Abstract
The extensive length, compaction, and interwound nature of DNA, together with its controlled and restricted movement in eukaryotic cells, create a number of topological issues that profoundly affect all of the functions of the genetic material. Topoisomerases are essential enzymes that modulate the topological structure of the double helix, including the regulation of DNA under- and overwinding and the removal of tangles and knots from the genome. Type II topoisomerases alter DNA topology by generating a transient double-stranded break in one DNA segment and allowing another segment to pass through the DNA gate. These enzymes are involved in a number of critical nuclear processes in eukaryotic cells, such as DNA replication, transcription and recombination, and are required for proper chromosome structure and segregation. However, because type II topoisomerases generate double-stranded breaks in the genetic material, they also are intrinsically dangerous enzymes that have the capacity to fragment the genome. As a result of this dualistic nature, type II topoisomerases are the targets for a number of widely prescribed anticancer drugs. This article will describe the structure and catalytic mechanism of eukaryotic type II topoisomerases and will go on to discuss the actions of topoisomerase II poisons, which are compounds that stabilize DNA breaks generated by the type II enzyme and convert these essential enzymes into “molecular scissors.” Topoisomerase II poisons represent a broad range of structural classes and include anticancer drugs, dietary components, and environmental chemicals.
Graphical Abstract
Introduction.
The genetic information required to encode the human genome is stored in approximately two meters of DNA, which is severely compacted within the nucleus of the cell (5-10 μm in diameter).1 The DNA contains two complementary polynucleotide strands that are plectonemically coiled, wrapped around a histone core to form nucleosomes, and further condensed into higher order chromatin and chromosome structures.1 Interactions with scaffolding proteins, together with the extreme length of the genome, hinder the free rotation of the DNA strands. These properties, together with the interwound nature and restricted rotation of DNA, create a number of topological issues that profoundly affect all of the functions of the genetic material.2–6 Topological relationships within DNA, including under- and overwinding (i.e., negative and positive supercoiling, respectively), tangling, and knotting can be altered only by breaking one or both strands of the double helix.4, 7
DNA Topology.
Human DNA is globally underwound by ~6%.2, 7, 8 This has important ramifications because duplex DNA acts as the storage form of the genetic information. In order to replicate or express this information, the two strands of DNA must be separated to gain access to the genetic material. DNA underwinding decreases the energy needed to break the hydrogen bonds between complementary bases, thereby facilitating strand separation.2, 8, 9
In contrast to basal levels of negative supercoiling, hyper-negative supercoiling that occurs behind transcription complexes promotes the formation of R-loops, which is a source of genomic instability.10–12 Furthermore, positive supercoiling creates significant problems for DNA systems that require helicases, such as replication and transcription.2, 4, 7, 8 Because helicases separate the two strands of DNA without unwinding them, they do not remove any of the turns of the double helix.2, 4, 8, 9 Consequently, these DNA turns are compressed ahead of the helicase, overwinding the double helix and generating severe torsional stress in the genetic material. Unless this torsional stress is alleviated, the replication and transcription machinery can no longer track along the DNA and these essential nuclear processes will stall rapidly.
DNA replication also generates intermolecular tangles (e.g., catenanes) between daughter chromosomes.2, 8 Unless these tangled DNA molecules are decatenated, daughter chromosomes cannot be segregated properly during mitosis or meiosis. Similarly, intramolecular knots formed within the same DNA molecule are generated during recombination.2, 8 Because DNA knots make it impossible to completely separate the two strands of the double helix, they block essential nucleic acid processes.4 Consequently, DNA tangles and knots can be lethal to cells if they are not resolved.2, 4, 7, 8
Topoisomerases.
Topoisomerases regulate the topological structure of the genetic material during cellular processes. These enzymes are essential for the survival of all organisms and alter DNA topology by generating transient breaks in the double helix.7, 8, 13–19
Topoisomerases can be separated into two major classes, type I and type II, based on the number of DNA strands that they cleave and ligate during their catalytic cycles.7, 8, 13–18, 20 Type I enzymes (denoted by odd numbers) transiently cleave a single strand of the DNA duplex, while type II enzymes (denoted by even numbers) cleave both strands. Because type I topoisomerases cleave only a single strand, they can modulate DNA supercoiling, but cannot remove knots or tangles from intact duplex DNA.4, 7, 8 In contrast, because type II topoisomerases cleave both strands of the double helix, they can regulate DNA under- and overwinding and can also remove tangles and knots from the genome.4, 7, 8, 13, 21
Type II Topoisomerases.
Eukaryotic type II topoisomerases function as homodimers and require ATP and divalent metal ions for overall catalytic activity.20–23 Briefly, they interconvert different topological structures by generating a transient double-stranded break in the DNA backbone, transporting a separate double helix through the nucleic acid gate, and resealing the break. Both of the protomer subunits of topoisomerase II contain an active site tyrosine residue, which cuts the DNA by a nucleophilic attack on the sugar-phosphate backbone.20, 21 In order to maintain the integrity of the genome and conserve the bond energy of the backbone while the DNA is cleaved, type II enzymes covalently attach to the newly generated 5’-terminal phosphates (4-base pairs apart on opposite strands).8, 13–17, 20, 24 This covalent enzyme-cleaved DNA complex, the cleavage complex, is a hallmark of all type II topoisomerases and contains a phosphotyrosine-linked protein and a free deoxyribose hydroxyl on the 5’- and 3’-termini, respectively.8, 13–17, 20
All living organisms encode at least one type II topoisomerase.7, 8, 17, 20 Lower eukaryotes encode a single type II enzyme, while vertebrates express two isoforms of topoisomerase II, α and β.16, 21, 25 Human topoisomerase IIα and IIβ are encoded by two separate genes (17q21–22 and 3p24, respectively) and differ in molecular mass (170 and 180 kDa, respectively). Both isoforms share extensive sequence identity (70%) and display similar enzymatic properties, but differ significantly in their expression, cellular regulation, and functions.14, 16, 17, 21, 25–27
Expression of topoisomerase IIα is linked to cellular growth and is essential for the survival of proliferating cells.4, 13, 14, 16, 21, 25 Although the α isoform is virtually non-existent in quiescent and differentiated tissues, rapidly proliferating cells contain ~500,000 molecules.18, 25, 27, 28 Topoisomerase IIα is associated with replication forks and remains tightly bound to chromosomes during mitosis. The enzyme plays roles in DNA transcription and recombination,18, 29 is important for fork convergence and termination of replication,30 and is required for proper chromosome organization and segregation.8, 18, 29, 30
In contrast to topoisomerase IIα, the β isoform is not required at the cellular level.25 However, it is essential for neuronal development.7, 16, 20, 25, 31 High levels of the β isoform are found in most cell types, independent of proliferation status.7, 18, 25, 32 Topoisomerase IIβ dissociates from chromosomes during mitosis, but appears to play an important role in the transcription of hormonally and developmentally regulated genes.25, 33–35 Despite the differences between the α and β isoforms of topoisomerase II, these enzymes are mechanistically and structurally similar and will be collectively referred to in this article as topoisomerase II, unless otherwise noted.
Structural Domain Organization of Eukaryotic Topoisomerase II.
Each topoisomerase II protomer contains three main regions: the N-terminal region, the catalytic core, and the C-terminal region (Figure 1).20, 21 The N-terminal region contains the N-gate (through which the DNA enters the enzyme), the GHKL (Gyrase, Hsp90, Histidine Kinase, MutL) domain (also known as the ATPase domain) that binds and hydrolyzes ATP, and the transducer domain that relays hydrolysis signals to the catalytic core of the enzyme.20, 21 The catalytic core contains the TOPRIM (topoisomerase/primase) domain that coordinates the active site divalent metal ions, the WHD (winged-helix domain) that includes the active site tyrosine, and the tower domain that makes polar and electrostatic contact with the DNA.20, 21 The C-terminal region (CTR) contains nuclear localization signals and post-transcriptional modification sites.20, 21, 37 The CTR is largely disordered in the protein when it is not bound to DNA and is poorly conserved among the type II enzymes.20, 21 It is not necessary for enzyme function, but it contains protein elements that allow topoisomerase IIα to discern the geometry of DNA supercoils and remove positive supercoils ~10 times faster than it does negative supercoils.8, 38, 39
Figure 1.
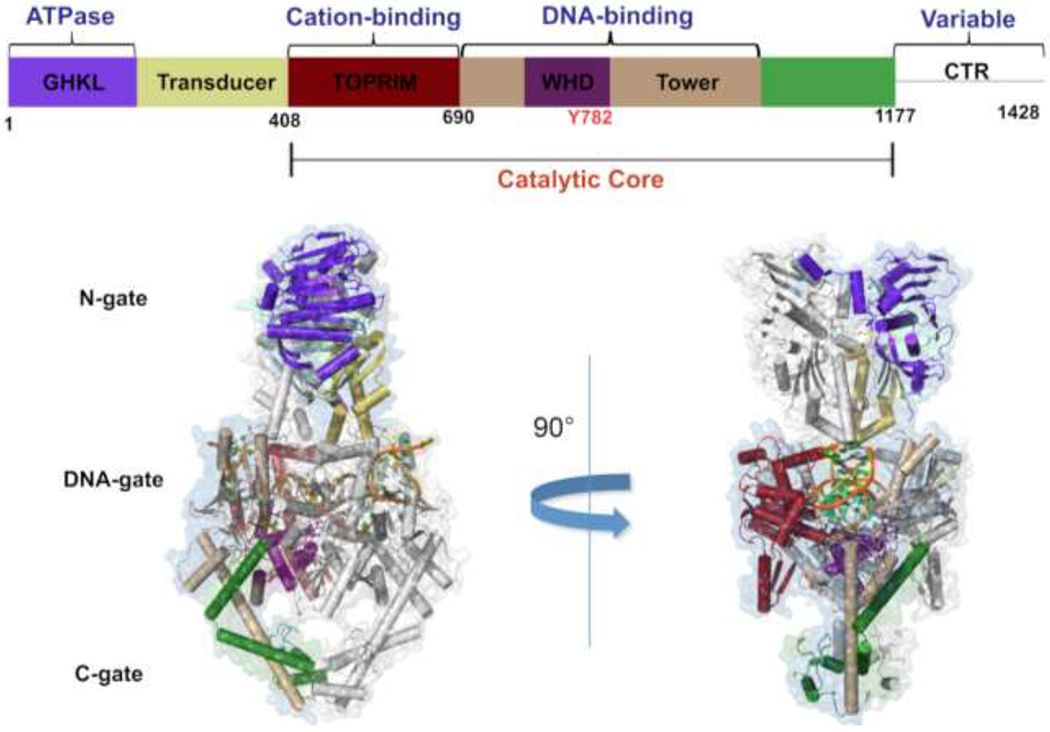
Domain organization and structure of eukaryotic topoisomerase II. A schematic depicting the functional regions of a topoisomerase II protomer is colored and labeled above. The structure of a functional eukaryotic topoisomerase II from yeast is shown below in a covalent complex with double-stranded DNA. The PDB model 4GFH is shown and the functional regions are highlighted according to the schematic above for visualization.36 One of the enzyme protomers is shaded gray to distinguish between the two protomers. The variable and unstructured C-terminal region (CTR) is not depicted in the structure.
Catalytic Mechanism of Topoisomerase II.
Topoisomerase II alters DNA topology using the double-stranded DNA passage reaction, which is shown as a series of discrete steps in Figure 2. 1) The enzyme starts its catalytic cycle by binding two DNA segments at a crossover.40 The first segment bound by the enzyme is the double helix that will be cleaved by the enzyme and is referred to as the “Gate-” or “G-segment.”41 The second segment is the double helix that will be transported through the transiently cleaved G-segment and is referred to as the “Transport-”or “T-segment.”40, 42, 43 2) In the presence of divalent metal ions (coordinated within the TOPRIM domain), the type II enzyme samples the G-segment for bendability.22, 44–46 DNA sequences that can be bent by the enzyme45, 47, 48 are distorted to an angle of ~150° and can be used as a site for enzyme action.41 3) Cleavage of the bent G-segment is catalyzed by the nucleophilic attack of the active site tyrosine residues to form the cleavage complex.41, 44 The two tyrosine residues act in a coordinated manner. Once the first strand has been cleaved, the second is cleaved >10-fold faster.49 4) Upon the binding of two ATP molecules, the enzyme closes the N-gate, forcing the T-segment through the open DNA-gate.13, 20, 21 The DNA strand passage event appears to take place more rapidly if one of the ATP molecules is hydrolyzed.50 5) The G-segment is then ligated, and the T-segment is released.13, 20, 21 6) Following hydrolysis of the second ATP molecule, the G-segment can be released and 7) the enzyme conformation is reset, allowing for the capture of another DNA crossover.13, 20, 21
Figure 2.
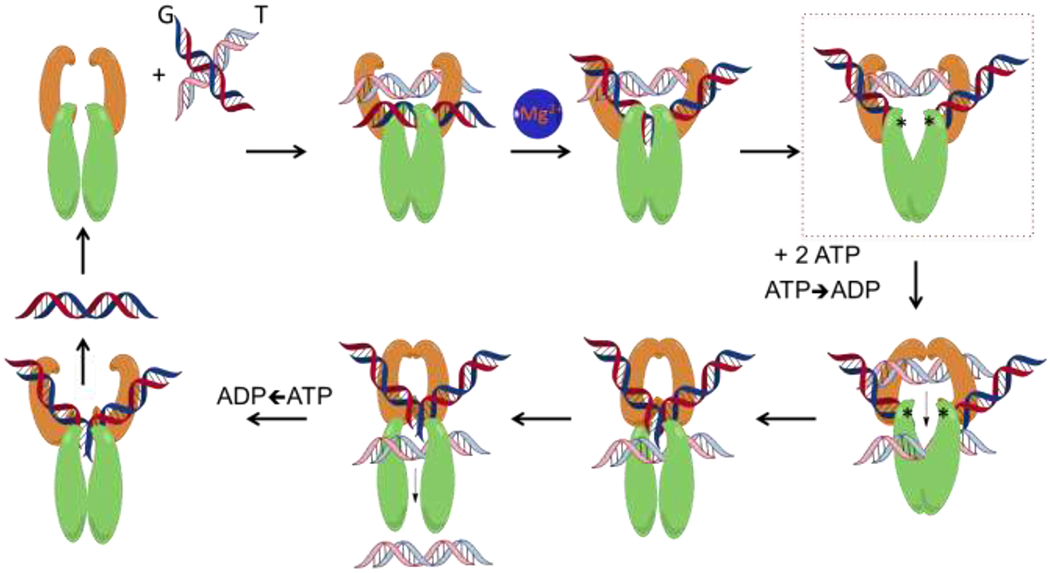
The catalytic cycle of topoisomerase II. Type II topoisomerases regulate DNA topology by a multi-step process illustrated in the schematic. 1) The homodimer binds to DNA crossovers, positioning the first double helix as the DNA Gate-segment (G-segment) and the second as the Transport-segment (T-segment). 2) Upon the coordination of divalent metal ions in the TOPRIM region of the enzyme, the G-segment is sampled for bendability and bent. 3) The bent G-segment is then cleaved by nucleophilic attacks on the DNA backbone by the catalytic tyrosine residues, covalently linking the enzyme to the cleaved DNA (depicted by the asterisks) and allowing for the DNA-gate to open. This intermediate step represents the “cleavage complex” (highlighted by the dotted box). 4) Following the binding of two molecules of ATP in the ATPase domain, the N-gate closes, and hydrolysis of one ATP transduces signals through the enzyme allowing for rapid passage of the T-segment through the DNA-gate. 5) The opened DNA-gate is then resealed, 6) and the T-segment is released. 7) After hydrolysis of the second ATP molecule, the enzyme releases the G-segment, and the enzyme is reset to capture another crossover.
Converting Type II Topoisomerases into Molecular Scissors.
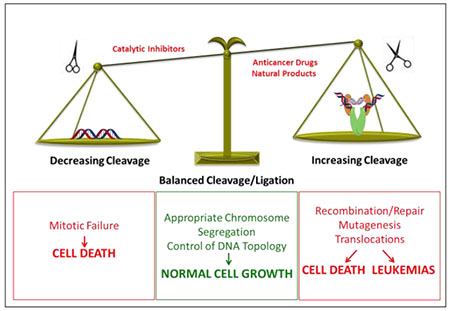
Because type II topoisomerases generate double-stranded DNA breaks as part of their reaction mechanism, they are intrinsically dangerous enzymes.13, 15, 18 Although essential to cell viability, type II topoisomerases also have the capacity to fragment the genome (Figure 3). As a result, levels of cleavage complexes are maintained in a critical balance that greatly favors ligation. Under normal circumstances, these complexes are short-lived and are readily reversible.13, 15, 18, 20 However, compounds that stabilize cleavage complexes have serious cellular consequences.13–18, 46
Figure 3.
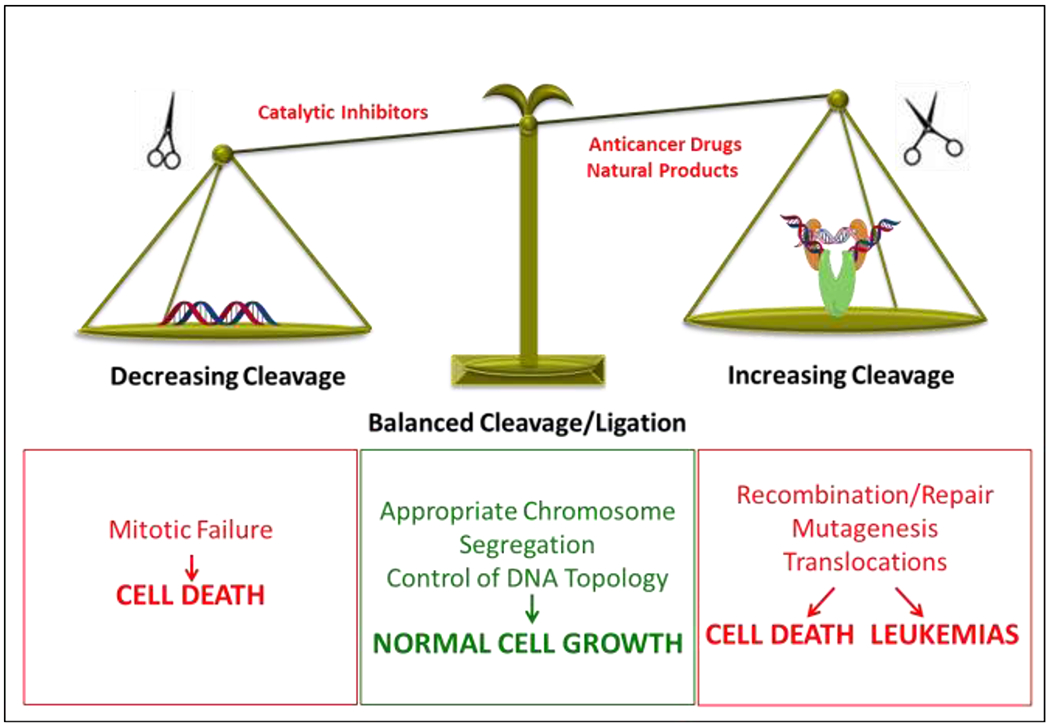
Balancing of topoisomerase II-mediated DNA cleavage. A balanced level of topoisomerase II-DNA cleavage complexes is required for the enzyme to perform its critical cellular functions (middle). If the level of topoisomerase II-DNA cleavage complexes falls too low (left), cells are unable to untangle daughter chromosomes after DNA replication and ultimately die of mitotic failure. If the level of cleavage complexes becomes too high (right), the actions of DNA tracking systems can convert these transient complexes to permanent double-stranded breaks. The resulting DNA breaks, as well as the inhibition of essential DNA processes, initiate recombination/repair pathways that can generate chromosome translocations and other DNA aberrations. If the strand breaks overwhelm the cell, they can trigger apoptosis. This is the basis for the actions of several widely prescribed anticancer drugs and natural products that target topoisomerase II. If the concentration of topoisomerase II-mediated DNA strand breaks is too low to overwhelm the cell, mutations or chromosomal aberrations may be present in surviving populations. In some cases, exposure to topoisomerase II poisons has been associated with the formation of specific types of leukemias.
When levels of cleavage complexes, reflecting overall levels of enzymatic activity, drop below threshold concentrations, daughter chromosomes remain entangled following replication. As a result, sister chromatids cannot properly segregate during mitosis and cells die due to catastrophic mitotic failure.8, 20 When levels of cleavage complexes rise to critical levels, cells also die, but for a different reason. In this instance, cell death is due to the conversion of transient DNA cleavage intermediates to strand breaks that can no longer be ligated by topoisomerase II and have to be rejoined by recombination/repair pathways.13, 15, 18, 20, 51, 52 These breaks are believed to be generated when replication forks, transcription complexes, or other DNA tracking enzymes approach or attempt to traverse the covalently bound protein ‘roadblock’ in the genetic material.13, 52–54 The mechanism that renders topoisomerase II incapable of resealing the DNA breaks has not yet been defined. It may be direct protein interactions with the type II enzyme or extreme DNA overwinding generated by oncoming DNA tracking systems. The conversion of trapped cleavage complexes to untethered DNA breaks is complex and involves the actions of DNA repair/processing enzymes.55–59 Ultimately, the resulting damage and induction of recombination/repair pathways can generate a variety of mutations and chromosomal aberrations. When topoisomerase II-generated breaks are present in sufficient numbers, they can overwhelm the cell and initiate cell death pathways.13, 52–54
Because of this potentially lethal property of topoisomerase II, the enzyme has become a prominent target for the development of anticancer drugs.13, 15, 54 However, in some cases, the use of these drugs initiates chromosomal translocations that trigger specific types of leukemias in ~2-3% of patients.18, 53, 60–64
Topoisomerase II Catalytic Inhibitors vs. Poisons.
Compounds that alter topoisomerase II activity can be divided into two categories: catalytic inhibitors and poisons (Figure 4). Topoisomerase II catalytic inhibitors are compounds that impair the overall catalytic activity without increasing the concentration of cleavage complexes.49, 54, 65, 66 Inhibitors have been described that act at a variety of different steps of the topoisomerase II catalytic cycle, including DNA binding, ATP binding, ATP hydrolysis, and DNA cleavage.65–69 Depending on which step of the catalytic cycle is targeted, inhibitors may have very different cellular effects. For example, an inhibitor that blocks topoisomerase II-DNA binding will rob the cell of all of the essential functions of the enzyme, including structural functions. Alternatively, an inhibitor that blocks ATP binding will rob the cell of the catalytic functions of topoisomerase II, while an inhibitor that blocks ATP hydrolysis will freeze the N-terminal gate in a closed conformation, trapping topoisomerase II on the DNA and blocking critical nucleic acid processes.65–69
Figure 4.
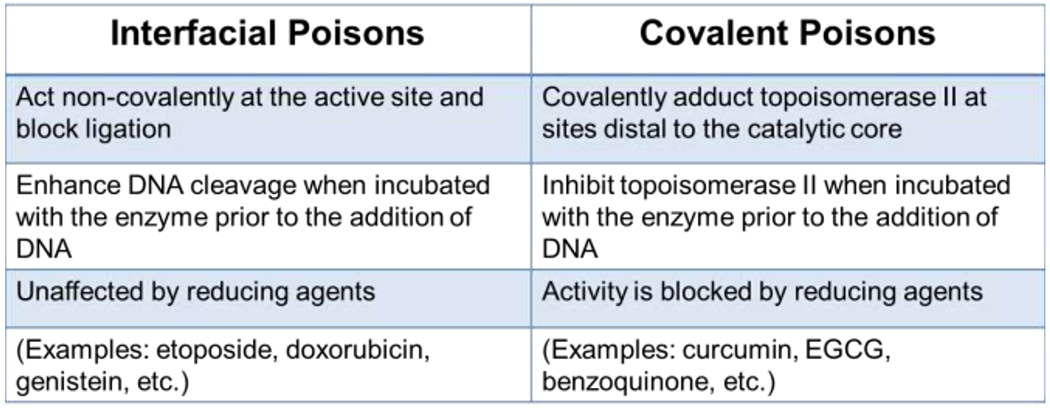
Mechanisms of topoisomerase II poisons. The two distinct mechanisms, interfacial and covalent, by which compounds can interact with the enzyme-DNA complex to enhance DNA cleavage are described.
Topoisomerase II poisons are compounds that increase levels of topoisomerase II-DNA cleavage complexes. The term “poison” was originally applied to these compounds because they convert the enzyme into a cellular toxin that initiates the mutagenic and lethal consequences described above.13, 15, 54, 68 Compounds that act in this manner are referred to as topoisomerase II poisons irrespective of whether or not they inhibit enzyme catalytic activity. This is because the increased levels of cellular DNA breaks induced by these compounds represent a gain-of-function, dominant phenotype.13, 15, 54, 68 All clinically relevant topoisomerase II-targeted drugs identified to date act as poisons and interfere with the ability of the enzyme to religate cleaved DNA molecules.13, 15, 54, 70, 71
Topoisomerase II inhibitors and poisons are often confused in the literature. However, considering the dramatically different effects that catalytic inhibitors and poisons exert on the type II enzyme and on the cell, it is important to understand the differences between the two classes of compounds and intellectually keep them separate.
It is also important to recognize that many topoisomerase II poisons can cure or prevent cancers. Therefore, despite the negative connotations associated with the term “poison,” these compounds have a considerable number of positive attributes. Put another way, just because something is called a poison does not mean that it is necessarily bad for you.
Mechanisms of Topoisomerase II Poisons.
Topoisomerase II poisons act by two distinct mechanisms: interfacial vs. covalent (Figure 4). Interfacial poisons interact in a non-covalent manner at the topoisomerase II-DNA interface within the vicinity of the active site tyrosyl residues.15, 17, 71 These compounds are bi-functional and must interact with both the protein and the DNA in order to increase levels of DNA scission. Upon DNA cleavage, interfacial poisons insert between the ends of the double helix at the cut scissile bonds on the G-segment.13, 15, 17, 54, 71 They distort the active site within the cleavage complex and appear to detach one of the catalytic magnesium ions from the phosphotyrosyl moiety.72 More importantly, interfacial poisons act as physical barriers to ligation, functioning as “molecular doorstops.”13, 54, 73 In order to stabilize double-stranded breaks, two molecules (one at each scissile bond) must be present in the active site of the type II enzyme. Etoposide (Figures 4 and 5) is a classic example of an interfacial topoisomerase II poison.
Figure 5.
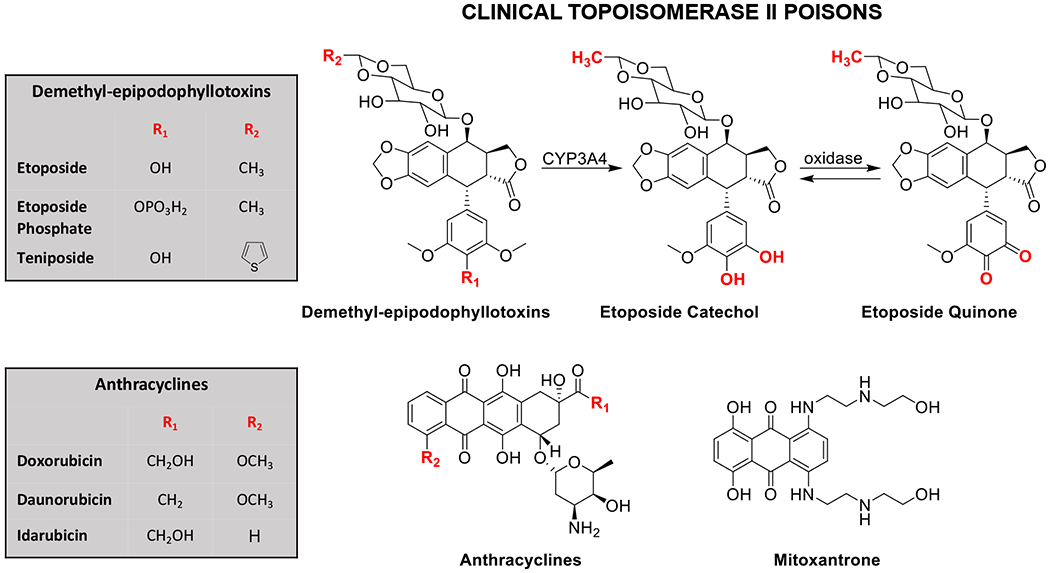
Clinical topoisomerase II poisons. The demethyl-epipodophyllotoxins (etoposide, etoposide phosphate, and teniposide), the anthracyclines (doxorubicin, daunorubicin, and idarubicin), and the anthracenedione mitoxantrone target topoisomerase II and are approved for clinical use in the United States. A scheme for the metabolism of etoposide by cytochrome P450 and oxidase is shown (top right).
Kinetic, drug-enzyme binding, and saturation transfer difference [1H]-nuclear magnetic resonance spectroscopy studies all indicate that interfacial poisons contact topoisomerase II in addition to the DNA.72–77 Despite the specific contacts that interfacial poisons make with both the protein and the cleaved DNA, they are surprisingly diverse in structure (Figure 5). Generally, but not always, they contain a multi-ring core with aromatic elements as well as a pendant ring.78, 79 However, based strictly on the structure of a compound, it is difficult to predict whether it will act as a topoisomerase II poison.
In contrast to interfacial topoisomerase II poisons, covalent poisons contain protein-reactive groups and covalently adduct the enzyme at amino acids outside of the active site.13, 80, 81 Most covalent poisons incorporate sulfhydryl-reactive groups such as quinones or maleimides and modify the enzyme through an acylation reaction.80–86 Benzoquinone (Figures 4 and 6) is a classic example of a covalent topoisomerase II poison.
Figure 6.
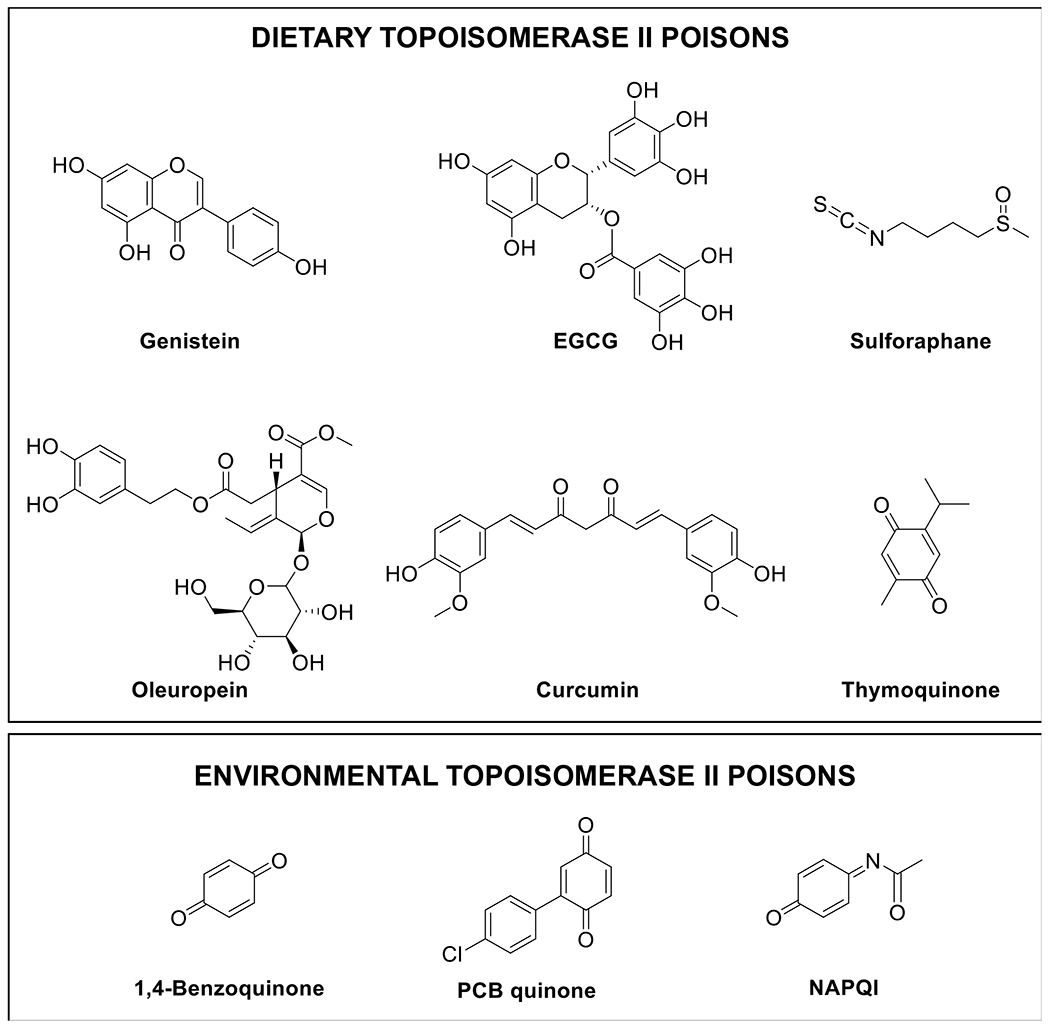
Dietary and environmental topoisomerase II poisons. A number of dietary and environmental compounds can act as topoisomerase II poisons. Included on the list of dietary natural products are: genistein (soy), EGCG [(−)-epigallocatechin gallate; green tea], sulforaphane (cruciferous vegetables), oleuropein (olives), curcumin (turmeric), thymoquinone (black seed). On the list of environmental chemicals are: 1,4-benzoquinone (benzene metabolite), PCB (polychlorinated biphenyl) quinone (industrial fluid), and NAPQI (N-acetyl-p-benzoquinone imine; acetaminophen metabolite).
Covalent topoisomerase II poisons can be distinguished from interfacial poisons by a number of unique properties (Figure 4). First, because covalent poisons contain reactive groups, their ability to poison topoisomerase II can be abolished by the presence of reducing or thiol-containing agents.86–88 Second, covalent poisons require the N-terminal region of the enzyme in order to exert their effects.43, 80, 84, 85 Studies suggest that these compounds increase topoisomerase II-mediated DNA cleavage, at least in part, by closing the N-terminal gate.84, 89 Consequently, in contrast to interfacial poisons, covalent poisons are unable to enhance DNA cleavage mediated by the catalytic core of topoisomerase II.43, 88 Third, although covalent poisons enhance DNA cleavage when added to the topoisomerase II-DNA complex, they display the distinguishing feature of inhibiting enzyme activity when incubated with the enzyme prior to the addition of DNA.80, 83, 84, 87, 88, 90–93 This feature may reflect the fact that closing the N-terminal protein gate prevents DNA binding.80, 89 However, there is also evidence that covalent poisons adduct an essential reactive residue that is exposed (or unprotected) in the absence of DNA.80, 85 Finally, while interfacial poisons always act by inhibiting DNA ligation, results with covalent poisons are more equivocal. Ultimately, it is still an open question as to the precise mechanism by which covalent poisons increase the levels of topoisomerase II-DNA cleavage complexes.
Topoisomerase II Poisons as Anticancer Drugs.
Topoisomerase II poisons represent a group of widely prescribed anticancer drugs.13, 15, 17, 18, 54 Currently, six topoisomerase II-targeted agents are approved for use in the United States.13, 15, 17, 18, 54 These drugs encompass a group of naturally derived and synthetic compounds and are used to treat a variety of human malignancies (Figure 5). Notably, etoposide, doxorubicin, and their derivatives are frontline therapies for a number of systemic cancers and solid tumors, including leukemias, lymphomas, sarcomas, breast cancers, lung cancers, neuroblastomas, and germ-cell malignancies.13, 15, 17, 18, 54 In addition, mitoxantrone is used to treat breast cancer, acute myeloid leukemia (AML), non-Hodgkin lymphoma, and multiple sclerosis. It is notable that all topoisomerase II-targeted anticancer drugs act as interfacial poisons.
Etoposide was one of the first topoisomerase II-targeted agents to be used clinically (Figure 5).13, 15, 17, 18, 54, 94, 95 The drug is a semisynthetic derivative of podophyllotoxin, which comes from Podophyllum peltatum (also known as the mayapple or American mandrake plant) and has been used as a folk remedy for over a thousand years. 94, 96
Etoposide is by far the best-characterized topoisomerase II poison.13, 15, 17–19, 54, 94, 95 Extensive research on this anticancer agent has provided a knowledge base that has paved the way for later drug studies. Etoposide was the first topoisomerase II poison that was demonstrated to inhibit the DNA ligation activity of the type II enzyme.70 Furthermore, the drug was shown to enter the binary enzyme-DNA complex primarily through interactions with the protein.74 Recent data has revealed structure-function relationships for the interaction of etoposide with topoisomerase II that are predictive with regard to new drug design.72, 73, 97, 98
Although topoisomerase II-targeted drugs represent important anticancer agents, several also appear to trigger chromosomal translocations that lead to the formation of specific leukemias. About 2-3% of patients treated with etoposide or doxorubicin eventually develop acute myeloid leukemia (AML) characterized by translocations with breakpoints in the mixed lineage leukemia (MLL) gene at chromosomal band 11q23.18, 53, 61, 62, 64, 95 Furthermore, the increased use of mitoxantrone to treat breast cancer and multiple sclerosis is associated with the development of acute promyelocytic leukemia (APL) characterized by chromosome 15:17 translocations involving the promyelocytic leukemia gene (PML) and the retinoic acid receptor α (RARA) genes.18, 60–62, 99 In all cases, chromosomal breakpoints are located in close proximity to sites of topoisomerase II-mediated DNA cleavage. In the case of APL patients, ~60% of the breakpoints that have been sequenced reside within an 8-bp region that centers on a mitoxantrone-induced topoisomerase II-DNA cleavage site.18, 60–62, 99
Etoposide quinone (Figure 5), a metabolite generated by phase I metabolism, has been implicated in the generation of etoposide-induced AMLs.18, 100–103 Etoposide is converted to a catechol by CYP3A4 in the liver, which is carried in the blood throughout the body. The catechol is converted to the quinone in the oxidizing environment of the hematopoietic system.18, 101–103 In contrast to the parent drug, which acts as an interfacial poison, etoposide quinone enhances topoisomerase II-mediated DNA cleavage by acting primarily as a covalent poison.92, 104, 105
Although topoisomerase IIα and topoisomerase IIβ are both targets for anticancer drugs, mounting circumstantial evidence indicates that topoisomerase IIβ is the isoform primarily responsible for initiating drug-induced leukemias.18, 54, 63, 64, 106 The initial evidence supporting this hypothesis came from a mouse skin carcinogenesis model in which the incidence of secondary malignancy was greatly diminished in skin-specific top2β–knockout mice.107 Moreover, etoposide-induced DNA sequence rearrangements in cellular models required topoisomerase IIβ.107
Later evidence implicating topoisomerase IIβ in the generation of leukemias came from translocation models. Translocations require the juxtaposition of chromosomal partners and genes are expressed in transcription factories that bring multiple chromosomes into close proximity.108, 109 Studies in cultured human cell lines indicate that MLL and its two most common translocation partners are found in the same transcription factory in multiple human cell lines63, 64, 110, 111 and the majority of MLL breaks generated by etoposide treatment in these factories are dependent on the β isoform.63 In addition, the genotoxic effects of etoposide in these cells appear to be mediated primarily by topoisomerase IIβ.63
Finally, because expression of topoisomerase IIα, but not topoisomerase IIβ, is proliferation dependent, differentiated tissues express the β isoform almost exclusively.18, 25 Thus, some of the off-target toxicities of topoisomerase II-targeted drugs, including cardiomyopathy, are attributed (at least in part) to the β isoform.18, 63, 106, 112 This has led to interest in the potential development of a drug that displays specificity for topoisomerase IIα, such as NK314 and ARN-21934.18, 113, 114
Dietary Topoisomerase II Poisons.
Topoisomerase II poisons are regularly consumed as part of the human diet. These include bioflavonoids (flavones, isoflavones, and flavonols), catechins, catechols, isothiocyanates, and quinones (Figure 6).81, 86, 90, 91, 115–118
Bioflavonoids are a diverse group of polyphenolic compounds that are constituents of many fruits, vegetables, legumes, and plant leaves.81, 119–122 Studies suggest that these compounds can help protect against cancer, cardiovascular disease, osteoporosis, age-related diseases, and inflammation.81, 119–122 Although bioflavonoids exert a range of effects on human cells, a number of them are potent topoisomerase II poisons in vitro and in cellulo.81, 100, 115, 119–124 Genistein, which is one of the most abundant isoflavones in soy, is perhaps the best-characterized bioflavonoid-based topoisomerase II poison (Figure 6).81, 100, 115, 124 It acts as an interfacial poison, and its activity is dependent on the presence of a 4’-hydroxyl moiety on the B ring.100, 115, 116 Genistein is highly active against both topoisomerase IIα and topoisomerase IIβ, and displays a potency and efficacy against the enzymes similar to that of etoposide.100, 115, 124 In humans, genistein reaches micromolar concentrations in plasma following ingestion.125
Catechins are another important class of bioflavonoids with activity towards topoisomerase II.100, 117 Green tea, which is one of the most commonly consumed beverages in the world, is a rich source of (−)-epigallocatechin gallate (EGCG) and related catechins (Figure 6). Diets rich in these compounds are associated with reduced risk of breast, prostate, colorectal, and lung cancer.116, 122, 126 In contrast to genistein, EGCG is a covalent topoisomerase II poison.116, 117 The mechanistic differences between bioflavonoids and catechins appear to be related to structural elements in the B and C rings. Although the C-4’ hydroxyl of the B ring is critical for bioflavonoids to act as interfacial poisons, the inclusion of two additional B ring hydroxyl groups increases redox activity and is required for catechins to act as covalent poisons. Because EGCG contains a non-aromatic C ring, it is unable to act as an interfacial topoisomerase II poison and functions exclusively as a covalent poison.116, 117 It is notable that the concentration of EGCG in human plasma and saliva is estimated to be as high as 4 and 48 μM, respectively, following consumption of ~3 cups of green tea.127
Olive plants are a rich source of catechols that are based on hydroxytyrosol (including oleuropein and verbascoside) (Figure 6).128, 129 These compounds are powerful antioxidants that are converted to reactive quinones under oxidizing conditions.81 Hydroxytyrosol, oleuropein, and verbascoside are found in all parts of the olive plant, including the leaves, bark, and fruit, as well as the oil pressed from the fruit. These metabolites are prevalent in the Mediterranean diet, which has great potential for cancer prevention.130–132 Similar to other compounds with the potential to form quinones, hydroxytyrosol, oleuropein, and verbascoside act as covalent topoisomerase II poisons.118 These compounds also enhance enzyme-mediated DNA cleavage in complex formulations intended for human dietary consumption, such as olive leaf extracts and olive oil.118 Olive oil and hydroxytyrosol are currently in clinical trials as preventative measures for breast cancer (Clinicaltrials.gov Identifier NCT04174391 and NCT02068092).
Isothiocyanates, such as sulforaphane, are found in cruciferous vegetables, such as broccoli, cabbage, cauliflower, and kale (Figure 6) and are believed to have chemopreventative properties.133 These compounds are reactive and also act as covalent topoisomerase II poisons.86
Curcumin is the principal flavor and color component of the spice turmeric (Figure 6).134 In aqueous solution at physiological pH, it undergoes a spontaneous and complex autoxidation reaction that generates a series of quinone methide intermediates.135 Although the parent compound and the stable bicyclopentadione end product display no activity toward topoisomerase II, the quinone oxidation intermediates are covalent poisons.90, 136 Even in the complex formulation of turmeric, curcumin intermediates function as topoisomerase II poisons.90 Beyond its culinary uses, curcumin is widely used in traditional Chinese herbal and Ayurvedic medicine to treat inflammation, bacterial infections, and cancers.137–139 More than 100 clinical research trials have been initiated to investigate curcumin and its metabolites as an anticancer agent [Clinincaltrials.gov Identifiers: NCT04731844 (prostate cancer), NCT02724202 (colon cancer), NCT04208334 (head and neck cancer)].
Thymoquinone is the major bioactive compound in Nigella sativa, also known as black seed (Figure 6).140 This Mediterranean plant has a rich history of medicinal use in Middle Eastern, Northern African, and Indian cultures that dates back more than 3,000 years.140 Thymoquinone is a covalent topoisomerase II poison.91 Black seed extract and oil, which are the medicinal form of the plant, also enhance topoisomerase II-mediated DNA cleavage.91 Historically, black seed has been used to treat a variety of illnesses associated with inflammation, including asthma, bronchitis, fever, arthritis, and rheumatism. Recently, it has been shown to display anticancer activity in cellular and animal models.140, 141 Following ingestion, its concentration in human plasma is in the micromolar range.142
6,6’-Dihydroxythiobinupharidine is the active ingredient in the Nuphar leutea, the yellow water lily. N. leutea has been used in traditional medicine by a variety of indigenous populations.143, 144 6,6’-Dihydroxythiobinupharidine is a covalent topoisomerase II poison and appears to display selectivity for the α over the β isoform.88
Besides the treatment-related leukemias associated with topoisomerase II-targeted drugs, the only other malignancies that display 11q23 translocations are infant AMLs or acute lymphoblastic leukemias (ALLs).18, 145 Approximately 80% of infants diagnosed with these leukemias display translocations in the MLL gene. The chromosomal translocations associated with these cancers have been observed in utero, indicating that infant leukemias are initiated during gestation.18, 145 Epidemiological studies indicate that the risk of developing these leukemias increases more than 3-fold when pregnant women consume foods/drinks that are rich in dietary topoisomerase II poisons.18, 53, 123, 146 Notably, treatment of cultured human cells with dietary bioflavonoids induces cleavage within the MLL gene.123 Thus, while the consumption of topoisomerase II poisons in the human diet appears to be chemopreventative, it has the capacity to trigger leukemias in developing embryos.
Environmental Topoisomerase II Poisons.
A variety of environmental quinone-based compounds that are damaging to human health are also potent topoisomerase II poisons (Figure 6).83, 84, 87, 89, 147–150 All of these compounds act as covalent poisons. For example, 1,4-benzoquinone, a major metabolite of benzene, is associated with the development of leukemia.150 Quinone metabolites of polychlorinated biphenyls (PCBs), which were used as industrial diluents, lubricants, and cooling fluids until the 1970s, are highly carcinogenic.149 N-acetyl-p-benzoquinone imine (NAPQI), which is the toxic metabolite of acetaminophen, is known to be a potent liver toxin.147 Finally, 1,2-naphthoquinone, a secondary metabolite of naphthalene, is an environmental pollutant found in diesel exhaust.93
Summary.
In order to carry out their essential nuclear functions, type II topoisomerases generate transient double-stranded breaks in the genetic material. Consequently, although they are required for cell survival, type II topoisomerases are intrinsically dangerous enzymes. Topoisomerase II poisons exploit this hazardous property and convert the type II enzyme into “molecular scissors” that fragment the genome. Topoisomerase II poisons are structurally and mechanistically diverse, and the only apparent feature that some of them have in common is the ability to increase levels of topoisomerase II-mediated DNA cleavage. Although several topoisomerase II poisons can cure or prevent cancer in humans, these same compounds have been linked to the generation of specific leukemias. Although the catalytic mechanism of topoisomerase II has been well-studied, we still have much to learn about how topoisomerase II poisons alter enzyme activity and generate DNA damage. Hopefully, further research in the field will lead to the development of more effective anticancer drugs that display fewer harmful consequences.
Funding
Work in the senior author’s laboratory is supported by National Institutes of Health (NIH) research grant 5R01 GM126363 (N.O.) and by United States Veterans Administration Merit Review award I01 Bx002198 (N.O.). K.R.V. was a trainee under NIH grants R25 GM062459 and T32 GM08320 and was supported in part by a Research Supplement to Grant 5R01 GM033944 to Promote Diversity in Health-Related Research. A.A.O. is a member of the Chemical and Physical Biology Program, was a trainee under NIH grant T32 CA009582, and is supported in part by NIH pre-doctoral fellowship F31 AI149863.
Footnotes
The authors declare no competing financial interests.
Contributor Information
Kendra R. Vann, Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, Tennessee 37232, United States.
Alexandria A. Oviatt, Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, Tennessee 37232, United States.
Neil Osheroff, Departments of Biochemistry and Medicine (Hematology/Oncology), Vanderbilt University School of Medicine, Nashville, Tennessee 37232, and VA Tennessee Valley Healthcare System, Nashville, TN 37212, United States.
REFERENCES
- [1].Gross DS, Chowdhary S, Anandhakumar J, and Kainth AS (2015) Chromatin, Curr. Biol. 25, R1158–1163. [DOI] [PubMed] [Google Scholar]
- [2].Bates AD, and Maxwell A (2005) DNA Topology, Oxford University Press, New York, USA. [Google Scholar]
- [3].Buck D (2009) DNA topology, In Proceedings of Symposia in Applied Mathematics: Applications of Knot Theory (Buck D, and Flapan E, Eds.), pp 47–80, American Mathematical Society, Providence. [Google Scholar]
- [4].Liu Z, Deibler RW, Chan HS, and Zechiedrich L (2009) The why and how of DNA unlinking, Nucleic Acids Res. 37, 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Irobalieva RN, Fogg JM, Catanese DJ Jr., Sutthibutpong T, Chen M, Barker AK, Ludtke SJ, Harris SA, Schmid MF, Chiu W, and Zechiedrich L (2015) Structural diversity of supercoiled DNA, Nat Commun 6, 8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Finzi L, and Olson WK (2016) The emerging role of DNA supercoiling as a dynamic player in genomic structure and function, Biophys. Rev. 8, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deweese JE, Osheroff MA, and Osheroff N (2008) DNA topology and topoisomerases: teaching a “knotty” subject, Biochem. Mol. Biol. Educ 37, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ashley RE, and Osheroff N (2016) Regulation of DNA topology by topoisomerases: mathematics at the molecular level, In Knots, Low-Dimensional Topology and Applications (Adams CC, Gordon CM, Jones VFR, Kauffman LH, Lambropoulou S, Millett KC, Przytycki JH, Ricca R, and Sazadanovic R, Eds.), Springer, Greece. [Google Scholar]
- [9].Travers A, and Muskhelishvili G (2007) A common topology for bacterial and eukaryotic transcription initiation?, EMBO Rep. 8, 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Santos-Pereira JM, and Aguilera A (2015) R loops: new modulators of genome dynamics and function, Nat Rev Genet 16, 583–597. [DOI] [PubMed] [Google Scholar]
- [11].Kuzminov A (2018) When DNA topology turns deadly - RNA polymerases dig in their R-loops to stand their ground: new positive and negative (super)twists in the replication-transcription conflict, Trends Genet. 34, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chakraborty P (2020) New insight into the biology of R-loops, Mutat. Res. 821, 111711. [DOI] [PubMed] [Google Scholar]
- [13].Deweese JE, and Osheroff N (2009) The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing, Nucleic Acids Res. 37, 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nitiss JL (2009) DNA topoisomerase II and its growing repertoire of biological functions, Nat. Rev. Cancer 9, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pommier Y, Leo E, Zhang H, and Marchand C (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs, Chem. Biol 17, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vos SM, Tretter EM, Schmidt BH, and Berger JM (2011) All tangled up: how cells direct, manage and exploit topoisomerase function, Nat. Rev. Mol. Cell Biol. 12, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pommier Y (2013) Drugging topoisomerases: lessons and challenges, ACS Chem. Biol 8, 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pendleton M, Lindsey RH Jr., Felix CA, Grimwade D, and Osheroff N (2014) Topoisomerase II and leukemia, Ann. N. Y. Acad. Sci. 1310, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McKie SJ, Maxwell A, and Neuman KC (2020) Mapping DNA topoisomerase binding and cleavage genome wide using next-generation sequencing techniques, Genes (Basel) 11, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen SH, Chan NL, and Hsieh TS (2013) New mechanistic and functional insights into DNA topoisomerases, Annu. Rev. Biochem 82, 139–170. [DOI] [PubMed] [Google Scholar]
- [21].Dalvie ED, and Osheroff N (2021) DNA topoisomerases: type II, In The Encyclopedia of Biological Chemistry (Jez JM, Ed.) in press ed., Elsevier, Inc. [Google Scholar]
- [22].Deweese JE, Burgin AB, and Osheroff N (2008) Human topoisomerase IIα uses a two-metal-ion mechanism for DNA cleavage, Nucleic Acids Res. 36, 4883–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stuchinskaya T, Mitchenall LA, Schoeffler AJ, Corbett KD, Berger JM, Bates AD, and Maxwell A (2009) How do type II topoisomerases use ATP hydrolysis to simplify DNA topology beyond equilibrium? Investigating the relaxation reaction of nonsupercoiling type II topoisomerases, J. Mol. Biol. 385, 1397–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bromberg KD, Hendricks C, Burgin AB, and Osheroff N (2002) Human topoisomerase IIα possesses an intrinsic nucleic acid specificity for DNA ligation. Use of 5’ covalently activated oligonucleotide substrates to study enzyme mechanism, J. Biol. Chem. 277, 31201–31206. [DOI] [PubMed] [Google Scholar]
- [25].Austin CA, Lee KC, Swan RL, Khazeem MM, Manville CM, Cridland P, Treumann A, Porter A, Morris NJ, and Cowell IG (2018) TOP2B: the first thirty years, Int. J. Mol. Sci 19, 2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Drake FH, Hofmann GA, Bartus HF, Mattern MR, Crooke ST, and Mirabelli CK (1989) Biochemical and pharmacological properties of p170 and p180 forms of topoisomerase II, Biochemistry 28, 8154–8160. [DOI] [PubMed] [Google Scholar]
- [27].Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, and Drake FH (1991) Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells, Cell Growth Differ. 2, 209–214. [PubMed] [Google Scholar]
- [28].Padget K, Pearson AD, and Austin CA (2000) Quantitation of DNA topoisomerase IIα and β in human leukaemia cells by immunoblotting, Leukemia 14, 1997–2005. [DOI] [PubMed] [Google Scholar]
- [29].Yu X, Davenport JW, Urtishak KA, Carillo ML, Gosai SJ, Kolaris CP, Byl JAW, Rappaport EF, Osheroff N, Gregory BD, and Felix CA (2017) Genome-wide TOP2A DNA cleavage is biased toward translocated and highly transcribed loci, Genome Res. 27, 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Heintzman DR, Campos LV, Byl JAW, Osheroff N, and Dewar JM (2019) Topoisomerase II is crucial for fork convergence during vertebrate replication termination, Cell Rep 29, 422–436 e425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Grue P, Grasser A, Sehested M, Jensen PB, Uhse A, Straub T, Ness W, and Boege F (1998) Essential mitotic functions of DNA topoisomerase IIα are not adopted by topoisomerase IIβ in human H69 cells, J. Biol. Chem 273, 33660–33666. [DOI] [PubMed] [Google Scholar]
- [32].Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, and Mielke C (2002) Dynamics of human DNA topoisomerases IIα and IIβ in living cells, J. Cell Biol 157, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yang X, Li W, Prescott ED, Burden SJ, and Wang JC (2000) DNA topoisomerase IIβ and neural development, Science 287, 131–134. [DOI] [PubMed] [Google Scholar]
- [34].Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, and Rosenfeld MG (2006) A topoisomerase IIβ-mediated dsDNA break required for regulated transcription, Science 312, 1798–1802. [DOI] [PubMed] [Google Scholar]
- [35].Chen W, Qiu J, and Shen YM (2012) Topoisomerase IIα, rather than IIβ, is a promising target in development of anti-cancer drugs, Drug Discov. Ther 6, 230–237. [PubMed] [Google Scholar]
- [36].Schmidt BH, Osheroff N, and Berger JM (2012) Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity, Nat. Struct. Mol. Biol 19, 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mirski SE, Gerlach JH, Cummings HJ, Zirngibl R, Greer PA, and Cole SP (1997) Bipartite nuclear localization signals in the C terminus of human topoisomerase IIα, Exp. Cell Res 237, 452–455. [DOI] [PubMed] [Google Scholar]
- [38].McClendon AK, Rodriguez AC, and Osheroff N (2005) Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks, J. Biol. Chem 280, 39337–39345. [DOI] [PubMed] [Google Scholar]
- [39].McClendon AK, Gentry AC, Dickey JS, Brinch M, Bendsen S, Andersen AH, and Osheroff N (2008) Bimodal recognition of DNA geometry by human topoisomerase IIα: preferential relaxation of positively supercoiled DNA requires elements in the C-terminal domain, Biochemistry 47, 13169–13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zechiedrich EL, and Osheroff N (1990) Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers, EMBO J. 9, 4555–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dong KC, and Berger JM (2007) Structural basis for gate-DNA recognition and bending by type IIA topoisomerases, Nature 450, 1201–1205. [DOI] [PubMed] [Google Scholar]
- [42].Roca J, and Wang JC (1992) The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases, Cell 71, 833–840. [DOI] [PubMed] [Google Scholar]
- [43].Lindsey RH Jr., Pendleton M, Ashley RE, Mercer SL, Deweese JE, and Osheroff N (2014) Catalytic core of human topoisomerase IIα: insights into enzyme-DNA interactions and drug mechanism, Biochemistry 53, 6595–65602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schmidt BH, Burgin AB, Deweese JE, Osheroff N, and Berger JM (2010) A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases, Nature 465, 641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lee S, Jung SR, Heo K, Byl JA, Deweese JE, Osheroff N, and Hohng S (2012) DNA cleavage and opening reactions of human topoisomerase IIα are regulated via Mg2+-mediated dynamic bending of gate-DNA, Proc. Natl. Acad. Sci. U. S. A 109, 2925–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bax BD, Murshudov G, Maxwell A, and Germe T (2019) DNA topoisomerase inhibitors: trapping a DNA-cleaving machine in motion, J. Mol. Biol 431, 3427–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lee I, Dong KC, and Berger JM (2013) The role of DNA bending in type IIA topoisomerase function, Nucleic Acids Res. 41, 5444–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jang Y, Son H, Lee SW, Hwang W, Jung SR, Byl JAW, Osheroff N, and Lee S (2019) Selection of DNA cleavage sites by topoisomerase II results from enzyme-induced flexibility of DNA, Cell Chem Biol 26, 502–511 e503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Deweese JE, and Osheroff N (2009) Coordinating the two protomer active sites of human topoisomerase IIα: nicks as topoisomerase II poisons, Biochemistry 48, 1439–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Harkins TT, and Lindsley JE (1998) Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 1. A DNA-dependent burst in ATP hydrolysis, Biochemistry 37, 7292–7298. [DOI] [PubMed] [Google Scholar]
- [51].Markovits J, Pommier Y, Kerrigan D, Covey JM, Tilchen EJ, and Kohn KW (1987) Topoisomerase II-mediated DNA breaks and cytotoxicity in relation to cell proliferation and the cell cycle in NIH 3T3 fibroblasts and L1210 leukemia cells, Cancer Res. 47, 2050–2055. [PubMed] [Google Scholar]
- [52].D’Arpa P, Beardmore C, and Liu LF (1990) Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons, Cancer Res. 50, 6919–6924. [PubMed] [Google Scholar]
- [53].Felix CA, Kolaris CP, and Osheroff N (2006) Topoisomerase II and the etiology of chromosomal translocations, DNA Repair (Amst.) 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- [54].Nitiss JL (2009) Targeting DNA topoisomerase II in cancer chemotherapy, Nat. Rev. Cancer 9, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability, Nat. Rev. Mol. Cell Biol. 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Morimoto S, Tsuda M, Bunch H, Sasanuma H, Austin C, and Takeda S (2019) Type II DNA topoisomerases cause spontaneous double-strand breaks in genomic DNA, Genes (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Riccio AA, Schellenberg MJ, and Williams RS (2020) Molecular mechanisms of topoisomerase 2 DNA-protein crosslink resolution, Cell. Mol. Life Sci 77, 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sun Y, Saha LK, Saha S, Jo U, and Pommier Y (2020) Debulking of topoisomerase DNA-protein crosslinks (TOP-DPC) by the proteasome, non-proteasomal and non-proteolytic pathways, DNA Repair (Amst) 94, 102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zagnoli-Vieira G, and Caldecott KW (2020) Untangling trapped topoisomerases with tyrosyl-DNA phosphodiesterases, DNA Repair (Amst) 94, 102900. [DOI] [PubMed] [Google Scholar]
- [60].Hasan SK, Mays AN, Ottone T, Ledda A, La Nasa G, Cattaneo C, Borlenghi E, Melillo L, Montefusco E, Cervera J, Stephen C, Satchi G, Lennard A, Libura M, Byl JA, Osheroff N, Amadori S, Felix CA, Voso MT, Sperr WR, Esteve J, Sanz MA, Grimwade D, and Lo-Coco F (2008) Molecular analysis of t(15;17) genomic breakpoints in secondary acute promyelocytic leukemia arising after treatment of multiple sclerosis, Blood 112, 3383–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Joannides M, and Grimwade D (2010) Molecular biology of therapy-related leukaemias, Clin. Transl. Oncol 12, 8–14. [DOI] [PubMed] [Google Scholar]
- [62].Joannides M, Mays AN, Mistry AR, Hasan SK, Reiter A, Wiemels JL, Felix CA, Coco FL, Osheroff N, Solomon E, and Grimwade D (2011) Molecular pathogenesis of secondary acute promyelocytic leukemia, Mediterr. J. Hematol. Infect. Dis 3, e2011045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cowell IG, Sondka Z, Smith K, Lee KC, Manville CM, Sidorczuk-Lesthuruge M, Rance HA, Padget K, Jackson GH, Adachi N, and Austin CA (2012) Model for MLL translocations in therapy-related leukemia involving topoisomerase IIβ-mediated DNA strand breaks and gene proximity, Proc. Natl. Acad. Sci. U. S. A 109, 8989–8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cowell IG, and Austin CA (2012) Mechanism of generation of therapy related leukemia in response to anti-topoisomerase II agents, Int. J. Environ. Res. Public Health 9, 2075–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Andoh T, and Ishida R (1998) Catalytic inhibitors of DNA topoisomerase II, Biochim. Biophys. Acta 1400, 155–171. [DOI] [PubMed] [Google Scholar]
- [66].Larsen AK, Escargueil AE, and Skladanowski A (2003) Catalytic topoisomerase II inhibitors in cancer therapy, Pharmacol. Ther 99, 167–181. [DOI] [PubMed] [Google Scholar]
- [67].Fortune JM, and Osheroff N (1998) Merbarone inhibits the catalytic activity of human topoisomerase IIα by blocking DNA cleavage, J. Biol. Chem 273, 17643–17650. [DOI] [PubMed] [Google Scholar]
- [68].Fortune JM, and Osheroff N (2000) Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice, Prog. Nucleic Acid Res. Mol. Biol 64, 221–253. [DOI] [PubMed] [Google Scholar]
- [69].Minniti E, Byl JAW, Riccardi L, Sissi C, Rosini M, De Vivo M, Minarini A, and Osheroff N (2017) Novel xanthone-polyamine conjugates as catalytic inhibitors of human topoisomerase IIα, Bioorg. Med. Chem. Lett 27, 4687–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Osheroff N (1989) Effect of antineoplastic agents on the DNA cleavage/religation reaction of eukaryotic topoisomerase II: inhibition of DNA religation by etoposide, Biochemistry 28, 6157–6160. [DOI] [PubMed] [Google Scholar]
- [71].Pommier Y, and Marchand C (2012) Interfacial inhibitors: targeting macromolecular complexes, Nat. Rev. Drug Discov 11, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wu CC, Li YC, Wang YR, Li TK, and Chan NL (2013) On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs, Nucleic Acids Res. 41, 10630–10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, and Chan NL (2011) Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide, Science 333, 459–462. [DOI] [PubMed] [Google Scholar]
- [74].Burden DA, Kingma PS, Froelich-Ammon SJ, Bjornsti MA, Patchan MW, Thompson RB, and Osheroff N (1996) Topoisomerase II-etoposide interactions direct the formation of drug-induced enzyme-DNA cleavage complexes, J. Biol. Chem 271, 29238–29244. [DOI] [PubMed] [Google Scholar]
- [75].Kingma PS, Burden DA, and Osheroff N (1999) Binding of etoposide to topoisomerase II in the absence of DNA: decreased affinity as a mechanism of drug resistance, Biochemistry 38, 3457–3461. [DOI] [PubMed] [Google Scholar]
- [76].Wilstermann AM, Bender RP, Godfrey M, Choi S, Anklin C, Berkowitz DB, Osheroff N, and Graves DE (2007) Topoisomerase II - drug interaction domains: identification of substituents on etoposide that interact with the enzyme, Biochemistry 46, 8217–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bender RP, Jablonksy MJ, Shadid M, Romaine I, Dunlap N, Anklin C, Graves DE, and Osheroff N (2008) Substituents on etoposide that interact with human topoisomerase IIα in the binary enzyme-drug complex: contributions to etoposide binding and activity, Biochemistry 47, 4501–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sinha BK, Politi PM, Eliot HM, Kerrigan D, and Pommier Y (1990) Structure-activity relations, cytotoxicity and topoisomerase II dependent cleavage induced by pendulum ring analogues of etoposide, Eur. J. Cancer 26, 590–593. [DOI] [PubMed] [Google Scholar]
- [79].Bailly C (2012) Contemporary challenges in the design of topoisomerase II inhibitors for cancer chemotherapy, Chem. Rev. 112, 3611–3640. [DOI] [PubMed] [Google Scholar]
- [80].Bender RP, Ham AJ, and Osheroff N (2007) Quinone-induced enhancement of DNA cleavage by human topoisomerase IIα: adduction of cysteine residues 392 and 405, Biochemistry 46, 2856–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ketron AC, and Osheroff N (2014) Phytochemicals as anticancer and chemopreventive topoisomerase II poisons, Phytochem. Rev 13, 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang H, Mao Y, Chen AY, Zhou N, LaVoie EJ, and Liu LF (2001) Stimulation of topoisomerase II-mediated DNA damage via a mechanism involving protein thiolation, Biochemistry 40, 3316–3323. [DOI] [PubMed] [Google Scholar]
- [83].Lindsey RH Jr., Bromberg KD, Felix CA, and Osheroff N (2004) 1,4-Benzoquinone is a topoisomerase II poison, Biochemistry 43, 7563–7574. [DOI] [PubMed] [Google Scholar]
- [84].Bender RP, Lehmler HJ, Robertson LW, Ludewig G, and Osheroff N (2006) Polychlorinated biphenyl quinone metabolites poison human topoisomerase IIα: altering enzyme function by blocking the N-terminal protein gate, Biochemistry 45, 10140–10152. [DOI] [PubMed] [Google Scholar]
- [85].Bender RP, and Osheroff N (2007) Mutation of cysteine residue 455 to alanine in human topoisomerase IIα confers hypersensitivity to quinones: enhancing DNA scission by closing the N-terminal protein gate, Chem. Res. Toxicol 20, 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lin RK, Zhou N, Lyu YL, Tsai YC, Lu CH, Kerrigan J, Chen YT, Guan Z, Hsieh TS, and Liu LF (2011) Dietary isothiocyanate-induced apoptosis via thiol modification of DNA topoisomerase IIα, J. Biol. Chem. 286, 33591–33600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lindsey RH Jr., Bender RP, and Osheroff N (2005) Effects of benzene metabolites on DNA cleavage mediated by human topoisomerase IIα: 1,4-hydroquinone is a topoisomerase II poison, Chem. Res. Toxicol 18, 761–770. [DOI] [PubMed] [Google Scholar]
- [88].Dalvie ED, Gopas J, Golan-Goldhirsh A, and Osheroff N (2019) 6,6’-Dihydroxythiobinupharidine as a poison of human type II topoisomerases, Bioorg. Med. Chem. Lett 29, 1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Mondrala S, and Eastmond DA (2010) Topoisomerase II inhibition by the bioactivated benzene metabolite hydroquinone involves multiple mechanisms, Chem. Biol. Interact 184, 259–268. [DOI] [PubMed] [Google Scholar]
- [90].Ketron AC, Gordon ON, Schneider C, and Osheroff N (2013) Oxidative metabolites of curcumin poison human type II topoisomerases, Biochemistry 52, 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ashley RE, and Osheroff N (2014) Natural products as topoisomerase II poisons: effects of thymoquinone on DNA cleavage mediated by human topoisomerase IIα, Chem. Res. Toxicol 27, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Smith NA, Byl JA, Mercer SL, Deweese JE, and Osheroff N (2014) Etoposide quinone is a covalent poison of human topoisomerase IIβ, Biochemistry 53, 3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Collins JA, and Osheroff N (2021) 1,2-Naphthoquinone as a poison of human type II topoisomerases, Chem. Res. Toxicol 34, 1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hande KR (1998) Etoposide: four decades of development of a topoisomerase II inhibitor, Eur. J. Cancer 34, 1514–1521. [DOI] [PubMed] [Google Scholar]
- [95].Baldwin EL, and Osheroff N (2005) Etoposide, topoisomerase II and cancer, Curr. Med. Chem. Anticancer Agents 5, 363–372. [DOI] [PubMed] [Google Scholar]
- [96].Loike JD, Brewer CF, Sternlicht H, Gensler WJ, and Horwitz SB (1978) Structure-activity study of the inhibition of microtubule assembly in vitro by podophyllotoxin and its congeners, Cancer Res. 38, 2688–2693. [PubMed] [Google Scholar]
- [97].Drwal MN, Marinello J, Manzo SG, Wakelin LP, Capranico G, and Griffith R (2014) Novel DNA topoisomerase IIα inhibitors from combined ligand- and structure-based virtual screening, PLoS One 9, e114904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Huang NL, and Lin JH (2014) Drug-induced conformational population shifts in topoisomerase-DNA ternary complexes, Molecules 19, 7415–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Mistry AR, Felix CA, Whitmarsh RJ, Mason A, Reiter A, Cassinat B, Parry A, Walz C, Wiemels JL, Segal MR, Ades L, Blair IA, Osheroff N, Peniket AJ, Lafage-Pochitaloff M, Cross NC, Chomienne C, Solomon E, Fenaux P, and Grimwade D (2005) DNA topoisomerase II in therapy-related acute promyelocytic leukemia, N. Engl. J. Med 352, 1529–1538. [DOI] [PubMed] [Google Scholar]
- [100].Austin CA, Patel S, Ono K, Nakane H, and Fisher LM (1992) Site-specific DNA cleavage by mammalian DNA topoisomerase II induced by novel flavone and catechin derivatives, Biochem. J 282, 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Felix CA, Walker AH, Lange BJ, Williams TM, Winick NJ, Cheung NK, Lovett BD, Nowell PC, Blair IA, and Rebbeck TR (1998) Association of CYP3A4 genotype with treatment-related leukemia, Proc. Natl. Acad. Sci. U. S. A 95, 13176–13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Lovett BD, Strumberg D, Blair IA, Pang S, Burden DA, Megonigal MD, Rappaport EF, Rebbeck TR, Osheroff N, Pommier YG, and Felix CA (2001) Etoposide metabolites enhance DNA topoisomerase II cleavage near leukemia-associated MLL translocation breakpoints, Biochemistry 40, 1159–1170. [DOI] [PubMed] [Google Scholar]
- [103].Atwal M, Lishman EL, Austin CA, and Cowell IG (2017) Myeloperoxidase enhances etoposide and mitoxantrone-mediated DNA damage: a target for myeloprotection in cancer chemotherapy, Mol. Pharmacol 91, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Jacob DA, Mercer SL, Osheroff N, and Deweese JE (2011) Etoposide quinone is a redox-dependent topoisomerase II poison, Biochemistry 50, 5660–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Jacob DA, Gibson EG, Mercer SL, and Deweese JE (2013) Etoposide catechol is an oxidizable topoisomerase II poison, Chem. Res. Toxicol 26, 1156–1158. [DOI] [PubMed] [Google Scholar]
- [106].Lyu YL, Kerrigan JE, Lin CP, Azarova AM, Tsai YC, Ban Y, and Liu LF (2007) Topoisomerase IIβ mediated DNA double-strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane, Cancer Res. 67, 8839–8846. [DOI] [PubMed] [Google Scholar]
- [107].Azarova AM, Lyu YL, Lin CP, Tsai YC, Lau JY, Wang JC, and Liu LF (2007) Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies, Proc. Natl. Acad. Sci. U. S. A 104, 11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, and Fraser P (2004) Active genes dynamically colocalize to shared sites of ongoing transcription, Nat. Genet 36, 1065–1071. [DOI] [PubMed] [Google Scholar]
- [109].Cook PR (2010) A model for all genomes: the role of transcription factories, J. Mol. Biol 395, 1–10. [DOI] [PubMed] [Google Scholar]
- [110].Strissel PL, Strick R, Tomek RJ, Roe BA, Rowley JD, and Zeleznik-Le NJ (2000) DNA structural properties of AF9 are similar to MLL and could act as recombination hot spots resulting in MLL/AF9 translocations and leukemogenesis, Hum. Mol. Genet 9, 1671–1679. [DOI] [PubMed] [Google Scholar]
- [111].Zhang Y, Strissel P, Strick R, Chen J, Nucifora G, Le Beau MM, Larson RA, and Rowley JD (2002) Genomic DNA breakpoints in AML1/RUNX1 and ETO cluster with topoisomerase II DNA cleavage and DNase I hypersensitive sites in t(8;21) leukemia, Proc. Natl. Acad. Sci. U. S. A 99, 3070–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zhang S, Liu X, Bawa-Khalfe T, Lu LS, Lyu YL, Liu LF, and Yeh ET (2012) Identification of the molecular basis of doxorubicin-induced cardiotoxicity, Nat. Med 18, 1639–1642. [DOI] [PubMed] [Google Scholar]
- [113].Toyoda E, Kagaya S, Cowell IG, Kurosawa A, Kamoshita K, Nishikawa K, Iiizumi S, Koyama H, Austin CA, and Adachi N (2008) NK314, a topoisomerase II inhibitor that specifically targets the alpha isoform, J. Biol. Chem 283, 23711–23720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ortega JA, Arencibia JM, Minniti E, Byl JAW, Franco-Ulloa S, Borgogno M, Genna V, Summa M, Bertozzi SM, Bertorelli R, Armirotti A, Minarini A, Sissi C, Osheroff N, and De Vivo M (2020) Novel, potent, and druglike tetrahydroquinazoline inhibitor that is highly selective for human topoisomerase II α over β, J. Med. Chem 63, 12873–12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Bandele OJ, and Osheroff N (2007) Bioflavonoids as poisons of human topoisomerase IIα and IIβ, Biochemistry 46, 6097–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Bandele OJ, Clawson SJ, and Osheroff N (2008) Dietary polyphenols as topoisomerase II poisons: B ring and C ring substituents determine the mechanism of enzyme-mediated DNA cleavage enhancement, Chem. Res. Toxicol 21, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Bandele OJ, and Osheroff N (2008) (−)-Epigallocatechin gallate, a major constituent of green tea, poisons human type II topoisomerases, Chem. Res. Toxicol. 21, 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Vann KR, Sedgeman CA, Gopas J, Golan-Goldhirsh A, and Osheroff N (2015) Effects of olive metabolites on DNA cleavage mediated by human type II topoisomerases, Biochemistry 54, 4531–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Kurzer MS, and Xu X (1997) Dietary phytoestrogens, Annu. Rev. Nutr 17, 353–381. [DOI] [PubMed] [Google Scholar]
- [120].Scalbert A, and Williamson G (2000) Dietary intake and bioavailability of polyphenols, J. Nutr 130, 2073S–2085S. [DOI] [PubMed] [Google Scholar]
- [121].Kandaswami C, Lee LT, Lee PP, Hwang JJ, Ke FC, Huang YT, and Lee MT (2005) The antitumor activities of flavonoids, In Vivo 19, 895–909. [PubMed] [Google Scholar]
- [122].Siddiqui IA, Adhami VM, Saleem M, and Mukhtar H (2006) Beneficial effects of tea and its polyphenols against prostate cancer, Mol. Nutr. Food Res 50, 130–143. [DOI] [PubMed] [Google Scholar]
- [123].Strick R, Strissel PL, Borgers S, Smith SL, and Rowley JD (2000) Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia, Proc. Natl. Acad. Sci. U. S. A 97, 4790–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Bandele OJ, and Osheroff N (2008) The efficacy of topoisomerase II-targeted anticancer agents reflects the persistence of drug-induced cleavage complexes in cells, Biochemistry 47, 11900–11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Setchell KD, Brown NM, Desai P, Zimmer-Nechemias L, Wolfe BE, Brashear WT, Kirschner AS, Cassidy A, and Heubi JE (2001) Bioavailability of pure isoflavones in healthy humans and analysis of commercial soy isoflavone supplements, J. Nutr 131, 1362S–1375S. [DOI] [PubMed] [Google Scholar]
- [126].Lopez-Lazaro M, Calderon-Montano JM, Burgos-Moron E, and Austin CA (2011) Green tea constituents (−)-epigallocatechin-3-gallate (EGCG) and gallic acid induce topoisomerase I- and topoisomerase II-DNA complexes in cells mediated by pyrogallol-induced hydrogen peroxide, Mutagenesis 26, 489–498. [DOI] [PubMed] [Google Scholar]
- [127].Bertram B, Bollow U, Rajaee-Behbahani N, Burkle A, and Schmezer P (2003) Induction of poly(ADP-ribosyl)ation and DNA damage in human peripheral lymphocytes after treatment with (-)-epigallocatechin-gallate, Mutat. Res. 534, 77–84. [DOI] [PubMed] [Google Scholar]
- [128].Agati G, Galardi C, Gravano E, Romani A, and Tattini M (2002) Flavonoid distribution in tissues of Phillyrea latifolia L. leaves as estimated by microspectrofluorometry and multispectral fluorescence microimaging, Photochem. Photobiol. 76, 350–360. [DOI] [PubMed] [Google Scholar]
- [129].Ayranci E, and Erkan N (2013) Radical scavenging capacity of methanolic Phillyrea latifolia L. extract: anthocyanin and phenolic acids composition of fruits, Molecules 18, 1798–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Hashim YZ, Eng M, Gill CI, McGlynn H, and Rowland IR (2005) Components of olive oil and chemoprevention of colorectal cancer, Nutr. Rev 63, 374–386. [DOI] [PubMed] [Google Scholar]
- [131].Couto E, Boffetta P, Lagiou P, Ferrari P, Buckland G, Overvad K, Dahm CC, Tjonneland A, Olsen A, Clavel-Chapelon F, Boutron-Ruault MC, Cottet V, Trichopoulos D, Naska A, Benetou V, Kaaks R, Rohrmann S, Boeing H, von Ruesten A, Panico S, Pala V, Vineis P, Palli D, Tumino R, May A, Peeters PH, Bueno-de-Mesquita HB, Buchner FL, Lund E, Skeie G, Engeset D, Gonzalez CA, Navarro C, Rodriguez L, Sanchez MJ, Amiano P, Barricarte A, Hallmans G, Johansson I, Manjer J, Wirfart E, Allen NE, Crowe F, Khaw KT, Wareham N, Moskal A, Slimani N, Jenab M, Romaguera D, Mouw T, Norat T, Riboli E, and Trichopoulou A (2011) Mediterranean dietary pattern and cancer risk in the EPIC cohort, Br. J. Cancer 104, 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Escrich E, Solanas M, and Moral R (2014) Olive oil and other dietary lipids in breast cancer, Cancer Treat. Res. 159, 289–309. [DOI] [PubMed] [Google Scholar]
- [133].Herr I, and Buchler MW (2010) Dietary constituents of broccoli and other cruciferous vegetables: implications for prevention and therapy of cancer, Cancer Treat. Rev. 36, 377–383. [DOI] [PubMed] [Google Scholar]
- [134].Jaffrey M (1994) Madhur Jaffrey’s spice kitchen: fifty recipes introducing indian spices and aromatic seeds, Clarkson Potter, New York. [Google Scholar]
- [135].Griesser M, Pistis V, Suzuki T, Tejera N, Pratt DA, and Schneider C (2011) Autoxidative and cyclooxygenase-2 catalyzed transformation of the dietary chemopreventive agent curcumin, J. Biol. Chem. 286, 1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Gordon ON, Luis PB, Ashley RE, Osheroff N, and Schneider C (2015) Oxidative transformation of demethoxy- and bisdemethoxycurcumin: products, mechanism of formation, and poisoning of human topoisomerase IIα, Chem. Res. Toxicol. 28, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Sharma RA, McLelland HR, Hill KA, Ireson CR, Euden SA, Manson MM, Pirmohamed M, Marnett LJ, Gescher AJ, and Steward WP (2001) Pharmacodynamic and pharmacokinetic study of oral Curcuma extract in patients with colorectal cancer, Clin. Cancer Res. 7, 1894–1900. [PubMed] [Google Scholar]
- [138].Mahady GB, Pendland SL, Yun G, and Lu ZZ (2002) Turmeric (Curcuma longa) and curcumin inhibit the growth of Helicobacter pylori, a group 1 carcinogen, Anticancer Res. 22, 4179–4181. [PubMed] [Google Scholar]
- [139].Patel VB, Misra S, Patel BB, and Majumdar AP (2010) Colorectal cancer: chemopreventive role of curcumin and resveratrol, Nutr. Cancer 62, 958–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Gali-Muhtasib H, Roessner A, and Schneider-Stock R (2006) Thymoquinone: a promising anti-cancer drug from natural sources, Int. J. Biochem. Cell Biol. 38, 1249–1253. [DOI] [PubMed] [Google Scholar]
- [141].Khader M, Bresgen N, and Eckl PM (2009) In vitro toxicological properties of thymoquinone, Food Chem. Toxicol. 47, 129–133. [DOI] [PubMed] [Google Scholar]
- [142].Elmowafy M, Samy A, Raslan MA, Salama A, Said RA, Abdelaziz AE, El-Eraky W, El Awdan S, and Viitala T (2016) Enhancement of bioavailability and pharmacodynamic effects of thymoquinone via Nanostructured Lipid Carrier (NLC) formulation, AAPS PharmSciTech 17, 663–672. [DOI] [PubMed] [Google Scholar]
- [143].Johnson LM (2006) Gitksan medicinal plants--cultural choice and efficacy, J Ethnobiol Ethnomed 2, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].El Beyrouthy M, Arnold N, Delelis-Dusollier A, and Dupont F (2008) Plants used as remedies antirheumatic and antineuralgic in the traditional medicine of Lebanon, J. Ethnopharmacol 120, 315–334. [DOI] [PubMed] [Google Scholar]
- [145].Spector LG, Xie Y, Robison LL, Heerema NA, Hilden JM, Lange B, Felix CA, Davies SM, Slavin J, Potter JD, Blair CK, Reaman GH, and Ross JA (2005) Maternal diet and infant leukemia: the DNA topoisomerase II inhibitor hypothesis: a report from the children’s oncology group, Cancer Epidemiol. Biomarkers Prev 14, 651–655. [DOI] [PubMed] [Google Scholar]
- [146].Ross JA, Potter JD, and Robison LL (1994) Infant leukemia, topoisomerase II inhibitors, and the MLL gene, J. Natl. Cancer Inst 86, 1678–1680. [DOI] [PubMed] [Google Scholar]
- [147].Bender RP, Lindsey RH Jr., Burden DA, and Osheroff N (2004) N-acetyl-p-benzoquinone imine, the toxic metabolite of acetaminophen, is a topoisomerase II poison, Biochemistry 43, 3731–3739. [DOI] [PubMed] [Google Scholar]
- [148].Lindsey RH, Bender RP, and Osheroff N (2005) Stimulation of topoisomerase II-mediated DNA cleavage by benzene metabolites, Chem. Biol. Interact. 153–154, 197–205. [DOI] [PubMed] [Google Scholar]
- [149].Ludewig G, Lehmann L, Esch H, and Robertson LW (2008) Metabolic activation of PCBs to carcinogens in vivo - a review, Environ. Toxicol. Pharmacol 25, 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Son MY, Deng CX, Hoeijmarkers JH, Rebel VI, and Hasty P (2016) A mechanism for 1,4-benzoquinone-induced genotoxicity, Oncotarget 7, 46433–46447. [DOI] [PMC free article] [PubMed] [Google Scholar]


