Abstract
Fermentative nitrogen-fixing bacteria have not yet been examined in detail in thermal environments. In the present study, we isolated the thermophilic fermentative bacterium, strain YA01 from a hot spring. This strain grew at temperatures up to 78°C. A phylogenetic analysis based on its 16S rRNA gene sequence indicated that strain YA01 belonged to the genus Caldicellulosiruptor, which are fermentative bacteria in the phylum Firmicutes, with 97.7–98.0% sequence identity to its closest relatives. Strain YA01 clearly exhibited N2-dependent growth at 70°C. We also confirmed N2-dependent growth in the relatives of strain YA01, Caldicellulosiruptor hydrothermalis 108 and Caldicellulosiruptor kronotskyensis 2002. The nitrogenase activities of these three strains were examined using the acetylene reduction assay. Similar activities were detected for all tested strains, and were slightly suppressed by the addition of ammonium. A genome analysis revealed that strain YA01, as well as other Caldicellulosiruptor, possessed a gene set for nitrogen fixation, but lacked the nifN gene, which encodes a nitrogenase iron-molybdenum cofactor biosynthesis protein that is commonly detected in nitrogen-fixing bacteria. The amino acid sequences of nitrogenase encoded by nifH, nifD, and nifK shared 92–98% similarity in Caldicellulosiruptor. A phylogenetic tree of concatenated NifHDK sequences showed that NifHDK of Caldicellulosiruptor was in the deepest clade. To the best of our knowledge, this is the first study to demonstrate the nitrogen-fixing ability of fermentative bacteria at 70°C. Caldicellulosiruptor may have retained an ancient nitrogen-fixing enzyme system.
Keywords: diazotroph, hot spring, thermophile, fermentation, Firmicutes
Nitrogen is one of the most abundant and important elements for life. Nitrogen-fixing microorganisms play significant roles in converting atmospheric N2 gas to ammonia in ecosystems. According to a review by Postgate (1998), the first nitrogen-fixing bacteria or diazotrophs were discovered by Winogradsky in 1893. Nitrogen-fixing microorganisms have been reported in 16 phyla in Bacteria and 1 phylum in Archaea from various environments (Mus et al., 2019). Aerobic free living and symbiotic Proteobacteria and phototrophs have been widely reported (Martinez-Romero, 2006; Flores et al., 2015; Wasai and Minamisawa, 2018), and the nitrogen-fixing ability of anaerobic respiratory bacteria, such as Anaeromyxobacter in soil, has recently been attracting increasing attention (Masuda et al., 2020). In 1988, fermentative nitrogen-fixing bacteria were reported in the genus Clostridium in Firmicutes (Leschine et al., 1988); however, limited information is currently available on fermentative nitrogen-fixing bacteria. Nitrogen fixation by fermentative metabolism utilizing polysaccharides (e.g., cellulose) has been suggested to play an important role in nitrogen cycles in soil and animal intestines (Monserrate et al., 2001; Yamada et al., 2007).
Nitrogen fixation is achieved by multiple proteins encoded by nif genes (Raymond et al., 2004). Phylogenetic examinations indicated that nitrogenase genes originated in archaea and were horizontally transferred to bacteria (Boyd et al., 2011a). The nifH gene encoding the nitrogenase reductase subunit of nitrogenase is widely regarded as an indicator of the existence of diazotrophs (Zehr et al., 2003). The diversity and distribution of nifH genes have been analyzed in natural ecosystems, including thermal environments (Mehta et al., 2003; Hamilton et al., 2011; Zehr, 2011; König et al., 2016; Pajares and Bohannan, 2016; Nishihara et al., 2018c). The nitrogen-fixing methanogenic archaeon, Methanocaldococcus FS406-22, was isolated from a deep-sea hyperthermal vent and its nitrogen-fixing ability was demonstrated at temperatures up to 92°C (Mehta and Baross, 2006). In 1986, nitrogen-fixing ability was reported in a thermophilic cellulose-degrading fermentative bacterium that grew at 60°C (Bogdahn and Kleiner, 1986a). Nishihara et al. (2018b) recently reported the nitrogen-fixing ability of H2-oxidizing aerobic bacteria in the genus Hydrogenobacter sp. in the deeply branching phylum Aquificae at 70°C; this is the highest temperature observed for N2 fixation in Bacteria. However, thermophilic isolates that grow at temperatures higher than 70°C are still limited.
In Nakabusa Hot Spring (Nagano, Japan), a sulfidic and slightly alkaline hot spring, chemosynthetic microbial communities develop well at temperatures higher than 70°C (Nakagawa and Fukui, 2002, 2003; Kimura et al., 2010; Nishihara et al., 2018a), and these communities are dominated by H2-/sulfur-oxidizing bacteria in Aquificae (Tamazawa et al., 2012, 2016). The nitrogenase activity of the communities was detected ex situ at 70°C under anaerobic conditions (Nishihara et al., 2018a). Nishihara et al. (2018c) also performed a nifH gene amplicon analysis of chemosynthetic microbial communities at temperatures between 72 and 77°C, and the findings obtained showed that the relative abundance of the nifH gene from Caldicellulosiruptor were 7.42, 48.97, 73.12, and 94.58% in the four samples analyzed. The genus Caldicellulosiruptor comprises thermophilic fermentative bacteria that exhibit cellulolytic activities (Zverlov et al., 1998; Blumer-Schuette et al., 2012; Brunecky et al., 2013) and is widely distributed in globally diverse thermal environments (Lee et al., 2018; Blumer-Schuette, 2020). Genes related to nitrogen fixation are found in the genomes of some species in this genus (CP002330.1, CP002326.1, CP002219.1, CP003001.1, CP000679.1, LACO01000001.1, LACM01000001.1, and LACN01000001.1); however, their nitrogenase activities and dinitrogen-dependent growth have not yet been demonstrated.
In the present study, we isolated thermophilic fermentative bacteria using a combined nitrogen-poor medium from microbial communities developed at approximately 80°C in Nakabusa Hot Spring and characterized their nitrogen-fixing abilities and genetic features in comparisons with their closest relatives.
Materials and Methods
Isolation of bacteria under nitrogen-fixing conditions
Pale tan-colored microbial mats developed in hot spring water at 78.3°C were collected at Nakabusa Hot Springs (36° 23′ 20″ N 137° 44′ 22″ E), Nagano, Japan on January 8th, 2018. Hot spring water was slightly alkaline (pH 8.5–8.9) and contained 5.0–6.1 μmol L–1 of ammonia (Kato et al., 2004), but not nitrate or nitrite (Kato et al., 2004; Kimura et al., 2010). Samples were immediately injected into the anoxic medium in glass vials (see below) with attempts to avoid oxygen contamination at the sampling site. The vials were stored in hot spring water at 60–75°C for 7 h during transportation to our laboratory and then incubated at 70°C.
Winogradsky’s nitrogen-poor mineral medium (Tchan and New, 1984) was prepared with a slight modification and used for the cultivation and isolation of bacteria (L–1): 0.28 g K2HPO4, 0.053 g KH2PO4, 0.12 g MgSO4·7H2O, 0.125 g NaCl, 0.05 g yeast extract, 0.01 g CaCl2·2H2O, 2.5 mg FeSO4·7H2O, 2.5 mg MnSO4·5H2O, 2.5 mg Na2MoO4·2H2O, 2.5 g glucose, 2.5 g sucrose, and 2.5 g Na-pyruvate. The pH of the medium was adjusted to 7.5. Twenty milliliters of the medium was placed into a 70-mL glass vial. The vial was sealed with a butyl rubber stopper and aluminum cap, and then autoclaved after the gas phase had been replaced with N2. In total, 0.5 mL of the culture was repetitively sub-cultured every week in fresh medium. After 10 sub-cultivations, an isolate was obtained by the twice dilution-to-extinction technique. The single morphology of microbial cells was confirmed under a phase-contrast microscope (Axio Imager 2; Carl Zeiss).
Cultivation and maintenance of bacteria
Caldicellulosiruptor hydrothermalis 108, Caldicellulosiruptor bescii DSM 6725, and Caldicellulosiruptor kronotskyensis 2002 were obtained from DSMZ (Germany) (Miroshnichenko et al., 2008; Yang et al., 2010). Bacterial strains were cultivated at 70°C under the N2:CO2 (8:2) gas phase in medium containing the following (L–1): 0.068 g KH2PO4, 0.087 g K2HPO4, 2.09 g MOPS, 0.33 g KCl, 0.25 g NH4Cl, 0.6 g MgSO4·7H2O, 0.4 g NaCl, 0.1 g CaCl2·2H2O, and 10 mL trace minerals. Twenty milliliters of the medium was prepared in 50-mL glass vials sealed with a butyl rubber stopper and aluminum cap. After autoclaving, 0.8 mL of a filter-sterilized 10% cellobiose solution, 0.2 mL of a 5% NaHCO3 solution, and 0.2 mL of a vitamin solution were injected into the vials. The trace minerals solution comprised the following (L–1): 1.5 g nitrilotriacetic acid, 3.0 g MgSO4·7H2O, 0.5 g MnSO4·5H2O, 1.0 g NaCl, 0.1 g FeSO4·7H2O, 0.1 g CaCl2·2H2O, 0.1 g CoCl2·6H2O, 0.13 g ZnCl2, 0.01 g CuSO4, 0.01 g AlK(SO4)·12H2O, 0.01 g H3BO3, 0.025 g Na2MoO4·2H2O, 0.024 g NiCl2·6H2O, and 0.025 g Na2WO4·H2O. The components of the vitamin solution were as follows (L–1): 2 mg biotin, 2 mg folic acid, 10 mg pyridoxine HCl, 5 mg riboflavin, 5 mg thiamine, 5 mg nicotinic acid, 5 mg pantothenic acid, 0.1 mg vitamin B12, p-aminobenzoic acid, and 5 mg thioctic acid.
16S rRNA gene sequence analysis
Bacterial cells were collected by centrifugation and total DNA was extracted according to a method reported by Noll et al. (2005). A DNA fragment of the 16S rRNA gene was PCR-amplified using the 27F and 1492R primers (Lane et al., 1985; Lane, 1991), and amplified DNA after purification by the LaboPass PCR purification Kit (CosmoGenetech) was directly sequenced using BigDye terminator kit v3.1 on an ABI3130 Genetic Analyzer (Applied Biosystems). Sequences were compared using the BLAST program (Altschul et al., 1997) with those available in the DDBJ/EMBL/GenBank databases.
Genome analysis
Total DNA was extracted from bacterial cells by Qiagen Genomic-tip 100/G for bacterial cells (Qiagen) and sequenced by Bioengineering Lab. using the combination of DNBSEQ-G400 (MGI Tech) and GridION with the flow cell-type R9.4.1 (Oxford Nanopore Technologies) platform. Regarding DNBSEQ-G400, DNA was fragmented using a Covaris S2 ultrasonicator (Covaris) to obtain 500-bp DNA fragments. The DNBseq DNA library was prepared according to the manufacturer’s instructions and sequenced using DNBSEQ-G400 (pair-end 150-bp reads). In GridION, the library was prepared using the Ligation Sequence Kit (SQK-LSK109) after barcoding using Native Barcoding Expansion (Oxford Nanopore Technologies EXP-NBD104) and was then sequenced. In the DNBSEQ-G400 analysis, 3,500,000 read pairs (1.05 Gbp) were sampled using Seqkit (v. 0.11.0) (Shen et al., 2016) and quality filtered using Sickle (v. 1.2.3) (Joshi and Fass, 2011) with the parameters -q 20 -l 127. In the GridION analysis, adaptors of the reads obtained were trimmed using Porechop (v. 0.2.3) (Wick et al., 2017) and quality filtered using Filtlong (v. 0.2.0) (https://github.com/rrwick/Filtlong) with the parameters --min_length 1000 --target_bases 250000000, and processed error-prone reads using Canu v1.8 (Koren et al., 2017). A total of 3,033,015 reads (DNBSEQ) and 67,859 reads (GridION) were obtained after quality filtering and subjected to use for hybrid assembly by Unicycler (v. 0.4.7) (Phillippy et al., 2017) with the default setting. The assembled genome was annotated using Prokka v1.14.0 (Seemann, 2014). Proteins involved in nitrogen fixation were visualized using ‘gggenes’ (https://CRAN.R-project.org/package=gggenes) with ‘ggplot2’ in R package (R Foundation, Vienna, Austria) (https://www.R-project.org/) (Seemann, 2014; Wickham, 2016).
Nitrogen fixation gene cluster and phylogenetic analysis
A concatenated phylogenetic tree of Nif/Anf/VnfHDK was constructed using 276 Nif/Anf/VnfHDK protein homologs, which were located in operons in 235 genomes including the genome newly analyzed in the present study. Genomes harboring Nif/Anf/VnfHDK homologues were examined using AnnoTree v1.2 (Boyd et al., 2011b; Mendler et al., 2019; Garcia et al., 2020), collected from the National Center for Biotechnology Information database, and annotated using Prokka v1.14.0 (Seemann, 2014). Amino acid sequences were aligned using Mafft v7.427 (Katoh et al., 2002). Maximum likelihood trees were constructed using RAxML-NG v. 0.9.0 with the LG+F+G4 model and 100 bootstrap replicates (Kozlov et al., 2019). Bootstrap support values were recalculated by BOOSTER (v0.1.2) (Lemoine et al., 2018). The MarHDK protein in Rhodospirillum rubrum ATCC11170 (WP_011388553.1, WP_011388552.1, and WP_011388551.1) was used as the outgroup (North et al., 2020).
Growth capability in nitrogen-poor media
Modified Winogradsky’s nitrogen-poor mineral medium (described above) was used to assess N2-dependent growth. Ten milliliters of medium was prepared in 32-mL glass test tubes sealed with butyl rubber stoppers and screw caps and the gas phase of the culture tube was filled with N2 or argon (Ar) gas. In total, 0.5 mL of bacterial cultures pre-cultivated in nitrogen-poor medium were inoculated into fresh nitrogen-poor medium. To test the growth capability of C. bescii DSM 6725, a pre-cultivation was conducted using medium supplemented with 2 mmol L–1 of NH4Cl. Growth in the culture was assessed by measurements of optical density (OD) at 660 nm (miniphoto 518R; Taitec). Cultivation medium containing 2 mmol L–1 NH4Cl was also used to compare N2-dependent growth with growth on ammonium.
Nitrogenase activity by the acetylene reduction assay
Nitrogenase activity was detected using the acetylene reduction assay method (Leschine et al., 1988). In total, 0.5 mL of bacterial pre-cultures in modified Winogradsky’s nitrogen-poor mineral medium was inoculated into 10 mL of the same medium in 25-mL glass vials and cultivated under a N2 gas atmosphere. At the exponential growth phase, a portion (0.5 mL) of the culture solution was removed and mixed with 0.05 mL of 10% Formalin Neutral Buffer Solution (pH 7.4–7.5, Fujifilm Wako Pure Chemical) to fix cells for the cell number count. The gas phase of culture vials was then replaced with N2 gas and 1.5 mL of 99.9999% acetylene gas was injected into each vial. Vials were incubated at 70°C and, after a 24-h incubation, 1 mL of 37% neutralized formaldehyde was added to stop the reaction. The production of ethylene by the reduction of acetylene was quantified using a GC-2014 gas chromatograph equipped with a flame ionization detector (Shimadzu) and 80/100 Porapak T (GL Science) column. Analysis conditions were as follows; carrier gas, N2 gas; column temperature, 70°C; injection temperature, 100°C; detector temperature, 100°C. Fresh medium containing no bacterial cells was prepared in the vial as a negative control to confirm abiotic ethylene production under the same conditions.
Nucleotide sequence accession number
The 16S rRNA gene sequence was deposited in the DDBJ/EMBL/GenBank databases with the accession number LC603168. The accession numbers of the genomic sequences of strain YA01 were AP024480 (chromosome) and AP024481 and AP024482 (two plasmids).
Results
Bacterial isolate from the hot spring under anaerobic nitrogen-fixing conditions
Pale tan-colored microbial mats collected at 78.3°C from Nakabusa Hot Spring were directly inoculated into glass vials with modified Winogradsky’s nitrogen-poor mineral medium and anaerobically incubated at 70°C. After several sub-cultivations at one-week intervals, a stable enrichment culture was obtained. A pure culture containing cells of a single morphotype, i.e., short rods (Fig. S1), was obtained by dilution-to-extinction and the isolate was designated as strain YA01. Strain YA01 grew at temperatures up to 78°C. The 16S rRNA gene sequence of strain YA01 (1,474 bp) showed 98.0, 97.7, and 97.7% identities to those of its closest relatives, C. hydrothermalis 108, C. bescii DSM 6725, and C. kronotskyensis 2002, respectively. This result indicated that strain YA01 was a species of the genus Caldicellulosiruptor.
Nitrogen-fixation related genes in Caldicellulosiruptor
DNBSEQ-G400 and GridION runs resulted in the generation of approximately 12,782,840 reads with a total of 1,917 Mbp and 155,919 reads with a total of 255 Mbp, respectively. The complete genome of strain YA01 consisted of a single chromosome with a length of 2,592,764 bp and two plasmids with lengths of 3,514 and 1,547 bp. The G+C content of the genome was 34.8%. Coding potential predictions identified 2,412 protein-coding genes, three rRNA operons, and 47 tRNA genes. Three 16S rRNA genes had the same sequence, which was identical to the 16S rRNA gene sequence amplified by PCR. Average nucleotide identity (ANI) between strain YA01 and its close relatives (C. hydrothermalis 108, C. bescii DSM 6725, and C. kronotskyensis 2002) calculated using an ANI calculator (http://enve-omics.ce.gatech.edu/ani/) (Rodriguez-R, L.M., and Konstantinidis, K.T. 2016 The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4: e1900v1) ranged between 90.31 and 91.10%.
Annotation results showed that the chromosome of strain YA01 contained at least seven proteins involved in nitrogen fixation: three genes for nitrogenase structural proteins (NifH, NifD, and NifK), two proteins involved in the biosynthesis of MoFe protein cofactors (NifB and NifE), and two proteins of the PII family involved in posttranslational nitrogenase regulation (NifI1 and NifI2) (Arcondéguy et al., 2001; Dodsworth et al., 2005; Burén et al., 2020) (Fig. 1). These seven genes were also identified in eight out of the 14 genomes in the genus Caldicellulosiruptor available in GenBank; C. morganii Rt8.B8 (accession no. LACO01000001.1), C. naganoensis NA10 (accession no. LACN01000001.1), C. danielii strain Wai35.B1 (accession no. LACM01000001.1), C. lactoaceticus 6A (accession no. CP003001.1), C. kronotskyensis 2002 (accession no. CP002330.1), C. kristjanssonii I77R1B (accession no. CP002326.1), C. hydrothermalis 108 (accession no. CP002219.1), and C. saccharolyticus DSM 8903 (accession no. CP000679.1) (Van De Werken et al., 2008; Kataeva et al., 2009; Blumer-Schuette et al., 2011; Wai et al., 2015; Blumer-Schuette, 2020). NifN was not found in the genomes based on the annotation using eggNOG-mapper v2 (Huerta-Cepas et al., 2017; 2019).
Fig. 1.
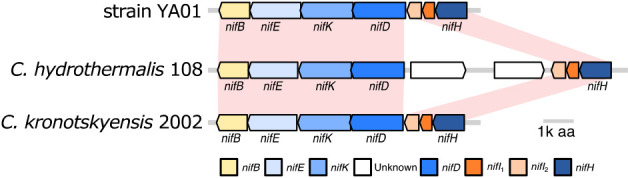
Nitrogen fixation gene clusters for strain YA01 and its closest relatives, Caldicellulosiruptor hydrothermalis 108 and Caldicellulosiruptor kronotskyensis 2002. The arrow indicates the transcriptional direction.
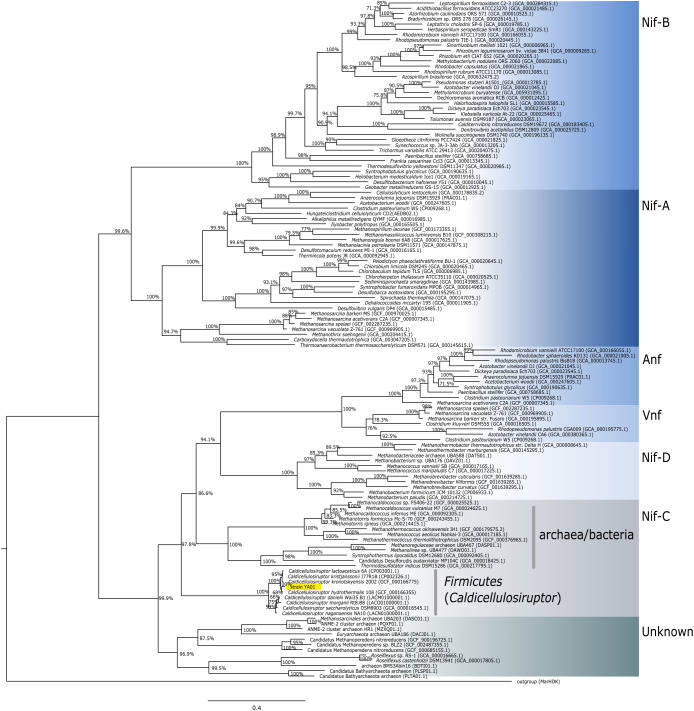
The amino acid sequences of the nitrogenase structural proteins were similar among the genus (NifH, 98.37±0.41%; NifD, 96.58±1.54%; NifK, 92.05±5.26%). A concatenated NifHDK phylogenetic tree was constructed for all Caldicellulosiruptor species possessing nitrogen fixation-related genes (Fig. 2). Strain YA01 clustered with all other members of the genus Caldicellulosiruptor within the cluster Nif-C (Fig. 2). The NifHDK sequences of Caldicellulosiruptor formed a monophyletic lineage, were placed in the deepest clade in the branch of the Anf/Vinf/Nif-D/Nif-C/Unknown lineage (Boyd and Peters, 2013; Poudel et al., 2018; Garcia et al., 2020), and were distantly related to those in other thermophilic bacteria, such as Thermoanaerobacterium thermosaccharolyticum (Nif-A lineage) and Hydrogenobacter sp. (Nif-B lineage) (Boyd and Peters, 2013). The thermophilic features of NifHDK did not appear to correlate with the primary structure.
Fig. 2.
Phylogenetic tree of concatenated NifHDK sequences
The phylogenetic tree was constructed by the Maximum Likelihood method with 100 bootstrap replicates. The newly isolated strain, strain YA01 is highlighted in yellow. Among the 276 sequences used, 130 representative taxa are shown. Bootstrap values of more than 50% are indicated at the respective nodes. MarHDK protein sequences in Rhodospirillum rubrum ATCC11170 were used as the outgroup. Abbreviations: Nif, Mo-nitrogenase; Anf, Fe-nitrogenase; Vnf, V-nitrogenase; Unknown, uncharacterized nitrogenase homologues. The cluster for Nif (Nif-A, B, C, and D) is shown according to the definition by Poudel et al., 2018.
Growth capability under nitrogen-fixing conditions
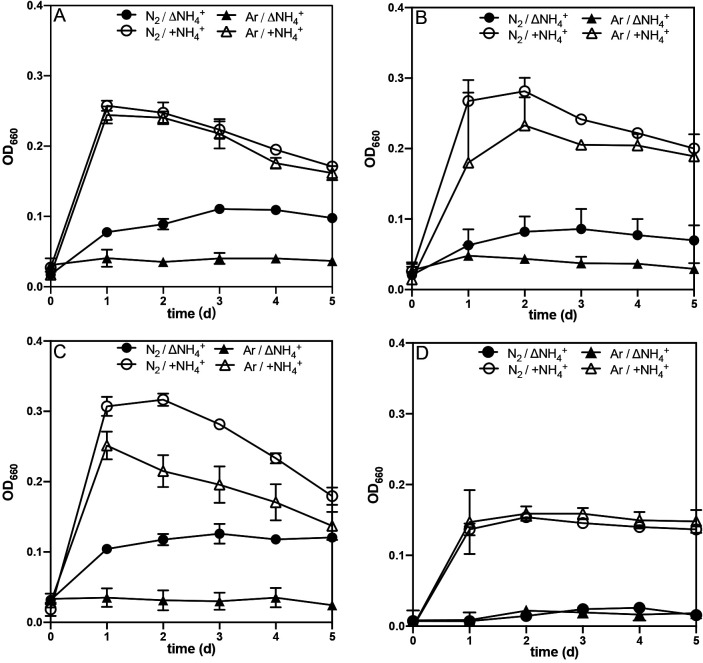
Strain YA01 and its three relatives, C. hydrothermalis 108, C. bescii DSM 6275, and C. kronotskyensis 2002, were cultivated in nitrogen-poor medium and ammonium-containing medium under the N2 or argon (Ar) gas phase (Fig. 3). C. bescii, which did not possess nitrogen-fixing genes, did not grow in nitrogen compound-free medium (Fig. 3D). Strain YA01, C. hydrothermalis 108, and C. kronotskyensis 2002 showed marked increases in OD in nitrogen compound-free medium under N2 gas, but not under the Ar gas phase (Fig. 3A, B, and C). These three strains reached the stationary phase within 2 to 3 days and final OD were 0.05 to 0.14 under N2-fixing conditions, corresponding to 1.06×107 to 2.27×107 cells mL–1. In ammonium-containing medium, the growth of all these strains was faster than in the absence of ammonium and final OD were 0.20 to 0.35. In C. hydrothermalis 108 and C. kronotskyensis 2002, growth yields in ammonium-containing medium were slightly higher under N2 gas than under Ar gas.
Fig. 3.
Growth of strain YA01 (A) and its closest species, Caldicellulosiruptor hydrothermalis 108 (B), Caldicellulosiruptor kronotskyensis 2002 (C), Caldicellulosiruptor bescii DSM 6275 (D) with (open symbols) or without (closed symbols) ammonium under a N2 or Ar atmosphere. Error bars indicate the standard deviation of three replicates.
Nitrogenase activity
To test nitrogenase activity, strain YA01 and its relatives C. hydrothermalis 108 and C. kronotskyensis 2002 were cultivated to the exponential growth phase in nitrogen-poor medium and ammonium-containing medium under the N2 gas phase. Acetylene was injected into the vials and incubated at 70°C. The results of ethylene production after a 24-h incubation are summarized in Table 1. Ethylene production was observed in all tested strains, even in the presence of ammonium. Strain YA01 showed the highest value among the three strains in the absence of ammonium. The amount of ethylene produced in the presence of ammonium for 24 h was lower than that in its absence in all strains. The suppressive effects of ammonium on C. kronotskyensis 2002 were weak (approximately 60% of that in its absence). Acetylene-reducing activities were also observed under an incubation at 78°C for all strains: 9.50±5.05, 8.37±5.53, and 15.0±12.1 nmol C2H4 106 cells–1 24 h–1 by strain YA01, C. hydrothermalis 108, and C. kronotskyensis 2002, respectively. Activities at 78°C were weaker than those at 70°C.
Table 1.
Acetylene-reducing activities of strain YA01 and its closest relatives, Caldicellulosiruptor hydrothermalis 108 and Caldicellulosiruptor kronotskyensis 2002 in the absence and presence of ammonium at 70°C.
| Strains | YA01 | C. hydrothermalis 108 | C. kronotskyensis 2002 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Conditions* | ΔNH4+ | +NH4+ | ΔNH4+ | +NH4+ | ΔNH4+ | +NH4+ | |||
| Acetylene-reducing activity (nmol C2H4 106 cells–1 24 h–1)** | 610±246 | 18.8±14.3 | 107±18.3 | 0.257±0.0694 | 363±74.4 | 216±156 | |||
*, ΔNH4+, modified Winogradsky’s nitrogen-poor mineral medium; +NH4+, ammonium-containing medium (2 mmol L–1 of NH4Cl).
**, values were obtained from three culture vials and shown with standard deviations.
Discussion
In the present study, we isolated a bacterial strain by cultivation in nitrogen-poor medium from Nakabusa Hot Spring, Japan. The results of the phylogenetic analysis based on the 16S rRNA gene sequence suggested that this isolate, strain YA01, is a new species in the genus Caldicellulosiruptor and is closely related to C. hydrothermalis, C. bescii, and C. kronotskyensis. The results of the genomic analysis indicated that strain YA01 as well as C. hydrothermalis 108 and C. kronotskyensis 2002 possess a set of nitrogen fixation-related genes (Fig. 1) and their NifHDK formed a monophyletic lineage in the deeply branching group of the NifHDK tree (Fig. 2). Growth capability with N2 gas as the sole nitrogen source and acetylene-reducing activity were successfully demonstrated for the new isolate, C. hydrothermalis 108, and C. kronotskyensis 2002 (Fig. 3 and Table 1). To the best of our knowledge, this is the first study to detect nitrogen-fixing ability in the genus Caldicellulosiruptor. The nitrogenase activities of bacteria were previously reported at temperatures up to 70°C by Nishihara et al. (2018b) in the chemolithoautotrophic bacteria, Hydrogenobacter sp. in the phylum Aquificae. The nitrogenase activities of Caldicellulosiruptor were detected at temperatures higher than 70°C, i.e., 78°C, which was the maximum growth temperature of strain YA01.
The nif gene operons of strain YA01 and its relatives, C. hydrothermalis 108 and C. kronotskyensis 2002 basically comprised nifHDKEB (Fig. 1). Commonly known nif gene operons contain the additional gene, nifN; however, a homologous gene to nifN was not identified in Caldicellulosiruptor. nifN encodes subunits of the tetrameric protein NifEN (2NifE 2NifN), which is required for the biosynthesis of the iron molybdenum co-factor of Mo-type nitrogenase (Hu et al., 2005, 2006, 2008; Corbett et al., 2006; Burén et al., 2020). Evolutionary studies based on molecular phylogram and comparative analyses of amino acid sequences suggested that nifN and nifK are paralogous genes that were derived through gene duplication (Raymond et al., 2004; Boyd et al., 2011b). Similar to Caldicellulosiruptor, the diazotrophic archaeon, Methanocaldococcus sp. FS406-22 also lacks nifN (Mehta and Baross, 2006). This finding indicates that the protein coded by nifK in these thermophiles performs the same function as NifN. Alternatively, NifE may work without an NifN subunit, as suggested by Garcia et al. (2020), because NifE in Caldicellulosiruptor showed low similarity with other known NifE (Fig. S2). The phylogenetic trees of NifHDK (Fig. 2) and NifE (Fig. S2) indicated that Caldicellulosiruptor has an ancient nitrogen-fixing enzyme system. As proposed in the phylogenetic study of nitrogen fixation-related genes by Garcia et al. (2020), Mo-nitrogenase in the genus Caldicellulosiruptor may have emerged earlier and then evolved into modern nitrogenases in wide lineages of prokaryotes.
The nitrogenase activities of the Caldicellulosiruptor strains were not completely suppressed by the addition of ammonium (Table 1); however, the inhibition of nitrogenase activity by ammonium has been traditionally reported in most diazotrophic bacteria (Dixon and Kahn, 2004). In cellulolytic fermentative diazotrophic bacteria, Clostridium sp. in Firmicutes, acetylene-reducing activity decreased under the detection limit when ammonium was added (Bogdahn and Kleiner, 1986a, 1986b). However, this activity was not suppressed by ammonium for the thermophilic relative, Clostridium thermocellum (now Hungateiclostridium thermocellum) (Bogdahn and Kleiner, 1986a; Tindall, 2019). Although the protein, NifA has been shown to regulate the expression of nif genes in nitrogen-fixing aerobes in Proteobacteria (Merrick, 1992), most anaerobic diazotrophs, including Caldicellulosiruptor and Clostridium, do not possess the nifA gene (Boyd et al., 2015). The evolution of Nif regulation systems from anaerobic to aerobic metabolism is still debatable (Boyd et al., 2015). Further transcriptional and enzymological studies are required to elucidate responses to ammonium in these thermophilic diazotrophs.
Caldicellulosiruptor are frequently detected from microbial mats in geothermal springs (Lee et al., 2018; Blumer-Schuette, 2020). Microbial mats are stratified communities of microorganisms with thicknesses of 3 to 5 mm and thermophilic microbial mats have been utilized as a model microbial community to investigate the development and maintenance of ecosystems (Taffs et al., 2009; Klatt et al., 2013; Kim et al., 2015; Lindemann et al., 2016; Bernstein et al., 2017; Haruta, 2020). Previous studies focused on primary production in communities in hot spring streams and reported a spatial and temporal distribution and the co-occurrence of carbon-fixing metabolism via oxygenic and anoxygenic photosynthesis, aerobic chemosynthesis (e.g., H2 and sulfide oxidation), and anaerobic chemosynthesis (e.g., sulfur disproportionation) (Rothschild and Mancinelli, 1990; Kimura et al., 2010; Kojima et al., 2016; Tamazawa et al., 2016; Sharrar et al., 2017; Gutiérrez-Preciado et al., 2018; Kawai et al., 2019). Dinitrogen fixation is also required for community development in spring waters that are poor in nitrogen compounds (Kato et al., 2004; Steunou et al., 2006; Kimura et al., 2010; Hamilton et al., 2011; Loiacono et al., 2012). However, possible thermophilic diazotrophs at temperatures higher than 70°C in terrestrial springs have not been clarified. The present results provide important insights into the development of micro-ecosystems in thermal environments. Caldicellulosiruptor may utilize organic compounds derived from primary producers at the anoxic layer of microbial mats and provide ammonium to the communities. Caldicellulosiruptor possessing the ancient type of nitrogenase may play important roles in carbon and nitrogen cycles not only in modern thermal springs, but also in the early Earth.
Citation
Chen, Y., Nishihara, A., and Haruta, S. (2021) Nitrogen-fixing Ability and Nitrogen Fixation-related Genes of Thermophilic Fermentative Bacteria in the Genus Caldicellulosiruptor. Microbes Environ 36: ME21018.
https://doi.org/10.1264/jsme2.ME21018
Supplementary Material
Acknowledgements
We wish to thank Mr. Takahito Momose, the president of Nakabusa Onsen Inn Inc., for allowing us to use the hot spring in the winter season. We are grateful to R. Craig Everroad for his critical reading of the manuscript and also for correcting the English. We would like to thank Dr. Shigeki Ehira for his useful discussions and providing acetylene. We also thank Dr. Masaru Konishi Nobu for his technical assistance with the phylogenetic analysis. A. N. was supported by a Grant-in-Aid for Early-Career Scientists (19K16234) from the Japan Society for the Promotion of Science (JSPS).
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcondéguy, T., Jack, R., and Merrick, M. (2001) PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev 65: 80–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, H.C., Brislawn, C., Renslow, R.S., Dana, K., Morton, B., Lindemann, S.R., et al. (2017) Trade-offs between microbiome diversity and productivity in a stratified microbial mat. ISME J 11: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer-Schuette, S.E., Ozdemir, I., Mistry, D., Lucas, S., Lapidus, A., Cheng, J.F., et al. (2011) Complete genome sequences for the anaerobic, extremely thermophilic plant biomass-degrading bacteria Caldicellulosiruptor hydrothermalis, Caldicellulosiruptor kristjanssonii, Caldicellulosiruptor kronotskyensis, Caldicellulosiruptor owensensis and Caldicellulosiruptor lactoaceticus. J Bacteriol 193: 1483–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer-Schuette, S.E., Giannone, R.J., Zurawski, J.V., Ozdemir, I., Ma, Q., Yin, Y., et al. (2012) Caldicellulosiruptor core and pangenomes reveal determinants for noncellulosomal thermophilic deconstruction of plant biomass. J Bacteriol 194: 4015–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer-Schuette, S.E. (2020) Insights into thermophilic plant biomass hydrolysis from Caldicellulosiruptor systems biology. Microorganisms 8: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdahn, M., and Kleiner, D. (1986a) Inorganic nitrogen metabolism in two cellulose-degrading Clostridia. Arch Microbiol 145: 159–161. [Google Scholar]
- Bogdahn, M., and Kleiner, D. (1986b) N2 fixation and NH4+ assimilation in the thermophilic anaerobes Clostridium thermosaccharolyticum and Clostridium thermoautotrophicum. Arch Microbiol 144: 102–106 [DOI] [PubMed] [Google Scholar]
- Boyd, E.S., Anbar, A.D., Miller, S., Hamilton, T.L., Lavin, M., and Peters, J.W. (2011a) A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9: 221–232. [DOI] [PubMed] [Google Scholar]
- Boyd, E.S., Hamilton, T.L., and Peters, J.W. (2011b) An alternative path for the evolution of biological nitrogen fixation. Front Microbiol 2: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, E.S., and Peters, J.W. (2013) New insights into the evolutionary history of biological nitrogen fixation. Front Microbiol 4: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, E.S., Garcia Costas, A.M., Hamilton, T.L., Mus, F., and Peters, J.W. (2015) Evolution of molybdenum nitrogenase during the transition from anaerobic to aerobic metabolism. J Bacteriol 197: 1690–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunecky, R., Alahuhta, M., Xu, Q., Donohoe, B.S., Crowley, M.F., Kataeva, I.A., et al. (2013) Revealing nature’s cellulase diversity : The digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342: 1513–1516. [DOI] [PubMed] [Google Scholar]
- Burén, S., Jiménez-Vicente, E., Echavarri-Erasun, C., and Rubio, L.M. (2020) Biosynthesis of nitrogenase cofactors. Chem Rev 120: 4921–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett, M.C., Hu, Y., Fay, A.W., Ribbe, M.W., Hedman, B., and Hodgson, K.O. (2006) Structural insights into a protein-bound iron-molybdenum cofactor precursor. Proc Natl Acad Sci U S A 103: 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R., and Kahn, D. (2004) Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2: 621–631. [DOI] [PubMed] [Google Scholar]
- Dodsworth, J.A., Cady, N.C., and Leigh, J.A. (2005) 2-Oxoglutarate and the PII homologues NifI1 and NifI2 regulate nitrogenase activity in cell extracts of Methanococcus maripaludis. Mol Microbiol 56: 1527–1538. [DOI] [PubMed] [Google Scholar]
- Flores, E., Lopez-Lozano, A., and Herrero, A. (2015) Nitrogen fixation in the oxygenic phototrophic prokaryotes (cyanobacteria): The fight against oxygen. In Biological Nitrogen Fixation. De Bruijn, F.J. (ed.) Hoboken, NJ: Wiley-Blackwell, pp. 879–889. [Google Scholar]
- Garcia, A.K., McShea, H., Kolaczkowski, B., and Kaçar, B. (2020) Reconstructing the evolutionary history of nitrogenases: evidence for ancestral molybdenum-cofactor utilization. Geobiology 18: 394–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Preciado, A., Saghaï, A., Moreira, D., Zivanovic, Y., Deschamps, P., and López-García, P. (2018) Functional shifts in microbial mats recapitulate early earth metabolic transitions. Nat Ecol Evol 2: 1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, T.L., Lange, R.K., Boyd, E.S., and Peters, J.W. (2011) Biological nitrogen fixation in acidic high-temperature geothermal springs in Yellowstone National Park, Wyoming. Environ Microbiol 13: 2204–2215. [DOI] [PubMed] [Google Scholar]
- Haruta, S. (2020) Thermophilic photosynthesis-based microbial communities -energy production and conversion. In Life in Extreme Environments -Biotechnological Applications of Extremophilic Microorganisms-. Lee, N.M. (ed.) Berlin, Germany: De Gruyter, pp. 153–162. [Google Scholar]
- Hu, Y., Fay, A.W., and Ribbe, M.W. (2005) Identification of a nitrogenase FeMo cofactor precursor on NifEN complex. Proc Natl Acad Sci U S A 102: 3236–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Corbett, M.C., Fay, A.W., Webber, J.A., Hodgson, K.O., Hedman, B., and Ribbe, M.W. (2006) FeMo cofactor maturation on NifEN. Proc Natl Acad Sci U S A 103: 17119–17124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Fay, A.W., Lee, C.C., Yoshizawa, J., and Ribbe, M.W. (2008) Assembly of nitrogenase MoFe protein. Biochemistry 47: 3973–3981. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas, J., Forslund, K., Coelho, L.P., Szklarczyk, D., Jensen, L.J., Von Mering, C., and Bork, P. (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34: 2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S.K., Cook, H., et al. (2019) eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47: D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, N.A., and Fass, J.N. (2011) Sickle: a sliding-window, adaptive, quality-based trimming tool for fastQ files (Version 1.33) [Software] URL https://github.com/najoshi/sickle
- Kataeva, I.A., Yang, S.J., Dam, P., Poole, F.L., Yin, Y., Zhou, F., et al. (2009) Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium ‘Anaerocellum thermophilum’ DSM 6725. J Bacteriol 191: 3760–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, K., Kobayashi, T., Yamamoto, H., Nakagawa, T., Maki, Y., and Hoaki, T. (2004) Microbial mat boundaries between chemolithotrophs and phototrophs in geothermal hot spring effluents. Geomicrobiol J 21: 91–98. [Google Scholar]
- Katoh, K., Misawa, K., Kuma, K.I., and Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai, S., Kamiya, N., Matsuura, K., and Haruta, S. (2019) Symbiotic growth of a thermophilic sulfide-oxidizing photoautotroph and an elemental sulfur-disproportionating chemolithoautotroph and cooperative dissimilatory oxidation of sulfide to sulfate. Front Microbiol 10: 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.M., Nowack, S., Olsen, M., Becraft, E.D., Wood, J.M., Thiel, V., et al. (2015) Diel metabolomics analysis of a hot spring chlorophototrophic microbial mat leads to new hypotheses of community member metabolisms. Front Microbiol 6: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H., Mori, K., Nashimoto, H., Hanada, S., and Kato, K. (2010) In situ biomass production of a hot spring sulfur-turf microbial mat. Microbes Environ 25: 140–143. [DOI] [PubMed] [Google Scholar]
- Klatt, C.G., Inskeep, W.P., Herrgard, M.J., Jay, Z.J., Rusch, D.B., Tringe, S.G., et al. (2013) Community structure and function of high-temperature chlorophototrophic microbial mats inhabiting diverse geothermal environments. Front Microbiol 4: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, H., Kazuhiro, U., and Manabu, F. (2016) Caldimicrobium thiodismutans sp. nov., a sulfur-disproportionating bacterium isolated from a hot spring, and emended description of the genus Caldimicrobium. Int J Syst Evol Microbiol 66: 1828–1831. [DOI] [PubMed] [Google Scholar]
- König, S., Gros, O., Heiden, S.E., Hinzke, T., Thürmer, A., Poehlein, A., et al. (2016) Nitrogen fixation in a chemoautotrophic lucinid symbiosis. Nat Microbiol 2: 16193. [DOI] [PubMed] [Google Scholar]
- Koren, S., Walenz, B.P., Berlin, K., Miller, J.R., Bergman, N.H., and Phillippy, A.M. (2017) Canu: scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov, A.M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, D.J., Pace, B., Olsen, G.J., Stahl, D.A., Sogin, M.L., and Pace, N.R. (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A 82: 6955–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, D.J. (1991) 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematic. Stackebrandt, E., and Goodfellow, M. (eds). New York, NY: John Wiley & Sons, pp. 115–175. [Google Scholar]
- Lee, L.L., Blumer-Schuette, S.E., Izquierdo, J.A., Zurawski, J.V., Loder, A.J., Conway, J.M., et al. (2018) Genus-wide assessment of lignocellulose utilization in the extremely thermophilic genus Caldicellulosiruptor by genomic, pangenomic, and metagenomic analyses. Appl Environ Microbiol 84: e02694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine, F., Domelevo-Entfellner, J.-B., Wilkinson, E., Correia, D., Davila Felipe, M., De Oliveira, T., and Gascuel, O. (2018) Renewing felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556: 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschine, S.B., Holwell, K., and Canale-Parola, E. (1988) Nitrogen fixation by anaerobic cellulolytic bacteria. Science 242: 1157–1159. [DOI] [PubMed] [Google Scholar]
- Lindemann, S.R., Bernstein, H.C., Song, H.-S., Fredrickson, J.K., Fields, M.W., Shou, W., et al. (2016) Engineering microbial consortia for controllable outputs. ISME J 10: 2077–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiacono, S.T., Meyer-Dombard, D.R., Havig, J.R., Poret-Peterson, A.T., Hartnett, H.E., and Shock, E.L. (2012) Evidence for high-temperature in situ nifH transcription in an alkaline hot spring of Lower Geyser Basin, Yellowstone National Park. Environ Microbiol 14: 1272–1283. [DOI] [PubMed] [Google Scholar]
- Martinez-Romero, E. (2006) Dinitrogen-fixing prokaryotes. In The Prokaryotes. New York, NY: Springer, pp. 793–817. [Google Scholar]
- Masuda, Y., Yamanaka, H., Xu, Z.X., Shiratori, Y., Aono, T., Amachi, S., et al. (2020) Diazotrophic anaeromyxobacter isolates from soils. Appl Environ Microbiol 86: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, M.P., Butterfield, D.A., and Baross, J.A. (2003) Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca ridge. Appl Environ Microbiol 69: 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, M.P., and Baross, J.A. (2006) Nitrogen fixation at 92°C by a hydrothermal vent archaeon. Science 314: 1783–1786. [DOI] [PubMed] [Google Scholar]
- Mendler, K., Chen, H., Parks, D.H., Lobb, B., Hug, L.A., and Doxey, A.C. (2019) AnnoTree: visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res 47: 4442–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick, M.J. (1992) Regulation of nitrogen fixation genes in free-living and symbiotic bacteria. In Biological Nitrogen Fixation. Stacey, G., Burris, R.H., and Evans, H.J. (eds). New York, NY: Chapman and Hall; pp. 835–876. [Google Scholar]
- Miroshnichenko, M.L., Kublanov, I.V., Kostrikina, N.A., Tourova, T.P., Kolganova, T.V., Birkeland, N.K., and Bonch-Osmolovskaya, E.A. (2008) Caldicellulosiruptor kronotskyensis sp. nov. and Caldicellulosiruptor hydrothermalis sp. nov., two extremely thermophilic, cellulolytic, anaerobic bacteria from Kamchatka thermal springs. Int J Syst Evol Microbiol 58: 1492–1496. [DOI] [PubMed] [Google Scholar]
- Monserrate, E., Leschine, S.B., and Canale-Parola, E. (2001) Clostridium hungatei sp. nov., a mesophilic, N2-fixing cellulolytic bacterium isolated from soil. Int J Syst Evol Microbiol 51: 123–132. [DOI] [PubMed] [Google Scholar]
- Mus, F., Colman, D.R., Peters, J.W., and Boyd, E.S. (2019) Geobiological feedbacks, oxygen, and the evolution of nitrogenase. Free Radical Biol Med 140: 250–259. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., and Fukui, M. (2002) Phylogenetic characterization of microbial mats and streamers from a Japanese alkaline hot spring with a thermal gradient. J Gen Appl Microbiol 48: 211–222. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., and Fukui, M. (2003) Molecular characterization of community structures and sulfur metabolism within microbial streamers in Japanese hot springs. Appl Environ Microbiol 69: 7044–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara, A., Haruta, S., McGlynn, S.E., Thiel, V., and Matsuura, K. (2018a) Nitrogen fixation in thermophilic chemosynthetic microbial communities depending on hydrogen, sulfate, and carbon dioxide. Microbes Environ 33: 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara, A., Matsuura, K., Tank, M., McGlynn, S.E., Thiel, V., and Haruta, S. (2018b) Nitrogenase activity in thermophilic chemolithoautotrophic bacteria in the phylum Aquificae isolated under nitrogen-fixing conditions from Nakabusa hot springs. Microbes Environ 33: 394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara, A., Thiel, V., Matsuura, K., McGlynn, S.E., and Haruta, S. (2018c) Phylogenetic diversity of nitrogenase reductase genes and possible nitrogen-fixing bacteria in thermophilic chemosynthetic microbial communities in Nakabusa hot springs. Microbes Environ 33: 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll, M., Matthies, D., Frenzel, P., Derakshani, M., and Liesack, W. (2005) Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ Microbiol 7: 382–395. [DOI] [PubMed] [Google Scholar]
- North, J.A., Narrowe, A.B., Xiong, W., Byerly, K.M., Zhao, G., Young, S.J., et al. (2020) A nitrogenase-like enzyme system catalyzes methionine, ethylene, and methane biogenesis. Science 369: 1094–1098. [DOI] [PubMed] [Google Scholar]
- Pajares, S., and Bohannan, B.J.M. (2016) Ecology of nitrogen fixing, nitrifying, and denitrifying microorganisms in tropical forest soils. Front Microbiol 7: 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillippy, A.M., Wick, R.R., Judd, L.M., Gorrie, C.L., and Holt, K.E. (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postgate, J. (1998) Nitrogen Fixation. New York, NY: Cambridge University Press. [Google Scholar]
- Poudel, S., Colman, D.R., Fixen, K.R., Ledbetter, R.N., Zheng, Y., Pence, N., et al. (2018) Electron transfer to nitrogenase in different genomic and metabolic backgrounds. J Bacteriol 200: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, J., Siefert, J.L., Staples, C.R., and Blankenship, R.E. (2004) The natural history of nitrogen fixation. Mol Biol Evol 21: 541–554. [DOI] [PubMed] [Google Scholar]
- Rothschild, L.J., and Mancinelli, R.L. (1990) Model of carbon fixation in microbial mats from 3,500 myr ago to the present. Nature 345: 710–712. [DOI] [PubMed] [Google Scholar]
- Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Sharrar, A.M., Flood, B.E., Bailey, J.V., Jones, D.S., Biddanda, B.A., Ruberg, S.A., et al. (2017) Novel large sulfur bacteria in the metagenomes of groundwater-fed chemosynthetic microbial mats in the Lake Huron Basin. Front Microbiol 8: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W., Le, S., Li, Y., and Hu, F. (2016) SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q File manipulation. PLoS One 11: e0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou, A.S., Bhaya, D., Bateson, M.M., Melendrez, M.C., Ward, D.M., Brecht, E., et al. (2006) In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic Cyanobacteria inhabiting hot spring microbial mats. Proc Natl Acad Sci U S A 103: 2398–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taffs, R., Aston, J.E., Brileya, K., Jay, Z., Klatt, C.G., McGlynn, S., et al. (2009) In silico approaches to study mass and energy flows in microbial consortia: a syntrophic case study. BMC Syst Biol 3: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamazawa, S., Takasaki, K., Tamaki, H., Kamagata, Y., and Hanada, S. (2012) Metagenomic and biochemical characterizations of sulfur oxidation metabolism in uncultured large sausage-shaped bacterium in hot spring microbial mats. PLoS One 7: e49793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamazawa, S., Yamamoto, K., Takasaki, K., Mitani, Y., Hanada, S., Kamagata, Y., and Tamaki, H. (2016) In situ gene expression responsible for sulfide oxidation and CO2 fixation of an uncultured large sausage-shaped aquificae bacterium in a sulfidic hot spring. Microbes Environ 31: 194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchan, Y.T., and New, P.B. (1984) Genus I. Azotobacter. In Bergey’s Manual of Systematic Bacteriology, vol. 1. Kreig, N.R., and Holt, J.G. (eds). Baltimore, UK: Williams and Wilkins, pp. 220–229. [Google Scholar]
- Tindall, B.J. (2019) The names Hungateiclostridium Zhang et al. 2018, Hungateiclostridium thermocellum (Viljoen et al. 1926) Zhang et al. 2018, Hungateiclostridium cellulolyticum (Patel et al. 1980) Zhang et al. 2018, Hungateiclostridium aldrichii (Yang et al. 1990) Zhang et al. 2018, Hungateiclostridium alkalicellulosi (Zhilina et al. 2006) Zhang et al. 2018, Hungateiclostridium clariflavum (Shiratori et al. 2009) Zhang et al. 2018, Hungateiclostridium straminisolvens (Kato et al. 2004) Zhang et al. 2018 and Hungateiclostridium saccincola (Koeck et al. 2016) Zhang et al. 2018 contravene Rule 51b of the International Code of Nomenclature of Prokaryotes and require replacement names in the genus Acetivibrio Patel et al. 1980. Int J Syst Evol Microbiol 69: 3927–3932. [DOI] [PubMed] [Google Scholar]
- Van De Werken, H.J.G., Verhaart, M.R.A., VanFossen, A.L., Willquist, K., Lewis, D.L., Nichols, J.D., et al. (2008) Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol 74: 6720–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai, S., Cottingham, R.W., Huntemann, M., Copeland, A., Chen, I-min A., Kyrpides, N., et al. (2015) Complete genome sequences of Caldicellulosiruptor sp. strain Rt8.B8. Genome Announc 3: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasai, S., and Minamisawa, K. (2018) Plant-associated microbes: from rhizobia to plant microbiomes. Microbes Environ 33: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, R.R., Judd, L.M., Gorrie, C.L., and Holt, K.E. (2017) Completing bacterial genome assemblies with multiplex MinION sequencing. Microb Genomics 3: e000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2016) ggplot2—Elegant Graphics for Data Analysis. New York, NY: Springer. [Google Scholar]
- Yamada, A., Inoue, T., Noda, S., Hongoh, Y., and Ohkuma, M. (2007) Evolutionary trend of phylogenetic diversity of nitrogen fixation genes in the gut community of wood-feeding termites. Mol Ecol 16: 3768–3777. [DOI] [PubMed] [Google Scholar]
- Yang, S.J., Kataeva, I., Wiegel, J., Yin, Y., Dam, P., Xu, Y., et al. (2010) Classification of ‘Anaerocellum thermophilum’ strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int J Syst Evol Microbiol 60: 2011–2015. [DOI] [PubMed] [Google Scholar]
- Zehr, J.P., Jenkins, B.D., Short, S.M., and Steward, G.F. (2003) Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5: 539–554. [DOI] [PubMed] [Google Scholar]
- Zehr, J.P. (2011) Nitrogen fixation by marine Cyanobacteria. Trends Microbiol 19: 162–173. [DOI] [PubMed] [Google Scholar]
- Zverlov, V., Mahr, S., Riedel, K., and Bronnenmeier, K. (1998) Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 144: 457–465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.