Abstract
Vaginal dysbiosis-induced by an overgrowth of anaerobic bacteria is referred to as bacterial vaginosis (BV). The dysbiosis is associated with an increased risk for acquisition of sexually transmitted infections. Women with symptomatic BV are treated with oral metronidazole (MET), but its effectiveness remains to be elucidated. This study used whole-genome sequencing (WGS) to determine the changes in the microbiota among women treated with MET. WGS was conducted on DNA obtained from 20 vaginal swabs collected at four time points over 12 months from five randomly selected African American (AA) women. The baseline visit included all women who were diagnosed with asymptomatic BV and were untreated. All subjects were tested subsequently once every 2 months and received a course of MET for each BV episode during the 12 months. The BV status was classified according to Nugent scores (NSs) of vaginal smears. The microbial and resistome profiles were analysed along with the sociodemographic metadata. Despite treatment, none of the five participants reverted to normal vaginal flora — two were consistently positive for BV, and the rest experienced episodic cases of BV. WGS analyses showed Gardnerella spp. as the most abundant organism. After treatment with MET, there was an observed decline of Lactobacillus and Prevotella species. One participant had a healthy vaginal microbiota based on NS at one follow-up time point. Resistance genes including tetM and lscA were detected. Though limited in subjects, this study shows specific microbiota changes with treatment, presence of many resistant genes in their microbiota, and recurrence and persistence of BV despite MET treatment. Thus, MET may not be an effective treatment option for asymptomatic BV, and whole metagenome sequence would better inform the choice of antibiotics.
Keywords: Lactobacillus, microbiota dysbiosis, 16s rRNA gene sequencing, bacteria, community
Introduction
Bacterial vaginosis (BV) is a polymicrobial condition with a dysbiosis of the vaginal microbiota where the levels of Lactobacilli decrease, while that of opportunistic, anaerobic bacteria increases [1]. BV is most common among women at reproductive age [2] and affects approximately 29 % of women in the United States [3]. It is associated with vaginal discharge, malodour, itching, and increased vaginal pH [3, 4]. However, about 50 % of cases of BV are asymptomatic [5]. BV has also been found to increase the risk of sexually transmitted infections (STIs) such as human immunodeficiency virus (HIV) [4, 6], herpes simplex virus type-2 [7], human papillomavirus (HPV) [8], Neisseria gonorrhoeae [9], Chlamydia trachomatis [9], and Trichomonas vaginalis [10]. Adverse reproductive sequelae, such as spontaneous abortion, preterm labour, and pelvic inflammatory disease, are associated with BV [2, 4, 6, 11]. Additionally, the aetiology of BV remains unknown, and the long-term effective treatment methods are yet to be determined [10, 12]. The longitudinal changes in the composition of the vaginal microbiota can allude to bacteria associated with healthy and unhealthy microbiota as well as the effect of treatment on their composition.
Lactobacillus species dominate a healthy vaginal microbiota [2, 13–15]. In general, the genus has been found to lower the vaginal pH through the production of lactic acid, which promotes an ideal vaginal ecosystem [14, 16]. Among the Lactobacillus species, L. crispatus and L. jensenii promote a healthy vaginal microbiota, whereas L. gasseri and L. iners do not [6, 14, 15]. In cases of BV, the vaginal microbiota is frequently dominated by Gardnerella spp., Prevotella spp., Mobiluncus spp., Atopobium vaginae, and Mycoplasma hominis [2, 6, 15]. The most typical antibiotics used to treat BV include metronidazole (MET) and clindamycin to inhibit bacterial growth [2, 10]. Though treatment often appears to be effective, there is an increased long-term recurrence (within 1 year) [2, 15]. Recurrent BV may probably be due to reinfection or increasing microbial resistance [15]. Demographic and behavioural characteristics further exacerbate the prevalence. For example, hormonal contraception has been shown to reduce the risk of BV [14]. An increased risk of BV can be attributed to race (African American), history of STIs, douching, and sexual behaviour [11].
BV diagnosis is based on either Amsel’s or Nugent tests [3]. Amsel’s criteria are widely used in clinical settings, whereas Nugent scores (NSs) are used extensively in research [10]. Amsel’s criteria are based on the presence of three of the four following signs: milky white discharge, presence of clue cells in wet mount microscopy, elevated vaginal pH, and whiff test [10, 17]. NSs range from 0 to 10 and are based on the relative abundance of bacteria in the Gram-staining of vaginal smears. A healthy vaginal microbiota is expected to have a NS of 0–3. Scores of 4–6 and 6–10 suggest intermediate flora and BV, respectively [18]. The vaginal microbiota can be further characterized by community state types (CSTs) [16]. There are five CSTs that describe the vaginal microbiota in terms of dominant bacteria; CSTs I, II, III and V are dominated by Lactobacillus species, especially L. crispatus, L. gasseri, L. iners, and L. jensenii , respectively; and the CST IV is highly heterogenous and dominated by strictly anaerobic bacteria [16]. Most early vaginal microbiota studies commonly used 16S rRNA sequencing to describe the composition [6, 13, 19]. In this study, we examined the change in the composition of the vaginal microbiota of women with asymptomatic BV using whole-genome sequencing (WGS) and investigated the longitudinal change in the vaginal microbiota among women treated with MET. The changes in the relative abundance of bacterial species, alpha diversity, CST, and BV status across time points were examined. Furthermore, the correlation between NS and identified bacterial species were assessed. We report the exploratory findings of a pilot study with five women (20 samples) who were followed for 1 year at four time points.
Methods
Clinical samples
Vaginal swabs in this study came from a previously completed clinical trial funded by the National Institutes for Health, known as the BRAVO study [20]. The BRAVO study sought to investigate the effect of home screening and treatment of asymptomatic BV on incident N. gonorrhoeae and C. trachomatis (NG/CT). This trial gathered data from women aged 15–25 years from five states in the US, namely Alabama, California, Maryland, North Carolina, and Pennsylvania. All women were diagnosed with asymptomatic BV at baseline using modified Amsel’s criteria. Women self-collected vaginal swabs at baseline and every 2 months over the next 12 months. Self-collected vaginal samples were shipped to a central laboratory in Pittsburg for testing. The vaginal swabs were tested for BV and NG/CT using NSs and the BD ProbeTec Amplified DNA Assay (Becton-Dickson, Inc. Sparks, MD), respectively.
Due to the pilot nature of the current study and limited funds, five women were randomly selected for WGS, which is known to provide a more detailed microbial profile than 16S sequencing. These five women were chosen from 124 subjects because they had the least possible confounding factors among the cohort. All five women chosen were African American, non-Hispanic, between the ages of 19–22, had never been pregnant before, did not have chlamydia or gonorrhoea, did not consume antibiotics during the survey, were sexually active, had prior BV exactly once, and stayed through the entire programme. The broader cohort of 124 subjects was not all African American; and some were Hispanic, many were pregnant during or prior to the study, and started with more severe BV at the start. Treatment was administered after baseline. Therefore, all baseline samples were untreated. Treatment was administered at each time point for which a woman tested positive for BV.
Epidemiological data
Baseline characteristics were assessed using a self-administered questionnaire in the parent study. Information on age, race (Black or African American/White or Caucasian/Hawaiian or Pacific Islander/Asian/Other), ethnicity (Hispanic/Latino, not Hispanic/Latino), and the highest level of education (eighth grade or less/Incomplete high school/High School graduate or GED/Incomplete college/Associate degree/College degree/Masters or advanced degree/Vocational certificate) were collected. Clinical and behavioural questions related to medical history explored prior antibiotic use (past 30 days), previous lifetime episodes of BV (never, once, 2–4 times, ≥5 times), prior lifetime treatment for BV (≤1 month ago, 1–6 months ago, 7–12 months, ≥12 months ago), prior pregnancy (yes/no), frequency of vaginal douching (daily/weekly/monthly/yearly/not in the past year/never), birth control methods used in the past year (pills, patch, Nuva-ring, condoms, a spermicide cream, Depo-Provera shot, intra-uterine device (IUD) and other), and the number of different sex partners and the types of sexual behaviour in the past year, (receptive oral sex, unprotected anal sex, unprotected vaginal sex, women who have sex with women, and new sex partners). The treatment arm assignment was also recorded.
BV assessment
BV was diagnosed using the NS [3]. A score of 0–3 is considered healthy vaginal microbiota, 4–6 intermediate flora, and 7–10 indicates BV. The vaginal microbiota can be further classified into five community state types (CSTs) [16]. The CSTs I, II, III, and V are dominated by L. crispatus , L. gasseri, L. iners, and L. jensenii, respectively, and are typically associated with a healthy vaginal ecosystem [16]. On the other hand, CST IV represents a diverse group with no individual dominant species but is associated with BV [16].
DNA preparation and whole genome sequencing
Vaginal swabs were processed and analysed with the ZymoBIOMICS Service (Zymo Research, Irvine, CA). Genomic DNA samples were profiled for shotgun metagenomic sequencing. Following the manufacturer’s protocol, sequencing libraries were prepared with KAPA HyperPlus (MA, US) with 100 ng DNA input. The libraries used internal 8 bp barcodes and TruSeq adapters. All libraries were quantified with TapeStation and pooled evenly. The final pool was quantified with quantitative PCR and sequenced with Illumina HiSeq. The operational taxonomic units were then counted, and the abundance of each taxon was normalized to a constant sum of one for each sample.
Bioinformatics and statistical analysis
Statistical tools
Data were analysed using the R statistical software package version 3.5.1 [21]. The plyr [22] and Hmisc [23] were used for matrix operations and data manipulation.
Microbial abundance analysis
The abundance profile for each subject was generated using in-house scripts. The resulting microbial profile of each sample was combined with clinical attributes of interest and labelled as treated/untreated correspondingly. Names of microbes were trimmed to focus only on the species level. Used the vegan [24] tool was to calculate the community diversity based on the Inverse Simpson Index [25].
Correlational analyses
Pearson and Spearman correlation coefficients [26] were calculated to find the taxa whose abundance values in the samples co-varied the most and the taxa that were most correlated with NSs, as described earlier [27].
Variance analysis
Variance analysis was performed on the sample microbial profiles. Principal component analysis (PCA), a dimensionality reduction tool, was used to assess the variance in the vaginal microbiota and to determine the taxa that contributed the most to the variance. It identifies the direction along which the samples display the most variance and also extracts the principal components that reduce the dimensionality of the data while preserving as much variance as possible. The balanced error rate of the Mahalanobis distance [28] was calculated for an increasing number of components. The mixomics R-package [29] was used for the PCA analyses.
Discriminant analysis
Discriminant analysis was performed on the sample microbial profiles to determine the taxa that contributed the most to the discrimination between the samples. The abundance of these taxa was then compared using Welch’s t-test [30], which was selected as there were unequal sample sizes, five samples untreated (baseline) versus 15 treated (follow-up). The Benjamini-Hochberg correction [31] was also used to decrease the false discovery rate. The Sparse Partial Least Squares Discriminant Analysis (sPLS-DA) [32] is similar to PCA, but it finds the direction along which the separation between the classes is maximized (discriminatory). The four classes for this analysis correspond to the sampling time points. Three-fold cross validation was repeated 100 times. The mixomics [29] was also used for the sPLS-DA analyses.
Analyses of ABR genes
The antimicrobial resistance profile for each subject was generated using scripts from PeTRi metagenomic pipeline [33]. First, accessed the list of 2786 antibiotic-resistance reference genes from the Comprehensive Antibiotic Resistance Database (CARD) version v3.1.1 [34]. Second, aligned metagenomic reads from each subject against the reference sequences using Bowtie2 [35]. Third, using the alignments computed ‘counts per million reads’ for each ABR gene, which served as an approximation of the metagenomic sample’s resistance ‘potential’. Since the CARD database does not contain nim genes associated with MET resistance, their presence was manually curated.
Visualization tools
The R libraries gplots [36], ggplot2 [37], and plotly [38] were used for the creation of all figures presented in this manuscript.
Results
Participant characteristics
The study sample included five AA women, who were less than 22 years of age (80%), did not have a college degree (80%) and lacked a history of pregnancy (100%). None of the women reported prior antibiotic use 30 days at the start of the study or a BV treatment history. Four of the five women reported douching in the past but did not douche regularly. These women actively used a form of birth control, most commonly condoms (75%), and had at least one sex partner in the past year. Most women had two or more oral sex partners in the past year (60%), engaged in vaginal sex without a condom (80%), and reported a new sex partner in the past year (80%). For this study, all five women received 1 week oral MET treatment after baseline assessment and every subsequent time point for which they tested positive for BV. Thus, at baseline, all vaginal samples were positive for BV and untreated. None of the participants had NG/CT.
Vaginal microbial profile
All the participants of this study had asymptomatic BV. Two participants (1 and 2) had BV (as per NSs) despite treatment for the entire year. Both participants reported having new and multiple sex partners as well as having received oral sex. In addition, Participant 2 also reported engaging in anal sex.
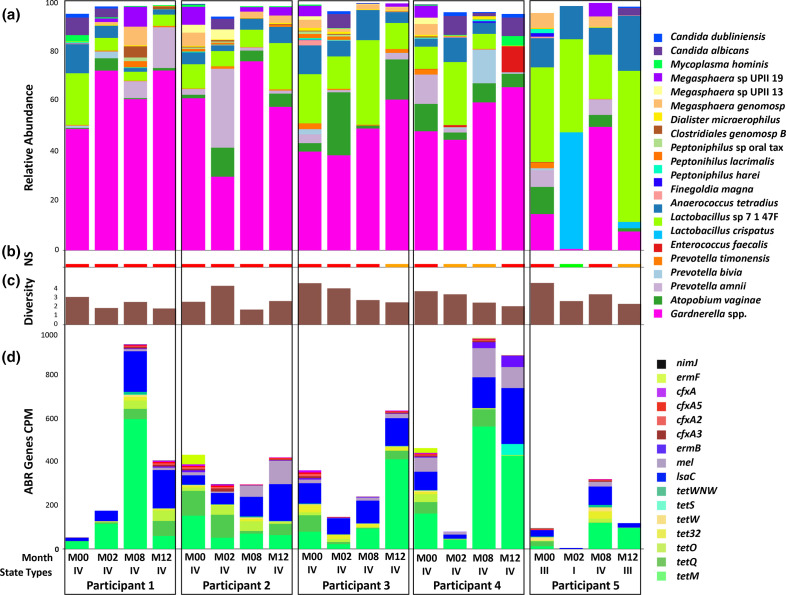
Fig. 1 describes the changes in the composition of vaginal microbiota over time for the five participants. Participants 1, 2, 3, and 4 were classified as having community state type (CST) V throughout the sampling period, even though they presented small variations in their microbial compositions. On the other hand, Participant 5 presented states III, I, V, and III in that order and was the only participant who transitioned to health-associated CSTs.
Fig. 1.
Vaginal microbial and resistome profile of five African American participants at four time points. Each column represents a specific time point for each participant beginning at baseline and followed by 4 month intervals, a total of four columns per participant ordered sequentially. Each column represents a specific time point for each participant beginning at baseline and followed by 4 month intervals, a total of four columns per participant ordered sequentially. The labels M00, M02, M08, and M12 correspond to baseline, and months four, eight, and twelve, respectively. The vaginal microbiota characterized by community state types I to IV are indicated in the x-axis. (a) Relative abundance of microbes. (b) The Nugent scores (NS) associated with the participant. The BV is indicated by red, intermediate vaginal flora by orange, and healthy vaginal microbiota by green. (c) Alpha diversity calculated using the Inverse Simpson Index. (d) Resistome profile is given as counts per million reads (CPM).
Participant 4 reportedly engaged in unprotected vaginal, oral, and anal sex throughout the study but also reported the use of condoms twice during the entire study. Participants 3 and 4 observed transitions between a BV state and intermediate state; the relative abundance of Gardnerella spp. increased as the study progressed. Gardnerella spp. consistently dominated the vaginal microbiota despite fluctuations in the relative abundance in the cases of consistent BV.
Participant 5 responded to treatment and had a healthy vaginal microbiota at month two but transitioned to BV at month eight, followed by intermediate BV at month 12. This participant consistently used condoms and had received Depo-Provera shots as forms of birth control throughout the study. She did not report oral, anal, or vaginal sex without a condom. Additionally, she had no new sex partners and had only one sex partner throughout the study period. This participant experienced the only instance of a healthy vaginal microbiota, which was dominated by L. crispatus . Markedly decreased levels of Gardnerella spp. were present in the healthy vaginal microbiota compared to intermediate and BV states. Once this participant transitioned from a healthy state to BV, the relative abundance of Gardnerella spp. increased and subsequently decreased after a transition from BV to the intermediate stage.
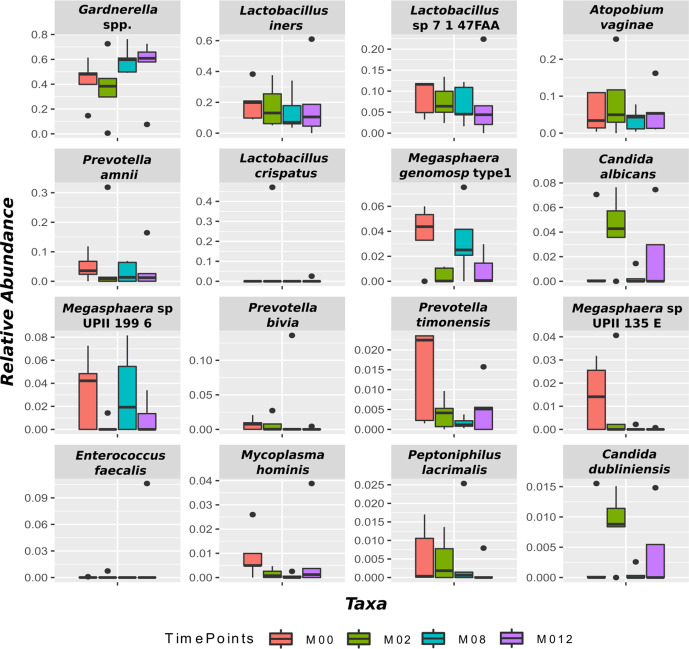
Fig. 2 shows the evolution of the most abundant bacterial species. We found the most abundant species associated with BV were Gardnerella spp., Atopobium vaginae , Prevotella amnii, and Prevotella bivia. Gardnerella spp. increased from baseline over time. The relative abundance of Prevotella bivia and Megasphaera sp. decreased from baseline to the end of the study period. The relative abundance of the remaining bacterial species fluctuated throughout the 12 months. Contrary to expectations, there was an increase in the abundance of Gardnerella spp. over time with MET treatment.
Fig. 2.
Evolution of the most abundant taxa throughout the study. The boxplots show the 16 taxa whose abundance was the highest over all subjects. The different colours represent different time points. None of the changes were statistically significant due to the size of the sample.
Variance in vaginal microbiota
Fig. S1 (available in the online version of this article) illustrates the results of PCA. Based on these results, the attributes (taxa) with maximum variance (Table 1) were determined and investigated further. Consistent with results presented earlier in this section, the vaginal microbiota of Participant 5 (only participant with a healthy vaginal microbiota at any time point) was sufficiently distinct from the rest. Based on the PCA analysis results, Gardnerella spp. and Megasphaera species are strongly associated with BV, while L. iners and L. crispatus were associated with a healthy vaginal microbiota (Table 1). This was also supported by the fact that all Lactobacillus species had a negative contribution in the last column of Table 1 (Principal component 1), while the BV-associated bacteria contributed to BV status.
Table 1.
Species determined by PCA to have maximum variance among five AA women
|
Taxa |
Principal component 1 |
|---|---|
|
Gardnerella spp. |
0.796033 |
|
−0.529284 |
|
|
−0.231106 |
|
|
Lactobacillus sp 7 1 47FAA |
−0.172205 |
|
Megasphaera sp UPII 199 6 |
0.032557 |
|
0.023059 |
Effect of metronidazole treatment
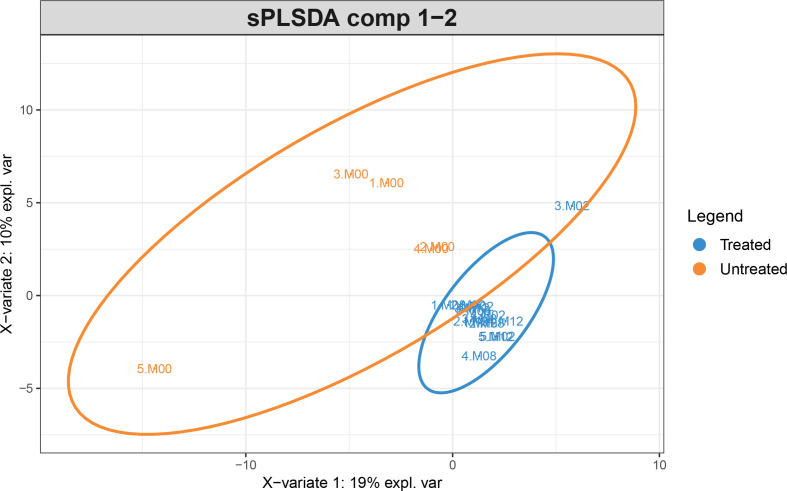
After the sPLS-DA analysis, the subjects’ data were plotted using their projections along with the first and second principal components (Fig. 3). Unlike PCA, sPLS-DA finds the direction that best separates the labelled classes. The samples were classified as untreated at time point 0 (baseline) and treated after receiving the treatment (months 2, 8 and 12). The treated group clusters together tightly, suggesting that treatment somehow ‘homogenizes’ the surviving microbiome. Subject 5 was a clear outlier. sPLS-DA determined that the genera, Peptoniphilus and Anaerococcus, were the most differentially abundant taxa between untreated and treated vaginal samples. However, this finding was likely due to overfitting the model as the Balanced Error Rate was 43%, close to the chance for a two-class classifier. Also, there was no statistically significant difference between treated and untreated samples for any of the taxa because of the small sample size. This argues that sPLS-DA findings should be interpreted with caution.
Fig. 3.
sPLS-DA projection of the samples into the first and second principal components (PC). Treated participants are coloured in blue while untreated ones are coloured in orange. Each sample is represented by X.MYY, where X means the participant number, and YY represents the month of data collection. The treated group clusters together in a tighter way than the heterogeneous untreated group (i.e. participants at baseline).
Resistome profile
The prevalence of resistance genes was analysed against the CARD database [34]. The metagenome of all the participants contained antibiotic resistance genes (Fig. 1d). The 15 most abundant genes analysis shows that tet genes that would confer resistance to tetracycline are most prevalent, with tetM being the most abundant, followed by tetQ. The next most abundant gene, lscA, is associated with clindamycin, lincomycin, dalfopristin, and tiamulin resistance. Analysis for the presence of nim genes related to the MET resistance showed the presence of nimJ in a few of the samples at extremely low levels.
Correlational studies
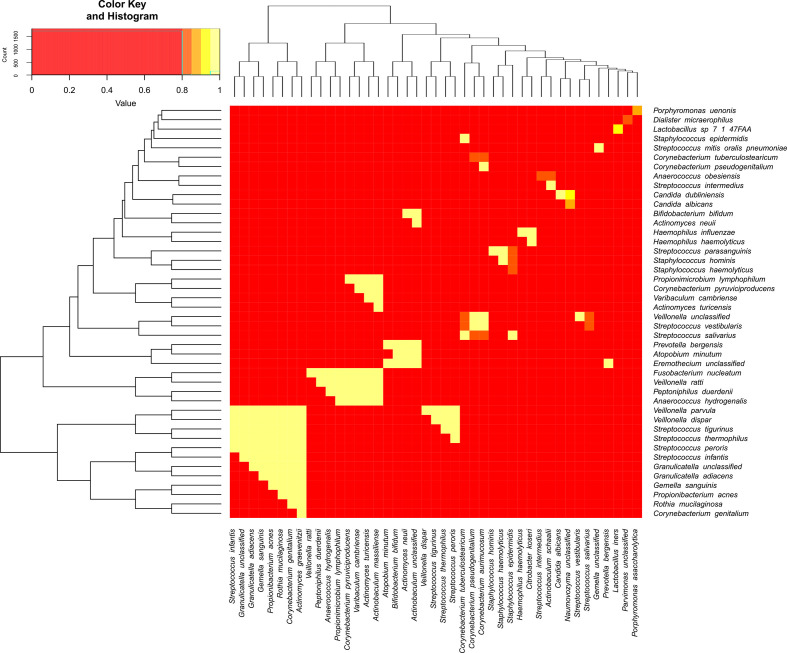
Fig. 4 shows the pairwise correlations between the taxa with the correlations displayed as a heatmap. Each row represents a different taxon. Rows and columns were ordered to depict the hierarchical clustering (obtained by iteratively grouping the most similar groups) shown as a dendrogram on the side of the heatmap. For readability, included only the strongest Spearman correlations (excluding self-correlations). A threshold of 0.8 (-0.8, resp.) was used for filtering positive (negative, resp.) correlation values. There were no correlations lower than −0.8. While these are not displayed in the figure, we note that the five strongest negative correlations were for the following taxa pairs: L. iners and Gardnerella spp. (−0.74), L. coleohominis and Prevotella amnii (−0.69); N. unclassified and Megasphaera sp. Type 1 (-0.70); C. dubliniensis and Megasphaera sp. Type 1 (-0.64); Lactobacillus sp. 7 47FAA and Gardnerella spp. (−0.62).
Fig. 4.
Top Spearman correlations for taxa pairs. Each interaction between a row and a column in this heatmap represents the Spearman correlation between the two taxa involved. Rows and columns are the different taxa ordered according to hierarchical clustering. For readability, only the top correlations were selected. Yellow colours indicate correlations closer to 1, while red colours imply lower correlations.
Streptococcus spp. and Staphylococcus spp. were highly correlated with many other species throughout the sample (Fig. 4). Streptococcus spp. was highly positively correlated with Staphylococcus spp. and Veillonella spp. Furthermore, Prevotella bergensis was highly positively correlated with Atopobium minutum , Bifidobacterium bifidum , Actinomyces neuii, and Actinobaculum unclassified. Of note is the correlation between Haemophilus haemolyticus and Streptococcus spp. as well as Haemophilus haemolyticus and Staphylococcus spp.
Table 2 shows the top Spearman correlations between taxa abundance and the NS. There was just one episode of a low NS (healthy vaginal microbiota), four episodes with intermediate scores, and 14 episodes with high NSs (BV). Even though the data were small and skewed, the strongest correlations highlight the NSs underlying logic. All Lactobacilli spp. were associated with a low NS. In other words, as the abundance of Lactobacilli spp. increased, the NSs tend to decrease (negative association). The abundance of BV-associated bacteria such as Mycoplasma , Prevotella , Megasphaera, and Porphyromonas were correlated with high NSs. Surprisingly, L. iners and Gardnerella spp. were absent from this top list, with Spearman correlation values in the range −0.35 through 0.34.
Table 2.
Spearman correlation between taxa and the Nugent score
|
Taxa name |
Spearman correlation |
|---|---|
|
−0.6347917188 |
|
|
−0.5947203609 |
|
|
−0.5345614474 |
|
|
Candida lusitaniae |
−0.5345614474 |
|
0.5236533323 |
|
|
0.5041102652 s |
|
|
−0.5000000000 |
|
|
Megasphaera sp. type 1 |
0.4949254787 |
|
0.4084817754 |
|
|
−0.3983923509 |
Discussion
Due to the study’s pilot nature, the limited sample size, findings should be interpreted with caution and should not be generalized. Since this study depended on the participant self-reporting, one cannot rule out the participants' failure to adhere to the antibiotic regimen. Another confounding factor might be when the samples were collected; even though specific guidelines were provided for the sample collection, we cannot guarantee it was the same for each participant. With these caveats, three important observations are noted in asymptomatic BV patients treated with MET – (i) specific changes in the vaginal microbiota with treatment, (ii) recurrence and persistence of BV despite treatment, (iii) presence of resistance genes.
According to previous studies, effective treatment of BV resulted in a decrease of the relative abundance of facultative and strict anaerobes in the vaginal microbiota accompanied by an increase in the relative abundance of Lactobacillus spp. [2, 39]. However, the current study showed a decrease over time in the relative abundance of Lactobacillus spp. Similar findings were observed in a study conducted by Hummelen et al. [40] with a Tanzanian population. The MET treatment decreased species diversity in their study, but did not shift it to a BV-associated vaginal microbial profile dominated by a Lactobacillus spp. [40]. The characteristic absence of L. jensenii among African populations compared to non-African could account for this observation [41].
Throughout the study, there was an observed increase in Gardnerella spp. for participants that did not maintain healthy vaginal flora. This increase may be due to the ineffectiveness of MET as a treatment option or increased Gardnerella spp. resistance. Xioa and colleagues reported findings that indicated Gardnerella spp. might be resistant to MET since the treatment exerts no influence on the bacteria at the molecular level [2]. Deng and colleagues also presented findings highlighting MET resistance in Gardnerella spp. [42]. Our resistome analyses demonstrated the presence of nimJ associated with MET resistance in few participants, albeit at extremely low level [43]. Though the nim alleles are widespread in both Gram-positive and -negative genera of aerobic and anaerobic bacteria and archaea, their presence in Gardnerella spp. has not been reported level [43]. Thus, it is not possible to infer if the presence of nimJ actually contributed to any resistance. In addition, majority of the samples harboured tetracycline resistance genes. The most abundant of the tet gene tetM has been associated with Gardnerella spp. [44]; and the gene lsaC associated with clindamycin resistance. Clindamycin, a lincosomide, has been used as an alternate to MET for BV treatment [10]. The participants of this study are likely to respond poorly to clindamycin.
At baseline, the only participant (Participant 5) with a relatively healthy vaginal microbiota had the lowest level of Gardnerella spp. which may have influenced treatment efficacy. This is also supported by the presence lowest level of tet genes. Furthermore, at baseline, all participants were classified as CST IV (Fig. 1), except for Participant 5, who was classified as CST III. Brooks and colleagues have determined that the ability to transition to another CST is determined by the current CST [45]. The difference in baseline CST could have facilitated the observed transition of Participant 5 to a healthy vaginal microbiota. Also, CST III is dominated by L. iners [45]. There was an inverse relationship between the abundance of Gardnerella spp. and L. iners, which was only observed for Participant 5 with a healthy vaginal microbiota (Fig. 1), another factor that could have potentially facilitated the transition to a healthy vaginal microbiota for this participant. It has also been determined that L. iners is the dominant species posttreatment [15]. Participant 5 was also the only participant to report the use of hormonal birth control, which has been shown to reduce the risk of BV [46]. The results of PCA and sPLS-DA further highlighted the outlier status of Participant 5 when compared to the other participants.
Gardnerella spp., L. iners, and L. crispatus were determined by PCA to contribute to maximum variance among the samples. The relative abundance of taxa was not homogeneous for all participants. This heterogeneity could have resulted from the use of different birth control methods and differences in sexual practices. The results of the Spearman correlation between taxa abundance and NSs were consistent with current literature [47]. The Lactobacillus species, L. crispatus, L. gasseri, and L. jensenii, were shown to be inversely associated with the NS, which suggests that they promote a healthy vaginal microbiota. Conversely, the relative abundance of Mobilincus hominis increased with the increase in NSs. Though Lactobacillus spp. promotes a healthy vaginal microbiota; not all Lactobacilli appeared to be protective. L. crispatus has been shown to protect against BV whereas L. iners has been found to be associated with BV [14]. Our studies showed a correlation between Haemophilus haemolyticus and Streptococcus spp. as well as Haemophilus haemolyticus and Staphylococcus spp. This observation is interesting because when BV was first identified, it was believed that the causative agent was Haemophilus vaginalis [21]. These three species appear highly correlated with many other species in the vaginal microbiota.
This study suggests MET may not be an effective course of treatment for women with a low relative abundance of Lactobacillus spp. and maybe more effective when the vaginal microbiota is dominated by L. iners instead of L. jensenii . Additionally, in cases of MET treatment, there is a high probability of long-term recurrence. The findings presented in this study warrant further research. For future research, the vaginal microbiota should be examined at shorter time intervals since the changes that occur are dynamic. Daily assessments may provide more useful information than using 4 month intervals. In addition to whole-genome sequencing, metabolomics should be conducted to determine whether BV may result from a specific pathogen, its metabolites, or a combination of the pathogen and its metabolites. Finally, research should also consider the contributing factors of a shift from a BV-associated vaginal microbial profile one that is dominated by Lactobacillus spp. after treatment with MET. Moreover, the resistome profile would better inform the treatment options.
Data availability
All sequence data generated in this study were deposited in NCBI under BioProject number: PRJNA669294 with Accession numbers SRR12830910 through SRR12830929.
Supplementary Data
Funding information
The parent study was funded by the Division of Microbiology and Infectious Diseases of the NIAID (contract HHSN26620040073C). The study, D.R.P., M.C., K.M., G.N., and P.M. were supported by NIH grant 1R15AI128714-01. D.R.P. was funded by the Dissertation Year Fellowship from Florida International University (FIU). M.C. was funded by a Research Assistantship from FIU.
Author contributions
The experiments were conceived and designed by K.K., H.K., K.M., G.N., P.M. The samples were processed by D.R.P., M.C., B.C., V.S. and H.K. and the experiments were performed by D.R.P. and M.C. Critical analyses of the data were done by D.R.P., M.C., V.S., K.M., G.N., P.M. The manuscript was prepared by D.R.P., M.C., K.M., G.N., P.M. Finally, all the authors were involved in the critical review of the paper.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The BRAVO study was registered with clinicaltrials.gov (clinical trials registration: NCT00667368). All sites included in the BRAVO study received Institutional Review Board (IRB) approval from their respective institutions. The current study was approved by the FIU IRB (IRB protocol number: IRB-17–0021-CR01).
Footnotes
Abbreviations: AA, African American; BV, bacterial vaginosis; CARD, comprehensive antibiotic resistance database; CST, community state type; CT, Chlamydia trachomatis; IUD, intra-uterine device; MET, metronidazole; NG, Neisseria gonorrhoeae; NS, Nugent score; PCA, principal component analysis; sPLS-DA, sparse partial least squares discriminant analysis; STI, sexually transmitted infection; TV, Trichomonas vaginalis; WGS, whole-genome sequencing.
One supplementary figure is available with the online version of this article.
References
- 1.Ferreira CST, Donders GG, Parada CMGdeL, Tristão AdaR, Fernandes T, et al. Treatment failure of bacterial vaginosis is not associated with higher loads of Atopobium vaginae and Gardnerella vaginalis . J Med Microbiol. 2017;66:1217–1224. doi: 10.1099/jmm.0.000561. [DOI] [PubMed] [Google Scholar]
- 2.Xiao B, Niu X, Han N, Wang B, Du P, et al. Predictive value of the composition of the vaginal microbiota in bacterial vaginosis, a dynamic study to identify recurrence-related flora. Sci Rep. 2016;6:26674. doi: 10.1038/srep26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ravel J, Brotman RM, Gajer P, Ma B, Nandy M, et al. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome. 2013;1:29. doi: 10.1186/2049-2618-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith WL, Hedges SR, Mordechai E, Adelson ME, Trama JP, et al. Cervical and vaginal flora specimens are highly concordant with respect to bacterial vaginosis-associated organisms and commensal Lactobacillus species in women of reproductive age. J Clin Microbiol. 2014;52:3078–3081. doi: 10.1128/JCM.00795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koumans EH, Sternberg M, Bruce C, McQuillan G, Kendrick J, et al. The prevalence of bacterial vaginosis in the United States, 2001-2004; associations with symptoms, sexual behaviors, and reproductive health. Sex Transm Dis. 2007;34:864–869. doi: 10.1097/OLQ.0b013e318074e565. [DOI] [PubMed] [Google Scholar]
- 6.Brotman RM. Vaginal microbiome and sexually transmitted infections: an epidemiologic perspective. J Clin Invest. 2011;121:4610–4617. doi: 10.1172/JCI57172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allsworth JE, Lewis VA, Peipert JF. Viral sexually transmitted infections and bacterial vaginosis: 2001-2004 National health and nutrition examination survey data. Sex Transm Dis. 2008;35:791–796. doi: 10.1097/OLQ.0b013e3181788301. [DOI] [PubMed] [Google Scholar]
- 8.Brotman RM, Shardell MD, Gajer P, Tracy JK, Zenilman JM, et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J Infect Dis. 2014;210:1723–1733. doi: 10.1093/infdis/jiu330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bautista CT, Wurapa E, Sateren WB, Morris S, Hollingsworth B, et al. Bacterial vaginosis: a synthesis of the literature on etiology, prevalence, risk factors, and relationship with Chlamydia and gonorrhea infections. Mil Med Res. 2016;3:4. doi: 10.1186/s40779-016-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coudray MS, Madhivanan P. Bacterial vaginosis-A brief synopsis of the literature. Eur J Obstet Gynecol Reprod Biol. 2020;245:143–148. doi: 10.1016/j.ejogrb.2019.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muzny CA, Sunesara IR, Kumar R, Mena LA, Griswold ME, et al. Characterization of the vaginal microbiota among sexual risk behavior groups of women with bacterial vaginosis. PLoS One. 2013;8:e80254. doi: 10.1371/journal.pone.0080254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balkus JE, Srinivasan S, Anzala O, Kimani J, Andac C, et al. Impact of periodic presumptive treatment for bacterial vaginosis on the vaginal microbiome among women participating in the preventing vaginal infections trial. J Infect Dis. 2017;215:723–731. doi: 10.1093/infdis/jiw622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitali B, Cruciani F, Picone G, Parolin C, Donders G, et al. Vaginal microbiome and metabolome highlight specific signatures of bacterial vaginosis. Eur J Clin Microbiol Infect Dis. 2015;34:2367–2376. doi: 10.1007/s10096-015-2490-y. [DOI] [PubMed] [Google Scholar]
- 14.Crucitti T, Hardy L, van de Wijgert J, Agaba S, Buyze J, et al. Contraceptive rings promote vaginal lactobacilli in a high bacterial vaginosis prevalence population: a randomised, open-label longitudinal study in Rwandan women. PLoS One. 2018;13:e0201003. doi: 10.1371/journal.pone.0201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert JA, John S, Sobel JD, Akins RA. Longitudinal analysis of vaginal microbiome dynamics in women with recurrent bacterial vaginosis: recognition of the conversion process. PLoS One. 2013;8:e82599. doi: 10.1371/journal.pone.0082599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammadzadeh F, Dolatian M, Jorjani M, Alavi Majd H. Diagnostic value of Amsel's clinical criteria for diagnosis of bacterial vaginosis. Glob J Health Sci. 2014;7:8–14. doi: 10.5539/gjhs.v7n3p8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amegashie CP, Gilbert NM, Peipert JF, Allsworth JE, Lewis WG, et al. Relationship between Nugent score and vaginal epithelial exfoliation. PLoS One. 2017;12:e0177797. doi: 10.1371/journal.pone.0177797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UME, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwebke JR, Lee JY, Lensing S, Philip SS, Wiesenfeld HC, et al. Home screening for bacterial vaginosis to prevent sexually transmitted diseases. Clin Infect Dis. 2016;62:531–536. doi: 10.1093/cid/civ975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 22.Wickham H. The Split-Apply-Combine strategy for data analysis. J Stat Softw. 2011;1:1–29. [Google Scholar]
- 23.Harrell Jr F Package ‘Hmisc’. 2018 https://hbiostat.org/R/Hmisc/
- 24.Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P. Vegan: community ecology package. R package version 2.5-3. https://cran.rproject.org/web/packages/vegan/index.html2018 . 2018
- 25.Somerfield PJ, Clarke KR, Warwick RM. Simpson index. Encyclopedia Ecol. 2008:3252–3255. [Google Scholar]
- 26.Schober P, Boer C, Schwarte LA. Correlation coefficients: appropriate use and interpretation. Anesth Analg. 2018;126:1763–1768. doi: 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez M, Riveros JD, Campos M, Mathee K, Narasimhan G. Microbial "social networks". BMC Genomics. 2015;16 Suppl 11:S6–13. doi: 10.1186/1471-2164-16-S11-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahalanobis PC. On the generalised distance in statistics. Proc Natl Inst Sci India. 1936;2:49–55. [Google Scholar]
- 29.Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 13:e10057522017. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch BL. The generalisation of student's problems when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 31.Haynes W. Benjamini–hochberg method. In: Dubitzky W, Wolkenhauer O, Cho K-H, Yokota H, editors. Encyclopedia of Systems Biology. New York: Springer; 2013. p. 78. p. [Google Scholar]
- 32.Ruiz-Perez D, Guan H, Madhivanan P, Mathee K, Narasimhan G. So you think you can PLS-DA? BMC Bioinformatics. 2020;21:207225. doi: 10.1186/s12859-019-3310-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stebliankin V, Sazal M, Valdes C, Mathee K, Narasimhan G. Novel approach for microbiome analysis using bacterial replication rates and causal inference with applications. bioRxiv. 2020 [Google Scholar]
- 34.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, et al. Card 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D25. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W. gplots: various R programming tools for plotting data. R package version 3.0.1. https://CRAN.R-project.org/package=gplots2016 . 2016
- 37.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: 2016. [Google Scholar]
- 38.Sievert C plotly for R. https://plotly-book.cpsievert.me . 2018
- 39.Mayer BT, Srinivasan S, Fiedler TL, Marrazzo JM, Fredricks DN, et al. Rapid and profound shifts in the vaginal microbiota following antibiotic treatment for bacterial vaginosis. J Infect Dis. 2015;212:793–802. doi: 10.1093/infdis/jiv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hummelen R, Fernandes AD, Macklaim JM, Dickson RJ, Changalucha J, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One. 2010;5:e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. Isme J. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 42.Deng Z-L, Gottschick C, Bhuju S, Masur C, Abels C, et al. Metatranscriptome analysis of the vaginal microbiota reveals potential mechanisms for protection against metronidazole in bacterial vaginosis. mSphere. 2018;3 doi: 10.1128/mSphereDirect.00262-18. 27 06 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alauzet C, Lozniewski A, Marchandin H. Metronidazole resistance and nim genes in anaerobes: A review. Anaerobe. 2019;55:40–53. doi: 10.1016/j.anaerobe.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Freeman CD, Klutman NE, Metronidazole LKC. A therapeutic review and update. Drugs. 1997;54:679–708. doi: 10.2165/00003495-199754050-00003. [DOI] [PubMed] [Google Scholar]
- 45.Brooks JP, Buck GA, Chen G, Diao L, Edwards DJ, et al. Changes in vaginal community state types reflect major shifts in the microbiome. Microb Ecol Health Dis. 2017;28:1303265. doi: 10.1080/16512235.2017.1303265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madden T, Grentzer JM, Secura GM, Allsworth JE, Peipert JF. Risk of bacterial vaginosis in users of the intrauterine device: a longitudinal study. Sex Transm Dis. 2012;39:217–222. doi: 10.1097/OLQ.0b013e31823e68fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdelmaksoud AA, Koparde VN, Sheth NU, Serrano MG, Glascock AL, et al. Comparison of Lactobacillus crispatus isolates from Lactobacillus-dominated vaginal microbiomes with isolates from microbiomes containing bacterial vaginosis-associated bacteria. Microbiology. 2016;162:466–475. doi: 10.1099/mic.0.000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data generated in this study were deposited in NCBI under BioProject number: PRJNA669294 with Accession numbers SRR12830910 through SRR12830929.