Highlights
-
•
The Brain/MINDS beyond project plans to collect multi-site/scanner brain MRI data.
-
•
Prospective harmonization of MRI was achieved by standardizing scanning protocols.
-
•
The preliminary data showed moderate reliability of brain connectome data.
-
•
Completing traveling subject plan will allow robust statistical harmonization.
-
•
Scanning protocols are publicly available and data will also be shared by 2024.
Keywords: Multi-site study, HCP-style brain imaging, Psychiatric disorders, Neurological disorders, Harmonization protocol, Traveling subjects
Abbreviations: DALYs, disability-adjusted life years; MRI, magnetic resonance imaging; HCP, Human Connectome Project; ABCD, Adolescent Brain Cognitive Development; BPD, bipolar disorder; MDD, major depressive disorder; DecNef, Decoded Neurofeedback; ASD, autism spectrum disorder; ADNI, Alzheimer’s Disease Neuroimaging Initiative; AD, Alzheimer’s disease; MCI, mild cognitive impairment; PPMI, Parkinson's Progression Markers Initiative; PD, Parkinson’s disease; T1w, T1-weighted; T2w, T2-weighted; rsfMRI, resting-state functional MRI; CRHD, Connectome Related to Human Disease; GLM, general linear model; TS, traveling subject; AMED, Japan Agency for Medical Research and Development; Brain/MINDS Beyond, Strategic International Brain Science Research Promotion Program; HARP, Harmonization protocol; DWI, diffusion-weighted imaging; QC, quality control; MNI, Montreal Neurological Institute; MSM, multi-modal surface matching; GLMM, general linear mixed model; CIFTI, Connectivity Informatics Technology Initiative; FEF, frontal eye field; PEF, premotor eye field; PSL, peri-sylvian language; STS, superior temporal sulcus; NODDI, nerite orientation and density imaging
Abstract
Psychiatric and neurological disorders are afflictions of the brain that can affect individuals throughout their lifespan. Many brain magnetic resonance imaging (MRI) studies have been conducted; however, imaging-based biomarkers are not yet well established for diagnostic and therapeutic use. This article describes an outline of the planned study, the Brain/MINDS Beyond human brain MRI project (BMB-HBM, FY2018 ~ FY2023), which aims to establish clinically-relevant imaging biomarkers with multi-site harmonization by collecting data from healthy traveling subjects (TS) at 13 research sites. Collection of data in psychiatric and neurological disorders across the lifespan is also scheduled at 13 sites, whereas designing measurement procedures, developing and analyzing neuroimaging protocols, and databasing are done at three research sites. A high-quality scanning protocol, Harmonization Protocol (HARP), was established for five high-quality 3 T scanners to obtain multimodal brain images including T1 and T2-weighted, resting-state and task functional and diffusion-weighted MRI. Data are preprocessed and analyzed using approaches developed by the Human Connectome Project. Preliminary results in 30 TS demonstrated cortical thickness, myelin, functional connectivity measures are comparable across 5 scanners, suggesting sensitivity to subject-specific connectome. A total of 75 TS and more than two thousand patients with various psychiatric and neurological disorders are scheduled to participate in the project, allowing a mixed model statistical harmonization. The HARP protocols are publicly available online, and all the imaging, demographic and clinical information, harmonizing database will also be made available by 2024. To the best of our knowledge, this is the first project to implement a prospective, multi-level harmonization protocol with multi-site TS data. It explores intractable brain disorders across the lifespan and may help to identify the disease-specific pathophysiology and imaging biomarkers for clinical practice.
1. Introduction
Psychiatric and neurological disorders are afflictions of the brain that can affect individuals throughout their lifespans. Using the disability-adjusted life years (DALYs), which is a measure of disease burden proposed by the World Health Organization Global Burden of Disease study, in 2010 mental and behavioral disorders accounted for 7.4% of the total DALYs and neurological disorders accounted for 3.0% (Murray et al., 2012), up from 5.4% and 1.9% in 1990, respectively. Since the 1990s, technical advances in magnetic resonance imaging (MRI) have allowed detailed analysis of the organization of brain function and structure in humans. Recent high-quality MRI studies with a large cohort are expected to provide neurobiological and life-span information in healthy subjects (Glasser et al., 2016b, Harms et al., 2018, Miller et al., 2016), which will hopefully provide diagnostic utility for patients with psychiatric and neurological disorders (Drysdale et al., 2017, Elliott et al., 2018b, Koutsouleris et al., 2015, Nunes et al., 2018). However, the diagnostic sensitivity of brain MRI for psychiatric disorders has not been well established, presumably because effect sizes tend to be small and overlap with variability in healthy individuals (Yamashita et al., 2019). Protocols of scanning and analysis have rarely been standardized across projects, though that has begun to change - especially for large projects such as the Human Connectome Project (HCP) (Van Essen et al., 2012), UK Biobank (Miller et al., 2016), and the Adolescent Brain Cognitive Development (ABCD) project (Casey et al., 2018). Designing multi-disorder study and individual’s unique connectome throughout development may provide a framework to establish specific, precise resting-state fMRI biomarkers in neuropsychiatry (Parkes et al., 2020).
In this article, we first review previous large-population or multi-site neuroimaging studies related to psychiatric disorders (Section 2). Then, we provide an overview and study design of our current study in progress in Japan, the Brain/MINDS Beyond human brain MRI project (BMB-HBM, FY2018 ~ FY2023), which aims to establish clinically relevant imaging biomarkers with multi-site/protocol harmonization by including travelling subjects in study design (Section 3). Sections 4 to 5 describe details on the BMB-HBM such as harmonized MRI scanning protocols (HARP), preprocessing and preliminary data, and conceptualization of traveling-subject harmonization, followed by Section 6 presenting ethical and data sharing aspects in this project. Finally, we discuss the future directions, and potential impacts of this project (Section 7).
2. Previous neuroimaging protocols
2.1. Previous multi-site neuroimaging studies for neuropsychiatric disorders
Several brain imaging projects have attempted to identify suitable biomarkers in neuropsychiatric diseases. Recent multi-site neuroimaging mega studies have revealed well-replicated and clinically applicable findings from structural images; the Enhancing NeuroImaging Genetics through Meta-Analysis Consortium in the U.S. (n = 4,568) and the Cognitive Genetics Collaborative Research Organization in Japan (n = 2,564) replicated findings that patients with schizophrenia have volumetric alterations of subcortical structures when compared to healthy controls (Okada et al., 2016, van Erp et al., 2016). The findings were partly evident in other psychiatric disorders, such as bipolar disorder (BPD) and major depressive disorder (MDD) (Hibar et al., 2018, Schmaal et al., 2017, Schmaal et al., 2016, van Erp et al., 2016). Using resting-state functional MRI (rsfMRI), a multi-site study successfully developed generalized classifiers for psychiatric disorders. The Decoded Neurofeedback (DecNef) Project (https://bicr.atr.jp/decnefpro/), a multi-site neuroimaging study in Japan (12 sites, n = 2,409), developed a generalized classifier for autism spectrum disorder (ASD) with a high accuracy—not only for the data in three Japanese sites (85%) but also for the Autism Brain Imaging Data Exchange dataset (75%) (Yahata et al., 2016). The project also quantified the spectrum of psychiatric disorders by applying the ASD classifier to other multi-disorder datasets (schizophrenia, MDD, and attention-deficit/hyperactivity disorder). Later studies in the Strategic Research Program for Brain science (SRPBS) consortium further combined high-quality harmonization with travelling subjects (TS) and achieved multi-site predictions of brain features in neuropsychiatric patients (Yamashita et al., 2019, Yamashita et al., 2020).
In the field of neurodegenerative disease, the Alzheimer’s Disease Neuroimaging Initiative (ADNI) is one of many major multi-site neuroimaging and biomarker studies of Alzheimer’s disease (AD) and mild cognitive impairment (MCI) that was started in 2005 in North America (Mueller et al., 2005, Weiner et al., 2015). It contributed to the development of blood and imaging biomarkers, the understanding of the biology and pathology of aging, and to date has resulted in over 1,800 publications. ADNI also impacted worldwide ADNI-like programs in many countries including Japan, Australia, Argentina, Taiwan, China, Korea, Europe, and Italy. The Japanese ADNI (J-ADNI) conducted a multi-site neuroimaging study on cognitively normal elderly patients, MCI, and mild AD (n = 537), which emphasized the harmonization of the protocol and procedures with the ADNI (Iwatsubo et al., 2018). J-ADNI also developed machine learning techniques using feature-ranking, a genetic algorithm, and a structural MRI-based atrophy measure to predict the conversion from MCI to AD (Beheshti et al., 2017). Inspired by the Parkinson's Progression Markers Initiative (PPMI; (Parkinson Progression Marker Initiative, 2011), the Japanese (J-) PPMI team has also started a cohort in patients with rapid eye movement sleep behavior disorder, which is regarded to be prodromal to Parkinson’s disease (PD) (Mukai and Murata, 2017).
These previous mega-studies have contributed to the discovery of potential mechanisms and biomarkers of multiple brain disorders. However, most of these imaging biomarkers have relatively small effect sizes and the study results were drawn from multi-site data which are often heterogeneous and used now outdated traditional low-resolution data acquisition protocols. In addition, there have been no human brain MRI studies that explore multiple psychiatric and neurological disorders that occur through the lifespan within the same cohort of subjects.
2.2. High-quality multi-modal MRI protocols and preprocessing pipelines
The HCP developed a broad approach to improving brain imaging data acquisition, preprocessing, analysis, and sharing (Glasser et al., 2016b). It includes: 1) high-quality multi-modal data acquisition; 2) in a large number of subjects; and 3) high-quality data preprocessing and has proven usefulness of MRI techniques for understanding the detailed organization of a healthy human brain (Elliott et al., 2018a, Glasser et al., 2016a, Smith et al., 2015). The HCP aimed to delineate the brain areas and characterize neural pathways that underlie brain function and behavior in 1,200 healthy young adults (Van Essen et al., 2012). HCP scans were performed by a single MR scanner (a customized 3T Skyra, Siemens Healthcare GmbH, Erlangen, Germany) in a total of 4-hour scan time for high-resolution multi-modal data, which included T1-weighted (T1w) images, T2-weighted (T2w) images, diffusion-weighted images (DWI), rsfMRI, and task fMRI (Glasser et al., 2016b, Glasser et al., 2013). The HCP also developed a set of preprocessing pipelines with improved cross-subject alignment that dramatically improves the spatial localization of brain imaging findings and also increasing statistical sensitivity (Coalson et al., 2018, Glasser et al., 2013, Robinson et al., 2018). For the Lifespan Developing and Aging HCP Projects (HCP-D and HCP-A) the original HCP protocol for healthy young adults was shortened, for children and the elderly (60 to 90 min scan time; (Bookheimer et al., 2019, Harms et al., 2018, Somerville et al., 2018), and for psychiatric and neurological disorders (the Connectomes Related to Human Disease [CRHD]), and adolescent development (the ABCD project; (Casey et al., 2018). The UK Biobank used an even more abbreviated scanning approach to collect a much larger number of cohorts (n = 100,000) to predict health conditions (Miller et al., 2016).
Many of these high-quality multimodal projects have been based on a single or small number of the same model scanners at different sites and thus did not fully address standardization of the data acquisition across different scanner models or vendors.
2.3. Traveling subjects
A harmonization approach is required for individual-based statistics using a multi-site dataset, because the data from each site has the bias from hardware and scanning protocol (measurement bias) and sampling variability (i.e. age, sex, handedness, and socioeconomic status). If measurement biases were correlated or anti-correlated with a specific disease state this would result in a positive or negative bias in a given measure, whereas uncorrelated biases would merely reduce sensitivity (i.e. SNR) of the measure. Sampling biases due to biological differences in the sampled populations should also be considered for both case and control groups. Data harmonization has been proposed to control for these biases, including a general linear model (GLM) with the site as the covariate, a Bayesian approach (Fortin et al., 2018, Fortin et al., 2017), and a meta-analytic approach (Okada et al., 2016, van Erp et al., 2016), but any statistical method is unable to distinguish between sampling and measurement biases as long as the subject’s data is obtained by different sites/scanners and protocols (Yamashita et al., 2019). Inter-site cross-validation by machine learning and deep learning techniques is a method that aims to remove bias without any specific preparation if large-sample datasets are available (Nunes et al., 2018). However, this method extracts stable characteristics across the images and is limited to using only a part of the information for further analysis. In addition, it is unclear whether the classifiers obtained by such methods can be applied to an independent new site of the initial multi-site project.
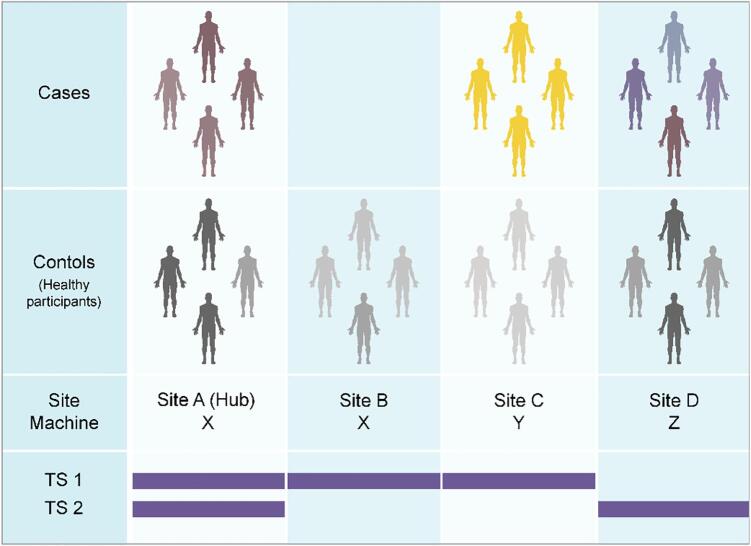
The traveling subject (TS) approach is a powerful research design to control for site differences (Fig. 1). This approach requires the images from the same participants at all the participating sites, but also requires significant effort from the sites and the participants when compared to other harmonization methods listed above, and the TS scans must be completed before the analysis starts. However, the TS approach can differentiate most of the sample variability from measurement bias in functional MRI (Yamashita et al., 2019), structure and diffusion MRI (Tong et al., 2020). In our prior study, by scanning nine TS participants repeatedly at all the twelve sites, Yamashita et al. achieved the high-quality harmonization of the functional connectivity obtained by functional MRI (Yamashita et al., 2019). Measurement and sampling biases for each group (schizophrenia, MDD, ASD, and healthy controls) were segregated from individual and disease-specific factors as the rest of sampling variability. Our analysis of functional connectivity indicated that the measurement bias was the largest effect, followed by sampling variability, and disease- or subject-specific variation was the smallest effect. With regard to measurement bias, differences in phase encoding direction had the biggest effect size when compared to those of the vendor, coil, and scanner within the same vendor. The harmonization method was estimated to reduce measurement bias by 29% and improve the signal-to-noise ratio by 40% (Yamashita et al., 2019). Further investigations are needed to determine the best approach for reducing sampling bias arising from biological differences in the sampled population.
Fig. 1.
Case-control studies and traveling subject approach. (Top) When we analyze multi-site data from a set of case-control MRI studies, we must consider machine and protocol-derived bias (measurement bias) as well as sampling bias (from biological differences in the sampled populations). Even if the scanner and protocol are the same between sites (e.g. Sites A and B), measurement bias may still occur because of slight differences in the magnetic or radiofrequency fields, etc. Sampling bias should be considered for patient groups as well as control groups, given that the control participants were recruited according to the demographics in the patient group. (Bottom) The traveling subject (TS) harmonization approach enables us to combine with case-control datasets by differentiating between measurement and sampling biases (Yamashita et al., 2019). To reduce the effort of TS participants and participating sites, the current project applies a hub-and-spoke design to the TS project. With this approach, multiple sets of participants, scanner, protocol data can be efficiently collected, and measurement bias is properly estimated using a GLMM for grouped and repeated datasets (TS 1 and 2).
3. Overview of BMB-HBM project
The Strategic International Brain Science Research Promotion Program (Brain/MINDS Beyond [BMB]; FY2018–FY2023) was funded by the Japan Agency for Medical Research and Development (AMED) to support global brain research by enhancing collaboration with the domestic projects of other countries. The human brain MRI project (BMB-HBM) is carried out by thirteen research institutions and universities among BMB project. A core part of the study design including MRI protocols, preprocessing, harmonization and data sharing is coordinated by the working group in this project.
Based on previous multi-site studies in Japan (Iwatsubo et al., 2018, Okada et al., 2016, Yahata et al., 2016, Yamashita et al., 2019), the overall goal of this project is expected to find altered brain imaging characteristics in psychiatric and neurological disorders that can be applied to future therapeutic investigations and clinical devices. Importantly, this project plans to establish the multi-level harmonization consisting of 1) travelling subject design and data collection, 2) harmonized MRI protocols, 3) harmonized preprocessing and preliminary data, and 4) statistical harmonization. Currently, first two items related to the core part of protocols are established and described in Section 3.1 and 3.2. The latter two are detailed in the next Section 4 and include a planned preprocessing pipeline and theorization of statistical harmonization.
3.1. Study design – multi-site, multi-disorder
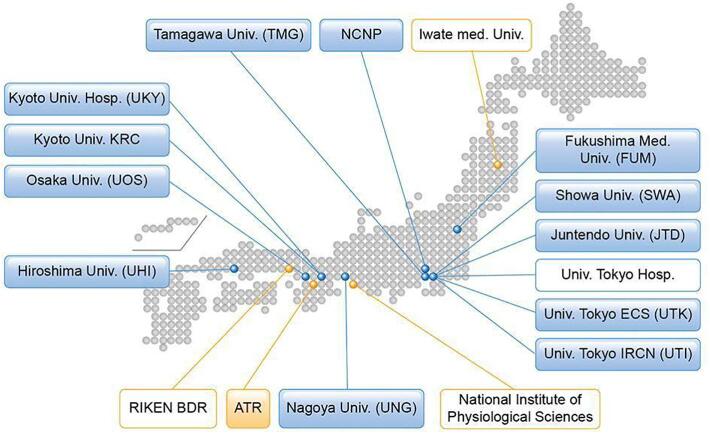
Over 2 k patients with various psychiatric and neurological disorders are scheduled to participate in the project. As of March 2020, 13 sites have approved this study project, received approval from their respective ethical review board(s), and obtained clinical and TS measurements using the appropriate MRI scanners (Fig. 2, Table 1). Of these, 5 sites mainly explore psychiatric disorders (schizophrenia, ASD, MDD, and BPD), 4 sites neurological disorders (AD, PD, multiple system atrophy, progressive supranuclear palsy, chronic pain disorder, and epilepsy), and 2 sites both categories. Two sites measure the general adolescent population to investigate brain development and recruit through advertisement and cohort studies (Ando et al., 2019, Okada et al., 2019). Each site intends to obtain brain images and demographic (and clinical) characteristics for clinical cases and match controls for age, sex, premorbid IQ or educational attainment, socio-economic status, and handedness (see Section 4.2). The exclusion criteria were set by each study purpose (i.e. low premorbid IQ, history of loss of consciousness for >5 min, illegal drug use, and alcohol dependency). Illegal drug use can be a major concern for disease onset and poor prognosis, especially for psychiatric disorders. However, there is far less illegal drug use in Japan compared to Western European countries (Degenhardt et al., 2008, Lee and Kwon, 2016), and most of the participating sites excluded those with a current illegal drug use or previous history of regular use (Koike et al., 2013).
Fig. 2.
Brain/MINDS Beyond human brain MRI project. Institutes in the blue boxes show measurement and analysis sites for neuropsychiatric disorders, and those in the orange boxes show analysis support sites. Institutes listed in boxes with a colored background represent participation in the traveling subject project. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Participating sites of the Brain/MINDS Beyond MRI project.
| Site | Research group | Role for the project | Role for TS | MRI scanner (System version) | Protocol | Main target population |
|---|---|---|---|---|---|---|
| UTK | G1-1D, G1-2 | Data acquisition/Analysis | Hub | Prisma (VE11C) | CRHD | Adolescent cohort, HP, ASD, Sch, MDD, Epilepsy |
| UTI | G1-1D, G1-2 | Data acquisition/Sharing | Hub | Prisma (VE11C) | CRHD | HP, ASD, Sch, MDD, BPD |
| ATR | G1-2, G3 | Data acquisition/Sharing/Analysis | Hub | Prisma (VE11C) | CRHD | HP |
| FUM | G1-1S | Data acquisition | Spoke | Skyra (VE11C) | HARP | HP, AD, PD |
| TMG | G1-1D | HARP setup/Data acquisition | Spoke | Trio (VB19A) | HARP | Adolescent cohort |
| SWA | G1-1D, G3, IR | HARP setup/Data acquisition | Spoke | Skyra (VE11E) | HARP | HP, ASD |
| NCNP | G1-1S | HARP setup/Data acquisition/Sharing/Analysis | Spoke | Verio Tim + Dot (VD13A) | HARP | HP, Sch, MDD, AD, PD |
| JTD | IR | Data acquisition | Spoke | Prisma (VE11C) | HARP | HP, PD, MSA, PSP |
| UOS | G2 | Data acquisition | Spoke | Prisma (VE11C) | HARP | HP, Chronic pain |
| UHI | G1-1A, G3 | HARP setup/Data acquisition | Spoke | Skyra (VE11C) | HARP | HP, MDD, BPD |
| UNG | BM | Data acquisition | Spoke | Verio (VB17A) | HARP | HP, Sch |
| UKY | G1-1S | HARP setup/Data acquisition | Spoke | Skyra (VE11C) | HARP | HP, AD, PD |
| KRC | G1-1A | Data acquisition | Spoke | Verio (VB17A) | HARP | HP, Sch, MDD, BPD |
| BDR | G1-2 | HARP setup/Data Analysis | Spoke | Prisma (VE11C) | HARP | NA |
Abbreviations: UTK, The University of Tokyo ECS (Komaba Campus); UTI, The University of Tokyo IRCN; FUM, Fukushima Medical University; TMG, Tamagawa Academy & University; SWA, Showa University; NCNP, National Center of Neurology and Psychiatry; JTD, Juntendo Hospital; ATR, Advanced Telecommunications Research Institute International; UOS, Osaka University; UHI, Hiroshima University; UNG, Nagoya University; UKY, Kyoto University; KRC, Kyoto University Kokoro Research Center; BDR, RIKEN Center for Biosystems Dynamics Research; IR, Innovative Research Group in Brain/MINDS Beyond; BM, Brain/MINDS project; CRHD, Human Connectome Studies Related To Human Disease protocol; HARP, harmonization protocol; HP, healthy participants; ASD, autism spectrum disorders; Sch, schizophrenia; MDD, major depressive disorder; BPD, bipolar disorder; AD, Alzheimer’s disease; PD, Parkinson disease; MSA, multiple system atrophy; PSP, progressive supranuclear palsy.
3.2. Travelling subject design
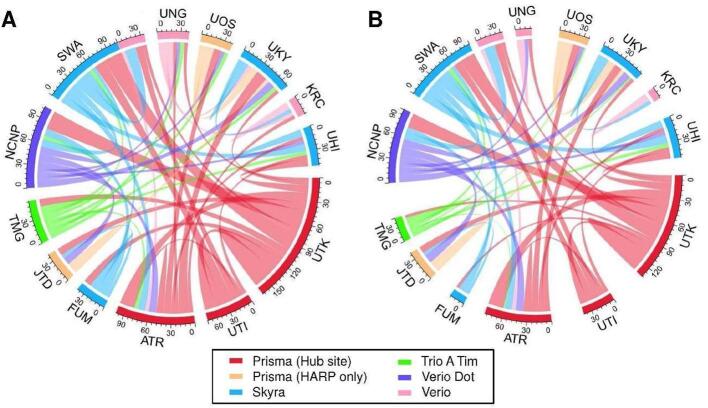
The traveling subject (TS) approach is a core part of the study design in this project. For designing TS, 75 healthy adults were planned to undergo 6 to 8 scans at three or more sites within 6 months. Five or more healthy participants were recruited at each site. Each participant received test–retest scans at the recruitment site and underwent scans at different sites including a hub site. We set up three hub sites, according to a hub-and-spoke model, in which all participants received scans using a MAGNETOM Prisma scanner and the CRHD and HARP protocols. The hub-and-spoke model planned is illustrated in Fig. 3. The details of sites, protocols in traveling subject are summarized in Supplementary Table S2.
Fig. 3.
Expected and current data connection of the traveling subjects (TS). Data connections in the traveling subject project (TS) that were initially planned (A) and the actual connections as of March 2020 (B). Hub sites using Prisma and other sites using Prisma, Skyra, Trio A Tim, Verio Dot, and Verio are illustrated in red, orange, blue, green, purple, and pink, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Each participant undergoes HARP and CRHD scans using the Prisma (~2 h) at one or more hub sites (UTK, UTI, and ATR) to harmonize the data within the Brain/MINDS Beyond project and other projects (e.g. Brain/MINDS, HCP, and ABCD) and test the difference in quality between the protocols. The other visiting sites were determined in consideration of the site locations, machine differences, and project similarities between the sites. Each participant receives multiple scans at the recruitment site to assess the test–retest reliability (1-hour × 2 sessions). At a part of sites, we also scan TS with the previous ‘SRPBS MRI protocol’ (termed SRPB in Brain/MINDS Beyond) used in the multi-site studies (Yamashita et al., 2019). The total number of scans and spokes between the sites are expected to be 455 and 465, respectively (Fig. 3A). As of March 2020, 74 participants were registered and 405 scans (89.0%) were completed and uploaded to the ATR XNAT server. The data provided 368 spokes (76.1%, Fig. 3B). The TS project will end in August 2020.
4. Protocols
4.1. HARP, CRHD and SRPB protocols
We developed protocols that minimize potential differences related to measurement and increase the MR image sensitivity to brain organization in psychiatric and neurological disorders. From a neurobiological perspective, the cerebral cortex is organized by a 2D sheet-like structure with an average thickness of 2.6 mm embedded and folded in the ~ 1300 mL of brain volume (Glasser et al., 2016b). From a neuroimaging perspective, the spatial resolution and homogeneity of the images are important factors that may induce bias and error during the image analysis; these include partial voluming, image distortion, errors in brain segmentation, and registration. Of these, respecting spatial fidelity of neuroanatomical structures is the most important approach for achieving unbiased imaging (Glasser et al., 2016b). Therefore, the spatial resolution of the imaging was determined based on cortical thickness and was matched across all scanners. The phase encoding direction of EPI-based functional and diffusion MRI is an important factor that relates to spatial distortion (and signal loss in fMRI) in association with the polarity of the direction, echo spacing, and B0 magnetic field homogeneity; therefore, we acquire a spin-echo filed map with opposite phase encoding directions to enable distortion correction (Andersson et al., 2003). Based on these strategies, two new MRI protocols were planned for use in the project: 1) a harmonized MRI protocol (HARP), which can be run on the multiple MRI scanners/sites within a period of 22 to 65 min; and 2) an ‘HCP style’ MRI protocol used by HCP CRHD for the high-performance 3 T MRI scanner such as a MAGNETOM Prisma (Siemens Healthcare GmbH, Erlangen, Germany). A legacy protocol, SRPB was used in sites which used former-generation MRI scanners (Yamashita et al., 2019).
The HARP was created to be used at multiple MRI scanners/sites, and it was designed to obtain high-quality and standardized brain MRI data in a ‘clinically’ practical window of time (Table 2 and Supplementary Table S1). The conditions of the MRI scanners were as follows: 1) static magnetic field strength of 3 T; 2) multi-array head coil with 32 or more channels; and 3) ability to perform a multi-band EPI sequence provided from Center for Magnetic Resonance Research, University of Minnesota with an acceleration factor of 6 (Moeller et al., 2010, Setsompop et al., 2012, Xu et al., 2013). The parameters of the HARP were extensively adjusted and harmonized across scanners, particularly those that may affect the spatial localization and quality of the metrics of cortical ribbon segmentation, including spatial resolution, temporal resolution and imaging contrast, such as an acquisition matrix, field of view, repetition & echo time, bandwidth, echo spacing, encoding directions an acceleration factor. This process required iterative optimizations between scanners, since available values of parameters are often limited across multiple, diverse systems provided by vendors. For example, the echo time value of the functional MRI (TE = 34.4 ms) was the only option that was allowed to be set in all the scanners. In 2021, the protocol was adapted for use with five MRI scanners/systems (MAGNETOM Prisma, Skyra, Trio A Tim, Verio, and Verio Dot; Siemens Healthcare GmbH, Erlangen, Germany), and we plan to expand it to different MRI scanners/vendors during the project period and in fact we are working on creating HARP protocol for GE scanners. The HARP was intended to perform the brain scan within a period of ~ 30 min using a high-resolution structural MRI scan (T1w and T2w, spatial resolution of 0.8 mm) and a set of two high-sensitive rsfMRI scans with opposing phase directions, a spatial resolution of 2.4 mm, and a temporal resolution of 0.8 s for a total of 10 min. Additional two sets of rsfMRI scans are performed if subjects agreed to be scanned. The protocols also include optional sequences for four additional rsfMRI scans, task fMRI (Emotion and CARIT) (Winter and Sheridan, 2014), two DWI scans with opposing phase encoding directions (b-value = 0, 700, and 2000 sec/mm2, and the number of diffusion direction = 96 in Prisma, 120 in other scanners), quantitative susceptibility mapping, and arterial spin labeling. The minimum and maximum scanning time of the HARP is 22 and 65 min, respectively (Table 2). The HARP protocols are publicly available at https://dx.doi.org/10.17605/OSF.IO/C49NP
Table 2.
Time table of CRHD and HARP protocols.
| Subset | Sequence | Duration |
Participant instruction | ||
|---|---|---|---|---|---|
|
Prisma |
Skyra, Trio, Verio Dot, Verio |
||||
| CRHD | HARP | HARP | |||
| rsfMRI 1 | SEF AP | 0:32 | 0:06 | Fixation | |
| BOLD AP | 5:46 | 5:08 | Fixation | ||
| SEF PA | 0:32 | 0:06 | Fixation | ||
| BOLD PA | 5:46 | 5:08 | Fixation | ||
| Structure | T1 MPR | 6:38 | 5:22 | Rest | |
| T2 SPC | 5:57 | 5:31 | 5:22–6:26 | Rest | |
| Subtotal | 25 min | 22 min | 22–23 min | ||
| ASL | NA | 2:45b | Rest | ||
| QSM | NA | 5:03c | Rest | ||
| DWI | AP | 6:07 | 3:29 | 4:50 | Rest |
| PA | 6:05 | 3:32 | 4:54 | Rest | |
| AP | 5:39 | NA | NA | Rest | |
| PA | 5:39 | NA | NA | Rest | |
| rsfMRI 2 | See rsfMRI 1a | 13 min | 11 min | Fixation | |
| rsfMRI 3 | See rsfMRI 1a | NA | 11 min | Fixation | |
| Task fMRI EMOTION | SEF AP | NA | 0.06 | Task | |
| SEF PA | NA | 0.06 | Task | ||
| BOLD PA | NA | 4.08 | Task | ||
| Task fMRI CARIT | SEF AP | NA | 0.06 | Task | |
| SEF PA | NA | 0.06 | Task | ||
| BOLD PA | NA | 4.08 | Task | ||
| Total | 61 min | 68 min | 59–68 min | ||
Abbreviations: rsfMRI, resting-state functional MRI; ASL, arterial spin labeling; QSM, quantitative susceptibility mapping; DWI, diffusion weighted imaging; SEF, spin echo field mapping; BOLD, blood oxygenation level dependent; T1 MPR, T1-weighted magnetization prepared rapid acquisition with gradient echo; T2 SPC, T2-weighted sampling perfection with application optimized contrasts using different flip angle evolutions.
a set of SEF AP, BOLD AP, SEF PA, and BOLD PA.
b Only for Prisma and Skyra.
c Only for Prisma, Skyra, and Verio Dot.
The CRHD protocol was planned for collaboration with the HCP CRHD for the Early Psychosis Project (Lewandowski et al., 2020). The HCP CRHD protocol included high-resolution structural MRI (spatial resolution of 0.8 mm), high-resolution resting-state fMRI with an opposing phase encoding direction and longer scan time (spatial resolution of 2 mm, TR = 0.8sec, 5.6 min × 4 scans), and high-resolution and high angular diffusion MRI (spatial resolution of 1.5 mm, b-value = 0, 200, 500, 1500, 3000 sec/mm2, number of diffusion direction = 194), publicly available at https://www.humanconnectome.org/storage/app/media/documentation/data_release/Appendix_1_HCP-EP_Release_Imaging_Protocols.pdf. The SRPB protocol is a legacy protocol used in the previous project (Yamashita et al., 2019) and includes T1w image (spatial resolution of 1.0 mm), a single resting-state fMRI scan (spatial resolution of 3.3 × 3.3 mm in plane, slice thickness and gap of 3.2 mm and 0.8 mm respectively, TR = 2.5sec, 10 min × 1 scan, phase encoding direction of posterior to anterior) and a gradient-echo field map (https://bicr.atr.jp/wp-content/uploads/2018/08/UnifiedProtocol-1–1.pdf).
The installation of the protocols in the MRI scanners was ensured by conducting hierarchical parameter checks and site visits at the beginning of the measurement period. After the protocol installation, each site sent XML files of the installed protocol from the MRI scanner to the protocol management site (UTK), and all the parameters were confirmed with a checksum algorithm using R (R Core Team, 2018). This process was useful for validating the protocols across sites/scanners because some of the MRI scanners actually underwent inappropriate installation and were set with different parameters. The results were then sent back to the collaborators, who edited the parameters. We also checked the DICOM files that are deposited in the ATR XNAT server. In this phase, we checked the parameters, slice numbers, and diffusion gradient information (bvec and bval).
The manuals were shared and used at the sites for protocol installation, demographic and clinical assessment before the scan (e.g. handedness), and the assessment of and instruction to participants during the scan (e.g. general instruction during the scan, fixation to the cross during rsfMRI scans, and the assessment of sleepiness during the rsfMRI).
4.2. Cognitive and behavioral assessment
Each participating site assesses demographic characteristics (i.e. age, sex, and socioeconomic status), clinical characteristics (i.e. diagnosis, symptom severity, cognitive function, and general functioning), and subjective social evaluations (i.e. quality of life and well-being) (Table 3). Each subgroup (G1-1D, G1-1A, G1-1S, and G1-2 TS) indicates standard scales, some of which are uniform across subgroups and easier to share and use when analyzing brain images.
Table 3.
Clinical and neuropsychological assessment.
| G1-1D | G1-1A | G1-1S | |
|---|---|---|---|
| Depression | K6 or BDI-II | BDI-II and PHQ-9 | PHQ-9 and BDI-II/GDS-15 |
| Anxiety | – | GAD-7 | STAI |
| Autism | AQ-10, AQ-50 or SRS-2 (for developmental disorders) | AQ-10 or AQ-50 | – |
| Psychosis | APSS | – | NPI-Q |
| Intellectual ability | JART-25 or WAIS-III (WISC at the age of 15 years or less) Information and Picture completion subtests | JART-25 | JART-25 |
| Cognitive function | CANTAB or BACS-J | CANTAB or BACS-J | ADAS-Cog11, CDT, CDR, FAB, HVLT-R, JLO, MMSE, MoCA-J, SDMT, TMT-A/B, WMS-R |
| General function and disability | GAF, mGAF or WHO-DAS 2.0 | GAF, mGAF or WHO-DAS 2.0 | Schwab & England ADL |
| Quality of life | EQ-5D | EQ-5D | PASE |
| Well-being | WHO-5 | WHO-5 | SHAPS |
| Handedness | EHRS or UTokyo | EHRS or UTokyo | UTokyo |
Abbreviations: K6, 6-item Kessler Screening Scale for Psychological Distress; BDI-II, Beck Depression Inventory-Second Edition; PHQ-9, Patient Health Questionnaire-9; GDS-15, Geriatric Depression Scale 15; GAD-7, General Anxiety Disorder-7; STAI, State-Trait Anxiety Inventory; AQ-10, 10-item short version of the Autism Spectrum Quotient; AQ-50, Autism Spectrum Quotient (original version); APSS, Adolescent Psychotic-like Symptom Screener; NPI-Q, Neuro Psychiatric Inventory-Brief Questionnaire Form; JART-25, 25-item short version of the Japanese Adult Reading Test; WAIS-III, Wechsler Adult Intelligence Scale-Third Edition; GAF, Global Assessment of Functioning; mGAF, modified GAF; WHO-DAS 2.0, the World Health Organization Disability Assessment Schedule II; Schwab & England ADL, Modified Schwab and England ADL (Activities of Daily Living) scale; CANTAB, Cambridge Neuropsychological Test Automated Battery; BACS-J, the Brief Assessment of Cognition in Schizophrenia Japanese version; ADAS-Cog, Alzheimer’s Disease Assessment Scale-cognitive component; CDT, Clock Drawing Test; CDR, Clinical Dementia Rating; FAB, Frontal Assessment Battery; HVLT-R, Hopkins Verbal Learning Test-Revised; JLO, Judgment of Line Orientation; MMSE, Mini-Mental State Examination; MoCA-J, Japanese version of Montreal Cognitive Assessment; SDMT, Symbol Digit Modality Test; TMT-A/B, Trail Making Test Parts A and B; WMS-R, Wechsler Memory Scale-Revised; EQ-5D, EuroQol 5 Dimension questionnaire; WHO-5, World Health Organization-Five Well-Being Index; PACE, Physical Activity Scale for Elderly; SHAPS, Snaith-Hamilton Pleasure Scale; EHI, Edinburgh Handedness Inventory; UTokyo, 14-item Rating Scale of Handedness for Biological Psychiatry Research among Japanese People.
4.3. Data logistics
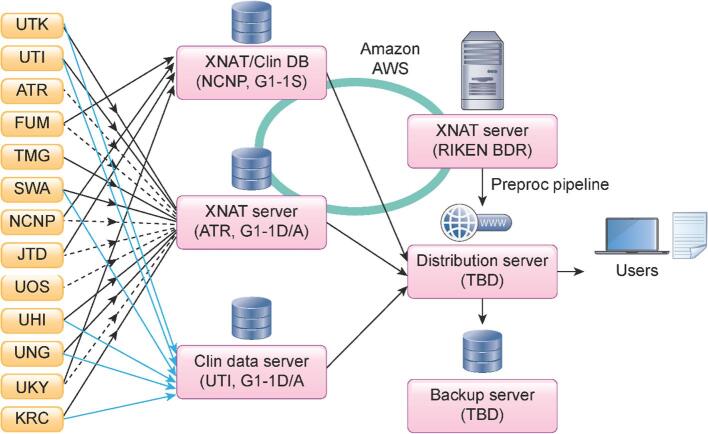
Brain MR images obtained using the CRHD and HARP protocols in this study project and related studies are stored, preprocessed, and distributed using the XNAT server system (https://www.xnat.org/) (Fig. 4). Due to the legacy of previous multi-site studies (Iwatsubo et al., 2018, Yahata et al., 2016, Yamashita et al., 2019), several data centers were already available for this project. The images obtained from the development and adult projects (G1-1D and G1-1A) will be sent to an XNAT server at ATR and the clinical data will be sent to UTI. For the senescent project (G1-1S), all the data will be sent to the NCNP (Iwatsubo et al., 2018). The TS data will also be sent to the ATR server shown in dashed lines. When uploading to the XNAT server, personal information (i.e. name and date of birth) contained in DICOM is automatically removed using an anonymization script of XNAT. A defacing procedure is performed for T1w and T2w images. These processes de-identify the MRI data. After manually checking whether the face images are completely obscured, all the anonymized MRI data are shared using Amazon AWS with RIKEN BDR, in which all image preprocessing is performed (see Section 5.1). Preprocessed data are sent back to the servers and can be seen with limited access (i.e. participating sites). After a quality control (QC) (see Section 5.3), cleaned imaging data with a demographic and clinical datasheet will be stored in the distribution server(s). All data will be also sent to the backup server(s).
Fig. 4.
Data storage, preprocessing, quality check, and data sharing. MRI (black line) and clinical (blue line) data from G1-1D and G1-1A sites are sent to the XNAT server and a data server at ATR and UTI, respectively. All data from G1-1S sites are sent to an XNAT server and a data server managed by NCNP, as this group applied a standard clinical assessment protocol to the project following a previous multi-site study. Traveling subject data from G1-1S sites are also sent to the XNAT server in ATR (dot line). XNAT servers at NCNP, ATR, and RIKEN BDR are linked by Amazon AWS to share the imaging data. NCNP manages a separate server for storing clinical data (Clin DB) being collected from the participants in this project. All MR images are preprocessed at RIKEN BDR. All MR images are preprocessed at RIKEN BDR. All the raw and preprocessed data will be stored and provided to the users in a distribution server. A backup server will be placed at a different site. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
5. Analysis and preliminary results
5.1. Preprocessing of HARP, CRHD, and SRPB MRI data
All neuroimaging data are preprocessed at RIKEN BDR for this project. The MR images are sent via Amazon S3 to a high-throughput parallel computing system at RIKEN BDR for preprocessing. The raw MRI data in DICOM format are converted to those in NIFTI using a conversion program, BCILDCMCONVERT (https://github.com/RIKEN-BCIL/BCILDCMCONVERT), by which folder structures are created and all the imaging parameters are read and stored including the type of gradient, k-space read out time in phase and read directions, phase encoding directions, to be used for preprocessing. The preprocessing is performed using the HCP pipeline 4.3.0 (Glasser et al., 2013) with modifications for adapting and harmonizing multiple scanners. In brief, the structural MRI (T1w and T2w) is first corrected for image distortions related to the gradient nonlinearity in each scanner type and the inhomogeneity of the B0 static magnetic field in each scan. The signal homogeneity is dealt with by prescan normalization and is also improved by a biasfield correction using T1w and T2w images (Glasser and Van Essen, 2011). The T1w and T2w images are fed into non-linear registration to the Montreal Neurological Institute (MNI) space and used for cortical surface reconstruction using FreeSurfer (Fischl, 2012) surface registration using multi-modal surface matching (MSM) (Robinson et al., 2018) and folding pattern (MSMsulc); this is followed by the creation of a myelin map using T1w divided by T2w and surface mapping (Glasser and Van Essen, 2011).
The functional MRI data is corrected for distortion (gradient nonlinearity and B0-inhomogeneity) and motion. The distortion from B0 static field inhomogeneity is corrected by means of opposite phase encoding spin echo fieldmap data using TOPUP (Andersson et al., 2003); it is then warped and resampled to MNI space at a 2 mm resolution and saved as a volume in the Neuroimaging Informatics Technology Initiative (NIFTI) format. The region of the cortical ribbon in the fMRI volume is further mapped onto the cortical surface and combined with voxels in the subcortical gray region to create 32 k greyordinates in the Connectivity Informatics Technology Initiative (CIFTI) format. Multiple runs of the fMRI data are merged and fed into independent component analyses (ICA) followed by an automated classification of noise components and the removal of noise components using FIX (Salimi-Khorshidi et al., 2014, Glasser et al., 2018). The automated classifier is trained using the data in this project and its accuracy is maximized. The denoised fMRI data, in combination with other cortical metrics (myelin, thickness; Fig. 4B and 4C, respectively), is further used for multi-modal registrations (MSMAll) over the cortical surface, followed by ‘de-drifting’ (removing registration bias after multimodal registration) (Glasser et al., 2016a).
The diffusion MRI is corrected for distortion and motion due to gradient nonlinearity, eddy current, motion, and B0 static field inhomogeneity using EDDY (Andersson and Sotiropoulos, 2016). The signal dropouts, susceptibility artefact, and their interaction with motion were also corrected (Andersson et al., 2018, Andersson et al., 2017). The resulting diffusion volumes are merged into a single volume and resampled in the subject’s real physical space aligned according to the ACPC convention. Diffusion modeling is performed using nerite orientation density imaging (NODDI) (Fukutomi et al., 2018, Zhang et al., 2012), and a Bayesian estimation of crossing fibers (Behrens et al., 2003, Sotiropoulos et al., 2016). Diffusion probabilistic tractography (Behrens et al., 2003) is also performed in a surface-based analysis (Donahue et al., 2016).
For analyzing the data of the SRPB protocol, we apply a legacy mode of HCP pipeline 4.3.0. The distortion of B0 field inhomogeneity of the T1w and the resting-state fMRI is corrected using a gradient field map. Since T2w image is not obtained, myelin map is not calculated and the MSMAll registration is optimized without using myelin contrast in future study.
5.2. Preliminary travelling subject results
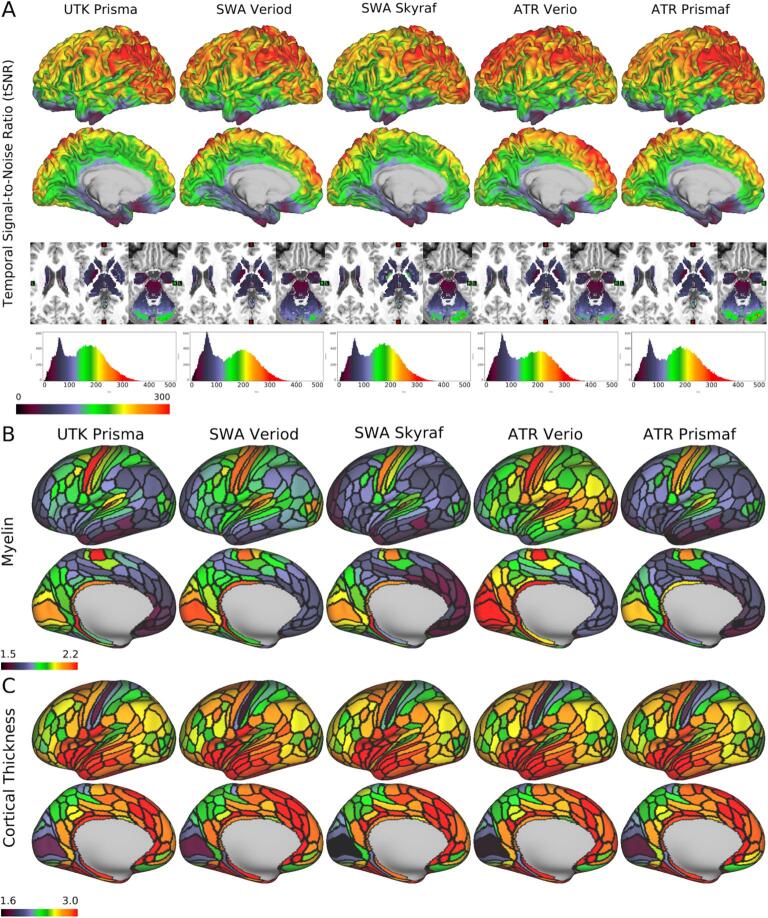
We first demonstrate the preliminary results of HARP in a single subject. Fig. 5A reveals the temporal signal-to-noise ratio (tSNR) in the same subject (ID = 9503), which was very high across scanners and multi-array coils. The mean ± standard deviation across 32 k greyordinates was 161 ± 80 in the Prisma at UTK, 155 ± 81 in the Verio Dot at SWA, 151 ± 72 in the Skyra fit at SWA, 151 ± 80 in the Verio at ATR, and 150 ± 74 in the Prisma fit at ATR; the values and their distributions were similar across scanners/sites. Fig. 5B shows cortical myelin map (not biasfield corrected [non BC]) in a single subject (ID = 9503) across scanners/sites, parcellated by HCP MMP v1.0 (Glasser et al., 2016a). It reveals the typical cortical distribution of the high myelin contrast in the primary sensorimotor (aeras 1, 3a, 3b, 4), auditory (A1), visual (V1), middle temporal, and ventral prefrontal (47 m) areas—as demonstrated previously (Glasser and Van Essen, 2011). The distributions over the cortex were comparable between scanners, although absolute values were slightly different suggesting the residual bias from transmit field across scans/scanners (see also 2.5.3).
Fig. 5.
Quality of MRI and preliminary cortical structures obtained by HARP in a single traveling subject across scanners/sites. A) Temporal signal-to-noise ratio (tSNR) obtained in a single subject (ID = 9503) across different scanners/sites by a harmonized MRI protocol (a sequence of functional MRI in HARP using a multi-band echo planar imaging with TR/TE = 800/34.4 ms; see Supplementary Table S1 for other details). The images from top to bottom show color-coded tSNR maps in 32 k greyordinates (see main text) overlaid on the lateral and medial surface of the mid-thickness surface of the left hemisphere, the subcortical sections of the T1w image, and the histogram of the tSNR values. B) Cortical myelin contrast (T1w/T2w ratio) across different scanners. The myelin contrast is not corrected for the biasfield and parcellated by the HCP MMP v1.0 (Glasser et al., 2016a). C) The map shows cortical thickness across different scanners. Cortical thickness is corrected by curvature and parcellated by the HCP MMP v1.0. The tSNR, myelin map and cortical thickness are comparable across scanners. Data at https://balsa.wustl.edu/7q4P9 and https://balsa.wustl.edu/6Vvqv.
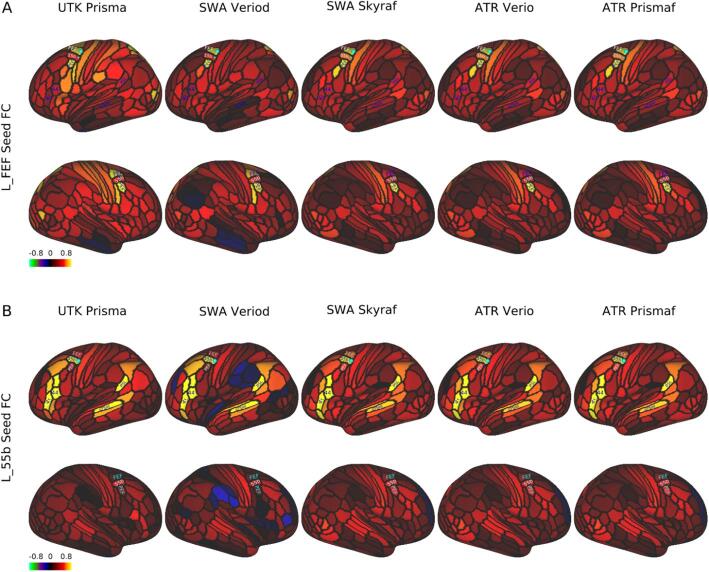
The resting-state seed-based functional connectivity in the same exemplar subject (ID = 9503) revealed a typical pattern over the cerebral cortex across scanners/sites; the left frontal eye field (FEF)-seed functional connectivity showed symmetric coactivation in the bilateral premotor eye field (PEF) (Fig. 6A), whereas the left area 55b-seed FC showed an asymmetric language network distributed in the perisylvian language (PSL) area, superior temporal sulcus (STS), and areas 44/45 predominantly in the left hemisphere (Fig. 6B).
Fig. 6.
Seed-based resting-state functional connectivity in a single traveling subject across scanners/sites. In a single subject (ID = 9503), the resting-state fMRI scans (5 min × 4) were collected using a scanning protocol of HARP across different scanners/sites (see Supplementary Table S1), preprocessed, and denoised by a surface-based analysis to generate parcellated functional connectivity (FC) using the HCP MMP v1.0 (Glasser et al., 2016a). A) FC seeded from the left frontal eye field (FEF), which was distributed symmetrically in the bilateral premotor eye field (PEF) and comparable across scanners/sites. B) FC seeded from the left area 55b, which showed an asymmetric language network predominant in the left hemisphere that was comparable across scanners/sites. The language network is distributed in the areas of 44/45, superior temporal sulcus, dorsal posterior part (STSdp), and perisylvian language (PSL). Data at https://balsa.wustl.edu/1B9VG and https://balsa.wustl.edu/5Xr71.
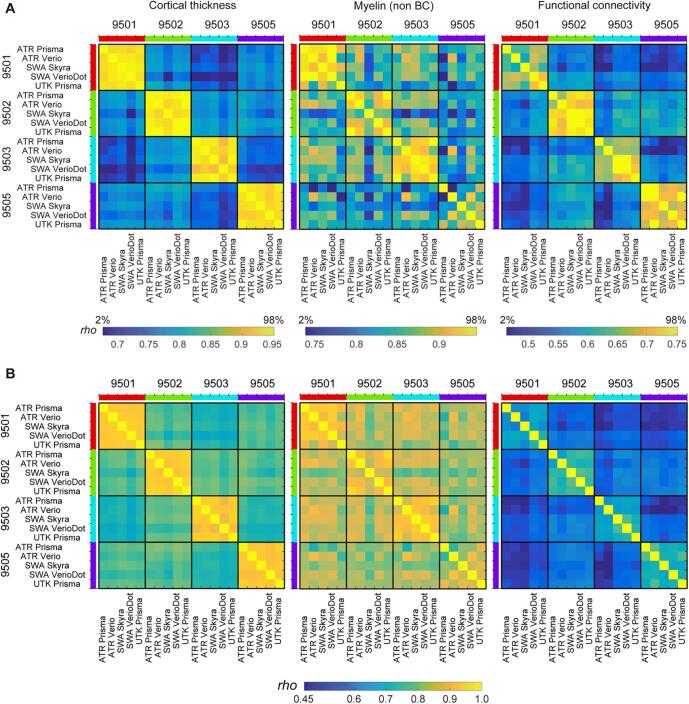
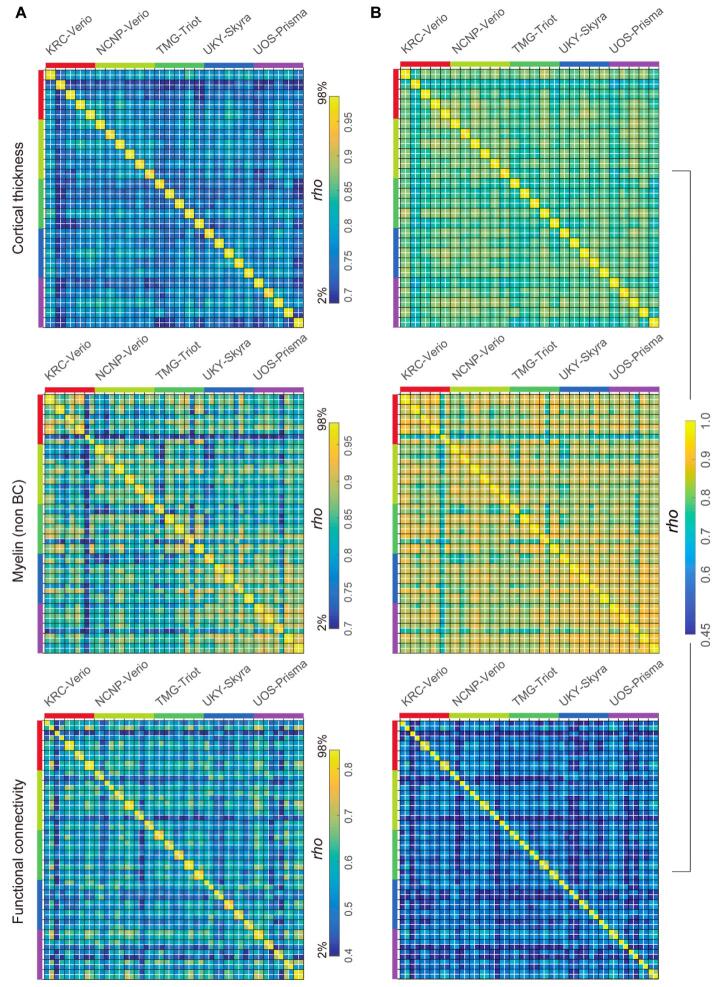
We further performed the preliminary preprocessing using the data from the initial TS study (N = 30), among which four healthy subjects participated and travelled across five sites and received MRI scanning with HARP in different scanners (4TS × 5S), and twenty-six subjects completed test–retest scans in any of 5 scanners (26TS × 2/5S). Datasets were analyzed with the current version of preprocessing (see Section 5.1) and each of the cortical thickness, myelin (non BC), and functional connectivity was parcellated using HCP MMP v1.0 (Glasser et al., 2016a) as described above (a part of the parcellated data in an exemplar subject [ID = 9503] was already shown in Fig. 5, Fig. 6). To investigate similarity of the data, each of the parcellated metrics was fed into an analysis of Spearman’s rank correlation across subjects and sites/scanners. Fig. 7 shows the resultant similarity matrices which demonstrate higher correlation coefficients of within-subjects & between scanners than those of cross-subjects & between scanners in all the meftrics of cortical thickness, myelin, and functional connectivity.
Fig. 7.
Similarity of the cortical metrics across subjects and sites/scanners in preliminary travelling subject study. From left to right shows the correlation matrices of the parcellated cortical thickness, myelin (non BC) and functional connectivity in four travelling subjects (TS), scanned by five scanners/sites. In the upper row (A), color ranges are scaled by the distribution of the correlation coefficients (2% to 98% of histogram) to highlight the contrast between ‘within-subject’ similarities and ‘across-subject’ similarities, while in the lower row (B) color ranges are scaled by the same absolute values across all three modalities. There were 360 parcellated values for thickness and myelin and 129,240 parcellated values for functional connectivity, which cover the cerebral cortex in both hemispheres. Spearman’s correlation coefficient (rho) is shown using a color bar placed at the bottom. Non BC: non biasfield corrected.
We also performed preliminary analysis for the validity of the quantitative measure such as cortical thickness. The variability (standard deviation) of global cortical thickness across subjects (n = 4) was < 0.14 mm (=5.0% of mean cortical thickness across the cerebral cortex and across subjects) whereas variability across scanners (n = 5) was < 0.058 mm (=2.1%). Indeed, statistical test using analysis of variance (ANOVA) revealed a significant effect of subjects (F3,12 = 17.5, p = 0.0001) but not of scanners (F4,12 = 2.8, p = 0.074). These values are consistent with the test–retest reproducibility of cortical thickness in other studies (Kharabian Masouleh et al., 2020). However, it will be important to reassess this issue critically once data collection is completed.
We also analyzed a different set of TS (N = 26), who received test–retest scanning with the HARP protocol in the same MRI sites/scanners. The results (Fig. 8) showed greater similarity of cortical thickness, myelin map, and functional connectivity between test–retest data within subjects as compared with those with different subjects and/or scanners. The correlation coefficients of within-subject & within-scanner were again moderately high and comparable with those of within-subject & between-scanners in Fig. 7.
Fig. 8.
Test-retest and cross-subject similarity of cortical thickness, myelin (non BC) and resting-state functional connectivity (FC). The matrices show the similarity for each brain metric (top cortical thickness; middle, myelin (no bias corrected, BC); bottom, functional connectivity) using a total N = 26 traveling subjects obtained at five sites/scanners. The black line separates different subjects’ data, while white line test–retest (within-subject) data. The left column in (A) shows matrices with color ranges scaled by the distribution of the correlation coefficients (2% to 98% of histogram) to highlight contrast of ‘test-retest’ similarities as compared with those of ‘between-scanner or between-subject’. The right column (B) shows those with color-range scaled by the same absolute values across three modalities. Note that 2x2 correlation matrix that is within a black square and is adjacent to the diagonal indicates similarity of a single subject’s test–retest data and is excellent in structure (thickness and myelin) and fairly good in FC. The different sites are colored along the left and top edges.
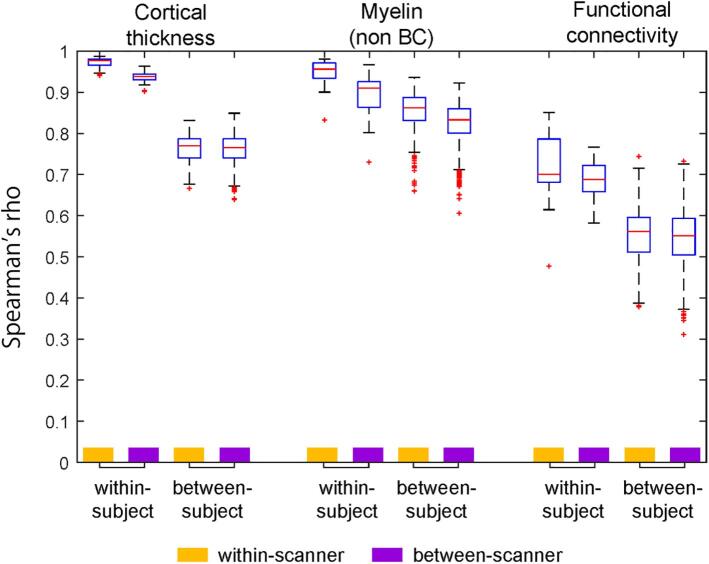
Fig. 9 summarizes the similarity values of all the TS30 data in Fig. 7, Fig. 8, classified into four types: within-subject & within-scanner, within-subject & between-scanner, between-subject & within-scanner, and between-subject & between-scanner. It is notable that the within-subject similarities are apparently higher than those of between-subject, indicating high sensitivity to subject-wise connectome. The between-subject similarities are smaller than within-subject and almost same across scanners, suggesting minimal bias between scanners and protocols. The within-subject & between-scanner similarity of the myelin map (0.89 ± 0.05) was slightly degraded as compared with within-subject & within-scanner (0.95 ± 0.03), suggesting the residual bias from transmit field across scans, for which we need to develop the correction method in future. That said, these preliminary datasets indicate that the HARP protocols and cortical parcellated analysis provide a similar and specific pattern of subject-wise connectome, which may effectively enhance statistical harmonization (see Section 5.4) once the data was fully collected in this project.
Fig. 9.
The summarized plots of similarity of cortical metrics in TS30. The plot summarizes the similarity (Spearman’s rho) of the cortical metrics. The similarity measures were from those in Fig. 7, Fig. 8, and classified into four types: within-subject & within-scanner (Subject N = 26, combination n = 16); within-subject & between-scanner (Subject N = 4, combination n = 40); between-subject & within-scanner (Subject N = 30, combination n = 250); and between-subject & between-scanner (Subject N = 30, combination n = 1200), where the combination n denotes a total number of similarity values used for statistics in the matrices in Fig. 7, Fig. 8. Non BC: non biasfield corrected.
5.3. Quality control
QC is implemented in several stages: 1) a brief image check during each scan; 2) an anomaly and abnormality inspection by the radiologists; 3) an assessment of raw data image quality when uploading data to the XNAT server; and 4) preprocessed image quality checks. QC 1 is conducted by site personnel and the participants are rescanned within the same session if scan time remains, if the images have major artifacts, such as those due to head movement. QC 2 is conducted by radiologists at the measurement site or other sites if any radiologist at the site is unable to check the images. QC 3 is manually conducted by researchers at the measurement sites before uploading the data to a server for all images in reference to the HCP QC manual (Marcus et al., 2013). After uploading the images to the XNAT servers, all images are first checked according to the DICOM file information as to whether the images are correctly updated. The researchers at each site are informed of missing DICOM files and any irregular parameters detected in the DICOM files. In QC 3, the T1w and T2w images are manually checked as to whether the face images are completely removed. Then, signal distributions of the myelin map are checked for outliers because of its sensitivity to several artifacts and errors such as motion, reconstruction of the images, and cortical surface reconstruction. Functional and diffusion images are automatically checked for outliers, and the images and data will be checked by visual inspection. In the QC 3 process, a QC pipeline will be implemented for checking the images (Marcus et al., 2013). QC 4 uses preprocessed CIFTI images that will be checked in several preprocessing steps. Any irregular scans and remarks are recorded in the clinical data servers and the information will be used when determining the eligibility criteria for each study.
5.4. Statistical harmonization using travelling subject data
Based on the previous study in the SRPBS consortium (Yamashita et al., 2019), we conduct a TS project in which subjects travels to multiple sites/scanners and receive MRI scans with multiple protocols (HARP, CRHD and SRPB). As described earlier, we expect that using the harmonized protocols may result in less contamination of measurement bias, and higher sensitivity to the neurobiological and/or disease-related change in clinical studies, because of high quality data acquisition and preprocessing across scanners. Nevertheless, statistical harmonization would be still valid to reduce any potential measurement bias and more importantly allow us to integrate our current project dataset with other datasets collected by different protocols. The statistical harmonization will be performed by using a model of the brain metrics as follows (Yamashita et al., 2019):
| (1) |
where the Subject variable is modeled by the subject-wise disease label and other neurobiological variables (age, sex, etc.); Sampling bias by the group-wise biological differences in participants between sites; and Measurement bias by the factor for scanner and protocol. The harmonized brain metrics can be estimated as follows:
| (2) |
However, it is difficult to separately estimate measurement and sampling biases using the multisite dataset alone (i.e. without TS) because these two types of bias covary across sites and not separable as long as different samples (participants) are scanned by different measurement variables (e.g. scanners and imaging protocols). In contrast, since the participants’ groups are considered to be fixed in the multisite TS design, the dataset in use may minimize the bias from sampling while holding the measurement bias constant across sites and protocols. A model of the brain metrics for TS datasets is as follows:
| (3) |
To solve this regression model, the previous SRPBS TS harmonization method applied a GLM and achieved equal estimation of measurement bias by requesting all the participants to be scanned at all the sites/scanners (Yamashita et al., 2019). In contrast, the present TS project was designed so that the participants travel to only some of the test sites/scanners in a hub-spoke design (Fig. 1) and statistical harmonization employs a general linear mixed model (GLMM) by treating the subject as a random effect. Use of GLMM may allow adaptation for grouped and repeated datasets like in the TS design and proper estimation by balancing inflation of type I error. It also makes it manageable to add the new TS or new scanners in the hub-spoke design in future. Our statistical harmonization will be more generalized by modeling measurement bias by sum of a scanner-type bias (e.g. Siemens Prisma, Skyra, Verio, Verio Dot, Trio) and a protocol bias (e.g. HARP, SRPB, CRHD) as follows:
| (4) |
For validations of our proposed harmonization method, we will compare our method with other established harmonization methods (e.g. ComBat), which employs an empirical Bayesian criterion. The validation will be done using several methods: clustering of the brain metrics for testing interpretability of variations in data acquisition/preprocessing (see Fig. 4 in Yamashita et al., 2019) as well as dimensionality reduction to confirm the effect of removing measurement bias from site differences and improvement of signal-to-noise ratio in harmonized brain metrics (see Fig. 5, Fig. 7 in Yamashita et al., 2019) and the number of important FCs discovered by the machine learning classifier that distinguish patients with psychiatric disorders from healthy controls based on brain metrics (Yamashita et al., 2020).
For harmonizing our data with those obtained with HCP/ABCD, although no subjects travel to the imaging site for HCP/ABCD, a similar statistical design can be applied to at least structural and functional MRI as the MRI scanning protocols of CRHD and HARP are either the same or similar to the HCP and ABCD, respectively, thus can be treated as the same in the term of the protocol bias in Eq. 4.
6. Ethics and data sharing
6.1. Ethical regulation
Sharing neuropsychiatric patient data, which may contain information linked to subjects’ privacy, requires special attention (Sadato et al., 2019). Therefore, the Brain/MINDS Beyond project put NCNP as the core site for supporting ethical considerations. Before participating in the project, all institutions are required to receive approval from their ethical review board regarding their research plans. This includes the following points and ethical documentation: 1) MR images and clinical data of the participants may be shared within the Brain/MINDS Beyond project or Japanese/International scientific institutions for collaboration. De-identified MR images with limited clinical data (see below) may become publicly accessible on an open database for research purposes. 2) MR images of the participants may be compared with non-human primate MRI data. 3) Intellectual property rights originating from the research of the Brain/MINDS Beyond project shall be attributed to the institutes of the researchers and not the participants. All participants must provide written informed consent to participate in this project after receiving a complete explanation of the experiment.
The Japanese regulations for the sharing of personal information used for research purposes requires attention in dealing with two types of data: “individual identification codes” and “special care-required personal information” (http://www.japaneselawtranslation.go.jp/law/detail/?id=2781&vm=04&re=01). Individual identification codes are direct identifiers—information sufficient to identify a specific individual. Special care-required personal information represents indirect identifiers needing special care in handling so as not to cause potential disadvantages to participants. In consideration of these regulations, data accompanied with the MR images are limited in the publicly accessible open database, and only include 5-year age bins, sex, diagnostic information, handedness, simple socioeconomic status, clinical scale scores, and sleepiness scale scores. In the Brain/MINDS Beyond project, we exclude the datasets of MR images containing facial information from the data in the publicly accessible open database.
6.2. Data sharing
In the current provisional plan of sharing the collected data, we have designated three types of data sharing:
-
1)
Access via an open database: de-identified MR images and limited clinical data are to become publicly accessible for research purposes after the research period ends. The initial release will be scheduled in 2024. Basic demographic and clinical characteristics such as 5-year age bin, sex, socioeconomic status, (premorbid) estimated intellectual quotient, main diagnosis, representative scale scores for each disease and sleepiness during rsfMRI scan will be shared.
-
2)
Application-based sharing: MR images and the clinical datasets are shared after receiving application approval for data usage by the Brain/MINDS Beyond human brain MRI study working group. Applicants are required to obtain approval of their research plan from the ethical review board of their institution and request the dataset type in the application form. The working group discusses the eligibility of the applicants, as well as the availability of the requested dataset, the ethical consideration in the Brain/MINDS Beyond site(s), and any conflict from other applications. Data is released from the distribution server of the Brain/MINDS Beyond project with limited access.
-
3)
Collaboration-based sharing: This form of sharing is used for individual collaborative studies. A research proposal collaborating with the institute(s) in the Brain/MINDS Beyond project is approved by the ethical review board of the institute(s). Data is shared from the relevant institute(s).
As of January 2021, the initial TS data was already transferred to the hub sites in a data logistics described in Section 4.3 and analyzed for preprocessing (Section 5) under ethical considerations (Section 6.1). The distribution to the researchers in the Brain/MINDS Beyond project has just started in February 2021.
7. Discussion
The Brain/MINDS Beyond human brain MRI (BMB-HBM) study expands upon research from previous multi-site neuroimaging studies in Japan and provides high quality brain images by standardizing multiple MRI scanners and protocols. An unbiased and quantitative assessment of cortical structure and function may be needed for sensitive and specific predictions of any dynamics, perturbations, or disorders of the brain system. Multi-modal cross-disease image datasets are systematically acquired, analyzed, and shared to enable investigation of common and disease-specific features for psychiatric and neurological disorders with a high sensitivity and specificity. A distinct feature of the BMB-HBM project is to include a study design including travelling subjects (TS) across sites/scanners/protocols and enables to harmonize the heterogenous data from lower (i.e. data acquisition, preprocessing) to higher levels (i.e. statistics). The latest information and resources of the BMB-HBM study is available at http://hbm.brainminds-beyond.jp
7.1. Prospective harmonization - scanning protocol and preprocessing
To date, several national projects have applied high-quality multimodal MRI protocols, in addition to a preprocessing pipeline, to a large cohort (e.g., HCP, UK biobank, and ABCD). Unlike these multi-site projects, we plan to investigate brain organization associated with brain disorders that occur throughout the lifespan and to develop imaging biomarkers that can be implemented in clinical trials. To facilitate the collection of a larger number of patients with different brain disorders, multiple clinical research sites are participating in this project and cooperating for standardized data acquisitions. The core of the project began from establishing a standardized protocol (i.e. HARP) based on five 3 T MRI scanners, but it will continue to develop a comparable protocol for other types of scanners/vendors. The protocol is designed not only for high-resolution structural MRI and high-quality resting-state fMRI, but also for diffusion MRI and other imaging—including scans for correcting distortions. The preprocessing is performed with a surface-based multi-modal analysis to minimize bias largely generated from the variability in cortical folding across subjects (Coalson et al., 2018, Glasser et al., 2016b). The preliminary data demonstrated high quality MRI images and the fidelity of structural and functional brain organizations across scanners/sites. The signal-to-noise ratio of MRI images was very high across scanners/sites (Fig. 5A). The cortical metrics of structure (myelin map, thickness) (Fig. 5B–C) were comparable to those previously reported in the literature (Fischl and Dale, 2000, Glasser and Van Essen, 2011), as well as the functional connectivity related to eye movements involving FEF and PEF (Fig. 6A) (Amiez and Petrides, 2009) and a language network involving left 55b, 44/45, STS, and PSL (Fig. 6B) (Glasser et al., 2016a). The initial trial with 30 TS also demonstrated the similar and specific pattern of subject-wise connectome across five scanners (Fig. 7, Fig. 8, Fig. 9), suggesting the reliability of our prospective harmonization (e.g. protocols and preprocessing) and promising future statistical harmonization. These findings suggest that a surface-based parcellated analysis may provide useful and reliable metrics concerning cortical structure, function, and connectivity, and may potentially contribute to the establishment of multi-modal imaging biomarkers of brain disorders.
7.2. Statistical harmonization - traveling-subject statistical model
The TS approach is a relatively new harmonization method for multi-site brain image data (Yamashita et al., 2019), which has proven that measurement bias from MRI equipment and protocols can be differentiated from sampling bias between sites. Instead of using a previously applied GLM, we plan to expand the statistical approach to a GLMM in this project. One of the obstacles of the GLMM approach is that it requires a larger number of total scans compared to those in a GLM approach; overlapping scans at hub sites are required for all TS participants to ensure the data connectivity; additionally, a larger number of TS participants is required in the TS project because the degree of freedom can be reduced in the GLMM. However, one of the benefits of the GLMM approach includes that it is flexible with the variability in data acquisition—such as the number of scans per participant and length of scan time per protocol; thus, is suitable for a larger multi-site project. Furthermore, this approach allows the addition of another site, scanner, and protocol to an existing TS network, which can deal with the future upgrades of scanners and protocols. In fact, the scanners at two sites (UHI and SWA) were upgraded to a MAGNETOM Skyra fit for institutional reasons, thus, we customized the design to include ‘time traveling’ subjects in two sites to ensure that the data are properly collected before and after the scanner upgrade. In case a new site will participate in the project afterwards, additional scanning of the TS between the new and the hub site may help data statistically harmonized in this network.
7.3. Future directions of technical validation and improvements
Although the data acquisition protocol was successfully harmonized across five scanners from the same vendor, the quality of the preprocessing and harmonization may need to be validated and further improved once collection of TS data is completed. First, the quality control and assurance of the brain metrics and error detection in the preprocessing needs to be established, and the preprocessing techniques need to be optimized or elaborated using the TS data. For example, the accuracy of the cortical reconstruction relies on the automatic tissue segmentation using a training dataset in FreeSurfer (Fischl, 2012) that may be vulnerable to the scanner differences, particularly related to variation in B1 transmission. Thus, segmentation algorithms may benefit from retraining using the current travelling subjects’ data. White matter lesion are often seen in eldery subjects and cause errors of the white matter segmentation and surface, and thus need to be addressed for accurate cortical surface reconstruction by combining automatic lesion prediction (Griffanti et al., 2016). The automatic tissue segmentation of subcortical structures may also be improved by using multi-modal data (Visser et al., 2016). Residual intensity biases in myelin maps present in the current study are largely due to differences of B1 transmit field across scans and scanners, which will be corrected in future preprocessing pipelines. Finally, validation of the scanning and prepressing protocols may require rigorous testing for the bias and reproducibility of the brain metrics, such as using an intraclass correlation analysis when completing the TS data collection.
Second, the accuracy and predictability of the functional MRI-derived metrics, FC, may be improved. In particular, denoising of fMRI data is critically important, since > 90% of variance in the resting-state fMRI signals is dominated by structured artifacts, motion and random noise (Marcus et al., 2013, Glasser et al., 2018). The current denoising classifier process may also have the bias in specificity and sensitivity across protocols, thus, automatic classifiers of ICA components may be improved by retraining using TS data across scanners for each protocol. The validation of the restrained automatic classification will be assessed using leave-one-out or cross validation. The automatic classification may be improved by combining and validating with the data of clinical patients. In addition, while the calculation of FC is commonly based on a Pearson’s correlation between the signals of remote brain areas, the accuracy or predictability of the FC may be potentially improved by using a Tiknov partial correlation or tangent space parametrization (Pervaiz et al., 2020). The optimization of the FC calculation may also benefit from a validation study with neural tracers in non-human primate studies, as is promoted in Non-Human Primate Neuroimaging & Neuroanatomy Project (NHP_NNP) (Hayashi et al., 2020). In addition, task fMRI scans using EMOTION and CARIT in HARP can be used to evaluate the validity, reliability and applicability of harmonization. The result of task fMRI may also be used for validation of the resting-state fMRI, although the TS project with task fMRI is now under planning and will be completed in coming years.
Finally, as for diffusion MRI, the specialized nature of dMRI signals and associated analysis methods has led to dMRI-specific harmonization methods, as recently proposed for harmonizing the multi-site & multi-shell data based on model-free approaches (see Pinto et al., 2020 for review). Ning et al. (2020) compared the effects of several dMRI harmonization methods using the multi-shell diffusion MRI data in the same subjects in multiple scanners. The algorithms used three approaches: rotational invariant spherical harmonics, deep neural networks and hybrid biophysical and statistical algorithms, and diffusion parametric maps, such as fractional anisotropy (FA), mean diffusibility (MD), and mean kurtosis (MK) were compared before and after the data harmonization. The results demonstrated that data harmonization reduced the variability of diffusion metrics across protocols. Hence, future studies should thoroughly evaluate neurobiological metrics like NODDI and tractography using the current TS data. As for QSM, reconstructed QSM may suffer from the incompleteness of the data acquisition and the bias depending on the head orientation, which could be reduced by applying a deep learning algorithm to solve the inverse problem of the magnetic dipole (Bollmann et al., 2019, Jung et al., 2020).
7.4. Potential applications of Brain/MINDS human brain MRI project
Because this project focuses on various brain disorders across the lifespan, we aim to identify common and disease-specific features of psychiatric and neurological disorders. While some case-control studies suggest possible neural mechanisms in a psychiatric disease, other studies suggest that the effects may not be specific to a single entity but instead may be shared across multiple neuropsychiatric disorders (Hibar et al., 2018, Schmaal et al., 2017, Schmaal et al., 2016, van Erp et al., 2016). Such non-specificity may be at least partly addressed by investigating diseases across the lifespan, since some of brain changes reported in psychiatric disorders also occur in aging or development in healthy subjects, e.g. volumetric changes in subcortical structures in schizophrenia (Okada et al., 2016, van Erp et al., 2016) and in healthy aging (O'Shea et al., 2016, Wang et al., 2019). We initially coordinated with 13 sites to explore various psychiatric and neurological disorders throughout the lifespan and to make use of a powerful harmonization method. Therefore, this project is expected to identify both the common and disease-specific pathophysiology features of psychiatric and neurological disorders, leading to identifying candidate imaging biomarkers for future clinical trials.
8. Conclusion
The Brain/MINDS Beyond human brain MRI project began with the participation of 13 clinical research sites—all of which have setup brain image scans using the standard MRI scanners and protocols, conducted TS scans, and will share acquired data with the project and the public in the future, and commit to the analysis and publication of the data. To the best of our knowledge, this is the first human brain MRI project to explore psychiatric and neurological disorders across the lifespan. The project aims to discover robust findings which may be directly related to the common or disease-specific pathophysiology features of such diseases and facilitate the development of candidate biomarkers for clinical application and drug discovery.
CRediT authorship contribution statement
Shinsuke Koike: Conceptualization, Project administration, Funding acquisition, Writing - original draft, Software, Resources. Saori C. Tanaka: Conceptualization, Project administration, Data curation, Software, Resources, Writing - original draft. Tomohisa Okada: Methodology, Investigation. Toshihiko Aso: Investigation, Formal analysis, Validation. Ayumu Yamashita: Conceptualization, Software, Writing - original draft. Okito Yamashita: Conceptualization, Software, Writing - original draft. Michiko Asano: Investigation, Data curation. Norihide Maikusa: Methodology, Resources. Kentaro Morita: Writing - original draft. Naohiro Okada: Investigation, Writing - original draft. Masaki Fukunaga: Methodology. Akiko Uematsu: Methodology, Investigation. Hiroki Togo: Methodology, Investigation. Atsushi Miyazaki: Methodology, Investigation. Katsutoshi Murata: Methodology. Yuta Urushibata: Methodology. Joonas Autio: Methodology. Takayuki Ose: Methodology. Junichiro Yoshimoto: Methodology. Toshiyuki Araki: Writing - original draft. Matthew F. Glasser: Software, Writing - review & editing. David C. Van Essen: Writing - review & editing. Megumi Maruyama: Project administration. Norihiro Sadato: Investigation, Funding acquisition, Project administration, Writing - review & editing. Mitsuo Kawato: Conceptualization, Funding acquisition, Project administration, Writing - review & editing, Supervision. Kiyoto Kasai: Investigation, Funding acquisition, Project administration, Supervision. Yasumasa Okamoto: Investigation, Funding acquisition. Takashi Hanakawa: Project administration, Funding acquisition, Methodology, Investigation, Resources, Writing - original draft. Takuya Hayashi: Conceptualization, Project administration, Funding acquisition, Software, Resources, Formal analysis, Writing - original draft, Writing - review & editing.
Acknowledgments
The data presented in Figure 5-6 are available at BALSA (https://balsa.wustl.edu/study/show/npD26). Harmonization protocols, preprocessing, data usage and other information of the project are available at the BrainMINDS-beyond human brain MRI site (http://hbm.brainminds-beyond.jp). For inquiry of the data usage, please contact to bmbhbm-info@nips.ac.jp. This research was supported by the Agency for Medical Research and Development (AMED) under Grant Numbers JP20dm0307001, JP20dm0307002, JP20dm0307003, JP20dm0307004, JP20dm0307008, JP20dm0307009, JP20dm‘0307020, JP20dm0207069 and JP20dm0307006. This study was also supported by the University of Tokyo Center for Integrative Science of Human Behaviour (CiSHuB) and the International Research Center for Neurointelligence (WPI-IRCN) at the University of Tokyo Institutes for Advanced Study (UTIAS).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2021.102600.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Amiez C., Petrides M. Anatomical organization of the eye fields in the human and non-human primate frontal cortex. Prog. Neurobiol. 2009;89:220–230. doi: 10.1016/j.pneurobio.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Andersson J.L., Skare S., Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20:870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- Andersson J.L.R., Graham M.S., Drobnjak I., Zhang H., Campbell J. Susceptibility-induced distortion that varies due to motion: correction in diffusion MR without acquiring additional data. Neuroimage. 2018;171:277–295. doi: 10.1016/j.neuroimage.2017.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J.L.R., Graham M.S., Drobnjak I., Zhang H., Filippini N., Bastiani M. Towards a comprehensive framework for movement and distortion correction of diffusion MR images: within volume movement. Neuroimage. 2017;152:450–466. doi: 10.1016/j.neuroimage.2017.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J.L.R., Sotiropoulos S.N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078. doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, S., Nishida, A., Yamasaki, S., Koike, S., Morimoto, Y., Hoshino, A., Kanata, S., Fujikawa, S., Endo, K., Usami, S., Furukawa, T.A., Hiraiwa-Hasegawa, M., Kasai, K., Scientific, T.T.C., Data Collection, T., 2019. Cohort Profile: The Tokyo Teen Cohort study (TTC). Int. J. Epidemiol. 48, 1414–1414g. [DOI] [PMC free article] [PubMed]
- Beheshti I., Maikusa N., Daneshmand M., Matsuda H., Demirel H., Anbarjafari G., Japanese-Alzheimer's Disease Neuroimaging I. Classification of Alzheimer's disease and prediction of mild cognitive impairment conversion using histogram-based analysis of patient-specific anatomical brain connectivity networks. J. Alzheimers Dis. 2017;60:295–304. doi: 10.3233/JAD-161080. [DOI] [PubMed] [Google Scholar]
- Behrens T.E., Woolrich M.W., Jenkinson M., Johansen-Berg H., Nunes R.G., Clare S., Matthews P.M., Brady J.M., Smith S.M. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn. Reson. Med. 2003;50:1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- Bollmann S., Rasmussen K.G.B., Kristensen M., Blendal R.G., Østergaard L.R., Plocharski M., O’Brien K., Langkammer C., Janke A., Barth M. DeepQSM - using deep learning to solve the dipole inversion for quantitative susceptibility mapping. Neuroimage. 2019;195:373–383. doi: 10.1016/j.neuroimage.2019.03.060. [DOI] [PubMed] [Google Scholar]
- Bookheimer S.Y., Salat D.H., Terpstra M., Ances B.M., Barch D.M., Buckner R.L., Burgess G.C., Curtiss S.W., Diaz-Santos M., Elam J.S., Fischl B., Greve D.N., Hagy H.A., Harms M.P., Hatch O.M., Hedden T., Hodge C., Japardi K.C., Kuhn T.P., Ly T.K., Smith S.M., Somerville L.H., Ugurbil K., van der Kouwe A., Van Essen D., Woods R.P., Yacoub E. The lifespan human connectome project in aging: an overview. Neuroimage. 2019;185:335–348. doi: 10.1016/j.neuroimage.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey B.J., Cannonier T., Conley M.I., Cohen A.O., Barch D.M., Heitzeg M.M., Soules M.E., Teslovich T., Dellarco D.V., Garavan H., Orr C.A., Wager T.D., Banich M.T., Speer N.K., Sutherland M.T., Riedel M.C., Dick A.S., Bjork J.M., Thomas K.M., Chaarani B., Mejia M.H., Hagler D.J., Jr., Daniela Cornejo M., Sicat C.S., Harms M.P., Dosenbach N.U.F., Rosenberg M., Earl E., Bartsch H., Watts R., Polimeni J.R., Kuperman J.M., Fair D.A., Dale A.M., Workgroup A.I.A. The adolescent brain cognitive development (ABCD) study: imaging acquisition across 21 sites. Dev Cogn Neurosci. 2018;32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coalson T.S., Van Essen D.C., Glasser M.F. The impact of traditional neuroimaging methods on the spatial localization of cortical areas. Proc. Natl. Acad. Sci. USA. 2018;115:E6356–E6365. doi: 10.1073/pnas.1801582115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt L., Chiu W.T., Sampson N., Kessler R.C., Anthony J.C., Angermeyer M., Bruffaerts R., de Girolamo G., Gureje O., Huang Y., Karam A., Kostyuchenko S., Lepine J.P., Mora M.E., Neumark Y., Ormel J.H., Pinto-Meza A., Posada-Villa J., Stein D.J., Takeshima T., Wells J.E. Toward a global view of alcohol, tobacco, cannabis, and cocaine use: findings from the WHO World Mental Health Surveys. PLoS Med. 2008;5:e141. doi: 10.1371/journal.pmed.0050141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue C.J., Sotiropoulos S.N., Jbabdi S., Hernandez-Fernandez M., Behrens T.E., Dyrby T.B., Coalson T., Kennedy H., Knoblauch K., Van Essen D.C., Glasser M.F. Using diffusion tractography to predict cortical connection strength and distance: a quantitative comparison with tracers in the monkey. J. Neurosci. 2016;36:6758–6770. doi: 10.1523/JNEUROSCI.0493-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale A.T., Grosenick L., Downar J., Dunlop K., Mansouri F., Meng Y., Fetcho R.N., Zebley B., Oathes D.J., Etkin A., Schatzberg A.F., Sudheimer K., Keller J., Mayberg H.S., Gunning F.M., Alexopoulos G.S., Fox M.D., Pascual-Leone A., Voss H.U., Casey B.J., Dubin M.J., Liston C. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat. Med. 2017;23:28–38. doi: 10.1038/nm.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott L.T., Sharp K., Alfaro-Almagro F., Shi S., Miller K.L., Douaud G., Marchini J., Smith S.M. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562:210–216. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott M.L., Romer A., Knodt A.R., Hariri A.R. A Connectome-wide functional signature of transdiagnostic risk for mental illness. Biol. Psychiatry. 2018;84:452–459. doi: 10.1016/j.biopsych.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Dale A.M. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.P., Cullen N., Sheline Y.I., Taylor W.D., Aselcioglu I., Cook P.A., Adams P., Cooper C., Fava M., McGrath P.J., McInnis M., Phillips M.L., Trivedi M.H., Weissman M.M., Shinohara R.T. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage. 2018;167:104–120. doi: 10.1016/j.neuroimage.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.P., Parker D., Tunc B., Watanabe T., Elliott M.A., Ruparel K., Roalf D.R., Satterthwaite T.D., Gur R.C., Gur R.E., Schultz R.T., Verma R., Shinohara R.T. Harmonization of multi-site diffusion tensor imaging data. Neuroimage. 2017;161:149–170. doi: 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutomi H., Glasser M.F., Zhang H., Autio J.A., Coalson T.S., Okada T., Togashi K., Van Essen D.C., Hayashi T. Neurite imaging reveals microstructural variations in human cerebral cortical gray matter. Neuroimage. 2018;182:488–499. doi: 10.1016/j.neuroimage.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser M.F., Coalson T.S., Bijsterbosch J.D., Harrison S.J., Harms M.P., Anticevic A., Van Essen D.C., Smith S.M. Using temporal ICA to selectively remove global noise while preserving global signal in functional MRI data. Neuroimage. 2018;181:692–717. doi: 10.1016/j.neuroimage.2018.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser M.F., Coalson T.S., Robinson E.C., Hacker C.D., Harwell J., Yacoub E., Ugurbil K., Andersson J., Beckmann C.F., Jenkinson M., Smith S.M., Van Essen D.C. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser M.F., Smith S.M., Marcus D.S., Andersson J.L., Auerbach E.J., Behrens T.E., Coalson T.S., Harms M.P., Jenkinson M., Moeller S., Robinson E.C., Sotiropoulos S.N., Xu J., Yacoub E., Ugurbil K., Van Essen D.C. The Human Connectome Project's neuroimaging approach. Nat. Neurosci. 2016;19:1175–1187. doi: 10.1038/nn.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser M.F., Sotiropoulos S.N., Wilson J.A., Coalson T.S., Fischl B., Andersson J.L., Xu J., Jbabdi S., Webster M., Polimeni J.R., Van Essen D.C., Jenkinson M., Consortium W.U.-M.H. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser M.F., Van Essen D.C. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J. Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffanti L., Zamboni G., Khan A., Li L., Bonifacio G., Sundaresan V., Schulz U.G., Kuker W., Battaglini M., Rothwell P.M., Jenkinson M. BIANCA (Brain Intensity AbNormality Classification Algorithm): A new tool for automated segmentation of white matter hyperintensities. Neuroimage. 2016;141:191–205. doi: 10.1016/j.neuroimage.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M.P., Somerville L.H., Ances B.M., Andersson J., Barch D.M., Bastiani M., Bookheimer S.Y., Brown T.B., Buckner R.L., Burgess G.C., Coalson T.S., Chappell M.A., Dapretto M., Douaud G., Fischl B., Glasser M.F., Greve D.N., Hodge C., Jamison K.W., Jbabdi S., Kandala S., Li X., Mair R.W., Mangia S., Marcus D., Mascali D., Moeller S., Nichols T.E., Robinson E.C., Salat D.H., Smith S.M., Sotiropoulos S.N., Terpstra M., Thomas K.M., Tisdall M.D., Ugurbil K., van der Kouwe A., Woods R.P., Zollei L., Van Essen D.C., Yacoub E. Extending the human connectome project across ages: imaging protocols for the lifespan development and aging projects. Neuroimage. 2018;183:972–984. doi: 10.1016/j.neuroimage.2018.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, T., Hou, Y., Glasser, M.F., Autio, J.A., Knoblauch, K., Inoue-Murayama, M., Coalson, T., Yacoub, E., Smith, S., Kennedy, H., Van Essen, D.C., 2020. The NonHuman Primate Neuroimaging & Neuroanatomy Project. arXiv:2010.00308 [q-bio]. [DOI] [PMC free article] [PubMed]
- Hibar D.P., Westlye L.T., Doan N.T., Jahanshad N., Cheung J.W., Ching C.R.K., Versace A., Bilderbeck A.C., Uhlmann A., Mwangi B., Kramer B., Overs B., Hartberg C.B., Abe C., Dima D., Grotegerd D., Sprooten E., Boen E., Jimenez E., Howells F.M., Delvecchio G., Temmingh H., Starke J., Almeida J.R.C., Goikolea J.M., Houenou J., Beard L.M., Rauer L., Abramovic L., Bonnin M., Ponteduro M.F., Keil M., Rive M.M., Yao N., Yalin N., Najt P., Rosa P.G., Redlich R., Trost S., Hagenaars S., Fears S.C., Alonso-Lana S., van Erp T.G.M., Nickson T., Chaim-Avancini T.M., Meier T.B., Elvsashagen T., Haukvik U.K., Lee W.H., Schene A.H., Lloyd A.J., Young A.H., Nugent A., Dale A.M., Pfennig A., McIntosh A.M., Lafer B., Baune B.T., Ekman C.J., Zarate C.A., Bearden C.E., Henry C., Simhandl C., McDonald C., Bourne C., Stein D.J., Wolf D.H., Cannon D.M., Glahn D.C., Veltman D.J., Pomarol-Clotet E., Vieta E., Canales-Rodriguez E.J., Nery F.G., Duran F.L.S., Busatto G.F., Roberts G., Pearlson G.D., Goodwin G.M., Kugel H., Whalley H.C., Ruhe H.G., Soares J.C., Fullerton J.M., Rybakowski J.K., Savitz J., Chaim K.T., Fatjo-Vilas M., Soeiro-de-Souza M.G., Boks M.P., Zanetti M.V., Otaduy M.C.G., Schaufelberger M.S., Alda M., Ingvar M., Phillips M.L., Kempton M.J., Bauer M., Landen M., Lawrence N.S., van Haren N.E.M., Horn N.R., Freimer N.B., Gruber O., Schofield P.R., Mitchell P.B., Kahn R.S., Lenroot R., Machado-Vieira R., Ophoff R.A., Sarro S., Frangou S., Satterthwaite T.D., Hajek T., Dannlowski U., Malt U.F., Arolt V., Gattaz W.F., Drevets W.C., Caseras X., Agartz I., Thompson P.M., Andreassen O.A. Cortical abnormalities in bipolar disorder: an MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Mol. Psychiatry. 2018;23:932–942. doi: 10.1038/mp.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsubo T., Iwata A., Suzuki K., Ihara R., Arai H., Ishii K., Senda M., Ito K., Ikeuchi T., Kuwano R., Matsuda H., Japanese Alzheimer's Disease Neuroimaging I., Sun C.K., Beckett L.A., Petersen R.C., Weiner M.W., Aisen P.S., Donohue M.C., Alzheimer's Disease Neuroimaging I. Japanese and North American Alzheimer's Disease Neuroimaging Initiative studies: harmonization for international trials. Alzheimers Dement. 2018;14:1077–1087. doi: 10.1016/j.jalz.2018.03.009. [DOI] [PubMed] [Google Scholar]
- Jung W., Yoon J., Ji S., Choi J.Y., Kim J.M., Nam Y., Kim E.Y., Lee J. Exploring linearity of deep neural network trained QSM: QSMnet+ NeuroImage. 2020;211:116619. doi: 10.1016/j.neuroimage.2020.116619. [DOI] [PubMed] [Google Scholar]
- Kharabian Masouleh S., Eickhoff S.B., Zeighami Y., Lewis L.B., Dahnke R., Gaser C., Chouinard-Decorte F., Lepage C., Scholtens L.H., Hoffstaedter F., Glahn D.C., Blangero J., Evans A.C., Genon S., Valk S.L. Influence of processing pipeline on cortical thickness measurement. Cereb. Cortex. 2020;30:5014–5027. doi: 10.1093/cercor/bhaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S., Takano Y., Iwashiro N., Satomura Y., Suga M., Nagai T., Natsubori T., Tada M., Nishimura Y., Yamasaki S., Takizawa R., Yahata N., Araki T., Yamasue H., Kasai K. A multimodal approach to investigate biomarkers for psychosis in a clinical setting: the Integrative Neuroimaging studies in Schizophrenia Targeting for Early intervention and Prevention (IN-STEP) project. Schizophr. Res. 2013;143:116–124. doi: 10.1016/j.schres.2012.11.012. [DOI] [PubMed] [Google Scholar]
- Koutsouleris N., Meisenzahl E.M., Borgwardt S., Riecher-Rossler A., Frodl T., Kambeitz J., Kohler Y., Falkai P., Moller H.J., Reiser M., Davatzikos C. Individualized differential diagnosis of schizophrenia and mood disorders using neuroanatomical biomarkers. Brain. 2015;138:2059–2073. doi: 10.1093/brain/awv111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.Y., Kwon J.S. Psychosis research in Asia: advantage from low prevalence of cannabis use. NPJ Schizophr. 2016;2:1. doi: 10.1038/s41537-016-0002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski K.E., Bouix S., Ongur D., Shenton M.E. Neuroprogression across the Early Course of Psychosis. J. Psychiatry Brain Sci. 2020;5 doi: 10.20900/jpbs.20200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus D.S., Harms M.P., Snyder A.Z., Jenkinson M., Wilson J.A., Glasser M.F., Barch D.M., Archie K.A., Burgess G.C., Ramaratnam M., Hodge M., Horton W., Herrick R., Olsen T., McKay M., House M., Hileman M., Reid E., Harwell J., Coalson T., Schindler J., Elam J.S., Curtiss S.W., Van Essen D.C., Consortium W.U.-M.H. Human Connectome Project informatics: quality control, database services, and data visualization. Neuroimage. 2013;80:202–219. doi: 10.1016/j.neuroimage.2013.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.L., Alfaro-Almagro F., Bangerter N.K., Thomas D.L., Yacoub E., Xu J., Bartsch A.J., Jbabdi S., Sotiropoulos S.N., Andersson J.L., Griffanti L., Douaud G., Okell T.W., Weale P., Dragonu I., Garratt S., Hudson S., Collins R., Jenkinson M., Matthews P.M., Smith S.M. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 2016;19:1523–1536. doi: 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S., Yacoub E., Olman C.A., Auerbach E., Strupp J., Harel N., Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson. Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.G., Weiner M.W., Thal L.J., Petersen R.C., Jack C., Jagust W., Trojanowski J.Q., Toga A.W., Beckett L. The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin. N. Am. 2005;15(869–877):xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai Y., Murata M. Japan Parkinson's Progression Markers Initiative (J-PPMI) Nihon Rinsho. 2017;75:151–155. [PubMed] [Google Scholar]
- Murray C.J., Vos T., Lozano R., Naghavi M., Flaxman A.D., Michaud C., Ezzati M., Shibuya K., Salomon J.A., Abdalla S., Aboyans V., Abraham J., Ackerman I., Aggarwal R., Ahn S.Y., Ali M.K., Alvarado M., Anderson H.R., Anderson L.M., Andrews K.G., Atkinson C., Baddour L.M., Bahalim A.N., Barker-Collo S., Barrero L.H., Bartels D.H., Basanez M.G., Baxter A., Bell M.L., Benjamin E.J., Bennett D., Bernabe E., Bhalla K., Bhandari B., Bikbov B., Bin Abdulhak A., Birbeck G., Black J.A., Blencowe H., Blore J.D., Blyth F., Bolliger I., Bonaventure A., Boufous S., Bourne R., Boussinesq M., Braithwaite T., Brayne C., Bridgett L., Brooker S., Brooks P., Brugha T.S., Bryan-Hancock C., Bucello C., Buchbinder R., Buckle G., Budke C.M., Burch M., Burney P., Burstein R., Calabria B., Campbell B., Canter C.E., Carabin H., Carapetis J., Carmona L., Cella C., Charlson F., Chen H., Cheng A.T., Chou D., Chugh S.S., Coffeng L.E., Colan S.D., Colquhoun S., Colson K.E., Condon J., Connor M.D., Cooper L.T., Corriere M., Cortinovis M., de Vaccaro K.C., Couser W., Cowie B.C., Criqui M.H., Cross M., Dabhadkar K.C., Dahiya M., Dahodwala N., Damsere-Derry J., Danaei G., Davis A., De Leo D., Degenhardt L., Dellavalle R., Delossantos A., Denenberg J., Derrett S., Des Jarlais D.C., Dharmaratne S.D., Dherani M., Diaz-Torne C., Dolk H., Dorsey E.R., Driscoll T., Duber H., Ebel B., Edmond K., Elbaz A., Ali S.E., Erskine H., Erwin P.J., Espindola P., Ewoigbokhan S.E., Farzadfar F., Feigin V., Felson D.T., Ferrari A., Ferri C.P., Fevre E.M., Finucane M.M., Flaxman S., Flood L., Foreman K., Forouzanfar M.H., Fowkes F.G., Fransen M., Freeman M.K., Gabbe B.J., Gabriel S.E., Gakidou E., Ganatra H.A., Garcia B., Gaspari F., Gillum R.F., Gmel G., Gonzalez-Medina D., Gosselin R., Grainger R., Grant B., Groeger J., Guillemin F., Gunnell D., Gupta R., Haagsma J., Hagan H., Halasa Y.A., Hall W., Haring D., Haro J.M., Harrison J.E., Havmoeller R., Hay R.J., Higashi H., Hill C., Hoen B., Hoffman H., Hotez P.J., Hoy D., Huang J.J., Ibeanusi S.E., Jacobsen K.H., James S.L., Jarvis D., Jasrasaria R., Jayaraman S., Johns N., Jonas J.B., Karthikeyan G., Kassebaum N., Kawakami N., Keren A., Khoo J.P., King C.H., Knowlton L.M., Kobusingye O., Koranteng A., Krishnamurthi R., Laden F., Lalloo R., Laslett L.L., Lathlean T., Leasher J.L., Lee Y.Y., Leigh J., Levinson D., Lim S.S., Limb E., Lin J.K., Lipnick M., Lipshultz S.E., Liu W., Loane M., Ohno S.L., Lyons R., Mabweijano J., MacIntyre M.F., Malekzadeh R., Mallinger L., Manivannan S., Marcenes W., March L., Margolis D.J., Marks G.B., Marks R., Matsumori A., Matzopoulos R., Mayosi B.M., McAnulty J.H., McDermott M.M., McGill N., McGrath J., Medina-Mora M.E., Meltzer M., Mensah G.A., Merriman T.R., Meyer A.C., Miglioli V., Miller M., Miller T.R., Mitchell P.B., Mock C., Mocumbi A.O., Moffitt T.E., Mokdad A.A., Monasta L., Montico M., Moradi-Lakeh M., Moran A., Morawska L., Mori R., Murdoch M.E., Mwaniki M.K., Naidoo K., Nair M.N., Naldi L., Narayan K.M., Nelson P.K., Nelson R.G., Nevitt M.C., Newton C.R., Nolte S., Norman P., Norman R., O'Donnell M., O'Hanlon S., Olives C., Omer S.B., Ortblad K., Osborne R., Ozgediz D., Page A., Pahari B., Pandian J.D., Rivero A.P., Patten S.B., Pearce N., Padilla R.P., Perez-Ruiz F., Perico N., Pesudovs K., Phillips D., Phillips M.R., Pierce K., Pion S., Polanczyk G.V., Polinder S., Pope C.A., 3rd, Popova S., Porrini E., Pourmalek F., Prince M., Pullan R.L., Ramaiah K.D., Ranganathan D., Razavi H., Regan M., Rehm J.T., Rein D.B., Remuzzi G., Richardson K., Rivara F.P., Roberts T., Robinson C., De Leon F.R., Ronfani L., Room R., Rosenfeld L.C., Rushton L., Sacco R.L., Saha S., Sampson U., Sanchez-Riera L., Sanman E., Schwebel D.C., Scott J.G., Segui-Gomez M., Shahraz S., Shepard D.S., Shin H., Shivakoti R., Singh D., Singh G.M., Singh J.A., Singleton J., Sleet D.A., Sliwa K., Smith E., Smith J.L., Stapelberg N.J., Steer A., Steiner T., Stolk W.A., Stovner L.J., Sudfeld C., Syed S., Tamburlini G., Tavakkoli M., Taylor H.R., Taylor J.A., Taylor W.J., Thomas B., Thomson W.M., Thurston G.D., Tleyjeh I.M., Tonelli M., Towbin J.A., Truelsen T., Tsilimbaris M.K., Ubeda C., Undurraga E.A., van der Werf M.J., van Os J., Vavilala M.S., Venketasubramanian N., Wang M., Wang W., Watt K., Weatherall D.J., Weinstock M.A., Weintraub R., Weisskopf M.G., Weissman M.M., White R.A., Whiteford H., Wiebe N., Wiersma S.T., Wilkinson J.D., Williams H.C., Williams S.R., Witt E., Wolfe F., Woolf A.D., Wulf S., Yeh P.H., Zaidi A.K., Zheng Z.J., Zonies D., Lopez A.D., AlMazroa M.A., Memish Z.A. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- Ning L., Bonet-Carne E., Grussu F., Sepehrband F., Kaden E., Veraart J., Blumberg S.B., Khoo C.S., Palombo M., Kokkinos I., Alexander D.C., Coll-Font J., Scherrer B., Warfield S.K., Karayumak S.C., Rathi Y., Koppers S., Weninger L., Ebert J., Merhof D., Moyer D., Pietsch M., Christiaens D., Gomes Teixeira R.A., Tournier J.-D., Schilling K.G., Huo Y., Nath V., Hansen C., Blaber J., Landman B.A., Zhylka A., Pluim J.P.W., Parker G., Rudrapatna U., Evans J., Charron C., Jones D.K., Tax C.M.W. Cross-scanner and cross-protocol multi-shell diffusion MRI data harmonization: algorithms and results. NeuroImage. 2020;221:117128. doi: 10.1016/j.neuroimage.2020.117128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes A., Schnack H.G., Ching C.R.K., Agartz I., Akudjedu T.N., Alda M., Alnaes D., Alonso-Lana S., Bauer J., Baune B.T., Boen E., Bonnin C.D.M., Busatto G.F., Canales-Rodriguez E.J., Cannon D.M., Caseras X., Chaim-Avancini T.M., Dannlowski U., Diaz-Zuluaga A.M., Dietsche B., Doan N.T., Duchesnay E., Elvsashagen T., Emden D., Eyler L.T., Fatjo-Vilas M., Favre P., Foley S.F., Fullerton J.M., Glahn D.C., Goikolea J.M., Grotegerd D., Hahn T., Henry C., Hibar D.P., Houenou J., Howells F.M., Jahanshad N., Kaufmann T., Kenney J., Kircher T.T.J., Krug A., Lagerberg T.V., Lenroot R.K., Lopez-Jaramillo C., Machado-Vieira R., Malt U.F., McDonald C., Mitchell P.B., Mwangi B., Nabulsi L., Opel N., Overs B.J., Pineda-Zapata J.A., Pomarol-Clotet E., Redlich R., Roberts G., Rosa P.G., Salvador R., Satterthwaite T.D., Soares J.C., Stein D.J., Temmingh H.S., Trappenberg T., Uhlmann A., van Haren N.E.M., Vieta E., Westlye L.T., Wolf D.H., Yuksel D., Zanetti M.V., Andreassen O.A., Thompson P.M., Hajek T., Group E.B.D.W. Using structural MRI to identify bipolar disorders - 13 site machine learning study in 3020 individuals from the ENIGMA Bipolar Disorders Working Group. Mol. Psychiatry. 2018 doi: 10.1038/s41380-018-0228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea A., Cohen R.A., Porges E.C., Nissim N.R., Woods A.J. Cognitive aging and the hippocampus in older adults. Front. Aging Neurosci. 2016;8:298. doi: 10.3389/fnagi.2016.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N., Ando S., Sanada M., Hirata-Mogi S., Iijima Y., Sugiyama H., Shirakawa T., Yamagishi M., Kanehara A., Morita M., Yagi T., Hayashi N., Koshiyama D., Morita K., Sawada K., Ikegame T., Sugimoto N., Toriyama R., Masaoka M., Fujikawa S., Kanata S., Tada M., Kirihara K., Yahata N., Araki T., Jinde S., Kano Y., Koike S., Endo K., Yamasaki S., Nishida A., Hiraiwa-Hasegawa M., Bundo M., Iwamoto K., Tanaka S.C., Kasai K. Population-neuroscience study of the Tokyo TEEN Cohort (pn-TTC): cohort longitudinal study to explore the neurobiological substrates of adolescent psychological and behavioral development. Psychiatry Clin. Neurosci. 2019;73:231–242. doi: 10.1111/pcn.12814. [DOI] [PubMed] [Google Scholar]
- Okada N., Fukunaga M., Yamashita F., Koshiyama D., Yamamori H., Ohi K., Yasuda Y., Fujimoto M., Watanabe Y., Yahata N., Nemoto K., Hibar D.P., van Erp T.G., Fujino H., Isobe M., Isomura S., Natsubori T., Narita H., Hashimoto N., Miyata J., Koike S., Takahashi T., Yamasue H., Matsuo K., Onitsuka T., Iidaka T., Kawasaki Y., Yoshimura R., Watanabe Y., Suzuki M., Turner J.A., Takeda M., Thompson P.M., Ozaki N., Kasai K., Hashimoto R. Abnormal asymmetries in subcortical brain volume in schizophrenia. Mol. Psychiatry. 2016;21:1460–1466. doi: 10.1038/mp.2015.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes L., Satterthwaite T.D., Bassett D.S. Towards precise resting-state fMRI biomarkers in psychiatry: synthesizing developments in transdiagnostic research, dimensional models of psychopathology, and normative neurodevelopment. Curr. Opin. Neurobiol. Whole-brain Interactions Between Neural Circuits. 2020;65:120–128. doi: 10.1016/j.conb.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervaiz U., Vidaurre D., Woolrich M.W., Smith S.M. Optimising network modelling methods for fMRI. NeuroImage. 2020;211:116604. doi: 10.1016/j.neuroimage.2020.116604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson Progression Marker Initiative The Parkinson Progression Marker Initiative (PPMI) Prog. Neurobiol. 2011;95:629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M.S., Paolella R., Billiet T., Van Dyck P., Guns P.-J., Jeurissen B., Ribbens A., den Dekker A.J., Sijbers J. Harmonization of brain diffusion MRI: concepts and methods. Front. Neurosci. 2020;14:396. doi: 10.3389/fnins.2020.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Robinson E.C., Garcia K., Glasser M.F., Chen Z., Coalson T.S., Makropoulos A., Bozek J., Wright R., Schuh A., Webster M., Hutter J., Price A., Cordero Grande L., Hughes E., Tusor N., Bayly P.V., Van Essen D.C., Smith S.M., Edwards A.D., Hajnal J., Jenkinson M., Glocker B., Rueckert D. Multimodal surface matching with higher-order smoothness constraints. Neuroimage. 2018;167:453–465. doi: 10.1016/j.neuroimage.2017.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadato N., Morita K., Kasai K., Fukushi T., Nakamura K., Nakazawa E., Okano H., Okabe S. Neuroethical issues of the brain/MINDS Project of Japan. Neuron. 2019;101:385–389. doi: 10.1016/j.neuron.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Salimi-Khorshidi G., Douaud G., Beckmann C.F., Glasser M.F., Griffanti L., Smith S.M. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L., Hibar D.P., Samann P.G., Hall G.B., Baune B.T., Jahanshad N., Cheung J.W., van Erp T.G.M., Bos D., Ikram M.A., Vernooij M.W., Niessen W.J., Tiemeier H., Hofman A., Wittfeld K., Grabe H.J., Janowitz D., Bulow R., Selonke M., Volzke H., Grotegerd D., Dannlowski U., Arolt V., Opel N., Heindel W., Kugel H., Hoehn D., Czisch M., Couvy-Duchesne B., Renteria M.E., Strike L.T., Wright M.J., Mills N.T., de Zubicaray G.I., McMahon K.L., Medland S.E., Martin N.G., Gillespie N.A., Goya-Maldonado R., Gruber O., Kramer B., Hatton S.N., Lagopoulos J., Hickie I.B., Frodl T., Carballedo A., Frey E.M., van Velzen L.S., Penninx B., van Tol M.J., van der Wee N.J., Davey C.G., Harrison B.J., Mwangi B., Cao B., Soares J.C., Veer I.M., Walter H., Schoepf D., Zurowski B., Konrad C., Schramm E., Normann C., Schnell K., Sacchet M.D., Gotlib I.H., MacQueen G.M., Godlewska B.R., Nickson T., McIntosh A.M., Papmeyer M., Whalley H.C., Hall J., Sussmann J.E., Li M., Walter M., Aftanas L., Brack I., Bokhan N.A., Thompson P.M., Veltman D.J. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol. Psychiatry. 2017;22:900–909. doi: 10.1038/mp.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L., Veltman D.J., van Erp T.G., Samann P.G., Frodl T., Jahanshad N., Loehrer E., Tiemeier H., Hofman A., Niessen W.J., Vernooij M.W., Ikram M.A., Wittfeld K., Grabe H.J., Block A., Hegenscheid K., Volzke H., Hoehn D., Czisch M., Lagopoulos J., Hatton S.N., Hickie I.B., Goya-Maldonado R., Kramer B., Gruber O., Couvy-Duchesne B., Renteria M.E., Strike L.T., Mills N.T., de Zubicaray G.I., McMahon K.L., Medland S.E., Martin N.G., Gillespie N.A., Wright M.J., Hall G.B., MacQueen G.M., Frey E.M., Carballedo A., van Velzen L.S., van Tol M.J., van der Wee N.J., Veer I.M., Walter H., Schnell K., Schramm E., Normann C., Schoepf D., Konrad C., Zurowski B., Nickson T., McIntosh A.M., Papmeyer M., Whalley H.C., Sussmann J.E., Godlewska B.R., Cowen P.J., Fischer F.H., Rose M., Penninx B.W., Thompson P.M., Hibar D.P. Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Mol. Psychiatry. 2016;21:806–812. doi: 10.1038/mp.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K., Gagoski B.A., Polimeni J.R., Witzel T., Wedeen V.J., Wald L.L. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn. Reson. Med. 2012;67:1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Nichols T.E., Vidaurre D., Winkler A.M., Behrens T.E., Glasser M.F., Ugurbil K., Barch D.M., Van Essen D.C., Miller K.L. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat. Neurosci. 2015;18:1565–1567. doi: 10.1038/nn.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville L.H., Bookheimer S.Y., Buckner R.L., Burgess G.C., Curtiss S.W., Dapretto M., Elam J.S., Gaffrey M.S., Harms M.P., Hodge C., Kandala S., Kastman E.K., Nichols T.E., Schlaggar B.L., Smith S.M., Thomas K.M., Yacoub E., Van Essen D.C., Barch D.M. The Lifespan Human Connectome Project in Development: a large-scale study of brain connectivity development in 5–21 year olds. Neuroimage. 2018;183:456–468. doi: 10.1016/j.neuroimage.2018.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos S.N., Hernandez-Fernandez M., Vu A.T., Andersson J.L., Moeller S., Yacoub E., Lenglet C., Ugurbil K., Behrens T.E.J., Jbabdi S. Fusion in diffusion MRI for improved fibre orientation estimation: an application to the 3T and 7T data of the Human Connectome Project. Neuroimage. 2016;134:396–409. doi: 10.1016/j.neuroimage.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Q., He H., Gong T., Li C., Liang P., Qian T., Sun Y., Ding Q., Li K., Zhong J. Multicenter dataset of multi-shell diffusion MRI in healthy traveling adults with identical settings. Sci. Data. 2020;7:157. doi: 10.1038/s41597-020-0493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp T.G., Hibar D.P., Rasmussen J.M., Glahn D.C., Pearlson G.D., Andreassen O.A., Agartz I., Westlye L.T., Haukvik U.K., Dale A.M., Melle I., Hartberg C.B., Gruber O., Kraemer B., Zilles D., Donohoe G., Kelly S., McDonald C., Morris D.W., Cannon D.M., Corvin A., Machielsen M.W., Koenders L., de Haan L., Veltman D.J., Satterthwaite T.D., Wolf D.H., Gur R.C., Gur R.E., Potkin S.G., Mathalon D.H., Mueller B.A., Preda A., Macciardi F., Ehrlich S., Walton E., Hass J., Calhoun V.D., Bockholt H.J., Sponheim S.R., Shoemaker J.M., van Haren N.E., Hulshoff Pol H.E., Ophoff R.A., Kahn R.S., Roiz-Santianez R., Crespo-Facorro B., Wang L., Alpert K.I., Jonsson E.G., Dimitrova R., Bois C., Whalley H.C., McIntosh A.M., Lawrie S.M., Hashimoto R., Thompson P.M., Turner J.A. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry. 2016;21:547–553. doi: 10.1038/mp.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen D.C., Ugurbil K., Auerbach E., Barch D., Behrens T.E., Bucholz R., Chang A., Chen L., Corbetta M., Curtiss S.W., Della Penna S., Feinberg D., Glasser M.F., Harel N., Heath A.C., Larson-Prior L., Marcus D., Michalareas G., Moeller S., Oostenveld R., Petersen S.E., Prior F., Schlaggar B.L., Smith S.M., Snyder A.Z., Xu J., Yacoub E., Consortium W.U.-M.H. The Human Connectome Project: a data acquisition perspective. Neuroimage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser E., Keuken M.C., Douaud G., Gaura V., Bachoud-Levi A.-C., Remy P., Forstmann B.U., Jenkinson M. Automatic segmentation of the striatum and globus pallidus using MIST: Multimodal Image Segmentation Tool. Neuroimage. 2016;125:479–497. doi: 10.1016/j.neuroimage.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu Q., Luo J., Hu M., Zuo C. Effects of age and sex on subcortical volumes. Front. Aging Neurosci. 2019;11:259. doi: 10.3389/fnagi.2019.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Cedarbaum J., Green R.C., Harvey D., Jack C.R., Jagust W., Luthman J., Morris J.C., Petersen R.C., Saykin A.J., Shaw L., Shen L., Schwarz A., Toga A.W., Trojanowski J.Q., Alzheimer's Disease Neuroimaging I. 2014 Update of the Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2015;11:e1–120. doi: 10.1016/j.jalz.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter W., Sheridan M. Previous reward decreases errors of commission on later 'No-Go' trials in children 4 to 12 years of age: evidence for a context monitoring account. Dev. Sci. 2014;17:797–807. doi: 10.1111/desc.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Moeller S., Auerbach E.J., Strupp J., Smith S.M., Feinberg D.A., Yacoub E., Ugurbil K. Evaluation of slice accelerations using multiband echo planar imaging at 3 T. Neuroimage. 2013;83:991–1001. doi: 10.1016/j.neuroimage.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata N., Morimoto J., Hashimoto R., Lisi G., Shibata K., Kawakubo Y., Kuwabara H., Kuroda M., Yamada T., Megumi F., Imamizu H., Nanez J.E., Sr., Takahashi H., Okamoto Y., Kasai K., Kato N., Sasaki Y., Watanabe T., Kawato M. A small number of abnormal brain connections predicts adult autism spectrum disorder. Nat. Commun. 2016;7:11254. doi: 10.1038/ncomms11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Yahata N., Itahashi T., Lisi G., Yamada T., Ichikawa N., Takamura M., Yoshihara Y., Kunimatsu A., Okada N., Yamagata H., Matsuo K., Hashimoto R., Okada G., Sakai Y., Morimoto J., Narumoto J., Shimada Y., Kasai K., Kato N., Takahashi H., Okamoto Y., Tanaka S.C., Kawato M., Yamashita O., Imamizu H. Harmonization of resting-state functional MRI data across multiple imaging sites via the separation of site differences into sampling bias and measurement bias. PLoS Biol. 2019;17(4):e3000042. doi: 10.1371/journal.pbio.3000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Sakai Yuki, Yamada Takashi, Yahata Noriaki, Kunimatsu Akira, Okada Naohiro, Itahashi Takashi, Hashimoto Ryuichiro, Mizuta Hiroto, Ichikawa Naho, Takamura Masahiro, Okada Go, Yamagata Hirotaka, Harada Kenichiro, Matsuo Koji, Tanaka Saori C, Kawato Mitsuo, Kasai Kiyoto, Kato Nobumasa, Takahashi Hidehiko, Okamoto Yasumasa, Yamashita Okito, Imamizu Hiroshi. Generalizable brain network markers of major depressive disorder across multiple imaging sites. PLoS Biol. 2020 doi: 10.1371/journal.pbio.3000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Schneider T., Wheeler-Kingshott C.A., Alexander D.C. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage. 2012;61:1000–1016. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.