Summary
Organoid models have been shown to be valuable tools for studying epithelial-mesenchymal crosstalk during biological and pathological settings. Our data identified ACTA2+ PDGFRα+ repair-supportive mesenchymal cells as an important component of the conducting airway niche. Here, we provide a detailed protocol for culturing airway organoids, or bronchiolospheres, which provide an assessment of the ability of mesenchymal cells to support club-cell growth.
For complete details on the use and execution of this protocol, please refer to Moiseenko et al. (2020).
Subject areas: Cell Biology, Cell culture, Cell isolation, Flow cytometry/mass cytometry, Stem cells, Organoids
Graphical abstract
Highlights
-
•
Bronchiolospheres are a useful tool to model epithelial-mesenchymal interactions
-
•
Different types of mesenchymal cells can be used to support club cell growth
-
•
Cell differentiation within bronchiolospheres can be assessed after 16 days of culture
-
•
Bronchiolospheres can be passaged multiple times
Organoid models have been shown to be valuable tools for studying epithelial-mesenchymal crosstalk during biological and pathological settings. Our data identified ACTA2+ PDGFRα+ repair-supportive mesenchymal cells (RSMCs) as an important component of the conducting airway niche. Here, we provide a detailed protocol for culturing airway organoids, or bronchiolospheres, which provide an assessment of the ability of mesenchymal cells to support club-cell growth.
Before you begin
Timing: 15 days
Tamoxifen administration and naphthalene injury
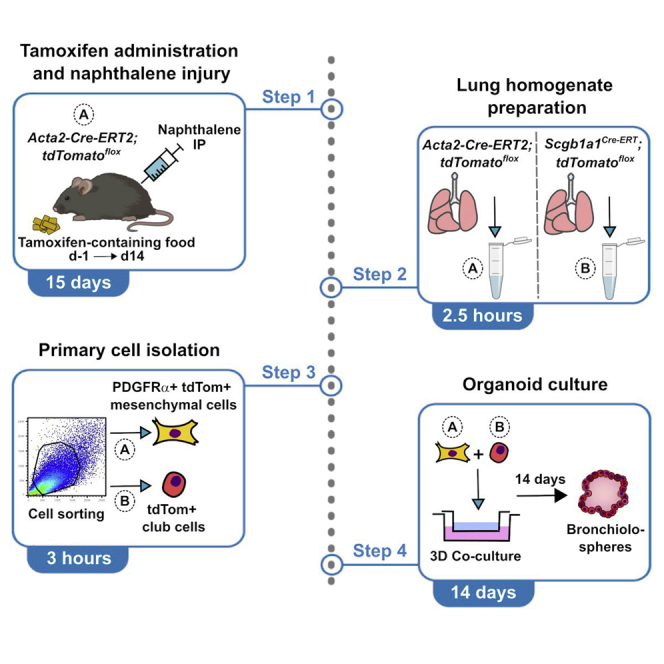
Lung mesenchymal cells used for the co-culture with club cells are obtained from 8–12-week-old Acta2-Cre-ERT2; tdTomatoflox mice 14 days after naphthalene-induced injury. Provide tamoxifen-containing food at a concentration of 0.4 g of tamoxifen per Kg of food (Altromin, Germany) starting at one day before naphthalene treatment for a period of 14 days. Administer naphthalene intraperitoneally at a concentration of 0.275 mg per gram of body weight.
Note: For naphthalene treatment, female mice are preferably used since they are more susceptible to injury.
Note: Naphthalene stock solution (20 mg/mL) is prepared by dissolving naphthalene powder in corn oil at 20°C–25°C while vortexing in the dark.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC Rat monoclonal anti-PDGFRα | BioLegend | Cat#135907; RRID: AB_2043969 |
| APC Rat IgG2a | BioLegend | Cat#400511; RRID: AB_1952838 |
| Chemicals, peptides, and recombinant proteins | ||
| Naphthalene | Sigma-Aldrich | Cat#84679 |
| Tamoxifen | Sigma-Aldrich | Cat#T5648 |
| Insulin-Transferrin-Selenium | Biozym | Cat#41400-045 |
| l-Glutamine | Gibco | Cat#11539876 |
| PBS | Gibco | Cat#10010023 |
| EDTA | Roth | Cat#6764.1 |
| Matrigel matrix | Corning | Cat#356231 |
| DMEM | Gibco | Cat#12491015 |
| αMEM | Gibco | Cat#41061029 |
| Dispase | Corning | Cat#354235 |
| Collagenase IV | Gibco | Cat # 17104019 |
| DNase I | Serva | Cat#18535.01 |
| Heparin | Stemcell Technologies | Cat#07980 |
| Y-27632 ROCK inhibitor | Stemcell Technologies | Cat#72304 |
| rmFGF10 | R&D | Cat#6224-FG-025/CF |
| rmHGF | R&D | Cat#2207-HG-025/CF |
| Ethanol | Merck | Cat#32305 |
| Hepes | Gibco | Cat#12509079 |
| Matrigel matrix | Corning | Cat #356231 |
| HBSS | Gibco | Cat#14175095 |
| FCS | Cell Biologics | Cat#6912 |
| Pen/Strep/L-Glut | Merck | Cat#G6784 |
| Corn oil | Sigma-Aldrich | Cat#C8267-500ML |
| Experimental models: organisms/strains | ||
| Mouse: Tg(Acta2-cre/ERT2)12Pcn | Pierre Chambon (Wendling et al., 2009) | MGI: 3831907 |
| Mouse: B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | Jackson Laboratory | RRID: IMSR_JAX:007905 |
| Mouse: B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh/J | Jackson Laboratory | RRID: IMSR_JAX:016225 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson Laboratory | RRID: IMSR_JAX:007914 |
| Software | ||
| FlowJo | FlowJo, LLC |
https://www.flowjo.com/solutions/flowjo RRID: SCR_008520 |
| Other | ||
| 5 mL polypropylene round-bottom tube | Corning | Cat#352063 |
| 5 mL polystyrene round-bottom tube with cell-strainer cap | Corning | Cat#352235 |
| 60-mm glass petri dishes | Sigma-Aldrich | Cat#CLS7016560 |
| Scalpel blades | Fisher Scientific | Cat# S95937A |
| Cell culture inserts 0.4 μm, 12 mm diameter | Millicell | Cat#PICM01250 |
| 24-Well cell culture plare | Greiner Bio-One | Cat#662160 |
| Cell strainer 100 μm | Sarstedt | Cat#833945100 |
| Cell strainer 70 μm | Sarstedt | Cat#833945070 |
| 0.2 μm filter | Sarstedt | Cat#831826001 |
| BD FACSAriaTMIII Cell Sorter | BD Biosciences | Cat#648282 |
| GentleMACS® Dissociator | MACS Miltenyi Biotec | Cat#130-093-235 |
| Laminar hood Class II | Heraeus | Cat#50033851 |
| Mettler-Toledo™ pH meter | Sigma-Aldrich | Cat#MT30130864 |
Materials and equipment
All the chemicals and reagents should be kept sterile. Media and buffers should be prepared under a Class II biological hood and filter-sterilized using a 0.2 μm filter (Sarstedt). For pH adjustment, use 1 M HCl or NaOH (Merck).
Digestion medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | n/a | 48.75 mL |
| HEPES | 25 mM | 1.25 mL |
| DNase I | 10 U/mL | 0.005 mL |
| Total | 50 mL |
Note: This medium should be freshly made and kept at 4°C for a maximum of 2 h.
MACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | n/a | 46.05 mL |
| EDTA | 2 mM | 3.7 mL |
| FCS | 0.5% | 0.25 mL |
| Total | 50 mL |
Note: Adjust PH to 7.2 and store at 4°C for no longer than 2 months.
Organoid medium
| Reagent | Final concentration | Amount |
|---|---|---|
| αMEM | n/a | 44.45 mL |
| FCS | 10% | 5 mL |
| Pen/Strep/L-Glut | 100 U/mL /0.1 mg/mL /2 mM | 0.5 mL |
| Insulin-transferrin-selenium | 10 μg/mL-5.5 μg/mL–6.7 ng/mL | 0.5 mL |
| Heparin | 4 μg/mL | 0.05 mL |
| Total | 50 mL |
Note: Store at 4°C for no longer than 2 weeks.
Note: For the initial culture, prepare 5 mL of organoid medium containing 10 μM of PBS-reconstituted ROCK inhibitor.
Alternatives: DMEM may also be used.
RBC lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| NH4Cl | 155 mM | 90 g |
| KHCO3 | 10 mM | 10 g |
| EDTA | 0.1 mM | 370 mg |
Note: Dissolve in 1 L of ddH2O and adjust PH to 7.4.
Note: Store at 20°C–25°C for no longer than 2 months.
Step-by-step method details
Lung homogenate preparation
Timing: 2.5 h
Two weeks after naphthalene treatment, prepare the lung homogenates needed for the co-culture from Acta2-Cre-ERT2; tdTomatoflox mice and Scgb1a1Cre-ERT; tdTomatoflox mice.
Note: Female 12- to 14-week-old Scgb1a1Cre-ERT; tdTomatoflox mice can be used without tamoxifen treatment.
-
1.
Euthanize animals by exposure to CO2 followed by cervical dislocation.
-
2.
Sterilize the mouse thorax and abdomen with 80% ethanol.
-
3.
Open the thoracic cavity to expose the lungs and heart using surgical scissors.
-
4.
Use a 25 G needle to perfuse the lungs with 20 mL of sterile Hank’s Balanced Salt Solution (HBSS, Life Technologies) through the right ventricle until the lungs are free of blood.
-
5.Lung harvest:
-
a.To obtain lung cell suspensions from Acta2-Cre-ERT2; tdTomatoflox mice:
-
i.Harvest the lungs and cut them into small pieces (∼2 mm) in a small petri dish using a blade.
-
ii.Place the lung pieces in 15 mL Falcon tubes and incubate in 5 mL of 0.5% collagenase type IV in HBSS for 45 min at 37°C while rotating.
-
i.
-
b.To obtain lung cell suspensions from Scgb1a1Cre-ERT; tdTomatoflox mice:
-
i.Make a small incision in the exposed trachea to insert a shortened 21 G cannula (firmly fixed).
-
ii.Instill 1.5 mL of dispase through the trachea.
-
iii.Remove the cannula and immediately tie the trachea using a surgical suture.
-
iv.Harvest the lungs and place them in separate Eppendorf tubes containing 2 mL of dispase
-
v.Incubate for 40 min at 20°C–25°C.
-
vi.Remove the trachea and heart using surgical scissors.
-
vii.Homogenize the lungs in 10 mL of digestion medium using GentleMACS® Dissociator (MACS Miltenyi Biotech).
-
i.
-
a.
CRITICAL: Be careful not to digest lungs longer than 45 min. This could lead to significant loss of cells.
-
6.
Filter the homogenates through 70 μm and 40 μm nylon cell strainers (BD Biosciences).
-
7.
Centrifuge at 500 ×g for 10 min at 4°C.
Note: Wash the filters using 5 mL of DMEM if necessary.
Note: The optimal yield expected at this stage for both cell types is between 15 ×106 to 30×106 cells per lung.
Primary cell isolation
Timing: 3 h
After the preparation of Acta2-Cre-ERT2; tdTomatoflox and Scgb1a1Cre-ERT; tdTomatoflox lung cell suspensions, use a cell sorter (For e.g., BD FACSAria™ III) to isolate mesenchymal and club cells, respectively.
-
8.Stain Acta2-Cre-ERT2; tdTomatoflox cells
-
a.Resuspend cells in 50 μL of anti-PDGFRα antibodies (1:100) or isotype control in MACS buffer for 20 min on ice in the dark.
-
b.Wash cells with 100 μL of MACS buffer.
-
c.Centrifuge cells at 500 × g for 10 min at 4°C.
-
d.Resuspend the cell pellet in 300 μL of MACS buffer.
-
a.
-
9.
Resuspend the cell suspension obtained from Scgb1a1Cre-ERT; tdTomatoflox mice in 300 μL of MACS buffer.
Filter cells with a final volume of 500 μL of MACS buffer using 40 μm mesh into FACS tubes for cell sorting.
-
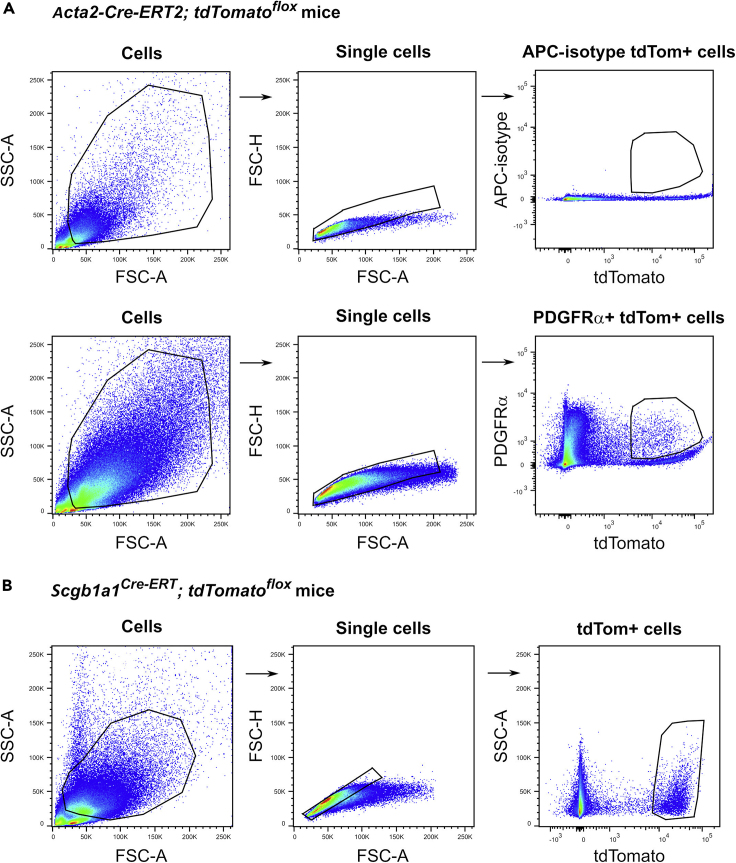
10.Set the appropriate gating strategy (Figure 1).
-
a.Isolate PDGFRα+ tdTom+ cells from the lung homogenates of Acta2-Cre-ERT2; tdTomatoflox mice (Figure 1A) (troubleshooting 1).
-
b.Isolate tdTom+ cells from the lung homogenates of Scgb1a1Cre-ERT; tdTomatoflox mice (Figure 1B) (troubleshooting 2).
-
a.
-
11.
Perform cell sorting into FACS tubes containing 350 μL organoid medium using an 85 μm nozzle (troubleshooting 3).
Note: The expected viability post-sort is around 96% for mesenchymal cells and 86% for club cells.
Figure 1.
Gating strategy for sorting mesenchymal and epithelial cell populations
(A) Representative flow cytometric dot plots of isotype control and PDGFRα and tdTomato expression in mesenchymal cells isolated from lung homogenates of Acta2-Cre-ERT2; tdTomatoflox mice 14 days after naphthalene injury.
(B) Representative flow cytometric dot plots of tdTomato expression in club cells isolated from lung homogenates of Scgb1a1Cre-ERT; tdTomatoflox mice.
Organoid culture
Timing: 14 days
Following cell isolation, co-culture mesenchymal and epithelial cells for 14 days to generate bronchiolospheres.
-
12.
Centrifuge flow-sorted cells for 5 min, 500 × g at 4°C.
-
13.Resuspend the cells in organoid medium.
-
a.Adjust cell concentrations as follows:
-
i.2×104 PDGFRα+ tdTom+ cells per 25 μL of medium per cell culture insert.
-
ii.1×103 SCGB1A1+ cells per 25 μL of medium per cell culture insert.
-
i.
-
a.
Note: The expected number of sorted cells per mouse is ∼15,000 PDGFRα+ tdTom+ mesenchymal cells and ∼100,000 SCGB1A1+ club cells.
Note: For each culture, between 15,000 and 20,000 mesenchymal cells and a minimum of 5,000 epithelial cells are required.
CRITICAL: It is advisable to perform a cell viability and quantification assay before the culture if the total number of isolated cells is lower than expected.
-
14.
Mix both cell populations to end up with a volume of 50 μL of organoid medium per insert.
-
15.
Dilute PDGFRα+ tdTom+ cells and SCGB1A1+ cells in 50 μL (per insert) with cold growth factor-reduced Matrigel at a 1:1 ratio as described before (Quantius et al., 2016) (troubleshooting 4).
CRITICAL: Mix well but carefully to avoid bubble formation.
CRITICAL: Work as fast as possible to avoid Matrigel polymerization in the tube.
-
16.
Add 100 μL of mixed cells in Matrigel on top of a 12-mm cell culture insert.
Note: Add the cell mixture drop by drop placing the pipette in the middle of the insert to ensure cell dispersion into the entire insert.
-
17.
Place inserts in a 24-well plate.
-
18.
Incubate for 15 min at 37°C to allow Matrigel polymerization.
-
19.
Remove the insert and add 350 μL of organoid medium into each well.
CRITICAL: Add ROCK inhibitor at a concentration of 10 μM to the organoid medium for 14–18 h, then change to regular organoid medium.
-
20.
Place back the insert into the well containing the organoid medium and incubate at 37°C with 5% CO2 for at least 2 weeks.
-
21.
Change medium three times per week for at least 14 days to generate bronchiolospheres (troubleshooting 5).
Note: Bronchiolospheres can be kept in culture for at least 4 weeks.
Expected outcomes
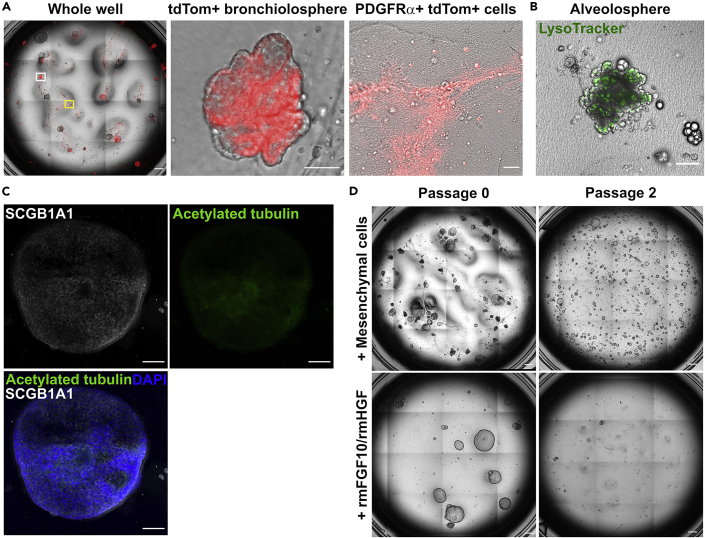
Using this detailed protocol for the isolation and 3D co-culture of lineage-labeled mesenchymal and epithelial cell populations, it is expected that tdTom+ bronchiolospheres will develop after 14 days of culture (Figure 2A). Notably, repair-supportive mesenchymal cells (RSMCs) classical spindle-shape morphology can also be observed around the bronchiolospheres (Figure 2A). The typical morphology of bronchiolospheres has been addressed in a previous publication (Vazquez-Armendariz et al., 2020). Besides bronchiolospheres, a low percentage of Lysotracker+ alveolospheres can also be detected (14%±3%) (Figure 2B). At day 14 of culture, these organoids mostly express the club cell marker SCGB1A1 with only a few acetylated tubulin+ ciliated cells (Figure 2C). Nevertheless, abundant beating cilia can be observed within bronchiolospheres at day 16 of culture, indicating the presence of mature ciliated cells (Methods video S1). At this stage, the supportive role of mesenchymal cells for club-cell growth can be assessed by quantification of bronchiolosphere colony-forming efficiency (CFE) and size, as well as identification of ciliated cell differentiation by immunofluorescence/live imaging. Bronchiolospheres can be passaged up to at least 5 times by using freshly isolated mesenchymal cells. Alternatively, the mesenchymal component can be substituted by a mixture of recombinant mouse (rm)FGF10 and rmHGF; however, the latter approach leads to significant reduction in bronchiolosphere CFE, lifespan until passaging and ciliated cell differentiation (Figure 2D).
Figure 2.
Bronchiolosphere formation after co-culture of mesenchymal and club cells
(A) Whole-well overview (left), representative image of a bronchiolosphere (white square; middle) and a mesenchymal cell cluster (yellow square; right) at day 14 of co-culture of PDGFRα+ tdTom+ cells and SCGB1A1+ club cells. Scale bars represent 100 μm.
(B) Representative confocal image of an alveolosphere stained for LysoTracker (stains lamellar bodies in AECII). Scale bar represents 100 μm.
(C) Representative confocal image of a bronchiolosphere stained for SCGB1A1 (club cells) and acetylated tubulin (ciliated cells). Scale bars represent 50 μm.
(D) Whole-well overview of bronchiolospheres grown in the presence or absence of mesenchymal cells before and after passaging. Note that bronchiolospheres grown in the absence of mesenchymal cells were supplemented with rmFGF10 and rmHGF. Scale bars represent 100 μm.
Limitations
Although most of the organoids are bronchiolospheres (∼86%), other types of organoids such as alveolospheres can also be generated (∼14%). This is due to the nature of isolated mesenchymal cells; in this case, RSMCs. If RSMCs are substituted by airway smooth muscle cell (ASMC)-enriched cells (PDGFRα- tdTom+), only bronchiolospheres will be generated albeit at significantly lower CFE (Moiseenko et al., 2020). Note that this bronchiolosphere model can also be used to test the ability of other mesenchymal cell populations to support club cell growth in vitro such as EpCAM- CD31- CD45- SCA-1+ resident mesenchymal cells.
Troubleshooting
Problem 1
There is no access to mesenchymal cells (step #10a).
Potential solution
Club cells can be cultured in organoid medium containing 50 ng/mL rmFGF10 and 30 ng/mL rmHGF. Bronchiolospheres will develop after 14 days albeit at significantly lower numbers (Figure 2D).
Problem 2
SCGB1A1+ club cell number is not enough for co-culturing with mesenchymal cells (step #10b).
Potential solution
SCGB1A1+ club cells can be expanded in vitro by culturing them in organoid medium containing 50 ng/mL rmFGF10 and 30 ng/mL rmHGF. The resulting bronchiolospheres can then be digested using 1 mg/mL dispase (BD Biosciences) for 10 min at 4°C, washed with MACS buffer twice by centrifugation at 300 ×g for 10 min and then passaged and expanded until co-culture with the mesenchymal cells can be performed (Figure 2D).
Problem 3
Slow cell sorting due to the presence of red blood cells (RBCs) in the sample (step #11).
Potential solution
Use an RBC lysis buffer as an additional step before sorting. Incubate cell suspensions in RBC lysis buffer for 90 s, wash with DMEM containing 10% FCS, centrifuge at 500 × g for 10 min at 4°C, and resuspend in 300 μL of MACS buffer.
Problem 4
Matrigel polymerization during co-culture preparation (step #15).
Potential solution
Keep Matrigel, organoid media and samples on ice during the entire culture preparation procedure. Keep a box of sterile 200 μL pipette tips at −20°C to use them exclusively for Matrigel mixing and transfer of the cells.
Problem 5
Bronchiolospheres do not develop even when the right number of cells per well was cultured (step #21).
Potential solution
Exclude dead cells by adding a live/dead cell marker before the sort of both cell types. For instance, add 1 μL of SYTOX™ Blue (Invitrogen) per mL of sample just before sorting.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Elie El Agha (Elie.El-Agha@innere.med.uni-giessen.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze any data sets or code.
Acknowledgments
This work was supported by the German Research Foundation (DFG, SFB 1021 C05 and Z02, SFB-TR84 B9; A6, SFB CRC1213 A04, KFO 309 P2/P7/P8/Z01, and EL 931/4-1, excellence cluster Cardio-Pulmonary Institute [CPI]), University Hospital Giessen and Marburg (FOKOOPV), Institute for Lung Health (ILH), and the German Center for Lung Research (DZL).
Author contributions
A.I.V.-A. and E.E.A. designed and wrote this protocol. W.S., S.H., and E.E.A. provided guidance during the experiments and manuscript preparation. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100594.
Contributor Information
Ana Ivonne Vazquez-Armendariz, Email: ana.i.vazquez-armendariz@innere.med.uni-giessen.de.
Elie El Agha, Email: elie.el-agha@innere.med.uni-giessen.de.
References
- Moiseenko A., Vazquez-Armendariz A.I., Kheirollahi V., Chu X., Tata A., Rivetti S., Günther S., Lebrigand K., Herold S., Braun T. Identification of a repair-supportive mesenchymal cell population during airway epithelial regeneration. Cell Rep. 2020;33:108549. doi: 10.1016/j.celrep.2020.108549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantius J., Schmoldt C., Vazquez-Armendariz A.I., Becker C., El Agha E., Wilhelm J., Morty R.E., Vadász I., Mayer K., Gattenloehner S. Influenza virus infects epithelial stem/progenitor cells of the distal lung: impact on Fgfr2b-driven epithelial repair. PLoS Pathog. 2016;12:e1005544. doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Armendariz A.I., Heiner M., El Agha E., Salwig I., Hoek A., Hessler M.C., Shalashova I., Shrestha A., Carraro G., Mengel J.P. Multilineage murine stem cells generate complex organoids to model distal lung development and disease. EMBO J. 2020;39:e103476. doi: 10.15252/embj.2019103476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling O., Bornert J.-M., Chambon P., Metzger D. Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis. 2009;47:14–18. doi: 10.1002/dvg.20448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze any data sets or code.