Abstract
Materials made of recombinant spider silk proteins are promising candidates for cardiac tissue engineering, and their suitability has so far been investigated utilizing primary rat cardiomyocytes. Herein, we expanded the tool box of available spider silk variants and demonstrated for the first time that human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes attach, contract, and respond to pharmacological treatment using phenylephrine and verapamil on explicit spider silk films. The hiPSC-cardiomyocytes contracted for at least 14 days on films made of positively charged engineered Araneus diadematus fibroin 4 (eADF4(κ16)) and three different arginyl-glycyl-aspartic acid (RGD)-tagged spider silk variants (positively or negatively charged and uncharged). Notably, hiPSC-cardiomyocytes exhibited different morphologies depending on the spider silk variant used, with less spreading and being smaller on films made of eADF4(κ16) than on RGD-tagged spider silk films. These results indicate that spider silk engineering is a powerful tool to provide new materials suitable for hiPSC-based cardiac tissue engineering.
Abbreviations: AFM, atomic force microscopy; APTES, (3-aminopropyl) triethoxysilane; ATR, attenuated total reflection; DPBS, Dulbecco's phosphate-buffered saline; eADF4, Engineered Araneus diadematus fibroin 4; EthHD1, ethidium homodimer 1; FT-IR, Fourier-transform infrared (spectroscopy); hiPSC, human-induced pluripotent stem cell; IPTG, isopropyl-β-D-thiogalactoside; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight
Graphical abstract
Highlights
-
•
hiPSC-cardiomyocytes attach and contract on positively charged and/or RGD-tagged spider silk variants.
-
•
hiPSC-cardiomyocytes exhibit spider silk variant-dependent morphology upon adhesion.
-
•
Explicit spider silk variants promote long-term contractility of hiPSC-cardiomyocytes.
-
•
hiPSC-cardiomyocytes grown on spider silk materials respond to pharmacological treatment.
1. Introduction
Cardiovascular disease is one of the most common causes of death worldwide [1]. Despite great advances in reducing the acute mortality of cardiovascular disease through identification of risk factors (e.g. physical inactivity, high fat diet, high blood pressure, and smoking), education and diagnosis, as well as minimizing cardiomyocyte loss (e.g. antiplatelet therapy, optimized hospitalization, acute percutaneous coronary intervention), the prevalence of heart failure is rising [1]. Thus, there is a great need for novel therapies to improve heart function. Moreover, it is becoming more apparent that data derived from animal models are often not translatable into clinical practice [[2], [3], [4]]. Therefore, researchers in the field are working toward developing predictive human engineered cardiac tissues to model cardiac diseases [[5], [6], [7]], screen for therapeutic drugs [7,8], as well as repair modalities for the injured heart [[9], [10], [11]]. A cornerstone of this development is the advent of human induced pluripotent stem cells (hiPSCs) and protocols for their efficient differentiation into cardiac lineages [[12], [13], [14], [15]]. The hiPSCs can be derived from patients’ own cells and easily modified by the genome editing technology, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/ CRISPR-associated protein 9 (Cas9) to introduce or correct specific mutations or to introduce reporters for monitoring specific processes [16,17].
The most advanced engineered cardiac tissues are based on cast collagen matrices [5,7], in which tissue organization can be enhanced by electrical and mechanical stimulation [18]. However, contractile forces generated by engineered heart tissues (0.05–2 mN/mm2 [19]) are still significantly smaller than forces generated by native tissue (approx. 50 mN/mm2 [20]). This might, for example, be due to suboptimal mechanical properties of the utilized matrix and the fabrication approach (casting). Only few other materials have been utilized for cardiac tissue engineering, such as fibrin [9,18,21] and gelatin methacrylate [22,23]. Expansion of the number of materials suitable for cardiac tissue engineering will be necessary to identify one with optimized mechanical properties and to enable fabrication approaches beyond casting to generate hierarchically structured tissues including, for example, a vasculature, which is required for the creation of thick heart tissues. Limited by the rate of diffusion, the maximum thickness of non-vascularized myocardial tissue in vitro generally ranges from 50 to 100 μm, but this can be increased to around 200 μm through perfusion and high oxygen content and even to 0.5 mm when using an advanced perfusion bioreactor [24,25].
In order to identify 3D printable new materials, suitable for tissue engineering, the ones made of silk proteins are promising candidates [[26], [27], [28], [29]]. We have previously demonstrated that materials made of natural silkworm silk and recombinant silk proteins are suitable for cardiac tissue engineering [[30], [31], [32]]. Recombinant spider silk proteins are non-cytotoxic, show no immunogenicity [27,33,34] and can be produced at a large scale with high purity and consistent quality [[35], [36], [37]]. Furthermore, their biological and mechanical properties can be engineered by genetic modifications, and they can be functionalized with growth factors or peptides specific for cardiomyocyte or cardiovascular needs [[38], [39], [40], [41], [42]], which confers a definite advantage over naturally derived (silk) proteins. A further advantage is the processability of recombinant spider silk proteins into 3D scaffolds without the need of additives or crosslinkers, such as hydrogels or foams, which can mimic the environment of natural tissue [43,44]. Our spider silk hydrogels show viscoelastic and shear-thinning behavior and could be 3D bioprinted in the presence or absence of encapsulated cells [38,45].
Our studies are based on the recombinant spider silk protein eADF4(C16), which is composed of 16 repeats of the C-module (Sequence: GSSAAAAAAAASGPGGYGPENQGPSGPGGYGPGGP) based on the Araneus diadematus fibroin (ADF4) of the European garden spider [37]. One glutamic acid residue (E) in each C-module yields a negative net charge in eADF4(C16) at neutral pH [37]. Although the material is highly biocompatible, previous studies showed that several cell types could not adhere to films made thereof [39,46], as most cells also have a negatively charged surface and prefer positively charged surfaces for attachment [47,48]. Further, the protein amino acid sequence of eADF4(C16) lacks any cell-binding motifs [39,46]. When eADF4(C16) is modified with the tripeptide arginyl-glycyl-aspartic acid (RGD) cell adhesion motif or the glutamic acid residues are replaced by lysine ones, yielding the positively charged variant eADF4(κ16), rat neonatal cardiomyocytes can attach to films made of both proteins [31,32]. These results suggested that materials made of specifically engineered spider silk proteins can be used for cardiac tissue engineering.
In this work, we expanded the number of spider silk variants tested. Further, we showed for the first time that hiPSC-cardiomyocytes can efficiently attach, spread, contract, and respond to pharmacological treatment on films made of explicit eADF4 variants.
2. Materials and methods
If not stated otherwise, all chemicals were purchased from Carl Roth, Germany.
2.1. Production of recombinant spider silk variants
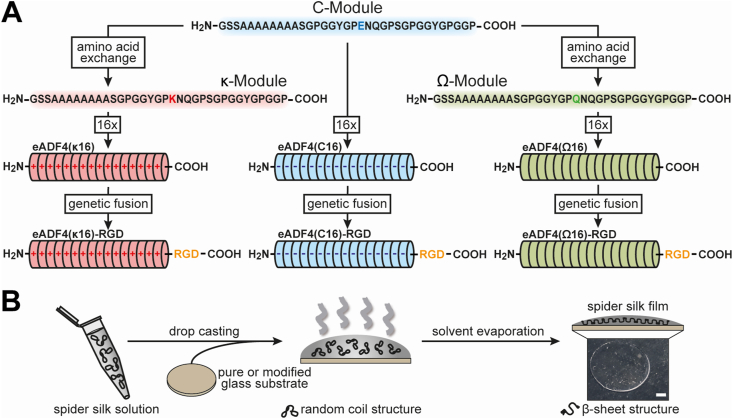
The cell-binding RGD-peptide was genetically fused to the C-terminus of eADF4(κ16) and the eADF4(Ω16) variant (Fig. 1A) using our established cloning strategy [37,39]. Therein, all recombinant eADF4-based spider silk variants contain an N-terminally fused T7 tag enabling detection by western blot analysis. While eADF4(C16) was purchased from AMSilk GmbH in a high quality, all other recombinant eADF4-based spider silk proteins were produced via time-dependent fed-batch fermentation in Escherichia coli BL21 gold (DE3) as described previously [37]. In brief, the protein synthesis was initialized by adding 0.5 mM isopropyl-β-D-thiogalactoside at 30 °C. While eADF4(C16)-RGD [39] was produced for 4 h, eADF4(κ16) [41] and eADF4(Ω16) [40] variants were expressed for 2 h at 37 °C due to faster protein degradation and inclusion body formation. After production, all recombinant spider silk proteins were purified as described previously [37,[39], [40], [41]]. After cell disruption, a heat step was used to denature residual E. coli proteins and an ammonium sulfate precipitation to yield spider silk particles. After several washing steps to remove residual E.coli DNA or salt, the spider silk particles were lyophilized and stored at −20 °C. The purity of the recombinant spider silk proteins was confirmed by ultraviolet visual (UV–Vis) spectroscopy, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western Blot, fluorescence spectroscopy, and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry to be >98% as described previously [37,39].
Fig. 1.
Design and processing of recombinant spider silk proteins based on ADF4.A The negatively charged glutamic acid (E) residue of the C-module, reflecting the consensus sequence of the core domain of the natural Araneus diadematus fibroin 4, was replaced by a positively charged lysine (K) or an uncharged glutamine (Q) residue to generate the κ- or the Ω-module, respectively. These modules were repeated 16 times to obtain the charged recombinant spider silk variants eADF4(C16), eADF4(κ16), or the uncharged eADF4(Ω16). Additionally, by genetically fusing an integrin-binding sequence RGD at the C-terminus, eADF4(C16)-RGD, eADF4(κ16)-RGD and eADF4(Ω16)-RGD were generated. B For film casting, the recombinant spider silk proteins were dissolved in a formic acid-water mixture (5:1 v/v). The spider silk solutions were drop-cast on glass substrates to obtain films with 0.5 mg protein per cm2. For negatively charged eADF4(C16) variants, the glass substrates were modified using (3-aminopropyl) triethoxysilane (APTES) before film casting to generate a positive surface. During solvent evaporation, β-sheets formed making the spider silk films water-insoluble. Scale bar: 2 mm.
2.2. Spider silk film casting
Recombinant spider silk protein films were cast on glass slides (Thermo Fisher Scientific, Germany) with an area of 1 cm2 by a drop-casting process (Fig. 1B). For negatively charged eADF4(C16) variants, glass slides were functionalized using (3-aminopropyl) triethoxysilane (APTES) (Sigma-Aldrich, Germany) before film casting to prevent film detachment due to electrostatic repulsion as described previously [31,32]. In brief, after washing using acetone (VWR, Germany) and water, the glass slides were incubated in ethanol (VWR, Germany)/APTES mixture (250:1 (v/v)) for 5 h at room temperature. Afterwards, the glass slides were incubated for 1 h at 70 °C to stabilize the APTES functionalization. Before film casting, the APTES-functionalized and the pure glass slides were washed using water and ethanol. All eADF4 variants were solved at a concentration of 5 mg/ml in a formic acid/water mixture (5:1 (v/v)) in a step-wise manner in which the protein powder was first solved in formic acid (VWR, Germany) for 30 min before water was added. 100 μL spider silk solution was dropped on glass slides. After evaporation of the solvent, spider silk films containing 0.5 mg spider silk protein per cm2 were obtained and stored at dry and dark conditions until further use. Formic acid was used for film casting, as the obtained films were already water-insoluble making a subsequent treatment afterwards unnecessary [[49], [50], [51]]. The resulting films were used as-cast for subsequent experiments without any further washing steps or treatment afterwards.
2.3. Fourier transform infrared spectroscopy
Fourier transform infrared (FT-IR) spectroscopy was performed using a Bruker Tensor 27 FT-IR-spectrometer with Mercury Cadmium Telluride (MCT)-detector (Bruker, Rheinstetten, Germany) coupled with a Hyperion 1000 FT-IR microscope (Bruker Rheinstetten, Germany) with an attenuated total reflection (ATR) lens (20x). The germanium crystal was brought into contact with the sample. The FT-IR spectra were recorded in reflectance mode in a wavenumber range of 4000 cm−1–800 cm−1 with a spectral resolution of 4 cm−1. One hundred scans were recorded for each spectrum. Atmospheric compensation and background spectra in air were applied to each spectrum. The spectra were smoothened with five points, and the baseline was corrected using the rubber band method with 64 baseline points and one iteration. Fourier self-deconvolution (FSD) and curve fitting were performed as described previously [52,53].
2.4. Atomic force microscopy
For surface topography characterization, the recombinant spider silk films on glass were analyzed in tapping mode using a Dimension Icon atomic force microscope (Bruker, Karlsruhe, Germany) with a resolution of 512 to 512 data points at 1 Hz. OTESPA-R3 silicon cantilevers with a spring constant of 26 Nm−1 were used. The scan size was 50 μm–50 μm. Images were processed with Nanoscope analysis 1.5 afterwards, plane fitted and flattened with first-order fits.
2.5. Water contact angle measurements
Water contact angle measurements were conducted on spider silk coatings as well as untreated and silanized glass slides at a Surftens-universal tensiometer (OEG GmbH, Germany) using the SCA-20 software and the sessile drop method. Therefore, water droplets were applied on the surface of recombinant spider silk films or appropriate glass substrates. After 10 s equilibration time at room temperature an image was taken, and the contact angle was analyzed. (Nine measurements per condition n = 9).
2.6. Human induced pluripotent stem cells culture and cardiac differentiation
Human induced pluripotent stem cells (hiPSCs) were cultured in StemMACS iPS-Brew XF (Miltenyi Biotec, Germany) on Matrigel™-coated multiwell culture plates. Medium change was performed daily. For maintenance culture, hiPSCs were passaged at 75%–80% confluency using ethylene diamine tetra acetic acid (EDTA, 0.5 mM, Thermo Fisher Scientific) and re-plated in StemMACS iPS-Brew XF supplemented with 10 μM rho-associated protein kinase (ROCK)-inhibitor (Y-27632, Selleck Chemicals). For subsequent differentiation, hiPSCs were dissociated into single cells using Accutase (Sigma-Aldrich) and re-plated in StemMACS iPS-Brew XF supplemented with ROCK-inhibitor at a density of ~21,000 cells/cm2. A cardiac differentiation protocol based on that described by Lian and coworkers was used [15]. At 80%–90% confluency, cardiac differentiation was initiated by exchanging the culture medium to differentiation medium (MDiff) comprising Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific) supplemented with 2% B-27 Minus Insulin (Thermo Fisher Scientific) and 100 μM l-ascorbic acid (Sigma-Aldrich). For the first 24 h (day 0–1), culture medium was further supplemented with 8–10 μM CHIR-99021 (Sigma Aldrich). Subsequently, medium was exchanged to MDiff without additional supplementation. On day 3 of differentiation, medium was exchanged to conditioned MDiff comprising equal parts of fresh MDiff and MDiff from days 1–3 of differentiation, and supplemented with 5 μM IWR-1 (Selleck Chemicals). Fresh MDiff without additional supplementation was added on day 5 of differentiation. Subsequently, on day 7 of differentiation culture medium was exchanged to maintenance medium (MMain) comprising RPMI 1640 supplemented with 2% B-27 Supplement (Thermo Fisher Scientific). Cardiomyocytes were purified by switching to lactate selection medium comprising Glucose-free RPMI 1640 (Thermo Fisher Scientific) supplemented with 5 mM sodium-lactate (Sigma-Aldrich) and 100 mM l-ascorbic acid. Metabolic selection was performed for 4–5 days starting on day 9–11 of differentiation, after which culture medium was changed back to MMain for continued maintenance.
2.7. Cardiomyocyte culture on recombinant spider silk films
Cardiomyocytes were dissociated on day 15–22 of differentiation by incubation with Accutase for 30 min at 37 °C. 200,000 cells in MMain supplemented with 1% RevitaCell (Thermo Fisher Scientific) were seeded per coverslip and left to attach for 24 h. Coverslips were then washed with Dulbecco's phosphate-buffered saline (DPBS, Thermo Fisher Scientific) and subsequently cultured in MMain, with medium changes every 2 days.
2.8. Live-dead-staining
Cardiomyocytes cultured on spider silk films for 72 h were incubated with 1 mM Calcein, 2 mM Ethidium Homodimer-1 (EthHD1) and 5 μg/mL Hoechst 33421 (all Thermo Fisher Scientific) in DPBS for 30 min at 37 °C. Cells were subsequently washed with fresh DPBS and directly mounted onto glass slides using Fluoromount-G (Thermo Fisher Scientific). Samples were imaged immediately on a Keyence BZ-9000 epifluorescence microscope equipped with a 20x Plan Apo objective (Nikon). Calcein+ and Ethidium Homodimer-1+ cells were counted manually using Hoechst to distinguish autofluorescent debris.
2.9. Immunofluorescence staining
Cardiomyocytes were washed with DPBS and fixed with 4% paraformaldehyde for 15 min at room temperature. Samples were washed and permeabilized (0.5% Triton X-100 in phosphate buffered saline [PBS]), followed by incubation with blocking buffer (5% bovine serum albumin [BSA], 0.2% Tween-20 in PBS). Samples were incubated with primary antibody overnight at 4 °C. The following antibodies were used at the indicated dilution: anti-sarcomeric α-actinin (abcam, ab9465, 1:500); anti-cardiac troponin-I (abcam, ab56357, 1:500); anti-connexin 43 (abcam, ab11370, 1:250). Secondary antibody incubation was performed for 1 h at room temperature: donkey anti-mouse Alexa Fluor 647 (1:500); donkey anti-rabbit Alexa Fluor 488 (1:500); donkey anti-goat Alexa Fluor 594 (1:500). All antibodies were diluted in blocking buffer. Samples were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted onto glass slides using Fluoromount G. Images were acquired using a laser scanning confocal microscope (LSM 800, Zeiss) equipped with a 20x Plan-APOCHROMAT and a 63x Plan-APOCHROMAT oil immersion objective (both Zeiss).
2.10. Quantification of immunofluorescence staining
For quantification of cardiomyocyte size, images (~320 × 320 μm) of hiPSC-cardiomyocytes, immunofluorescently stained for sarcomeric α-actinin and counterstained with DAPI, were used. Intensity thresholds were set for each image individually, and the area of α-actinin signal was measured. Nuclei were counted and outlined manually, and their size was recorded. All analyses were performed using the Fiji image processing package of ImageJ [54].
2.11. Contraction analysis
Videos of beating cardiomyocytes were recorded at 15 frames per second using a Keyence BZ-9000 microscope equipped with a 20x Plan Fluor objective (Nikon). The number of contractions in each video (10 s) was extrapolated to obtain beats per minute (bpm). The beating frequency calculated from three to four videos were averaged for each sample. Contraction profiles were generated using the MUSCLEMOTION plugin [55] for the image processing software ImageJ/FIJI [54]. For analysis of drug response, baseline contraction behavior was recorded first. Then, 50 μM phenylephrine was added to the culture medium. Videos were recorded after incubating the samples for 10 min at 37 °C. Subsequently, 1 μM verapamil was added, followed by an additional incubation for 10 min at 37 °C and a final recording.
2.12. Statistics
Statistical analyses were conducted in PRISM 5 (GraphPad). Statistical significance was determined by one-way analysis of variance (ANOVA), followed by a post hoc test according to Tukey, assuming normal distribution, or two-way ANOVA when indicated. P-values of <0.05 were considered to indicate significant differences between the groups tested.
3. Results and discussion
3.1. Analysis of spider silk variant films as scaffolds for hiPSC cardiomyocytes
Negatively charged eADF4(C16) is based on 16 repeats of the consensus sequence of ADF4 and has been established almost two decades ago [37]. In recent years, modifications of this recombinant protein comprised the introduction of an RGD-tag in eADF4(C16)-RGD [39]. Furthermore, the exchange of all glutamic acid residues with lysine or glutamine ones, yielded positively charged eADF4(κ16) [41], and uncharged eADF4(Ω16) [40], respectively. Herein, to further extend the range of modified spider silk proteins, RGD-fusions with eADF4(κ16) and eADF4(Ω16) were generated (Fig. 1A).
Spider silk films were processed on untreated and 3-aminopropyltriethoxysilane (APTES)-functionalized (i.e. silanized) glass substrates at a density of 0.5 mg protein per cm2. The differently charged and RGD-modified spider silk proteins were successfully processed into flat films. The positively charged eADF4(κ16) and uncharged eADF4(Ω16) variants were stably fixed on the negatively charged glass substrates without treatment beforehand. In contrast, silanization was used to immobilize films made of the negatively charged eADF4(C16) and eADF4(C16)-RGD variants. All films were stable during all experiments, including long-term cell cultivation. No apparent degradation or detachment was detected, confirming results from previous experiments using spider silk films [31,32,34,56].
The secondary structure of recombinant spider silk films was analyzed using FT-IR spectroscopy. In the herein used approach, a subsequent treatment of the spider silk proteins afterwards was not necessary [[49], [50], [51]], since the initial β-sheet content was sufficiently high, after processing from the very beginning, turning the recombinant spider silk films water-insoluble [51,52]. All recombinant spider silk variants showed nearly the same broad curve shape of amide I (1,600 cm−1 – 1,700 cm−1) and amide II (1,570 cm−1 – 1,540 cm−1) bands (Supplementary Fig. 1A).
Fourier self-deconvolution (FSD) and curve fitting were used to evaluate the secondary structure content of the amide I region (Supplementary Fig. 1B, Table 1) [53].
Table 1.
Secondary structure content [%] of recombinant spider silk films determined using FSD analysis according to Hu et al. [53].
| Secondary structure | eADF4(C16) | eADF4(C16)-RGD | eADF4(κ16) | eADF4(κ16)-RGD | eADF4(Ω16) | eADF4(Ω16)-RGD |
|---|---|---|---|---|---|---|
| β-sheets | 31.0 ± 0.4 | 34.4 ± 0.9 | 24.7 ± 7.9 | 28.1 ± 1.0 | 27.4 ± 2.0 | 26.9 ± 1.4 |
| random coils | 35.4 ± 0.5 | 34.4 ± 0.3 | 36.9 ± 3.7 | 34.1 ± 0.7 | 34.5 ± 0.3 | 34.9 ± 1.3 |
| α-helices | 11.5 ± 0.3 | 10.9 ± 0.1 | 12.2 ± 0.7 | 11.1 ± 0.4 | 11.2 ± 0.3 | 11.5 ± 0.5 |
| β-turns | 19.7 ± 0.3 | 18.8 ± 0.3 | 23.4 ± 1.5 | 20.1 ± 0.9 | 20.1 ± 1.5 | 20.4 ± 0.5 |
| others | 2.5 ± 1.5 | 1.5 ± 0.8 | 2.7 ± 2.2 | 6.6 ± 0.6 | 6.7 ± 0.4 | 6.4 ± 0.8 |
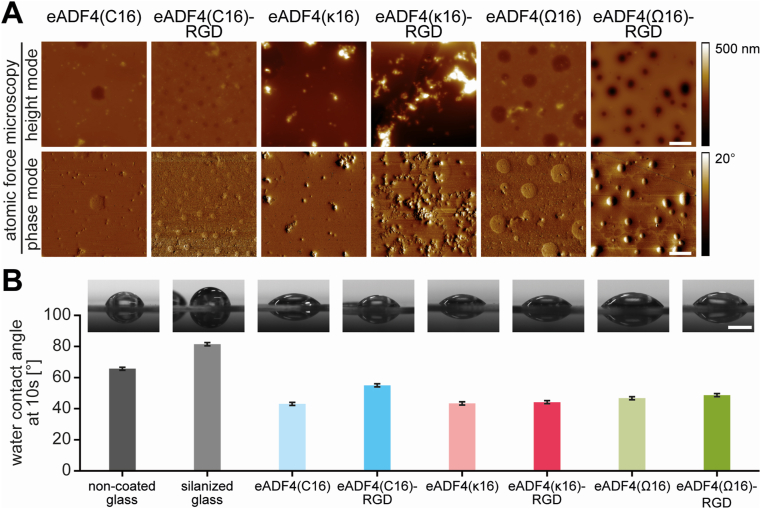
Primary cell attachment to biomaterial surfaces is influenced by surface topography and, in most cases, by the surface roughness of a material. The surface topography of flat 2D spider silk films was determined without further washing using AFM (Fig. 2A). The height images (Fig. 2A, AFM pictures, upper row) showed a smooth surface topography for eADF4(C16) and eADF4(Ω16) variants. However, the height AFM pictures indicated circular patches in the cases of eADF4(C16)-RGD, eADF4(Ω16) and eADF4(Ω16)-RGD films, which were also visible in the related phase images (Fig. 2A, lower row). These phase images represent the delay of the oscillation of the cantilever in tapping mode. The phase signal is sensitive to different material-related properties like composition, stiffness/softness and viscoelastic properties [57]. These patches on eADF4(C16)-RGD, eADF4(Ω16) and eADF4(Ω16)-RGD films (Fig. 2A, lower row) indicated different viscoelastic properties of the patches in comparison to the rest of the film. The patches could result from microphase separation of hydrophobic and hydrophilic parts of the protein as published previously [51,52]. Thereby, during film assembly the hydrophilic parts (GGX and GPGX motifs) of the eADF4-variants separate from the hydrophobic parts (polyalanine stretches), leading to structural patches with crystalline areas embedded in an amorphous matrix. The coupled RGD sequence affected the size of these patches in case of the negatively and uncharged variants, as they were smaller for eADF4(C16)-RGD and eADF4(Ω16)-RGD compared to eADF4(Ω16). In contrast, both positively charged films, eADF4(κ16) and eADF4(κ16)-RGD, showed a smooth film surface without any patches despite salt crystals distributed over the entire surface. These salt crystals were increasing the apparent total surface roughness.
Fig. 2.
Spider silk film surface characterization. A Film surface topography was determined using atomic force microscopy (AFM) in tapping mode in air. Scale bar: 10 μm B Water contact angles on spider silk films and non-coated and silanized glass materials were determined after 10 s using the sessile drop method. Scale bar: 1 mm.
Finally, water contact angle measurements were performed to evaluate the surface hydrophobicity of the films (Fig. 2B). Surfaces with a water contact angle below 90° are characterized as hydrophilic, while water contact angles above this threshold are indicative of hydrophobic surfaces. Untreated and APTES-functionalized (silanized) glass slides showed contact angles around 66° and 81°, respectively, and are per se defined as hydrophilic surfaces. Nevertheless, spider silk surfaces were more hydrophilic, indicated by a better spread water droplet (Fig. 2B). All spider silk variants, except eADF4(C16)-RGD, showed contact angles between 43° and 49°. The slightly higher contact angle of 55° for eADF4(C16)-RGD in comparison to eADF4(C16) confirmed results of a former study [31]. The presence of salt crystals on eADF4(κ16)-based spider silk films showed no influence on the water contact angle and surface hydrophobicity [32].
3.2. Human induced pluripotent stem cell-cardiomyocytes attach to films of distinct spider silk variants
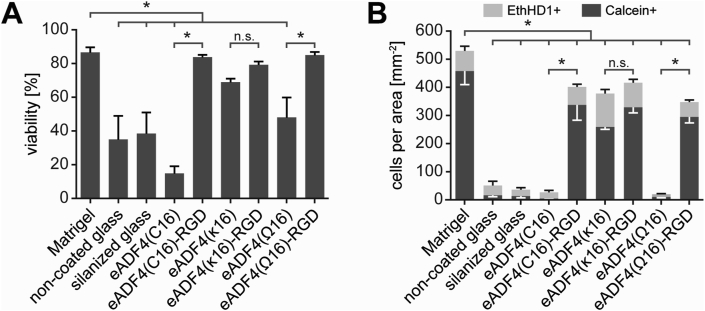
In order to evaluate the spider silk variants’ suitability for human cardiac tissue engineering applications, human induced pluripotent stem cell (hiPSC)-cardiomyocytes were seeded on silk films, and the viability of attached cells was determined after 3 days using Calcein and Ethidium Homodimer-1 (EthHD1) staining. As a control adhesion matrix, Matrigel™ was used, representing a laminin-rich mixture of extracellular matrix (ECM) proteins. While Matrigel™ is typically used to culture hiPSC-cardiomyocytes in vitro, it is not suitable for clinical translation, due to its tumor-related origin and associated safety concerns. In addition, the composition of Matrigel™ is not fully defined and is also subject to lot-to-lot variability [58].
High viability was found on films made of RGD-modified spider silk variants, irrespective of their charge, comparable to the viability of hiPSC-cardiomyocytes on Matrigel™ (Fig. 3A). Similarly, cells on eADF4(κ16) films exhibited high viability. In contrast, the degree of viability was significantly reduced on eADF4(C16) and eADF4(Ω16) films (similar to glass controls). Considering the non-cytotoxic nature of spider silk proteins [27,33,34], the reduced viability is probably due to a lack of proper cell attachment. Note, our experience shows also that non-proliferative cardiomyocytes undergo cell death when seeded at very low densities. Importantly, part of the seeded cells is dead as a result of metabolic selection during differentiation and cell dissociation. These dead cells stick to surfaces (in our case also non-coated glass) independently of adhesion motifs and thus are overrepresented on films to which hiPSC-derived cardiomyocytes adhere only inefficiently.
Fig. 3.
hiPSC-cardiomyocytes adhere to films made of several spider silk variants. A Percentage of viable cells on spider silk films made of different variants 3 days after seeding as determined by Calcein and Ethidium Homodimer-1 (EthHD1) staining. Data are mean ± standard error (SE) of the mean. ∗: p < 0.05, one-way ANOVA followed by a post hoc test according to Tukey. B Count of viable (Calcein+) and dead (EthHD1+) cells per area on spider silk films 3 days after seeding. N = 4 independent experiments. Data are mean ± standard error of the mean. ∗: p < 0.05, one-way ANOVA followed by a post hoc test according to Tukey.
To further examine the interaction of hiPSC-cardiomyocytes with the different surfaces, the absolute numbers of live (Calcein+) and dead (EthidiumHD1+) cells were assessed (Fig. 3B). The number of cells on unmodified eADF4(C16) and eADF4(Ω16) films were very low and comparable to glass controls, where only a few viable cells were found. In contrast, the amounts were markedly higher on RGD-modified spider silk variants, as well as eADF4(κ16). Importantly, the number of viable cells on eADF4(κ16) films was comparable to that observed for eADF4(κ16)-RGD ones. No significant differences were observed between the different RGD-modified variants. The observed higher number of attached cells on Matrigel™, compared to RGD-variant and eADF4(κ16) films, might be explained by the greater variety of cell adhesion motifs and integrin recognition sites, as Matrigel™ comprises a multitude of extracellular matrix (ECM) proteins [59]. In addition, growth factors present in Matrigel™ might promote attachment.
Attachment on eADF4(κ16) films appeared to be mediated by its charge, since the primary amino acid sequence of this spider silk sequence contains no cell-binding motif [46], and the cell membrane of mammalian cells is mainly negatively charged and interacts preferentially with polycationic surfaces [32,47,48]. The attachment efficiency to eADF4(κ16) films was comparable with the efficiencies of eADF4(C16)-RGD or eADF4(Ω16)-RGD ones. Interestingly, the RGD-modification of eADF4(κ16) did not further improve cell attachment. This suggested that the peptide motif of eADF4(κ16)-RGD has no additional, synergistic effect on initial adhesion to the positively charged surface.
3.3. hiPSC-cardiomyocytes exhibit different morphologies depending on the spider silk variant
Immunofluorescent staining of sarcomeric α-actinin revealed hiPSC-cardiomyocyte spreading on RGD-modified variants, similar to Matrigel™-controls, 3 days after seeding (Fig. 4A). In contrast, on eADF4(C16) and eADF4(Ω16) films, hiPSC-cardiomyocytes remained rounded and did not spread. Notably, while being comparable in number (Fig. 3B), the hiPSC-cardiomyocytes on eADF4(κ16) films appeared less spread and smaller than those on eADF4(κ16)-RGD films (Fig. 4A). To obtain an estimation of cardiomyocyte size, the area of α-actinin staining was determined as well as the number of nuclei of a given field of view (Fig. 4B). Our analyses showed that the few cardiomyocytes attached to unmodified variants, irrespective of charge, exhibited the smallest α-actinin areas/nuclei with values comparable to those of cells on glass controls. The analyses further revealed that cardiomyocytes spread most on films made of RGD-variants, with α-actinin/nuclei levels highest for cells grown on Matrigel™, eADF4(Ω16)-RGD and eADF4(C16)-RGD and slightly smaller for cells on eADF4(κ16)-RGD. Notably, the analyses confirmed that cells on eADF4(κ16) films exhibited a significantly lower α-actinin area per nuclei compared with cells on eADF4(κ16)-RGD.
Fig. 4.
hiPSC-cardiomyocytes display different morphologies depending on the spider silk variant used as scaffold.A Immunofluorescence staining for hiPSC-cardiomyocyte marker protein sarcomeric α-actinin (red) 3 days after seeding. Scale bar: 100 μm. B Quantification of sarcomeric α-actinin immunofluorescence signal area, normalized to nuclei count 3 days after seeding. Data from four independent experiments are presented as mean ± standard error of the mean. ∗: p < 0.05, one-way ANOVA followed by a post hoc test according to Tukey. C Violin plots of nuclear size 3 days after seeding. Data from four independent experiments are presented (n = 101 to 1944). Red lines: median; dashed lines: 1st and 3rd quartile. ▲: number of data points outside of the depicted range.
To validate these results, nuclear size was quantified as an indirect indicator of cardiomyocyte cell size. Notably, cells usually maintain a roughly constant nuclear-to-cytoplasmic volume ratio [60]. It has been previously shown that during maturation, the increase in cardiomyocyte size is accompanied by an increase in size of the nuclei [61].
As shown in Fig. 4C, nuclear size data were in agreement with α-actinin area/nuclei data, whereby the average nuclear size was largest on eADF4(Ω16)-RGD, surpassing sizes on both eADF4(C16)-RGD and eADF4(κ16)-RGD as well as Matrigel™.
In summary, hiPSC-cardiomyocytes attachment did not correlate with cell size, as hiPSC-cardiomyocytes attached on eADF4(κ16) films were markedly smaller than those attached on films made of RGD-modified variants. These findings are in agreement with reports that RGD-mediated integrin signaling is required for hypertrophic responses in cardiomyocytes [62,63].
3.4. Long-term cultivation of hiPSC-cardiomyocytes on spider silk films
To evaluate the possibility of long-term cultivation of hiPSC-cardiomyocytes on spider silk films, cultures were analyzed for up to 14 days. Immunofluorescent staining for α-actinin revealed a striated pattern, typical for hiPSC-cardiomyocytes, on all RGD-modified spider silk variants, similar to that on Matrigel™ (Fig. 5A). hiPSC-cardiomyocytes cultured on eADF4(κ16) remained smaller, even after this prolonged culture period. Additionally, cardiac troponin I (cTnI), an isoform of troponin I associated with hiPSC-cardiomyocytes maturation, was found in a striated pattern on RGD-modified variants, whereas it appeared more disorganized on eADF4(κ16) (Fig. 5A). Further, the gap junction protein connexin 43 could be identified at cell-to-cell contacts, suggesting electrical coupling (Fig. 5A). In accordance, spontaneous contractions of hiPSC-cardiomyocytes were observed on films made of RGD-modified variants and eADF4(κ16) as early as 24 h after seeding, and consistently by day 3 of culture. Connexin 43 was not specifically localized to longitudinal termini of cardiomyocytes (i.e. intercalated discs) on either of the matrices investigated, but distributed around the entire cells, suggesting that hiPSC-derived cardiomyocytes did not undergo advanced maturation on spider silk films compared to Matrigel™.
Fig. 5.
Long-term cultivation of hiPSC-cardiomyocytes on spider silk films. A Immunofluorescence staining of the hiPSC-cardiomyocyte marker proteins sarcomeric α-actinin (red), cardiac troponin I (magenta) and connexin 43 (green) 14 days after seeding. White scale bar: 50 μm; magenta scale bar: 10 μm. B Beating frequency of hiPSC-cardiomyocytes at 3, 7 and 14 days after seeding. Data from 3 independent experiments are presented as mean ± standard error of the mean.
Of note, hiPSC-cardiomyocytes on RGD-modified variants and Matrigel™ contracted synchronously across each field of view, while contractions on eADF4(κ16) were asynchronous (Supplementary Fig. 2), which appears to result from the smaller cell size and, therefore, lower coverage and cell-to-cell contact impairing electrical coupling.
To investigate hiPSC-cardiomyocyte contractility on spider silk variant films over time, cultures were analyzed for up to 14 days, and the beating behavior was recorded (Fig. 5B). An overall reduction in beating frequency was observed with increasing culture time (influence of culture time: p < 0.05 determined by two-way ANOVA). However, no significant differences between RGD-modified variants, eADF4(κ16) or Matrigel™ were observed at either time point (day 3, 7 or 14). These findings indicated that films made of these spider silk variants supported long-term culture and did not impair hiPSC-cardiomyocyte contractility.
3.5. hiPSC-cardiomyocytes on spider silk films respond to pharmacological stimulation
Next, cells were treated with phenylephrine, an adrenergic agonist, and subsequently verapamil, an inhibitor of L-type calcium channels, and the contractions were recorded (Fig. 6A). Phenylephrine induced a significant increase in beating frequency of hiPSC-cardiomyocytes on all spider silk variants tested, while verapamil abolished beating (Fig. 6B and C). A 1.7–2.9-fold increase in beating frequency was detected upon phenylephrine treatment.
Fig. 6.
hiPSC-cardiomyocytes on selected spider silk films are responsive to drug treatment. A Scheme of workflow and exemplary contraction profiles of hiPSC-cardiomyocytes at baseline and in response to adrenergic stimulation (50 μM phenylephrine) and calcium channel blockade (1 μM verapamil). B Beating frequencies of hiPSC-cardiomyocytes upon treatment with phenylephrine and verapamil on spider silk films. Individual samples (n = 7 to 10) are shown in black; means are shown in red. C Quantification of the increase in beating frequencies upon phenylephrine treatment. Data from 4 (3 for eADF4(Ω16)-RGD) independent experiments are presented as individual data points (black dots) and mean ± standard error of the mean (gray bars). ∗: p < 0.05, one-way ANOVA followed by a post hoc test according to Tukey.
4. Conclusion
Two Previous studies have established that spider silk materials made of eADF4 variants are very suitable and promising materials for tissue engineering and biomedical applications, as spider silk materials are highly biocompatible and cause no unspecific immune response [27,33,34]. From the findings presented here, we deduce that these materials are also promising scaffolds for cardiac tissue engineering. Interestingly, while hiPSC-cardiomyocytes could not adhere to eADF4(C16) without additional modification, they could be cultivated on specifically engineered spider silk variants. We conclude that specific modified spider silk variants, tailored to tissue-specific requirements, can be utilized to engineer human cardiac tissues aiming at modeling human diseases, screening for therapeutic drugs, and improving heart function in patients. This is an important observation, as it might allow to design strategies for selective growth of cardiomyocytes in the future, and thus might help in designing hierarchically structured cardiac tissues. Further, hiPSC-cardiomyocytes exhibited selective cell behavior on films made of the different spider silk variants, which might be of advantage for certain applications. The smaller cell size in contact with eADF4(κ16) surfaces might allow the generation of cardiac tissues with a higher cardiomyocyte density, which might, therefore, exhibit a higher contractile force. Such a construct could then also be enlarged by subsequent stimulation with pro-hypertrophic agents. In the context of 3D printing, a smaller cell size might also be of advantage, for example, regarding shear stress.
Credit author statement
TUE, VTT, and SL carried out experimental work and analyzed data; TUE, VTT, FBE, and TS wrote the original draft of the manuscript; FBE and TS conceptualized and supervised the study; all authors revised and approved the final version of the manuscript.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Prof. Scheibel is co-founder and shareholder of AMSilk GmbH.
Acknowledgments
The authors would like to thank Isabel Schoenauer for assistance in confirming stem cell pluripotency, Jana Petzold for technical advice, as well as Annika Döbl, Kaveh Roshanbinfar and Ingo Thievessen for critical discussions. Support from the Elite Network of Bavaria is also acknowledged. This work was supported by the Deutsche Forschungsgemeinschaft [DFG, German Research Foundation, grant numbers: Projektnummer 326998133 – TRR 225 (subproject C01 to F.B.E. and T.S.) and INST 410/91-1 FUGG (to F.B.E.)].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2021.100114.
Contributor Information
F.B. Engel, Email: felix.engel@uk-erlangen.de.
T. Scheibel, Email: thomas.scheibel@bm.uni-bayreuth.de.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Virani S.S., Alonso A., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Delling F.N., Djousse L., Elkind M.S.V., Ferguson J.F., Fornage M., Khan S.S., Kissela B.M., Knutson K.L., Kwan T.W., Lackland D.T., Lewis T.T., Lichtman J.H., Longenecker C.T., Loop M.S., Lutsey P.L., Martin S.S., Matsushita K., Moran A.E., Mussolino M.E., Perak A.M., Rosamond W.D., Roth G.A., Sampson U.K.A., Satou G.M., Schroeder E.B., Shah S.H., Shay C.M., Spartano N.L., Stokes A., Tirschwell D.L., VanWagner L.B., Tsao C.W., E. American Heart Association Council on, C. Prevention Statistics, S. Stroke Statistics Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- 2.Lecour S., Botker H.E., Condorelli G., Davidson S.M., Garcia-Dorado D., Engel F.B., Ferdinandy P., Heusch G., Madonna R., Ovize M., Ruiz-Meana M., Schulz R., Sluijter J.P., Van Laake L.W., Yellon D.M., Hausenloy D.J. ESC working group cellular biology of the heart: position paper: improving the preclinical assessment of novel cardioprotective therapies. Cardiovasc. Res. 2014;104:399–411. doi: 10.1093/cvr/cvu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clauss S., Bleyer C., Schuttler D., Tomsits P., Renner S., Klymiuk N., Wakili R., Massberg S., Wolf E., Kaab S. Animal models of arrhythmia: classic electrophysiology to genetically modified large animals. Nat. Rev. Cardiol. 2019;16:457–475. doi: 10.1038/s41569-019-0179-0. [DOI] [PubMed] [Google Scholar]
- 4.Perry C.J., Lawrence A.J. Hurdles in basic science translation. Front. Pharmacol. 2017;8:478. doi: 10.3389/fphar.2017.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiburcy M., Hudson J.E., Balfanz P., Schlick S., Meyer T., Chang Liao M.L., Levent E., Raad F., Zeidler S., Wingender E., Riegler J., Wang M., Gold J.D., Kehat I., Wettwer E., Ravens U., Dierickx P., van Laake L.W., Goumans M.J., Khadjeh S., Toischer K., Hasenfuss G., Couture L.A., Unger A., Linke W.A., Araki T., Neel B., Keller G., Gepstein L., Wu J.C., Zimmermann W.H. Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation. 2017;135:1832–1847. doi: 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinson J.T., Chopra A., Nafissi N., Polacheck W.J., Benson C.C., Swist S., Gorham J., Yang L., Schafer S., Sheng C.C., Haghighi A., Homsy J., Hubner N., Church G., Cook S.A., Linke W.A., Chen C.S., Seidman J.G., Seidman C.E. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y., Rafatian N., Feric N.T., Cox B.J., Aschar-Sobbi R., Wang E.Y., Aggarwal P., Zhang B., Conant G., Ronaldson-Bouchard K., Pahnke A., Protze S., Lee J.H., Davenport Huyer L., Jekic D., Wickeler A., Naguib H.E., Keller G.M., Vunjak-Novakovic G., Broeckel U., Backx P.H., Radisic M. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell. 2019;176:913–927. doi: 10.1016/j.cell.2018.11.042. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemme M., Ulmer B.M., Lemoine M.D., Zech A.T.L., Flenner F., Ravens U., Reichenspurner H., Rol-Garcia M., Smith G., Hansen A., Christ T., Eschenhagen T. Atrial-like engineered heart tissue: an in vitro model of the human atrium. Stem Cell Rep. 2018;11:1378–1390. doi: 10.1016/j.stemcr.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menasche P., Vanneaux V., Hagege A., Bel A., Cholley B., Parouchev A., Cacciapuoti I., Al-Daccak R., Benhamouda N., Blons H., Agbulut O., Tosca L., Trouvin J.H., Fabreguettes J.R., Bellamy V., Charron D., Tartour E., Tachdjian G., Desnos M., Larghero J. Transplantation of human embryonic stem cell-derived cardiovascular progenitors for severe ischemic left ventricular dysfunction. J. Am. Coll. Cardiol. 2018;71:429–438. doi: 10.1016/j.jacc.2017.11.047. [DOI] [PubMed] [Google Scholar]
- 10.Madonna R., Van Laake L.W., Botker H.E., Davidson S.M., De Caterina R., Engel F.B., Eschenhagen T., Fernandez-Aviles F., Hausenloy D.J., Hulot J.S., Lecour S., Leor J., Menasche P., Pesce M., Perrino C., Prunier F., Van Linthout S., Ytrehus K., Zimmermann W.H., Ferdinandy P., Sluijter J.P.G. ESC Working Group on Cellular Biology of the Heart: position paper for Cardiovascular Research: tissue engineering strategies combined with cell therapies for cardiac repair in ischaemic heart disease and heart failure. Cardiovasc. Res. 2019;115:488–500. doi: 10.1093/cvr/cvz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao L., Gregorich Z.R., Zhu W., Mattapally S., Oduk Y., Lou X., Kannappan R., Borovjagin A.V., Walcott G.P., Pollard A.E., Fast V.G., Hu X., Lloyd S.G., Ge Y., Zhang J. Large cardiac muscle patches engineered from human induced-pluripotent stem cell–derived cardiac cells improve recovery from myocardial infarction in swine. Circulation. 2018;137:1712–1730. doi: 10.1161/circulationaha.117.030785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cyganek L., Tiburcy M., Sekeres K., Gerstenberg K., Bohnenberger H., Lenz C., Henze S., Stauske M., Salinas G., Zimmermann W.-H., Hasenfuss G., Guan K. Deep phenotyping of human induced pluripotent stem cell–derived atrial and ventricular cardiomyocytes. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 2017;21:179–194. doi: 10.1016/j.stem.2017.07.003. e4. [DOI] [PubMed] [Google Scholar]
- 14.Protze S.I., Liu J., Nussinovitch U., Ohana L., Backx P.H., Gepstein L., Keller G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2016;35:56–68. doi: 10.1038/nbt.3745. [DOI] [PubMed] [Google Scholar]
- 15.Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long C., Li H., Tiburcy M., Rodriguez-Caycedo C., Kyrychenko V., Zhou H., Zhang Y., Min Y.L., Shelton J.M., Mammen P.P.A., Liaw N.Y., Zimmermann W.H., Bassel-Duby R., Schneider J.W., Olson E.N. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y.H., Kao H.K.J., Chang C.W., Merleev A., Overton J.L., Pretto D., Yechikov S., Maverakis E., Chiamvimonvat N., Chan J.W., Lieu D.K. Human induced pluripotent stem cell line with genetically encoded fluorescent voltage indicator generated via CRISPR for action potential assessment post-cardiogenesis. Stem Cells. 2020;38:90–101. doi: 10.1002/stem.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirt M.N., Hansen A., Eschenhagen T. Cardiac tissue engineering: state of the art. Circ. Res. 2014;114:354–367. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]
- 20.Hasenfuss G., Mulieri L.A., Blanchard E.M., Holubarsch C., Leavitt B.J., Ittleman F., Alpert N.R. Energetics of isometric force development in control and volume-overload human myocardium. Comparison with animal species. Circ. Res. 1991;68:836–846. doi: 10.1161/01.res.68.3.836. [DOI] [PubMed] [Google Scholar]
- 21.Breckwoldt K., Letuffe-Breniere D., Mannhardt I., Schulze T., Ulmer B., Werner T., Benzin A., Klampe B., Reinsch M.C., Laufer S., Shibamiya A., Prondzynski M., Mearini G., Schade D., Fuchs S., Neuber C., Kramer E., Saleem U., Schulze M.L., Rodriguez M.L., Eschenhagen T., Hansen A. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017;12:1177–1197. doi: 10.1038/nprot.2017.033. [DOI] [PubMed] [Google Scholar]
- 22.Lee J., Manoharan V., Cheung L., Lee S., Cha B.H., Newman P., Farzad R., Mehrotra S., Zhang K., Khan F., Ghaderi M., Lin Y.D., Aftab S., Mostafalu P., Miscuglio M., Li J., Mandal B.B., Hussain M.A., Wan K.T., Tang X.S., Khademhosseini A., Shin S.R. Nanoparticle-based hybrid Scaffolds for deciphering the role of multimodal cues in cardiac tissue engineering. ACS Nano. 2019;13:12525–12539. doi: 10.1021/acsnano.9b03050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin S.R., Zihlmann C., Akbari M., Assawes P., Cheung L., Zhang K., Manoharan V., Zhang Y.S., Yuksekkaya M., Wan K.T., Nikkhah M., Dokmeci M.R., Tang X.S., Khademhosseini A. Reduced graphene oxide-GelMA hybrid hydrogels as Scaffolds for cardiac tissue engineering. Small. 2016;12:3677–3689. doi: 10.1002/smll.201600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patra C., Boccaccini A.R., Engel F.B. Vascularisation for cardiac tissue engineering: the extracellular matrix. Thromb. Haemost. 2015;113:532–547. doi: 10.1160/TH14-05-0480. [DOI] [PubMed] [Google Scholar]
- 25.Esser T.U., Roshanbinfar K., Engel F.B. Promoting vascularization for tissue engineering constructs: current strategies focusing on HIF-regulating scaffolds. Expet Opin. Biol. Ther. 2019;19:105–118. doi: 10.1080/14712598.2019.1561855. [DOI] [PubMed] [Google Scholar]
- 26.Song Y., Wang H., Yue F., Lv Q., Cai B., Dong N., Wang Z., Wang L. Silk-based biomaterials for cardiac tissue engineering. Adv. Healthc. Mater. 2020 doi: 10.1002/adhm.202000735. [DOI] [PubMed] [Google Scholar]
- 27.Salehi S., Koeck K., Scheibel T. Spider silk for tissue engineering applications. Molecules. 2020;25 doi: 10.3390/molecules25030737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland C., Numata K., Rnjak-Kovacina J., Seib F.P. The biomedical use of silk: past, present, future. Adv. Healthc. Mater. 2019;8 doi: 10.1002/adhm.201800465. [DOI] [PubMed] [Google Scholar]
- 29.Kundu B., Kurland N.E., Bano S., Patra C., Engel F.B., Yadavalli V.K., Kundu S.C. Silk proteins for biomedical applications: bioengineering perspectives. Prog. Polym. Sci. 2014;39:251–267. doi: 10.1016/j.progpolymsci.2013.09.002. [DOI] [Google Scholar]
- 30.Patra C., Talukdar S., Novoyatleva T., Velagala S.R., Muhlfeld C., Kundu B., Kundu S.C., Engel F.B. Silk protein fibroin from Antheraea mylitta for cardiac tissue engineering. Biomaterials. 2012;33:2673–2680. doi: 10.1016/j.biomaterials.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 31.Kramer J.P.M., Aigner T.B., Petzold J., Roshanbinfar K., Scheibel T., Engel F.B. Recombinant spider silk protein eADF4(C16)-RGD coatings are suitable for cardiac tissue engineering. Sci. Rep. 2020;10:8789. doi: 10.1038/s41598-020-65786-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petzold J., Aigner T.B., Touska F., Zimmermann K., Scheibel T., Engel F.B. Surface features of recombinant spider silk protein eADF4(kappa 16)-made materials are well-suited for cardiac tissue engineering. Adv. Funct. Mater. 2017;27 doi: 10.1002/adfm.201701427. [DOI] [Google Scholar]
- 33.Steiner D., Lang G., Fischer L., Winkler S., Fey T., Greil P., Scheibel T., Horch R.E., Arkudas A. Intrinsic vascularization of recombinant eADF4(C16) spider silk matrices in the arteriovenous loop model. Tissue Eng. 2019;25:1504–1513. doi: 10.1089/ten.tea.2018.0360. [DOI] [PubMed] [Google Scholar]
- 34.Zeplin P.H., Maksimovikj N.C., Jordan M.C., Nickel J., Lang G., Leimer A.H., Römer L., Scheibel T. Spider silk coatings as a bioshield to reduce periprosthetic fibrous capsule formation. Adv. Funct. Mater. 2014;24:2658–2666. doi: 10.1002/adfm.201302813. [DOI] [Google Scholar]
- 35.DeFrancesco L. Hanging on a thread. Nat. Biotechnol. 2017;35:496–499. doi: 10.1038/nbt.3894. [DOI] [PubMed] [Google Scholar]
- 36.Heidebrecht A., Scheibel T. Recombinant production of spider silk proteins. Adv. Appl. Microbiol. 2013;82:115–153. doi: 10.1016/b978-0-12-407679-2.00004-1. [DOI] [PubMed] [Google Scholar]
- 37.Huemmerich D., Helsen C.W., Quedzuweit S., Oschmann J., Rudolph R., Scheibel T. Primary structure elements of spider dragline silks and their contribution to protein solubility†. Biochemistry. 2004;43:13604–13612. doi: 10.1021/bi048983q. [DOI] [PubMed] [Google Scholar]
- 38.Schacht K., Jungst T., Schweinlin M., Ewald A., Groll J., Scheibel T. Biofabrication of cell-loaded 3D spider silk constructs. Angew Chem. Int. Ed. Engl. 2015;54:2816–2820. doi: 10.1002/anie.201409846. [DOI] [PubMed] [Google Scholar]
- 39.Wohlrab S., Muller S., Schmidt A., Neubauer S., Kessler H., Leal-Egana A., Scheibel T. Cell adhesion and proliferation on RGD-modified recombinant spider silk proteins. Biomaterials. 2012;33:6650–6659. doi: 10.1016/j.biomaterials.2012.05.069. [DOI] [PubMed] [Google Scholar]
- 40.Kumari S., Lang G., DeSimone E., Spengler C., Trossmann V.T., Lücker S., Hudel M., Jacobs K., Krämer N., Scheibel T. Engineered spider silk-based 2D and 3D materials prevent microbial infestation. Mater. Today. 2020 doi: 10.1016/j.mattod.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doblhofer E., Scheibel T. Engineering of recombinant spider silk proteins allows defined uptake and release of substances. J. Pharm. Sci. 2015;104:988–994. doi: 10.1002/jps.24300. [DOI] [PubMed] [Google Scholar]
- 42.Saric M., Scheibel T. Engineering of silk proteins for materials applications. Curr. Opin. Biotechnol. 2019;60:213–220. doi: 10.1016/j.copbio.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Schacht K., Scheibel T. Controlled hydrogel formation of a recombinant spider silk protein. Biomacromolecules. 2011;12:2488–2495. doi: 10.1021/bm200154k. [DOI] [PubMed] [Google Scholar]
- 44.Schacht K., Vogt J., Scheibel T. Foams made of engineered recombinant spider silk proteins as 3D Scaffolds for cell growth. ACS Biomater. Sci. Eng. 2016;2:517–525. doi: 10.1021/acsbiomaterials.5b00483. [DOI] [PubMed] [Google Scholar]
- 45.DeSimone E., Schacht K., Pellert A., Scheibel T. Recombinant spider silk-based bioinks. Biofabrication. 2017;9 doi: 10.1088/1758-5090/aa90db. [DOI] [PubMed] [Google Scholar]
- 46.Leal-Egaña A., Lang G., Mauerer C., Wickinghoff J., Weber M., Geimer S., Scheibel T. Interactions of fibroblasts with different morphologies made of an engineered spider silk protein. Adv. Eng. Mater. 2012;14:B67–B75. doi: 10.1002/adem.201180072. [DOI] [Google Scholar]
- 47.Blau A. Cell adhesion promotion strategies for signal transduction enhancement in microelectrode array in vitro electrophysiology: an introductory overview and critical discussion. Curr. Opin. Colloid Interface Sci. 2013;18:481–492. doi: 10.1016/j.cocis.2013.07.005. [DOI] [Google Scholar]
- 48.De Rosa M., Carteni M., Petillo O., Calarco A., Margarucci S., Rosso F., De Rosa A., Farina E., Grippo P., Peluso G. Cationic polyelectrolyte hydrogel fosters fibroblast spreading, proliferation, and extracellular matrix production: implications for tissue engineering. J. Cell. Physiol. 2004;198:133–143. doi: 10.1002/jcp.10397. [DOI] [PubMed] [Google Scholar]
- 49.Spiess K., Ene R., Keenan C.D., Senker J., Kremer F., Scheibel T. Impact of initial solvent on thermal stability and mechanical properties of recombinant spider silk films. J. Mater. Chem. 2011;21:13594–13604. doi: 10.1039/c1jm11700a. [DOI] [Google Scholar]
- 50.Um I.C., Kweon H.Y., Lee K.G., Park Y.H. The role of formic acid in solution stability and crystallization of silk protein polymer. Int. J. Biol. Macromol. 2003;33:203–213. doi: 10.1016/j.ijbiomac.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 51.Wohlrab S., Spiess K., Scheibel T. Varying surface hydrophobicities of coatings made of recombinant spider silk proteins. J. Mater. Chem. 2012;22:22050–22054. doi: 10.1039/c2jm35075k. [DOI] [Google Scholar]
- 52.Borkner C.B., Lentz S., Müller M., Fery A., Scheibel T. Ultrathin spider silk films: insights into spider silk assembly on surfaces. ACS Appl. Polym. Mater. 2019;1:3366–3374. doi: 10.1021/acsapm.9b00792. [DOI] [Google Scholar]
- 53.Hu X., Kaplan D., Cebe P. Determining beta-sheet crystallinity in fibrous proteins by thermal analysis and infrared spectroscopy. Macromolecules. 2006;39:6161–6170. doi: 10.1021/ma0610109. [DOI] [Google Scholar]
- 54.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sala L., van Meer B.J., Tertoolen L.G.J., Bakkers J., Bellin M., Davis R.P., Denning C., Dieben M.A.E., Eschenhagen T., Giacomelli E., Grandela C., Hansen A., Holman E.R., Jongbloed M.R.M., Kamel S.M., Koopman C.D., Lachaud Q., Mannhardt I., Mol M.P.H., Mosqueira D., Orlova V.V., Passier R., Ribeiro M.C., Saleem U., Smith G.L., Burton F.L., Mummery C.L. MUSCLEMOTION: a versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ. Res. 2018;122:e5–e16. doi: 10.1161/CIRCRESAHA.117.312067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borkner C.B., Wohlrab S., Möller E., Lang G., Scheibel T. Surface modification of polymeric biomaterials using recombinant spider silk proteins. ACS Biomater. Sci. Eng. 2016;3:767–775. doi: 10.1021/acsbiomaterials.6b00306. [DOI] [PubMed] [Google Scholar]
- 57.Giurlani W., Berretti E., Innocenti M., Lavacchi A. Measuring the thickness of metal coatings: a review of the methods. Coatings. 2020;10 doi: 10.3390/coatings10121211. [DOI] [Google Scholar]
- 58.Hughes C.S., Postovit L.M., Lajoie G.A. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 59.Benton G., Arnaoutova I., George J., Kleinman H.K., Koblinski J. Matrigel: from discovery and ECM mimicry to assays and models for cancer research. Adv. Drug Deliv. Rev. 2014;79–80:3–18. doi: 10.1016/j.addr.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 60.Vukovic L.D., Jevtic P., Edens L.J., Levy D.L. New insights into mechanisms and functions of nuclear size regulation. Int Rev Cell Mol Biol. 2016;322:1–59. doi: 10.1016/bs.ircmb.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Gerdes A.M., Morales M.C., Handa V., Moore J.A., Alvarez M.R. Nuclear size and DNA content in rat cardiac myocytes during growth, maturation and aging. J. Mol. Cell. Cardiol. 1991;23:833–839. doi: 10.1016/0022-2828(91)90216-9. [DOI] [PubMed] [Google Scholar]
- 62.Ross R.S., Pham C., Shai S.Y., Goldhaber J.I., Fenczik C., Glembotski C.C., Ginsberg M.H., Loftus J.C. Beta1 integrins participate in the hypertrophic response of rat ventricular myocytes. Circ. Res. 1998;82:1160–1172. doi: 10.1161/01.res.82.11.1160. [DOI] [PubMed] [Google Scholar]
- 63.Brancaccio M., Hirsch E., Notte A., Selvetella G., Lembo G., Tarone G. Integrin signalling: the tug-of-war in heart hypertrophy. Cardiovasc. Res. 2006;70:422–433. doi: 10.1016/j.cardiores.2005.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.