Abstract
Clonal hematopoiesis (CH) is common in older persons and is associated with an increased risk of hematologic cancer. Here, we review studies establishing an association between CH and hematopoietic malignancy, discuss features of CH that are predictive of leukemic progression, and explore the role of hematopoietic stressors in the evolution of CH to acute myeloid leukemia or myelodysplastic syndrome. CH due to point mutations or structural variants such as copy-number alterations is associated with an ∼10-fold increased risk of hematopoietic malignancy. Although the absolute risk of hematopoietic malignancy is low, certain features of CH may confer a higher risk of transformation, including the presence of TP53 or spliceosome gene mutations, a variant allele fraction >10%, the presence of multiple mutations, and altered red blood indices. CH in the setting of peripheral blood cytopenias carries a very high risk of progression to a myeloid malignancy and merits close observation. There is emerging evidence suggesting that hematopoietic stressors contribute to both the development of CH and progression to hematopoietic malignancy. Specifically, there is evidence that genotoxic stress from chemotherapy or radiation therapy, ribosome biogenesis stress, and possibly inflammation may increase the risk of transformation from CH to a myeloid malignancy. Models that incorporate features of CH along with an assessment of hematopoietic stressors may eventually help predict and prevent the development of hematopoietic malignancies.
Visual Abstract
Introduction
The accumulation of mutations in hematopoietic stem cells (HSCs) with age results in the production of a genetically heterogeneous cell population, with each HSC possessing its own unique set of private mutations.1 HSCs that acquire somatic mutations conferring a competitive fitness advantage relative to their normal counterparts may expand, resulting in clonal hematopoiesis (CH). Genes commonly mutated in CH are also recurrently mutated in myeloid malignancies. Thus, it is not surprising that CH is associated with the development of hematopoietic malignancy. However, the absolute risk of hematopoietic malignancy development in persons with CH is low, and there is a need to identify persons most at risk for transformation. The mechanisms contributing to leukemic transformation from CH are unclear, limiting the development of strategies to prevent progression. Here, we review studies establishing an association between CH and hematopoietic malignancy, discuss features of CH that are predictive of leukemic progression, and explore the role of hematopoietic stressors in the progression of CH to acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS).
CH is associated with an increased risk of hematologic malignancy
Several large cohort studies in persons unselected for prior history of hematopoietic disorder have established an association between CH and the development of hematopoietic malignancies. For purposes of this review, unless otherwise stated, CH is defined as the presence of 1 or more somatic mutations with a variant allele fraction (VAF) of at least 0.02. Genovese et al performed whole-exome sequencing on 12 380 Swedish persons, identifying CH in 455.2 The presence of CH conferred an increased risk for subsequent development of hematologic cancer with a hazard ratio of 12.9. Jaiswal et al performed panel sequencing on 3341 persons with longitudinal follow-up: 134 had CH.3 The hazard ratio for the development of hematologic malignancy in patients with CH was 11.1. Zink et al analyzed whole-genome sequencing data from 11 262 Icelanders.4 In this study, CH was defined as the presence of a high number (>20) of somatic mutations, regardless of coding or noncoding. Although this method identified a larger total peak prevalence of CH, there was still a significantly increased risk for development of hematologic malignancy with a hazard ratio of 2.43. Altogether, these studies show that the risk of developing a hematopoietic malignancy in persons with CH is increased. However, the incidence of hematopoietic malignancies is low. For example, with a median follow-up of nearly 8 years, only 4% of persons with CH in the Jaiswal study developed a hematopoietic malignancy. Thus, the predictive value of CH per se to identify patients at risk for hematopoietic malignancy is low.
CH due to large chromosomal anomalies also is associated with an increased risk of hematopoietic malignancy. Jacobs et al used single-nucleotide polymorphism (SNP) arrays to identify copy-number alterations and copy-number neutral loss of heterozygosity in the peripheral blood of 57 583 individuals.5 This technique, which is able to detect chromosomal abnormalities (>2 Mb in size) with a frequency of >7% of cells, identified CH in 517 individuals (1.2%). Of note, the frequency of CH was 20% or 22% for individuals who subsequently developed myeloid or lymphoid leukemia, respectively, compared with 0.74% in cancer-free controls. Laurie et al used a similar approach to identify CH due to chromosomal abnormalities in 149 of 8562 persons (1.7%) with longitudinal follow-up.6 Compared with persons without CH, the risk of hematopoietic malignancy in persons with CH was increased ∼10-fold after adjusting for age. Finally, Loh et al also used SNP arrays to interrogate 151 202 UK Biobank participants.7 They included long-range phasing of the SNP data to increase sensitivity, allowing them to detect chromosomal abnormalities at cell fractions as low as 1%. Using this approach, they identified CH in 7484 cases (4.9%). Similar to the other studies, an association of CH with hematopoietic malignancy was observed. In all 3 studies, the strongest association of CH was with the development of chronic lymphocytic leukemia (CLL). Indeed, chromosomal abnormalities frequently found in CLL, such as trisomy 12 or deletions of 13q, are associated with the subsequent development of CLL.
CH following cytotoxic therapy is associated with an increased risk of therapy-related myeloid malignancy
There is considerable evidence suggesting that CH is also associated with an increased risk of developing a therapy-related myeloid neoplasm (tMN). Exposure to cytotoxic therapy is associated with an increase in CH due to mutations in genes, such as TP53 and PPM1D, which are frequently mutated in tMNs.8-10 Two retrospective case-control studies showed that CH at the time of primary cancer treatment is associated with an ∼10-fold increased risk of tMN.11,12 Gibson et al showed that CH at the time of autologous stem cell transplantation for lymphoma conferred an increased risk of tMN, with a cumulative incidence of 14.1% for those with CH vs 4.3% for those without.10 In the largest series reported to date, Bolton et al reported on 9549 persons with cancer who received “oncologic” therapy (includes chemotherapy, radiation therapy, or immunotherapy), of whom 75 developed a tMN.13 CH was strongly associated with tMN (hazard ratio, 6.9), with a median time to progression to tMN of 26 months.
Clonal evolution from CH to hematopoietic malignancy
In all of the large cohort sequencing studies, lymphoid malignancies were more common than myeloid malignancies, despite the fact that the mutations identified in the CH samples were mostly associated with myeloid malignancies.2,3 Moreover, Zink et al showed that CH without any identifiable driver mutation is still associated with an increased risk of hematopoietic malignancy.4 These observations raise the possibility that hematopoietic malignancies may not always derive from the expanded hematopoietic stem and progenitor cell (HSPC) clone present at the time of CH. Instead, the presence of CH may represent a biomarker of a stressed hematopoietic system that is prone to malignant transformation. As summarized later in this section, there is evidence to support both pathways.
Accumulating evidence indicates that the majority of tMNs are derived from the expanded HSPC clone present in CH. We reported 4 cases of tMNs in which the exact TP53 mutations found at diagnosis were also detectable at a low level in the blood or bone marrow 3 to 6 years prior to tMN development.14 Bolton et al reported on 35 cases of tMN in which prior peripheral blood samples were available. One or more disease-defining mutations present in the tMN sample were identified in the CH sample in 32 cases (94%).13 Similar results were reported in studies by Gillis et al and Takahashi et al.11,12 Collectively, these 2 studies identified tMN mutations in the prior CH sample in 14 of 19 cases (74%). Of note, a recent report showed that specific somatic chromosomal abnormalities present in patients with tMNs were detectable in prior blood samples in 3 of 14 cases.15 Collectively, these data suggest that the majority of tMN cases develop from an expanded HSPC clone carrying tMN-associated mutations that are present years prior to tMN development.
With respect to de novo hematopoietic malignancies, with the exception of clonal somatic chromosomal abnormalities and the development of CLL, data to make firm conclusions are lacking. Several large case-control studies have confirmed the association of CH with de novo AML.16,17 Indeed, AML-associated mutations were detected in the CH sample in the majority of cases. However, because the AML was not sequenced, a direct relationship between CH mutations and AML could not be established.
There is also evidence that CH may serve as a biomarker of prior hematopoietic stress, particularly genotoxic stress. As discussed in the next section, prior exposure to chemotherapy is associated with an increased frequency of CH in a dose-dependent fashion. Interestingly, Gibson et al showed that the presence of CH in patients with relapsed lymphoma undergoing autologous hematopoietic cell transplantation is associated with increased nonrelapse mortality.10 They also showed that the CH mutations detectable at the time of transplantation are not always the ones that give rise to tMN. Together, these observations suggest that, in the setting of prior cytotoxic therapy, the presence of CH may reflect a perturbed hematopoietic state, which might include a reduced HSPC pool and/or altered bone marrow microenvironment.
Specific features of CH that are predictive of hematopoietic malignancy development
Current evidence suggests that the great majority of patients with CH do not develop a hematopoietic malignancy, making the positive predictive value low. Thus, there is a need to better stratify persons with CH who are at risk of developing a hematopoietic malignancy. Indeed, recent data suggest that certain features of CH may confer a higher risk of malignant transformation, including the specific gene mutated, clone size, and the presence of multiple mutations (Table 1).
Table 1.
CH characteristics increasing risk for leukemia transformation
| Definition | References |
|---|---|
| Total no. of mutations | |
| >1 gene | 10,13,16,17 |
| >250 somatic SNPs | 4 |
| VAF | |
| >10% | 3,17 |
| RDW | |
| >14% | 13,16 |
| Specific gene | |
| Study specific: | 13,16,17 |
| TP53, U2AF1/spliceosome | |
| IDH1/2, RUNX1, PHF6 | 17 |
| Specific variant | 18 |
| DNMT3A (R882C/H, R729W, R326C, R320*, R736H/C, Y735C, W860R, R771*, R598*, P904L) | |
| SRSF2 P95R/H/L | |
| SF3B1 K700E, K666N | |
| JAK2 V617F | |
| IDH2 R140Q | |
| GNB1 K57E |
RDW, red cell distribution width.
Case-control studies of patients with CH who developed AML show that mutations in certain genes confer a higher risk of AML development. In the study by Abelson et al, mutations of TP53 and the splicing factor U2AF1 conferred the highest risk of AML development with hazard ratios of 12.5 and 7.9, respectively.16 This contrasts with hazard ratios for DNMT3A and TET2 of 1.4 and 1.6, respectively. In the study by Desai et al, mutations of TP53 and spliceosome genes (U2AF1, SF3B1, or SRSF2) also conferred a higher risk of AML development, with odds ratios of 47.2 and 7.4, respectively.17 They also identified IDH1, IDH2, and RUNX1 with PHF6 mutations as high-risk mutations. Remarkably, in this study, all persons with TP53 mutations (n = 21) or IDH1/2 mutations (n = 15) developed AML a median of 5 to 6 years from CH sampling. Due to the case-control nature of this study, caution should be used in extrapolating these data to the general population to conclude that all CH cases with TP53 and IDH1/2 mutations will inevitably undergo malignant transformation. In the Bolton study of CH and the development of tMN in persons receiving prior oncologic therapy, the strongest associations were observed for TP53 and spliceosome gene mutations.13 Of note, in this study, CH due to IDH1/2 mutations had only a modest association with tMN development. Taking this analysis 1 step further, Watson et al looked beyond the gene level to assess the impact of specific variants on the development of CH.18 They identified 20 high-risk variants that had a strong fitness advantage and conferred a higher risk of developing AML (Table 1). Although additional data are needed to confirm these findings, it is likely that incorporating specific gene variants into predictive algorithms will improve our ability to identify persons with CH at highest risk for malignant transformation.
The size of a hematopoietic clone can be inferred from the VAF, with some caveats such as the presence of homozygous mutations or germline variants. In the study by Jaiswal et al, the hazard ratio for the development of hematologic malignancy increased to nearly 50 for those persons with a VAF of 10% or greater (corresponding to a clone representing 20% of cells in the blood).3 Case-control studies of patients with CH who developed AML also suggested that clone size correlates with leukemic progression for most gene mutations. For example, in the study by Desai et al, multivariate analysis showed that the odds ratio of AML development increased from 2.5 for persons with DNTM3A mutations with a VAF <10% to 4.8 for persons with a VAF ≥10%.17 However, both studies suggested that the impact of clone size on AML development is reduced for mutations in certain genes, including TP53, U2AF1, and IDH1. Clone size was also predictive of the development of tMN, with the odds ratio for tMN development increasing more than threefold comparing CH with a VAF of 2% to 5% to cases with a VAF >10%.13
The risk of transformation to AML also correlates with the total number of mutations detected in the CH sample. In the Gibson et al study of tMN development in persons undergoing autologous stem cell transplantation for lymphoma, the presence of >1 CH mutation was associated with a higher risk of tMN than those with only 1 mutation (16.5% vs 4% at 5 years).10 In the studies by Abelson et al and Desai et al, CH with >1 mutation was associated with a higher risk of AML development.16,17 In particular, certain combinations of gene mutations, such as DNTM3A plus a spliceosome gene mutation, conferred a high risk of leukemic transformation. Strikingly, in the Zink et al study, the total number of somatic single-nucleotide variants (SNVs) dramatically increased the risk for hematologic malignancy development; the hazard ratio for cases with <180 SNVs was 1.98 vs a hazard ratio of 42.2 for cases with >250 SNVs.4 Finally, in the Bolton et al study, the presence of >1 CH mutation conferred an increased risk of tMN development, with a hazard ratio of 1.8.13 Of note, in all of these studies, whether the multiple mutations identified in the CH sample are present in the same clone is unknown, but are likely to be relevant.
Along with these clone-specific parameters, there are data suggesting that certain red blood cell parameters, including red cell distribution width and mean corpuscular volume, are associated with AML development.13,16 Bolton et al incorporated CH mutation type, maximum clone size, number of mutations, and peripheral blood count indices into a model to identify a high-risk group with an absolute 10-year risk of tMN of 4% to 12%. Large prospective studies are needed to validate and refine this model, but these data suggest that we may eventually be able to identify persons with CH at high-enough risk to be actionable.
CH with cytopenia
Prior studies had identified CH in persons with peripheral blood cytopenia, a condition termed clonal cytopenia of undetermined significance (CCUS).19,20 In a prospective study of 154 persons with idiopathic blood cytopenia, panel sequencing identified 1 or more mutation in 56 (36%).21 The 5-year probability of progression to a myeloid neoplasm in this cohort of persons with CCUS was 82%. This compares to the 5-year probability of transformation of only 9% in persons with idiopathic cytopenia without CH. Mutations in certain genes, such as SF3B1, or multiple mutations confer an even higher risk, with over 90% of persons with CH carrying these mutations progressing to a myeloid neoplasm by 5 years. Thus, CCUS appears to be a special category of CH with a very high risk of transformation to a myeloid malignancy.
Role of hematopoietic stressors in leukemic transformation from CH
Accumulating evidence suggests that hematopoietic stressors play an important role in the development of CH, which is discussed in further detail elsewhere in this review series. For example, there is strong clinical and preclinical evidence that cytotoxic therapy selects for hematopoietic progenitor cells carrying mutations in DNA damage response genes, including TP53, PPM1D, and CHEK2.8,11,12,14 Animal studies have confirmed that HSPCs carrying TP53 or PPM1D mutations have a competitive advantage in vivo after exposure to chemotherapy.14,22-24 Intriguingly, it appears that there may be chemotherapy-specific effects on the development of CH. For example, Hsu et al showed that expansion of PPM1D-mutated clones was strongly associated with exposure to platinum agents and etoposide, but not to other cytotoxic agents or transplant.24 This is consistent with recent data showing that platinum agents and etoposide, but not alkylator agents, are associated with an increased frequency of CH.13 There is also emerging evidence that inflammatory cytokines may contribute to the expansion of hematopoietic stem/progenitor clones carrying mutations in epigenetic modifiers, such as TET2.25-27
Whether hematopoietic stressors also contribute to the progression from CH to hematopoietic malignancy is less clear. Our recent study of CH in persons with Shwachman-Diamond syndrome (SDS) provides insight into this question. SDS is a congenital bone marrow failure syndrome characterized by blood cytopenias, exocrine pancreatic insufficiency, bony abnormalities, and a high rate of transformation to MDS or AML. We identified CH due to TP53 mutations in 13 of 27 persons with SDS (43%; median age, 6.3 years) vs 0 of 17 age-matched healthy controls.28 SDS is caused in most cases by biallelic loss-of-function mutations of SBDS,29 which results in impaired ribosome biogenesis.30-32 Of note, there is evidence that ribosome biogenesis stress induces p53 expression, which in turn results in a growth arrest.33,34 Together, these observations suggest a model in which elevated p53 expression due to ribosome biogenesis stress in SDS HSPCs results in impaired HSPC growth and/or survival (Figure 1). Mutation of TP53 in HSPCs is predicted to attenuate this growth arrest, resulting in their selective expansion in patients with SDS.
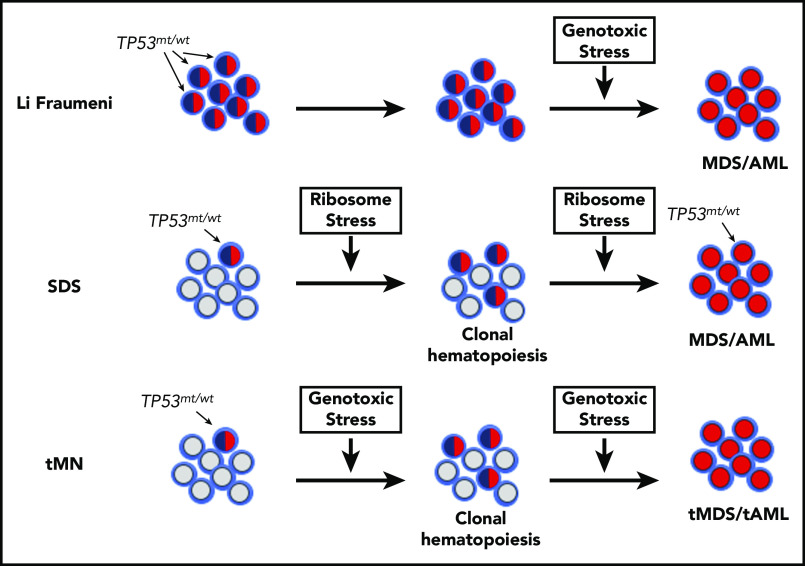
Figure 1.
Model of MDS/AML pathogenesis in SDS, Li Fraumeni syndrome, and tMN. In Li Fraumeni syndrome, all of the HSPCs carry heterozygous TP53 mutations (indicated by blue/red shaded cells). In the absence of genotoxic stress (eg, chemotherapy), there is no selective pressure to mutate the second allele. Thus, MDS/AML in Li Fraumeni syndrome is uncommon. In SDS, mutations of SBDS lead to chronic ribosome stress, which can select for HSPCs that have mutated 1 allele of TP53 and result in TP53-mutated CH. Continued ribosome stress then selects for HSPCs in which both TP53 alleles are mutated (red shaded cells), which ultimately leads to MDS/AML. Patients undergoing treatment with cytotoxic chemotherapy or radiation experience significant genotoxic stress, resulting in selection of HSPCs carrying age-related TP53 mutations. In this model, repeated exposure to cytotoxic therapy selects for HSPCs carrying biallelic TP53 mutations, which ultimately leads to MDS/AML. Other hematopoietic stressors, such as chronic inflammation, may promote progression from CH to myeloid malignancy in a similar fashion by providing a fitness advantage to HSPCs carrying mutations in other genes besides TP53. tAML, therapy-related AML; tMDS, therapy-related MDS.
However, simply having a TP53 mutation within the hematopoietic compartment is clearly not sufficient to drive leukemogenesis. Li Fraumeni is a cancer predisposition syndrome caused by heterozygous germline mutations of TP53.35 Despite 100% of HSPCs carrying a heterozygous TP53 mutation, the risk of MDS/AML development in Li Fraumeni syndrome is low (<5%),36 especially compared with SDS, where the cumulative incidence of MDS/AML is >20%.37 So why is the risk of leukemic progression higher in SDS? We suggest that continued ribosome stress in SDS HSPCs carrying a heterozygous TP53 mutation selects for clones that inactivate the second TP53 allele (Figure 1). Mouse studies show that development of hematopoietic malignancy is faster and more penetrant in HSPCs carrying biallelic loss-of-function TP53 mutations compared with heterozygous TP53-mutated cells.38-40 However, biallelic loss of TP53 alone is not sufficient to induce AML or MDS in mice, and other cooperating mutations are required for transformation.41,42 In humans, there is a high frequency of chromosome 5 and 7 abnormalities in TP53-mutated AML/MDS. When these chromosomal abnormalities are acquired in relation to the biallelic loss of TP53 is unknown. Nonetheless, these observations suggest a model for myeloid malignancy development in SDS in which ribosome stress contributes not only to the development of TP53-mutated CH, but also to its progression to a myeloid malignancy. Consistent with this conclusion, a recent study showed that the majority of MDS or AML cases arising in the setting of SDS carry biallelic TP53 mutations.43 In contrast, in Li Fraumeni syndrome, there is no selective pressure to mutate the second TP53 allele and the risk of MDS/AML is low. Of note, a recent case series of persons with Li Fraumeni showed that 5 of 5 patients with myeloid malignancy developed tMN after being treated with cytotoxic chemotherapy for a first malignancy.44 Thus, in Li Fraumeni syndrome, genotoxic stress may contribute to tMN development by selecting for HSPCs that have inactivated the second TP53 allele.45,46
There is evidence that other hematopoietic stressors contribute to clonal evolution from CH to hematopoietic malignancy in certain disorders. For example, CH due to activating mutations of CSF3R (encoding the granulocyte colony-stimulating factor [G-CSF] receptor) is frequent in patients with severe congenital neutropenia.47-50 Of note, circulating levels of G-CSF are elevated in patients with severe congenital neutropenia, either due to pharmacologic administration, or increased endogenous production.51 This high level of G-CSF, in combination with the activating CSF3R mutations, provides a fitness advantage to HSPCs52 that likely contributes to the high rate of transformation to MDS/AML seen in this disorder. Another example of hematopoietic stressors shaping clonal evolution is in patients with germline mutations of SAMD9 or SAMD9L, which are on chromosome 7 (clinical syndromes reviewed in Davidsson et al53). These gain-of-function mutations impair HSPC proliferation, which may be promoted by inflammation-induced SAMD9/SAMD9L expression.54,55 In this setting, HSPCs that have lost the copy of chromosome 7 containing the mutant SAMD9 or SAMD9L allele have a fitness advantage, resulting in their clonal expansion and, in some cases, progression to MDS/AML with monosomy 7.54-56 Finally, there is evidence that inflammation may contribute to leukemic progression from TET2-mutated CH. Loss of Tet2 in mice results in enhanced HSC self-renewal and a myeloproliferative-like syndrome.57 Meisel et al showed that this myeloproliferative phenotype is abrogated when Tet2−/− mice are housed under germ-free conditions.58 They provided evidence that Tet2 loss disrupts intestinal integrity, leading to increased translocation of intestinal bacteria, which in turn, results in increased inflammatory cytokine production that contributes to the myeloproliferative phenotype.
It is worth noting that not all clonal adaptions to hematopoietic stressors are deleterious. For example, in SDS, the most common somatic chromosomal abnormality is isochromosome 7.59 Studies show that the duplicated region of chromosome 7 includes the SBDS allele capable of producing full-length protein, effectively increasing SBDS expression,60 and is associated with a lower risk of transformation to myeloid malignancy. Thus, considering the context in which CH arises to determine whether close monitoring or therapeutic intervention is needed will be important.
Conclusions, open questions, and future directions
In summary, current evidence shows that CH confers an increased risk of myeloid and lymphoid malignancies. However, these cancers are uncommon, and the absolute risk of developing a hematopoietic malignancy is small. There are emerging data that certain features of CH may confer a higher risk of transformation, including the presence of TP53 or spliceosome gene mutations, a VAF >10%, the presence of multiple mutations, and altered red blood indices. There is also emerging evidence that specific variants within a given gene confer differing degrees of risk for transformation. Prospective trials are needed to validate these finding and establish the absolute risk of hematopoietic malignancy development. CH in the setting of peripheral blood cytopenias (CCUS) carries a very high risk of progression to a myeloid malignancy. Prospective clinical studies are needed to determine whether early intervention in persons with CCUS prevents or delays the development of MDS/AML. Nonetheless, we suggest that current evidence supports testing for CH in persons with unexplained persistent blood cytopenias. Persons with CCUS should receive frequent follow-up for the development of MDS/AML.
There are several open questions in the field. Sequencing and array-based studies show that CH due to point mutations and structural variants (eg, copy-number alterations) are each associated with an increased risk of hematopoietic malignancy. However, to date, no studies have tested for both types of mutations in the same study population. Whether the cooccurrence of structural variants with point mutations increases the risk of hematopoietic malignancy is uncertain. Improved methods to detect structural variants using next-generation sequencing data should allow investigators to address this issue in the near future.
For persons with CH containing multiple mutations, are these mutations in the same cell? Longitudinal studies in persons with CH carrying multiple mutations show that, in some cases, 1 of the mutations is lost.9,13 Thus, in these cases, the mutations likely arose in distinct HSPC clones. Is the likelihood of leukemic progression different between CH due to a single HSPC clone containing multiple mutations and CH in which multiple HSPCs carry single mutations? Single-cell sequencing techniques with improved mutation calling will allow the field to better address this question.
There is accumulating evidence suggesting that the cell of origin in which a mutation occurs contributes to the emergence of the tumor phenotype.61 For example, the phenotype of MLL-AF9–induced leukemia in mice differs when HSCs are the cells of origin as compared with granulocyte-monocyte progenitors.62,63 Studies are needed to determine whether the cell of origin in CH impacts the risk of malignant transformation.
The role of hematopoietic stressors in the development of CH and subsequent development of hematopoietic malignancy need further investigation. Our study of SDS suggests that ribosomal stress plays a key role both in the development of CH carrying heterozygous TP53 mutations and in the progression to myeloid malignancy carrying biallelic TP53 mutations. Does repeated exposure to cytotoxic therapy likewise promote progression from heterozygous TP53-mutated CH to biallelic TP53-mutated tMN? Does inflammation play a role in the development and leukemic progression of CH due to epigenetic modifier genes, such as TET2, DNMT3A, or ASXL1? Ultimately, identifying hematopoietic stressors that contribute to leukemic progression in persons with CH may suggest strategies to minimize its development.
Finally, and perhaps most importantly, strategies to prevent the development of hematopoietic malignancies in persons with CH and high-risk features are needed. The identification of stressors that contribute to leukemic progression in persons with CH may suggest strategies to minimize its development. For example, smoking has been linked to CH, suggesting that smoking cessation may reduce its incidence.8 Aggressive treatment of inflammation may also reduce the incidence of CH and/or its progression to hematopoietic malignancy. In persons with TP53-mutated CH, minimizing genotoxic therapy may reduce the rate of leukemic transformation. Finally, treatment with agents that target specific high-risk gene mutations may provide an approach to suppress the HSPC clone carrying that mutation. For example, treatment with enasidenib, an IDH2 inhibitor, has been shown to suppress HSPCs carrying IDH2 mutations in patients with AML.64 As more targeted therapies become available, their use to suppress CH or prevent its progression could be tested.
Authorship
Contribution: J.T.W. and D.C.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel C. Link, Division of Oncology, Department of Internal Medicine, School of Medicine, Washington University in St. Louis, 660 S. Euclid Ave, Campus Box 8007, St. Louis, MO 63110; e-mail: danielclink@wustl.edu.
REFERENCES
- 1.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zink F, Stacey SN, Norddahl GL, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130(6):742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs KB, Yeager M, Zhou W, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44(6):651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laurie CC, Laurie CA, Rice K, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44(6):642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh P-R, Genovese G, Handsaker RE, et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature. 2018;559(7714):350-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombs CC, Zehir A, Devlin SM, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21(3):374-382.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong TN, Miller CA, Jotte MRM, et al. Cellular stressors contribute to the expansion of hematopoietic clones of varying leukemic potential. Nat Commun. 2018;9(1):455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson CJ, Lindsley RC, Tchekmedyian V, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol. 2017;35(14):1598-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillis NK, Ball M, Zhang Q, et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol. 2017;18(1):112-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi K, Wang F, Kantarjian H, et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol. 2017;18(1):100-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolton KL, Ptashkin RN, Gao T, et al. Oncologic therapy shapes the fitness landscape of clonal hematopoiesis. bioRxiv. 2020; doi:10.1101/848739. [DOI] [PMC free article] [PubMed]
- 14.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Wang F, Kantarjian H, et al. Copy number alterations detected as clonal hematopoiesis of indeterminate potential. Blood Adv. 2017;1(15):1031-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai P, Mencia-Trinchant N, Savenkov O, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24(7):1015-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson CJ, Papula AL, Poon GYP, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367(6485):1449-1454. [DOI] [PubMed] [Google Scholar]
- 19.Cargo CA, Rowbotham N, Evans PA, et al. Targeted sequencing identifies patients with preclinical MDS at high risk of disease progression. Blood. 2015;126(21):2362-2365. [DOI] [PubMed] [Google Scholar]
- 20.Kwok B, Hall JM, Witte JS, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126(21):2355-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malcovati L, Gallì A, Travaglino E, et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood. 2017;129(25):3371-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6(4):309-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahn JD, Miller PG, Silver AJ, et al. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood. 2018;132(11):1095-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu JI, Dayaram T, Tovy A, et al. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell. 2018;23(5):700-713.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abegunde SO, Buckstein R, Wells RA, Rauh MJ. An inflammatory environment containing TNFα favors Tet2-mutant clonal hematopoiesis. Exp Hematol. 2018;59:60-65. [DOI] [PubMed] [Google Scholar]
- 26.Cai Z, Kotzin JJ, Ramdas B, et al. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell. 2018;23(6):833-849.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuster JJ, Walsh K. Somatic mutations and clonal hematopoiesis: unexpected potential new drivers of age-related cardiovascular disease. Circ Res. 2018;122(3):523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia J, Miller CA, Baty J, et al. Somatic mutations and clonal hematopoiesis in congenital neutropenia. Blood. 2018;131(4):408-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boocock GRB, Morrison JA, Popovic M, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33(1):97-101. [DOI] [PubMed] [Google Scholar]
- 30.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118(16):4305-4312. [DOI] [PubMed] [Google Scholar]
- 31.Menne TF, Goyenechea B, Sánchez-Puig N, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39(4):486-495. [DOI] [PubMed] [Google Scholar]
- 32.Burwick N, Coats SA, Nakamura T, Shimamura A. Impaired ribosomal subunit association in Shwachman-Diamond syndrome. Blood. 2012;120(26):5143-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451(7176):335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dutt S, Narla A, Lin K, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols KE, Malkin D, Garber JE, Fraumeni JF Jr., Li FP. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol Biomarkers Prev. 2001;10(2):83-87. [PubMed] [Google Scholar]
- 36.Bougeard G, Renaux-Petel M, Flaman J-M, et al. Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol. 2015;33(21):2345-2352. [DOI] [PubMed] [Google Scholar]
- 37.Freedman MH, Bonilla MA, Fier C, et al. Myelodysplasia syndrome and acute myeloid leukemia in patients with congenital neutropenia receiving G-CSF therapy. Blood. 2000;96(2):429-436. [PubMed] [Google Scholar]
- 38.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215-221. [DOI] [PubMed] [Google Scholar]
- 39.Harvey M, McArthur MJ, Montgomery CA Jr., Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5(3):225-229. [DOI] [PubMed] [Google Scholar]
- 40.Lang GA, Iwakuma T, Suh Y-A, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119(6):861-872. [DOI] [PubMed] [Google Scholar]
- 41.Zuber J, Radtke I, Pardee TS, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23(7):877-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Z, Zuber J, Diaz-Flores E, et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev. 2010;24(13):1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy AL, Myers KC, J Bowman, et al. Distinct genetic pathways define pre-leukemic and compensatory clonal hematopoiesis in Shwachman-Diamond syndrome. bioRxiv. 2020; doi:10.1101/2020.06.04.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swaminathan M, Bannon SA, Routbort M, et al. Hematologic malignancies and Li-Fraumeni syndrome. Cold Spring Harb Mol Case Stud. 2019;5(1):a003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zebisch A, Lal R, Müller M, et al. Acute myeloid leukemia with TP53 germ line mutations. Blood. 2016;128(18):2270-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulz E, Valentin A, Ulz P, et al. Germline mutations in the DNA damage response genes BRCA1, BRCA2, BARD1 and TP53 in patients with therapy related myeloid neoplasms. J Med Genet. 2012;49(7):422-428. [DOI] [PubMed] [Google Scholar]
- 47.Dong F, Dale DC, Bonilla MA, et al. Mutations in the granulocyte colony-stimulating factor receptor gene in patients with severe congenital neutropenia. Leukemia. 1997;11(1):120-125. [DOI] [PubMed] [Google Scholar]
- 48.Dong F, Brynes RK, Tidow N, Welte K, Löwenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333(8):487-493. [DOI] [PubMed] [Google Scholar]
- 49.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: results of a long-term survey. Blood. 2007;109(1):93-99. [DOI] [PubMed] [Google Scholar]
- 50.Klimiankou M, Uenalan M, Kandabarau S, et al. Ultra-sensitive CSF3R deep sequencing in patients with severe congenital neutropenia. Front Immunol. 2019;10:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mempel K, Pietsch T, Menzel T, Zeidler C, Welte K. Increased serum levels of granulocyte colony-stimulating factor in patients with severe congenital neutropenia. Blood. 1991;77(9):1919-1922. [PubMed] [Google Scholar]
- 52.Liu F, Kunter G, Krem MM, et al. Csf3r mutations in mice confer a strong clonal HSC advantage via activation of Stat5. J Clin Invest. 2008;118(3):946-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davidsson J, Puschmann A, Tedgård U, Bryder D, Nilsson L, Cammenga J. SAMD9 and SAMD9L in inherited predisposition to ataxia, pancytopenia, and myeloid malignancies. Leukemia. 2018;32(5):1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tesi B, Davidsson J, Voss M, et al. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood. 2017;129(16):2266-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong JC, Bryant V, Lamprecht T, et al. Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight. 2018;3(14):e121086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buonocore F, Kühnen P, Suntharalingham JP, et al. Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans. J Clin Invest. 2017;127(5):1700-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20(1):11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meisel M, Hinterleitner R, Pacis A, et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 2018;557(7706):580-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimamura A. Shwachman-Diamond syndrome. Semin Hematol. 2006;43(3):178-188. [DOI] [PubMed] [Google Scholar]
- 60.Minelli A, Maserati E, Nicolis E, et al. The isochromosome i(7)(q10) carrying c.258+2t>c mutation of the SBDS gene does not promote development of myeloid malignancies in patients with Shwachman syndrome. Leukemia. 2009;23(4):708-711. [DOI] [PubMed] [Google Scholar]
- 61.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314-322. [DOI] [PubMed] [Google Scholar]
- 62.Stavropoulou V, Kaspar S, Brault L, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30(1):43-58. [DOI] [PubMed] [Google Scholar]
- 63.Krivtsov AV, Figueroa ME, Sinha AU, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27(4):852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quek L, David MD, Kennedy A, et al. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nat Med. 2018;24(8):1167-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]