Abstract
Cytochrome P4501 (CYP1) and CYP3A proteins are primarily responsible for the metabolism of 17β-estradiol (E2) in mammals. We have cloned and heterologously expressed CYP1A, CYP1B1, CYP1C1, CYP1C2, CYP1D1, and CYP3A65 from zebrafish (Danio rerio) to determine the CYP-mediated metabolism of E2 in a non-mammalian species. Constructs of each CYP cDNA were created using a leader sequence from the bacterial ompA gene to allow appropriate expression in Escherichia coli without 5′ modification of the gene. Membrane vesicles were purified, and functional CYP protein was verified using carbon monoxide difference spectra and fluorescent catalytic assays with the substrates 7-ethoxyresorufin and 7-benzyloxy-4-(trifluoromethyl)-coumarin. Rates of in vitro E2 metabolism into 4-hydroxyE2 (4-OHE2), 2-hydroxyE2 (2-OHE2), and 16α-hydroxyE1 (16α-OHE1) metabolites were determined by gas chromatography/mass spectrometry. The 2-OHE2 metabolite was produced by all CYPs tested, while 4-OHE2 was only detected following incubation with CYP1A, CYP1B1, CYP1C1, and CYP1C2. The 16α-OHE1 metabolite was only produced by CYP1A. The highest rates of E2 metabolism were from CYP1A and CYP1C1, followed by CYP1C2. CYP1B1, CYP1D1, and CYP3A65 had low rates of E2 metabolism. E2 metabolism by zebrafish CYP1A, CYP1C1, and CYP1C2 produced similar ratios of 4-OHE2 to 2-OHE2 as previous studies with mammalian CYP1As. CYP1B1 formed the highest ratio of 4-OHE2 to 2-OHE2 metabolites. Contrary to mammals, these results suggest that fish CYP1A and CYP1C proteins are primarily responsible for E2 metabolism, with only minor contributions from CYP3A65 and CYP1B1. Similar to mammals, 2-OHE2 is the predominant metabolite from CYP-mediated E2 metabolism in fish, suggesting that all vertebrate species produce the same major E2 metabolite.
Introduction
The major estrogen in vertebrates is 17β-estradiol (E2). The first step in the metabolism of E2 is NADPH-dependant oxidative metabolism catalyzed by hepatic cytochrome P450 (CYP) enzymes (Zhu & Conney 1998). Mammalian CYP enzymes are critical for E2 metabolism, including those belonging to the CYP1A, CYP1B, and CYP3A subfamilies; yet, there are distinct differences in the CYP1 and CYP3 families across vertebrates. The CYP3A subfamily includes multiple genes in mammals; humans have four CYP3As (Qiu et al. 2008). Most fish contain only one or two CYP3As; zebrafish (Danio rerio) have a single CYP3A gene, CYP3A65 (Qiu et al. 2008). The CYP1 family contains more subfamilies in non-mammalian vertebrates. There are two CYP1A genes (CYP1A1 and CYP1A2) in mammals, and a single CYP1A gene in fish, with the exception of some polyploid species such as the salmonids (Goldstone et al. 2007). CYP1B1 is the single CYP1B gene found in all vertebrate lineages. CYP1Cs are found in all non-mammalian vertebrate lineages (Goldstone et al. 2007). Fish have CYP1C1 and CYP1C2 paralogs arising from a gene duplication event (Godard et al. 2005, Goldstone et al. 2007). The CYP1D subfamily, a non-mammalian subfamily which has been recently identified, contains a single CYP1D1 gene (Goldstone et al. 2009). The liver is the major site of E2 metabolism; all CYP1s and CYP3As are known to be expressed in the liver of mammals (Bieche et al. 2007) and fish (Jonsson et al. 2007, Goldstone et al. 2009).
In mammals, the major product of E2 oxidation is the 2-hydroxyE2 (2-OHE2) metabolite, with limited production of 4-hydroxyE2 (4-OHE2) and small levels of metabolites hydroxylated at either the 6β, 16β, or 16α positions (Lee et al. 2001). The regioselective hydroxylation of E2 by purified CYPs has been well studied in mammals (Lee et al. 2003). CYP1A1, CYP1A2, and CYP3A4 show the highest rates of metabolism in humans, mostly as a result of high 2-hydroxylation activity (Lee et al. 2003). Consequently, these three CYPs have very low ratios of 4-OHE2 to 2-OHE2 formation (Lee et al. 2003). On the other hand, E2 metabolism by mammalian CYP1B1 produces similar amounts of 4-OHE2 to CYP1As and CYP3As, but very little 2-OHE2, such that three times more 4-OHE2 is formed in relation to 2-OHE2 (Lee et al. 2003).
The two catecholestrogens, 2-OHE2 and 4-OHE2, are further metabolized by catechol-O-methyltransferase (COMT) into the major urinary estrogen metabolites: 2-methoxyE2 (2-MeOE2) and 4-methoxyE2 (4-MeOE2) respectively (Creveling 2003). However, the catecholestrogens can also be oxidized to form reactive semiquinones and quinines, which can form adducts with purine bases (Dawling et al. 2001, Belous et al. 2007). COMT has a higher affinity for 2-OHE2 over 4-OHE2, resulting in an increased genotoxicity of the 4-OHE2 metabolite (Tsuchiya et al. 2005). High concentrations of 4-OHE2 metabolites have been associated with human breast cancers (Rogan et al. 2003), and elevated CYP1B1 mRNA transcripts have been identified in many cancerous tissues including breast, testis, and ovary (McKay et al. 1995, Murray et al. 1997) suggesting that elevated levels of CYP1B1-mediated E2 metabolism can lead to carcinogenesis. 2-OHE2 metabolites appear to be tumor inhibitors (Tsuchiya et al. 2005).
In fish liver microsomes, the presence of E2 2-hydroxylase activity was identified in channel catfish (Butala et al. 2004), English sole (Stein et al. 1991), scup, and winter flounder (Snowberger & Stegeman 1987), and recently, 4-OH activity was detected in channel catfish (Butala et al. 2004). The only reported E2 metabolism by purified fish CYPs has been from Japanese eel showing 2-OH activity by CYP1A9 but not by CYP1C1; however, neither CYP produced 4-OHE2 metabolites (Uno et al. 2008). Since E2 metabolism predominantly occurs in the liver, and the dominant metabolic products are 2-OHE2 with smaller amounts of 4-OHE2 for both mammals and fish, similar CYP families could be responsible for E2 metabolism in mammalian and non-mammalian vertebrates.
In the present study, we describe our findings on the rates of formation of E2 metabolites by bacterially expressed zebrafish CYPs. Zebrafish CYP1A, CYP1B1, CYP1C1, CYP1C2, CYP1D1, and CYP3A65 were each cloned and expressed in bacteria to produce catalytically active proteins used for in vitro assessment of E2 metabolism. As fish have similar CYP content in the CYP1 and CYP3 families as other non-mammalian vertebrates, our data will serve as a model for non-mammalian vertebrate CYP-mediated E2 metabolism.
Materials and Methods
Cloning of zebrafish CYPs
RNA was extracted with TRIzol reagent (Invitrogen) from pools of liver or gill tissue taken from female wild type zebrafish (D. rerio; n=8). cDNA was synthesized using cloned AMV RT (Invitrogen) and an oligo (dT)20 primer. Zebrafish CYP1A, CYP1C1, CYP1C2, CYP1D1, and CYP3A65 were amplified from liver cDNA, and CYP1B1 was amplified from gill cDNA using Platinum Taq Polymerase (Invitrogen) following the manufacturer’s protocol and using gene-specific primers and annealing temperatures as listed in Table 1. PCR products were gel purified, cloned into pGEM-T Easy vectors (Promega), and transformed into competent Escherichia coli JM109 cells (Promega). Plasmids were purified by QIAprep Spin Miniprep Kits (Qiagen) and sequenced by MobixLab (McMaster University, Hamilton, ON, USA).
Table 1.
Primers and annealing temperatures used for (A) cloning of zebrafish cytochrome P450 (CYPs) and (B) fusion of the ompA(+2) segment to the zebrafish CYPs. All primers are listed in the 5′–3′ direction
| A | Forward primer |
Reverse primer |
Annealing temperature (°C) |
GenBank accession number |
|---|---|---|---|---|
| Construct | ||||
| CYP1A | TGGAGCTAATTGGCACTGGT | CCTGGATTTCAGGAGCTCAA | 53 | NM_131879 |
| CYP1B1 | CCACCCGAACTCTGAAACTC | GTTTTCTTAGCCGCCTTCATTT | 53 | NM_001013267 |
| CYP1C1 | GGAGGCTGAGTTTGGACTGA | CCCATTCGACTGGATGTTTT | 53 | NM_001020610 |
| CYP1C2 | TGAGCCATCCTCCGGTAA | GCAGTGGGTTAGACAGCACA | 53 | XM_686678 |
| CYP1D1 | GCGATTTGCCAACACTGATA | TGCCAACATTAGCTTGATGC | 54 | NM_001007310 |
| CYP3A65 | GGAGCTTCATCATCTTCAGCA | CCTCCTCCTCCTCCTCAGAC | 53 | NM_001020797 |
| B | Reverse primer | Linker primer | Annealing temperature (°C)b |
|---|---|---|---|
| Constructa | |||
| ompA-CYP1A | TCTAGAGCGGCCGCGAATTCACTAc | TGGAAGAATAGCCAGAGCCATXXXd | 50, 55 |
| ompA-CYP1B1 | TCTAGAGCGGCCGCGAATTCACTA | AGCCAGCAGGACATCCATCATXXX | 50, 55 |
| ompA-CYP1C1 | TCTAGAGCGGCCGCGAATTCACTA | CTGTCCGCTCCATTCCCGCATXXX | 50, 55 |
| ompA-CYP1C2 | TCTAGAGCAGTGGGTTAGACAGCACA | CTCTGAATCCGACTGCGCCATXXX | 50, 55 |
| ompA-CYP1D1 | TCTAGATGGAACACTGAGAGAATGATGG | GGAGATATTCTCAAGATTCATXXX | 55, 53 |
| ompA-CYP3A65 | TCTAGAGCGGCCGCGAATTCACTA | TGTTTCTGCCGAGAAGAACATXXX | 50, 55 |
The forward primer used for ompA(+2)-CYP fusions was always 5′-GGAATTCCATATGAAAAAGACAGCTATCGCG-3′ with an NdeI restriction site underlined.
The first temperature was used to anneal the forward and linker primer to the ompA(+2) template during step 1 of the PCR. The second annealing temperature is used during the second step of PCR to attach the ompA(+2) segment to the full-length CYP using the forward and reverse primers. See Materials and Methods section for details.
The XbaI restriction site is underlined.
XXX represents 5′-CGGGACGGCCTGCGCTACGGTAGCGA-3′, which primes against the 3′ end of the ompA(+2) segment.
To facilitate proper membrane targeting of expressed CYPs in bacteria, an ompA(+2) sequence consisting of 69 nucleotides was attached in frame with the CYP start site by PCR using the methods of Pritchard et al. (2006). The source for the ompA(+2) was purified genomic JM109 DNA for CYP1A, CYP1B1, CYP1C1, and CYP3A65. The ompA-CYP1A and ompA-CYP1C1 constructs were used as ompA templates for attachment to CYP1C2 and CYP1D1 respectively. The ompA(+2) sequence was attached using two separate PCRs using a high fidelity polymerase, Accuprime Pfx (Invitrogen). The first PCR used a forward primer against the ompA segment and a linker primer that primed against the first 21 bases of the CYP and the last 21 bases of the ompA segment (Table 1). This generated a ~90 nucleotide fragment containing the first 21 bases of the appropriate CYP gene, and was gel purified and used in a second PCR with the same forward primer and a CYP-specific reverse primer against the 3′ end (Table 1) of the zebrafish CYP cloned gene. The forward and reverse primers contained NdeI and XbaI restriction sites respectively. PCR fragments containing the complete CYP gene and the ompA(+2) sequence were digested, gel purified, ligated into the pCW vector, and co-transfected with the pACYC vector containing human NADPH–CYP reductase (CPR) into JM109 cells. Final sequences were confirmed by sequencing prior to expression.
Expression and purification of zebrafish CYPs
Overnight cultures of E. coli were diluted 1:100 into TB with ampicillin (50 μg/ml) and chloramphenicol (25 μg/ml) and shaken at 30 °C and 200 r.p.m. Isopropyl β-d-1-thiogalacto-pyranoside (1 mM; Fisher Scientific, Pittsburgh, PA, USA) was added when cultures reached an OD600 of between 0·7 and 1·0. Expression was optimized with the addition of 0–1 mM δ-aminolevulinic acid (Ala; MP Biomedicals, Solon, OH, USA) based on maximizing the catalytic activity of purified membranes (see Catalytic assays). Ala was added to a final concentration of 0·1 mM for all CYP constructs except CYP1A and CYP1B1, where Ala was added at 0·5 and 1·0 mM respectively. Expression was allowed to proceed for another 20–24 h. Cells were harvested, and bacterial membranes were purified using previously published methods (Pritchard et al. 2006). Total protein content was measured using a bicinchoninic acid assay kit (Pierce protein research products) according to the manufacturer’s protocols (Smith et al. 1985).
Total P450 analysis and cytochrome c activity
Total P450 content was measured by diluting membranes into P450 spectrum buffer (Pritchard et al. 1997) and measuring the carbon monoxide (CO) difference spectra using the peak absorbance difference between 450 and 490 nm and an extinction coefficient of 92/mM/cm (Omura & Sato 1964). Cytochrome c reductase activity of membranes was determined by measuring the absorbance change at 450 nm of a reaction mixture containing 1 mg/ml of cytochrome c and 0·4 mM NADH in 0·2 M potassium phosphate buffer (pH 7·7) at 37 °C using an extinction coefficient of 21·1/mM per cm (Massey 1959).
Catalytic assays
Ethoxyresorufin-O-deethylase activity was measured at 30 °C using methods from Hahn et al. (1993). The rate of conversion of 7-benzyloxy-4-(trifluoromethyl)coumarin (BFC; BD Biosciences, San Jose, CA, USA) to 7-hydroxy-4-(trifluoromethyl)coumarin by purified membranes was measured in the presence of 1 mM BFC and 1·33 mM NADPH in 0·5 M potassium phosphate buffer (pH 7·4) at 30 °C, as optimized from Crespi & Stresser (2000). Catalytic assays containing cytochrome b5 (Calbiochem, EMD Chemicals Inc., Gibbstown, NJ, USA) had 50 pmol added. All catalytic assays were normalized for total P450 in the reaction mixture. Optimization of CYP expression was based on maximizing 7-ethoxyresorufin (7-ER; CYP1A, 1B1, 1C1, 1C2, 1D1) or BFC (CYP3A65) metabolism with assays normalized for total protein.
Zebrafish liver microsomal preparation
Zebrafish (D. rerio) were purchased from DAP International (Etobicoke, ON, Canada). Fish were kept in a semi-recirculating system at 28–30 °C and fed three times per day, alternating between tropical flake food and fresh brine shrimp (Artemia sp.). Adult zebrafish were removed from our regular breeding stock, and livers were collected and pooled (male n=57; female n=38) and flash frozen in liquid nitrogen. Fish were ~1 year of age at collection, and were still actively breeding. Microsomal fractions were collected according to Stegeman et al. (1979). All samples were kept at −80 °C until use. Total protein concentrations were determined, and the remaining microsomal pools were used to determine hepatic E2 metabolism.
E2 metabolism
E2 (Sigma–Aldrich) was incubated with 0·5 mg membrane or microsomal protein at 28 °C for 2 h according to Spink et al. (1990) with and without cytochrome b5 (100 pmol). The incubation time was chosen based on data collected during the optimization of the assay for studies with fish (Butala et al. 2004). Reactions were terminated with 20 mM ascorbic acid (Sigma–Aldrich); the deuterated estradiol internal standard was added, and metabolites were extracted according to Butala et al. (2004). The samples were derivatized with N,O-bis(trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane (Sigma–Aldrich), evaporated under N2 and resuspended in iso-octane (Sigma–Aldrich) prior to analysis. Samples were analyzed for the presence of estrone (E1), estriol, E2, 4-OHE2, 2-OHE2, 4-MeOE2, 2-MeOE2, 16α-hydroxyE1 (16α-OHE1), and the deuterated standard by GC/MS according to Butala et al. (2004). All samples were run in duplicate.
Materials
NADPH, leupeptin, aprotonin, phenylmethanesulphonylfluoride, and cytochrome c were purchased from Sigma–Aldrich. Ampicillin and chloramphenicol were purchased from Fisher Scientific. The deuterated estradiol internal standard was prepared as described in Dehennin et al. (1980) and kindly provided by Dr John Rimoldi (University of Mississippi). The pCW vector (Muchmore et al. 1989) was a kind gift from Dr FW Dahlquist (University of California Santa Barbara, Santa Barbara, CA, USA), and the pACYC expression vector containing human CPR was a kind gift from Dr T Friedberg (University of Dundee, Dundee, UK).
Results
Expression of zebrafish CYPs
The CO difference spectra were analyzed for each expressed CYP (Fig. 1), and peaks were seen at 420 and 450 nm, indicating the presence of significant amounts of both inactive and active P450. Total P450 content was between 0·139 and 0·655 nmol/mg protein (Table 2). Cytochrome c activity was measured to determine the levels of NADPH–CPR in each sample. There were similar levels of reductase activity in each sample, except for cells expressing human CPR only which showed approximately two- to three-fold higher activity than those co-expressed with a CYP gene (Table 2).
Figure 1.
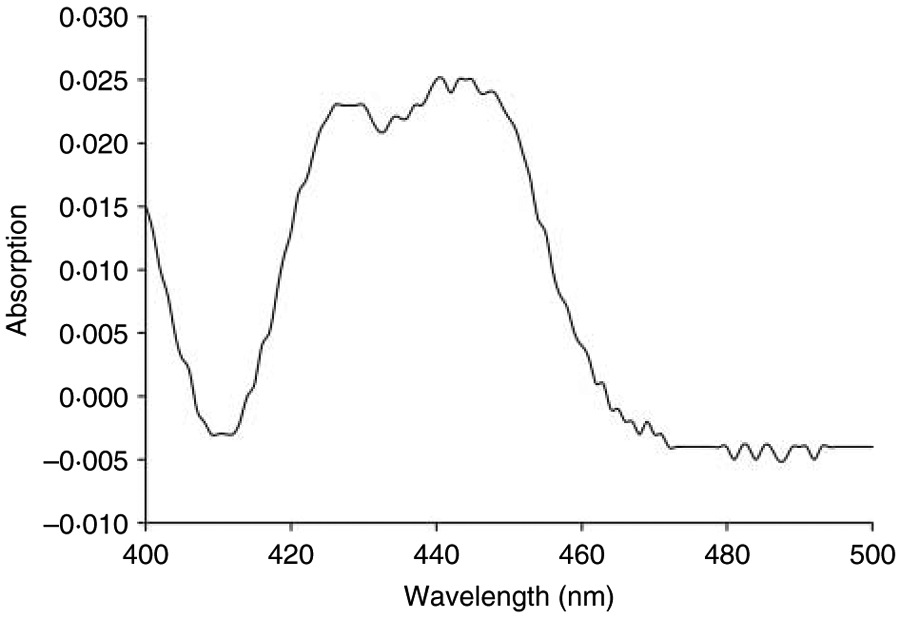
Representative CO difference spectrum of recombinant zebrafish CYP purified membrane vesicles. Example shown is for membrane vesicles containing CYP1C2.
Table 2.
Total protein, P450 content, and cytochrome c activity in membrane vesicles purified from bacteria co-expressing zebrafish cytochrome P450 (CYP) genes and human NADPH–CYP reductase (CPR), or expressing CPR alone. Cytochrome c activity expressed as mean activity ±s.e.m. of reactions done in triplicate
| Total protein (mg/ml) |
Total P450 content (nmol/mg) |
Cytochrome c activity (nmol/mg per min) |
|
|---|---|---|---|
| Construct | |||
| CPRa | 11·65 | –b | 29·68±3·2 |
| CYP1A | 16·42 | 0·655 | 16·77±2·1 |
| CYP1B1 | 19·20 | 0·278 | 16·27±0·68 |
| CYP1C1 | 22·25 | 0·395 | 13·93±0·37 |
| CYP1C2 | 20·50 | 0·473 | 17·52±1·0 |
| CYP1D1 | 22·77 | 0·139 | 15·50±0·11 |
| CYP3A65 | 17·04 | 0·282 | 18·74±0·43 |
Human NADPH–cytochrome P450 reductase.
– Total P450 content not determined.
The catalytic activity of each recombinant protein was assessed using 7-ER and BFC as substrates to verify that the proteins were functionally active. All CYP1s showed catalytic activity for 7-ER and BFC except for CYP1D1 and CYP3A65, which showed no metabolism of BFC and 7-ER respectively (Fig. 2). CYP1D1 showed detectable but low catalytic activity for a broad range of other substrates tested (unpublished data). The addition of cytochrome b5 significantly increased the ability of CYP1A, CYP1B1, CYP1C1, and CYP1C2 to metabolize 7-ER. However, cytochrome b5 did not affect the metabolism of 7-ER by CYP1D1 or the metabolism of BFC by CYP3A65 (data not shown).
Figure 2.
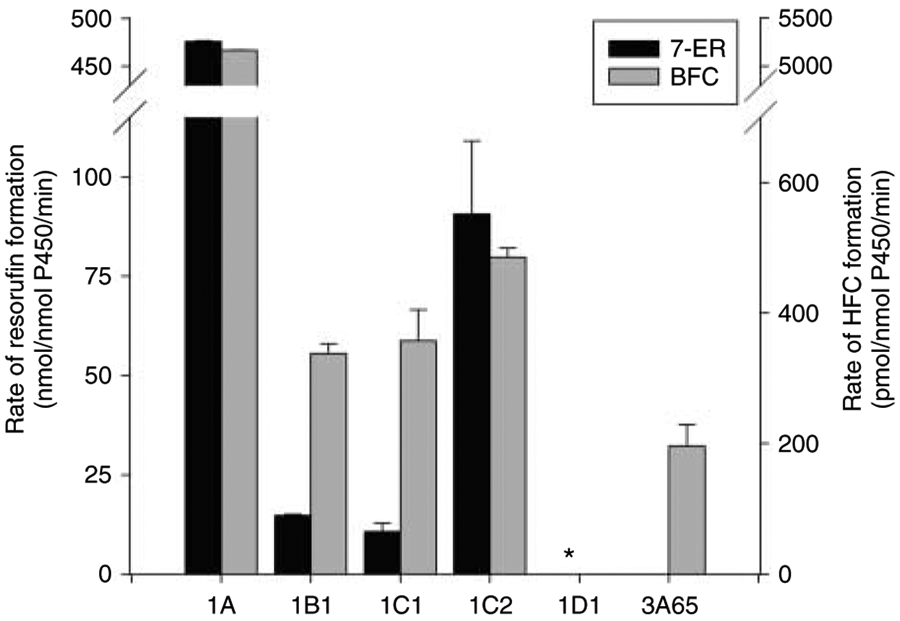
Catalytic activity of recombinant zebrafish CYPs co-expressed with human NADPH–CYP reductase. Data are shown for the metabolism of 7-ethoxyresorufin (7-ER) and 7-benzyloxy-4-(trifluoromethyl)coumarin (BFC), typical CYP1 and CYP3A substrates in mammals (Lake et al. 2009) and fish (Kashiwada et al. 2005). 7-ER and BFC are metabolized to resorufin and HFC respectively. Results are mean±s.e.m., for duplicate reactions. Catalytic activity was below the limits of detection for CYP3A65 (7-ER) and CYP1D1 (BFC). Values represent Mean±s.e.m. of reactions done in duplicate. *Resorufin was detected in CYP1D1 at a rate of 24±5 pmol/nmol P450/min.
E2 metabolism
E2 metabolism data are expressed as mean±s.e.m. of reactions done in duplicate. There was no 2-MeOE2 or 4-MeOE2 detected in any of the samples, and no E2 metabolites were detected in reactions with the control membranes (expressing CPR only). Since E. coli do not express COMT, the enzyme responsible for the methylation of hydroxylated E2 metabolites, the measurement of 2-MeOE2 and 4-MeOE2 was additional negative controls for our assay. 16α-OHE1 was only detected in reactions with CYP1A at a rate of 6·8±1·6 pmol/nmol P450 per min.
CYP1A and CYP1C1 showed the highest rates of E2 metabolism and a strong preference for 2-OHE2 over 4-OHE2 (Table 3). CYP1C2 metabolized moderately less 2-OHE2, and 4-OHE2, than CYP1A and CYP1C1, but metabolism at the 2 position was highest (Table 3). All CYP1A, CYP1C1, and CYP1C2 had percent ratios of 4-OHE2/2-OHE2 between 3 and 8. CYP1B1, CYP1D1, and CYP3A65 had the lowest rates of E2 metabolism in general (<40 pmol/nmol P450 per min), and 2-OHE2 formation predominated. CYP1B1 had the highest proportion of 4-OHE2 production (4-OHE2/2-OHE2=32%) of all the CYPs tested, but still formed twice as much 2-OHE2 than 4-OHE2. There was no 4-OHE2 produced by CYP1D1 or CYP3A65 without cytochrome b5.
Table 3.
Rate of formation of 2-hydroxyestradiol (2-OHE2) and 4-hydroxyestradiol (4-OHE2) by heterologously expressed zebrafish with and without cytochrome b5. Data from human and eel are from published studies for comparison with the zebrafish data
| 2-OHE2 (pmol/nmol P450/min) |
4-OHE2 (pmol/nmol P450/min) |
Ratio of 4-OHE2/ 2-OHE2 (%) |
|
|---|---|---|---|
| CYP | |||
| Zebrafisha | |||
| CYP1A | 425·2±85·0 | 21·8±5·4 | 5 |
| CYP1A+b5 | 892·6±81·1 | NDb | NAc |
| CYP1B1 | 27·1±10·7 | 8·8±5·7 | 32 |
| CYP1B1+b5 | 18·0±0·1 | 4·7±0·1 | 26 |
| CYP1C1 | 510·0±1·1 | 40·9±3·9 | 8 |
| CYP1C1+b5 | 1083±33 | 35·2±0·7 | 3 |
| CYP1C2 | 155·4±1·5 | 4·2±0·2 | 3 |
| CYP1C2+b5 | 57·0±1·4 | 1·8±0·3 | 3 |
| CYP1D1 | 31·4±1·9 | ND | NA |
| CYP3A65 | 25·6±5·3 | ND | NA |
| CYP3A65+b5 | 23·0 | 3·2 | 14 |
| Humand | |||
| CYP1A1 | 2523±208 | 163±22 | 7 |
| CYP1A2 | 4065±156 | 343±24 | 9 |
| CYP1B1 | 108±3 | 371±16 | 344 |
| CYP3A4 | 355±41 | 78±11 | 22 |
| CYP3A4+b5 | 3093±91 | 497±27 | 16 |
| CYP3A5 | 125±22 | 67±5 | 53 |
| CYP3A7+b5 | 146±17 | 55±3 | 58 |
| Japanese eele | |||
| CYP1A9 | 177 | ND | NA |
| CYP1C1 | ND | ND | NA |
Data from this study, using 50 μM E2 at 28 °C for 2 h; values represent mean±s.e.m. of duplicate reactions.
ND – metabolite not detected.
NA – not applicable; one or both metabolites not detected.
Human data from Lee et al. (2003), using 20 μM E2 at 37 °C for 20 min; values represent mean±s.d. of triplicate reactions.
Japanese eel data calculated based on Uno et al. (2008), using 100 μM E2, at 37 °C assuming constant rate over 8-h reaction.
Addition of cytochrome b5 approximately doubled the rate of formation of 2-OHE2 by CYP1A and CYP1C1, but reduced the amount of 4-OHE2 produced (Table 3). Cytochrome b5 decreased the rate of E2 metabolism by CYP1B1 and CYP1C2, while metabolism by CYP3A65 was not affected. Small amounts of estrone were produced, at rates below 3·0 pmol/nmol P450/min, by CYP1A, CYP1B1, CYP1C1, and CYP1C2 with cytochrome b5, but CYP3A65 with cytochrome b5 produced estrone at a rate of 8·2±5·3 pmol/nmol P450/min. Estriol was detected only in samples with CYP1C1 or CYP1C2 that included cytochrome b5. Estriol was produced at a rate of 223±19 pmol/nmol P450/min by CYP1C1 and 7±0·24 pmol/nmol P450/min by CYP1C2.
Zebrafish liver microsomes produced low, but detectable amounts of 2-OHE2 (Table 4) and estrone. Only one replicate reaction of each produced 2-OHE2, while both replicates produced estrone at 0·62±0·03 pmol/mg/min from female liver and 0·23±0·02 pmol/mg/min from male liver.
Table 4.
Rate of formation of 2-hydroxyestradiol (2-OHE2) and 4-hydroxyestradiol (4-OHE2) by liver microsomes from zebrafish. Data from other fish and mammalian species are from published studies for comparison to the zebrafish data
| Sex |
N |
2-OHE2 (pmol/mg/min) |
4-OHE2 (pmol/mg/min) |
Ratio of 4-OHE2/2-OHE2 (%) |
|
|---|---|---|---|---|---|
| Species | |||||
| Zebrafisha | Male | 1 | 0·068 | NDb | NAc |
| Female | 1 | 0·269 | ND | NA | |
| Channel catfishd | Male | 3 | 20·3±1·1 | 0·748±0·032 | 3·7 |
| Scupe | Male | 5 | 362±141 | –f | NA |
| Female | 6 | 157±37 | – | NA | |
| Winter floundere | Male | 13 | 296±79 | – | NA |
| Female | 13 | 89±39 | – | NA | |
| English soleg | Juvenile | 5 | 260±17 | – | NA |
| Humanh | Male | 21 | 46·6±25·7 | 7·1±3·4 | 15 |
| Female | 12 | 47·7±28·2 | 8·2±4·4 | 17 | |
| Rati | Male | 3–4 | 1710±180 | 390±50 | 23 |
| Female | 3–4 | 240±20 | 70±20 | 29 |
Data from this study, using 50 μM E2 at 28 °C.
ND – metbolite not detected.
NA – not applicable; one or both metabolites not detected.
Channel catfish data from Butala et al. (2004), using 50 μM E2 at 30 °C; values represent mean±s.e.m.
Data from Snowberger & Stegeman (1987), using 25 μM E2 at 25 °C for flounder, and 30 °C for scup; values represent mean±s.d.
– not investigated in this study.
English sole data from Stein et al. (1991), using 100 μM E2 at 25 °C; values represent mean±s.e.m.
Human data from Lee et al. (2001), using 20 μM E2 at 37 °C; values represent mean±s.d.
Rat data from Dannan et al. (1986), using 200 μM E2 at 37 °C; values represent mean±s.e.m.
Discussion
E2 metabolism in mammals is primarily catalyzed by CYP1 and CYP3A proteins (Lee et al. 2003), important CYP enzymes that are highly expressed in mammalian liver, the primary site of E2 metabolism (Lee et al. 2001). Because E2 metabolism also occurs predominantly in the liver of fish (Butala et al. 2004), we chose to express zebrafish CYP1s and CYP3A65 to investigate their capacity for E2 metabolism in vitro.
Bacterial expression of recombinant eukaryotic CYP proteins is possible, but often involves the modification of the 5′ end to allow appropriate targeting to membranes (Gillam et al. 1995, Waterman et al. 1995). Instead of 5′ modification, we used an ompA strategy that involves the attachment of a leader sequence to the desired CYP gene, to target the native CYP to the bacterial outer membrane (Pritchard et al. 1997). The leader sequence is cleaved off after targeting, allowing expression of unmodified, native CYP protein in bacteria. The expression conditions were optimized for each CYP by expressing the proteins in cultures with a range of Ala concentrations (0·1–1 mM). CYP1B1 was the only CYP to show a large increase in activity with an increase in Ala to 1 mM (data not shown); this was consistent with previous bacterial expressed human CYP1B1 (Jansson et al. 2000). All other CYPs required less, but some, Ala for optimal function. CO difference spectra confirmed that a peak around 450 nm was present indicating that active CYP heme-protein was present in the preparations (See Fig. 1 for a representative CO difference spectra). Another major peak at 420 nm represented other heme-containing proteins or inactive CYP at levels which are quite similar to other CYPs overexpressed in the same system (Pritchard et al. 2006).
In addition to CO difference spectra, functional protein was confirmed by fluorescent catalytic assays using 7-ER and BFC. 7-ER is a widely used specific substrate for CYP1 activity in mammals (CYP1A1, 1A2, 1B1; Shimada et al. 1998) and fish (CYP1A, 1B1; Hegelund et al. 2004). BFC is selective for CYP3A activity in mammals (Crespi & Stresser 2000), and was considered as a suitable substrate for fish CYP3A proteins based on studies with fish hepatic microsomes (Hegelund et al. 2004) and expressed fish CYP3A genes (Kashiwada et al. 2005). All CYP1s metabolized 7-ER to some degree. CYP1A catalyzed 7-ER metabolism at a rate 30 times greater than that of CYP1B1 (Fig. 2), a ratio comparable to that reported for human CYP1A1 and CYP1B1 (Murray et al. 2001). Unlike the CYP1As and CYP1Bs, there is no previous catalytic data available for CYP1Cs or CYP1Ds, but clearly the expressed CYP1C1, CYP1C2, and CYP1D1 are functional proteins as each were able to metabolize at least 7-ER (Fig. 2). While CYP1D1 had low activity for 7-ER and did not metabolize BFC, the function of the protein is completely unknown and it is likely that the substrates used are not optimal or specific for CYP1D1 activity. CYP3A65 did not metabolize 7-ER but did metabolize BFC (Fig. 2), similar to that reported for expressed medaka CYP3A proteins (Kashiwada et al. 2005). However, unlike medaka (Kashiwada et al. 2005), zebrafish CYP3A65 metabolism of BFC was not affected by cytochrome b5. Collectively, the presence of P450 peaks in the CO difference spectrum, and the ability of these proteins to metabolize 7-ER and/or BFC confirm that the expressed proteins are functional. Overall, three points favor the differences seen between these expressed CYP proteins for E2 metabolism as representative of true in vivo functional differences: 1) the expressed proteins are functional, 2) the CYP3A65, CYP1A, and CYP1B1 expressed proteins function as expected, and 3) there were similar activity levels of CPR present for each expressed protein.
CYP1A and CYP1C1 demonstrated the highest overall activity for E2 compared with the other fish CYP1s. They were also the only two zebrafish CYPs to show increased metabolism of E2 when supplemented with cytochrome b5. The rate of overall E2 metabolism of CYP1B1 compared with CYP1A was similar to that of the metabolism of human CYP1B1 to CYP1A1 (Lee et al. 2003). Surprisingly, both CYP1C1 and CYP1C2 metabolize E2 to a higher degree than CYP1B1 and CYP3A65. Phylogenetically, vertebrate CYP1Cs are clustered in a clade with the CYP1Bs, which is distinct from the CYP1A/CYP1D clade (Goldstone et al. 2009). That mammalian CYP1B1 is capable of E2 metabolism but in fish the CYP1Cs have much higher, and likely, more biologically relevant E2 metabolism than CYP1B1, suggests that E2 metabolism was present in the ancestor to the CYP1B and CYP1C clade, and that the fish CYP1Cs have retained this ancestral function while fish CYP1B1 has acquired novel function. This might suggest that both the tunicate CYP1Es and CYP1Fs, homologs to the vertebrate CYP1B/1C and CYP1A clade respectively (Goldstone et al. 2007), may be functionally capable of E2 metabolism.
Mammalian CYP genes show distinct regioselectivity in E2 metabolism. Like mammals, fish CYPs produced 2-OHE2, 4-OHE2, and 16α-OHE1 metabolites. In mammals, CYP1A1 was the only CYP1 able to hydroxylate at the 16α position, and does so in very low concentrations (Lee et al. 2003) similar to what was seen by zebrafish CYP1A. 2-OHE2 was the predominant metabolite formed in both zebrafish (this study) and mammalian studies (Lee et al. 2003), and the 4-OHE2 metabolite was produced to a much lower extent in both groups. In addition, Japanese eel CYP1A9 (one of two CYP1As in this species) also predominantly formed 2-OHE2 (Uno et al. 2008), though at a lower rate than seen here, possibly due to an 8 h reaction time in that study.
Zebrafish CYP1Cs and CYP1A have a similar 4-OHE2 to 2-OHE2 ratio compared with mammalian CYP1As (Lee et al. 2003). Zebrafish CYP1B1 was similar to mammalian CYP1B1 in its regioselectivity, as it metabolized the largest proportion of 4-OHE2 relative to 2-OHE2 formation compared with any other tested zebrafish CYP. However, unlike human CYP1B1 (Lee et al. 2003), zebrafish CYP1B1 still metabolized more 2-OHE2 than 4-OHE2, and overall, the rates of metabolism were much less than the mammalian ortholog. Distinct from the other CYP1s, CYP1D1 did not produce any 4-OHE2 metabolites. The lower ratio of 4-OHE2 to 2-OHE2 metabolism seen here from zebrafish CYP1B1 compared with human CYP1B1, and the preference of all other CYPs tested for 2-OH regioselectivity, agrees with the lower in vivo proportion of 4-OHE2 seen in channel catfish (Butala et al. 2004) compared with human (Lee et al. 2001) and rat (Dannan et al. 1986; Table 4). Our zebrafish microsomal protein had only low activity for E2, and we did not detect any 4-OHE2 metabolites. We also detected the production of estrone, which along with the parent compound, E2, are the primary estrogens in fish that are either glucuronidated or sulfated for elimination (Stein et al. 1991). Owing to the small size of zebrafish, a large sample size was required to obtain sufficient microsomal material for the present study; in fact, our microsomal pools required ~40–60 animals each and precluded a large number of replicates. To obtain sufficient numbers of animals, we collected livers from 1-year-old animals that were slated for removal from our breeding stock, and were already in reproductive decline. This likely led to an under-representation of the activity of zebrafish microsomal E2 activity, and may account for the lower levels of E2 metabolism that were seen from our zebrafish compared with other fish (Table 4). Additionally, there may be hormonal differences between species that could be responsible for the lower E2 metabolism by zebrafish. The relatively low amounts of 4-OHE2 detected in our samples suggest that though fish are susceptible to genotoxic damage by increased levels of E2 (Teles et al. 2006), they may be less sensitive than mammals.
Unlike mammalian liver where CYP1A2 is the dominant CYP1, fish lack the CYP1A2 gene and CYP1A is the dominant hepatic CYP1 (Jonsson et al. 2007). All CYP1C1, CYP1C2, and CYP1B1 are constitutively expressed at similar low levels in the liver (Jonsson et al. 2007). CYP1A, CYP1B1, CYP1C1, and CYP1C2 are inducible through the AHR pathway (Jonsson et al. 2007), and their role in E2 metabolism suggests a potential mechanism for AHR control of E2 concentrations. Our data suggests that CYP1A would be responsible for the majority of CYP1-mediated hepatic E2 metabolism in fish. First, CYP1A has higher in vitro activity for E2 metabolism compared with all other CYP1s and CYP3A65 (Table 3). Secondly, liver expression of CYP1A is significantly higher than all other CYP1s (Jonsson et al. 2007). Thirdly, CYP1A has a much higher induction via AHR agonists (Jonsson et al. 2007) suggesting that even when induced, CYP1A-mediated E2 metabolism is likely to predominate in vivo. The relatively high production of estrone compared with 2-OHE2 using zebrafish (this study), channel catfish (Butala et al. 2004), English sole (Stein et al. 1991), winter flounder, and scup (Snowberger & Stegeman 1987) microsomes suggests that CYP3A65, a very highly expressed hepatic CYP with the highest estrone production of our expressed proteins, may also be important for in vivo E2 metabolism in spite of its lower overall enzymatic activity in vitro.
CYP1D1 is constitutively expressed in the liver but, unlike other CYP1s, is not inducible through the AhR pathway (Goldstone et al. 2009). CYP1D1 expression was significantly less than CYP1A in liver, gut, gill, and kidney but higher in brain of zebrafish (Goldstone et al. 2009). Coupled with the lack of AHR-mediated induction of CYP1D1 (Goldstone et al. 2009), compared with all other CYP1 genes (Jonsson et al. 2007), this data suggest that CYP1D1 does not share a primary functional role with CYP1A1 in xenobiotic metabolism. Basal expression of CYP1A was threefold higher than CYP1D1 in liver (Goldstone et al. 2009); the low overall activity of CYP1D1 in vitro for E2 metabolism, and the much lower expression in liver compared with CYP1A, strongly suggests that CYP1D1 is unlikely to play an important in vivo role in E2 metabolism. It is possible that the low activities seen from CYP1D1 were due to the presence of predominantly non-functional protein in the preparation, because we did not see high catalytic capacity in the fluorescent assays. However, all CYPs were similarly prepared, a P450 peak was present for this preparation and functional reductase activity was similar to the other CYP preparations. As we do not know the function of this protein, it is more likely, given the different metabolic capacity documented in this study, the distinct expression profile and lack of induction of CYP1D1 by typical AhR ligands in other studies (Goldstone et al. 2009), that CYP1D1 has a function that is different and distinct from the other CYP1 genes, including E2 metabolism.
Mammalian species have more CYP3A genes than fish, as a result of more recent local gene duplications (Thomas 2007, Qiu et al. 2008). Zebrafish CYP3A65 (Tseng et al. 2005) and human CYP3A4, CYP3A5, and CYP3A7 (Maruyama et al. 2007) show similar induction patterns in the liver. In humans, CYP3A4 efficiently metabolizes E2 similar to the CYP1As (Lee et al. 2003), while zebrafish CYP3A65 poorly metabolizes E2 at rates that are similar to human CYP3A5 and CYP3A7 (Lee et al. 2003). Zebrafish CYP3A65 only produced 4-OHE2 metabolites, when supplemented with cytochrome b5, while mammalian CYP3As do produce 4-OHE2 regardless of whether cytochrome b5 is present (Lee et al. 2003). Although the mammalian CYP3A subfamily is phylogenetically clustered together with other vertebrate CYP3As, the CYP3A genes from the Actinopterygii class of ray-finned fishes, of which zebrafish are a member, cluster with fish CYP3B, CYP3C, and CYP3D genes (Qiu et al. 2008). As such, fish CYP3As share less of a common evolutionary history with mammalian CYP3As than they do with fish CYP3Bs, CYP3Cs, and CYP3Ds. In the case of fish and mammalian CYP3As, common nomenclature may not be reflective of common function. Fish CYP3As have some similar functions compared with mammalian CYP3As; zebrafish CYP3A65 can metabolize E2, and both zebrafish (this study) and medaka (Kashiwada et al. 2005) CYP3A proteins can metabolize mammalian CYP3A fluorescent substrates such as BFC; however, there are clear differences in the total activity of fish CYP3As as compared with mammalian CYP3As.
Ours is the first study involving the cloning, heterologous expression, and metabolic analysis of purified membrane preparations of both CYP1 and CYP3A isoforms from a non-mammalian vertebrate species. Purified membrane preparations resulted in concentrated levels of catalytically active CYP proteins. Using these preparations, we found that CYP1A and CYP1C1 have the highest rate of E2 metabolism, followed by more modest E2 metabolism by CYP1C2. The dominant metabolite formed was 2-OHE2, similar to studies seen in other fish (Uno et al. 2008) and humans (Lee et al. 2003). Similar to human E2 metabolism by CYP1B1 (Lee et al. 2003), the ratio of 4-OHE2 to 2-OHE2 was highest in zebrafish CYP1B1, yet the overall rates of metabolism by zebrafish CYP1B1 were lower than expected. CYP3A65 had low E2 metabolism comparable with human CYP3A5 and CYP3A7 (Lee et al. 2003), formed no 4-OHE2, and may not contribute greatly to E2 metabolism in vivo. CYP1D1, which had a different developmental and tissue expression pattern from CYP1A and was not inducible by AhR agonists in zebrafish (Goldstone et al. 2009), had functional differences compared with other CYP1s. CYP1D1 has been suggested to have an endogenous function (Goldstone et al. 2009); our study suggests that E2 metabolism is not a likely function of this protein. While overall E2 metabolism in fish and other vertebrates is similar to mammalian E2 metabolism, in non-mammalian vertebrates, the primary E2 metabolizers in vivo are likely CYP1A and CYP1C enzymes. The contributions by CYP1B1 and CYP3A are likely less than in mammalian species.
Acknowledgements
We would like to thank Dr Thomas Friedberg for supplying the human NADPH–cytochrome P450 reductase construct, Dr Frederick W Dahlquist for supplying the pCW vector, and Dr John Rimoldi for supplying the deuterated estradiol standard. We thank Emily Smith and Eric Hirsch for the optimization of fluorescent assays. This research was conducted in accordance with the accepted standards of humane animal care and approved under the McMaster University Animal Ethics Research Board, Animal Use Protocol #06-08-47.
Funding
This work was supported by the National Sciences and Engineering Research Council of Canada (NSERC) Discovery (grant number 328204) to JYW and the National Institute of Environmental Health Sciences (NIH; grant number R01ES012710) to KLW. Funding for MLS was partially provided by the Department of Biology, McMaster University.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that can be perceived as prejudicing the impartiality of the research reported.
References
- Belous AR, Hachey DL, Dawling S, Roodi N & Parl FF 2007. Cytochrome P450 1B1-mediated estrogen metabolism results in estrogen-deoxyribonucleoside adduct formation. Cancer Research 67 812–817. [DOI] [PubMed] [Google Scholar]
- Bieche I, Narjoz C, Asselah T, Vacher S, Marcellin P, Lidereau R, Beaune P & de Waziers I 2007. Reverse transcriptase-PCR quantification of mRNA levels from cytochrome (CYP)1, CYP2 and CYP3 families in 22 different human tissues. Pharmacogenetics and Genomics 17 731–742. [DOI] [PubMed] [Google Scholar]
- Butala H, Metzger C, Rimoldi J & Willett KL 2004. Microsomal estrogen metabolism in channel catfish. Marine Environmental Research 58 489–494. [DOI] [PubMed] [Google Scholar]
- Crespi CL & Stresser DM 2000. Fluorometric screening for metabolism-based drug–drug interactions. Journal of Pharmacological and Toxicological Methods 44 325–331. [DOI] [PubMed] [Google Scholar]
- Creveling CR 2003. The role of catechol-O-methyltransferase in the inactivation of catecholestrogen. Cellular and Molecular Neurobiology 23 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannan GA, Porubek DJ, Nelson SD, Waxman DJ & Guengerich FP 1986. 17beta-estradiol 2- and 4-hydroxylation catalyzed by rat hepatic cytochrome P-450: roles of individual forms, inductive effects, developmental patterns, and alterations by gonadectomy and hormone replacement. Endocrinology 118 1952–1960. [DOI] [PubMed] [Google Scholar]
- Dawling S, Roodi N, Mernaugh RL, Wang X & Parl FF 2001. Catechol-O-methyltransferase (COMT)-mediated metabolism of catechol estrogens: comparison of wild-type and variant COMT isoforms. Cancer Research 61 6716–6722. [PubMed] [Google Scholar]
- Dehennin L, Reiffsteck A & Scholler R 1980. Simple methods for the synthesis of 20 different, highly enriched deuterium labeled steroids, suitable as internal standards for isotope-dilution mass-spectrometry. Biomedical Mass Spectrometry 7 493–499. [Google Scholar]
- Gillam EM, Guo Z, Ueng YF, Yamazaki H, Cock I, Reilly PE, Hooper WD & Guengerich FP 1995. Expression of cytochrome P450 3A5 in Escherichia coli: effects of 5′ modification, purification, spectral characterization, reconstitution conditions, and catalytic activities. Archives of Biochemistry and Biophysics 317 374–384. [DOI] [PubMed] [Google Scholar]
- Godard CA, Goldstone JV, Said MR, Dickerson RL, Woodin BR & Stegeman JJ 2005. The new vertebrate CYP1C family: cloning of new subfamily members and phylogenetic analysis. Biochemical and Biophysical Research Communications 331 1016–1024. [DOI] [PubMed] [Google Scholar]
- Goldstone JV, Goldstone HM, Morrison AM, Tarrant A, Kern SE, Woodin BR & Stegeman JJ 2007. Cytochrome P450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): origin and diversification of the CYP1 gene family. Molecular Biology and Evolution 24 2619–2631. [DOI] [PubMed] [Google Scholar]
- Goldstone JV, Jonsson ME, Behrendt L, Woodin BR, Jenny MJ, Nelson DR & Stegeman JJ 2009. Cytochrome P450 1D1: a novel CYP1A-related gene that is not transcriptionally activated by PCB126 or TCDD. Archives of Biochemistry and Biophysics 482 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn ME, Lamb TM, Schultz ME, Smolowitz RM & Stegeman JJ 1993. Cytochrome-P4501a induction and inhibition by 3,3′,4,4′-tetrachlorobiphenyl in an Ah receptor-containing fish hepatoma-cell line (Plhc-1). Aquatic Toxicology 26 185–208. [Google Scholar]
- Hegelund T, Ottosson K, Radinger M, Tomberg P & Celander MC 2004. Effects of the antifungal imidazole ketoconazole on CYP1A and CYP3A in rainbow trout and killifish. Environmental Toxicology and Chemistry/SETAC 23 1326–1334. [DOI] [PubMed] [Google Scholar]
- Jansson I, Stoilov I, Sarfarazi M & Schenkman JB 2000. Enhanced expression of CYP1B1 in Escherichia coli. Toxicology 144 211–219. [DOI] [PubMed] [Google Scholar]
- Jonsson ME, Orrego R, Woodin BR, Goldstone JV & Stegeman JJ 2007. Basal and 3,3′,4,4′,5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B and 1C genes in zebrafish. Toxicology and Applied Pharmacology 221 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada S, Hinton DE & Kullman SW 2005. Functional characterization of medaka CYP3A38 and CYP3A40: kinetics and catalysis by expression in a recombinant baculovirus system. Comparative Biochemistry and Physiology. Part C, Pharmacology, Toxicology & Endocrinology 141 338–348. [DOI] [PubMed] [Google Scholar]
- Lake BG, Price RJ, Giddings AM & Walters DG 2009. In vitro assays for induction of drug metabolism. Methods in Molecular Biology 481 47–58. [DOI] [PubMed] [Google Scholar]
- Lee AJ, Kosh JW, Conney AH & Zhu BT 2001. Characterization of the NADPH-dependent metabolism of 17beta-estradiol to multiple metabolites by human liver microsomes and selectively expressed human cytochrome P450 3A4 and 3A5. Journal of Pharmacology and Experimental Therapeutics 298 420–432. [PubMed] [Google Scholar]
- Lee AJ, Cai MX, Thomas PE, Conney AH & Zhu BT 2003. Characterization of the oxidative metabolites of 17beta-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology 144 3382–3398. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Matsunaga T, Harada E & Ohmori S 2007. Comparison of basal gene expression and induction of CYP3As in HepG2 and human fetal liver cells. Biological and Pharmaceutical Bulletin 30 2091–2097. [DOI] [PubMed] [Google Scholar]
- Massey V 1959. The microestimation of succinate and the extinction coefficient of cytochrome c. Biochimica et Biophysica Acta 34 255–256. [DOI] [PubMed] [Google Scholar]
- McKay JA, Melvin WT, Ah-See AK, Ewen SW, Greenlee WF, Marcus CB, Burke MD & Murray GI 1995. Expression of cytochrome P450 CYP1B1 in breast cancer. FEBS Letters 374 270–272. [DOI] [PubMed] [Google Scholar]
- Muchmore DC, McIntosh LP, Russell CB, Anderson DE & Dahlquist FW 1989. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods in Enzymology 177 44–73. [DOI] [PubMed] [Google Scholar]
- Murray GI, Taylor MC, McFadyen MC, McKay JA, Greenlee WF, Burke MD & Melvin WT 1997. Tumor-specific expression of cytochrome P450 CYP1B1. Cancer Research 57 3026–3031. [PubMed] [Google Scholar]
- Murray GI, Melvin WT, Greenlee WF & Burke MD 2001. Regulation, function, and tissue-specific expression of cytochrome P450 CYP1B1. Annual Review of Pharmacology and Toxicology 41 297–316. [DOI] [PubMed] [Google Scholar]
- Omura T & Sato R 1964. The carbon monoxide-binding pigment of liver microsomes. Ii. Solubilization, purification, and properties. Journal of Biological Chemistry 239 2379–2385. [PubMed] [Google Scholar]
- Pritchard MP, Ossetian R, Li DN, Henderson CJ, Burchell B, Wolf CR & Friedberg T 1997. A general strategy for the expression of recombinant human cytochrome P450s in Escherichia coli using bacterial signal peptides: expression of CYP3A4, CYP2A6, and CYP2E1. Archives of Biochemistry and Biophysics 345 342–354. [DOI] [PubMed] [Google Scholar]
- Pritchard MP, McLaughlin L & Friedberg T 2006. Establishment of functional human cytochrome P450 monooxygenase systems in Escherichia coli. Methods in Molecular Biology 320 19–29. [DOI] [PubMed] [Google Scholar]
- Qiu H, Taudien S, Herlyn H, Schmitz J, Zhou Y, Chen G, Roberto R, Rocchi M, Platzer M & Wojnowski L 2008. CYP3 phylogenomics: evidence for positive selection of CYP3A4 and CYP3A7. Pharmacogenetics and Genomics 18 53–66. [DOI] [PubMed] [Google Scholar]
- Rogan EG, Badawi AF, Devanesan PD, Meza JL, Edney JA, West WW, Higginbotham SM & Cavalieri EL 2003. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: potential biomarkers of susceptibility to cancer. Carcinogenesis 24 697–702. [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamakazi H, Foroozesh M, Hopkins NE, Alworth WL & Guengerich FP 1998. Selectivity of polycyclic inhibitors for human cytochrome P450s 1A1, 1A2, and 1B1. Chemical Research in Toxicology 11 1048–1056. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ & Klenk DC 1985. Measurement of protein using bicinchoninic acid. Analytical Biochemistry 150 76–85. [DOI] [PubMed] [Google Scholar]
- Snowberger EA & Stegeman JJ 1987. Patterns and regulation of estradiol metabolism by hepatic microsomes from two species of marine teleosts. General and Comparative Endocrinology 66 256–265. [DOI] [PubMed] [Google Scholar]
- Spink DC, Lincoln DW II, Dickerman HW & Gierthy JF 1990. 2,3,7,8-Tetrachlorodibenzo-p-dioxin causes an extensive alteration of 17 beta-estradiol metabolism in MCF-7 breast tumor cells. PNAS 87 6917–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman JJ, Binder RL & Orren A 1979. Hepatic and extrahepatic microsomal electron transport components and mixed-function oxygenases in the marine fish Stenotomus versicolor. Biochemical Pharmacology 28 3431–3439. [DOI] [PubMed] [Google Scholar]
- Stein JE, Hom T, Sanborn HR & Varanasi U 1991. Effects of exposure to a contaminated-sediment extract on the metabolism and disposition of 17-beta-estradiol in English sole (Parophrys-Vetulus). Comparative Biochemistry and Physiology Part C, Pharmacology, Toxicology & Endocrinology 99 231–240. [Google Scholar]
- Teles M, Pacheco M & Santos MA 2006. Biotransformation, stress and genotoxic effects of 17beta-estradiol in juvenile sea bass (Dicentrarchus labrax L.). Environment International 32 470–477. [DOI] [PubMed] [Google Scholar]
- Thomas JH 2007. Rapid birth-death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genetics 3 e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng HP, Hseu TH, Buhler DR, Wang WD & Hu CH 2005. Constitutive and xenobiotics-induced expression of a novel CYP3A gene from zebrafish larva. Toxicology and Applied Pharmacology 205 247–258. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M & Yokoi T 2005. Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Letters 227 115–124. [DOI] [PubMed] [Google Scholar]
- Uno T, Okamoto S, Masuda S, Imaishi H, Nakamura M, Kanamaru K, Yamagata H, El-Kady MA, Kaminishi Y & Itakura T 2008. Bioconversion by functional P450 1A9 and P450 1C1 of Anguilla japonica. Comparative Biochemistry and Physiology. Part C, Pharmacology, Toxicology & Endocrinology 147 278–285. [DOI] [PubMed] [Google Scholar]
- Waterman MR, Jenkins CM & Pikuleva I 1995. Genetically engineered bacterial cells and applications. Toxicology Letters 82–83 807–813. [DOI] [PubMed] [Google Scholar]
- Zhu BT & Conney AH 1998. Functional role of estrogen metabolism in target cells: review and perspectives. Carcinogenesis 19 1–27. [DOI] [PubMed] [Google Scholar]


