Summary
The necromenic nematode Pristionchus entomophagus has been frequently found in nests of the invasive European ant Myrmica rubra in coastal Maine, United States, and may contribute to ant mortality and collapse of colonies by transferring environmental bacteria. Paenibacillus and several other bacterial species were found in the digestive tracts of nematodes harvested from collapsed ant colonies. Serratia marcescens, Serratia nematodiphila, and Pseudomonas fluorescens were collected from the hemolymph of nematode-infected wax moth (Galleria mellonella) larvae. Virulence against waxworms varied by the site of origin of the nematodes. In adult nematodes, bacteria were highly concentrated in the digestive tract with none observed on the cuticle. In contrast, juveniles had more on the cuticle than in the digestive tract. Host species was the primary factor affecting bacterial community profiles, but Spiroplasma sp. and Serratia marcescens sequences were shared across ants, nematodes, and nematode-exposed G. mellonella larvae.
Subject areas: Entomology, Microbiology, Microbiome
Graphical abstract
Highlights
-
•
Serratia and Pseudomonas isolated from nematodes emerged from M. rubra ant cadavers
-
•
Nematodes acquired a strain of Pseudomonas from the environment
-
•
Strains were shared in bacterial communities of field-collected ants and nematodes
-
•
Exposure to nematodes transferred bacteria to G. mellonella larvae
Entomology; Microbiology; Microbiome
Introduction
Myrmica rubra (Linnaeus) (Hymenoptera: Formicidae) is commonly known as the European fire ant because of its painful sting; however, it is only distantly related to fire ants in the genus Solenopsis (Pitts et al., 2018). Myrmica rubra is native to much of the Palearctic ecozone in Europe and Asia, stretching from Ireland in the west to Western Siberia in the east (Czechowski et al., 2002) and from approximately the 25°N latitude in the south to the 66°N latitude in the Arctic Circle (Elmes et al., 1999). M. rubra has been introduced into regions where it is non-native through unintentional human transport, including in North America where it is considered invasive (Arevalo and Groden, 2007; National Biological Information Infrastructure (NBII) & IUCN/SSC Invasive Species Specialist Group (ISSG), 2009). Since the early 1900s, established populations have been reported along the east coast in the U.S. in Maine, Massachusetts, New York, Pennsylvania, New Jersey, Washington D.C., Rhode Island, and New Hampshire and in Canada in Ontario, Québec, New Brunswick, Prince Edward Island, Newfoundland, and Nova Scotia (Wetterer and Radchenko, 2011). The ants were first observed in Maine in the late 1960s to early 1970s (Groden et al., 2004; Ouellette et al., 2010), and since 1998, reports of M. rubra have increased dramatically (Groden et al., 2005) in humid regions along Maine's coast (Groden et al., 2004), including in Acadia National Park on Mount Desert Island (Ouellette et al., 2010); however, colonies established inland suggest that the ant is able to survive in other environments throughout the state (Arevalo and Groden, 2007). These ants have been demonstrated to have potentially negative impacts on biological community dynamics through suppression of native ant species (Garnas et al., 2014; Naumann and Higgins, 2015), exacerbation of plant-feeding Homoptera populations (McPhee et al., 2012), and lowering arthropod abundance (Verble-Pearson and Pearson, 2016) and diversity (Naumann and Higgins, 2015). In addition, with their aggressive stinging behavior, they are a significant nuisance for home and business owners and public land managers in infested areas where density frequently exceeds an average of 1.5 nests/m2.
Surveys of pathogens and parasitoids associated with invasive M. rubra populations were conducted between 2002 and 2010 on Mount Desert Island in Maine. Collections of both live and moribund workers from nests and middens, respectively, revealed several species of entomopathogenic fungi as well as a nematode emerging from incubated dead individuals. The nematode was identified as Pristionchus entomophagus, and subsequent bioassays demonstrated that exposure of M. rubra workers to this nematode resulted in infection and mortality significantly greater than that observed in controls (Groden and Stock, personal communication). Nematodes in the genus Pristionchus are characterized as necromenic (Herrmann et al., 2006a, 2006b; Rae et al., 2008). Necromenic nematodes are usually free-living microbivores that associate in a tight phoretic (temporary and commensal) relationship with insect hosts, feeding on them after they die of natural causes (Dillman et al., 2012). Juvenile Pristionchus spp. enter their insect host through natural openings such as the mouth or anus (Poinar, 1972) and persist in dauer diapause, in which larvae are in stasis until conditions become favorable for growth. Once the host dies from other causes, dauer-stage nematode larvae detect favorable conditions and re-engage the reproductive life cycle, becoming J4 juveniles. Inside the intestines, the nematodes mature to adults and proliferate, rupturing the digestive tract wall and entering into the hemolymph (Poinar, 1969) and eventually emerging from the host to continue their life cycle in soil (Rae et al., 2008; Sudhaus, 2008).
Adult Pristionchus nematodes feed selectively on bacteria and fungi proliferating in and on the insect carcass, during which ingestion of pathogenic bacteria can negatively impact nematode fitness or cause death (Rae et al., 2008). Unlike entomopathogenic types, these nematodes do not have an obligate relationship with specific bacteria (Dillman et al., 2012) and may host a diverse bacterial community of common inhabitants of soil, water, and other insects (Dumont, 2011; Rae et al., 2008). Members of the nematode family Diplogasteridae, such as Pristionchus spp., have specialized stoma morphology, including shorter, broader mouthparts with no grinder, which allow them to ingest whole bacteria without crushing them and prevent them from being regurgitated after ingestion (Poinar, 1969; von Lieven and Sudhaus, 2000). Pristionchus nematodes may sporadically infect insect hosts with intact pathogenic bacteria picked up from the environment (Al Own et al., 2010; Dieterich et al., 2008; Rae et al., 2008). Facultative virulence (Dillman et al., 2012; Poinar, 1969) and infection of non-beetle hosts (Wahab, 1962) have been reported previously for Pristionchus species.
Field-collected M. rubra colonies that experienced considerable worker mortality when returned to the laboratory showed high levels of P. entomophagus nematode infection. Virulence of the nematodes was verified via reinfection bioassays (Groden and Stock, personal communications). However, nematodes gathered from M. rubra cadavers collected from various sites on Mount Desert Island, Maine, and used to inoculate M. rubra in reinfection assays revealed differential induced mortality, suggesting that there are inherent differences in the pathogenicity of P. entomophagus. We hypothesize that this difference is likely due to their site of origin and local soil bacteria at the sites (Groden and Stock, 2010). Pristionchus entomophagus may have caused mortality of these invasive ants by actively transporting pathogenic bacterial species from the environment into the ants (Groden and Stock, 2010). To explore whether P. entomophagus nematodes may be impacting M. rubra ant colony mortality on Mount Desert Island, in coastal Maine, we conducted a series of experiments to investigate the bacterial community associated with the nematodes and their potential transfer of pathogenic bacteria species from the environment to the ant. We isolated bacteria cultured from nematodes emerging from M. rubra cadavers and assessed the ability of the nematodes to acquire environmental bacteria and their subsequent transfer to an insect host using the greater wax worm, Galleria mellonella, a common model used for insect pathology research. We also identified bacteria which were potentially transferred from nematodes to infected ant nests on the island using bacterial community similarity and sequence tracking methods.
Results
Identification of bacteria from nematodes emerged from M. rubra ant cadavers
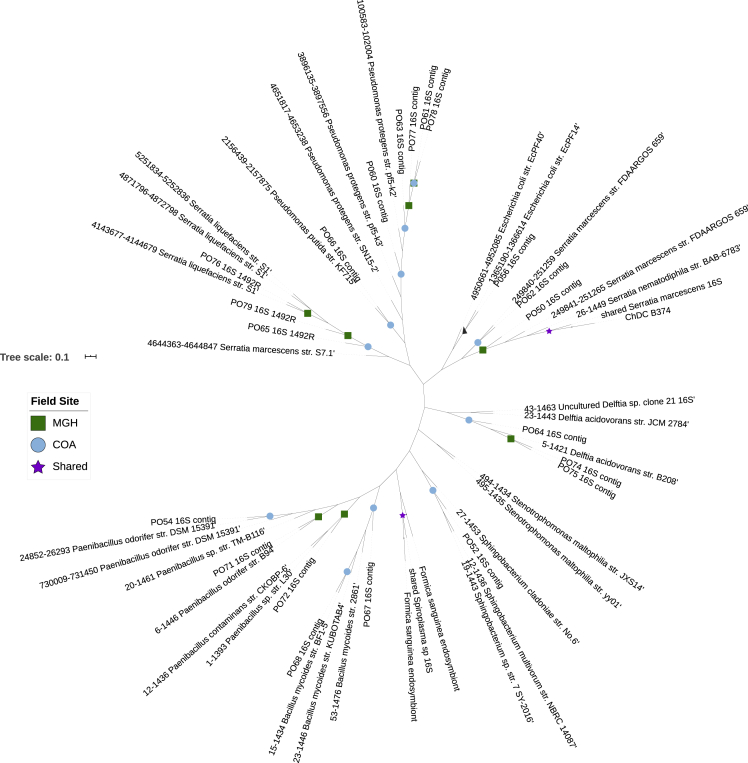
Ants were collected from colonies at the coastal Mount Desert Island sites Eden St. South (COA) and Otter Cliff Road (MGH) (Table 1), as these sites exhibited high mortality and emergence of nematodes from ant cadavers. Emerged nematodes were used to culture a total of 45 bacterial isolates (Table S1; Figure S1). Twenty-eight isolates were recovered from the nematodes' cuticle and were grouped into 12 unique bacterial morphotypes based on culture and cell morphology. Eight isolates were recovered from the nematodes' digestive tract resulting in two distinct morphotypes. Nine bacterial isolates yielding five unique morphotypes were identified from the hemolymph of Galleria mellonella larvae exposed to nematodes harvested from ant cadavers. Of these, 32 isolates, representing all the observed morphotypes, were selected for molecular identification (Table S2). Nearly full-length 16S rRNA gene sequences were obtained for 24 isolates. Genetic distance comparison of query sequences with those in the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) database revealed 13 species in eight genera (Table 2), visualized with additional related sequences in Figure 1. Various species of cultured bacteria were recovered from the cuticle of nematodes. Three sequenced isolates collected from the nematodes' digestive tract were identified as Paenibacillus spp (Table 2). Five isolates were identified from the hemolymph of nematode-infected Galleria mellonella larvae (Table 2). These bacteria were identified as Serratia marcescens, Serratia nematodiphila, and Pseudomonas fluorescens.
Table 1.
GPS coordinates of Myrmica rubra colony collection sites.
| Location | GPS coordinates | Experiments these samples were used in |
|---|---|---|
| Mount Desert Island, Maine | ||
| Woodchip (WC) | 68.2564 W 44.3769 N | 1 |
| Otter Cliff Road (MGH) | 68.2003 W 44.3292 N | 1 |
| Visitors' Center (VC) | 68.248 1 W 44.4103 N | 2 |
| Sports Park (SP) | 68.2033 W 44.3811 N | 1,3 |
| Eden St. South (COA) | 68.2225 W 44.3947 N | 1,3 |
| Old Farm Road (OFR) | 68.1950 W 44.3733 N | 1,3 |
| Breakneck Road (BNR) | 68.2561 W 44.3775 N | 2 |
| Orono, Maine | ||
| Orono (OR) | 68.6675 W 44.8872 N | 2 |
Table 2.
Taxonomic identification for bacteria isolated from the external cuticle and digestive tract of Pristionchus entomophagus and from the hemolymph of Galleria mellonella larvae co-cultured with this nematode
| Isolate ID | Length (nt) | Best match | Coverage (%) | Identity (%) | Accession of best match |
|---|---|---|---|---|---|
| Isolated from external cuticle of Pristionchus entomophagus | |||||
| PO52 | 1436 | Sphingobacterium multivorum str. NBRC 14087 | 99 | 99.9 | AB680559 |
| PO64 | 1420 | Delftia acidovorans str. B208 16S | 99 | 100 | KJ781879 |
| PO65 | 959 | Stenotrophomonas maltophilia str. yy01 | 100 | 100 | MN177222 |
| PO66 | 1437 | Pseudomonas putida str. JCM 13063 | 100 | 100 | LC507960 |
| PO67 | 1441 | Bacillus mycoides str. BF1-5 | 99 | 99.9 | MT078667 |
| PO68 | 1423 | Bacillus mycoides str. 2861 | 100 | 99.9 | MT586023 |
| PO74 | 1420 | Delftia lacustris str. MB38 | 100 | 99.9 | MH675503 |
| PO75 | 1419 | Delftia lacustris str, MB38 | 99 | 99.9 | MH675503 |
| PO76 | 1025 | Serratia quinivorans str. 5619 | 100 | 99.8 | MT256279 |
| PO77 | 1428 | Pseudomonas protegens str. SN15-2 | 100 | 100 | CP043179 |
| PO78 | 1426 | Pseudomonas protegens str. SN15-2 | 100 | 100 | CP043179 |
| PO79 | 872 | Serratia sp. RS7 | 100 | 99.9 | MN006027 |
| Isolated from digestive tract of Pristionchus entomophagus | |||||
| PO54 | 1445 | Paenibacillus odorifer str. DSM 15391 | 99 | 99.7 | CP009428 |
| PO71 | 1443 | Paenibacillus sp. FSL H7-0737 | 100 | 99.5 | CP009279 |
| PO72 | 1429 | Paenibacillus contaminans str. CKOBP-6 | 99 | 99.9 | NR044325 |
| Isolated from hemolymph of Galleria mellonella larvae co-cultured with nematodes | |||||
| PO50 | 1420 | Serratia marcescens str. FY | 100 | 100 | CP053378 |
| PO60 | 1449 | Pseudomonas protegens str. SN15-2 | 100 | 100 | CP043179 |
| PO61 | 1445 | Pseudomonas protegens str. SN15-2 | 100 | 100 | CP043179 |
| PO62 | 1424 | Serratia nematodiphila str. BAB-6783 | 100 | 99.9 | MF319860 |
| PO63 | 1422 | Pseudomonas protegens str. SN15-2 | 100 | 100 | CP043179 |
| Stock | |||||
| PO56 | 1424 | Escherichia coli str. EcPF5 | 100 | 99.9 | CP054236 |
These sequences are visualized as a phylogenetic tree in Figure 1.
Figure 1.
Phylogenetic tree of bacteria isolated from Pristionchus entomophagus nematodes collected from two field sites (MGH, COA) in Maine, along with closest BLAST matches
BLAST match information can be found in Table 2. Tree generated using the maximum likelihood algorithm and is presented unrooted.
Nematode uptake of environmental bacteria and vectoring to other insect hosts
Insect virulence varied by site of origin of nematode isolates
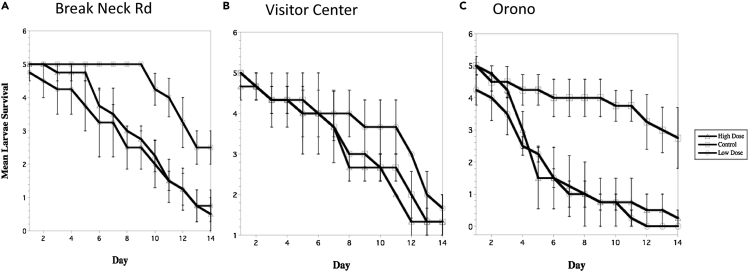
Nematode cultures similarly sourced from ants at three collected sites (Table 1), Breakneck Road (BNR) and Visitors' Center (VC) in Mount Desert Island (coastal) and Orono (OR, inland), exhibited varying virulence against Galleria mellonella larvae after 14 days (Figure 2). Virulence significantly varied by site (X22,8 = 64.91, p = 0.02); overall, nematodes collected from OR had the highest virulence, followed by BNR, and VC did not have significantly higher mortality compared to controls. There was not a significant difference in mortality between two tested doses of nematode exposure for any field site.
Figure 2.
Mortality in Galleria mellonella larvae over 14 days of exposure to Pristionchus entomophagus nematodes that emerged from Myrmica rubra colonies
Sites collected include (A) Breakneck Road in Acadia National Park, in Bar Harbor, ME (BNR); (B) Acadia National Park Visitors Center in Hulls Cove, Bar Harbor, ME (VC); and (C) Orono, ME (OR). The mean survival indicates the average quantity of G. mellonella larvae surviving out of the initial five. Error bars depict standard error (SE) for 4 replicates of 5 larvae per data point.
Nematodes may uptake bacteria from their environment with varying survival
Five percent of live adult nematodes (2/40) were able to uptake red fluorescent protein (RFP)-labeled Escherichia coli HB101 from culture media, i.e., environmental acquisition (Figure S2). This E. coli strain is known to be non-pathogenic to nematodes. No bacteria were observed in dead nematodes, positive controls (nematodes exposed to non-labeled Paenibacillus sp. previously isolated from nematodes), or negative controls (no bacterial exposure). Nematode survival rates were not significantly different (p > 0.05) at two days after exposure (87% survival ± 4 standard error [SE]) and when compared to controls exposed to no bacteria. However, after 5 days after exposure, survival fell to 69% ± 16 SE for E. coli-treated nematodes. There was no change in survival for nematodes exposed to Paenibacillus sp. (87% survival ± 0.1 SE) when compared to controls (86% survival ± 0.4 SE). Five days after exposure, survival was at 68% ± 05 SE for Paenibacillus sp.-treated nematodes and 80% ± 0.3 SE for control nematodes. Overall, mortality at day 5 was significantly higher in both bacterial treatment groups than in no bacterial controls (F1,6 = 7.08, p = 0.04).
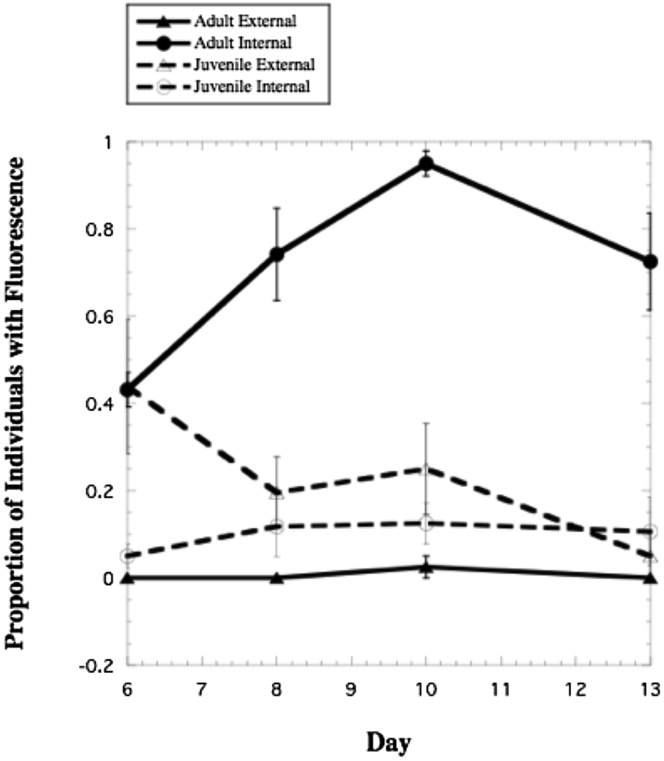
The uptake of RFP-labeled Pseudomonas aeruginosa strain PA14 from culture media by nematodes was visually confirmed (Figures S2 and 3) and was highest in the digestive tract of adult nematodes. No bacteria were observed on the cuticle of adults. Conversely, bacteria were confirmed on the external cuticle of juveniles, with a small amount of fluorescence observed in their digestive tract (Figure 3). The greatest difference in the proportion of overall fluorescence, as well as peak fluorescence, was between the digestive tract of adults (95%) and the cuticle of juveniles (30%) on day 10 (p < 0.001; Figure 3). Fluorescence overall varied over time (p < 0.001), with a general increase in the digestive tract of adults but a steady and significant decrease in the proportion of juveniles with fluorescence (external and internal) over time (p = 0.038). Survival in nematodes exposed to RFP-labeled P. aeruginosa strain PA14, known to be virulent to nematodes, was not significantly different (44.9% ± 7 SE survival) when compared to nematodes with their natural microflora (50.8% ± 4 SE survival) after two days of exposure (t7 = 0.75, p > 0.05).
Figure 3.
Proportion of adult and juvenile Pristionchus entomophagus nematodes showing fluorescence from RFP-labeled Pseudomonas aeruginosa bacteria in their digestive tract (internal) or on their cuticle (external)
Error bars depict standard error (SE) with N = 4.
Nematode vectoring of P. aeruginosa to G. mellonella larvae
Moth larval mortality increased when exposed to juvenile nematodes carrying the RFP-labeled P. aeruginosa strain PA14 (92% mortality; X21,11 = 53.32, p < 0.001), in comparison to control larvae not exposed to nematodes (36% mortality). Larvae mortality likewise increased when exposed to juvenile nematodes reared on nematode growth agar without bacteria (96% mortality; X21,11 = 65.77, p < 0.001), in comparison to control larvae. Consequently, Galleria mellonella larvae exposed to nematodes carrying bacteria did not exhibit significantly higher mortality compared to larvae exposed to nematodes without bacteria (X21,11 = 1.92, p > 0.05). Microscopic observations did not reveal the presence of RFP-labeled P. aeruginosa in G. mellonella larvae hemolymph, indicating a lack of bacterial transference from juvenile nematodes to larvae despite larval mortality.
Microbial community profiling of host-associated bacteria
Host species was the primary factor affecting bacterial communities
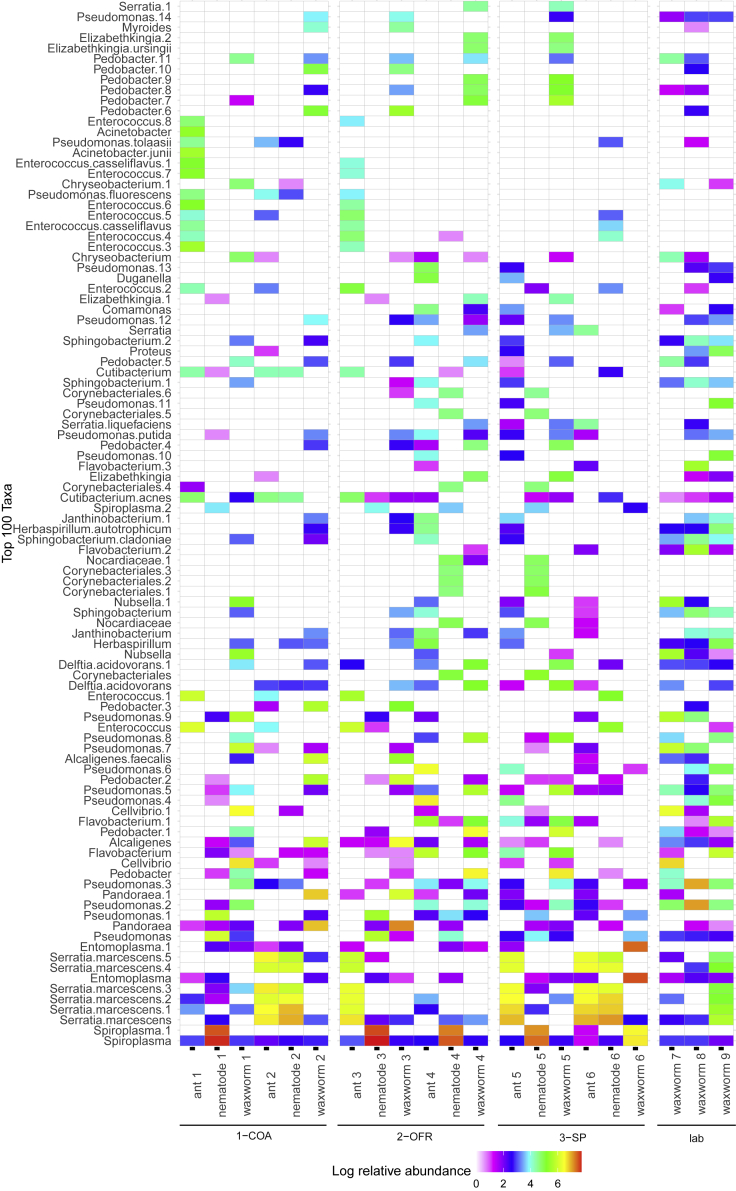
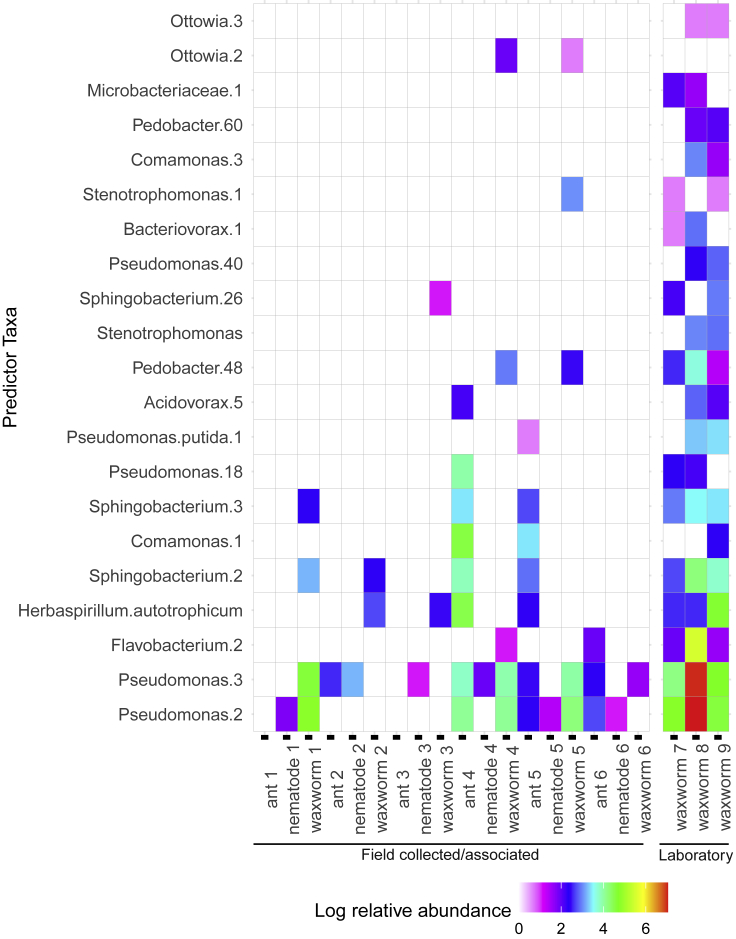
A total of 317 bacterial sequence variants (SVs) were present in field-collected ant samples that harbored nematodes in the wild, including multiple SVs identified as Spiroplasma sp., Serratia marcescens, Entomoplasma sp., Enterococcus sp., and Cutibacterium acnes, among others (Figure 4). A total of 274 unique SVs were present in field-collected nematodes which emerged from ant cadavers, including multiple SVs identified as Spiroplasma sp., Serratia marcescens, Entomoplasma sp., Pandorea sp., Flavobacterium sp., and Cutibacterium acnes, among others (Figure 4). Four SVs were shared between ant and nematode samples at an abundance of 1% and a prevalence of 70% (Table 3). Three of these SVs were variants of Serratia marcescens. A random forest analysis (77% accuracy) identified 13 SVs with differential abundance between ant and nematode samples (Figure S3) and demonstrated multiple SVs identified as Spiroplasma or Pandorea that were host species specific.
Figure 4.
Top 100 most abundant genera represented in the bacterial communities in field-collected Myrmica rubra ants and Pristionchus entomophagus nematodes from three field sites and Galleria mellonella larvae exposed to nematodes or not
Field sites on Mount Desert Island, Maine include Eden St. South (COA, #1–2), Old Farm Road (OFR, #3–4), and Sports Park (SP, #5–6) or laboratory for control larvae (#7–9).
Table 3.
Core bacterial sequence variants (SVs) between Myrmica rubra ants Pristionchus entomophagus nematodes and Galleria mellonella larvae co-cultured with nematodes
| Shared between | Core bacteria | Number of SVs |
|---|---|---|
| Ants and nematodes | Spiroplasma | 1 |
| Serratia marcescens | 3 | |
| Nematodes and nematode-exposed Galleria larvae | Spiroplasma | 1 |
| Serratia marcescens | 1 | |
| Pandoraea | 1 | |
| Pedobacter | 1 | |
| Ants and nematode-exposed Galleria larvae | Spiroplasma | 1 |
| Serratia marcescens | 1 | |
| Bacillus pumilus | 1 | |
| Pseudomonas | 2 | |
| Delftia acidovorans | 1 |
Core was defined at 70% prevalence across samples, with 1% minimum relative abundance for ant:nematodes or nematode:larvae comparisons or 1% and prevalence of 60% for ant:larvae comparisons.
Observed SV richness (Figure S6) did not vary significantly between ants and nematodes (F1,10 = 0.375, p = 0.554), whereas ant samples had higher evenness than nematode samples (F1,10 = 7.81, p = 0.019). Neither observed richness or evenness varied significantly between field sites for ant (F2,3 = 0.035, p = 0.966; F2,3 = 3.57, p = 0.161, respectively) or nematode samples (F2,3 = 0.37, p = 0.963; F2,3 = 0.527, p = 0.636, respectively).
A total of 563 bacterial SVs were present across all G. mellonella samples, and many commonly occurring bacterial SVs were identified as members of the genera Alcaligenes, Cellvibrio, Entomoplasma, Flavobacterium, Pandoraea, Pedobacter, Pseudomonas, Serratia marcescens, and Spiroplasma (Figure 4). G. mellonella larvae, including controls and those exposed to field-collected nematodes, shared 4 SVs at an abundance of 1% and prevalence of 70%, including three Pseudomonas and two Pedobacter species. There was no consistent pattern differentiating the most abundant taxa found in nematode-exposed versus control samples. Within the nematode-exposed G. mellonella larvae, there appears to be little consistency between abundant bacterial taxa by field site.
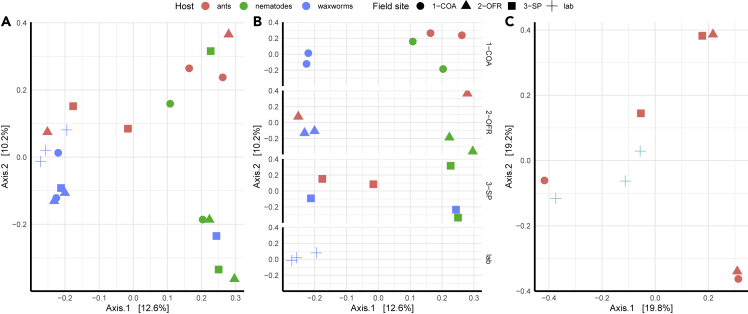
Across all samples, bacterial communities clustered (Figure 5A) based on host species (permANOVA, F2,20 = 1.68859, R2 = 0.16129, p < 0.001) but not by field site (p > 0.05; Figure 5B). For the field-collected ants and nematodes and the nematode-exposed moth larvae, there was no host x field site interaction, nor was the effect of the host stronger when compared within the field site (Figure 5B).
Figure 5.
Principal coordinate analysis of bacterial community similarity across field-collected Myrmica rubra ants, field-collected Pristionchus entomophagus nematodes, and laboratory-based Galleria mellonella larvae co-cultured with nematodes or not
Comparisons are provided for (A) all samples, (B) all samples by field collection site or laboratory (control), and (C) comparing nematode-exposed and control G. mellonella waxworm larvae. Field sites on Mount Desert Island, Maine include Eden St. South (COA), Old Farm Road (OFR), and Sports Park (SP), or laboratory for control waxworm larvae. Distance calculated using Jaccard unweighted similarity.
Testing the hypothesis of bacterial transfer by nematodes
There were two SVs shared across all ants, nematodes, and nematode-exposed G. mellonella larvae (n = 18), at an abundance of 1% and prevalence of 70%: Spiroplasma sp. and Serratia marcescens (Figure 1). When this was disaggregated by field site of the original ant colonies, hosts at COA (n = 6), as well as at Old Farm Road (OFR) (n = 6), shared only the same S. marcescens SV, and hosts at SP (n = 6) shared both those same Spiroplasma sp. and Serratia marcescens SVs, at an abundance of 1% and prevalence of 99% per site.
Random forest feature selection comparing bacterial communities between ants and nematode-exposed G. mellonella larvae identified 21 SVs with differential abundance between the sample groups, with a model accuracy of 75% (Figure S4). Several ant samples were characterized by a high abundance of taxa within the genus Serratia. In comparison, most nematode-exposed larvae were characterized by the genera Pedobacter and Pseudomonas. Six SVs were identified as shared between ant and larvae samples at an abundance of 1% and prevalence of 60% (Table 3). Two of these SVs were variants within the genus Pseudomonas.
Random forest feature selection comparing nematode to nematode-exposed G. mellonella larvae identified 20 SVs differentiating the sample groups, with an accuracy of 83% (Figure S5). Four SVs were shared between nematode and nematode-exposed larvae at an abundance of 1% and prevalence of 70% (Table 3). Each of these SVs was from different genera.
Nematode-exposed G. mellonella larvae were not significantly different from controls (p > 0.05), although samples were visually trending toward separate clustering (Figure 5C). Random forest feature prediction similarly showed little alteration of the collective bacterial state, with a model accuracy of only 66%.
Testing the hypothesis of bacterial community disruption by nematodes
Alternative to the idea that nematodes transfer bacteria from the environment to their ant host, we considered whether nematode exposure might disorder or depauperate the endobiotic community of insect hosts. Random forest feature selection identified 9 SVs which were more abundant in G. mellonella larvae controls than in any sample which was field collected or field associated (i.e., nematode-exposed larvae), with a model accuracy of 86% (Figure 6).
Figure 6.
Abundance of taxa identified as important community members associated with being in field-collected or associated hosts compared to laboratory-based control hosts
Samples are numbered by the transfer set (see Figure 4). The permutational random forest model had an 86% accuracy rate, and only statistically significant (p < 0.05) taxa are shown.
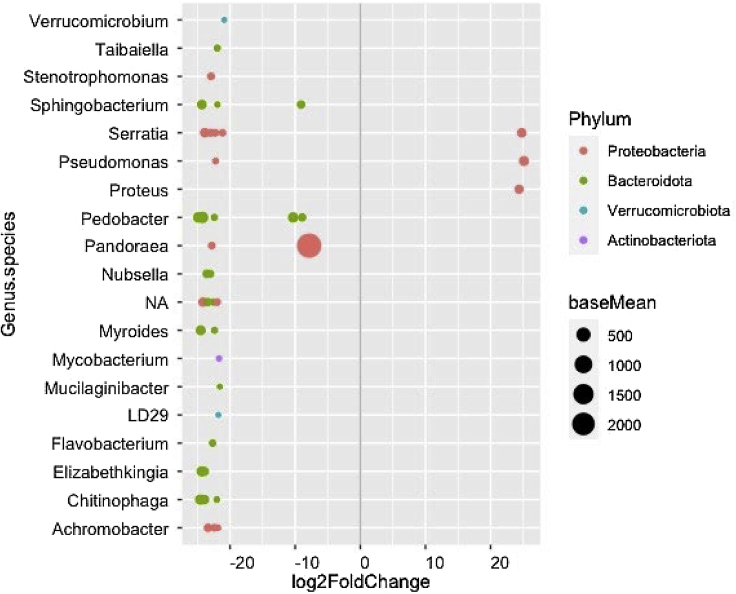
While total bacterial diversity visually appeared to be lower in nematode-exposed larvae than in controls (Figure S6), this was not significant for bacterial SV observed richness (W = 2, p = 0.092), evenness (W = 7, p = 0.714), or Shannon diversity (W = 3, p = 0.167). However, 16 bacterial SVs were found in lower abundance (p < 0.05) in nematode-exposed larvae than in controls (Figure 7), while three SVs identified as Serratia, Pseudomonas, and Proteus were found in higher abundance, all belonging to the Proteobacteria phylum.
Figure 7.
Bacterial taxa which were differentially abundant when comparing Galleria mellonella larvae co-cultured with Pristionchus entomophagus nematodes to control larvae
Positive fold changes indicate increased bacterial abundance (shown as baseMean) in nematode-exposed G. mellonella larvae, and negative fold changes indicate a reduction in bacterial abundance in nematode-exposed larvae.
Discussion
We examined the bacterial community associated with the necromenic nematode, P. entomophagus, and the potential for these nematodes to vector bacteria and contribute to mortality of their insect host. Demonstration that infection by P. entomophagus leads to M. rubra mortality would indicate that this nematode could represent an avenue for the biological control of this invasive ant in coastal Maine. While the use of nematodes as a biological control vector has clear logistical constraints, as discussed below, it represents a more targeted, safer, and perhaps self-sustaining approach than direct application of a chemical agent.
Nematode-carried bacteria and their acquisition from the environment
In the present study, only two (BNR, OR) of the three isolated nematode cultures resulted in significant insect mortality (Figure 2). This suggests differences between P. entomophagus populations from different sites, with one hypothesis being that bacteria associated with P. entomophagus vary between sites due to localized environmental selection on bacterial communities. Unlike many entomopathogenic nematodes (Steinernematidae and Heterorhabditidae), Pristionchus species are not associated with specific symbiotic bacteria but are capable of ingesting and carrying a diversity of bacteria (Poinar, 1969; von Lieven and Sudhaus, 2000), including species of Pseudomonas, Serratia, Enterobacter, and Bacillus (Rae et al., 2008). It has been demonstrated that Pristionchus nematodes sporadically infect insect hosts with pathogenic bacteria acquired from the environment (Al Own et al., 2010; Dieterich et al., 2008; Rae et al., 2008).
In the present study, Paenibacillus was isolated from the digestive tract of adult nematodes that emerged from ant cadavers (discussed more thoroughly in (Dumont, 2011)), and most adult nematodes readily consumed fluorescently labeled Pseudomonas aeruginosa from culture plates and maintained them in their gut environment (Figure 3). Adults that emerged from ant cadavers carried a variety of bacterial species on their cuticles but did not acquire and vector P. aeruginosa (Figure 3).
Conversely, juvenile nematodes appeared to seldom consume P. aeruginosa but harbored them on their cuticles (Figure 3). In dauer third stage juveniles, the second stage cuticle is retained in the third instar dauer juvenile state (Bellows and Fisher, 1999); thus, RFP-labeled P. aeruginosa may have been contained between the two cuticle layers and potentially remained between the cuticles during sampling. Adult and juvenile P. entomophagus rarely acquired RFP-labeled E. coli HB 101 from culture media, suggesting this strain is not an ideal bacterial transfer candidate as it is infrequently consumed by nematodes. This may be because of a lack of chemoattraction or perhaps the bacteria were unable to adhere to the cuticle. These nematodes can only infect ant hosts during the nematode's juvenile stage. Even if juvenile nematodes could frequently carry bacteria on the external cuticle, without their internal harborage, the bacteria may not be able to survive and multiply, thus making them poor vectors.
The life stages and morphology of nematodes may play a role in the attachment and retention of environmental bacteria to nematode surfaces as the cuticle is shed from one stage to the next and of insect hosts to void nematode juveniles before they establish and mature. For example, Poinar showed that dauer stage Pristionchus juveniles caused significantly fewer lethal infections when applied to G. mellonella larvae in comparison with samples containing a mix of dauer and juvenile stages (Poinar, 1969). Once established within the insect host, P. uniformis caused mortality in all subjects, yet many of the inoculated insect larvae were able to successfully void the nematodes, presumably passing dauer stage larvae through their digestive tracts before they could establish and mature (Poinar, 1969). In addition, the proteins and other secretions of other species of parasitic nematodes are known to be cytotoxic to their hosts (Chang et al., 2019). In our study, we found that G. mellonella mortality was significantly higher when they were exposed to nematodes, regardless of whether those nematodes were carrying the RFP-labeled P. aeruginosa strain PA14 and an apparent lack of transfer of these bacteria to larvae hemolymph. It is possible that wax moth larval mortality (Figure 2) was driven by other nematode-specific factors.
While bacteria have been isolated from the space between the two cuticular layers of the dauer stage juveniles, Pristionchus dauer stage juveniles do not retain the J2 cuticle as a protective sheath for the free-living stage (Gaugler, 2002). Collectively, these morphological differences between nematode species and life stages may moderate the transfer of bacteria from nematodes to insects and the subsequent mortality of the insect host, potentially explaining why Pristionchus nematode infections in ant colonies are not necessarily fatal.
Olfaction and chemoattraction profiles of Pristionchus spp. are highly diverse, allowing nematodes to find and colonize specific insect hosts (Hong and Sommer, 2006) and to find or avoid particular microorganisms. It is possible that carriage of certain bacteria by ants may draw nematode infection, rather than nematode infection of ants being the source of transfer of infectious bacteria to ants. In chemotaxis assays, P. entomophagus nematodes have been demonstrated to be strongly attracted to P. vulgaris bacteria (Rae et al., 2008). This may be a critical factor when designing biological control measures for M. rubra ants. In the present study, Proteus mirabilis and another Proteus sp. were not differentially abundant by host or by site (data not shown).
Pristionchus entomophagus was significantly more attracted to non-pathogenic specific soil- and beetle-derived bacteria than E. coli OP50, an artificial strain used in laboratory culturing of nematodes (Rae et al., 2008). Similarly, nematodes can recognize and avoid some entomopathogenic bacteria that negatively impact their fitness, including Bacillus thuringiensis and other bacilli, that are nematocidal or reduce fecundity in Pristionchus spp. (Rae et al, 2008, 2010; van Frankenhuyzen, 2009; Wei et al., 2003). P. entomophagus nematodes have been shown to be moderately attracted to Serratia marcescens and variably attracted to Pseudomonas strains (Rae et al., 2008). In the present study, Serratia and Pseudomonas were isolated from nematodes that emerged from M. rubra ant cadavers; nematodes acquired a strain of Pseudomonas from the environment; and strains were shared in the bacterial community of field-collected ants and nematodes, as well as G. mellonella larvae exposed to these nematodes in the lab.
Capacity to transfer bacteria from nematodes to insects
Several strains of Serratia marcescens, S. nematodiphila, and Pseudomonas protegens were isolated from the hemolymph of G. mellonella exposed to P. entomophagus cultures (Table 2; Figure 1). We were unable to visually determine if nematode-transferred Pseudomonas aeruginosa contributed to G. mellonella larvae mortality (discussed more thoroughly in (Michaud, 2013)). The high virulence of strain PA14 in G. mellonella and other insects (Jander et al., 2000) indicates it would have likely contributed to mortality if transfer were successful.
There are many potential reasons for a failure to transfer pathogenic bacteria, including community competition in the host, insufficient conditions for growth or survival, or insufficient bacteria present on the cuticle of juveniles and a low rate of transference. In this study, fluorescent microscopy suggested that P. aeruginosa in the digestive tract of nematode juveniles was rare. Thus, successful transfer of bacteria to insect larvae relied on the presence of the bacteria on the external cuticle of the nematodes. If labeled bacteria were present between the second and third stage juvenile cuticles rather than externally, its physical constraint may preclude transfer to larvae. Additionally, P. aeruginosa may have been washed off or mechanically removed as the nematodes moved through the sand in the experimental arena.
Serratia marcescens has previously been detected as part of the bacterial communities of other ant species such as Camponotus japonica (He et al., 2011), Formica cinerea (Sirviö and Pamilo, 2010), Anoplolepis gracilipes (Cooling et al., 2017), and Myrmica scabrinodis (Di Salvo et al., 2019). In the present study, we observed Serratia marcescens in the hemolymph of nematode-infected G. mellonella larvae and S. quinivorans from the external cuticles of nematodes collected from ant cadavers. Serratia marcescens has been shown to have insecticidal (Nishiwaki et al., 2007) and nematocidal (Rae et al., 2008) activity. Furthermore, S. marcescens has been used in the biological control of A. gracilipes ant colonies in Australia (Cooling et al., 2017). Serratia marcescens has also been shown to cause mortality in G. mellonella larvae but only when transferred via entomopathogenic nematodes and not by the bacteria alone (Ortega-Estrada et al., 2012). Sequencing of the bacterial community revealed that S. marcescens was among the top 10 SVs present in most ant and nematode bacterial communities, and three variants of S. marcescens were detected in the core microbiome shared by ant and nematode samples.
While our results suggest the possibility of transfer of S. marcescens to ants via nematode infection, ants may acquire this pathogen from elsewhere as they occur frequently in the environment and at low abundances in healthy insects (Grimont and Grimont, 1978). In some systems, ant acquisition of Serratia is hypothesized to occur through ingestion of aphids that have secondary endosymbioses with Serratia (He et al., 2011). Our results showed that ants' bacterial communities were characterized by a higher abundance of S. marcescens than nematode bacterial communities (Figure 1). Infecting nematodes may also provide a means of ingress for bacteria harbored outside the hemocoel but inside the ant's body. Ants are known to collect a diversity of microorganisms acquired with feeding and grooming in pellets within the buccal chamber in their heads (Hölldobler and Wilson, 1990). These pellets are held for a period of time before being expelled into refuse areas outside the nests (Eisner and Happ, 1962). Bacteria recovered within these pellets include Serratia and Pseudomonas species among others (Hansen et al., 1999; Zhang et al., 2018).
Results from our bacterial culturing experiment identified Pseudomonas fluorescens in the hemolymph of infected G. mellonella larvae and on the external cuticle of the nematodes. Bacterial community sequencing revealed that Pseudomonas SVs occurred in the top 10 genera of both ant and nematode samples but were in low abundances in all but one ant sample from site OFR. Pseudomonas have previously been reported to be pathogenic to insects (Chapuisat et al., 2007; Khan et al., 2016; dos Santos et al., 2009), as it produces the secondary metabolite hydrogen cyanide (Devi and Kothamasi, 2009), which inhibits the function of the enzyme cytochrome c oxidase, an integral part of the respiratory chain of the test organism Odontotermes obesus and other insects. Infection by Pseudomonas species can be fatal to other genera of nematodes (Khan et al., 2016) but did not impact survival of Pristionchus nematodes (Rae et al., 2008). Further, Pristionchus were attracted to Pseudomonas in culture (Rae et al., 2008). Pseudomonas species are commonly found in soil (Raaijmakers et al., 1997), so it is unclear if the occurrence of Pseudomonas in ant and nematode samples is related to intra-specific transfer of the bacteria or due to acquisition of the bacteria from the environment.
Other patterns in bacterial community structure in the field-collected samples include the common occurrence of Spiroplasma in nematode samples (Figure 4). Spiroplasma are often found in association with insect hosts (Duron et al., 2008) and can be a maternally transferred symbiont of M. rubra (Ballinger et al., 2018). A specific strain of Spiroplasma poulsonii has been shown to confer resistance of Drosophila neotestacea to infection by the parasitic nematode Howardula aoronymphium (Jaenike et al., 2010). In this study, nematodes had higher occurrences of Spiroplasma than ants had, but the core microbial community of ants and nematodes did include a Spiroplasma SV. Given the known relationship between Spiroplasma and M. rubra and the defensive role that Spiroplasma can take against nematodes, this study's finding of Spiroplasma in higher abundance in nematodes than in ants is unexpected. However, the presence of Spiroplasma in the bacterial communities of both ant and nematode samples suggests the potential transfer of these taxa from ants to nematodes.
Delftia sp., which was cultured in the present study from the cuticle of nematodes and the hemolymph of nematode-exposed G. mellonella larvae, are common soil-dwelling bacteria that have not been demonstrated to cause insect mortality. Delftia sp. has been identified previously in the hemolymph of insects (Hail et al., 2011). It is possible that Delftia sp. may be ingested by the insect host, where they are consumed by nematodes, and stick to the cuticle of the nematode as it exits the host.
Suitability of Pristionchus entomophagus nematodes as vectors
Our data suggest that Pristionchus entomophagus nematodes are potential candidates owing to their ability to acquire and transfer bacteria and their association with ants. Pristionchus nematodes are necromenic and proliferate inside insect cadavers, and a previous theory suggests that these nematodes are on the evolutionary path to parasitism (Weller et al., 2010). Pristionchus nematodes demonstrated differential chemoattraction to bacteria based on host association and have been shown to be able to suppress spore germination in their intestine, as well as using pathogenic spore-forming (e.g. Bacillus) and non-spore-forming pathogenic bacteria (e.g. Pseudomonas) as a food source (Rae et al., 2010). Pristionchus nematodes can be functionally resistant to insecticidal crystal proteins. However, there are several considerations and limitations to their use.
Dissemination of bacteria from nematode to host likely occurs through ingestion and defecation of intact bacteria (Chantanao and Jensen, 1969) and potentially through transfer of bacteria adhered to the cuticle or in the hemolymph (Dumont, 2011). The lack of bacterial transfer to the insect host in this study suggests the importance of internal harborage, as bacteria adhered to the external cuticle may be a more serendipitous and/or transient association. The absence of RFP-labeled P. aeruginosa, an experimental proxy for environmental bacteria, in the digestive tract of juveniles and high prevalence in the digestive tract of adults suggests different nutritional preferences, but there is currently no information on this. This provides a potential explanation for the decrease in juveniles with external fluorescence over time, as fewer environmentally located bacteria would be present to adhere to the cuticle as adults consume them.
Conclusions
The work presented demonstrates the complexity of the interactions between nematodes, insects, and bacteria. The experiments and field observations reported here suggest that the transfer of bacteria from Pristionchus entomophagus nematodes to ants is feasible and was possibly a contributor to ant mortality and death in wild colonies in Maine. While a single, clear, common causative agent of colony mortality did not emerge, Serratia marcescens and Spiroplasma SVs were present in ants, nematodes, and waxworm larvae, and Serratia were in increased abundance in waxworms infected by nematodes, relative to controls. Ants live in a microbially rich environment in soil, and infection by these and other bacteria species requires a vector, such as nematodes to breach the cuticular barrier that protects them externally as well as their buccal cavity, foregut, and hindgut regions. The use of an engineered biological control against invasive ant species, such as nematodes carrying specifically seeded bacterial species, is highly desirable, especially if the pathogenic bacteria are normally found in soil ecosystems and represents a low risk for biosafety control. Further studies are needed to confirm the feasibility and consistency of such an approach.
Limitations of the study
In the present study, we were unable to visually determine if nematode-transferred Pseudomonas aeruginosa contributed to G. mellonella larvae mortality. The putative lack of consumption of P. aeruginosa by juveniles would pose a challenge to using P. entomophagus for biological control, as the juvenile stage is needed for host infection to occur. The low sample numbers in the bacterial transfer group were a result of difficulty in sequencing, and this would need to be repeated with more power. Moreover, several mechanistic questions remain to be answered, including the dynamics and repeatability of bacterial acquisition from the environment, repeatability and dosage of successful vectoring to ants, and whether suitable pathogens to ants could be identified which would not introduce contaminating bacteria into the environment. Further laboratory experimentation is needed to validate and expand the scope of these findings, including transfer of nematodes to ants from different sites to assess mortality and validation of environmental bacteria acquisition of nematodes and vectoring into ants using tagged bacterial strains.
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli HB101 (p6TT1) bacteria expressing a red fluorescent protein | (Singer et al., 2010) | |
| Pseudomonas aeruginosa (strain PA14) | (Jander et al., 2000) | |
| Chemicals, peptides, and recombinant proteins | ||
| MoBio Soil Extraction kit | MoBio Laboratories, Inc., US | Catalog No. 12888-50 |
| QIAmp DNA Micro Kit | QIAgen | Cat. No. / ID: 56304. |
| Promega Wizard® Genomic DNA Ourification Kit | Promega, USA | Cat no. A1120 |
| Deposited data | ||
| Proofed 16S rRNA gene sequences | NCBI Genbank | GenBank accessions MT797825 - MT797845 |
| Raw 16S rRNA V3/V4 gene sequences from bacterial communities | NCBI Sequence Read Archive | BioProject Accession PRJNA646935 |
| Oligonucleotides | ||
| 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) | Integrated DNA Technologies, (Lane, 1991) | 27F |
| 1492R (5’-ACGGGCGGTGTGTAC-3’) | Integrated DNA Technologies, (Lane, 1991) | 1492R |
| 519R (5′-GTNTTACNGCGGCKGCTG-3’) | Integrated DNA Technologies, (Myer et al., 2016) | 519R |
| Software and algorithms | ||
| Nanodrop 1000 with version 3.3 software | ThermoScientific, USA | Version 3.3 |
| Interactive Tree of Life | (Letunic and Bork, 2019) | |
| MEGA | MEGA (Hall, 2013) | Version X |
| JMP | SAS Institute Inc. | Version 2012 |
| R Studio | (RCoreTeam, 2020) | R version 3.6.3 “Holding the Windsock” |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Suzanne Ishaq, sue.ishaq@maine.edu.
Materials availability
This study did not generate new unique reagents or insect lines.
Data and code availability
Proofed 16S rRNA gene sequences from cultured bacterial isolates can be accessed on NCBI (Bioproject PRJNA646935, GenBank accessions MT797825 - MT797845).
Raw 16S rRNA V3/V4 gene sequences from bacterial communities are publicly available under BioProject Accession PRJNA646935, from the NCBI Sequence Read archive (SRA).
Code is available upon request to Suzanne Ishaq (sue.ishaq@maine.edu) and can generally be found on the DADA2 (https://benjjneb.github.io/dada2/tutorial.html) and phyloseq (https://joey711.github.io/phyloseq/) tutorial pages.
Experimental model and subject details
Myrmica rubra colonies were collected at multiple sites in Acadia National Park Mount Desert Island, Maine (Table 1) in September 2010. Nests were located under natural (logs, rocks, etc.) and human (boards, plant pots, etc.) debris and exposed colonies were collected with an aspirator and transferred into plastic nest boxes (11 cm x 26 cm x 9 cm) or 19 L buckets with their nest soil. Colonies collected in buckets were stored at 4°C until used for experiments, when they would be transferred into nest boxes and held at 21°C. Each nest box contained a small portion of a cardboard egg carton covering a 3 x 3 x 3 cm3 piece of moistened sponge. The sponge was remoistened, and ants were fed ca. 2 g of sugar and tuna diet mix every 2 - 4 days.
Collection of nematodes: Ant colonies were inspected for dead individuals every 2-3 days. Dead worker ants were removed using sterile tweezers and surface sterilized by submerging in a 0.1% zephiran chloride solution for 30 sec (Roberts, 1973) , followed by two rinses in sterile dH2O, and drying on clean absorbent paper. After drying, cadavers from each nest were held in individual wells of 48-well microtiter plates, with cadavers from each colony maintained in their own separate plate. Plates were stored at ambient temperature (ca. 21°C) inside plastic bags containing damp paper towels to maintain high humidity (Evans et al., 2010). Cadavers were monitored for nematode emergence every other day. As nematodes emerged, they were transferred with tweezers from the wells and transferred to 50 mL centrifuge tubes with dH2O held at 10° C.
New M. rubra colonies were collected from field sites for each set of experiments to assure that the bacterial communities associated with the nematodes did not drift with repeated culturing. Nematode infections levels and successful recovery from different sites varied between collections. Nematodes recovered from colonies and locations that resulted in moderate to high ant mortality and high emergence of nematodes were chosen for each set of experiments.
G. mellonella infections and hemolymph collection: Last instar Galleria mellonella (Lepidoptera: Pyraidae) larvae were purchased from Petco (Bangor, ME) or Grubco Inc. (Fairfield, OH), and held at 5-10°C until used for nematode infections and/or bleeding experiments.
Method details
For infection, Galleria mellonella larvae were placed in 100 mL cups or 100 mm diameter Petri dishes with 20 g sterilized sand moistened with sterile dH20. Nematodes in dH20 solution were pipetted directly onto the sand and/or the dorsal cuticle of the larvae, which were incubated without food at ambient temperature (approximately 21°C) and monitored daily for signs of infection and death. White traps (Lacey, 2012; Stock and Goodrich-Blair, 2012) were used to harvest nematodes from dead larvae.
The hemolymph of larvae, infected and non-infected, was sampled for the presence of bacteria by bleeding caterpillars under aseptic conditions in a laminar flow hood. Caterpillars were removed from rearing/ infection cups, rinsed with dH20, and surface sterilized with 0.1% zephrin chloride as described previously (Roberts, 1973) or with 70% ethanol and 1% sodium hypochlorite (NaClO) as described previously (Lacey, 2012). After surface sterilization, a single proleg was cut with sterile, micro-dissecting scissors, and a microcapillary tube was used to draw the hemolymph that welled up from the wound. Slight pressure was applied to the body with soft forceps to exude as much sample as possible. Hemolymph samples were stored briefly at 4°C until bleeding of all specimens was completed, with plating or DNA extractions immediately following this process.
Isolation and identification of associated bacteria from nematodes emerged from M. rubra ant cadavers (Experiment 1)
Additional detail for experiment 1 can be found elsewhere (Dumont, 2011). The conceptual schematic for this work is visualized in Figure S7, as well as in the graphical abstract image created with BioRender.com.
Preparation of ant colonies
Multiple M. rubra colonies collected from different sites (Table 1) in September 2010, were maintained at 4°C for approximately 2 weeks prior to being transferred to nest boxes for use in bacterial culturing experiments. Colonies were maintained and monitored for dead ants and emergence of nematodes from ant cadavers. Colonies from two sites experiencing high levels of mortality and emergence of many nematodes were selected for use in this study, and nematodes were harvested from their corresponding cadaver plates.
Collection of externally located bacteria from nematodes
Approximately 50 μL of sterile distilled water was added to wells containing M. rubra cadavers and emerged nematodes. Nematodes from each well were pipetted into a 1.5 mL microcentrifuge tube filled with 1 mL of 1% Tween. Tubes were vortexed gently to mix and centrifuged for 10 sec at 13,000 RPM to concentrate the nematodes in the bottom of the tube. This stock solution was serial diluted four times, using a 1:10 ratio of rinse solution to distilled water. Three 300 μL aliquots of each 1×10-4 dilution were plated onto Trypticase Soy Agar (TSA). Plates were incubated at 29°C for 48 hr, after which, bacterial colonies were observed at 1-100X under a dissecting microscope and unique morphotypes were identified based on colony size, color, shape, and surface characteristics (Table S1; Figure S1A). Individual colony forming units of all unique morphotypes within a sample were transferred to fresh TSA plates. After two days of growth, monoculture plates were stored at 4°C.
Collection of digestive tract bacteria from nematodes (internal)
A total of 50 μL of sterile distilled water was added to selected wells in the 48-well plates housing the M. rubra cadavers and emerged nematodes. After gently mixing, three 5 μL aliquots were taken from each well and plated onto a contrasting black surface for counting. The number of nematodes in each aliquot was counted and averaged across each of the three aliquots for each site. The remaining 40 μL of each nematode solution was equilibrated to ½ the concentration of the least concentrated solution by adding sterile distilled water to each solution in the appropriate amount.
After standardization of nematode concentration, a 50 μL aliquot of nematode suspension was gently loaded onto a concavely folded piece of vacuum filter paper. The filter paper was loaded into an appropriately sized Buchner Funnel and attached to the laboratory vacuum system. Nematodes were continuously surfaced sterilized for 2-3 minutes by pipetting 1% bleach solution onto them, making sure not to spill the nematodes off the filter paper. After surface sterilization, the nematodes were rinsed with sterile distilled water in the same manner for 2-3 minutes. The filter paper was then loaded into a small Petri dish and flooded with sterile distilled water to dislodge the nematodes from the filter paper. After nematode presence was confirmed using a dissecting microscope, 300 μL aliquots of the nematode suspension were plated onto TSA agar. To assure inoculation of a sufficient number of nematodes, individual nematodes were pipetted out of the remaining solution using a 200 μL pipette tip and added to the TSA plate with the nematode suspension. It was determined that holding nematodes at room temperature until pipetting was best, as nematodes tended to stick to the surface of the Petri dish if refrigerated for long periods. Nematodes feeding and tunneling on the agar gave rise to colonies of bacteria excreted from the nematode digestive tract (Figure S1B). Plates were incubated at 29°C for 48 hr, after which trails of feeding nematodes and bacterial colonies were observable. Morphotypes were identified and individual colony forming units of unique morphotypes were transferred, grown, and stored as above.
Collection of bacteria in infected waxworm hemolymph
A total of 60 last instar G. mellonella larvae were inoculated with nematodes from the well plates of the two selected ant colonies/sites. Nematodes were harvested from six individual ants from each of the two colonies and transferred to individual infection dishes containing five larvae. Inoculated larvae were monitored daily, and the dead were collected and surface sterilized in 0.1% zephiran chloride solution. Sterilized cadavers were placed in individually marked Petri dishes for each set of five larvae. All cadavers were stored for 24 - 72 hours at 4°C until caterpillars could be bled for hemolymph en masse. Hemolymph collected from each set of five larvae was pooled for one sample and placed into a 1.5 mL microcentrifuge tube filled with 1mL of sterile dH2O. Four-fold serial dilutions were made and quadrant streaks of each 1×10-4 dilution were plated onto two Trypticase Soy Agar (TSA) plates for each of the 12 samples. Plates were incubated at 29°C for 48 hr, after which morphotypes were identified and individual colony forming units of unique morphotypes were transferred, grown, and stored as above.
DNA extraction and sequencing of bacterial isolates
Of the 45 bacterial isolates cultured (Table S1), 32 were selected for further evaluation via 16S rRNA sequencing. Due to the morphological similarities of isolates derived from samples from the same location and culture source (internal, external, hemolymph), samples originally derived from one well from each location were selected as representative samples of the external bacterial associates for sequencing. Fewer isolates were obtained from the internal and hemolymph samples and all were prepared for sequencing (Table S2).
All isolates were grown for 14-18 hr in 3 mL LB Broth, with a salt concentration of 5 g/L (Formedium), in 14 mL test tubes (VWR, USA) at 29°C and 90 rotations per minute (RPM). Broth cultures were centrifuged to pellet cells. Bacterial DNA was extracted using the Promega Wizard® Genomic DNA Purification Kit Cat no. A1120 (Promega, USA). DNA extract was stored at -20°C. Gel electrophoresis was used to assess presence of DNA extract using 0.8% Agarose gels made using 30 mL of TAE Buffer (tris base, acetic acid, and EDTA), 3 μL GelStar® GelStain (Lonza, USA) and 0.24 g Agarose (VSB Company, Cleveland, OH, USA). Samples were loaded using 5 μL of DNA and 1 μL of 6x loading dye (Gilbert, USA). The Lambda HindIII ladder (Promega) was used as the standard and samples were run at 90V for approximately 1 hr. Gels were visualized on an ultraviolet (UV) transilluminator and recorded with a remote shooting camera.
Polymerase chain reaction (PCR) amplification of bacterial 16s rRNA was conducted using the primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-ACGGGCGGTGTGTAC-3’) (Lane, 1991). Thermocycler conditions were as follows: 3 min at 95°C followed by 35 cycles of 15 s at 95°C, 30 s at 55°C, 1.5 min at 72°C, and a final step of 6.5 min at 72°C (Rae et al., 2008). Reactions were carried out at a volume of 20 μL. The PCR master mix recipe was derived from the original reaction conditions listed by Rae et al. (2008). After modifications, the final reaction mix included 9.6 μL H2O, 4 μL 5x PCR Buffer, 1.2 μL 25 mM MgCl2, 2 μL 2 mM dNTPs, 1 μL 10 μM 27f, 1 μL 10 μM 1492r, 0.2 μL GoTaq DNA Polymerase (Promega), and 1 μL of bacterial DNA.
PCR amplification was verified using 1.2% Agarose gels (30 μL of TAE Buffer, 3 μL GelStar® GelStain (Lonza, USA), and 0.36 g agarose). Invitrogen Low DNA Mass Ladder Cat. # 10068-013 (Invitrogen, USA) was used as a standard to detect the expected fragment length of 1465 bp; 5 μL of low mass ladder and 1 μL of 6x loading dye. Three microliters of PCR product and 2 μL of 2x loading dye was mixed and loaded for each sample. Gels were visualized on a UV transilluminator and recorded with a remote shooting camera. A fragment at the correct base pair size was taken to be a successful PCR run. PCR products were purified using a QiaQuick PCR Purification Kit (Qiagen, Venlo, Netherlands) following the Qiagen QiaQuick protocol, and stored at -20°C.
Purified PCR product quality was determined on a Nanodrop 1000 with version 3.3 software (ThermoScientific, USA). DNA concentrations were adjusted to 10 ng/μL using sterile water, as needed. Sanger sequencing was performed by the University of Maine Sequencing Facility (Orono, Maine, USA), using primers 27f and 1492r primers and ABI model 3730 DNA Sequencer with the XL Upgrade.
Transfer potential of pathogenic bacteria from environment to nematodes to other insect hosts (Experiment 2)
Additional detail for all methods used in experiment 2 can be found elsewhere (Michaud, 2013).
Harvest and storage of Pristionchus entomophagus nematodes
In September 2012, approximately 200 specimens were collected from each of 16 Myrmica rubra colonies located in eight sites in Acadia National Park and Orono, ME (Table 1) where Pristionchus entomophagus infection has previously been confirmed. Colonies were maintained and nematodes were collected from all colonies.
Virulence of P. entomophagus nematode populations
Galleria mellonella larvae were exposed to nematodes collected from ant cadavers from different colonies and monitored for mortality to assay nematode virulence. For each nematode population, larvae were exposed to low (20 - 25) and high (200 - 250) numbers of nematodes and compared to a control group exposed to no nematodes. Per site and dose assay, four replicate plates with 5 larvae per plate were exposed. The larvae were inoculated and maintained in 100 mm diameter Petri dishes and monitored daily for mortality for 14 days. Survival analysis was conducted using a general parametric model based on the Weibull distribution to examine difference in time to death between sites and nematode treatments (JMP, SAS Institute Inc. 2012). Dead larvae were removed and placed into white traps (Lacey, 2012), and nematodes were harvested once per week over 10 weeks by pipetting from the white traps into 50 mL centrifuge tubes. These nematode stocks were stored at 10° C. To replace old and dying nematode stocks 12 weeks after initial harvesting, nematode populations from the BNR site were exposed to G. mellonella as described above and collected via white traps (Figure S8).
Assessing uptake of RFP-labeled bacteria by P. entomophagus nematodes
To assess the ability of P. entomophagus to ingest and carry bacteria from their environment (Figure S2), nematodes from the BNR were used in the following experiments in which the nematodes were transferred to and held on culture media plates containing various bacteria treatments. For the first experiment, bacteria treatments included: 1) Escherichia coli HB101 (p6TT1) bacteria expressing a red fluorescent protein (Singer et al., 2010), and which have not been documented to be pathogenic towards nematodes; 2) bacterial-control plates containing non-labeled Paenibacillus sp. previously isolated from P. entomophagus; and 3) no-bacterial control plates without any additional bacteria added. For the second experiment bacteria treatments included: 1) the bacterial-control, non-labeled Paenibacillus plates; 2) no-bacterial control plates; and 3) plates containing a red fluorescent protein (p66TT1 plasmid d-tomato) labeled Pseudomonas aeruginosa (strain PA14) shown to be highly virulent in Caenorhabditis elegans worms and G. mellonella larvae with an LD50 of fewer than 10 bacteria (Jander et al., 2000), but not documented to be virulent in P. entomophagus nematodes.
To create treatment plates, 2.5 mL aliquots of nematode growth media (NGM; Carolina Biological, US) were poured into 45 mm diameter Petri plates under sterile conditions, streaked with bacteria, and incubated at 37° C for 48 - 72 hr to allow for bacterial growth. Four replicates were produced per treatment for a total of 12 plates. Approximately 200 - 400 nematodes in 140 - 150 μL of dH2O were pipetted onto each plate and cultured at 20°C for 48 hr to allow nematode grazing, after which nematodes were sampled for mortality and the evidence of acquiring the fluorescent protein.
To determine mortality, total dead (indicated by straight, stiff, or disintegrating nematodes) and living were counted in the entire plate (E. coli) or a subsample of ca. 50% of the plate sampling variable fields of view at 100X magnification under a dissecting microscope. To determine prevalence of uptake of environmental bacteria, 10 - 12 live juveniles and 10 - 12 live adults per plate with labeled E. coli or P. aeruginosa and unlabeled Paenibacillus sp. treatments were manually transferred to well slides and viewed on a Zeiss SteREO Discovery.V12 microscope with the Texas Red fluorescent filter (excitation 596/emission 615). The presence of external (cuticle) and internal fluorescence (digestive tract) was determined for adults and juveniles by visual observation (Figure S9). Repeated measures analysis of variance was conducted to examine the difference in the proportion survival and proportion with fluorescence between treatments over time in the E. coli trial, and a one-way ANOVA was used to examine treatment differences in the P. aeruginosa trial (JMP, SAS Institute Inc. version 2012). Proportion survival or fluorescence was transformed by arcsine square root for analysis (JMP, SAS Institute Inc. version 2012).
Transference of RFP-labeled bacteria from P. entomophagus nematodes to G. mellonella larvae
To determine if nematodes can transfer newly associated bacteria into their insect hosts, G. mellonella larvae were exposed to P. entomophagus nematodes carrying RFP-labeled bacteria. Three different treatments were established: 1) no nematode controls, 2) nematodes grown on plates with their naturally associated bacteria, and 3) nematodes grown on plates with RFP-labeled bacteria. There were 4 replicates per treatment for a total of 12 plates. Nine larvae were placed into each inoculation dish (100 mm2 diameter Petri dish with moistened autoclaved sand. Thirty-five live juvenile nematodes were transferred from their respective nematode growth agar to dish using a probe. Nematodes were added to a 50 μL sterile dH2O droplet within areas cleared of sand. Sand was gently pushed back into place to introduce nematodes into the moist sand. The petri dishes were stored in a humid chamber at ambient temperature (approximately 21°C) for 11 d, and larvae were monitored daily for mortality.
To assess bacterial transfer from nematodes, larvae were surface sterilized with a 1% sodium hypochlorite solution, which had been confirmed as non-fluorescing via direct observation using the Zeiss SteREO Discovery V12 microscope. One to two microliters of hemolymph were collected from three live larvae per plate 3 days after introduction of the nematodes, and then from each of the remaining larvae as they died over the 11-day duration of the experiment. Hemolymph was added to 20 μL of sterile dH2O on a standard microscope slide, covered with a 25×25 mm coverslip, and the edges were sealed with clear nail polish to prevent desiccation. The samples were viewed at 100X magnification on a Zeiss SteREO Discovery.V12 with the Texas Red fluorescent filter (excitation 543/emission 610) to determine presence/absence of fluorescence.
Microbial community profiling of bacteria associated with ants, nematodes, and infected G. mellonella larvae to determine in situ bacterial transfer between hosts
The design for this experiment is visualized in Figure S10, image created with BioRender.com. Three invasive ant colonies were collected from different sites on Mount Desert Island, Maine, in September 2015, held in rearing boxes in the laboratory. Ant colonies were maintained with regular watering and food, and dead ants were removed every 2-3 days and set up for emergence of nematodes as described above. Any adult nematodes that emerged were collected from wells and transferred to Petri dishes with Pasteur pipettes, reared in Baby Food (BF) agar according to procedures described previously (Stock and Nadler, 2006), and identified via molecular and morphological assessment (Lacey, 2012; Stock et al., 2009).
Last instar G. mellonella larvae were inoculated with nematode cultures originating from each of the three ant colonies. Twenty-μL aliquots of nematode solution (7 nematodes/mL dH2O) were applied directly onto the dorsal surface of each of five larvae per inoculation dish with 5 replicate dishes per nematode culture and 5 dishes of untreated larvae. Larvae were monitored daily and two dishes per nematode culture plus two dishes with untreated larvae were selected for hemolymph sampling 3 days post exposure when treated larvae showed signs of septicemia, but had not yet died (bloated, discolored, with only minor movement when prodded). Larvae were surface-sterilized, and hemolymph was collected and pooled from the five larvae per dish to yield 75-100 μL samples for DNA extraction.
Whole ants and nematodes were sampled from their original colonies and cultures for bacterial community analysis. Two samples of 5 ants were randomly collected from each nest box, transferred to sterile 1.5 mL snap tubes, and frozen for 15 min at -80°C prior to transfer to extraction tubes. Nematodes were collected by pipetting and manual transfer with a bent probe until two tubes with concentrations of 100 mixed aged nematodes in 500 μL dH2O were collected for each of the three nematode cultures. Neither ants nor nematode samples were surface sterilized.
DNA extraction and sequencing of bacterial communities
DNA was extracted from the whole ants, and whole nematodes samples, as well as from four individual whole G. mellonella larvae using the MoBio Soil Extraction kit (MoBio Laboratories, Inc., US) per the manufacturer’s protocol. DNA was extracted from hemolymph samples using QIAmp DNA Micro Kit for small sample volumes following the manufacturer’s protocols for Isolation of Genomic DNA from Small Volumes of Blood. A total of six nematode samples (2 per field site), six ant samples (2 per field site), and 13 G. mellonella larvae samples were selected for sequencing. Of the larvae samples, two were single whole larvae, two were single surface-sterilized larvae, and the remainder were hemolymph samples collected as described above from both nematode inoculated (infected, n = 6) and control (not inoculated, n = 2). Amplification of the 16S rRNA gene was done using eubacterial primers 27F (5’-AGRGTTTGATCMTGGCTCAG-3’) (Lane, 1991) and 519R (5′-GTNTTACNGCGGCKGCTG-3’) (Myer et al., 2016), and a 30-cycle PCR protocol per Molecular DNA Lab (MR DNA, Stillwater, TX). PCR products were pooled in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, US). Resulting library preparations were sequenced using the Illumina platform following the manufacturer’s protocols (MR DNA). Sequence data are publicly available under BioProject Accession PRJNA646935, from the NCBI Sequence Read archive (SRA).
Quantification and statistical analysis
Bacterial 16S rRNA gene identification (experiment 1)
Quality-edited bacterial 16S rRNA gene sequences were obtained from the University of Maine Sequencing Facility and identified using the Basic Local Alignment Search Tool (BLAST) online database provided by the National Center for Biological Information (NCBI, accessed 2011). Proofed sequences can be accessed on NCBI (Bioproject PRJNA646935, GenBank accessions MT797825 - MT797845). A phylogenetic tree was generated by aligning isolates and their closest matches in MEGA ver X (Hall, 2013) using the MUSCLE algorithm, and calculating branch lengths with Maximum Likelihood algorithm. Tree was visualized in the Interactive Tree of Life (Letunic and Bork, 2019).
Survival analysis (experiment 2)
Survival analysis was conducted using a general parametric model based on the Weibull distribution to examine difference in time to death between nematode treatments and control (JMP, SAS Institute Inc. version 2012).
Sequence data processing (experiment 3)
Raw data were denoised, barcodes were removed, and forward and reverse reads merged into contiguous sequences (i.e., ‘contigs’) by MR DNA. Resulting FASTQ files were processed in RStudio using R version 3.6.3 “Holding the Windsock” (RCoreTeam, 2020), following the DADA2 pipeline (Callahan et al., 2016). Sequence quality was measured and plotted using the ShortRead package (Morgan et al., 2009) and base R plotting. The first 15 bases and last 50 bases of reads were removed due to low sequence quality (sequence quality scores < 20). Error rates for each sequencing run were calculated, sequence variants identified, and chimeric reads were removed using DADA2. Taxonomy was assigned using the Silva ver. 138 (Pruesse et al., 2007) taxonomic training database for DADA2, and any sequences which matched as eukaryotic mitochondria were removed using the dplyr package (Wickham et al., 2015). Sequencing runs, along with taxonomy and metadata, were merged into one object using the phyloseq package (McMurdie and Holmes, 2013). This phyloseq object was then subset into ant, nematode, and G. mellonella larvae groups, as needed for analytical comparisons. All subset groups were pruned to remove null samples or taxa (phyloseq) and rarefied to 5,000 bacterial sequence variants (SV)s per sample for data analysis that included larvae only, or 10,000 SVs per sample for data analysis that included ants and nematodes.
Initial exploration of the data showed that two whole larvae samples that were prepared using surface sterilization prior to microbial DNA extraction had bacterial profiles that were distinct from the remainder of the larvae samples. These two samples were removed from the dataset along with a positive control sample that had been spiked with Bacillus bacteria.
Observed SV richness and evenness were calculated for all ant, nematode, and G. mellonella larvae samples in the package phyloseq. A Shapiro-Wilkes test was used to test if diversity data were normally distributed. Differences in observed richness and evenness between sample groups were tested using an ANOVA for normally distributed data, and a Wilcoxon test for those data that were not normally distributed. To determine differential abundance of bacteria SVs across sample groups, random forest classification algorithms were run using the rfpermute package (Archer, 2020). Principal Coordinate Analysis (PCoA) was performed using Jaccard’s distance to explore similarity of samples based on host species identity and host infection status. PCoA was visualized for all samples, then separately for G. mellonella larvae samples. The taxa belonging to the core microbial community between ant and nematode samples at an abundance of at least 1% and a prevalence of 70% -90%, noted in the respective results, were identified using the microbiome package (Lahti and Shetty, 2020).
Acknowledgments
We thank our scientific technicians, Tamara Levistsky, Jennifer Lund, and Elissa Ballman for their invaluable assistance with sampling, laboratory maintenance and experimentation, and oversight of students; the National Park Service at Acadia National Park and Park biologists, David Manski, Bruce Connery, and Judy Hazen-Connery for their permission and assistance with sample acquisitions; Drs. Michelle Goody and the late John Singer for their assistance with fluorescent microscopy; Dr. Frank Drummond for guiding students with the statistical analyses of bioassay data; and Dr. Amanda Klemmer for her assistance with initial visualization of bacterial community data. This project was supported by the University of Maine: Maine Agricultural and Forest Experiment Station (MAFES), the Honors College, and the Center for Undergraduate Research Fellowships.
Author contributions
S.P.S. and E.G. conceived of the original project and performed the molecular and morphological characterization of the nematodes and provided guidance for all subsequent work on this project. E.G. oversaw and directed sample collection and processing, isolations and maintenance of cultures, and infections and bioassays with students and technical staff in her laboratory. J.D.M. and E.G. oversaw infection and DNA extraction for community analysis. J.E.D. performed the culturing, bacterial isolation, and culture identification. A.M. performed virulence bioassays and visualization of labeled bacteria on and in nematodes and assessed transfer during infection, and S.L.I. taught and oversaw DNA data analysis and contributed to writing and reviewing the manuscript. S.S. and A.H. performed data analysis and contributed to writing the manuscript. All authors reviewed and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102663.
Contributor Information
Suzanne L. Ishaq, Email: sue.ishaq@maine.edu.
Eleanor Groden, Email: groden@maine.edu.
Supplemental information
References
- Al Own, F., Feil, E., Stock, S.P., Waterfield, N.R. 2010. Serratia proteamaculans strains associated with novel insect pathogenic nematodes in UK soils. Presented at the 2nd NEMASYM Workshop, International Symbiosis Society.
- Archer, E., 2020. rfPermute. CRAN. 2.1, 2020. https://cran.r-project.org/web/packages/rfPermute/rfPermute.pdf.
- Arevalo H.A., Groden E. University of Florida; 2007. European Fire Ant - Myrmica rubra Linnaeus (No. EENY-410) [Google Scholar]
- Ballinger M.J., Moore L.D., Perlman S.J. Evolution and diversity of inherited Spiroplasma symbionts in Myrmica ants. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02299-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellows T.S., Fisher T.W. A perspective on pathogens as biological control agents for insect pests. In: Fisher T.W., Bellows T.S., Caltagirone L.E., Dahlsten D.L., Huffaker C.B., Gordh G., editors. Handbook of Biological Control: Principles and Applications of Biological Control. Academic Press; 1999. pp. 517–545. [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.Z., Serra L., Lu D., Mortazavi A., Dillman A.R. A core set of venom proteins is released by entomopathogenic nematodes in the genus Steinernema. PLoS Pathog. 2019;15:e1007626. doi: 10.1371/journal.ppat.1007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantanao A., Jensen H.J. Saprozoic nematodes as carriers and disseminators of plant pathogenic bacteria. J. Nematol. 1969;1:216–218. [PMC free article] [PubMed] [Google Scholar]
- Chapuisat M., Oppliger A., Magliano P., Christe P. Wood ants use resin to protect themselves against pathogens. Proc. Biol. Sci. 2007;274:2013–2017. doi: 10.1098/rspb.2007.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooling M., Gruber M.A.M., Hoffmann B.D., Sébastien A., Lester P.J. A metatranscriptomic survey of the invasive yellow crazy ant, Anoplolepis gracilipes, identifies several potential viral and bacterial pathogens and mutualists. Insectes Soc. 2017;64:197–207. [Google Scholar]
- Czechowski W., Radchenko A., Czechowska W. Museum and Institute of Zoology Warszawa; 2002. The Ants (Hymenoptera, Formicidae) of Poland. [Google Scholar]
- Devi K.K., Kothamasi D. Pseudomonas fluorescens CHA0 can kill subterranean termite Odontotermes obesus by inhibiting cytochrome c oxidase of the termite respiratory chain. FEMS Microbiol. Lett. 2009;300:195–200. doi: 10.1111/j.1574-6968.2009.01782.x. [DOI] [PubMed] [Google Scholar]
- Dieterich C., Clifton S.W., Schuster L.N., Chinwalla A., Delehaunty K., Dinkelacker I., Fulton L., Fulton R., Godfrey J., Minx P. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 2008;40:1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman A.R., Chaston J.M., Adams B.J., Ciche T.A., Goodrich-Blair H., Stock S.P., Sternberg P.W. An entomopathogenic nematode by any other name. PLoS Pathog. 2012;8:e1002527. doi: 10.1371/journal.ppat.1002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Salvo M., Calcagnile M., Talà A., Tredici S.M., Maffei M.E., Schönrogge K., Barbero F., Alifano P. The microbiome of the <iMaculinea-Myrmica host-parasite interaction. Sci. Rep. 2019;9:8048. doi: 10.1038/s41598-019-44514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos P.F., Fonseca A.R., Sanches N.M. [Ants (Hymenoptera: Formicidae) as vectors for bacteria in two hospitals in the municipality of Divinópolis, state of Minas Gerais] Rev. Soc. Bras. Med. Trop. 2009;42:565–569. doi: 10.1590/s0037-86822009000500016. [DOI] [PubMed] [Google Scholar]
- Dumont J.E. University of Maine; 2011. Characterization of the Microbial Associates of Nematodes Pathogenic to Myrmica Rubra (Bachelor’s of Science ) [Google Scholar]
- Duron O., Bouchon D., Boutin S., Bellamy L., Zhou L., Engelstädter J., Hurst G.D. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008;6:27. doi: 10.1186/1741-7007-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner T., Happ G.M. The infrabuccal pocket of a Formicine ant: a social filtration device. Psyche. 1962;69:107–116. [Google Scholar]
- Elmes G.W., Wardlaw J.C., Nielsen M.G., Kipyatkov V.E., Lopatina E.B., Radchenko A.G., Barr B., Others Site latitude influences on respiration rate, fat content and the ability of worker ants to rear larvae: a comparison of Myrmica rubra (Hymenoptera: Formicidae) populations over their European range. Eur. J. Entomol. 1999;96:117–124. [Google Scholar]
- Evans H.C., Groden E., Bischoff J.F. New fungal pathogens of the red ant, Myrmica rubra, from the UK and implications for ant invasions in the USA. Fungal Biol. 2010;114:451–466. doi: 10.1016/j.funbio.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Garnas J., Groden E., Drummond F.A. Mechanisms of competitive displacement of native ant fauna by invading Myrmica rubra (Hymenoptera: Formicidae) populations. Environ. Entomol. 2014;43:1496–1506. doi: 10.1603/EN14079. [DOI] [PubMed] [Google Scholar]
- Gaugler R. CABI; 2002. Entomopathogenic Nematology. [Google Scholar]
- Grimont P.A., Grimont F. The genus Serratia. Annu. Rev. Microbiol. 1978;32:221–248. doi: 10.1146/annurev.mi.32.100178.001253. [DOI] [PubMed] [Google Scholar]
- Groden, E., Stock, S.P., 2010. Establishment of a new host relationship with an invasive? Or hitchhiker across the Atlantic? Presented at the Second NEMASYM Workshop, International Symbiosis Society.
- Groden E., Drummond F.A., Stack L.B., Graham C. University of Maine Cooperative Extension; 2004. European Fire Ant: A New Invasive Insect in Maine (No. 2550) [Google Scholar]
- Groden E., Drummond F.A., Garnas J., Franceour A. Distribution of an invasive ant, Myrmica rubra (Hymenoptera: Formicidae), in Maine. J. Econ. Entomol. 2005;98:1774–1784. doi: 10.1093/jee/98.6.1774. [DOI] [PubMed] [Google Scholar]
- Hail D., Lauzìere I., Dowd S.E., Bextine B. Culture independent survey of the microbiota of the glassy-winged sharpshooter (Homalodisca vitripennis) using 454 pyrosequencing. Environ. Entomol. 2011;40:23–29. doi: 10.1603/EN10115. [DOI] [PubMed] [Google Scholar]
- Hall B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013;30:1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- Hansen L.D., Spangenberg W.J., Gaver M.M. The infrabuccal chamber of Camponotus modoc (Hymenoptera: Formicidae): ingestion, digestion, and survey of bacteria. In: Robinson W.H., Rettich F., Rambo G.W., editors. Proceedings of the 3rd International Conference on Urban Pests. Presented at the International Conference on Urban Pests. 1999. pp. 211–219. [Google Scholar]
- He H., Chen Y., Zhang Y., Wei C. Bacteria associated with gut lumen of Camponotus japonicus Mayr. Environ. Entomol. 2011;40:1405–1409. doi: 10.1603/EN11157. [DOI] [PubMed] [Google Scholar]
- Herrmann M., Mayer W.E., Sommer R.J. Nematodes of the genus Pristionchus are closely associated with scarab beetles and the Colorado potato beetle in Western Europe. Zoology. 2006;109:96–108. doi: 10.1016/j.zool.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Herrmann M., Mayer W.E., Sommer R.J. Sex, bugs and Haldane’s rule: the nematode genus Pristionchus in the United States. Front. Zool. 2006;3:14. doi: 10.1186/1742-9994-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölldobler B., Wilson E.O. Belknap Press; 1990. The Ants. [Google Scholar]
- Hong R.L., Sommer R.J. Chemoattraction in Pristionchus nematodes and implications for insect recognition. Curr. Biol. 2006;16:2359–2365. doi: 10.1016/j.cub.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Jaenike J., Unckless R., Cockburn S.N., Boelio L.M., Perlman S.J. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science. 2010;329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- Jander G., Rahme L.G., Ausubel F.M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.R., Mohidin F.A., Khan U., Ahamad F. Native Pseudomonas spp. suppressed the root-knot nematode in in vitro and in vivo, and promoted the nodulation and grain yield in the field grown mungbean. Biol. Control. 2016;101:159–168. [Google Scholar]
- Lacey L.A., editor. Manual of Techniques in Invertebrate Pathology. 2nd ed. Academic Press; 2012. pp. 1–484. [Google Scholar]
- Lahti, L., Shetty, S., 2020. Introduction to the microbiome R package. CRAN. 2020. https://www.bioconductor.org/packages/release/bioc/vignettes/microbiome/inst/doc/vignette.html.
- Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee K., Garnas J., Drummond F., Groden E. Homopterans and an invasive red ant, Myrmica rubra (L.), in Maine. Environ. Entomol. 2012;41:59–71. doi: 10.1603/EN11046. [DOI] [PubMed] [Google Scholar]
- Michaud A.M. University of Maine; 2013. Alteration of Microflora of the Facultative Parasitic Nematode Pristionchus entomophagus and its Potential Application as a Biological Control Agent (Bachelor’s of Science) [Google Scholar]
- Morgan M., Anders S., Lawrence M., Aboyoun P., Pagès H., Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer P.R., Kim M., Freetly H.C., Smith T.P.L. Evaluation of 16S rRNA amplicon sequencing using two next-generation sequencing technologies for phylogenetic analysis of the rumen bacterial community in steers. J. Microbiol. Methods. 2016;127:132–140. doi: 10.1016/j.mimet.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Naumann K., Higgins R.J. The European fire ant (Hymenoptera: Formicidae) as an invasive species: impact on local ant species and other epigaeic arthropods. Can. Entomol. 2015;147:592–601. [Google Scholar]
- National Biological Information Infrastructure (NBII) & IUCN/SSC Invasive Species Specialist Group (ISSG) Ecology of Myrmica rubra [WWW Document]. Global Invasive Species Database. 2009. http://issg.org/database/species/ecology.asp?si=1014&fr=1&sts=&lang=EN
- Nishiwaki H., Ito K., Shimomura M., Nakashima K., Matsuda K. Insecticidal bacteria isolated from predatory larvae of the antlion species Myrmeleon bore (Neuroptera: Myrmeleontidae) J. Invertebr. Pathol. 2007;96:80–88. doi: 10.1016/j.jip.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Ortega-Estrada M.D.J., Rincón-Castro M.C.D., Basurto-Ríos R., Toledo J., Ibarra J.E. Phoresis between Serratia marcescens and Steinernema carpocapsae (Rhabditida: Steinernematidae) during infection of Galleria mellonella (Lepidoptera: Pyralidae) larvae. Florida Entomologist. 2012;91:120–127. [Google Scholar]
- Ouellette G.D., Drummond F.A., Choate B., Groden E. Ant diversity and distribution in Acadia National Park, Maine. Environ. Entomol. 2010;39:1447–1456. doi: 10.1603/EN09306. [DOI] [PubMed] [Google Scholar]
- Pitts J.P., Camacho G.P., Gotzek D., Mchugh J.V., Ross K.G. Revision of the fire ants of the Solenopsis saevissima species-group (Hymenoptera: Formicidae) Proc. Entomol. Soc. Wash. 2018;120:308–411. [Google Scholar]
- Poinar G.O. Diplogasterid nematodes (Diplogasteridae: Rhabditida) and their relationship to insect disease. J. Invertebr. Pathol. 1969;13:447–454. [Google Scholar]
- Poinar G.O. Nematodes as facultative parasites of insects. Annu. Rev. Entomol. 1972;17:103–122. [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J.M., Weller D.M., Thomashow L.S. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 1997;63:881–887. doi: 10.1128/aem.63.3.881-887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae R., Riebesell M., Dinkelacker I., Wang Q., Herrmann M., Weller A.M., Dieterich C., Sommer R.J. Isolation of naturally associated bacteria of necromenic Pristionchus nematodes and fitness consequences. J. Exp. Biol. 2008;211:1927–1936. doi: 10.1242/jeb.014944. [DOI] [PubMed] [Google Scholar]
- Rae R., Iatsenko I., Witte H., Sommer R.J. A subset of naturally isolated Bacillus strains show extreme virulence to the free-living nematodes Caenorhabditis elegans and Pristionchus pacificus. Environ. Microbiol. 2010;12:3007–3021. doi: 10.1111/j.1462-2920.2010.02278.x. [DOI] [PubMed] [Google Scholar]
- RCoreTeam . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing.https://www.r-project.org/ [Google Scholar]
- Roberts L.S. Modifications in media and surface sterilization methods for in vitro cultivation of Hymenolepis diminuta. J. Parasitol. 1973;59:474–479. [PubMed] [Google Scholar]
- Singer J.T., Phennicie R.T., Sullivan M.J., Porter L.A., Shaffer V.J., Kim C.H. Broad-host-range plasmids for red fluorescent protein labeling of gram-negative bacteria for use in the zebrafish model system. Appl. Environ. Microbiol. 2010;76:3467–3474. doi: 10.1128/AEM.01679-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirviö A., Pamilo P. Multiple endosymbionts in populations of the ant Formica cinerea. BMC Evol. Biol. 2010;10:335. doi: 10.1186/1471-2148-10-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock S.P., Glazer I., Boemare N., Vandenberg J., editors. Insect Pathogens: Molecular Approaches and Techniques. CABI Publishing; 2009. pp. 1–440. [Google Scholar]
- Stock S.P., Goodrich-Blair H. Nematode parasites, pathogens and associates of insects and invertebrates of economic importance. In: Lacey Lawrence., editor. Manual of Techniques in Invertebrate Pathology. Vol. 2. Academic Press; San Diego, CA: 2012. pp. 373–426. (Manual of Techniques in Invertebrate Pathology). [Google Scholar]
- Stock S.P., Nadler S. Morphological and molecular characterisation of Panagrellus spp. (Cephalobina: Panagrolaimidae): taxonomic status and phylogenetic relationships. Nematology. 2006;8:921–938. [Google Scholar]
- Sudhaus W. Vol. 12. 2008. Evolution of insect parasitism in rhabditid and diplogastrid nematodes; pp. 143–161. (Advances in Arachnology and Developmental Biology). [Google Scholar]
- van Frankenhuyzen K. Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertebr. Pathol. 2009;101:1–16. doi: 10.1016/j.jip.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Verble-Pearson R., Pearson S. European fire ant presence decreases native Arboreal insect abundance in Acadia National Park, Maine, USA. naar. 2016;36:162–165. [Google Scholar]
- von Lieven A.F., Sudhaus W. Comparative and functional morphology of the buccal cavity of Diplogastrina (Nematoda) and a first outline of the phylogeny of this taxon. J. Zoolog. Syst. 2000;38:37–63. [Google Scholar]
- Wahab A. Untersuchungen über Nematoden in den drüsen des kopfes der Ameisen (Formicidae) Z. für Morphol. Ökol. Tiere. 1962;52:33–92. [Google Scholar]
- Wei J.-Z., Hale K., Carta L., Platzer E., Wong C., Fang S.-C., Aroian R.V. Bacillus thuringiensis crystal proteins that target nematodes. Proc. Natl. Acad. Sci. U S A. 2003;100:2760–2765. doi: 10.1073/pnas.0538072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller A.M., Mayer W.E., Rae R., Sommer R.J. Quantitative assessment of the nematode fauna present on Geotrupes dung beetles reveals species-rich communities with a heterogeneous distribution. J. Parasitol. 2010;96:525–531. doi: 10.1645/GE-2319.1. [DOI] [PubMed] [Google Scholar]
- Wetterer J.K., Radchenko A.G. Worldwide spread of the ruby ant, Myrmica rubra (Hymenoptera: Formicidae) Myrmecol. News. 2011;14:87–96. [Google Scholar]
- Wickham H., Francois R., Henry L., Müller K. R Found. Stat. Comput.; 2015. Dplyr: A Grammar of Data Manipulation. R Package Version 0.4. 3.https://CRAN.R-project.org/package=dplyr [Google Scholar]
- Zhang K., Wei C., Nan X., Wang Y., He H. Composition and diversity of microbes in the infrabuccal pocket of Camponotus japonicus (Hymenoptera: Formicidae) Acta Entomol. Sin. 2018;61:686–697. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proofed 16S rRNA gene sequences from cultured bacterial isolates can be accessed on NCBI (Bioproject PRJNA646935, GenBank accessions MT797825 - MT797845).
Raw 16S rRNA V3/V4 gene sequences from bacterial communities are publicly available under BioProject Accession PRJNA646935, from the NCBI Sequence Read archive (SRA).
Code is available upon request to Suzanne Ishaq (sue.ishaq@maine.edu) and can generally be found on the DADA2 (https://benjjneb.github.io/dada2/tutorial.html) and phyloseq (https://joey711.github.io/phyloseq/) tutorial pages.