Abstract
Bovine tuberculosis (bTB) is endemic in cattle in Ethiopia, a country that hosts the largest national cattle herd in Africa. The intensive dairy sector, most of which is peri-urban, has the highest prevalence of disease. Previous studies in Ethiopia have demonstrated that the main cause is Mycobacterium bovis , which has been investigated using conventional molecular tools including deletion typing, spoligotyping and Mycobacterial interspersed repetitive unit-variable number tandem repeat (MIRU-VNTR). Here we use whole-genome sequencing to examine the population structure of M. bovis in Ethiopia. A total of 134 M . bovis isolates were sequenced including 128 genomes from 85 mainly dairy cattle and six genomes isolated from humans, originating from 12 study sites across Ethiopia. These genomes provided a good representation of the previously described population structure of M. bovis , based on spoligotyping and demonstrated that the population is dominated by the clonal complexes African 2 (Af2) and European 3 (Eu3). A range of within-host diversity was observed amongst the isolates and evidence was found for both short- and long-distance transmission. Detailed analysis of available genomes from the Eu3 clonal complex combined with previously published genomes revealed two distinct introductions of this clonal complex into Ethiopia between 1950 and 1987, likely from Europe. This work is important to help better understand bTB transmission in cattle in Ethiopia and can potentially inform national strategies for bTB control in Ethiopia and beyond.
Keywords: Bovine tuberculosis, Ethiopia, livestock, transmission, whole genome sequencing
Data Summary
Files S1 and S2 (available in the online version of this article)
Raw sequencing data for all sequenced isolates have been deposited in the European Nucleotide Archive (ENA) under project PRJEB32192. Accession numbers are detailed in File S1.
The R code used to analyse the data in this manuscript is available on Github: https://github.com/avantonder/Ethiopia2020.
Impact Statement.
Bovine tuberculosis (bTB) is endemic in cattle in Ethiopia, a country that hosts the largest national cattle herd in Africa. Tracing the source of M. bovis infections of livestock is an important tool for understanding the epidemiology of bTB and defining control/eradication strategies. This study performed the first WGS-based analysis of M. bovis isolates from Ethiopia and attempted to include isolates from multiple herds in different parts of the country. Our study highlighted the diversity of M. bovis in Ethiopia, identified within- and between-herd transmission often over long distances and showed that there had been at least two introductions of clonal complex European 3 (Eu3) into Ethiopia in the early 1970s. This work is important as it helps to better understand bTB transmission in cattle in Ethiopia and will potentially inform national strategies for bTB control in Ethiopia and beyond.
Introduction
Mycobacterium bovis is one of several highly related subspecies of the Mycobacterium tuberculosis complex (MTBC) which causes tuberculosis (TB) in humans and a range of domesticated and wild animals [1]. While the main human pathogen of this complex, Mycobacterium tuberculosis sensu stricto, causes approximately ten million new TB cases yearly [2], M. bovis is the main causative agent of TB in cattle, also known as bovine TB (bTB). The actual global prevalence of bTB in cattle is not known but recent estimates suggest that around 7 % of all cattle around the world are likely affected, having a significant impact on their productivity [3]. M. bovis is also capable of being transmitted to humans and it is believed that at least 1.5 % of all human TB cases are due to zoonotic TB [2].
Bovine TB is present across all continents and many countries in Europe, Australasia and the Americas have been able to eliminate or control bovine TB in their national herds, often through costly test-and-slaughter programmes [4, 5]. In countries without adequate resources to control the disease, bTB is frequently endemic [2]. This is the case for Ethiopia which has the fifth largest national cattle herd in the world with over 60 million animals, of which the vast majority are local zebu breeds reared in extensive farming systems [6]. bTB is endemic at a very low level in these extensive husbandry systems but thrives in the intensive dairy sector, with mainly high milk-yield Holstein-Friesian (H-F) or H-F/zebu cross-bred dairy cattle, where the prevalence is high [7]. This is particularly true in the well-established dairy belt in central Ethiopia where several studies have recorded over 25 % bTB prevalence in animals [8, 9] (Almaw et. al. under review). With the current increase in urbanization in Ethiopia, demand for milk is increasing around urban centres [10] and consequently, the intensive dairy sector is expanding and emerging in new centres across the country. Such expansion is associated with trading of dairy cattle between herds and regions, leading to increased risk of disease transmission in this sector.
To address questions around transmission and control of bTB in the highly affected dairy sector, a project named Ethiopia Control of Bovine TB Strategies (ETHICOBOTS) was funded by the Zoonoses and Emerging Livestock Systems programme (UK). One aim of this multi-disciplinary project was to investigate the population structure of M. bovis in the Ethiopia dairy sector and to explore epidemiological links. Previous studies have used strains from across Ethiopia to establish a basic picture of the M. bovis population through conventional methods including spoligotyping and Mycobacterial interspersed repetitive unit-variable number tandem repeat (MIRU-VNTR) [8, 11–19]; this contributed to the definition of the African 2 clonal complex of M. bovis (Af2) confined to Ethiopia and East Africa [20]. However, these techniques have their limitations: both MIRU-VNTR and spoligotyping have less discriminatory power and identical spoligotype patterns can be found in phylogenetically unrelated strains [21–24]. Whole-genome sequencing (WGS) and SNP-based analyses can now provide more detailed and discriminating phylogenetic analyses [25, 26]. Also, due to a lack of recombination in the MTBC, SNPs exhibit very low degrees of homoplasy [22, 24].
A small number of M. bovis isolates (n=14) from neighbouring Eritrea have previously been analysed using WGS and established the presence of an undescribed clonal complex since named Eu3 [27, 28]. In this study we performed genomic analyses of 134 Ethiopian M. bovis isolates collected from dairy cattle, humans and a dromedary from several study sites across the country, giving the first WGS-based picture of the population structure of M. bovis in Ethiopia. We further analysed a collection of 107 Eu3 genomes, including 40 from Ethiopia, to date the most recent common ancestor (MRCA) of this lineage and to estimate the date of introduction into Ethiopia.
Methods
Isolate selection, culturing, and preparation of genomic DNA
Metadata on the host species of the 134 M . bovis isolates selected for this study are summarized in File S1. The vast majority (n=121) originated from 76 dairy cattle (12 H-F, 8 Zebu and 56 H-F/Zebu cross-bred dairy cattle), collected from known dairy farms in Mekele, Gondar and Alage (near Hawassa) as well as from the large dairy belt in central Ethiopia (Addis Ababa, Holeta, Sebeta, Bishoftu, Sendafa and Sululta) while six isolates were collected from zebu cattle and one from a dromedary slaughtered at Gondar, Addis Ababa, Butajira, Negele and Filtu abattoirs, respectively (Fig. 1a, File S1). The latter six, the dromedary sample, plus 29 isolates from dairy cattle from central Ethiopia were collected in previous studies [8, 11, 14] and frozen stocks (archived at −80 °C) were used for re-culturing and subsequent genomic DNA preparation. Ninety-two of the isolates from dairy cattle were collected as part of the ETHICOBOTS project. Eighty-six of these isolates originated from tuberculin reactor animals that were slaughtered and tissue samples of suspected TB lesions were processed and cultured for mycobacteria according to a previously described protocol [29]. The remaining six isolates originated from milk collected from tuberculin reactors. These milk samples were processed based on published methodology [30–32] and cultured as described for the tissue samples. The six M. bovis isolates of human origin were isolated from pulmonary TB cases (sputum) or from TB lymphadenitis cases (fine-needle aspirates), from study sites overlapping with the cattle samples (File S1) [33, 34] (H. Taye, in preparation). All M. bovis isolates included in this study were isolated between 2006 and 2018 (File S1). Genomic DNA of each isolate was extracted from heat-inactivated M. bovis cells using the DNeasy Blood and Tissue Kit (Qiagen). Before submission for WGS, all genomic DNA samples were confirmed as M. bovis by RD4 deletion typing [11].
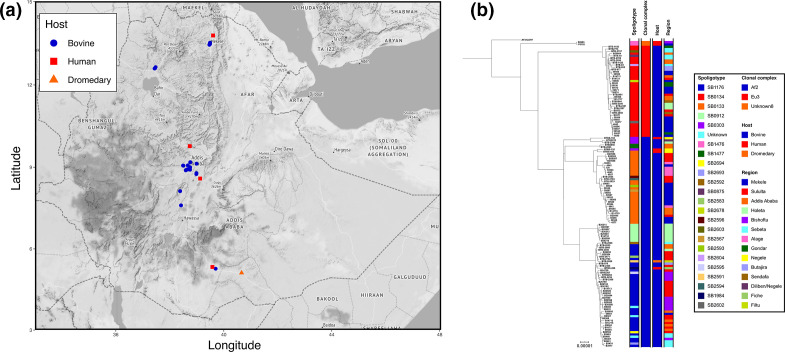
Fig. 1.
(a) Map of Ethiopia showing the location of isolation for each sequenced isolate coloured by the host .(b) Maximum-likelihood phylogenetic tree of 134 Ethiopian genomes rooted using M. bovis AF2122/97.
Whole genome sequencing and sequence analysis
WGS was performed in four rounds using the Illumina HiSeq 2500, MiSeq, NextSeq 500, and HiSeqX platforms to produce paired-end reads of between 50 and 150 base-pairs in length. Raw sequencing reads were deposited at the European Nucleotide Archive (ENA) under project PRJEB32192; all accessions used in this project are listed in File S1. FastQC v0.11.9 [35] was used to generate basic quality control metrics for the raw sequence data. Sequence reads were classified using Kraken v0.10.6 [36] and the abundance of the classification was refined to a single level using Bracken v1.0 [37]. Samples with less than 70 % of all reads assigned to the Mycobacterium genus were excluded. In silico spoligotyping was performed using SpoTyping v2.1 [38] and the binary spoligotype representations were queried against the M. bovis spoligotype database (www.mbovis.org) to extract spoligotypes named in the format of SBXXXX. Clonal complexes were assigned to samples using RD-analyzer v1.0 [39] with samples not identified as belonging to previously described M. bovis clonal complexes [European 1 (Eu1), European 2 (Eu2), African 1 (Af1), and African 2 (Af2)] designated as ‘Other’ [20, 40–42]. Further assignment to clonal complex was based on the phylogenetic lineages recently identified by Loiseau et al. [43].
Sequence reads were mapped to the M. bovis AF2122/97 reference genome (NC0002945) using BWA v0.7.17 (Burrow-Wheeler Aligner) (minimum and maximum insert sizes of 50 and 1000, respectively) [44]. SNPs were called using SAMtools v1.2 mpileup and BCFtools v1.2 (minimum base call quality of 50 and minimum root squared mapping quality of 30) as previously described [44, 45]. Samples with reads mapping to less than 90 % of the AF2122/97 reference were excluded. Genomic regions consisting of GC-rich sequences such as PPE proteins and PE-PGRS repeats were masked in the resulting alignment using previously published coordinates [46]. Variable sites were extracted from the masked alignment using snp-sites v2.5.1 [47]. Maximum-likelihood phylogenetic trees were constructed using IQ-tree v1.6.5 [48] (constant sites added to alignment, extended model selection and 1000 bootstraps) and the resulting trees were annotated and rooted in iTOL [49]. Pairwise SNP distances for all genomes were calculated using pairsnp (https://github.com/gtonkinhill/pairsnp).
To provide wider context for the isolates sequenced in this study, a global representative M. bovis dataset (n=352; File S1) was assembled. Genomes were selected based on the following criteria: they were previously published [27, 43, 50–54] and represented the currently understood M. bovis diversity. All previously characterized clonal complexes (Af1, Af2, Eu1, Eu2 and Eu3) and uncharacterized lineages (Unknown1 to Unknown8) from Loiseau et al. [43] were included. Due to the large number of available sequences for clonal complexes Eu1 and Eu2, a random selection of 100 genomes was chosen for each. Sequence data was downloaded from the ENA and trimmed using Trimmomatic v0.33 [55]. Sample QC, spoligotype assignment, mapping and phylogenetic tree construction were performed as above. The tree was rooted with a Mycobacterium caprae isolate.
Spatial analysis
A map of the geographical locations of isolate collection (latitude and longitude) was constructed using R and the ggmap library [56, 57]. The spatial position of genomes from each clonal complex was plotted and a convex hull (the smallest polygon incorporating a given set of points), calculated using the getConvexHull function from the R library contoureR [58], was drawn around genomes from each clonal complex. Pairwise geographic distances between each isolate in kilometres (km) were calculated using the distHaversine function from the R library geosphere [59]. The associations between genetic and spatial distance for clonal complexes Af2 and Eu3 were tested with a Mantel test (1000 permutations to assess significance) implemented using the R library ade4 [60].
Putative transmission clusters
Two different maximum inter-isolate pairwise SNP thresholds were used to define transmission clusters: 15 SNPs to capture older transmission events and five SNPs to capture recent transmission events. The transmission clusters were calculated using the R library iGRAPH [61] and plotted and annotated using the R library ggraph [62].
Molecular dating of Eu3 clonal complex
beast v1.8.4 [63] was run on a SNP alignment of all Ethiopian (n=40) and published (n=67) Eu3 genomes (File S1), using tip sampling dates for calibration. Three runs of 108 Markov chain Monte Carlo (MCMC) iterations were performed using a HKY substitution model, strict or constant molecular clock and constant or exponential population size and growth (12 separate runs). The performance of each model was assessed through the comparison of posterior marginal likelihood estimates (MLE) and the model with the highest Bayes factor [64] (strict clock/constant population size) was selected (Table S1). These three MCMC runs were combined using LogCombiner v1.8.4 (10 % burnin) and convergence was assessed [posterior effective sample size (ESS) > 200 for each parameter] using Tracer v1.6. A maximum clade creditability tree summarizing the posterior sample of trees in the combined MCMC runs was produced using TreeAnnotator v1.8.4. The resulting tree was annotated using ggtree [65]. The R library TIPDATINGBEAST [66] was used to confirm a temporal signal in the dataset; briefly, the dates for each sample were resampled to generate 20 new datasets with randomly assigned dates. beast was then run on these new datasets using the same strict constant priors.
Results
Study sites and population
A total of 134 M . bovis isolates were successfully sequenced and passed QC; these included 127 isolates collected from 85 cattle, six isolates from six human TB patients and a single isolate from a dromedary. The origin of isolation and WGS-based genotypes of these isolates are listed in Table 1 (the complete set of metadata can be found in File S1). The majority of the isolates were collected from the large dairy belt in central Ethiopia, in the surroundings of Addis Ababa (Fig. 1a). In silico spoligotype assignment revealed a total of 22 different spoligotypes with the most prevalent types being SB0134 (n=36), SB1176 (n=36) and SB0133 (n=28). Three different clonal complexes were observed in the dataset (Fig. 1b): African 2 (Af2; n=92), European 3 (Eu3; n=40) and Unknown8 (n=2).
Table 1.
Metadata of 134 sequenced M. bovis isolates included in this study, assigned to the African 2, the European 3, and the Unknown8 clonal complexes
|
Clonal complex |
Samples (n) |
Spoligotype (n) |
Study site (n) |
Host (n) |
|---|---|---|---|---|
|
African 2 |
92 |
SB1176 (36), SB0133 (28), SB0912 (8), SB0303 (4), Unknown (3), SB1477 (2), Singletons (11) |
Sululta (26), Mekele (15), Bishoftu (11), Addis Ababa (11), Holeta (10), Sebeta (6), Alage (6), Negele (3), Others (4) |
Bovine (87), human (4), dromedary (1) |
|
European 3 |
40 |
SB0134 (35), Singletons (5) |
Mekele (14), Addis Ababa (6), Sebeta (5), Sululta (4), Gondar (4), Holeta (3), Butajira (2), Sendafa (2) |
Bovine (40) |
|
Unknown8 |
2 |
SB1476 |
Mekele (1), Bishoftu (1) |
Human (2) |
Comparison of sequenced genotypes with previously published genotypes
Spoligotype information for previously published M. bovis isolates from animals, mainly cattle, was collated and the likely clonal complex inferred [67] (Table 2). The most frequent Af2 and Eu3 spoligotypes (SB1176, SB0133, SB0912, SB1477 and SB0134) previously identified in Ethiopia were also represented amongst the spoligotypes of the sequenced isolates in this study at similar frequencies. SB1476 (Unknown8), previously found exclusively in Ethiopian cattle, was found in two of the six isolates collected from humans.
Table 2.
Genotyping data and inferred clonal complex of previously published M. bovis isolates from Ethiopia
|
Clonal complex |
Samples (n) |
Spoligotype (n) |
Study site (n) |
Host (n) |
Reference (n) |
|---|---|---|---|---|---|
|
African 2 |
190 |
SB1176 (104), SB0133 (50), SB1477 (7), SB0912 (6), SB1490 (4), SB0933 (3), SB1491 (2), SB1942 (2), SB1983 (2), Singletons (10) |
Addis Ababa (64), Holeta (41), Kombolcha (26), Negele (24), Jinka (12), Adama (7), Woldiya (4), Gondar (3), Fiche (3), Ghimbi (2), Butajira (2), Melga-Wondo (1), Hawassa (1) |
Bovine (188), camel (2) |
Berg 2009 [11](48), Ameni 2007 [13](41), Biffa 2010 [15](27), Gumi 2012 [31](24), Firdessa 2012 [8](19), Ameni 2010 [12](13), Tsegaye 2010 [18](7), Mekibeb 2013 [17](6), Ameni 2013 [14](3), Mamo 2011 [19](2) |
|
European 3 |
24 |
SB0134 (19), SB1517 (3), Singletons (2) |
Addis Ababa (21), Yabello (2), Gondar (1) |
Bovine (24) |
Firdessa 2012 [8](12), Biffa 2010 [15](6), Berg 2009 [11](5), Tsegaye 2010 [18](1) |
|
Unknown8 |
18 |
SB1476 |
Ghimbi (8), Gondar (7), Butajira (2), Jinka (1) |
Bovine (18) |
Berg 2009 [11](18) |
Phylogenetic relationships of M. bovis genomes
The phylogenetic tree of 134 Ethiopian genomes, rooted with the Eu1 reference AF2122/97, shows a distinct phylogenetic structure with the three included clonal complexes clearly segregating in the phylogeny (Fig. 1b). The Unknown8 clonal complex is basal followed by a split leading to Eu3 and Af2. Three distinct sub-lineages were observed for Af2 and each was associated with a predominant spoligotype (SB0303, SB0133 and SB1176).
Spatial distribution of M. bovis clonal complexes
Fig. 2a shows a spatial distribution of the 134 isolates overlaid with the smallest polygon incorporating the isolates from each clonal complex. There appears to be a distinct geographical distribution with Eu3 isolates more likely to be distributed to the north and west of the country whilst Af2 isolates have a more southern and eastern distribution. There is a significant overlap between these two clonal complexes in central Ethiopia around Addis Ababa, likely reflecting a greater density of infected cattle from across the country.
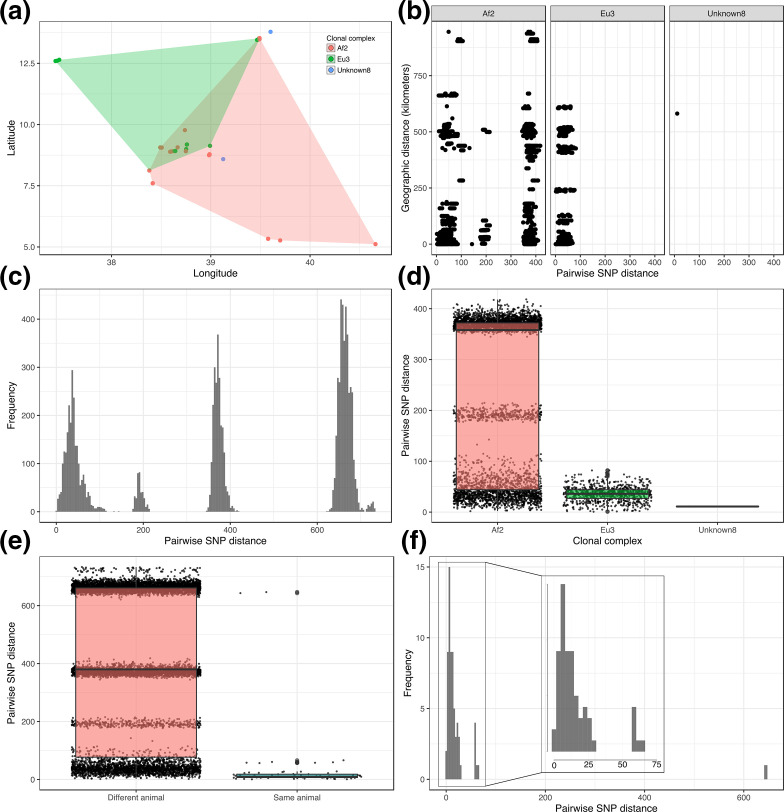
Fig. 2.
(a) Spatial analysis of distribution of isolates coloured by clonal complex. Each polygon represents the minimum convex polygon of the sampled locations of the isolates from each clonal complex. (b) Scatterplot of SNP distance against geographic distance for all pairs of genomes. (c) Histogram of all pairwise SNP distances. (d) Boxplot of all pairwise SNP distances separated by clonal complex. (e) Boxplot of all pairwise SNP distances separated by within- and between-animal. (f) Histogram of within-animal pairwise SNP distance. The insert shows the 0–75 SNP range zoomed in.
A comparison of pairwise genetic distance (SNPs) and geographic distance (kilometres) for all pairs of genomes is shown in Fig. 2b. The results of the Mantel tests for Eu3 and Af2 (abattoir isolates removed) showed that there was no association between genetic and spatial distance (Eu3 observation=0.10; simulated P-value: 0.001; Af2 observation=0.03; simulated P-value: 0.001). Instead, the pattern observed reflects the large geographic distribution of these genetic lineages in Ethiopia.
Genetic diversity and putative transmission
The distribution of pairwise SNP distances within the dataset was multimodal reflecting the population structure of the dataset with distinct modes observed for the three clonal complexes as well as the distinct sub-structure of Af2 (Fig. 2c). Categorizing the pairwise SNP distances by clonal complex showed that there is considerably more diversity within Af2 compared to Eu3 and Unknown8 with the maximum pairwise SNP distance within Af2 being 418 SNPs (median 358 SNPs), compared to 82 SNPs (median 36 SNPs) and 11 SNPs (median 11 SNPs) for Eu3 and Unknown8, respectively (Fig. 2d). The within-host pairwise SNP diversity had a median of ten SNPs (range 1–647 SNPs) whilst the between-host pairwise SNP diversity had a median of 380 SNPs (range 2–733 SNPs; Fig. 2e). Further examination of the within-host diversity revealed three distinct peaks within the distribution of pairwise SNP distances with the majority of within-host isolates being within 1 and 30 SNPs of each other (Fig. 2f). The maximum within-host pairwise SNP distance of 647 SNPs was observed in an animal (ser-002) with three isolates, two from Af2 and one from Eu3.
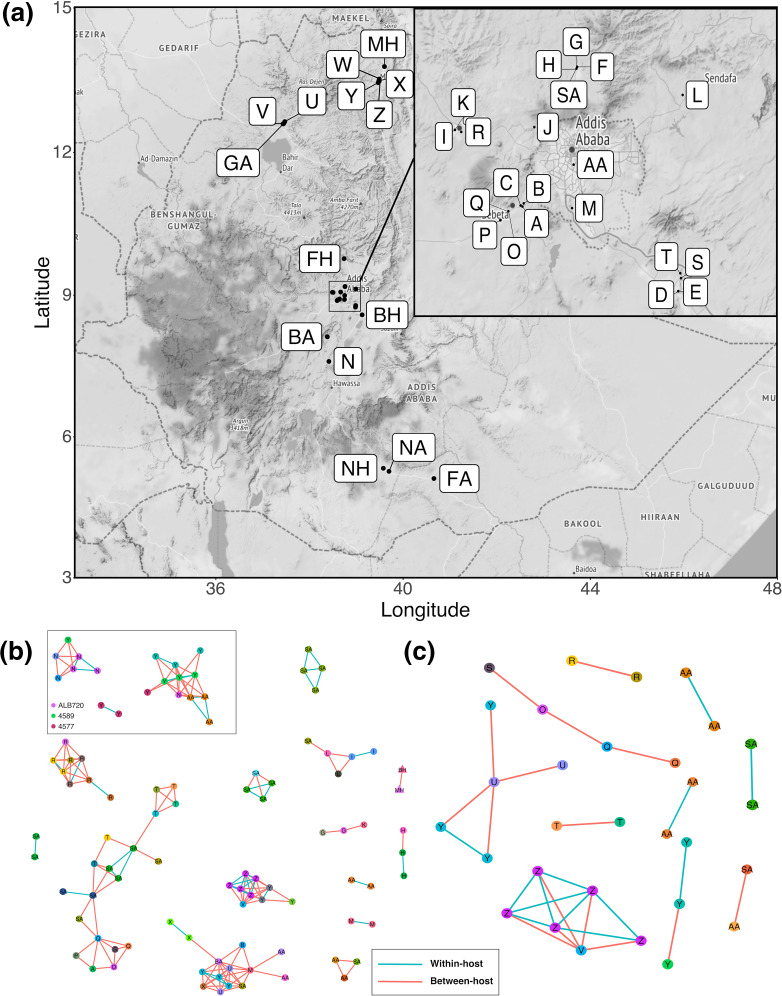
A total of 17 putative transmission clusters were defined using the conservative pairwise SNP threshold of 15 SNPs. The clusters varied in size between two and 19 isolates, and there were 35 isolates that were not assigned to a cluster (singletons; n=35; Fig. 3b). Extensive within- and between-herd transmission was observed with both short- and long-distance transmission occurring. Examples of isolates from the same animal being in different transmission clusters were observed; for instance, isolates from animal ALB720 were found in two different transmission clusters along with isolates from animal 4589 (Fig. 3b). These animals were from herds N (Alage) and Y (Mekele), which are approximately 662 km apart. The largest transmission cluster, containing 19 isolates from 16 animals, showed clear epidemiological links between the Sululta abattoir (SA) and herds from around Sebeta (A, O, P, Q) and Bishoftu (S, T), whilst the second largest transmission cluster, containing 14 isolates from 11 animals showed links to abattoirs in and around Addis Ababa (AA, BA, SA) and herds from as far away as Gondar (U) and Mekele (X, Y).
Fig. 3.
(a) Map of Ethiopia showing the locations of the herds, abattoirs and hospitals from which the isolates were sourced (letter coding from File S1). The region around Addis Ababa was magnified in the insert. (b) Putative transmission clusters defined using a pairwise SNP threshold of 15 SNPs. (c) Putative transmission clusters defined using a pairwise SNP threshold of five SNPs. Nodes are coloured by animal and labelled with the herd, abattoir or hospital of isolation. Edges coloured in blue represent within-host links whilst edges coloured in red represent between animal links. For simplicity, clusters where n <2 are not shown.
When using the more stringent threshold of five pairwise SNPs, ten putative transmission clusters comprising between two and six isolates were defined; 104 isolates were not assigned to a cluster (Fig. 3c). Transmission clusters with both within- and between-herd transmission were defined. Two of the transmission clusters showed evidence of long-distance transmission (>230 km apart) between herds in Gondar (U, V) and Mekele (Y, Z).
Global and European 3 specific phylogenetics
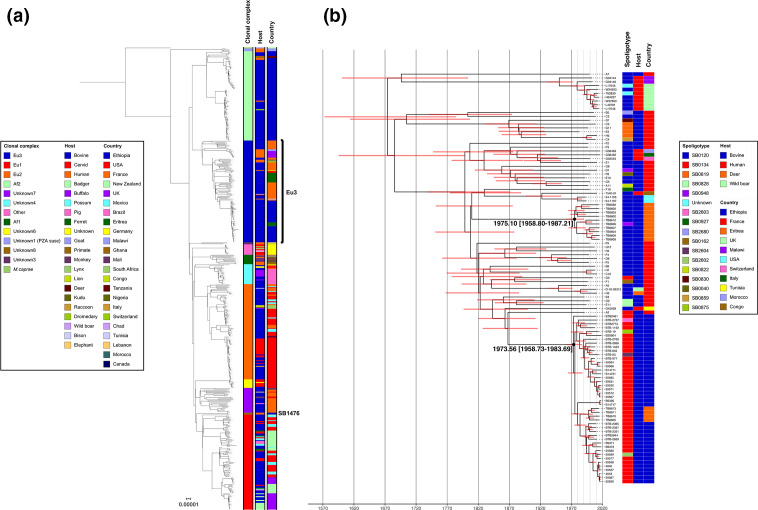
Fig. 4a shows a maximum-likelihood phylogeny of 485 M . bovis genomes from 22 countries (including the genomes from this study), collected from 20 different host species between 1983 and 2018. All previously described clonal complexes along with one undescribed clonal complex (labelled as ‘Other’) are represented in the collection. The structure of the phylogeny is consistent with recently published work [43] and shows that pyrazinamide (PZA)-susceptible M. bovis from Malawi are basal to the other clonal complexes, which are all PZA-resistant. The first of the next two ancestral splits in the phylogeny leads to the sister clades Af2 and Eu3, where the majority (n=132) of the Ethiopian genomes are contained, and the clade containing all other clonal complexes (Other, Af1, Eu1, Eu2 and Unknown3-8). The remaining two Ethiopian genomes (SB1476/Unknown8) are the outgroup to the globally distributed Eu1 clonal complex (Fig. 4a).
Fig. 4.
(a) Global maximum-likelihood phylogeny of 485 M. bovis genomes rooted using M. caprae . (b) Time‐calibrated maximum clade credibility tree of European 3 genomes.
The 40 Ethiopian Eu3 genomes were combined with 67 other Eu3 genomes from ten countries and the resulting phylogeny was dated using beast (Fig. 4b). The median date of the MRCA for this clade was estimated to be 1674 [95 % CI: 1557–1769]. The dated phylogeny shows two potential Eu3 introduction events into Ethiopia (Eritrea was part of Ethiopia until 1993) around the same time period: 1975 [95 % CI: 1959–1987] and 1974 [95 % CI: 1959–1984] (Fig. 4b). The Eu3 phylogeny also contains nine BCG vaccine isolates collected in Malawi and the UK and these have a MRCA dating back to 1950 [95 % CI: 1926–1968]. The results of the dated tip randomization analysis are shown in Fig. S1. The substitution rate in the observed data did not overlap with the estimated substitution rates in the randomized datasets showing that the temporal signal observed was not obtained by chance.
Discussion
This is the first study to use WGS to examine the population structure of M. bovis in Ethiopia. A total of 134 isolates from 12 study sites across Ethiopia were successfully sequenced and three distinct clonal complexes, which appeared to be mostly geographically segregated, and 22 different spoligotypes were observed in the dataset. The in-depth genome analysis showed there was no association between geographic and genetic distance and differing levels of genetic diversity were observed amongst the three clonal complexes. A range of within-host diversity was also observed with isolates from the same animal often being in different transmission clusters, as well as evidence of both short- and long-distance transmission with isolates in transmission clusters collected hundreds of kilometres apart. Molecular dating of a collection of Eu3 genomes which included the Ethiopian genomes in this study estimated the date of introduction of different Eu3 sub-lineages into Ethiopia and Eritrea during the same time period around the early 1970s (CI: 1958–1987).
To get a better understanding of how well the genomes analysed in this study represented the known population structure of M. bovis in Ethiopia, we collated spoligotype information for the vast majority of Ethiopian M. bovis isolates from animals (mainly cattle) published to date [67]. When comparing the spoligotypes in the current study (Table 1) with those previously published (Table 2), it was clear that the most frequent spoligotypes of Af2 and Eu3 identified in Ethiopia (SB1176, SB0133, SB0912, SB1477 and SB0134) were also represented in our dataset with their prevalence observed at a similar frequency. Spoligotype SB1476 has also been frequently observed in cattle in previous studies [11] but we provide the first evidence of this spoligotype in humans, possibly as a result of zoonotic transmission. On this basis, the isolates sequenced in this study appear to be representative of the population structure of M. bovis in Ethiopia.
Examination of the geographical distribution of the clonal complexes, in particular Af2 and Eu3, contained within the dataset showed a potential geographic divide between their distributions with an overlap in the herds around Addis Ababa. The number of isolates from outside this region is small but given the expected movement, often long-distance, of animals between different herds, the geographic separation of clonal complexes is still distinct. The reason for this pattern is unclear but may be due to historical isolation of different parts of the country due to geographic distance; it is interesting that Af2, the indigenous lineage in Ethiopia, was not found in the north west of the country but this may simply be due to a lack of sampling. We could find no evidence of a relationship between genetic and geographic distance for the different clonal complexes implying that the M. bovis population in Ethiopia is well-mixed and maintained by both short- and long-distance cattle movements. There was a large difference observed in genetic diversity between clonal complexes Af2 and Eu3; this is likely due to the long-term endemic nature of Af2 in Ethiopia which has allowed for significant genetic divergence over time with the emergence of clear sub-lineages within this clonal complex observed. Conversely, the comparatively recent introduction of Eu3 into Ethiopia (see below), has not allowed enough time for more genetic diversity to emerge. As expected, considerable genetic diversity was observed between isolates from different animals with some isolates, from different clonal complexes, being as much as 733 SNPs apart. Less expected was the range of within-host diversity observed. The majority of isolates from the same animal were up to 30 SNPs (median=10 SNPs) apart from each other; given the previously estimated mutation rate of M. bovis of 0.15–0.53 SNPs per genome per year [68, 69] it is likely infections are being maintained over a long period of time. Less closely related isolates from the same animal were also found showing that multiple infections by different strains was taking place amongst our samples.
With bTB being endemic in Ethiopia and the intensive dairy sector highly affected, there is considerable interest from stakeholders to explore control strategies for this disease. One of the aims of the ETHICOBOTS project was to use WGS to identify potential transmission of bTB in Ethiopian dairy cattle. There is currently no clear consensus as to what the most appropriate SNP threshold use for defining transmission clusters in either bTB or M. tuberculosis sensu stricto with thresholds of 5, 12 and 15 SNPs having been previously applied [70–72]. Thus, we defined transmission clusters both using a conservative threshold of 15 SNPs (Fig. 3b), which would allow for both the identification of older transmission events and account for varying rates of mutation within the animals sampled, and a stricter threshold of five SNPs (Fig. 3c), which would identify more recent transmission. Using either threshold, we could find clear evidence of transmission occurring between animals from the same herd as well as animals from different herds and from animals sampled at local abattoirs. Using the more conservative threshold of 15 SNPs, we found evidence of isolates from the same animal in different transmission clusters, showing that animals are likely being re-infected with different strains. There was also strong evidence of both short- and long-distance transmission with isolates in some transmission clusters being hundreds of kilometres apart. Despite the sparse sampling, the fact that we were able to identify various types of transmission events even with a strict threshold of five SNPs suggests that there is considerable ongoing transmission amongst cattle in Ethiopia. These findings are not surprising as very few farms in Ethiopia have the ability to control bTB in their herds, e.g. by test-and-slaughter based on the tuberculin skin-test and there is considerable long-distance trade between affected herds. Instead, this uncontrolled chronic disease results in cattle infected with M. bovis being likely to survive for a long enough time to be subject to exposure to multiple bTB strains and/or cattle trade (short- or long-distance), increasing the risk of disease transmission and reinfection.
Depending on the SNP threshold used, there is considerable heterogeneity with respect to the size of defined transmission clusters and contribution from individual farms. With the more conservative threshold of 15 SNPs, genomes isolated from Farm Y (Fig. 3a) were part of three clusters suggesting epidemiological links both locally and nationally. Although we must be cautious in interpreting patterns within such a sparse sample and higher SNP threshold, this pattern is consistent with so-called ‘super-spreading’ behaviour where a small number of herds contribute disproportionately to transmission. The potential for such super-spreading, with an opportunity to target herds for control, was evident in our movement-based network analysis [73].
Additionally, we wanted to examine the possibility of zoonotic transmission between humans and cattle in Ethiopia. The minimum pairwise SNP distance between any pairs of human and animal isolates was 41 SNPs (median 658 SNPs) providing no evidence of recent zoonotic transmission from the cattle or herds that were sampled (potential transmission events would be highlighted by small pairwise SNP distances). This was unsurprising, given there were only six isolates from humans included in this study. However, two of the human isolates (of type SB1476) were only 11 SNPs apart from each other suggesting a potential epidemiological link; these isolates were from Bishoftu and Mekele, approximately 581 km apart, suggestive of long-distance travel of infected individuals or animals. Overall, these data provide clear evidence of considerable transmission between cattle but denser sampling will be required to establish specific evidence of zoonotic transmission. Denser, more targeted sampling would also allow for more sophisticated transmission analyses using tools such as TransPhylo [74], which account for within-host diversity and can estimate unsampled cases within transmission networks.
Recent work has suggested an East African origin for M. bovis [43]. Given its position in the global phylogeny and confined geographical distribution [20], Af2 has likely been circulating in East Africa for hundreds of years; unfortunately, the lack of genomes from other parts of the world to provide context to the Ethiopian Af2 genomes, due to the small number sequenced, means confirming this in Ethiopia is currently not possible. However, we can show that there is a clear phylogenetic structure in this clonal complex with the Ethiopian genomes dividing into three distinct lineages. The position of Eu3 as the sister group to Af2 also implies a likely East African origin; however, based on the samples available for this study, the basal position of French isolates (Fig. 4b) suggests that the currently circulating Eu3 lineage in Ethiopia may in fact be European in origin. One possible explanation for this is that the ancestor of Eu3 was brought to Europe a few hundred years ago and that the modern Ethiopian and Eritrean genomes are descendants of that population, not of the ancestral population that may or may not still be circulating somewhere in East Africa. What is reasonably clear, given the long branches and subsequent expansion, is that there were two introductions of Eu3 sub-lineages, consistent with the study that first analysed the Eritrean WGS data [27], into Ethiopia between 1958 and 1987 (which also included Eritrea during that time) with the median estimate for introduction being in the early 1970s. Given the very similar dates, it is possible that these were part of the same series of cattle imports. In terms of likely origin of these imports, France should be viewed as a proxy for the M. bovis diversity seen in mainland Europe (these samples were chosen to represent the diversity seen in France [50]), so the actual origin may be elsewhere in Europe. There are historical records of the first dairy cattle being imported into Ethiopia around 1950 as part of the United Nations Relief and Rehabilitation Administration (UNRRA) [75] with further subsequent imports from Kenya in 1959 [76]. Several livestock and dairy development projects that took place in the 1950–70s and funded by Sweden and the World Bank may have brought in dairy cattle of exotic breeds from overseas [3].
The other clonal complex found in Ethiopia, Unknown8, represented by two human genomes with the spoligotype SB1476, has thus far only been found in Ethiopia [11]. The position of this lineage in the global tree, and the hypothesized East African origin of M. bovis [43], suggests that this clonal complex may be the ancestor of Eu1, the most prevalent and geographically distributed M. bovis lineage known to date. However, further work would need to be done to confirm this through the collection of larger numbers of isolates with spoligotype SB1476.
This study performed the first WGS-based analysis of M. bovis isolates from Ethiopia and attempted to include isolates from multiple herds in different parts of the country. There is considerable genetic diversity amongst M. bovis in Ethiopia with multiple clonal complexes circulating and that they were likely to have been introduced in the country at different time-points. This work is important as it helps to better understand bTB transmission in cattle in Ethiopia and will potentially inform national strategies for bTB control in Ethiopia and beyond.
Supplementary Data
Funding information
This research was financially supported by the Ethiopia Control of Bovine Tuberculosis Strategies (ETHICOBOTS) project funded by the Biotechnology and Biological Sciences Research Council, the Department for International Development, the Economic and Social Research Council, the Medical Research Council, the Natural Environment Research Council and the Defence Science and Technology Laboratory, under the Zoonoses and Emerging Livestock Systems (ZELS) program, ref: BB/L018977/1. Stefan Berg was also funded by Defra, UK, ref: TBSE3294. Glyn Hewinson holds a Sêr Cymru II Research Chair funded by the European Research Development Fund and Welsh Government.
Acknowledgements
The members of the ETHICOBOTS consortium are: Abraham Aseffa, Adane Mihret, Bamlak Tessema, Bizuneh Belachew, Eshcolewyene Fekadu, Fantanesh Melese, Gizachew Gemechu, Hawult Taye, Rea Tschopp, Shewit Haile, Sosina Ayalew, Tsegaye Hailu, all from Armauer Hansen Research Institute, Ethiopia; Rea Tschopp from Swiss Tropical and Public Health Institute, Switzerland; Adam Bekele, Chilot Yirga, Mulualem Ambaw, Tadele Mamo, Tesfaye Solomon, all from Ethiopian Institute of Agricultural Research, Ethiopia; Tilaye Teklewold from Amhara Regional Agricultural Research Institute, Ethiopia; Solomon Gebre, Getachew Gari, Mesfin Sahle, Abde Aliy, Abebe Olani, Asegedech Sirak, Gizat Almaw, Getnet Mekonnen, Mekdes Tamiru, Sintayehu Guta, all from National Animal Health Diagnostic and Investigation CenterCenter, Ethiopia; James Wood, Andrew Conlan, Alan Clarke, all from Cambridge University, UK; Henrietta L. Moore and Catherine Hodge, both from University College London, UK; Constance Smith at University of Manchester, UK; R. Glyn Hewinson from Aberystwyth University, UK; Stefan Berg, Martin Vordermeier, Javier Nunez-Garcia, all from Animal and Plant Health Agency, UK; Gobena Ameni, Berecha Bayissa, Aboma Zewude, Adane Worku, Lemma Terfassa, Mahlet Chanyalew, Temesgen Mohammed, Miserach Zeleke, all from Addis Ababa University, Ethiopia. We thank the Ethiopian Ministry of Agriculture and the Ethiopian Ministry of Health for their support to the ETHICOBOTS project. We thank NAHDIC for their logistical support. James Wood is supported by The ALBORADA Trust.
Author contributions
Conceptualization: J.L.N.W., S.B., A.J.K.C., A.A., R.G.H., G.A. Data curation: S.B., A.J.v.T., G.A.M., G.A. Formal analysis: S.B., A.J.T., E.P., J.N. Investigation: G.A., G.A.M., H.T., A.Z., M.T., A.O., A.A., M.L., M.S., C.D., M.H., A.F., B.M. Methodology: G.A., G.A.M., J.L.N.W., S.B., J.P., A.J.v.T., R.J.E. Project administration: S.G., A.M., A.A., S.B., J.L.N.W. Supervision: A.M., T.A., G.A., S.G., S.B., J.P., A.J.v.T. Writing – original draft: S.B., A.J.v.T., G.A.M., G.A. Writing – review and editing: A.J.K.C., A.M., A.A., T.A., G.A., B.G., J.P., J.L.N.W., E.P., R.J.E., R.G.H.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Ethical approvals to implement the research of ETHICOBOTS were granted by the National Research Ethics Review Committee (NRERC No. 3.10/800/07), and by the institutional review boards at Aklilu Lemma Institute of Pathobiology, Addis Ababa University (Reference No. IRB/ALIPB/2018), and at AHRI-ALERT (Proj. Reg. No. P046/14). Enrolment of human study participants was done after written informed consent was secured and signed agreements taken from all eligible participants.
Footnotes
Abbreviations: Af1, African 1; Af2, African 2; bTB, bovine tuberculosis; BWA, burrow-wheeler aligner; ENA, European Nucleotide Archive; ESS, effective sample size; ETHICOBOTS, Ethiopia Control of Bovine Tuberculosis Strategies; Eu1, European 1; Eu2, European 2; Eu3, European 3; H-F, Holstein-Friesian; HKY, Hasegawa, Kishino & Yano; km, kilometre; MCMC, Markov chain Monte Carlo; MIRU-VNTR, Mycobacterial interspersed repetitive unit-variable number tandem repeat; MLE, maximum likelihood estimates; PE, ProGlu; PE-PGRS, ProGlu-polymorphic GC-rich repetitive sequences; QC, quality control; SNP, single nucleotide polymorphism; TB, tuberculosis; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure, one supplementary table and two supplementary files are available with the online version of this article.
References
- 1.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO, FAO, OIE, The Union Roadmap for zoonotic tuberculosis 2017. ISBN: (WHO) 978-92-4-151304-3
- 3.Grace D, Mutua F, Ochungo P, Kruska R, Jones K, et al. Mapping of poverty and likely zoonoses hotspots. 2012.
- 4.Reviriego Gordejo FJ, Vermeersch JP, Gordejo FJR. Towards eradication of bovine tuberculosis in the European Union. Vet Microbiol. 2006;112:101–109. doi: 10.1016/j.vetmic.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 5.Cousins DV, Roberts JL. Australia's campaign to eradicate bovine tuberculosis: the battle for freedom and beyond. Tuberculosis. 2001;81:5–15. doi: 10.1054/tube.2000.0261. [DOI] [PubMed] [Google Scholar]
- 6.Vol I Report on livestock and livestock characteristics. Statistical Bulletin. 570 [Google Scholar]
- 7.Sibhat B, Asmare K, Demissie K, Ayelet G, Mamo G, et al. Bovine tuberculosis in Ethiopia: a systematic review and meta-analysis. Prev Vet Med. 2017;147:149–157. doi: 10.1016/j.prevetmed.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firdessa R, Tschopp R, Wubete A, Sombo M, Hailu E, et al. High prevalence of bovine tuberculosis in dairy cattle in central Ethiopia: implications for the dairy industry and public health. PLoS One. 2012;7:e52851. doi: 10.1371/journal.pone.0052851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elias K, Hussein D, Asseged B, Wondwossen T, Gebeyehu M. Status of bovine tuberculosis in Addis Ababa dairy farms. Rev Sci Tech Off Int Epiz. 2008;27:915–923. doi: 10.20506/rst.27.3.1850. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro B, Gebru G, Desta S, Negassa A, Nigussie K, et al. LRI Project Report. Nairobi, Kenya: International Livestock Research Institute (ILRI); 2015. Ethiopia livestock master plan. [Google Scholar]
- 11.Berg S, Firdessa R, Habtamu M, Gadisa E, Mengistu A, et al. The burden of mycobacterial disease in Ethiopian cattle: implications for public health. PLoS One. 2009;4:e5068. doi: 10.1371/journal.pone.0005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ameni G, Desta F, Firdessa R. Molecular typing of Mycobacterium bovis isolated from tuberculosis lesions of cattle in North eastern Ethiopia. Vet Rec. 2010;167:138–141. doi: 10.1136/vr.b4881. [DOI] [PubMed] [Google Scholar]
- 13.Ameni G, Erkihun A. Bovine tuberculosis on small-scale dairy farms in Adama town, central Ethiopia, and farmer awareness of the disease. Rev Sci Tech. 2007;26:711–719. [PubMed] [Google Scholar]
- 14.Ameni G, Tafess K, Zewde A, Eguale T, Tilahun M, et al. Vaccination of calves with Mycobacterium bovis Bacillus Calmette-Guerin reduces the frequency and severity of lesions of bovine tuberculosis under a natural transmission setting in Ethiopia. Transbound Emerg Dis. 2018;65:96–104. doi: 10.1111/tbed.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biffa D, Skjerve E, Oloya J, Bogale A, Abebe F, et al. Molecular characterization of Mycobacterium bovis isolates from Ethiopian cattle. BMC Vet Res. 2010;6:28. doi: 10.1186/1746-6148-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gumi B, Schelling E, Firdessa R, Erenso G, Biffa D, et al. Low prevalence of bovine tuberculosis in Somali pastoral livestock, Southeast Ethiopia. Trop Anim Health Prod. 2012;44:1445–1450. doi: 10.1007/s11250-012-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mekibeb A, Fulasa TT, Firdessa R, Hailu E. Prevalence study on bovine tuberculosis and molecular characterization of its causative agents in cattle slaughtered at Addis Ababa municipal abattoir, central Ethiopia. Trop Anim Health Prod. 2013;45:763–769. doi: 10.1007/s11250-012-0287-x. [DOI] [PubMed] [Google Scholar]
- 18.Tsegaye W, Aseffa A, Mache A, Mengistu Y, Stefan B. Ameni G: Conventional and molecular epidemiology of bovine tuberculosis in dairy farms in addis ababa city, the capital of ethiopia. J Appl Res Vet Med. 2010;8:143. [Google Scholar]
- 19.Mamo G, Bayleyegn G, Sisay Tessema T, Legesse M, Medhin G, et al. Pathology of camel tuberculosis and molecular characterization of its causative agents in pastoral regions of Ethiopia. PLoS One. 2011;6:e15862. doi: 10.1371/journal.pone.0015862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berg S, Garcia-Pelayo MC, Muller B, Hailu E, Asiimwe B, et al. African 2, a clonal complex of Mycobacterium bovis epidemiologically important in East Africa. J Bacteriol. 2011;193:670–678. doi: 10.1128/JB.00750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Raoult D, Fournier P-E. Bacterial strain typing in the genomic era. FEMS Microbiol Rev. 2009;33:892–916. doi: 10.1111/j.1574-6976.2009.00182.x. [DOI] [PubMed] [Google Scholar]
- 22.Kanduma E, McHugh TD, Gillespie SH. Molecular methods for Mycobacterium tuberculosis strain typing: a users guide. J Appl Microbiol. 2003;94:781–791. doi: 10.1046/j.1365-2672.2003.01918.x. [DOI] [PubMed] [Google Scholar]
- 23.Allix C, Walravens K, Saegerman C, Godfroid J, Supply P. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J Clin Microbiol. 2006;44:3471. doi: 10.1128/JCM.01349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith NH, Gordon SV, de la Rua-Domenech R, Clifton-Hadley RS, Hewinson RG. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis . Nat Rev Microbiol. 2006;4:670–681. doi: 10.1038/nrmicro1472. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Campos S, Aranaz A, de Juan L, Saez-Llorente JL, Romero B, et al. Limitations of spoligotyping and variable-number tandem-repeat typing for molecular tracing of Mycobacterium bovis in a high-diversity setting. J Clin Microbiol. 2011;49:3361–3364. doi: 10.1128/JCM.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahlstrom C, Barkema HW, Stevenson K, Zadoks RN, Biek R, et al. Limitations of variable number of tandem repeat typing identified through whole genome sequencing of Mycobacterium avium subsp. paratuberculosis on a national and herd level. BMC Genomics. 2015;16:161. doi: 10.1186/s12864-015-1387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghebremariam MK, Hlokwe T, Rutten VPMG, Allepuz A, Cadmus S, et al. Genetic profiling of Mycobacterium bovis strains from slaughtered cattle in Eritrea. PLoS Negl Trop Dis. 2018;12:e0006406. doi: 10.1371/journal.pntd.0006406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Branger M, Loux V, Cochard T, Boschiroli ML, Biet F, et al. The complete genome sequence of Mycobacterium bovis Mb3601, a SB0120 spoligotype strain representative of a new clonal group. Infect Genet Evol. 2020;82:104309. doi: 10.1016/j.meegid.2020.104309. [DOI] [PubMed] [Google Scholar]
- 29.Roberts GD, Koneman EW, Kim YK. Mycobacterium. Edited by Barlow et al. Manual of clinical microbiology. Washington, DC: American Society for Clinical Microbiology; 1991. pp. 304–339. [Google Scholar]
- 30.Zumárraga MJ, Soutullo A, García MI, Marini R, Abdala A, et al. Detection of Mycobacterium bovis -infected dairy herds using PCR in bulk tank milk samples. Foodborne Pathog Dis. 2012;9:132–137. doi: 10.1089/fpd.2011.0963. [DOI] [PubMed] [Google Scholar]
- 31.Medeiros L, Marassi CD, Duarte RS, da Silva MG, Lilenbaum W. Comparison of decontamination methods for primary isolation of Mycobacterium bovis in paucibacillary bovine tissues. Lett Appl Microbiol. 2012;54:182–186. doi: 10.1111/j.1472-765X.2011.03185.x. [DOI] [PubMed] [Google Scholar]
- 32.Machado A, Rito T, Ghebremichael S, Muhate N, Maxhuza G, et al. Genetic diversity and potential routes of transmission of Mycobacterium bovis in Mozambique. PLoS Negl Trop Dis. 2018;12:e0006147. doi: 10.1371/journal.pntd.0006147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Firdessa R, Berg S, Hailu E, Schelling E, Gumi B, et al. Mycobacterial lineages causing pulmonary and extrapulmonary tuberculosis, Ethiopia. Emerg Infect Dis. 2013;19:460–463. doi: 10.3201/eid1903.120256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gumi B, Schelling E, Berg S, Firdessa R, Erenso G, et al. Zoonotic transmission of tuberculosis between pastoralists and their livestock in south-east Ethiopia. Ecohealth. 2012;9:139–149. doi: 10.1007/s10393-012-0754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.FastQC A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- 36.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3:e104. doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 38.Xia E, Teo Y-Y, Ong RT-H. SpoTyping: fast and accurate in silico Mycobacterium spoligotyping from sequence reads. Genome Med. 2016;8:19. doi: 10.1186/s13073-016-0270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faksri K, Xia E, Tan JH, Teo Y-Y, Ong RT-H. In silico region of difference (RD) analysis of Mycobacterium tuberculosis complex from sequence reads using RD-Analyzer. BMC Genomics. 2016;17:847. doi: 10.1186/s12864-016-3213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith NH, Berg S, Dale J, Allen A, Rodriguez S, et al. European 1: A globally important clonal complex of Mycobacterium bovis . Infection Genetics and Evolution. 2011;11:1340–1351. doi: 10.1016/j.meegid.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 41.Müller B, Hilty M, Berg S, Garcia-Pelayo MC, Dale J, et al. African 1, an epidemiologically important clonal complex of Mycobacterium bovis dominant in Mali, Nigeria, Cameroon, and Chad. J Bacteriol. 2009;191:1951–1960. doi: 10.1128/JB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez-Campos S, Schürch AC, Dale J, Lohan AJ, Cunha MV, et al. European 2--a clonal complex of Mycobacterium bovis dominant in the Iberian Peninsula. Infect Genet Evol. 2012;12:866–872. doi: 10.1016/j.meegid.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Loiseau C, Menardo F, Aseffa A, Hailu E, Gumi B, et al. An African origin for Mycobacterium bovis . Evol Med Public Health. 2020;2020:49–59. doi: 10.1093/emph/eoaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price-Carter M, Brauning R, de Lisle GW, Livingstone P, Neill M, et al. Whole Genome Sequencing for Determining the Source of Mycobacterium bovis Infections in Livestock Herds and Wildlife in New Zealand. Front Vet Sci. 2018;5:272. doi: 10.3389/fvets.2018.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ, Nguyen L-T. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letunic I, Bork P. Interactive tree of life (iTOL) V4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hauer A, Michelet L, Cochard T, Branger M, Nunez J, et al. Accurate phylogenetic relationships among Mycobacterium bovis strains circulating in france based on whole genome sequencing and single nucleotide polymorphism analysis. Front Microbiol. 2019;10:955. doi: 10.3389/fmicb.2019.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guerra-Assuncao JA, Houben R, Crampin AC, Mzembe T, Mallard K, et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J Infect Dis. 2015;211:1154–1163. doi: 10.1093/infdis/jiu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Branger M, Hauer A, Michelet L, Karoui C, Cochard T, et al. Draft genome sequence of Mycobacterium bovis strain D-10-02315 isolated from wild boar. Genome Announc. 2016;4:e01268-16. doi: 10.1128/genomeA.01268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malm S, Linguissi LSG, Tekwu EM, Vouvoungui JC, Kohl TA, et al. New Mycobacterium tuberculosis Complex Sublineage, Brazzaville, Congo. Emerg Infect Dis. 2017;23:423–429. doi: 10.3201/eid2303.160679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Votintseva AA, Pankhurst LJ, Anson LW, Morgan MR, Gascoyne-Binzi D, et al. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol. 2015;53:1137–1143. doi: 10.1128/JCM.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kahle D, Wickham H. ggmap: spatial visualization with ggplot2. R J. 2013;5:144–161. doi: 10.32614/RJ-2013-014. [DOI] [Google Scholar]
- 57.Team RC R: a Language and Environment for Statistical Computing. Vienna, Austria: 2013. [Google Scholar]
- 58.contoureR Contouring of Non-Regular three-dimensional data
- 59.Geosphere Spherical trigonometry
- 60.Dray S, Dufour A-B. The ade4 package: Implementing the duality diagram for ecologists. J Stat Softw. 2007;22:1–20. doi: 10.18637/jss.v022.i04. [DOI] [Google Scholar]
- 61.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal, Complex Systems. 2006 [Google Scholar]
- 62.Pedersen T. ggraph: an implementation of grammar of graphics for graphs and networks. 2020.
- 63.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the beast 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. doi: 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
- 65.GC Y, Smith DK, Zhu HC, Guan Y. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 66.Rieux A, Khatchikian CE. tipdatingbeast: an R package to assist the implementation of phylogenetic tip-dating tests using beast. Mol Ecol Resour. 2017;17:608–613. doi: 10.1111/1755-0998.12603. [DOI] [PubMed] [Google Scholar]
- 67.Romha G, Gebru G, Asefa A, Mamo G. Epidemiology of Mycobacterium bovis and Mycobacterium tuberculosis in animals: transmission dynamics and control challenges of zoonotic TB in Ethiopia. Prev Vet Med. 2018;158:1–17. doi: 10.1016/j.prevetmed.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 68.Biek R, O'Hare A, Wright D, Mallon T, McCormick C, et al. Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathog. 2012;8:e1003008. doi: 10.1371/journal.ppat.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crispell J, Zadoks RN, Harris SR, Paterson B, Collins DM, et al. Using whole genome sequencing to investigate transmission in a multi-host system: bovine tuberculosis in New Zealand. Bmc Genomics. 2017:18. doi: 10.1186/s12864-017-3569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker TM, Ip CLC, Harrell RH, Evans JT, Kapatai G, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu Y, Cancino-Munoz I, Torres-Puente M, Villamayor LM, Borras R, et al. High-Resolution mapping of tuberculosis transmission: whole genome sequencing and phylogenetic modelling of a cohort from Valencia region, Spain. PLoS Med. 2019;16:e1002961. doi: 10.1371/journal.pmed.1002961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hatherell HA, Colijn C, Stagg HR, Jackson C, Winter JR, et al. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 2016;14:21. doi: 10.1186/s12916-016-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mekonnen GA, Ameni G, Wood JLN, Berg S, et al. ETHICOBOTS consortium Network analysis of dairy cattle movement and associations with bovine tuberculosis spread and control in emerging dairy belts of Ethiopia. BMC Vet Res. 2019;15:262. doi: 10.1186/s12917-019-1962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Didelot X, Fraser C, Gardy J, Colijn C. Genomic infectious disease epidemiology in partially sampled and ongoing outbreaks. Mol Biol Evol. 2017;34:msw075–1007. doi: 10.1093/molbev/msw275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hopkirk CSM The veterinary service of Ethiopia. N Z Vet J. 1954 [Google Scholar]
- 76.Ameni G, Aseffa A, Sirak A, Engers H, Young DB, et al. Effect of skin testing and segregation on the prevalence of bovine tuberculosis, and molecular typing of Mycobacterium bovis, in Ethiopia. Vet Rec. 2007;161:782–786. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.