Abstract
Cancer metastasis is largely incurable and accounts for 90% of breast cancer deaths, especially for the aggressive basal-like or triple negative breast cancer (TNBC). Combining patient database analyses and functional studies, we examined the association of integrin family members with clinical outcomes as well as their connection with previously identified microRNA regulators of metastasis, such as miR-206 that inhibits stemness and metastasis of TNBC. Here we report that the integrin receptor CD49b-encoding ITGA2, a direct target of miR-206, promotes breast cancer stemness and metastasis. ITGA2 knockdown suppressed self-renewal related mammosphere formation and pluripotency marker expression, inhibited cell cycling, compromised migration and invasion, and therefore decreased lung metastasis of breast cancer. ITGA2 overexpression reversed miR-206-caused cell cycle arrest in G1. RNA sequencing analyses revealed that ITGA2 knockdown inhibits genes related to cell cycle regulation and lipid metabolism, including CCND1 and ACLY as representative targets, respectively. Knockdown of CCND1 or ACLY inhibits mammosphere formation of breast cancer cells. Overexpression of CCND1 rescues the phenotype of ITGA2 knockdown-induced cell cycle arrest. ACLY-encoded ATP citrate lyase is essential to maintain cellular acetyl-CoA levels. CCND1 knockdown further mimics ITGA2 knockdown in abolishing lung colonization of breast cancer cells. We identified that the low levels of miR-206 as well as high expression levels of ITGA2, ACLY and CCND1 are associated with an unfavorable relapse-free survival of the patients with estrogen receptor-negative or high grade breast cancer, especially basal-like or TNBC, possibly serving as potential biomarkers of cancer stemness and therapeutic targets of breast cancer metastasis.
Keywords: ACLY, Breast cancer, CCND1, CD49b, Integrins, ITGA2, Metastasis, Stemness
Introduction
Metastasis causes a majority of cancer related deaths, estimated 9.6 million globally in 2018.1 Breast cancer is the most common cancer in women other than skin cancer and its five-year survival rate decreases to 27% when distant metastases develop.2,3 Among all breast cancers, one of the most aggressive subtypes with early metastases is triple negative breast cancer (TNBC) which largely overlaps with basal-like breast cancer, and has most limited options for targeted therapy so is associated with a poor prognosis.4 Therefore, it is imperative to identify metastasis drivers and novel targets of TNBC.
Over the last two decades, human tumor cells with enriched stem cell properties (stemness) have been identified to initiate tumor growth, and demonstrated to accelerate progression with key attributes of resistance to therapies and evasion to immune attacks.5, 6, 7, 8, 9, 10, 11, 12 Nevertheless, comparing to its importance in primary tumor growth, cancer stemness might be more indispensable for metastasis initiation,13,14 including clustered circulating tumor cells mediated polyclonal metastasis.15,16 Genetic and epigenetic aberrations are known to enable tumor plasticity and stemness.9,17, 18, 19 However, targeting of cancer stemness and metastasis has yet to be widely achieved due to the lack of comprehensive understanding of potentially heterogeneous stemness drivers and modulators. In this study we focus on identifying and linking metastasis-driving stemness factors and pathways to targetable surface receptors. We report here a new pathway coupling integrin CD49b to its upstream and downstream regulation mechanisms.
Integrins include a large family of over twenty subunit molecules forming αβ heterodimers with binding affinity for various extracellular membrane components. Many integrins are key players in physiological and pathological processes.20 However, the intrinsic roles and signaling pathways of many integrins in breast cancer metastasis are not fully elucidated. Our previous studies have demonstrated that microRNAs (miRs) regulate breast cancer chemoresistance and progression in TNBC.21, 22, 23, 24 Specifically, miR-206 suppresses breast cancer stemness and lung metastasis.21 Microarray-based global transcriptome analyses revealed that ITGA2, which encodes a surface integrin receptor CD49b (integrin α2), is one of the genes suppressed by miR-206. Further clinical correlation studies identified that ITGA2 expression is one of the few integrin alpha genes associated with unfavorable of relapse-free survival (RFS) of basal-like breast cancer patients. CD49b is relatively known to be expressed in immune cells especially in regulatory T cells (Treg).25 However, its tumor-intrinsic role and regulatory signaling in breast cancer progression are largely elusive.
To test the hypothesis that CD49b is stemness driver for metastasis of breast cancer, we utilized multiple TNBC model systems in vitro and in vivo to assess the functional importance of CD49b (ITGA2), and to identify its signaling pathways and targets. In this study, we found that CD49b enhances breast cancer stemness and metastasis, requiring the genes involved in lipid metabolism and cell cycle pathways, such as ACLY and CCND1, respectively.
Materials and methods
Animal studies
All mice used in this study were kept in specific pathogen-free facilities in the Animal Resources Center at Northwestern University and Case Western Reserve University. All animal procedures complied with the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the respective Institutional Animal Care and Use Committees. Animals were randomized by age and weight. The exclusion criterion of mice from experiments was sickness or conditions unrelated to tumors. Sample sizes were determined based on the results of preliminary experiments. NSG and NOD-Scid mice were used for tail vein injection experiments with MDA-MB-231 cells and 4T1 cells expressing luciferase2-fused eGFP (pFU-L2G) or tdTomato (pFU-L2T).14 For spontaneous metastasis experiments the murine mouse cell lines (4T1) were injected in the mammary fat pad of BALB/c mice. Imaging of all in vivo experiments was performed using a Perkin Elmer IVIS.
Human databases and cell line expression analyses
Relapse free survival plots were obtained from KM-Plot (www.Kmplot.com/analysis).26 This tool downloads gene expression data, relapse free and overall survival information from GEO (Affymetrix microarrays only), EGA and TCGA. It uses a PostgreSQL server, which integrates gene expression and clinical data simultaneously. KM-Plot analyzes the prognostic value of a particular gene by splitting the patient samples into two groups according to various quantile expressions of the proposed biomarker and the best cut off was chosen for each analysis. The hazard ratio with 95% confidence intervals and logrank p values were calculated.
Breast Cancer Miner was used for the analyses of any event free survival of basal like and/or TNBC (IHC-based) patients. This tool uses GSE studies as well others independent of KM plot.27,28 The total number of patients analyzed was 876 (filtered from 5861). For the RNA sequencing-based gene expression correlation analyses, the updated number of patients with basal-like and/or TNBC was 276.
UCSF Xena browser was used to study the ITGA2 gene expression through different breast cancer cell lines according to their subtype.29 The profiles of the set of breast cancer cell lines are seen in the study by Heiser 2012.30 The single cell RNA sequencing data of human circulating tumor cells isolated from breast cancer patients and xenografts were retrieved from the published databases GSE109761 and GSE11106516,31 and compared using Wilcox t test.
RNA sequencing and data analysis
Total RNA extraction was performed using Trizol and phase separation by chloroform, then extraction by alcohol. Samples were submitted to the RNA core for deep sequencing analysis. RNA sequencing was performed on a HiSeq 4000 at Northwestern University's Center for Genetic Medicine Sequencing core facility and a library was made using TruSeq Total RNA-Seq Library Prep. The raw data files were submitted to the GEO repository (GSE132961) of the NCBI tracking system (scheduled release on Feb 01, 2020). Sequencing data was aligned using STAR32 and analyzed using HTSeq33 and DESeq2.34 Differentially expressed genes were subject to a cutoff of FDR <0.05 and Log2 (Fold Change) > 0.48 or < −0.48. Pathway analysis was performed on significantly-differentially expressed genes in Metascape (http://metascape.org).35
Cell culture
MDA-MB-231 and BT20 cells were cultured in DMEM high glucose, supplemented with 10% FBS and 1% Pen/step antibiotic. 4T1 cells were cultured in RPMI media supplemented with 10% FBS and 1% Pen/strep antibiotic. MDA-MB-231 cells were obtained from ATCC, and BT20 and 4T1 cells were originally from ATCC and expanded by Dr. Dai Horiuchi. Transcriptome and phenotypic analyses of these cells matched the published profiles.
Cell transfections
Transient transfection of breast cancer cells with Dharmacon small interfering RNAs (siRNAs), or microRNA mimics was performed once or in tandem using Dharmafect transfection reagents (siRNA control:D-001810-01-05, ITGA2siRNA: L-004566-00-0005, CCND1: L-003210-00-0020; set of 4 ITGA2 siRNA: LU-004566-00-0005; miR control: CN-001000-01-05). Cells were at least 70% confluent at the moment of transfection, siRNA concentration is 100 nM per transfection. Most phenotypic analyses were conducted 2 days after the last transfection. Mammosphere formation in vitro and metastasis assays in vivo were done on the following day after the second transfection. For rescue experiments, cells were co-transfected with GFP empty vector or GFP ITGA2 cDNA in addition to either miR control or miR-206; alternatively OFP vector control, OFP-CCND1 cDNA vector in addition to either siRNA control or siITGA2 (Sino Biological, CV025, CV026, HG13024-ACG, HG10905-ACR). Transfection of vectors were made in serum free media, using PolyJet (Signa Gen Laboratories, SL100688).
Mammosphere assay
Cells were plated at a low density of 1000 cells in suspension in a 6- well plate covered with poly-HEMA in PRIME-XV® Tumorsphere serum-free medium (Irvine scientific, 91130). 8 days after plating a total number of spheres (diameter >50 μm) were counted for each well and pictures were taken.
Scratch wound assays of cell migration and invasion
Plates were coated with collagen type I (Corning 354231) and incubated at RT for at least an hour prior to cell plating. Cells were plated in an image lock 96 well plate overnight at a confluency of 20K cells per well. On next day morning a scratch was created using the IncuCyte wound maker. After washing of the floating cells, the remaining adherent cells were monitored for migratory filling of the wound by Incucyte. For the cell invasion assay, the washed wells of adherent cells with the scratch wound was covered with another layer of growth factor-reduced Matrigel for 1 h at 37 °C or room temperature. Then the plate was washed again and added with culture medium. The filling of the scratch wound was monitored real-time by IncuCyte over 24–48 h.
Cell cycle analysis
Cells were collected, then fixed with 70% alcohol, washed with PBS, incubated with RNAse A for 1 h then propidium iodide dye was added. Some cells were stained with DAPI and the RNAse A incubation was skipped. Cells were kept in the dark and at 4°C until flow cytometry analysis on LSRII (PE and UV channels).
3’UTR luciferase assay
Vectors expressing luciferase with the 3’UTR region of ITGA2 as well as luciferase assay reagents were obtained through Active Motif and their protocol was followed (Switchgear, Lightswitch luciferase assay kit, 32031; GAPDH control, 32014; ITGA2 3'utr: pLS_3UTR).
Flow cytometry
Cells were detached using 2 mM EDTA in PBS and cell surface expression of CD49b was measured (Biolegend 359310). Annexin V staining was performed using FITC Annexin V apoptosis detection Kit (BD Biosciences 556547).
Western blotting
Cells were lysed by RIPA buffer supplemented with Amresco protease inhibitor cocktail (1:100 diluted) and centrifuged for 10 min at 4°C and 10,000 RPM. Protein concentration was measured through a Bradford assay and 20-40ug of protein were loaded for each sample. Antibodies used: Cd49b (Rabbit pAb, Thermo Fisher Scientific PA5-26061), pFAK, total FAK (Rabbit, Cell Signaling, mAb 8556 and pAB 3285), b-actin (Mouse mAb, Abcam ab8224). Secondary antibodies where HRP conjugated (Promega, Rabbit W401B, Mouse W402B) for ECL detection using Pierce ECL2 solution (Thermo Fisher Scientific, 1896433A).
Immunohistochemistry
Tissue was first deparaffinized by xylene incubation followed by rehydration of tissue obtained through alcohol incubation in decreasing concentrations. Heat induced antigen retrieval was obtained using a decloaking chamber in decloaking solution for 20 min (Biocare Medical, RD913L). Further staining of primary antibody was performed following Dako envision plus kit and DAB staining (Anti-ITGA2, Sigma HPA063556). Hematoxolin and eosin staining was performed following dehydration of the tissue by incubation with increasing concentrations of alcohol. Tissues were later mounted with permount.
Acetyl-CoA measurement
Samples were fixed then stored at −80°C freezer, then dried with N2 blowing. MeOH:water mix with an internal standard (2C13 Ace-CoA) in each sample. Samples were diluted with buffer before SPE on water HLB plate. Sample was eluted with MeOH. The LC/MS analysis was performed on AB Sciex Qtrap 5500 coupled to Agilent UPLC/HPLC system. All samples are analyzed by Agilent proshell 120 SB-C18 column 100 Å, 2.7 μm, 2.1 mm × 100 mm coupled to an Agilent UPLC/HPLC system, which was operated at a flow rate of 450 uL/min.
Statistical analysis
Student's t-test was performed for most of the comparisons except for the ones specified such as in human database analyses. Probabilities under 0.05 were considered significant and represented with one star (*). Probabilities under 0.01 and 0.001 were represented with two stars (**) and three stars (***), respectively.
Results
Identification of ITGA2 as an oncogenic promotor of breast cancer metastasis
To identify potential phenotype drivers of breast cancer progression and metastasis, we first analyzed the association of all integrin encoding genes with clinical outcomes using Breast Cancer Miner with multiple GSE databases containing 2997 unique breast cancer samples.26 We found that high expression levels of eight integrin genes correlated with unfavorable RFS of basal-like breast cancer, including ITGA2, ITGA5, ITGA6, ITGA7, ITGAV, ITGB1, ITGB3, and ITGB6, whereas the rest of 17 integrin genes were associated with favorable outcomes (Fig. S1), implicating heterogeneous functions and relevance of integrin genes in breast cancer.
In order to narrow down the candidate screening of integrin regulators in TNBC stemness and metastasis, we examined possible association of these integrins with subtypes of breast cancer cell lines and known stemness regulators of breast cancer, such as microRNAs we have identified in previous studies.21 Based on the gene expression profiles of over 50 established breast cancer cell lines, high ITGA2 expression levels are mainly observed in basal A subtype cells (Fig. S2). Our previous work revealed that miR-206 suppresses TNBC stemness and metastasis.21 Based on our microarray dataset GEO-GSE 59751, we found that the expression of CD49b-encoding ITGA2 was specifically inhibited by miR-206 whereas other integrin genes such as ITGB1 levels remained unaffected (Fig. S3a). Consistently, we observed that the miR-206 over-expressing breast cancer cells caused minimal, residual lung metastases along with reduced CD49b levels, compared to the significant lung metastases mediated by control cells upon tail vein infusion (Fig. S3b). Therefore, we hypothesized that ITGA2 promotes cancer stemness and metastasis in TNBC as an oncogenic target of miR-206.
To examine the importance of ITGA2 in TNBC metastasis, we set out to determine if ITGA2 knockdown (KD) mimics miR-206 overexpression in regulating breast cancer metastasis in vivo. Upon tail vein injection, MDA-MB-231 cells with reduced CD49b levels due to ITGA2-KD had significantly decreased lung colonization in NOD/SCID mice (Fig. 1A–C). While equivalent cells of the control and ITGA2-KD groups homed to the lungs immediately after tail vein injection, cells with ITGA2-KD had a reduced capacity to colonize the lungs as measured from 24 h to 4 days post injection.
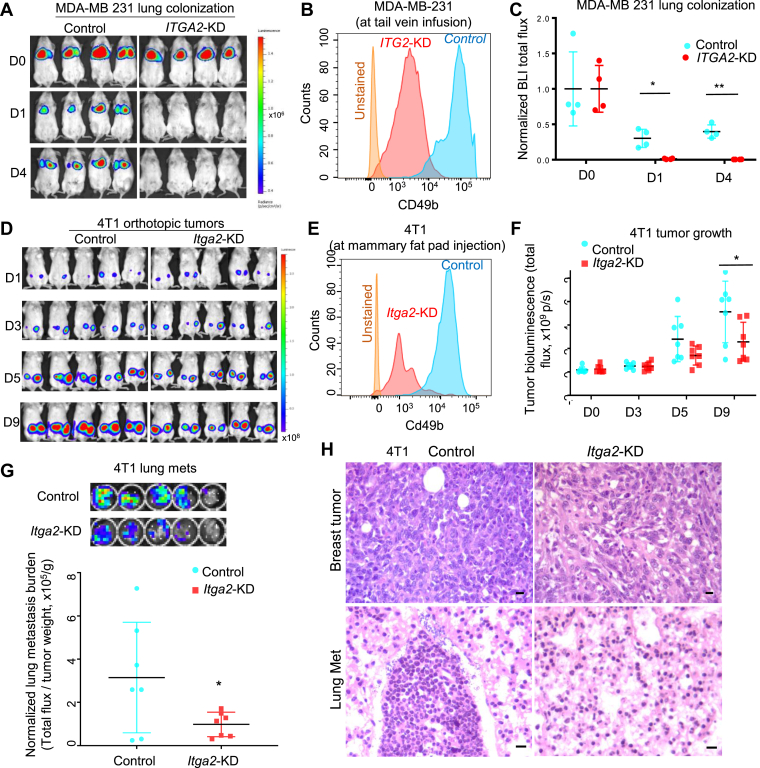
Figure 1.
ITGA2 knockdown inhibits lung colonization and metastasis. (A-C) Bioluminescence images (a), CD49b flow analyses (b), and normalized lung colonization signals (c, ratio of the total flux compared to D0) of NOD/Scid mice on day 0 (D0), 1 (D1), and 4 (D4) post tail vein infusion of L2T/L2G-labeled MDA-MB-231 cells, control and ITGA2-KD. n = 4, *P = 0.02 (D1) and **P = 0.004 (D4) for paired comparisons (t-test). Error bars represent S.D. values. (D) Bioluminescence images of tumor growth signals of mouse 4T1 cells, the control and Itga2 KD, on day 1 (D1), 3 (D3), 5 (D5), and 9 (D9) post orthotopic implantation into the 4th mammary fat pads of BALB/c mice. (E) Flow analyses of mouse Cd49b levels in 4T1 cells ± Itga2-KD prior to implantation. (F) 4T1 tumor growth analyses (tumor burden) (total flux p/s, n = 7, *t-test P< 0.01). Error bars represent S.D. values. (G) Bioluminescence images (top panels) and normalized lung metastasis by primary tumor burden (total flux/tumor weight) of the mouse lungs bearing 4T1 spontaneous metastases on D9, n = 7, t-test P = 0.05. Error bars represent S.D. values. (H) H & E staining images of 4T1 breast tumors and spontaneous lung metastases (present in the control group and absent in the Itga2 KD group) on day 9 post orthotopic implantations shown in d. Scale bars = 20 μm.
In addition, the murine 4T1 TNBC cells were utilized to investigate the effects of Itga2 KD on spontaneous metastasis in immune-competent Balb/c mice. When these cells were orthotopically transplanted at large numbers (106 cells per injection), 4T1 cells could metastasize from the mammary fat pads to the lungs within a week.36 Using IVIS imaging of luciferase-labeled cells, we found that silencing Itga2 gene with reduced Cd49b expression only modestly compromised primary tumor growth, as measured by BLI (Fig. 1D–F), without a significant reduction on tumor weight (Fig. S3c, t-test P = 0.17, n = 7). However, Itga2 silencing dramatically suppressed lung metastasis after normalization on tumor weight (Fig. 1G). Reduced Cd49b levels in engrafted tumors and inhibited lung metastases of Itga2-KD 4T1 cells were validated by immunohistochemistry and H & E staining (Fig. S3d and Fig. 1H). In contrast to the lungs, metastasis of 4T1 to the liver and other organs was not detected in these mice within the short observation window (data not shown).
ITGA2 is a target of miR-206 enhancing mammosphere formation and cancer stemness
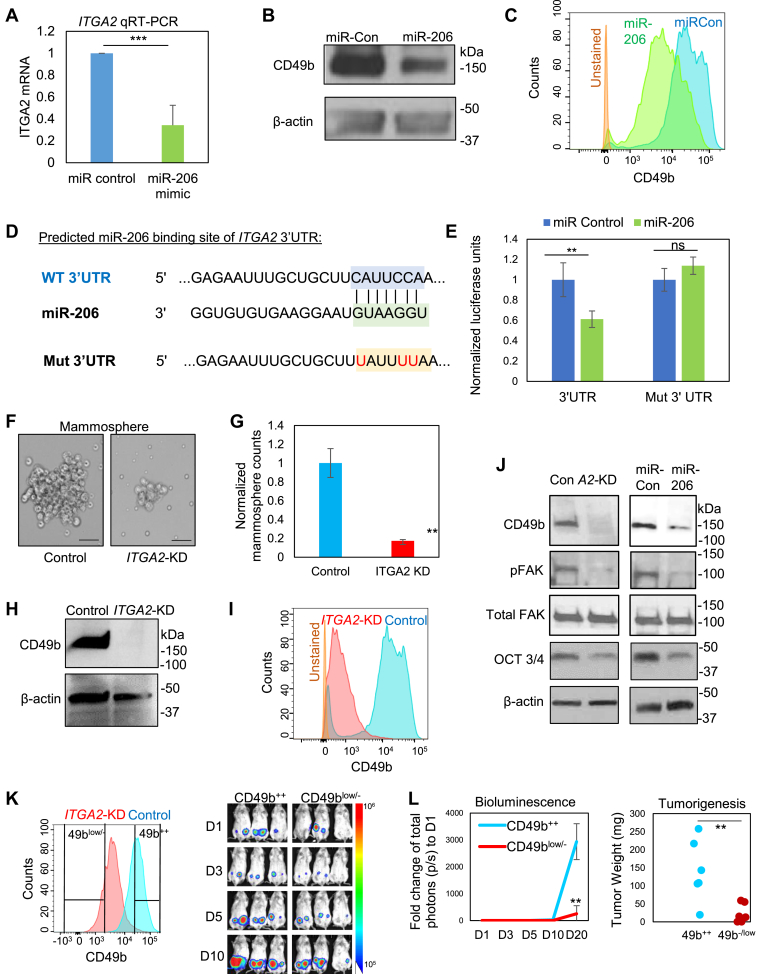
The inhibitory effects of miR-206 on ITGA2 mRNA and CD49b protein expression (both cellular and surface protein levels) were further validated in MDA-MB-231 cells upon transient transfection of miR-206 mimics, measured by qRT-PCR, immunoblotting, and flow cytometry analyses (Fig. 2A–C and S3a). To determine if ITGA2 is a direct target of miR-206, we utilized multiple algorithms including TARGET SCAN and others to assess the ITGA2 sequence and identified a predicted miR-206 binding site within its 3’UTR region (Fig. 2D). The ITGA2 3’UTR region (2 kb) was then cloned downstream of a luciferase reporter gene in pLS_3UTR. The luciferase reporter expression/activity was inhibited by co-transfected miR-206 and the inhibition was reversed when the three nucleotides were mutated from C to T (U) within the predicted site for miR-206 binding (Fig. 2D and E). These data suggest that ITGA2 is inhibited by miR-206 through a direct binding to its 3’UTR site.
Figure 2.
ITGA2 is a target of miR-206 and promotes mammosphere formation and stemness factor expression. (A–B) ITGA2 mRNA (top panel) and protein (bottom panel) levels are reduced by transfected miR-206 in MDA-MB-231 cells, measured by qRT-PCR and immunoblotting, respectively. ***t-test P < 0.001. Error bars represent S.D. values. (C) CD49b surface protein expression inhibited by miR-206 in MDA-MB-231 cells, evaluated by flow cytometry. (D) Predicted binding sites between the complementary sequences of ITGA2 3’UTR (WT) and miR-206. The bottom line shows the mutated C to T (U, in red) within the interaction site of 3’UTR. (E) 3’UTR luciferase assay shows direct regulation of ITGA2 by miR-206, n = 3, **t-test P = 0.005. (F-G) Images (F) and quantified counts (G) of breast cancer cell-derived mammospheres upon siITGA2 knockdown. **t-test P < 0.01. Error bars represent S.D. values. Scale bars = 25 μm (H-I) siITGA2 transfection depletes CD49b protein expression, measured by immunoblotting (H) and flow cytometry analyses (I). (J) Immunoblots to detect reduced expression of CD49b, phosphorylated FAK (pFAK), and OCT3/4 levels without affecting total FAK levels upon ITGA2 KD or miR-206 upregulation. (K) Left panel: Flow histogram of CD49b expression in control and siITGA2-transfected cells and gated sorting of CD49b++ and CD49blow/- populations, respectively. Right panel: Bioluminescent images of CD49b++ and CD49blow/- implants of 100–1000 cells from day 1–10 (D1-D10). (L) Left panel: Fold change of tumor burden (total photons) to Day 1 (D1) from D1-D20. ** t-test P = 0.007 on D20. Error bars represent S.D. values. Right panel: Weight of tumors derived from CD49b++ and CD49b−/low cells, orthotopically implanted at 100–1000 cells (t-test **P = 0.008).
We continued to examine the importance of ITGA2 in regulating cancer stemness. First, using the online Breast Cancer Gene Expression Miner v4.0, we observed a positive correlation between expression of IGTA2 and the breast cancer stem cell marker CD44 in human basal-like and TNBC, measured by RNA sequencing (Fig. S4A). We also detected a higher ITGA2 mRNA expression and protein levels in CD44+ versus CD44- tumor cells isolated from TNBC PDXs (Fig. S4B). Notably, siITGA2-mediated knockdown (KD) did not alter CD44 protein levels (Fig. S4C), suggesting a possible independent role of ITGA2 in TNBC. Mimicking the mammosphere-inhibitory phenotype of miR-206 as we previously observed,21 ITGA2 KD suppressed mammosphere formation of MDA-MB-231 cancer cells (Fig. 2F and G). The effectiveness of ITGA2 knockdown in MDA-MB-231 cells was confirmed by both immunoblotting and flow analyses (Fig. 2H and I). Furthermore, we investigated the effects of miR-206 upregulation and ITGA2 KD on pluripotency transcription factors and focal adhesion kinase (FAK) signaling. FAK is known to be activated by many integrins37 and the FAK signaling pathway promotes stemness in breast cancer cells.38 Indeed, both ITGA2 KD and miR-206 upregulation similarly decreased the protein levels of pluripotency marker OCT 3/4, and inhibited the phosphorylation/activation of FAK, without altering the total FAK protein levels (Fig. 2J). To determine the importance of CD49b in tumorigenicity in vivo, we sorted CD49high (++) and CD49 low/- cells from the control and ITGA2 KD cells, respectively. ITGA2 KD significantly inhibited the tumor growth and tumorigenesis from 100-1000 cells upon orthotopic implantation into the mammary fat pads of NSG mice (tumor weight 0 for 33% implantations of CD49b negative cells) (Fig. 2K and L and S4d). ITGA2 KD further inhibited mRNA levels of SOX2, measured by RT-PCR (Fig. S4e). These results demonstrate that ITGA2/CD49b is intrinsically required to enhance cancer stemness in TNBC.
ITGA2 knockdown reduces cell cycle progression
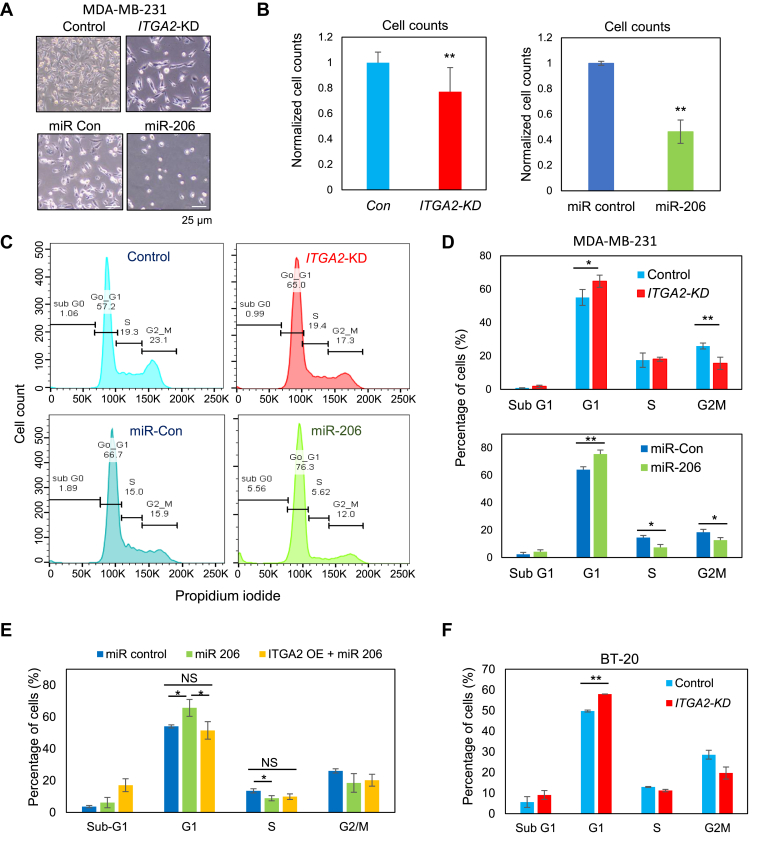
We further investigated whether CD49b regulates cancer stemness by modulating cell cycle or cellular viability. Similar to the miR-206-induced effects, ITGA2 KD reduced confluence (Fig. 3A and S4f) and counts of MDA-MB-231 breast cancer cells (Fig. 3B). We then examined if alterations in cell cycle and cell death contribute to the phenotypic changes. Mimicking the effects of miR-206 upregulation, ITGA2 KD in MDA-MB-231 cells increased the percentage of cells in the G1 phase and decreased populations in cycling phases such as G2/M (Fig. 3C and D). As expected, exogenous cDNA-mediated overexpression of CD49b rescued the cell cycle arrest caused by miR-206 in MDA-MB-231 cells a (Fig. 3E). Consistently, in a different TNBC line BT-20, ITGA2 KD also resulted in an accumulation of G1 arrested cells (Fig. 3E and S4g-h).
Figure 3.
ITGA2 knockdown inhibits cell cycle. (A-B) Images (A) and cell counts (B) of MDA-MB-231 cells 48 h after transfections with siITGA2 (ITGA2-KD) or miR-206 mimic. Cell number was measured via hemocytometer counting (B), n = 12, P = 0.002 (ITGA2-KD), and n = 3, P = 0.0012 (miR-206) (t-test). Error bars represent S.D. values. (C) Representative flow cytometry analyses of cell cycle with phases of subG1, G1, S, and G2M using propidium iodide upon ITGA2-KD and miR-206 overexpression in MDA-MB-231 cells. (D) Quantified percentage of MDA-MB-231 cells in each cell cycle phase as shown in (C) Top panel: siITGA2 mediated ITGA2-KD and control, n = 4 biological replicates; P = 0.03 (G1); P = 0.004 (G2M) (t-test). Bottom panel: miR-206 overexpression and control, n = 3 biological replicates; P = 0.01 (G1), 0.02 (S), 0.04 (G2M) (t-test). Error bars represent S.D. values. (E) Percentage of MDA-MB-231 cells in each cell cycle phase upon ITGA2 cDNA-mediated rescue of G1 arrest in miR-206 transfected cells (gated GFP+ cells with ITGA2 overexpression). P < 0.05 (t-test). NS = not significantly different. Error bars represent S.D. values. (F) Percentage of BT20 cells in each cell cycle phase upon ITGA2-KD. P < 0.01 (t-test). Error bars represent S.D. values.
We then measured cell death in breast cancer cells upon altering CD49b expression. From the cell cycle analyses, ITGA2 KD did not cause significant changes of subG1 phase (Fig. 3C–E), and differences in levels of cell death were not detectable in either Annexin V or propidium iodide (PI)-stained cells (Fig. S5a-b). A DNA binding dye with red fluorescence only marked a marginal increase of cytotoxicity (from 2% to 5%) of ITGA2 KD cells during cell migration in a scratch wound healing assay (Fig. S5c-d). Furthermore, in order to ensure minimal off-target effects of siRNAs, we employed four individual ITGA2 siRNAs #1–4 for additional validation studies. Most of the siITGA2s had minimal or marginal effects on cell death, measured by Annexin V and PI-staining, as well as subG1 populations (Fig. S5e and S6a-c) whereas three siITGA2s significantly altered G1 and/or G2/M phases of the cell cycle, in consistency with their knockdown efficiency on ITGA2 (Fig. S6a-c). Therefore, cell death was excluded as a major factor driving cell count differences which are likely to be primarily caused by cell cycle arrest. These data demonstrate that ITGA2 promotes cell cycle which is required for proliferation and self-renewal of TNBC.
ITGA2 siRNAs inhibit tumor cell migration and invasion
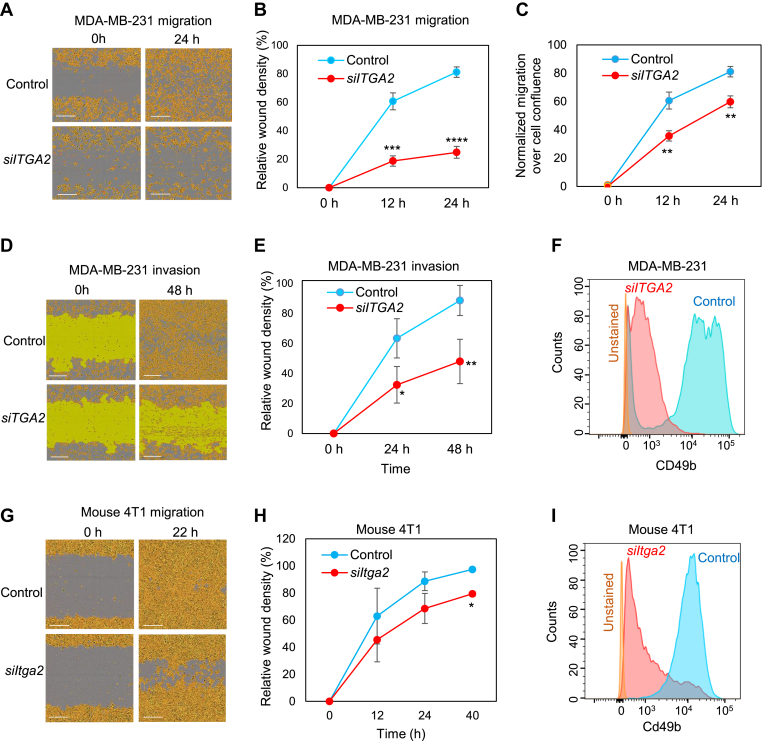
We subsequently investigated the functions of ITGA2/CD49b in regulating metastasis-related phenotypes, such as migration and invasion in multiple human and mouse TNBC cell lines. Collagen I is one of the primary ligands of integrins, which can bind extracellularly through the MIDAS motif and activate the integrin signaling cascade.39 To assess the impact of CD49b suppression on Collagen I binding, migration and invasion assays were conducted on collagen I-coated dishes. Transfection of a smart pool of four siITGA2s significantly suppressed migration of MDA-MB-231 cells (Fig. 4A–C). In addition, the siRNAs that conveyed the most effective reduction of CD49 levels as measured by flow cytometry also led to the most dramatic decrease in cell density and cell migration (Fig. S7a-b). Subsequently, cell invasion was measured when migrating cells were overlaid with Matrigel after the wound scratch was made. Upon ITGA2 siRNA transfection, MDA-MB-231 cells exhibited a dramatic decrease of invasion along with reduced cell density over time compared to the controls (Fig. 4D–F). Cell migration was also inhibited in mouse TNBC 4T1 cells when transfected with siRNAs targeting mouse Itga2 (Fig. 4G–I).
Figure 4.
ITGA2 knockdown reduces migration and invasion of TNBC cells in the presence of collagen. (A) Images of migratory MDA-MB-231 cells with human siITGA2 smart pool-mediated knockdown (ITGA2-KD) and siRNA controls (Con), at 0 and 24 h (h) following scratched wounding. Cells were plated on collagen I-coated plates. (B) Quantification of relative cell density filled by ITGA2-KD and control cells in wounded areas, n = 3; P = 0.0002 (12 h), 0.0001 (24 h) (t-test). Error bars represent S.D. values. (C) Normalized migration over confluence of MDA-MB-231 cells during wound healing. P < 0.01 (t-test). Error bars represent S.D. values. (D-E) Images (D) and relative cell density (E) of invasive MDA-MB-231 cells (A2-KD and control) covered by Matrigel, at 0 and 48 h after scratched wounding. n = 3, P = 0.02 (24 h), 0.007 (48 h) (t-test). Error bars represent S.D. values. (F) Flow cytometry plots of CD49b levels in MDA-MD-231 cells upon ITGA2 KD. (G-H) Images (G) and quantified wound density (H) of murine 4T1 mammary cancer cells upon Itga2 KD via siItga2, within 12, 22 (24), and 40 h following scratch wounding. *t-test P < 0.05 (n = 3). Error bars represent S.D. values. (I) Flow cytometry analyses of reduced murine Cd49b levels upon Itga2 KD in 4T1 cells.
Supporting the potential for CD49b serving as a new therapeutic target to block cancer progression, we further found that an anti-CD49b neutralizing antibody blocked the migration of MDA-MB-231 cells during wound healing (Fig. S7c). Depletion of CD49b specifically abolished the adhesion of MDA-MB-231 cells to its extracellular matrix ligand protein Collagen I but had minimal effects on adhesion to laminin and fibronectin (Fig S7d). However, the wound-healing migration of these cells was significantly altered on any of these three substrates upon ITGA2 KD (Fig S7e), suggesting that the CD49b-mediated tumor cell motility might be intrinsic and independent of its binding to extracellular matrix.
ITGA2 knockdown inhibits the expression of genes in lipid metabolism and cell cycle pathways
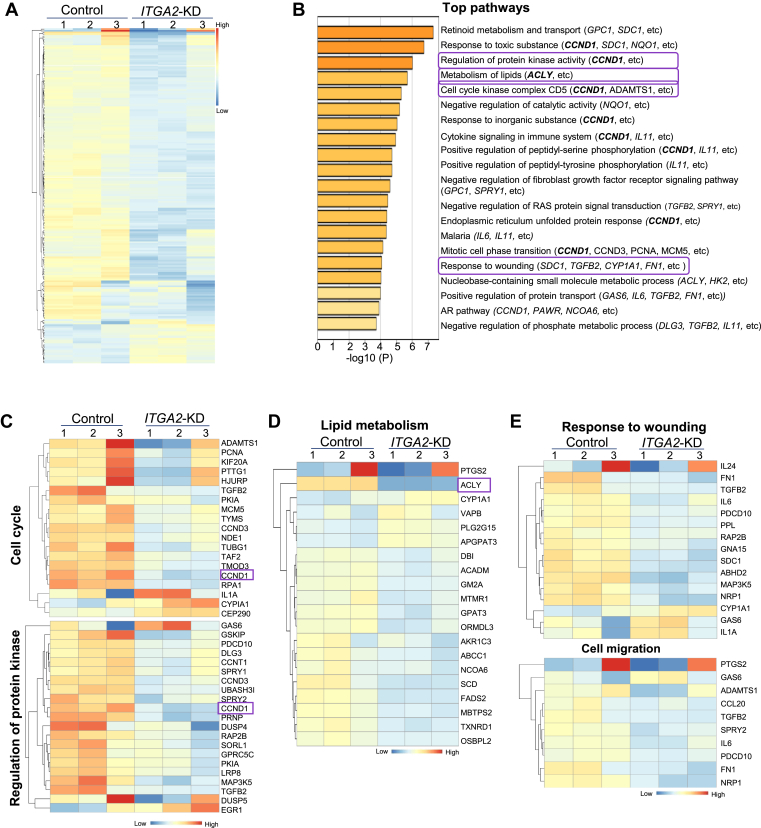
To further understand the downstream pathways regulated by ITGA2 in breast cancer, we performed RNA sequencing to compare the transcriptomes of ITGA2 KD and control MDA-MB-231 cells and identified the top 195 differentially expressed genes, based on a cutoff of FDR <0.05 and Log2 (Fold Change) > 0.48 or < −0.48 (Fig. 5A and Table S1). Notably, based on Metascape analysis (http://metascape.org), ITGA2 KD altered the pathways of lipid metabolism, retinoid metabolism and transport, cell cycle, response to wounding, regulation of protein kinase activity, and other pathways, including representative genes such as ACLY, CCND1, CCND3, MCM5, TGFB2, PCNA, ADAMTS1, MAP3K5 (Fig. 5B–E and S8a-b). ACLY-mediated acetyl-CoA production for lipid metabolism was recently shown to promote tumorigenesis in pancreatic cancer.40 CCND1 appeared in multiple pathways regulated by ITGA2/CD49b, such as cell cycle, response to stress (toxic substance), regulation of protein kinase activity (serine/tyrosine phosphorylations), cytokine signaling, unfolded protein response, mitotic cell phase transition, androgen receptor (AR) pathway, and other pathways (Fig. 5B and S8a-b). Meta-analysis of several breast cancer gene expression datasets using bc-GenExMiner further demonstrated a significant positive correlation of ITGA2 with both CCND1 and ITGB1 levels (Fig. S8c-d). We chose ACLY and CCND1 for further functional studies to determine their importance in CD49b-mediated phenotypes in TNBC stemness.
Figure 5.
ITGA2 downregulation-altered genes and clusters. (A) Heatmap of differentially expressed 195 genes >1.5 fold change upon ITGA2 KD via siITGA2 (see Table S1), with most being downregulated (top and middle rows of 169 genes) and a small portion of upregulated genes (bottom rows of 26 genes). (B) Top 20 gene clusters enriched by ITGA2 KD, colored by p-values. The Metascape gene list analyses were carried out with the ontology sources of KEGG Pathway, GO, Reactome, Canonical Pathways, and CORUM. (C-E) Heatmap of genes in representative pathways of cell cycle and regulation of protein kinase activity (C), lipid metabolism (D), and wound healing and cell migration (E), with representative genes CCND1 and ACLY shown in C and D, respectively.
ACLY and cyclin D1 are CD49b targets enhancing cancer stemness and cell cycling
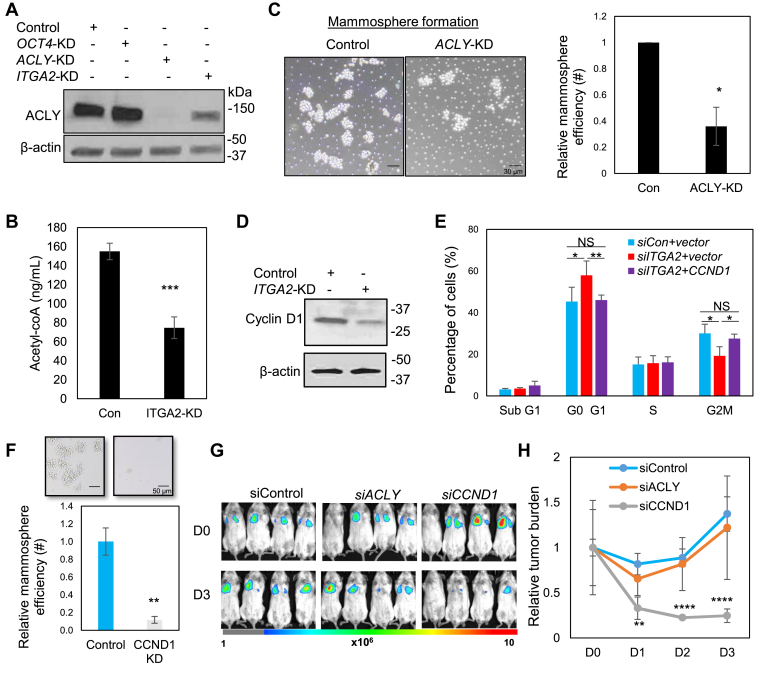
ACLY was one of the top genes downregulated in ITGA2 KD cells and we hypothesized that it is a regulator of cancer stemness in breast cancer. We first confirmed a decreased protein level of ACLY in siRNA-mediated ITGA2 KD cells as well as ACLY KD cells (Fig. 6A). Using LC/mass spectrometry analysis with a labeled standard control, we detected a reduced concentration of cellular Acetyl-CoA levels upon ITGA2 KD (Fig. 6B). ACLY KD also mimicked ITGA2 KD in inhibiting mammosphere formation (Fig. 6C), without significantly altering total cell counts or cell density in wound healing (Fig. S9a-b), suggesting a role of ACLY in stemness regulation independent of cell cycle effects.
Figure 6.
ACLY and Cyclin D1 are CD49b targets enhancing self-renewal and cell cycle. (A) Immunoblot of downregulated ACLY proteins in MDA-MB-231 cells transfected with siACLY and siITGA2 in comparison to siRNA control and siOCT4. (B) Reduced cellular acetyl-coA levels in the ITGA2-knock down MDA-MB-231 cells, as measured by LC/MS (t-test P = 0.001, n = 3 biological replicates). Error bars represent S.D. values. (C) Compromised mammosphere formation in ACLY KD cells, shown representative images (right panels) and quantified histograms (left panel) (t-test P < 0.05, n = 3 biological replicates). Error bars represent S.D. values. Scale bars = 30 μm. (D) Immunoblot of downregulated cyclin D1 proteins in MDA-MB-231 cells transfected with siITGA2 compared to control. (E) siITGA2-induced G1 arrest and altered G2/M phase were rescued by CCND1 overexpression in MDA-MB-231 cells. *P < 0.05, **P < 0.01 (t-test), n = 3 biological replicates. Error bars represent S.D. values. (F) Representative images (top panels) and qualification (bottom panel) of mammosphere formation of MDA-MB-231 cancer cells upon siITGA2 mediated KD. ** t-test P < 0.01, n = 3 biological replicates. Error bars represent S.D. values. Scale bars = 50 μm. (G) Bioluminescence images of siControl, siACLY, siCCND1-transfected MDA-MB-231 tumor cells upon tail vein infusion (n = 4 mice each group). (H) Fold change of lung metastasis signals (total photons, p/s), normalized to day 0 (D0). t-test P = 0.003 (**), 2e-5 (****), and 7e-5 (****), for D1, D2, and D3 comparisons between the control and siCCND1 groups, respectively. Error bars represent S.D. values.
We then continued to determine if CD49b-regulated cell cycle attributes to CCND1. ITGA2 KD also decreased the protein levels of CCND1-encoded cyclin D1 (Fig. 6D), which is an important driver of the G1-S phase transition during the cell cycle.41 Furthermore, enforced expression of CCND1 rescued the cell cycle defects in ITGA2 KD cells (Fig. 6E). Small RNA mediated silencing of CCND1 (KD) also mimicked ITGA2 KD in inhibiting the efficiency of mammosphere formation (Fig. 6F), suggesting cell cycle-coupled regulation of cancer stemness. To evaluate the effects of ACLY and CCND1 in lung colonization, we continued to inoculate these three groups of cells (control, ACLY-KD, CCND1-KD) into mice via the tail vein. The bioluminescence imaging showed that CCND1 KD instead of ACLY KD mimicked the ITGA2 KD in inhibiting lung colonization (Fig. 6G and H), suggesting the essential role of CCND1 in enhancing metastasis of breast cancer.
ITGA2 pathway components are associated with poor survival of breast cancer patients
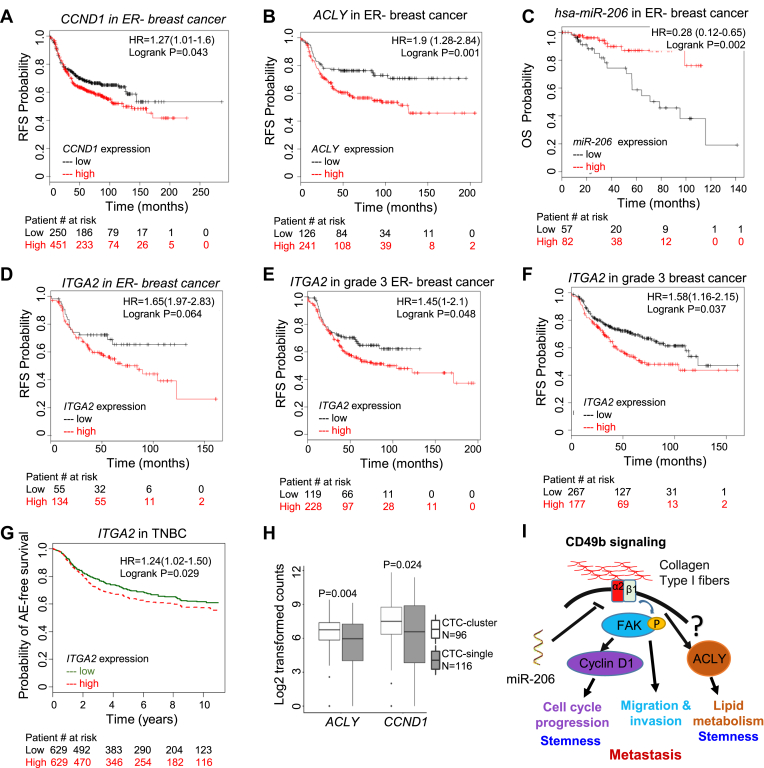
Lastly, we examined the pathological and clinical relevance of ITGA2 targets and upstream miR-206 in breast cancer. Similar to ITGA2-based KM Plots, CCND1 and ACLY expression show a similar pattern of correlation with a short RFS in ER− breast cancer patients (Fig. 7A and B), whereas miR-206 expression indicates an extended RFS in ER− breast cancer (Fig. 7C). High expression levels of ITGA2 are not only associated with reduced RFS probability in patients with ER- or the intrinsic basal subtype of breast cancer (Fig. 7D and S1), but also in patients with all grade 3 breast cancer patients independent of ER expression (Fig. 7E and F, and S9c-d). Consistently, the analysis of another database also demonstrated the correlation of high ITGA2 expression with poor event free survival of the patients with TNBC (Fig. 7G). Our recent discoveries and other reports demonstrated that circulating tumor cell (CTC) clusters are enriched with cancer stemness and proliferation compared to single CTCs.15,16,31,42 Notably, shown by single cell RNA sequencing from the published databases,16,31 both ACLY and CCND1 expression levels are upregulated in CTC clusters versus single CTCs isolated from patients with breast cancer and patient-derived xenografts (Fig. 7H), correlating with their effects on improved self-renewal and cell cycle progression respectively.
Figure 7.
ITGA2 pathway is associated with outcomes of the patients with breast cancer. (A-C) Breast Cancer Kaplan–Meier Plotter analyses of RFS correlated with CCND1 expression (a, Log rank P = 0.043, n = 701), and ACLY expression (b, Log rank P = 0.001, n = 407), and OS associated with miR-206 expression (c, Log rank P = 0.002, n = 159) in ER-breast cancer with best cut off. (D-F) KM plots of RFS correlation with ITGA2 expression in ER- (Log rank p = 0.0406, n = 189) (D), grade 3 ER- (Log rank P = 0.048, n = 347) (E), and all grade 3 (Log rank P = 0.037, n = 444) (F) breast cancer, with best cut off selected in Breast Cancer Kaplan–Meier Plotter analyses. (G) Breast cancer miner plot of any-event (AE) free survival association with ITGA2 expression in TNBC. Log rank P = 0.02, n = 1258, median cut off. (H) Box plot for differential ACLY and CCND1 expression levels between clustered and single CTCs of breast cancer patients and PDXs (GSE109761 and GSE111065).16,31 Wilcox t test P = 0.004 and 0.024 for ACLY and CCND1 comparisons, respectively. (I) Schematic CD49b signaling pathway induced by extracellular matrix factors such as collagen I fibers that result in the phosphorylation of FAK and upregulation of CCND1 and ACLY levels to promote proliferation, stemness, and metastasis of TNBC.
Since FAK has been known to promote CCND1 expression43 and both ITGA2 KD and miR-206 could inhibit FAK phosphorylation (Fig. 2J), our data suggest a regulation network that miR-206 inhibits ITGA2/CD49b-mediated signaling cascade through compromised FAK phosphorylation and cyclin D1 expression as well as reduced ACLY expression and metastasis-regulating pathways (Fig. 7I). The miR-206/ITGA2/ACLY-CCND1 signaling cascade is a clinically relevant target for both biomarker identification and therapeutic development to potentially improve patient outcomes.
Discussion
Our findings have identified that ITGA2/CD49b as a direct target of miR-206 in promoting breast cancer stemness and metastasis. The surface integrin protein CD49b can be potentially targeted by neutralizing antibodies and small molecules, serving as a new therapeutic target for metastatic TNBC. FAK, a non-receptor tyrosine kinase is activated by CD49b signaling and has been identified as a key mediator of intracellular signaling by integrins.44,45 Our studies have also identified several other integrins which expression levels are associated with an unfavorable RFS of breast cancer, such as cancer stem cell markers CD49f (integrin α6) and integrin α7.46, 47, 48 Both integrins and FAK can play crucial roles in the maintenance of breast cancer stem cells.38,49 It might be worth pursing if CD49b coordinates the pluripotency transcription factor OCT 3/4 with the FAK signaling pathway to regulate the breast cancer stemness phenotype.49 As FAK is a known mediator of the downstream signaling of many integrins in cell cycle regulation,50 we speculate that FAK activation may be involved in CD49b signaling-enhanced transactivation of CCND1 in TNBC. However, the transcriptional regulation of ACLY has not been well studied and the connection with FAK and/or other regulators will need to be further elucidated.
CD49b appears to activate cell cycling of breast cancer stem and progenitor cells in cancer progression. Our global transcriptome analyses reveal that CD49b regulates cell cycle, wound healing, and protein kinase pathways related to self-renewal and migration. CCND1 is amplified and overexpressed in one of the high risk of relapse subgroup -Integrative Cluster 2 of ER negative breast cancer with a very poor prognosis.51 Cell cycle progression through G1 phase in mammalian cells is precisely regulated by integrin-mediated adhesion to the extracellular matrix growth and growth factor binding to receptor tyrosine kinases (RTKs).52,53 These two classes of receptors coordinately promote activation of the G1 phase cyclin-dependent kinases (CDKs), such as CDK4/6, and increase expression of G1 phase cyclins, especially Cyclin D1/Cyclin E, which then mediate S phase entry. Our results show that transient ITGA2 knockdown inhibited TNBC proliferation by inducing G1 arrest. Previous studies have shown that the adaptor Shc might be involved in integrin α5 ligation and activation of mitogen-activated protein kinase (MAPK) activity.54,55 Elucidation of the molecular mechanisms underlying the upregulation of CCND1 is clinically relevant, and new targeting strategies might be useful to strengthen and broaden the application of the CDK4/6 inhibitors for treatment of metastatic hormone receptor positive breast cancer in the clinic.56
Altered metabolism has been connected to cellular reprograming. In addition to cell cycle and stemness-related pathways, our studies have also discovered that there are multiple metabolic pathways that are impacted by ITGA2, such as ACLY-involved lipid metabolism with acetyl CoA as a notable component. ACLY has recently been revealed to positively promote tumorigenesis and cell plasticity via elevated acetyl-CoA abundance and global histone acetylation.57 Moreover, recent findings show a contribution of de novo fatty acid synthesis (in which ACLY is involved) in intestinal tumorigenesis of adenomatous polyposis coli (APC) mutant mice.58 Our studies confirm a modulation of ACLY expression by CD49b signaling pathway and a role of ACLY in breast cancer self-renewal, showing an unprecedented connection of integrin signaling to ACLY-mediated lipid metabolism and cancer stemness. Lastly, it will be important to determine if this is specific to CD49b or shared with other integrin subunits.
In addition to the tumor autonomous role of CD49 in promoting cancer stemness, its expression in immune cells especially in regulatory T cells (Treg)25 needs to be taken into consideration when it serves as a potential cancer target. Future studies with conditional Itga2 knockout in mammary epithelial cells and immune cells respectively will assist clarifying the extent of impact of Itga2 on tumor cells versus stromal cells in the metastatic process.
Conflict of Interests
The authors declare no conflict of interests.
Funding
This manuscript has been partially supported by NIH/NCI grants R00CA160638 and R01CA245699 (H.L.), and Supplement for Diversity (V.A.), T32 CA080621-15 (R.T.), and R01CA213843 (R.A.K), American Cancer Society grant ACS127951-RSG-15-025-01-CSM (H.L.); the Susan G. Komen Foundation CCR15332826 (H.L.) and CCR18548501 (X.L.); the Department of Defense W81XWH-16-1-0021 (H.L.); the Lynn Sage Cancer Research Foundation (X.L. and H. L.); Northwestern University’s Endocrinology Training Grant T32DK007169-39 (A.H.); and start-up funds from Case Western Reserve University and at Northwestern University (H.L.).
Acknowledgements
V.A-C. conducted literature search, designed and performed experiments, collected data, analyzed data, made figures, interpreted data, and wrote the manuscript. A.D.H and X.L. assisted with experimental performance, data analyses, figure presentation, and manuscript writing. N.K.D and RT assisted with experimental performance. B.W. assisted bioinformatic analyses and figures, R.A.K provided cell lines, designed the experiments, and interpreted data. H.L. conducted literature search, designed experiments, analyzed data, interpreted data, and revised the manuscript.
We would like to thank Huiping Liu lab members particularly, Dhwani Patel, Wenjing Chen, and Natalia Brokate for assisting this manuscript. We are grateful to Dr. Dai Horiuchi for sharing cell lines as well as equipment with our laboratory; Dr. Thomas J. Hope laboratory for sharing the IVIS instrument to image immune competent animals; and Dr. Marc L. Mendillo laboratory for sharing the Incucyte imaging system. We appreciate the Animal Imaging Center as well as the Center for Advanced Microscopy and Nikon Imaging Center at Northwestern University for assisting analysis of IVIS data and for providing equipment; the NUSeq Core Facility for the RNA sequencing and the Bioinformatics Core for the analysis and the Mass Spectrometry Core in research Resources Center of University of Illinois at Chicago.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.01.015.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cancer IAfRo . The Glocal Cancer Observatory; 2018. All cancers. [online] [Google Scholar]
- 2.Howlader N.N.A., Noone A.M., Krapcho M. National Cancer Institute; 2016. SEER Cancer Statistics Review. [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA A Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Anders C.K., Carey L.A. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Canc. 2009;9(Suppl 2):S73–S81. doi: 10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapidot T., Sirard C., Vormoor J. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehn M., Cho R.W., Lobo N.A. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zabala M., Lobo N.A., Qian D., van Weele L.J., Heiser D., Clarke M.F. Chapter 2 - overview: cancer stem cell self-renewal A2 - Liu, Huiping. In: Lathia J.D., editor. Cancer Stem Cells. Academic Press; Boston: 2016. pp. 25–58. [Google Scholar]
- 9.Ramos E.K., Hoffmann A.D., Gerson S.L., Liu H. New opportunities and challenges to defeat cancer stem cells. Trends Canc. 2017;3(11):780–796. doi: 10.1016/j.trecan.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu J.M., Xia W., Hsu Y.H. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Danes A., Larsimont J.C., Liagre M. A slow-cycling LGR5 tumour population mediates basal cell carcinoma relapse after therapy. Nature. 2018;562(7727):434–438. doi: 10.1038/s41586-018-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao Y., Yang H., Levorse J. Adaptive immune resistance emerges from tumor-initiating stem cells. Cell. 2019;177(5):1172–1186. doi: 10.1016/j.cell.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Sousa e Melo F., Kurtova A.V., Harnoss J.M. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543(7647):676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Patel M.R., Prescher J.A. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107(42):18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X., Taftaf R., Kawaguchi M. Homophilic CD44 interactions mediate tumor cell aggregation and polyclonal metastasis in patient-derived breast cancer models. Canc Discov. 2019;9(1):96–113. doi: 10.1158/2159-8290.CD-18-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gkountela S., Castro-Giner F., Szczerba B.M. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176(1–2):98–112. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreso A., Dick J.E. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Mu P., Zhang Z., Benelli M. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355(6320):84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ku S.Y., Rosario S., Wang Y. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355(6320):78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Canc. 2010;10(1):9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samaeekia R., Adorno-Cruz V., Bockhorn J. miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin Canc Res. 2017;23(4):1091–1103. doi: 10.1158/1078-0432.CCR-16-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bockhorn J., Dalton R., Nwachukwu C. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4 doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bockhorn J., Prat A., Chang Y.F. Differentiation and loss of malignant character of spontaneous pulmonary metastases in patient-derived breast cancer models. Canc Res. 2014;74(24):7406–7417. doi: 10.1158/0008-5472.CAN-14-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bockhorn J., Yee K., Chang Y.F. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Canc Res Treat. 2013;137(2):373–382. doi: 10.1007/s10549-012-2346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gagliani N., Magnani C.F., Huber S. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med. 2013;19(6):739–746. doi: 10.1038/nm.3179. [DOI] [PubMed] [Google Scholar]
- 26.Gyorffy B., Lanczky A., Eklund A.C. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Canc Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 27.Jezequel P., Campone M., Gouraud W. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Canc Res Treat. 2012;131(3):765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 28.Jezequel P., Frenel J.S., Campion L. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database. 2013;2013 doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldman M., Craft B., Hastie M. The UCSC Xena Platform for cancer genomics data visualization and interpretation. bioRxiv. 2018 [Google Scholar]
- 30.Heiser L.M., Sadanandam A., Kuo W.L. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szczerba B.M., Castro-Giner F., Vetter M. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566(7745):553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 32.Dobin A., Davis C.A., Schlesinger F. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12) doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tripathi S., Pohl M.O., Zhou Y. Meta- and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yori J.L., Seachrist D.D., Johnson E. Kruppel-like factor 4 inhibits tumorigenic progression and metastasis in a mouse model of breast cancer. Neoplasia. 2011;13(7):601–610. doi: 10.1593/neo.11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitra S.K., Schlaepfer D.D. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18(5):516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Luo M., Fan H., Nagy T. Mammary epithelial-specific ablation of the focal adhesion kinase suppresses mammary tumorigenesis by affecting mammary cancer stem/progenitor cells. Canc Res. 2009;69(2):466–474. doi: 10.1158/0008-5472.CAN-08-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adorno-Cruz V., Liu H. Regulation and functions of integrin α2 in cell adhesion and disease. Genes Dis. 2018;6(1):16–24. doi: 10.1016/j.gendis.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrer A., Trefely S., Zhao S. Acetyl-CoA metabolism supports multistep pancreatic tumorigenesis. Canc Discov. 2019;9(3):416–435. doi: 10.1158/2159-8290.CD-18-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stacey D.W. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15(2):158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 42.Aceto N., Bardia A., Miyamoto D.T. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adorno-Cruz V., Liu H. Regulation and functions of integrin alpha2 in cell adhesion and disease. Genes Dis. 2019;6(1):16–24. doi: 10.1016/j.gendis.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan J.L., Shalloway D. Regulation of focal adhesion-associated protein tyrosine kinase by both cellular adhesion and oncogenic transformation. Nature. 1992;358(6388):690–692. doi: 10.1038/358690a0. [DOI] [PubMed] [Google Scholar]
- 45.Schlaepfer D.D., Hanks S.K., Hunter T., Geer Pvd. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature. 1994;372(6508):786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 46.Ming X.Y., Fu L., Zhang L.Y. Integrin α7 is a functional cancer stem cell surface marker in oesophageal squamous cell carcinoma. Nat Commun. 2016;7 doi: 10.1038/ncomms13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bierie B., Pierce S.E., Kroeger C. Integrin-beta4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc Natl Acad Sci USA. 2017;114(12):E2337–E2346. doi: 10.1073/pnas.1618298114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krebsbach P.H., Villa-Diaz L.G. The role of integrin alpha6 (CD49f) in stem cells: more than a conserved biomarker. Stem Cell Dev. 2017;26(15):1090–1099. doi: 10.1089/scd.2016.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan J.L. Integrin signaling through FAK in the regulation of mammary stem cells and breast cancer. IUBMB Life. 2010;62(4):268–276. doi: 10.1002/iub.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J.H., Reiske H., Guan J.L. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol. 1998;143(7):1997–2008. doi: 10.1083/jcb.143.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rueda O.M., Sammut S.J., Seoane J.A. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature. 2019;567(7748):399–404. doi: 10.1038/s41586-019-1007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz M.A., Assoian R.K. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J Cell Sci. 2001;114(Pt 14):2553–2560. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]
- 53.Moreno-Layseca P., Streuli C.H. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 2014;34:144–153. doi: 10.1016/j.matbio.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 54.Wary K.K., Mainiero F., Isakoff S.J., Marcantonio E.E., Giancotti F.G. The adaptor protein Shc couples a class of integrins to the control of cell cycle progression. Cell. 1996;87(4):733–743. doi: 10.1016/s0092-8674(00)81392-6. [DOI] [PubMed] [Google Scholar]
- 55.Roovers K., Davey G., Zhu X., Bottazzi M.E., Assoian R.K. Alpha5beta1 integrin controls cyclin D1 expression by sustaining mitogen-activated protein kinase activity in growth factor-treated cells. Mol Biol Cell. 1999;10(10):3197–3204. doi: 10.1091/mbc.10.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Im S.A., Lu Y.S., Bardia A. Overall survival with ribociclib plus endocrine therapy in breast cancer. N Engl J Med. 2019;381(4):307–316. doi: 10.1056/NEJMoa1903765. [DOI] [PubMed] [Google Scholar]
- 57.Carrer A., Trefely S., Zhao S. Acetyl-CoA metabolism supports multi-step pancreatic tumorigenesis. Cancer Discov. 2019;9(3):416–435. doi: 10.1158/2159-8290.CD-18-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goncalves M.D., Lu C., Tutnauer J. High-fructose corn syrup enhances intestinal tumor growth in mice. Science. 2019;363(6433):1345–1349. doi: 10.1126/science.aat8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.