Abstract
CircRNAs are a large class of endogenous single-stranded RNA that is different from other linear RNA, which are produced by back-splicing and fusion of either exons, introns, or both exon-intron into covalently closed loops. CircRNAs are found in almost all living organisms and have emerged as potentially important players effecting on all life activities. It was characterized by stable structure, resistant to RNA degradation, highly abundance and conservation and tissue-specific expression. Early circRNAs were ignored as a by-product of meaningless abnormally cut RNA and had little biological function. Currently, circRNAs have become a research hotspot due to its special characteristics. CircRNAs could function as miRNA sponges, interfere with splicing and bind to protein to regulate the expression of parental genes and so on. In recent years, an increasing number of studies have revealed that circRNAs are closely related to a series of physiological and pathological processes. Additionally, circRNAs play an important role in the occurrence and development of a variety of diseases, suggesting circRNAs may be as novel indicators or biomarkers for cancer and other diseases with which they are associated. In this article, we review the biogenesis, biological functions of circRNAs and recent advances in circRNAs research in human diseases. Results will provide new insights on the roles and new ideas of circRNAs for the diagnosis and treatment of diseases and possible directions and approach for future circRNA applications.
Keywords: Biogenesis, Cancer, CircRNAs, Diseases, Function, Regulation
Introduction
CircRNAs are non-coding RNAs that have a special covalent loop structure with no 5′ caps or 3′ ploy (A), making them resistant to RNA exonucleases or RNaseR and more stable than linear RNAs.1 CircRNAs were identified more than 40 years ago, which firstly were discovered as a viroid in plant in 1976 and were shown to encode subviral agents.2 Later on, a handful of such circRNAs were found in yeast mitochondrial RNAs and hepatitis delta virus.3,4 However, at that time, circRNAs have long been considered to be as by-products of pre-mRNAs rare mis-splicing. Thanks to the rapid development and breakthroughs of RNA sequencing and bioinformatics, the value of circRNAs in scientific field have been renewed starting about 7 years ago5,6 and numerous circRNAs have been successfully identified in various cell lines and as well as in model organisms such as mice, rice, fly and worm.7, 8, 9 Studies have revealed that circRNAs are abundantly and widely expressed in eukaryotes, and often show tissue/developmental stage-specific manners.10, 11, 12, 13 Furthermore, several researchers have confirmed that some circRNAs preferentially localize to exert their function in the cytoplasm, while the others circRNAs were localized in the nucleus.14,15 Although their biological functions are still rudimentary, a select number of circRNAs are functioning as miRNA sponges, transcriptional regulators, protein translation, and so on. Growing evidence and existing reports has indicated the expression of circRNAs is usually abnormal in many diseases such as cancers.16,17 In this review, we briefly review introduction the biogenesis, functions of circRNAs and emphasis on the currently progress of researches concerning circRNAs in different human diseases.
Categories and biogenesis of circRNAs
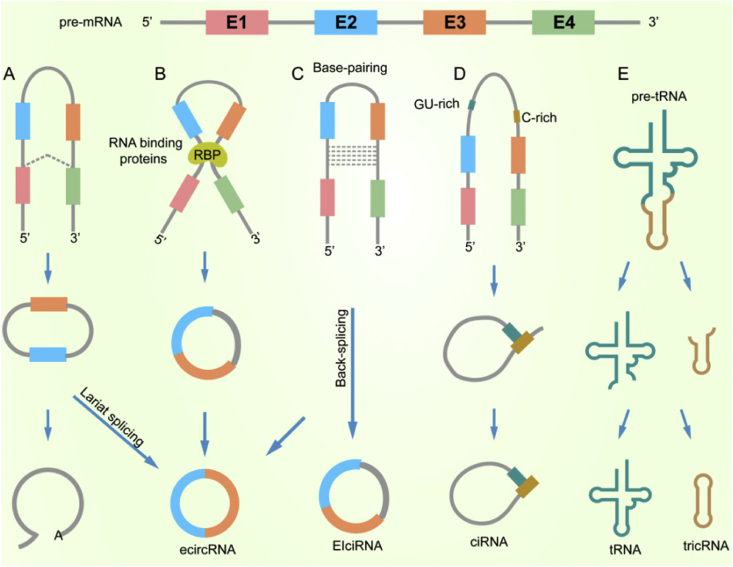
Understanding the structure features of circRNAs is critical for insight into their function. CircRNAs are produced from protein-coding genes in a non-canonical splicing event called back-splicing, in which a 5′ splice site (donor) attacks an upstream 3′ splice site (acceptor) generating a circular RNA molecule. Studies demonstrated that circularization and pre-mRNA splicing compete against each other via inhibiting the spliceosome by depleting components increases the ratio of circular to linear RNAs.18 When pre-mRNA processing events are slowed down, nascent RNA can be directed to alternative pathways that facilitate back-splicing. Based on their composition by competition of RNA pairing across the flanking sequences,19,20 circRNAs can be divided into four categories: exon-circRNAs (ecircRNAs), circular intron RNAs (ciRNAs), exon-intron circRNAs (EIciRNAs) and tRNA intronic circRNAs (tricRNAs)21 (Fig. 1). Although the mechanisms of circRNA biogenesis are still unclear, there are three models of circRNAs biogenesis that we possible have an agreement including lariat-driven circularization, base-pairing-driven circularization, and RNA-binding proteins (RBP)-mediated circularization.22 In lariat-driven circularization, a circRNA is formed by the joining of splice sites of the exons that are correlated with exon skipping during linear RNA formation, such as ecircRNAs and EIciRNAs.23,24 Base-pairing-driven circularization is also known as the back splicing mechanism, which depends on inverse-repeating or complementary base-pairing sequences across different introns to generate circRNA including ciRNA and EIciRNAs.13,25 Moreover, ciRNA is formed depending on a consensus motif containing an 11-nt C-rich element near the branch point and a 7-nt GU rich element near the 5′ splice site to escape disbranching. The other way that circRNA is derived from RBP-mediated circularization, which is the binding of RBP to the introns near splice sites to promote the circularization including the formation of ecircRNAs and EIciRNAs.22,26 The pre-tRNAs is recognized and spliced by the tRNA splicing nuclease complex, and then release and ligate the end to form tRNA and tricRNA.27
Figure 1.
CircRNA biogenesis mechanisms.
Biological functions of circRNAs
CircRNAs act as miRNA and protein (RBP) sponges
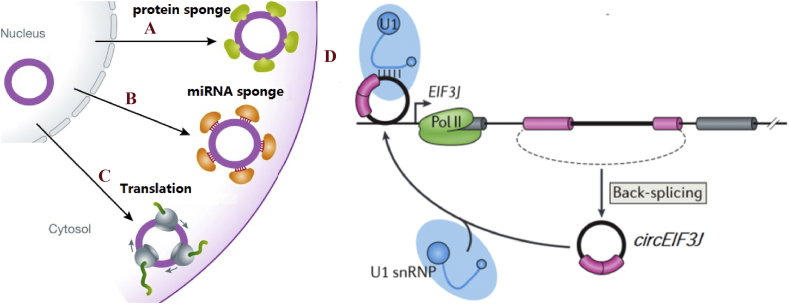
Accumulating studies has shown that circRNAs can bind miRNAs as the competitive endogenous RNA (ceRNA) affecting miRNA activities to bind its target mRNA, resulting in the increased expression of targets9,14 (Fig. 2A). This regulatory mechanism has become most common manner for cricRNAs. An illustrative example is that circRNA named CDR1as sponge for miR-7, which has been experimentally verified to act as a miR-7 sponge over 70 conserved miR-7 binding sites associating with Argonaute (AGO) proteins by a miR-7-dependent manner.14 Another typically example is that a highly expressed Circsry in mouse testis possessing 16 binding sites for miR-138.1 This is firstly demonstrated circRNAs act as miRNA sponges. In addition to above two circRNAs, more and more studies have confirmed that circRNAs can function as a ceRNA preventing their combinations or interactions with target mRNAs.28 Liu et al revealed that circCER regulated MMP13 expression by sponging miR-136 in human cartilage degradation.29 Chen and his group found that circFGFR2 could regulate skeletal muscle proliferation and differentiation in fibroblast cell lines of chicken embryo via sponging miR-133a-5p and miR-29b-1-5p.30
Figure 2.
Functions of circRNAs. (A) miRNA sponge; (B) protein sponge; (C) translation and (D) parental gene regulation.
In addition, circRNAs may be used as protein or RBP sponges to interact with proteins to regulate their function (Fig. 2B). A convincing example is circFoxo3, which interacts with p21 and CDK2 to form a complex of the ternary complex circFoxo3-p21-CDK2 giving rise to block cell cycle progression.31 CircMBL is also the best example, which sequesters muscleblind (MBL) protein and prevents it from binding to other targets.25 In a recent research of Qin et al they reported that hsa_circ_0001649 harbor six potential protein binding sites for U2 auxiliary factor 65 kDa subunit (U2AF65), 5 potential protein binding sites for eukaryotic initiation factor 4A-III, and one potential protein binding site for regulator of nonsense transcripts1.32 However, functions of hsa_circ_0001649 interacting with these proteins is still unknown and further studies should be investigated.
CircRNAs can be translated into proteins or peptides
As a conventional viewpoint, circRNAs are generally believed having no ability to encode proteins due to lacking 5′ and 3′ ends. Emerging evidence indicated that circRNAs function is as a template for translation or synthesis of proteins or peptides (Fig. 2C). For example, a study confirmed that a circRNA with 220 nts length from the rice yellow mottle virus can encode a highly basic, 16-kDa protein.33 Zhong's group reported that not only proteins can be translated from circRNAs but it can also perform its potential function. They demonstrated that circ-ZNF609 could translate proteins in murine myoblasts when driven by internal ribosome entry site (IRES), which functions in myogenesis.34 From a research by Yang et al showed that N6-methyladenosine (m6A) is sufficient to drive translation initiation of protein from circRNAs in human cells.35 Subsequently, further studies confirmed the prevalence of m6A in circRNAs.36 More recently, Yang et al revealed that circFBXW7 can directly encode protein FBXW7-185aa, which could regulate the stability of c-Myc leading to repress the progress of malignant glioma.37
CircRNAs regulate the expression of parental gene
Recent advances have revealed that circRNAs could regulate the expression of parental genes. Researcher has reported that knockdown of circRNAs decrease the transcription of their host genes. As described in early 2013 from Molecular Cell published a study, they reported that three circRNAs (ci-ankrd52, ci-mcm5 and ci-sirt7) from intron can also function as positive regulators of their parental gene transcription by interacting with RNA polymerase II (Pol II) and knockdown of their led to the reduced expression of their parental genes.38 In addition, circRNAs can interact with U1 small nuclear ribonucleoproteins (U1snRNP) to regulate the translations of the transcript from its parental gene by binding to RNA Pol II complex (Fig. 2D). For instance, both circEIF3J and circPAIP2 can bind to U1snRNP and RNA Pol II in a cis-acting form to elevate their parental gene transcription.39 In addition, a study showed that circPABPN1 positive regulates PABPN1 mRNA to inhibit the target HuR protein, and subsequently suppress the translation of its parental gene.40 Due to the unique structure and high stability, circRNAs may exert a series of biological functions through different mechanisms. In addition to above defined functions, other functions of circRNAs are summarized in Table 1. However, more detailed studies are needed to explore and determine the functions of circRNAs in future.
Table 1.
Putative functions of circRNAs.
CircRNAs and cancer or tumor
A vast number of evidence has uncovered a potential role of circRNAs which involved in a variety of human diseases, especially in the occurrence and development of cancer. It is also increasingly understood that circRNAs functions associating with cancer are being recognized. Cancerogenesis involves a number of causal molecular alterations, including genetic mutations and changes of gene expression states. Here, we focus mainly on the recent progress of circRNAs in different cancers.
CircRNAs and breast cancer
Breast cancer (BC) is one of the most ordinary malignant tumors with the highest and increasing morbidity and mortality affects more than 10% of females over 100 countries.48 According to the latest data from Global Cancer Statistics 2018, there are over 2 million females who are newly diagnosed with BC. It is well acknowledged that circRNAs were often unconventionally expressed in BC, which may have potential functions. Currently, many BC-related circRNAs had been identified. Related studies have suggested that circ_0067934 functions as an oncogene in BC by targeting Mcl-1.49 Study showed that circ_0067934 expression was significantly higher in BC tissues compared with that in adjacent non-tumor samples. After knockdown of circ_0067934 in vitro, cell proliferation in BC was inhibited, thereby effecting cell cycle in BC. Results implied that Mcl-1 was down-regulated via the knockdown of circ_0067934 in BC. The expression levels of hsa-circRNA-0005795 and hsa-circRNA-0088088 were significantly different both in serum exosomes and tissues and might function as ceRNA and play vital roles in BC development suggesting circRNAs hold significant diagnostic value for the prediction of BC by examining the expression of the circRNAs in serum exosomes between BC patients with healthy donors.50 Hsa_circ_0008039 can be used as a sponge of miR-432-5p to promote proliferation and invasion of the BC cells.51 More recently, Li et al investigated the differentially expressed (DE) profiles of circRNAs between plasma of patients with BC and normal human. Among DE circRNAs, hsa_circ_0069094, hsa_circ_0079876, hsa_circ_0017650, and hsa_circ_0017536 showed significant differences in expression levels between plasma of patients with BC and normal human.52 Results will provide new directions for the prediction, diagnosis and treatment for BC.
Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths and accounts for 90% of primary liver carcinomas in the word.53 It is estimated that HCC has high morbidity and mortality rates in humans, which give rise to over over 750,000 deaths each year.54 A recent study suggests that circRNAs are important for the initiation, migration, and invasion of HCC. Liu et al55 confirmed that circRNA-5692 overexpression effectively attenuated the malignant behaviors of HCC. Furthermore, circRNA-5692 sponge oncogenic miR-328-5p to enhance tumor suppressor DAB2IP expression, decreasing the malignant behaviors of HCC in vitro and in vivo. The investigations by Zhan et al showed that hsa_circRNA_103809 could promote HCC development via regulation axis of miR-377-3p/FGFR1/ERK.56 Studies by Su et al57 revealed that CDR1as could promote HCC progression by sponging miR-1270, which is directly transferred from HCC cells to surrounding normal cells via exosomes to further mediate the biological functions of surrounding cells. Zhang et al58 found that circTRIM33-12 is down-regulated in HCC which promoted HCC progression including cell proliferation, invasion, and immune evasion. According to Han's researches, they reported that circMTO1 repressed HCC progression through sponging miR-9.59 Therefore, the implication is that circMTO1 may become new targets in treating HCC.
Gastric cancer
Gastric cancer (GC), alias stomach adenocarcinoma, is one of the most frequent cancers and poses a serious threat to human life and leading causes of cancer-induced death worldwide, with geographically high morbidity in East Asia.60 In 2018, there are estimated 1,033,701 new cases of GC and 782,685 cases of GC-related death, which ranks the fifth and third in cancer incidence rates and mortality rates, respectively.61 Sun et al uncovered a regulatory network of circCOL6A3/miR-3064e5p/COL6A3 mRNA, which regulated malignant behaviors of GC cells by sequestration of miR-3064e-5p and dis-inhibition of COL6A3.62 CircRNA_100269 is down-regulated in GC and further inhibits cell growth in GC by targeting miR-630.63 Other study showed that overexpressed circRNA_001569 promoted cell viability of GC via suppressing the expression of miR-145, which was mediated by NR4A2.64 Wei et al analyzed 30 patients and found that hsa_circRNA_102958 exhibited highly expression in GC tissues compared with paired adjacent normal tissues. A Clinic Pathological and experiment analysis showed that hsa_circRNA_102958 level in GC tissues was positively associated with TNM stage (P = 0.032). The area under the ROC curve was 0.74.65 Hsa_circ_0042881, derived from exons 2–8 of tumor-suppressive gene NF1, serves as a novel oncogenic circRNA through functioning as miR-16 sponge in GC.66 For example, hsa_circ_0000096 regulates the proliferation and migration of GC cells by modulating the gene expression of cyclin D1, CDK-6, MMP-2, MMP-9, and E-cadherin.67 These discoveries will provide new therapeutic strategies for the prevention and treatment of GC in future. In a recent research reported that cir-cYAP1 was down-regulated in GC, and acted as a tumor suppressor by sponging miR-367-5p to further repress expression of target p27, thereby result in inhibition of GC cell proliferation and invasion.68
Lung cancer
Lung cancer (LC) is a global problem of cancer-associated deaths with high incidence and mortality for both sexes. A total of 85% of LC cases are diagnosed as non-small-cell lung cancer (NSCLC).69 With ongoing environmental degradation and pollution, the morbidity and mortality rate of NSCLC is increasing.70 Recently, circRNAs were reported to be closely associated with NSCLC. From the investigation by Zhao et al found that hsa_circ_404833 sponges miR-149-5p and inhibits cell motility, resulting in the acquired gefitinib resistance in LC.71 A research group used human circRNAs microarray analysis technology to screen circRNAs expression profiles in NSCLC oncogenesis. They identified a total of 957 abnormally expressed circRNAs in NSCLC tissue compared with normal tissue.72 Further experiment revealed that hsa_circ_0007385 was significantly up regulated in NSCLC tissue and cells. Moreover, the knockdown of hsa_circ_0007385 could significantly suppress the proliferation, migration and invasion of NSCLC cells. Dai et al analyzed the expression of circ0006916 and its parental gene HOMER1 in normal and LC bronchial epithelial cells and in LC tissues in relation to prognosis.73 Subsequently, the author also further examined the effect of circ0006916 on cell proliferation and demonstrated an important role for circ0006916 in anti-BPDE-induced carcinogenesis. CircRNA-100876 was found to up-regulate in NSCLC and closely associated with tumor staging and lymph node metastasis, and could serve as a potential prognostic biomarker for NSCLC.74 Zhang et al revealed the expression profiles of circRNAs in NSCLC tissues and their adjacent lung tissue and found that a circPIP5K1A expression was higher, indicating it mays be as a new circular RNA biomarker in NSCLC.75 Additionally, lung adenocarcinoma (LUAD) is a common pathological type of lung cancer, which belongs to non-small cell carcinoma. CDR1as is an extensively investigated circRNA, which has been reported to be involved in various cancers. A study indicated that the expression of CDR1as was significantly up-regulated in LUAD tissues in comparison to the matched controls. Moreover, the high-expression of CDR1as is related with the pemetrexed and cisplatin insensitivity of LUAD patients.76 The same trend is that circ100146 is also highly expressed in NSCLC cell lines and the chemically induced malignant transformed bronchial cell line, 16HBE-T, as well as 40 paired tissue samples of NSCLC. Subsequently, circ100146 inhibited the proliferation and invasion of cells and promoted apoptosis by sponging miR-361-3p and miR-615-5p.77 The functional mechanism of these identified circRNAs in LC may be not limited to miRNA sponges and others functions would be explored for LC diagnosis and therapy in future.
Bladder cancer
Bladder cancer (BC) is one of the ten most common tumors, which commonly diagnosed malignancy of the urinary system worldwide.78 Growing evidence has suggested that circRNAs have been found own great correlation with BC. In BC cells, circHIPK3, derived from Exon2 of the HIPK3 gene, was significantly down-regulate and targeted miR-558 to inhibit the growth and metastasis of BC in vivo.79 And then, Xie et al reported that circHIPK3 islow-expressed in BC and its over-expression will promotes gemcitabine sensitivity in BC.80 Results suggested that circHIPK3 could be as an independent prognostic biomarker for BC patients. Zhong et al performed a DE profile of circTCF25/miR-103a-3p/miR-107/CDK6 regulatory pathway in BC. They found that over-expression of circTCF25 could promote proliferation and migration of EJ and T24 BC cell lines.81 The other study by Su et al showed that the expression of circTFRC could elevate the proliferation of BC cell line, which is related to the low tumor stage and survival rate.82 Yu et al found that circPDSS1 was up-regulated in urothelial BC and promoted BC progress by sponging miR-16. Moreover, overexpression of miR-16 not only led to suppress proliferation, invasion and migration of urothelial BC cells, but also attenuated the effects the overexpression of circPDSS1.83 Research found that CDR1as was low expressed in BC tissues compared with adjacent normal tissues. Further, overexpression of CDR1as can inhibit the proliferation and invasion of BC cells by sponging miR-135a.84
Esophageal carcinoma
Esophageal carcinoma (ESCC) is a highly malignant digestive tract tumor in the word, which is divided into esophageal adenocarcinoma and ESCC. Shi et al found that circ-PRKCI is obviously up-regulated in ESCC, which as a ceRNA to regulate AKT3 expression by sponging miR-3680-3p in ESCC.85 For instance, a study has showed that circ_100876 is highly expressed in ESCC, which can promote cell proliferation, incursion, and distal metastasis, as well as the progress of EMT.86 Circ-DLG1 was observed to be increased in ESCC tissues, cell lines, and plasma and can significantly promote cell proliferation.87 Cir-ITCH was expressed at low levels in ESCC compared with peritumoral tissue, and may inhibit ESCC by regulating the Wnt signaling pathway.88 However, the specific regulatory function of cir-ITCH requires further study. Another research shows that the expression of circ_0067934 is significantly up-regulated in ESCC than normal tissues, suggesting its expression is concerned with ESCC differentiation and proliferation.89
Renal cell carcinoma
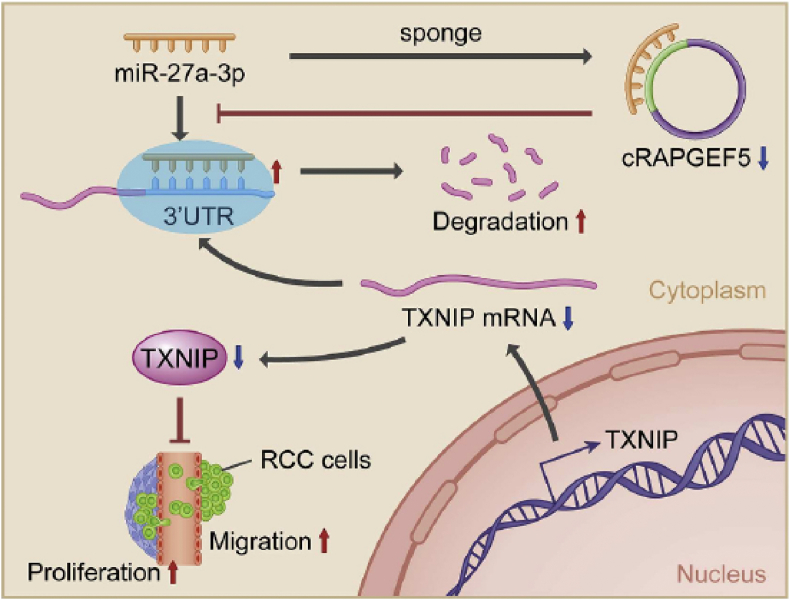
Renal cell carcinoma (RCC) has been increasing, accounting for 3% of cancer-related incidence rate, and there are over 400,000 as new cases and more than 170,000 deaths in 2018 worldwide.61 circRAPGEF5 is derived from exons 2–6 of the RAPGEF5 gene, and its down-regulation decreases the adsorption of miR-27a-3p, resulting in increasing the degradation of TXNIP and thus promoting RCC cells proliferation and migration90 (Fig. 3). Previous studies have indicated that circ-ZNF609 can promote retinal glial cell proliferation.91 Recently, Xiong and his colleagues investigated the effect of circ-ZNF609 on cell proliferation or invasion ability, and found that circ-ZNF609 is significantly increased in RCC lines by sponging miR-138-5p mediated FOXP4 expression to modulate tumor growth.92 They demonstrated that the high expression of circ-ZNF609 promotes cell proliferation and invasion ability.
Figure 3.
Schematic diagram of the circRAPGEF5-mediated pathway in RCC cells.
Osteosarcoma
Osteosarcoma (OS) is a common malignant bone tumor and mainly affects young adults. Accumulating studies indicated that circRNAs play a critical role in the pathogenesis of OS. As reported by Li et al circGLI2 is up-regulated in OS tissue and cells, and promotes OS cell proliferation, migration, and invasion by targeting miR-125b-5p.93 It was discovered by a research team that in chemoresistant OS patients, hsa_circ 0004674 was the most up-regulated circRNA, and was negatively correlated with prognosis.94 Another research group found that circ-HIPK3 was down-regulated in OS and correlated with poor prognosis, up-regulation of circ-HIPK3 suppressed cell proliferation, migration and invasion of OS cells.95
Prostate cancer
Prostate cancer (PC) is a common malignant tumor of the male genitourinary system, and its incidence ranks is the highest among man malignancies in worldwide, especially in Western countries. It were reported that there were 1,276,106 new cases of prostate cancer in 2018 from the United States data, and the disease resulted in 358,989 deaths.96 Feng et al investigated that circ0005276 increased migration and proliferation in prostate cancer.97 Additionally, Kong et al revealed circ-SMARCA5 exhibited high expression levels in PC samples than in matched noncancerous prostate tissues, implying that circ-SMARCA5 could be an oncogenic circRNA.98
Cervical cancer
Cervical cancer (CC) is also one of the common gynecologic cancers effecting female health in worldwide, accounting for a large proportion of death cancer-related throughout the world. Zheng et al investigated the expression profiles of circRNAs by transfecting E7 siRNA in CC Caski cells with high-throughput microarray technology.99 A total of 526 dys-regulated circRNAs were identified. Further bioinformatics analysis indicated that DE circRNAs might implicate in the mTOR signaling pathway, suggesting these DE circRNAs have the relationship between dys-regulation and CC progression. A recently functional investigation by Ji et al100 revealed that knockdown of circSLC26A4 repressed the proliferation, invasion, and tumor growth in cervical cancer tissue and cells by sponging miR-1287-5p implying circSLC26A4 may be as an oncogene in CC tumorigenesis.
Oral squamous cell carcinoma
Oral cancer has become a severe public issue worldwide and threat to people's health.101 Currently, some studies on circRNAs in Oral squamous cell carcinoma (OSCC) have been reported. A study showed that hsa_circ_0008309 was down-regulated in OSCC tissues compared with adjacent normal control tissues by application of RNA sequencing and qRT-PCR analysis.102 Besides, circRNA can be as a cancer suppressor. For example, cir-cDOCK1 could inhibit cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC.103
Ovarian cancer
Ovarian cancer (OC) is one of the most common malignant tumors and threatening female reproductive system causing over 25,000 deaths annually.104 Actually, accumulating researches indicated that dys-regulated expression of circRNAs played a significant role in the occurrence and progression of OC. Xu et al105 identified 5917 circRNAs and performed their DE analysis between ovarian normal and ovarian cancer tissues. They found that circ0004390 could promote ovarian cancer cells proliferation by sponging miR-198. The similar trend has been reported that hsa_circ_0013958 was up-regulated in OC tissues and knockdown of hsa_circ_0013958 inhibited proliferation, migration, and invasion of OC cells.106 Sheng et al reported that circRNAUBAP2 was highly expressed in OC tissues and promoted the progression of OC by sponging miRNA-144.107 These results might provide a valuable reference for clinical diagnosis and treatment of OC.
Colorectal cancer
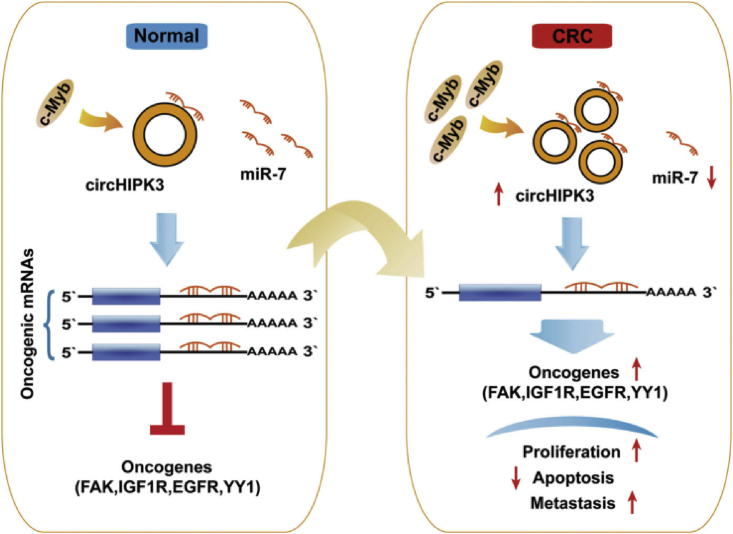
Colorectal cancer (CRC) is also most commonly diagnosed cancer, which led to 9.2% of all cancer-related deaths each year. Study found that CiRS-122, exosome-delivered circRNA, can promotes glycolysis to induce chemoresistance by miR-122 sponging and PKM2 up-regulation in CRC.108 Recently, Li et al109 verified that circVAPA was significantly up-regulated and promoted CRC progression by sponging miR-101, indicating circVAPA may be as a promising biomarker to predict chemoradiation resistance in CRC. Xie et al110 found that has-circ-001569 could promote the proliferation and invasion of CRC cells via sponging miR-145 to enhance the expression of E2F5, BAG4, and FMNL2. Another report demonstrated that circHIPK3 could promote CRC cells growth and metastasis by directly sponging miR-7.111 They further found that circHIPK3 is frequently up-regulated partly caused by the overexpression of transcription factor c-Myb, then circHIPK3 sequester and inhibit miR-7 activity via sponging miR-7, thereby result in enhancing the expression proto-oncogenes including FAK, IGF1R, EGFR, and YY1, which promoting CRC development and progression (Fig. 4).
Figure 4.
The schematic diagram of the c-Myb/circHIPK3/miR-7 axis in CRC.
CircRNAs and other human diseases
Age-related cataract
Age-related cataract (ARC) is the most cause of vision loss among the aged population.112 Liu et al113 revealed that circHIPK3 expression was down-regulated in all three subtypes of ARC compared with the control. Moreover, circHIPK3 silencing in ARC cases stimulated the human lens epithelial cells apoptosis mediated by oxidative stress by the circHIPK3/miR-193a/CRYAA axis, suggesting the role of circHIPK3 as a functional regulator and providing a novel insight into the pathogenesis of ARC.
Psoriasis
Psoriasis is one of the most common chronic inflammatory skin conditions, with 1–3% of the adult population affected worldwide.114 Moldovan et al firstly performed circRNA profiling of paired lesional and non-lesional skin from patients with psoriasis by RNA sequencing. This research provided fundamental information regarding DE circRNAs. However, whether circRNA is a cause or a consequence of the disease, the specific roles of circRNA is need to be further revealed.115
System lupus erythematosus
System lupus erythematosus (SLE) is a prototypic autoimmune disease. The study of Li et al firstly screened circRNAs profiles in SLE patients’ peripheral blood plasma. Some circRNA candidates were selected for validation and potential circRNA/miRNA interaction networks were also constructed.116 Result implied that circRNAs may associate with SLE. In another study, Luan et al investigated circRNA profiles in renal samples from SLE patients and health controls.117 They spotted circHLA-C as a probable regulator for miR-150 because of circHLA-C and miR-150 displaying a negative correlation, indicating the potential roles of circHLA-C in SLE. Future studies should focus on their regulatory mechanism between circRNAs and SLE.
Pulmonary tuberculosis
Pulmonary tuberculosis (PT), caused by Mycobacterium tuberculosis, is still remaining severe threat to public health. Recent study has revealed that circRNAs are intensively associated with PT.118 A report by Zhang et al showed 170 dys-regulated circRNAs in PT compared with healthy control. The findings indicated an important role of circRNA mediating gene regulation in the pathogenesis of PT.119 Yi et al performed the plasma circRNA expression profiles of active PT patients and found that hsa_circRNA_103571 was significantly decreased in active PT patients.120 At present, study on the regulatory mechanism of circRNAs in PT is at drawn. These circRNAs potential function in PT will be explained in future.
Duchenne muscular dystrophy
Duchenne muscular dystrophy (DMD) is a severe muscular disorder involving a progressive deterioration of muscle function and is caused by frame-shifting deletions or nonsense mutations in the DMD gene resulting in the absence or reduced production of the dys-trophin protein.121 Study has shown that two circRNAs (circQKI and circBNC2) were up-regulated during in vitro differentiation of normal myoblasts as described above and were down-regulated in the DMD conditions, well correlating with the notion that dystrophic cells have altered progression into the differentiation process.122 In addition, circ-ZNF609 was found to be down-regulated during myogenesis in control myoblasts, which was highly expressed in differentiated DMD myoblasts.34
Nervous system diseases
CircRNAs involved in nervous system diseases have been largely reported. Hong's research group has reported that circDLGAP4 participated in the progression of Parkinson's disease.123 Zhao et al found that sporadic Alzheimer's disease is associated with an aberrant miRNA/circRNA system in the hippocampal CA1 region.124 Another study showed that Alzheimer's disease mediated by CDR1as-miR-7 axis. Moreover, they found that CDR1as is significantly reduced in Alzheimer patients indicating that CDR1as may have a function in Alzheimer disease.125 Several studies demonstrated that circRNAs associated with the process of stroke. For example, Dong et al identified different expression of circRNAs in patients with acute ischemic stroke and healthy control by RNA sequencing.126 Wu et al127 also found that circTLK1 can aggravate neuronal injury via miR-335-3p/TIPARP resulting in neurological deficits and AIS. The great an effort is needed to develop new approaches to treat nervous system diseases complications in future. In addition to above diseases, more and more circRNAs associated with various disease are gradually being discovered. A brief summary of some other human disease-related circRNAs were summarized in Table 2. It was believed that more dys-regulated circRNAs will be identified in various types of cancer with development of cancer specific algorithms for RNA-seq data and their detailed functions are waiting to be elaborated in near future.
Table 2.
Summary of circRNAs involving in other human diseases.
| Disease types | Representative circRNA | Putative functions and mechanisms | Reference |
|---|---|---|---|
| Thyroid cancer | cRAPGEF5 | miR-198/FGFR1 pathway | 128 |
| Laryngeal cancer | circ-CCND1 | miRNA sponge (miR-646) | 129 |
| Pancreatic cancer | circRNA_0007334 | miRNA sponge (miR-144-3p and miR-577) | 130 |
| Kidney cancer | circ-HIAT1 | miR-29a-3p/29c-3p/195-5p pathway | 131 |
| Glioma | cZNF292 | Inhibits cell proliferation | 132 |
| Melanoma | circRNA_0084043 | miRNA sponge (miR-153-3p) | 133 |
| Cholangiocarcinoma | circ_0001649 | Induces cell tumor apoptosis | 134 |
| Hematological malignancy | f-circM9 and f-circPR | Induces cell tumor apoptosis | 135 |
| Leukemia | circAF4 | miRNA sponge (miR-128) | 136 |
| Cardiovascular disease | cZNF609 | miRNA sponge (miR-615-5p) | 137 |
| Osteoarthritis | circRNA-CER | miRNA sponge (miR-136) | 29 |
| Heart failure | cZNF292 | Induces tube formation and endothelial cells sprouting | 138 |
Conclusions and perspectives
With the advancement of RNA sequencing technology and the rapid development of bioinformatics, a large number of circRNAs were discovered. Moreover, emerging as a new regulatory RNA molecules, circRNAs have been widely focused on RNA research world, especially in biomedical fields. It was proved that circRNAs play multiple roles in gene expression regulation involving in life processes and initiation and development of diverse diseases. Furthermore, circRNAs function as oncogenic factors or tumor suppressors that participate in cancer occurrence and progression through multiple mechanisms. CircRNAs often had expressed in a tissue and developmental stage specific manner with abundance in certain specific pathological tissue. It has partially proven that circRNAs could be used as biomarkers or regulators for the diagnosis and prognosis of tumors. Currently, some methods have been developed to identify circRNAs, for example, RNA sequencing, circRNAs microarray, northern blotting, circRNA sequencing, PCR-based analyses, RNA fluorescence in situ hybridization.139,140 With the prompt advance of molecular biology techniques, these methods may be more accurate for detecting or quantifying circRNA although each technique might has specific advantages and disadvantages in this area. Up to now, the most important and most widely studied function of circRNAs is in their role as miRNA sponges. However, miRNAs sponge mechanism does not represent all the functions of circRNAs. Understanding the full aspect of functions for these molecules is only the tip of the iceberg and other functions of circRNAs require further study. Taken together, more challenging functional studies of circRNAs needed to be uncovered in both physiological and pathological conditions. It is widely believed that the regulatory function of a large number of circRNAs will be revealed gradually in human diseases.
Authors contribution
Yong Huang: conceptualization, design of the study, writing original draft.
Cai Zhang: resources, writing-review and editing, supervision.
Jianli Xiong: writing-review and editing, supervision, data curation.
Hongtao Ren: writing, review and editing.
Conflict of Interests
The authors declare that they have no conflict of interest.
Funding
This work was jointly supported by the Natural Science Foundation of China [grant numbers 1471971, 31402263 and 31872537].
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Memczak S., Jens M., Elefsinioti A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 2.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kos A., Dijkema R., Arnberg A.C., van der Meide P.H., Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 4.Arnberg A.C., Van Ommen G.J., Grivell L.A., Van Bruggen E.F., Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19(2):313–319. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z., Xie L., Han L. Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics. 2017;7(12):3106–3117. doi: 10.7150/thno.19016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L., Wang C., Sun H. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. 2021;22(2):1706–1728. doi: 10.1093/bib/bbaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P.L., Bao Y., Yee M.C. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0090859. e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westholm J.O., Miura P., Olson S. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 10.Ouyang H., Chen X., Wang Z. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 2018;25(1):71–86. doi: 10.1093/dnares/dsx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7) doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9) doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rybak-Wolf A., Stottmeister C., Glazar P. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 14.Hansen T.B., Jensen T.I., Clausen B.H. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 15.Xiao M.S., Ai Y., Wilusz J.E. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020;30(3):226–240. doi: 10.1016/j.tcb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu M., Xie F., Tang X., Wang T., Wang S. Insights into the role of circular RNA in macrophage activation and fibrosis disease. Pharmacol Res. 2020;156 doi: 10.1016/j.phrs.2020.104777. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Sun D., Pu W., Wang J., Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Liang D., Tatomer D.C., Luo Z. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell. 2017;68(5):940–954 e943. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X., Cai Y., Xu J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int J Mol Sci. 2019;20(16) doi: 10.3390/ijms20163926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zang J., Lu D., Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 23.Jeck W.R., Sorrentino J.A., Wang K. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilusz J.E. A 360 degrees view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9(4):e1478. doi: 10.1002/wrna.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashwal-Fluss R., Meyer M., Pamudurti N.R. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Wu J., Qi X., Liu L. Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids. 2019;16:589–596. doi: 10.1016/j.omtn.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt C.A., Giusto J.D., Bao A., Hopper A.K., Matera A.G. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res. 2019;(12):6452–6465. doi: 10.1093/nar/gkz311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aufiero S., Reckman Y.J., Pinto Y.M., Creemers E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16(8):503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 29.Liu Q., Zhang X., Hu X. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 'sponge' in human cartilage degradation. Sci Rep. 2016;6 doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., Ouyang H., Wang Z., Chen B., Nie Q. A novel circular RNA generated by FGFR2 gene promotes myoblast proliferation and differentiation by sponging miR-133a-5p and miR-29b-1-5p. Cells. 2018;7(11) doi: 10.3390/cells7110199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin M., Liu G., Huo X. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 33.AbouHaidar M.G., Venkataraman S., Golshani A., Liu B., Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci USA. 2014;111(40):14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legnini I., Di Timoteo G., Rossi F. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y., Fan X., Mao M. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou C., Molinie B., Daneshvar K. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20(9):2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Gao X., Zhang M. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3):304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Zhang X.O., Chen T. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 39.Li Z., Huang C., Bao C. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 40.Abdelmohsen K., Panda A.C., Munk R. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conn V.M., Hugouvieux V., Nayak A. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3 doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 42.Liu J., Liu T., Wang X., He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1) doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holdt L.M., Stahringer A., Sass K. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7 doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du W.W., Fang L., Yang W. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Yang F., Fang E. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26(7):1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng W.L., Marinov G.K., Chin Y.M., Lim Y.Y., Ea C.K. Transcriptomic analysis of the role of RasGEF1B circular RNA in the TLR4/LPS pathway. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-12550-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12) doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeSantis C.E., Ma J., Goding Sauer A., Newman L.A., Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi: 10.3322/caac.21412. [DOI] [PubMed] [Google Scholar]
- 49.Wang J.M., Li X.J., Wang J. Circular RNA circ_0067934 functions as an oncogene in breast cancer by targeting Mcl-1. Eur Rev Med Pharmacol Sci. 2019;23(13):9499–9505. doi: 10.26355/eurrev_201911_19444. [DOI] [PubMed] [Google Scholar]
- 50.Yang S.J., Wang D.D., Zhou S.Y. Identification of circRNA-miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics. 2020;12(2):101–125. doi: 10.2217/epi-2019-0058. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Lu C., Zhou Y., Zhang Z., Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502(3):358–363. doi: 10.1016/j.bbrc.2018.05.166. [DOI] [PubMed] [Google Scholar]
- 52.Li Z., Chen Z., Hu G. Profiling and integrated analysis of differentially expressed circRNAs as novel biomarkers for breast cancer. J Cell Physiol. 2020;235(11):7945–7959. doi: 10.1002/jcp.29449. [DOI] [PubMed] [Google Scholar]
- 53.Venook A.P., Papandreou C., Furuse J., de Guevara L.L. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;15(Suppl 4):5–13. doi: 10.1634/theoncologist.2010-S4-05. [DOI] [PubMed] [Google Scholar]
- 54.Sayiner M., Golabi P., Younossi Z.M. Disease burden of hepatocellular carcinoma: a global perspective. Dig Dis Sci. 2019;64(4):910–917. doi: 10.1007/s10620-019-05537-2. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z., Yu Y., Huang Z. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12) doi: 10.1038/s41419-019-2089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhan W., Liao X., Chen Z. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 2020;235(2):1733–1745. doi: 10.1002/jcp.29092. [DOI] [PubMed] [Google Scholar]
- 57.Su Y., Lv X., Yin W. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY) 2019;11(19):8182–8203. doi: 10.18632/aging.102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang P.F., Wei C.Y., Huang X.Y. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-1031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han D., Li J., Wang H. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 60.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 61.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 62.Sun X., Zhang X., Zhai H., Zhang D., Ma S. A circular RNA derived from COL6A3 functions as a ceRNA in gastric cancer development. Biochem Biophys Res Commun. 2019;515(1):16–23. doi: 10.1016/j.bbrc.2019.05.079. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y., Liu H., Li W. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY) 2017;9(6):1585–1594. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen F., Liu P., Xu Z. CircRNA_001569 promotes cell proliferation through absorbing miR-145 in gastric cancer. J Biochem. 2019;165(1):27–36. doi: 10.1093/jb/mvy079. [DOI] [PubMed] [Google Scholar]
- 65.Wei J., Wei W., Xu H. Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomark. 2020;27(2):139–145. doi: 10.3233/CBM-182029. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z., Ma K., Pitts S. Novel circular RNA circNF1 acts as a molecular sponge, promoting gastric cancer by absorbing miR-16. Endocr Relat Cancer. 2019;26(3):265–277. doi: 10.1530/ERC-18-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li P., Chen H., Chen S. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633. doi: 10.1038/bjc.2016.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu H., Liu Y., Bian Z. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 (Kip1) axis. Mol Cancer. 2018;17(1):151. doi: 10.1186/s12943-018-0902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 70.Zhang W., Wu X., Hu L. Overexpression of human papillomavirus type 16 oncoproteins enhances epithelial-mesenchymal transition via STAT3 signaling pathway in non-small cell lung cancer cells. Oncol Res. 2017;25(5):843–852. doi: 10.3727/096504016X14813880882288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao J., Li L., Wang Q., Han H., Zhan Q., Xu M. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44(6):2138–2146. doi: 10.1159/000485953. [DOI] [PubMed] [Google Scholar]
- 72.Jiang M.M., Mai Z.T., Wan S.Z. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144(4):667–674. doi: 10.1007/s00432-017-2576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai X., Zhang N., Cheng Y. RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA circ0006916, with important functions in lung cancer cells. Carcinogenesis. 2018;39(8):981–992. doi: 10.1093/carcin/bgy061. [DOI] [PubMed] [Google Scholar]
- 74.Yao J.T., Zhao S.H., Liu Q.P. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 75.Zhang S., Zeng X., Ding T. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mao Y., Xu R. Circular RNA CDR1-AS contributes to pemetrexed and cisplatin chemoresistance through EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed Pharmacother. 2020;123 doi: 10.1016/j.biopha.2019.109771. [DOI] [PubMed] [Google Scholar]
- 77.Chen L., Nan A., Zhang N. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 79.Li Y., Zheng F., Xiao X. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie F., Zhao N., Zhang H., Xie D. Circular RNA CircHIPK3 promotes gemcitabine sensitivity in bladder cancer. J Cancer. 2020;11(7):1907–1912. doi: 10.7150/jca.39722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhong Z., Lv M., Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6 doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Su H., Tao T., Yang Z. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18(1) doi: 10.1186/s12943-019-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu Q., Liu P., Han G., Xue X., Ma D. CircRNA circPDSS1 promotes bladder cancer by down-regulating miR-16. Biosci Rep. 2020;40(1) doi: 10.1042/BSR20191961. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Li P., Yang X., Yuan W. CircRNA-Cdr1as exerts anti-oncogenic functions in bladder cancer by sponging MicroRNA-135a. Cell Physiol Biochem. 2018;46(4):1606–1616. doi: 10.1159/000489208. [DOI] [PubMed] [Google Scholar]
- 85.Shi N., Shan B., Gu B., Song Y., Chu H., Qian L. Circular RNA circ-PRKCI functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p in esophageal squamous cell carcinoma. J Cell Biochem. 2019;120(6):10021–10030. doi: 10.1002/jcb.28285. [DOI] [PubMed] [Google Scholar]
- 86.Cao S., Chen G., Yan L., Li L., Huang X. Contribution of dysregulated circRNA_100876 to proliferation and metastasis of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11:7385–7394. doi: 10.2147/OTT.S177524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rong J., Wang Q., Zhang Y. Circ-DLG1 promotes the proliferation of esophageal squamous cell carcinoma. OncoTargets Ther. 2018;11:6723–6730. doi: 10.2147/OTT.S175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li F., Zhang L., Li W. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia W., Qiu M., Chen R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6 doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Q., Liu T., Bao Y. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77. doi: 10.1016/j.canlet.2019.10.017. [DOI] [PubMed] [Google Scholar]
- 91.Wang J.J., Liu C., Shan K. Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics. 2018;8(12):3408–3415. doi: 10.7150/thno.25156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiong Y., Zhang J., Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR-138-5p in renal carcinoma. J Cell Physiol. 2019;234(7):10646–10654. doi: 10.1002/jcp.27744. [DOI] [PubMed] [Google Scholar]
- 93.Li J.F., Song Y.Z. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biol. 2017;39(7) doi: 10.1177/1010428317709991. [DOI] [PubMed] [Google Scholar]
- 94.Kun-Peng Z., Xiao-Long M., Lei Z., Chun-Lin Z., Jian-Ping H., Tai-Cheng Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. 2018;10(10):1327–1346. doi: 10.2217/epi-2018-0023. [DOI] [PubMed] [Google Scholar]
- 95.Xiao-Long M., Kun-Peng Z., Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–1862. doi: 10.7150/jca.24619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rawla P. Epidemiology of prostate cancer. World J Oncol. 2019;10(2):63–89. doi: 10.14740/wjon1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng Y., Yang Y., Zhao X. Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis. 2019;10(11):792. doi: 10.1038/s41419-019-2028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kong Z., Wan X., Zhang Y. Androgen-responsive circular RNA circSMARCA5 is up-regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493(3):1217–1223. doi: 10.1016/j.bbrc.2017.07.162. [DOI] [PubMed] [Google Scholar]
- 99.Zheng S.R., Zhang H.R., Zhang Z.F. Human papillomavirus 16 E7 oncoprotein alters the expression profiles of circular RNAs in Caski cells. J Cancer. 2018;9(20):3755–3764. doi: 10.7150/jca.24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ji F., Du R., Chen T. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 Axis. Mol Ther Nucleic Acids. 2020;19:413–420. doi: 10.1016/j.omtn.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang H., Feng C., Wang M., Yang S., Wei F. Circular RNAs: diversity of functions and a regulatory nova in oral medicine: a pilot review. Cell Transplant. 2019;28(7):819–830. doi: 10.1177/0963689719837917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li B., Wang F., Li X., Sun S., Shen Y., Yang H. Hsa_circ_0008309 may Be a potential biomarker for oral squamous cell carcinoma. Dis Markers. 2018;2018 doi: 10.1155/2018/7496890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang L., Wei Y., Yan Y. CircDOCK1 suppresses cell apoptosis via inhibition of miR196a5p by targeting BIRC3 in OSCC. Oncol Rep. 2018;39(3):951–966. doi: 10.3892/or.2017.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 105.Xu F., Ni M., Li J. Circ0004390 promotes cell proliferation through sponging miR-198 in ovarian cancer. Biochem Biophys Res Commun. 2020;526(1):14–20. doi: 10.1016/j.bbrc.2020.03.024. [DOI] [PubMed] [Google Scholar]
- 106.Pei C., Wang H., Shi C., Zhang C., Wang M. CircRNA hsa_circ_0013958 may contribute to the development of ovarian cancer by affecting epithelial-mesenchymal transition and apoptotic signaling pathways. J Clin Lab Anal. 2020;34(7) doi: 10.1002/jcla.23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sheng M., Wei N., Yang H.Y., Yan M., Zhao Q.X., Jing L.J. CircRNA UBAP2 promotes the progression of ovarian cancer by sponging microRNA-144. Eur Rev Med Pharmacol Sci. 2019;23(17):7283–7294. doi: 10.26355/eurrev_201909_18833. [DOI] [PubMed] [Google Scholar]
- 108.Wang X., Zhang H., Yang H. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li X.N., Wang Z.J., Ye C.X., Zhao B.C., Huang X.X., Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother. 2019;112 doi: 10.1016/j.biopha.2019.108611. [DOI] [PubMed] [Google Scholar]
- 110.Xie H., Ren X., Xin S. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zeng K., Chen X., Xu M. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4) doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 112.Abdelkader H., Alany R.G., Pierscionek B. Age-related cataract and drug therapy: opportunities and challenges for topical antioxidant delivery to the lens. J Pharm Pharmacol. 2015;67(4):537–550. doi: 10.1111/jphp.12355. [DOI] [PubMed] [Google Scholar]
- 113.Liu X., Liu B., Zhou M. Circular RNA HIPK3 regulates human lens epithelial cells proliferation and apoptosis by targeting the miR-193a/CRYAA axis. Biochem Biophys Res Commun. 2018;503(4):2277–2285. doi: 10.1016/j.bbrc.2018.06.149. [DOI] [PubMed] [Google Scholar]
- 114.Lebwohl M. Psoriasis. Lancet. 2003;361(9364):1197–1204. doi: 10.1016/S0140-6736(03)12954-6. [DOI] [PubMed] [Google Scholar]
- 115.Moldovan L.I., Hansen T.B., Veno M.T. High-throughput RNA sequencing from paired lesional- and non-lesional skin reveals major alterations in the psoriasis circRNAome. BMC Med Genomics. 2019;12(1) doi: 10.1186/s12920-019-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li H., Li K., Lai W. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta. 2018;480:17–25. doi: 10.1016/j.cca.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 117.Luan J., Jiao C., Kong W. circHLA-C plays an important role in lupus nephritis by sponging miR-150. Mol Ther Nucleic Acids. 2018;10:245–253. doi: 10.1016/j.omtn.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang J., Zhu M., Pan J., Chen C., Xia S., Song Y. Circular RNAs: a rising star in respiratory diseases. Respir Res. 2019;20(1) doi: 10.1186/s12931-018-0962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang X., Zhu M., Yang R., Zhao W., Hu X., Gan J. Identification and comparison of novel circular RNAs with associated co-expression and competing endogenous RNA networks in pulmonary tuberculosis. Oncotarget. 2017;8(69):113571–113582. doi: 10.18632/oncotarget.22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yi Z., Gao K., Li R., Fu Y. Dysregulated circRNAs in plasma from active tuberculosis patients. J Cell Mol Med. 2018;22(9):4076–4084. doi: 10.1111/jcmm.13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 122.Cazzella V., Martone J., Pinnaro C. Exon 45 skipping through U1-snRNA antisense molecules recovers the Dys-nNOS pathway and muscle differentiation in human DMD myoblasts. Mol Ther. 2012;20(11):2134–2142. doi: 10.1038/mt.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Feng Z., Zhang L., Wang S., Hong Q. Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson's disease. Biochem Biophys Res Commun. 2020;522(2):388–394. doi: 10.1016/j.bbrc.2019.11.102. [DOI] [PubMed] [Google Scholar]
- 124.Zhao Y., Alexandrov P.N., Jaber V., Lukiw W.J. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer's disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7) Genes. 2016;7(12) doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang Q., Qu L., Chen X., Zhao Y.H., Luo Q. Progress in understanding the relationship between circular RNAs and neurological disorders. J Mol Neurosci. 2018;65(4):546–556. doi: 10.1007/s12031-018-1125-z. [DOI] [PubMed] [Google Scholar]
- 126.Dong Z., Deng L., Peng Q., Pan J., Wang Y. CircRNA expression profiles and function prediction in peripheral blood mononuclear cells of patients with acute ischemic stroke. J Cell Physiol. 2020;235(3):2609–2618. doi: 10.1002/jcp.29165. [DOI] [PubMed] [Google Scholar]
- 127.Wu F., Han B., Wu S. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J Neurosci. 2019;39(37):7369–7393. doi: 10.1523/JNEUROSCI.0299-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu W., Zhao J., Jin M., Zhou M. circRAPGEF5 contributes to papillary thyroid proliferation and metastatis by regulation miR-198/FGFR1. Mol Ther Nucleic Acids. 2019;14:609–616. doi: 10.1016/j.omtn.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 129.Zang Y., Li J., Wan B., Tai Y. circRNA circ-CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR-646. J Cell Mol Med. 2020;24(4):2423–2433. doi: 10.1111/jcmm.14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang J., Cong X., Ren M. Circular RNA hsa_circRNA_0007334 is predicted to promote MMP7 and COL1A1 expression by functioning as a miRNA sponge in pancreatic ductal adenocarcinoma. J Oncol. 2019;2019 doi: 10.1155/2019/7630894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang K., Sun Y., Tao W., Fei X., Chang C. Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 2017;394:1–12. doi: 10.1016/j.canlet.2016.12.036. [DOI] [PubMed] [Google Scholar]
- 132.Yang P., Qiu Z., Jiang Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7(39):63449–63455. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Luan W., Shi Y., Zhou Z., Xia Y., Wang J. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502(1):22–29. doi: 10.1016/j.bbrc.2018.05.114. [DOI] [PubMed] [Google Scholar]
- 134.Xu Y., Yao Y., Zhong X. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496(2):455–461. doi: 10.1016/j.bbrc.2018.01.077. [DOI] [PubMed] [Google Scholar]
- 135.Guarnerio J., Bezzi M., Jeong J.C. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 136.Huang W., Fang K., Chen T.Q. circRNA circAF4 functions as an oncogene to regulate MLL-AF4 fusion protein expression and inhibit MLL leukemia progression. J Hematol Oncol. 2019;12(1) doi: 10.1186/s13045-019-0800-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu C., Yao M.D., Li C.P. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7(11):2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boeckel J.N., Jae N., Heumuller A.W. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ Res. 2015;117(10):884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 139.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 140.Pandey P.R., Munk R., Kundu G., De S., Abdelmohsen K., Gorospe M. Methods for analysis of circular RNAs. Wiley Interdiscip Rev RNA. 2020;11 doi: 10.1002/wrna.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]