1). To determine the level of Zeocin resistance in individual strains, different cell numbers (104 / 105) were spotted on TAP agar plates, containing distinct concentrations of Zeocin (5, 60, 100, 150, 200 μg mL−1) and their growth was analysed. After UV mutagenesis, colonies were selected based on an elevated tolerance towards Zeocin (400, 600, 800, 1000, 1200 μg mL−1).
2.5. UV mutagenesis
The selected low resistance strain (hereafter named ‘40p’) was grown in 25 mL of TAP medium to a cell density of 2.4 × 106 cells mL−1. Cells were harvested by centrifugation (1,000 × g, 5 min) and the pellet was resuspended in 5 mL of TAP medium. For each sample, 0.25 mL corresponding to 3 × 106 cells were spread over agar plates containing either 150 μg/mL or 200 μg/mL Zeocin. Agar plates were then placed on a transilluminator with distance between cell suspension and UV lamp of 2 cm. Cell suspensions were directly exposed to UV light (312 nm) and different exposure times applied (1, 3, 5, 10, 15, 20, 25, 30, 40 min). Following mutagenesis, cells were transferred to darkness for 24 h. Finally, UV treated cells were incubated for 3 weeks on TAP agar plates at 100–150 μE m−2 s−1 continous light illumination and regenerated colonies were tested for an elevated Zeocin tolerance by assessing growth on TAP agar plates containing different concentrations of Zeocin (400, 600, 800, 1000, 1200 μg mL−1).
2.6. Fluorescence microplate reader assay
Cells were grown in TAP medium until late log phase and 100 μl of culture transferred to wells of a black 96-well plate (Greiner 96 Flat Bottom Black Polystyrol (GRE96fb chimney). For fluorescence readings, a microplate reader Tecan Infinite M200 (Tecan, Männedorf, Switzerland) was used and readings acquired by using the following excitation wavelengths and emission filters: 480 / 515 nm for the detection of Clover fluorescence and 559 / 600 nm for mRuby2-fluorescence detection. As a blank, TAP medium was used. Reporter protein fluorescence was normalized against the chlorophyll absorbance at 680 nm. Normalized fluorescence values were also determined for the wildtype, which were used as a reference (set to 1).
2.7. Protein quantification via Western Blot analyses
The strains of interest were grown to an OD750nm of 0.9–1 in 20 mL TAP medium and then cells were harvested by centrifugation (3000 x g for 3 min). After that, proteins were extracted by resuspending pelleted cells in 200 μl of lysis buffer (60 mM Tris pH 6.8, 2% (w/v) SDS, 10 % (v/v) glycerol, 100 mM DTT). Protein concentrations were quantified using the amido black assay [24]. Samples containing 10 μg of total protein fractions were denatured at 95 °C for 5 min, separated via Tris-glycine-SDS-PAGE in 8% (w/v) polyacrylamide gels and transferred to 0.45 μm Protran Nitrocellulose membranes (Amersham) using transfer buffer (25 mM Tris-HCl, 192 mM glycine and 20 % (v/v) methanol). Immunochemical protein detection was performed with HA-tag polyclonal antibody (Thermo Fisher Scientific) using the Thermo Scientific Pierce ECL Western blotting substrate. Signals were visualized using the FUSION-FX7 detection system (Peqlab, Germany).
2.8. Fluorescence microscopy
The randomly selected colonies harboring plasmids carring the reporter genes were grown on TAP plates with appropriate antibiotic during 1–2 days and then fluorescence signals were analysed with Leica M205 FA fully motorized fluorescence stereo microscope as previously described [25]. The visualization of the Clover fluorescence was performed at an excitation of 450–490 nm and at an emission of 505–515 nm. The following parameters were used to detect mRuby2 fluorescence: excitation at 540−580 nm and emission at 589−599 nm.
2.9. Algal growth in HSM medium and assessment of cadaverine production
Strains were grown in HSM medium under constant illumination with a light intensity of 100–200 μE m−2 s−1 and vigorous (vvm = 2) bubbling with carbon dioxide-enriched air (3% (v/v) CO2) for two weeks, with samples taken at days 1, 2, 3, 4, 5, 7, 11 and 14. For each sample, biomass, cell numbers and the optical density at 680 nm were determined. Polyamines were extracted and quantified, as previously described [9].
2.10. Mating and tetrad separation of the UV-mutagenized CC-1690
Mating experiments were performed as previously described [26]. For mating experiments, strains were grown on TAP agar plates for 2–3 days and gametogenesis was induced by nitrogen depletion and formation of dykaryons was observed microscopically.
3. Results and discussion
3.1. UV-mutagenesis and Zeocin selection yield in a Chlamydomonas strain with increased reporter expression
To add the capablility of expressing transgenes at a high level to the robust C.reinhardtii wildtype strain CC-1690 [17], the following strategy was applied (Fig. 1). First, strain CC-1690 was transformed with a ShBle containing plasmid [20] conferring resitance against the antibiotic Zeocin [27,28] and a transformant displaying only a low resistance level was isolated. This strain was transformed with a nuclear expression construct for the expression of a Clover green fluorescent protein reporter alone or a red fluorescent (mRuby2) reporter protein fused to the l-lsyine decarboxylase CadA. To obtain mutant strains expressing transgenes at a higher level by inactivating or circumventing the transgene silencing mechanisms inherent to Chlamydomonas [[29], [30], [31]], UV-mutagenesis was applied. UV mutagenized strains were then tested for an elevated resitance towards Zeocin. The Zeocin resistance level is proportional to the amount of expressed Zeocin-binding protein, since it forms a 1:1 complex with the antibiotic, which prevents DNA cleavage [32]. Zeocin resitance can be thus used as a proxy for the transgene expression capacity of UV mutants. UV mutants showing an elevated antibiotic resistance level were then also transformed with the fluorescent Clover reporter and transformants screened for high reporter expression levels. After obtaining a strain with retained robust phototrophic growth and high transgene expression capacity, this strain was transformed with a lysine decarboxylase (CadA)-mRuby2 reporter fusion construct as a proof-of-concept application for phototrophic cadaverine production.
Fig. 1.
Workflow chart depicting the different steps which led to the isolation of novel CC-1690 strains with desired properties. Created with BioRender.com.
To isolate the starting strain with a low Zeocin resitance level, 95 Zeocin-resitant transformants were subjected to growth assays on agar plates containing Zeocin concentrations in the range of 5−200 μg/mL of the antibiotic. Among all transformants, 11 showed little growth on 100 μg/mL Zeocin and could not grow at all on 150 μg/mL of the antibiotic (Fig. S1).
Among them, strain 40p showed the lowest resitance level and was selected as the starting strain. To this starting strain, which is further on designated “low resistance strain”, we applied UV-mutagenesis to obtain mutants with an altered transgene expression capacity.
Among the isolated UV mutants seven could survive on up to 1200 μg/mL Zeocin and we focussed on one of these mutants (Fig. S2; 7a; CC-1690***), designated “CC-1690***” in further experiments.
Along with UVM4, CC-1690*** was subjected to growth analyses under phototrophic conditions in a flat panel bioreactor (Fig. 2A) with vigorous bubbling (gas flow rate of 2.5 vvm, 1.7 % (v/v) CO2) to test the shear force resitance of both strains. The final biomass yield noted for the cell wall-containing strain CC-1690*** was about two-fold higher compared to the cell-wall deficient [11] UVM4 strain (1.46 ± 0.03 g L−1 for CC1690*** vs. 0.70 ± 0.07 g L−1 for UVM4 at day 5).
Fig. 2.
UV mutagenesis and Zeocin selection yields in a strain with improved reporter expression.
A) Analysis of phototrophic biomass accumulation for strain CC-1690*** and UVM4 grown in 400 mL flat panel photobioreactors with HS medium. The mean value and standard deviations (error bars) of two biological replicates is shown.
B) Scheme showing the tandem DNA construct, which was used for transformation of CC-1690 derivative strains (low resistance strain and UV strain). Nuclear expression of the fluorescent Clover gene and the paromomycin resitance cassette (AphVIII – ParoR) are driven by HSP70A and RBCS2 promoters (red and black arrows, respectively), RBCS2 3‘UTR and PSAD 5` UTR regulatory elements. RBCS2 intron 1 (yellow) was incorporated into the RBCS2 promoter region and RBCS2 intron 2 (orange) was incorporated into the Clover fluorescence gene. Strep-tag II is a synthetic peptide / epitope tag for immunodetection.
C) Fluorescence in Clover-expressing transformants derived from either the low resistance strain (white box) or the UV-mutagenized low resistance strain (CC-1690***; dark grey box).
For analysis, 25 out of 133 mutants from low resistance strain and 25 out of 294 mutants from UV strain have been used. Fluorescence was normalized to chlorophyll absorption (OD680nm) and is given relative to wild type values (background fluorescence set to 1). An asterisk indicates a P value lower than 0.05 according to Student’s t-test.
It is well-known, that cell wall-reduced C. reinhardtii strains cannot resist the high shear forces resulting from vigorous mixing in bioreactors [33]. The much better performance of CC-1690*** compared to UVM4 should therefore result from the fact that CC1690*** contains a wildtype-like robust cell wall, whereas UVM4 does not.
CC-1690*** and the low resistance strain were transformed with a construct for the expression of a Clover green fluorescent protein (Fig. 2B). For each transformation 25 distinct transformants were analysed in regard to their Clover expression based on fluorescence emission (Figs. 2C and S3). The Clover fluorescence emitted by UV strain-derived transformants was significantly (P < 0.05) higher than in those derived from the low resitance strain.
3.2. The novel UV strain displays a UVM4-like capacity to express nuclear transgenes
The UV strain, its progenitor the low resitance strain and UVM4 were then transformed with a nuclear expression construct (Fig. 3A) encoding a fusion of the lysin decarboxylase CadA [9,15,16] and the fluorescent reporter mRuby2 [34].
Fig. 3.
Expression of a mRuby2-CadA fusion protein in the UV strain.
A) The DNA construct, which was used for transformation in CC-1690 derivative strains (wild type, low resistance strain, CC-1690***) and UVM4. The hygromycin resistance gene (AphVII – HygroR) was placed under control of a PSAD promoter and FDX 3‘UTR. The lysine decarboxylase gene was regulated by HSP70A, RBCS2 and FDX1 3‘UTR regulatory elements and the open reading frame of CadA fused to the ORF of the mRuby 2 fluorescence reporter. Yellow vertical bars indicate RBCS2 intron 1, which was inserted into the RBCS2 promoter region as well as lysine decarboxylase and mRuby2 genes. HA indicates the human influenza hemagglutinin (HA) epitope tag.
B) Distribution of mRuby2 fluorescence in strains CC-1690 before and after UV mutagenesis in comparison to UVM4. All strains were transformed with the plasmid shown in panel A). For analysis, 12 out of 600 mutants from low resistance strain, 11 out of 200 mutants from CC-1690*** strain and 11 out of 288 mutants from UVM4 were used. mRuby2 fluorescence in CC-1690 lines was normalized first to OD680 and then to wild type background fluorescence. In the case of UVM4 mutants the mRuby2 fluorescence was normalized first to OD680nm and then to UVM4 background fluorescence.
After transformation, transformants were randomly picked, obtaining a broad spectrum of transgene expression levels (Fig. 3B), which is due to positioning effects, as a result from random integration of expression constructs into the nuclear genome of C. reinhardtii [35]. Fluorescence levels in transformants derived from CC-1690*** (11 out of 200 transformants analysed) and UVM4 (11 out of 288 transformants) were significanty (P < 0.05) higher when compared to those derived from the low resistance strain (Fig. 3B). Although, the median fluorescence and the upper quartile range was higher in UVM4-derived transformants, fluorescence differences between the two transformant populations were insignificant according to a Student`s t-test (p < 0.01, two-tailed hypothesis).
Therefore the UV strain displays a high transgene expression capacity, comparable to that of strain UVM4, a strain which along with UVM11 was for many years the only option, if high level transgene expression in C. reinhardtii was required. The nuclear genomes of both strains, UVM4 and UVM11 carry mutations in a gene encoding the Sir2-type histone deacetylase SRTA [36], which were shown to be causative for the high transgene expression phenotype. Sequencing of the SRTA gene in CC-1690*** confirmed that UV mutagenesis did not cause mutations in its coding sequence, pointing at mutations in other genes as the cause for higher transgene expression in this strain. Future experiments will aim at identifying the genotype responsible for the phenotype of CC-1690***.
3.3. The novel UV strain efficiently produces the diamine cadvarine under photoautotrophic conditions
Among the analysed UV strain-derived transformants UV_6, UV_10 and UV_3 displayed the highest mean mRuby2 fluorescence (Fig. 4A) and were subjected to further analyses (Fig. 4B) together with UV_1 and UV_2, as strains with an intermediate fluorescence. UV_6, UV_3 and UV_2 showed the highest cadaverin titers in the range of 3.5–3.7 mg L−1 after 7 days of cultivation in mixotrophic TAP medium (Fig. 4B) and were further analysed regarding their cadaverine production capacity under phototrophic conditions (Fig. 4C) and using the novel 6xP medium [9], developed for high cell density cultivation of C. reinhardtii. Under these conditions UV_6 displayed the highest biomass (20.49 g L−1 biomass dry weight) and cadaverine accumulation (22.5 mg L−1) after 14 days of phototrophic cultivation.
Fig. 4.
Selection of strain UV_6 for further analyses.
A) Analysis of the mean mRuby2 fluorescence in transformants derived from strain CC-1690*** (grey and coloured bars) or UVM4 (black bars). Fluorescence readings were normalized to chlorophyll absorption determined at 680 nm. Values are given relative to the normalized fluorescence recorded for the wildtype (set to 1). Error bars indicate the standard error derived from three biological replicates (n = 3).
B) Cadaverine titers reached in different UV strain-derived transformants after 7 days of cultivation in TAP medium containing acetate for mixotrophic growth.
C) Biomass dry weight (left y-axis, lines) and cadaverine titers (right y-axis, bars) determined for strains UV_2 (red), UV_10 (blue) and UV_6 (green) after 14 days of cultivation in 6xP medium for high cell density cultivation under photoautotrophic conditions.
In order to compare the cadaverine production capacity of UV_6 to that present in the best UVM4-derived strain (UVM4_3; Fig. 4A), cultivations in photoautotrophic HSM medium were performed (Fig. 5), which, in contrast to 6xP medium, contains ammonium instead of nitrate as the nitrogen source [9,37].
Fig. 5.
Phototrophic cadaverine production in strains UV_6 and UVM4_3.
A) Biomass dry weight determined for strains UV_6 (green) and UVM_3 (black) after 14 days of cultivation in phototrophic HSM medium. Error bars indicate the standard error derived from three biological replicates (n = 3).
B) Cadaverine accumulation in UV_6 (green) and UVM_3 (black) during 14 days of cultivation in phototrophic HSM medium. Error bars indicate the standard error derived from three biological replicates (n = 3).
C) Immunodetection of the CadA-mRuby2 fusion in UV_6 and UVM4_3 using an antibody raised against HA-tag and whole cell extracts derived from samples taken at day 2 of the cultivation shown in A) and B). Three distinct biological replicates (#1-#3) were analysed for each strain and protein loading assessed by a Coomassie Brilliant Blue (CBB) stain of whole cell extracts.
Strain UV_6 accumulated about 20 % more biomass than strain UVM4_3 (1.63 g L−1 in UV_6 vs. 1.38 g L−1) at day five, where biomass accumulation peaked for both strains (Fig. 5A). Cadaverine titers (Fig. 5B) showed a comparable increase in both strains, reaching a similar level (23.2 ± 1.6 mg L−1 in UVM4_3 vs. 22.3. ± 2.8 mg L−1 in UV_6) at day 11, before cadaverine further accumulated until day 14 (34.2 ± 3.1 mg L−1 in UVM4_3 vs. 21.4 ± 6.7 mg L−1 in UV_6). Immunodetection of the CadA-reporter fusion protein after 3 days of cultivation (Fig. 5C) showed that in UVM4_3 the lysine decarboxylase enzyme accumulated to higher levels, explaining the higher cadaverine titer in UVM4_3 at day 3. The higher biomass accumulation in UV_6 at day 11 seems to compensate for the lower transgene expression, resulting in comparable cadaverine titers.
Based on the observed traits of the UV strain including high transgene expression capacity (Figs. 2C, 3 B, 4 A and 5 C) and robust growth (Figs. 2A and 5 A), it was tested if this strain could be crossed with other C. reinhardtii strains of the opposite mating type (Fig. 6).
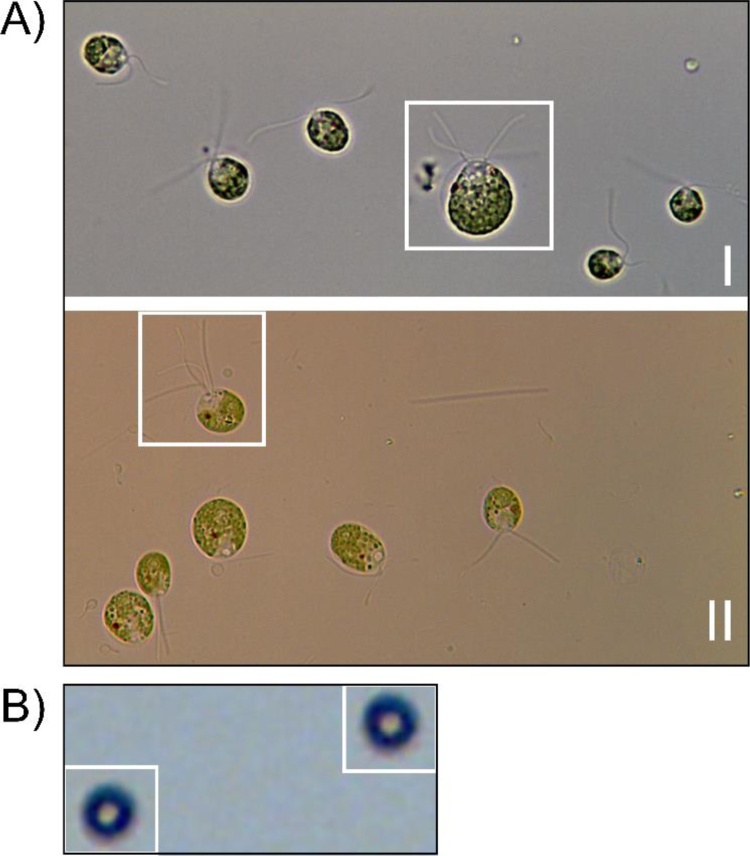
Fig. 6.
CC-1690*** is mating-competent.
A) Microscopic images taken in two distinct mating experiments with the novel strain CC-1690*** (mt+) and CC-1691 (mt−). Zygote-forming dikaryons possessing four flagella are highlighted by white boxes.
B) Magnification of two representative zygotes derived from CC-1690***.
3.4. The novel UV strain is mating-competent
As expected from the presence of long flagella and the motility of the UV strain, dikaryons (white boxes) possessing four flagella could be identified in microscopic images taken after mixing UV strain (derived from CC1690 mt+) with strain CC1691 (mt−). It is well established that almost every dikaryon (>99.9 %; [38]) becomes a zygote, and indeed these dykaryons turned into zygotes (Fig. 6B), showing the typical thick cell wall [39]. The cell-wall deficiency and immotility of UVM4/11 precluded mating experiments [11,36] representing a major drawback for their use a bioproduction chassis strain. Only recently, UVM11 could be crossed with wildtype strain CC-124 and the progeny retained the capacity to express transgenes at a high level [36]. The mating-competence of the UV strain will facilitate future map-based cloning or whole genome re-sequencing experiments [40] to identify the causative mutation.
4. Conclusion
A new Chlamydomonas reinhardtii cell wall-containing strain derived from the robust wildtype CC-1690 was generated by combining UV mutagenesis and subsequent candidate selection based on an increased Zeocin antibiotic tolerance. This novel strain (UV strain) will be further on designated CC-1690*** (spoken CC-1690 triple star) because it possess three key traits for industrial production processes. Besides possessing a cell wall-mediated high shear force resistance, it efficiently expresses nuclear transgenes, which are both prerequistes for industrial bioproduction schemes. As am third trait, CC-1690*** can be crossed to other strains, thus enabling fast trait combination and outcrossing of antibiotic resistance cassettes.
Author contributions
OK, BW, TB and LW: conceptualization. PD and KR: Investigation. PD: Methodology, and formal analysis. BW and OK: Supervision and funding acquisition. LW, TB, OK and PD: Writing – Review and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors gratefully acknowledge support from the state of North Rhine-Westphalia (NRW) and the “European Regional Development Fund (EFRE)”, Project “Cluster Industrial Biotechnology (CLIB) Kompetenzzentrum Biotechnologie (CKB)” (34.EFRE-0300095/1703FI04). The authors would like to thank Prof. Dr. Ralph Bock for sharing strain UVM4. We are grateful to the Center for Biotechnology (CeBiTec) at Bielefeld University for access to the Technology Platforms.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2021.e00644.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Lauersen K.J. Eukaryotic microalgae as hosts for light-driven heterologous isoprenoid production. Planta. 2019;249:155–180. doi: 10.1007/s00425-018-3048-x. [DOI] [PubMed] [Google Scholar]
- 2.Wichmann J., Lauersen K.J., Biondi N., Christensen M., Guerra T., Hellgardt K. Engineering biocatalytic solar fuel production: the PHOTOFUEL consortium. Trends Biotechnol. 2021;39 doi: 10.1016/j.tibtech.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Fernández F.G.A., Reis A., Wijffels R.H., Barbosa M., Verdelho V., Llamas B. The role of microalgae in the bioeconomy. N. Biotechnol. 2021;61:99–107. doi: 10.1016/j.nbt.2020.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Wichmann J., Lauersen K.J., Kruse O. Green algal hydrocarbon metabolism is an exceptional source of sustainable chemicals. Curr. Opin. Biotechnol. 2020;61:28–37. doi: 10.1016/j.copbio.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Naduthodi M.I.S., Claassens N.J., D’Adamo S., van der Oost J., Barbosa M.J. Synthetic biology approaches to enhance microalgal productivity. Trends Biotechnol. 2021 doi: 10.1016/j.tibtech.2020.12.010. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Salomé P.A., Merchant S.S. A series of fortunate events: introducing Chlamydomonas as a reference organism. Plant Cell. 2019;31:1682–1707. doi: 10.1105/tpc.18.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wichmann J., Baier T., Wentnagel E., Lauersen K.J., Kruse O. Tailored carbon partitioning for phototrophic production of (E)-α-bisabolene from the green microalga Chlamydomonas reinhardtii. Metab. Eng. 2018;45:211–222. doi: 10.1016/j.ymben.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Lauersen K.J., Wichmann J., Baier T., Kampranis S.C., Pateraki I., Møller B.L., Kruse O. Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng. 2018;49:116–127. doi: 10.1016/j.ymben.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Freudenberg R.A., Baier T., Einhaus A., Wobbe L., Kruse O. High cell density cultivation enables efficient and sustainable recombinant polyamine production in the microalga Chlamydomonas reinhardtii. Bioresour. Technol. 2021;323 doi: 10.1016/j.biortech.2020.124542. [DOI] [PubMed] [Google Scholar]
- 10.Ng I.-S., Tan S.-I., Kao P.-H., Chang Y.-K., Chang J.-S. Recent developments on genetic engineering of microalgae for biofuels and bio-based chemicals. Biotechnol. J. 2017;12 doi: 10.1002/biot.201600644. [DOI] [PubMed] [Google Scholar]
- 11.Neupert J., Karcher D., Bock R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009;57:1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- 12.Baier T., Kros D., Feiner R.C., Lauersen K.J., Müller K.M., Kruse O. Engineered fusion proteins for efficient protein secretion and purification of a human growth factor from the green microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018;7:2547–2557. doi: 10.1021/acssynbio.8b00226. [DOI] [PubMed] [Google Scholar]
- 13.Baier T., Jacobebbinghaus N., Einhaus A., Lauersen K.J., Kruse O. Introns mediate post-transcriptional enhancement of nuclear gene expression in the green microalga Chlamydomonas reinhardtii. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C., Lan C.Q. Effects of shear stress on microalgae – a review. Biotechnol. Adv. 2018;36:986–1002. doi: 10.1016/j.biotechadv.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Kind S., Neubauer S., Becker J., Yamamoto M., Völkert M., von Abendroth G. From zero to hero – production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab. Eng. 2014;25:113–123. doi: 10.1016/j.ymben.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Kind S., Wittmann C. Bio-based production of the platform chemical 1,5-diaminopentane. Appl. Microbiol. Biotechnol. 2011;91:1287–1296. doi: 10.1007/s00253-011-3457-2. [DOI] [PubMed] [Google Scholar]
- 17.Pröschold T., Harris E.H., Coleman A.W. Portrait of a species: chlamydomonas reinhardtii. Genetics. 2005;170:1601–1610. doi: 10.1534/genetics.105.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris E.H. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. 1st ed. Academic Press; San Diego [u.a.]: 1989. Culture and storage methods; pp. 25–63. [DOI] [PubMed] [Google Scholar]
- 19.Sueoka N. Mitotic replication of deoxyribonucleic acid in chlamydomonas reinhardi. Proc. Natl. Acad. Sci. U. S. A. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crozet P., Navarro F.J., Willmund F., Mehrshahi P., Bakowski K., Lauersen K.J. Birth of a photosynthetic chassis: a MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018;7:2074–2086. doi: 10.1021/acssynbio.8b00251. [DOI] [PubMed] [Google Scholar]
- 21.Baier T., Wichmann J., Kruse O., Lauersen K.J. Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res. 2018;46:6909–6919. doi: 10.1093/nar/gky532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berthold P., Schmitt R., Mages W. An engineered Streptomyces hygroscopicus aph 7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153:401–412. doi: 10.1078/14344610260450136. [DOI] [PubMed] [Google Scholar]
- 23.Yamano T., Iguchi H., Fukuzawa H. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 2013;115:691–694. doi: 10.1016/j.jbiosc.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 24.Popov N., Schmitt M., Schulzeck S., Matthies H. Eine störungsfreie Mikromethode zur Bestimmung des Proteingehaltes in Gewebehomogenaten. [Reliable micromethod for determination of the protein content in tissue homogenates] Acta Biol. Med. Ger. 1975;34:1441–1446. [PubMed] [Google Scholar]
- 25.Lauersen K.J., Kruse O., Mussgnug J.H. Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl. Microbiol. Biotechnol. 2015;99:3491–3503. doi: 10.1007/s00253-014-6354-7. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X., Stern D. Mating and tetrad separation of Chlamydomonas reinhardtii for genetic analysis. J. Vis. Exp. 2009;30 doi: 10.3791/1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens D.R., Rochaix J.D., Purton S. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 1996;251:23–30. doi: 10.1007/BF02174340. [DOI] [PubMed] [Google Scholar]
- 28.Lumbreras V., Stevens D.R., Purton S. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 1998;14:441–447. doi: 10.1046/j.1365-313X.1998.00145.x. [DOI] [Google Scholar]
- 29.Cerutti H., Johnson A.M., Gillham N.W., Boynton J.E. A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: integration into the nuclear genome and gene expression. Genetics. 1997;145:97–110. doi: 10.1093/genetics/145.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerutti H., Johnson A.M., Gillham N.W., Boynton J.E. Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell. 1997;9:925–945. doi: 10.1105/tpc.9.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroda M. Good news for nuclear transgene expression in Chlamydomonas. Cells. 2019;8:1534. doi: 10.3390/cells8121534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dumas P., Bergdoll M., Cagnon C., Masson J.M. Crystal structure and site-directed mutagenesis of a bleomycin resistance protein and their significance for drug sequestering. EMBO J. 1994;13:2483–2492. doi: 10.1002/j.1460-2075.1994.tb06535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duman E.T., Kose A., Celik Y., Oncel S.S. Design of a horizontal-dual bladed bioreactor for low shear stress to improve hydrodynamic responses in cell cultures: a pilot study in Chlamydomonas reinhardtii. Biochem. Eng. J. 2021;169 doi: 10.1016/j.bej.2021.107970. [DOI] [Google Scholar]
- 34.Lam A.J., St-Pierre F., Gong Y., Marshall J.D., Cranfill P.J., Baird M.A. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mussgnug J.H. Genetic tools and techniques for Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2015;99:5407–5418. doi: 10.1007/s00253-015-6698-7. [DOI] [PubMed] [Google Scholar]
- 36.Neupert J., Gallaher S.D., Lu Y., Strenkert D., Segal N.’a, Barahimipour R. An epigenetic gene silencing pathway selectively acting on transgenic DNA in the green alga Chlamydomonas. Nat. Commun. 2020;11:6269. doi: 10.1038/s41467-020-19983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris E.H., editor. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. 1st ed. Academic Press; San Diego [u.a.]: 1989. [DOI] [PubMed] [Google Scholar]
- 38.Dutcher S.K. Chapter 76 mating and tetrad analysis in chlamydomonas reinhardtii. In: Dentler W., Witman G., editors. Methods in Cell Biology. Academic Press; 1995. pp. 531–540. [DOI] [PubMed] [Google Scholar]
- 39.Minami S.A., Goodenough U.W. Novel glycopolypeptide synthesis induced by gametic cell fusion in Chlamydomonas reinhardtii. J. Cell Biol. 1978;77:165–181. doi: 10.1083/jcb.77.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schierenbeck L., Ries D., Rogge K., Grewe S., Weisshaar B., Kruse O. Fast forward genetics to identify mutations causing a high light tolerant phenotype in Chlamydomonas reinhardtii by whole-genome-sequencing. BMC Genomics. 2015;16:57. doi: 10.1186/s12864-015-1232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.