Abstract
Metagenomics is a valuable diagnostic tool for enhancing microbial food safety because (i) it enables the untargeted detection of pathogens, (ii) it is fast since primary isolation of micro-organisms is not required, and (iii) it has high discriminatory power allowing for a detailed molecular characterization of pathogens. For shotgun metagenomics, total nucleic acids (NAs) are isolated from complex samples such as foodstuff. Along with microbial NAs, high amounts of matrix NAs are extracted that might outcompete microbial NAs during next-generation sequencing and compromise sensitivity for the detection of low abundance micro-organisms. Sensitive laboratory methods are indispensable for detecting highly pathogenic foodborne bacteria like Brucella spp., because a low infectious dose is sufficient to cause human disease through the consumption of contaminated dairy or meat products. In our study, we applied shotgun metagenomic sequencing for the identification and characterization of Brucella spp. in artificially and naturally contaminated raw milk from various ruminant species. With the depletion of eukaryotic cells prior to DNA extraction, Brucella was detectable at 10 bacterial cells ml−1, while at the same time microbiological culture and isolation of the fastidious bacteria commonly failed. Moreover, we were able to retrieve the genotype of a Brucella isolate from a metagenomic dataset, indicating the potential of metagenomics for outbreak investigations using SNPs and core-genome multilocus sequence typing (cgMLST). To improve diagnostic applications, we developed a new bioinformatics approach for strain prediction based on SNPs to identify the correct species and define a certain strain with only low numbers of genus-specific reads per sample. This pipeline turned out to be more sensitive and specific than Mash Screen. In raw milk samples, we simultaneously detected numerous other zoonotic pathogens, antimicrobial resistance genes and virulence factors. Our study showed that metagenomics is a highly sensitive tool for biological risk assessment of foodstuffs, particularly when pathogen isolation is hazardous or challenging.
Keywords: Brucella, detection limits, host depletion, metagenomics, raw milk
Data Summary
Raw data generated by next-generation sequencing have been uploaded to the European Nucleotide Archive (ENA) under study accession number ERP121102 (https://www.ebi.ac.uk/ena/browser/view/PRJEB37772). The bioinformatics analysis pipeline developed in our study is publicly available at GitLab (https://gitlab.com/bfr_bioinformatics/refsnper).
Impact Statement.
Foodborne infections pose a major threat to public health, with 600 million cases reported annually worldwide. The monitoring of food is vitally important to prevent transmission of pathogens to humans through food consumption. Currently, next-generation sequencing is most commonly used to generate genomic data of bacterial isolates from patients and foodstuffs, which can be compared to trace foodborne outbreaks. Culture-independent metagenomics can speed up results, because time-consuming bacterial isolation is not required. Instead, the entire DNA of a food sample is analysed, but the relatively small proportion of pathogen DNA as compared to food-matrix DNA represents a challenge. As a result, pathogens are not detected or only small parts of their genomes can be recovered from the food sample, which significantly reduces the diagnostic value of this untargeted approach. To improve diagnostic applications in food control, the background signal from the food matrix has to be reduced to increase the amount of DNA retrieved from the micro-organisms. In our study, the advantage of eukaryotic cell depletion in food microbiology was proven for highly pathogenic zoonotic Brucella spp. in raw milk, which enabled the detection and molecular characterization of the pathogen without prior isolation.
Introduction
Brucella spp. are fastidious, slow-growing zoonotic bacteria, often transmitted from animals to humans through the consumption of raw animal products, predominantly unpasteurized milk and cheese [1]. Bacterial isolation from food products is challenging, because brucellae are frequently overgrown by other bacteria of the food microbiome using classical culture methods. In addition, infected animals often shed the pathogen in low concentrations [<103 c.f.u. (ml milk)−1] [2] and the infectious dose for humans is also very low, ranging from 10 to 100 bacteria. Therefore, genus-specific quantitative PCR (qPCR) is considered an adequate alternative diagnostic method to reliably identify Brucella spp. in foodstuff [3], but it does not allow for a detailed characterization of the pathogen, which is essential for risk assessment.
In contrast, whole-genome sequencing (WGS) of bacterial isolates provides a high resolution for the characterization of micro-organisms. Applying metagenomics as a culture-independent method to food affected by microbial contamination reveals a large number of sequencing reads originating from matrix material, which poses a challenge for the detection of low-abundance pathogens [4]. Sequencing of the food matrix can be avoided by amplification and sequencing of the bacterial 16S rDNA. However, with the so-called metabarcoding approach, various bacterial species cannot be resolved, making this method unsuitable for the universal detection of pathogenic bacteria [5]. Furthermore, non-bacterial taxa are not recorded and a more detailed characterization of pathogens, including genotyping or verification of virulence factors and antibiotic resistance genes, is impossible. Compared to the WGS commonly used for pathogen surveillance, the application of whole-metagenome sequencing (WMS) in food safety is still in the fledging stage [6]. Most of the present studies struggle with an insufficient percentage of microbial sequence reads, which are necessary to describe the relevant properties of a pathogen [5, 7–10].
Consequently, a drastic increase of sequencing depth is needed to get genome coverage sufficient for the detection of low-abundance species. According to Ottesen et al., a 250-fold increase in sequencing depth is necessary to reach a coverage of one for all genomes in the sample [9]. Several studies improved detection limits by enrichment culture before metagenomics [7, 9, 10]. Enrichment for at least 8 h decreased the limit of detection from 10 000 c.f.u. Shiga toxin-producing Escherichia coli in 100 g fresh spinach to 10 c.f.u. [7]. However, enrichment culture only works for bacterial pathogens with high replication rates, such as E. coli , that are not overgrown by the food microbiome. Viruses and parasites remain unnoticed, as well as bacteria that require other growth conditions. In general, enrichment culture may alter microbial composition [7, 10]. Therefore, protocols for the enrichment of microbial DNA have been established, e.g. by using CpG-methylation in eukaryotic DNA [11] or differential centrifugation [12]. An alternative method to reduce undesired matrix signals is the selective lysis of eukaryotic cells based on the different membrane properties of eukaryotic and prokaryotic cells. In various studies, the depletion of human DNA in clinical samples increased the sensitivity of metagenomic analysis for the detection of microbes [13–19]. Enrichment of bacterial DNA by depletion of eukaryotic cells only slightly modifies the microbial composition of a broad range of bacteria compared to conventionally extracted DNA [13, 16, 17].
The goal of our study was to apply metagenomic shotgun sequencing for direct detection and detailed characterization of zoonotic pathogens in food matrices, with Brucella spp. in raw milk as a model. We used both artificially contaminated raw cow’s milk, and milk from naturally infected sheep, goats, buffalos and cattle, to establish metagenomic analysis in food samples and to prove the concept, respectively.
Methods
Collection of raw milk samples
A total of 151 midstream milk samples (2 ml each) were collected from 100 dairy cattle, 25 buffalos, 13 goats and 13 sheep following aseptic and standardized milking procedures. The animals were reared by 20 rural farming communities in Meet El-Amel, Aga District in Dakahlia Governorate, Delta region, Egypt. The animals included in our study either suffered from health disorders, such as subclinical mastitis (n=33), clinical mastitis (n=27), reproductive disorders (n=7) and abortion (n=31), or were apparently healthy (n=50) and gave normal birth (n=3). All samples were kept in a cooler until transport to the laboratory within 1 h after sample collection.
Inoculation of raw cow’s milk
Raw milk was sampled from dairy cows kept on the experimental farm of the German Federal Institute for Risk Assessment (BfR), officially free of bovine brucellosis, and was inoculated with Brucella abortus bv. 1 (strain 544, NCTC 10093) with exponentially increasing concentrations ranging from 10 to 107 cells (ml milk)−1. The bacterial solutions prepared for inoculation experiments were grown on BBL Brucella agar with 5 % horse blood (Becton Dickinson) for 3 days at 37 °C, to determine the actual number of cells by counting c.f.u., following the general assumption that one bacterial cell equates to one c.f.u. Three independent experiments were performed, resulting in three biological replicates with three technical replicates each. The artificially contaminated milk samples were inactivated by adding 100 % (v/v) ethanol to obtain a final concentration of 75 % (v/v), followed by an incubation at room temperature for at least 15 min and were stored at −80 °C until further use.
Isolation of Brucella from milk
qPCR-positive milk samples were subjected to culture for bacterial isolation. In brief, 1 ml raw milk was diluted 1 : 10 in Brucella selective broth (Oxoid) [20] and incubated in a 25 ml cell culture flask at 37 °C and 5 % CO2. Over 6 weeks, 1 µl each was plated weekly on Brucella agar (Becton Dickinson) and selective agar (Oxoid) [20]. Suspicious isolates were identified as Brucella spp. using MALDI-TOF MS [21] and further characterized with classical microbiological methods [22].
DNA extraction
DNA was extracted from Brucella milk isolates and raw milk samples (500 µl) using the DNeasy mericon food kit (Qiagen) according to the manufacturer’s protocol for 200 mg starting material. For eukaryotic cell depletion, 1 ml raw milk was centrifuged at 16 000 g and the pellet was resuspended in 200 µl PBS. The HostZERO microbial DNA kit (Zymo) was used to deplete eukaryotic cells and to extract DNA according to the manufacturer’s instructions, with a 3 min bead-beating step in a precooled TissueLyser LT (Qiagen) at 50 Hz. DNA concentration and purity were determined using the Qubit dsDNA HS assay kit with a Qubit 2.0 fluorometer (Thermo Fisher Scientific) and a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies).
Detection of Brucella using qPCR
The genus-specific marker sequences bcsp31 and IS711 found in all Brucella spp. [23, 24] were amplified in a total volume of 25 µl including 5 µl template DNA using the QuantiFast pathogen PCR +IC kit (Qiagen) according to the manufacturer’s instructions. All samples were analysed in triplicates using a CFX96 Touch real-time PCR detection system (Bio Rad). C t values ≤40 in both qPCR assays were empirically considered as a Brucella -positive test result.
Next-generation sequencing
DNA libraries of Brucella milk isolates were generated with the Nextera XT DNA library prep kit (Illumina), and DNA libraries of raw milk samples with the TruSeq Nano DNA library prep kit, both according to the manufacturer’s instructions. In case the DNA concentration of a sample was below 2 ng µl–1, a fixed volume of 50 µl instead of 100 ng was used as input material. DNA was sheared using the M220 focused-ultrasonicator (Covaris). Next-generation sequencing was performed with NextSeq 500 and MiSeq (Illumina) in paired-end mode with 2×151 cycles and 2×251 cycles, respectively.
Bioinformatics analysis
Quality control and read extraction
Raw reads were trimmed using fastp version 0.20.0 [25] with a mean phred-score of 30. For taxonomic classification, Kraken2 version 2.0.8-beta [26] with the RefSeq95 database and KrakenUniq version 0.5.8 [27] with the RefSeq bacteria database (downloaded on December 2, 2019) were applied on trimmed reads. Read extraction and blast confirmation were executed as previously described [5].
Species and strain prediction
Species and/or strain prediction based on extracted reads were performed with Mash Screen version 2.2 [28] using a winner-takes-all strategy or with the RefSNPer pipeline version 1.0.0 (https://gitlab.com/bfr_bioinformatics/refsnper). In the latter case, extracted reads were mapped to a set of complete or draft genomes available in the RefSeq database using bowtie2 version 2.3.5 [29], followed by the calculation of the reference genome coverage and SNP calling with SAMtools version 1.10 [30] and BEDTools version 2.29.0 [31]. All complete genomes of the genus Brucella were used for species prediction, and all complete and draft genomes of B. abortus for strain prediction. Subsampling of reads was performed with seqtk version 1.2-r94 using different seeds for each replicate (https://github.com/lh3/seqtk). SNP analysis of publicly available B. abortus assemblies was carried out with parSNP version 1.2 [32].
Assembly and genotyping
Metagenomic assemblies were generated using megahit version 1.1.3 [33]. Shovill version 1.0.4 (https://github.com/tseemann/shovill) was used to assemble WGS data and SPAdes version 3.10.0 for the assembly of extracted reads from WMS data. The quality and completeness of the resulting assemblies were analysed with quast 4 version 4.6.1 [34]. The Brucella melitensis core-genome multilocus sequence typing (cgMLST) analysis was carried out with chewBBACA version 2.0.16 [35] using a scheme of 2704 target genes [36]. SNP calling with the complete reference genome and phylogenetic analysis were conducted using snippy version 4.4.3 (https://github.com/tseemann/snippy) with mincov=3. For pairwise comparison of isolates and WMS data, bowtie2 and BCFtools were used. The community analysis was performed with R package vegan.
Prediction of virulence factors and antimicrobial resistance (AMR) genes
Virulence factors and AMR genes were identified by srst2 version 0.2.0 [37] with default parameters for WGS data (minimum coverage=90 %, minimum depth=5) and more relaxed criteria for WMS data (minimum coverage=30 %, minimum depth=1) using the Virulence Factor Database (VFDB, set A_nt) as downloaded on March 22, 2018 and National Center for Biotechnology Information (NCBI) AMRfinder database version 2019-10-30.1, respectively.
Graphical representation
Plots were generated in R with ggplot2 version 3.2.1, pheatmap version 1.0.12, plotly version 4.9.1 and vegan version 2.5-6. For graphic representation of phylogenetic trees, iTOL [38] and grapetree [39] were used.
Results
Detection of Brucella spp. in raw milk using metagenomic sequencing
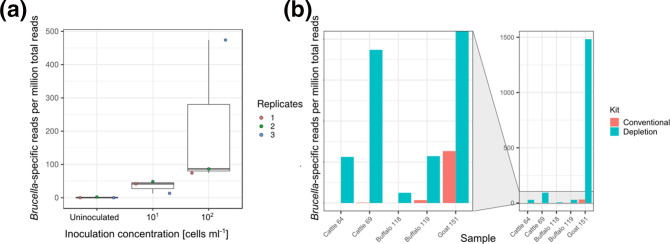
We determined the molecular detection limits of two different genus-specific qPCRs targeting IS711 and bcsp31. Independent of the target sequence, Brucella was reliably detected above 103 bacterial cells (ml milk)–1 (Fig. S1, available in the online version of this article). Based on preliminary experiments (data not shown), the detection limit of metagenomics was tested at concentrations of 101 and 102 cells ml−1. Metagenomic sequencing of foodstuff generates high numbers of reads (>95 %) deriving from the food matrix, which results in a tremendous reduction of sensitivity for the detection of pathogenic micro-organisms. To enhance the fraction of bacterial sequences, we extracted DNA from inoculated and non-inoculated raw milk samples depleted of eukaryotic cells. In three independent sequencing experiments, we found 0.02–2.15 reads specific for Brucella per million reads in non-inoculated milk, 13–49 Brucella reads per million reads in milk samples inoculated at a concentration of 101 cells ml−1 and 74–474 Brucella reads per million reads in milk samples inoculated at a concentration of 102 cells ml−1 (Fig. 1a). Remarkably, with depletion of eukaryotic cells we were able to detect Brucella at very low concentrations between 22 and 31 bacterial cells (ml raw milk)–1, which corresponds to a 100-fold lower detection limit than the ones observed for qPCR assays testing the same samples.
Fig. 1.
Eukaryotic cell depletion improves the detection of Brucella in raw milk by metagenomic analysis. (a) Detection of Brucella -specific reads per million total reads in artificially contaminated raw cow’s milk after eukaryotic cell depletion in three replicates. (b) Brucella -specific reads per million reads in milk samples from different dairy animals detected after conventional DNA extraction and DNA extraction with eukaryotic cell depletion.
In addition to artificially contaminated cow’s milk, we analysed raw milk samples from various ruminants potentially infected with Brucella spp. using qPCR and metagenomic sequencing. While 10 out of 151 raw milk samples were tested positive by the two genus-specific qPCR assays, B. melitensis was only isolated from a single goat's milk sample (no. 151). Three raw milk samples from buffalo (no. 118 and no. 119) and goat (no. 151) were selected on the basis of low C t values in both qPCRs for metagenomic sequencing. Additionally, two samples with a C t value >40 in the bcsp31 qPCR were chosen from cattle (no. 64 and no. 69) (Table 1). The DNA of all samples was sequenced with and without previous eukaryotic cell depletion.
Table 1.
Raw milk samples of potentially Brucella -infected animals selected for metagenomics
|
Sample no. |
Origin |
Health status |
Mean C t value for IS711 (n=2) |
C t value for bcsp31 |
|---|---|---|---|---|
|
64 |
Cattle |
Parturition |
35.86 |
41.75 |
|
69 |
Cattle |
Apparently healthy (q fever) |
35.03 |
40.39 |
|
118 |
Buffalo |
Abortion |
35.25 |
39.07 |
|
119 |
Buffalo |
Abortion |
32.65 |
36.30 |
|
151 |
Goat |
Apparently healthy |
25.64 |
30.48 |
Using Kraken2 classification and blastn verification, we found 0–32 Brucella -specific reads per million reads with the conventional DNA extraction method and 6–1483 Brucella -specific reads per million reads with the eukaryotic cell depletion approach. Two (no. 64 and no. 118) out of five milk samples were considered Brucella -negative with 0 and 0.02 Brucella -specific reads per million reads, when conventional DNA extraction was applied. In contrast, all five samples were Brucella -positive when DNA extraction was conducted with eukaryotic cell depletion (Fig. 1b). Hence, Brucella -specific reads could be enriched 16- to 1400-fold by depleting eukaryotic cells.
In summary, metagenomic sequencing of total sample DNA after eukaryotic cell depletion led to an improved detection of Brucella in inoculated raw cow’s milk, with a 100-fold higher sensitivity than qPCR. Eukaryotic cell depletion also improved pathogen detection in naturally contaminated raw milk samples and revealed the presence of Brucella in two samples that were primarily considered as Brucella -negative, when conventional DNA extraction was applied.
Brucella species and strain prediction from metagenomic datasets
Since qPCR does not allow for the differentiation of Brucella strains, we assessed whether metagenomic analysis will enable species and strain identification. We used Mash Screen and the newly developed bioinformatics pipeline RefSNPer for this purpose. RefSNPer executes read mapping to reference genomes followed by SNP calling and outputs the coverage, depth and number of SNPs for each reference genome (Fig. 2a). The objective behind this approach was to identify the most closely related genome exhibiting the smallest density of SNPs when compared to the sequences generated by next-generation sequencing (NGS).
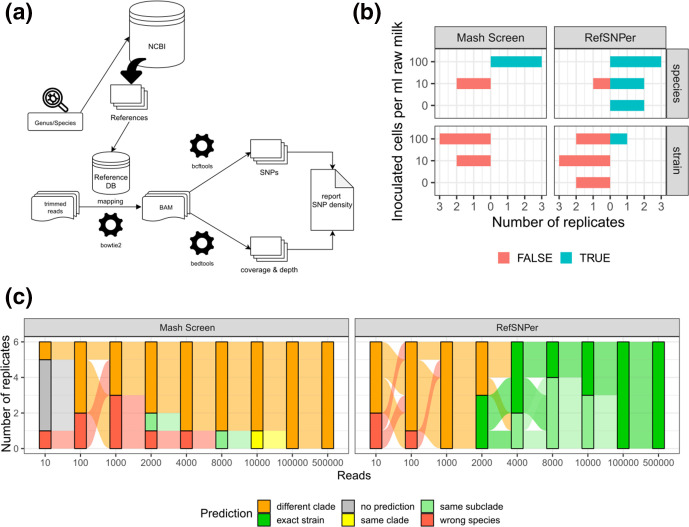
Fig. 2.
Brucella species and strain prediction in metagenomic samples. (a) RefSNPer: a workflow for the identification of the closest reference strains. The input (a set of isolate or metagenomic sequences provided as paired fastq files) is mapped to a reference database. The reference database can be automatically generated for user-defined genera or species from draft and/or complete genome sequences available from the NCBI RefSeq database. Coverage, coverage depth (BEDTools) and number of putative SNPs (BCFtools) are determined for each reference genome. A summary file is generated that outputs the SNP density in each reference genome. (b) Species and strain prediction in raw milk inoculated with different numbers of B. abortus (strain 544) cells using Mash Screen and RefSNPer. Results of three replicates are given, excluding replicates without any prediction. (c) Species and strain prediction by Mash Screen and RefSNPer for an increasing number of in silico randomly subsampled reads from WGS data of B. abortus (strain 544) in six replicates.
First, we tested the ability of the two pipelines to predict the correct species and strain from the artificially contaminated raw milk samples, which were inoculated with B. abortus strain 544 (NCTC 10093). Brucella -specific reads classified by Kraken2 were extracted, analysed with Mash Screen and RefSNPer, and the correctness of predictions was evaluated (Fig. 2b, Table S1). With Mash Screen, we detected B. abortus in every replicate at a concentration of 100 bacterial cells (ml milk)−1. In milk samples inoculated at lower bacterial concentrations (10 cells ml−1), the correct species was only identified in one out of three replicates, and reported either not at all or incorrectly as B. melitensis in the other replicates. Strain prediction was always incorrect independent of bacterial concentrations. No results were obtained for the control samples, not inoculated with Brucella . In contrast, RefSNPer predicted the correct species in two out of three replicates at the lower concentration of 10 cells ml−1 and for all replicates at the higher concentration of 100 cells ml−1. The correct strain was predicted in one sample inoculated with the higher bacterial concentration.
Since unreliable strain prediction might be caused by a small number of reads, we tested the minimum number of reads needed for correct strain prediction using both bioinformatics tools. Increasing numbers of reads from WGS data of B. abortus strain 544 were randomly subsampled in silico in replicates (n=6) and analysed with both tools to define the phylogenetic distance to B. abortus strain 544 after core-genome SNP analysis with all publicly available complete and draft genomes of B. abortus (Figs 2c and S2, Table S1). We observed wrong species prediction with Mash Screen up to 4000 reads, whereas the correct species was already reliably predicted with 1000 subsampled reads using RefSNPer. Mash Screen analysis was not able to predict the correct strain, and resulted in the prediction of strains that locate to phylogenetically more distant clades (Fig. S2, Table S1). In contrast, RefSNPer predicted the expected strain or a very closely related strain in 50 % of the replicates using only 2000 subsampled reads for analysis and in all replicates using 4000 reads.
Mash Screen and RefSNPer analyses were also conducted on metagenomic data from naturally contaminated raw milk samples in order to determine the Brucella species after DNA extraction with and without previous eukaryotic cell depletion (Table 2). For most samples with less than 1000 Brucella -specific reads, a reliable prediction failed. Furthermore, the predictions could not be verified because we did not recover Brucella isolates from any of these samples. The result of species prediction changed from B. abortus to B. melitensis and from B. abortus to Brucella ovis in sample numbers 69 and 119, respectively, when a higher number of bacteria-specific reads was available due to eukaryotic cell depletion. While the identification of B. melitensis in cattle from Egypt is very likely, B. ovis , which is usually isolated from sheep, might be a false prediction for buffalo milk. Mash Screen analysis was not able to predict a species or most frequently Brucella suis was found in raw milk samples. Mash Screen did not predict the correct species from 2787 Brucella -specific reads in sample number 151 after conventional DNA extraction. In contrast, RefSNPer accurately identified the correct species as well as the closest complete reference genome B. melitensis strain 2008724259 (GCF_001715485.1) from the NCBI RefSeq database.
Table 2.
Brucella reads and species prediction from naturally contaminated raw milk samples
|
Sample no. |
DNA extraction kit* |
blast confirmed Brucella -specific reads |
Predicted species |
|
|---|---|---|---|---|
|
Mash Screen |
RefSNPer |
|||
|
64 |
c |
2 |
np |
|
|
64 |
d |
155 |
||
|
69 |
c |
24 |
np |
|
|
69 |
d |
810 |
||
|
118 |
c |
0 |
np |
np |
|
118 |
d |
56 |
np |
|
|
119 |
c |
50 |
||
|
119 |
d |
225 |
||
|
151 |
c |
2787 |
||
|
151 |
d |
112 627 |
||
np, No prediction.
*c, Conventional DNA extraction; d, DNA extraction after eukaryotic cell depletion.
In summary, species and strain prediction were more reliable when RefSNPer was used. Mash Screen analysis never resulted in the expected strain. Using RefSNPer ≥1000 Brucella -specific reads were needed for reliable species prediction and ≥4000 reads for correct strain prediction.
Comparison of genomic sequences from a B. melitensis isolate and metagenomic sequences from related goat's milk
To evaluate the potential of WMS for pathogen characterization in food matrices, we compared WGS data of three isolates from raw goat's milk with the Brucella-specific data from the WMS of the related sample (no. 151). We first conducted a RefSNPer analysis with the WGS reads and Brucella -specific reads extracted from WMS data, and consequently identified the genome sequence of B. melitensis strain 2008724259 (GCF_001715485.1) as the closest complete reference genome for comparison. The number of Brucella -specific sequencing reads was about 12-fold higher in the WGS than in the WMS dataset, also resulting in a higher coverage depth of 39- to 55-fold after WGS of isolates in comparison to 5.3-fold on average after WMS of the contaminated raw milk sample. WGS and WMS datasets covered the reference genome almost completely with 99.99 and 99.05 %, respectively. Pairwise comparison of the three isolate sequences resulted in 2–5 SNPs and pairwise comparison of the metagenome to the assemblies from WGS of the isolates resulted in 43–44 SNPs, but only 14–16 SNPs were supported by ≥3 reads as shown in a SNP matrix (Fig. S3).
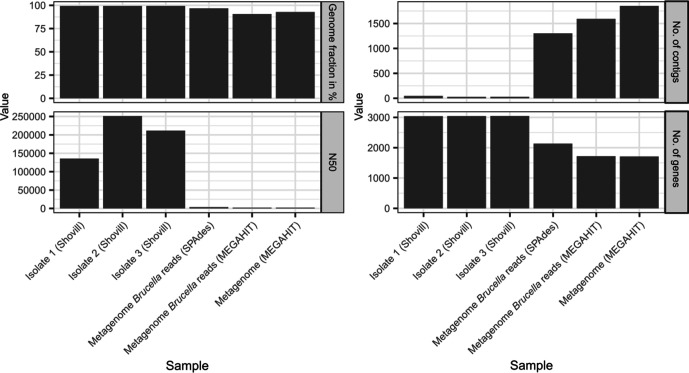
In a next step, we tested two different assembly strategies: assemblies generated from all metagenomic sequences (metagenome) and from Brucella -specific reads extracted from the metagenome (metagenome Brucella reads) with SPAdes and megahit. These assemblies were evaluated with B. melitensis strain 2008724259 (GCF_001715485.1) as a reference genome (Fig. 3). A metagenomic assembly of the WMS data resulted in 1857 contigs covering 92.9 % of the reference genome. Brucella read extraction and assembly resulted in fewer contigs (n SPAdes=1303, n MEGAHIT=1593) and a higher recovery of the reference genome (96.8 %) when SPAdes was used. When WGS data of isolates were assembled, 99.4 % of the genome could be recovered and the number of contigs varied between 29 and 48.
Fig. 3.
Statistics for different assembly strategies. Assemblies were generated from the whole metagenome (metagenome), from Brucella -specific reads extracted from the metagenome (metagenome Brucella reads), and from WGS data of isolates originating from the related raw milk sample. The genome fraction, N50, number of contigs and number of genes recovered from these assemblies were analysed using B. melitensis strain 2008724259 (GCF_001715485.1) for reference.
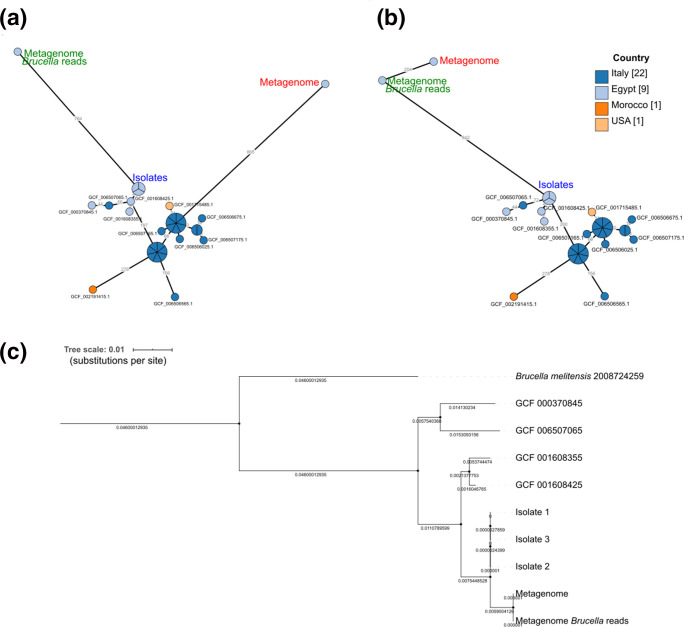
A cgMLST analysis was conducted with all publicly available draft and complete B. melitensis genomes (n=336) retrieved from the RefSeq database, the assembled genome sequences of isolates, the Brucella -specific reads (extracted from metagenomes) assembled with SPAdes and the metagenomic assemblies of WMS data. The isolates and the assembly of Brucella -specific reads extracted from WMS data clustered in the same subclade of a minimum spanning tree based on cgMLST allelic profiles (Fig. S4), whereas the metagenomic assembly of WMS data was found in a more distant clade (Fig. 4a). The isolates analysed in our study and the assembled Brucella -specific reads from WMS data clustered more closely together with three other isolates from Egypt (GCF_000370845.1, GCF_001608425.1, GCF_001608355.1) and one isolate from Italy (GCF_006507065.1) than with the metagenomic assembly from WMS data. The phylogenetic distance of the metagenomic assembly resulted from the relative low number of matching alleles, with 1105 and 682 when isolate 2 was compared to the assembled Brucella -specific reads and the assembled total metagenomic dataset, respectively. To test the impact of the assembler chosen for analysis, we also assembled the WGS data of isolates and Brucella -specific reads (extracted from the metagenome) with megahit and conducted a cgMLST analysis. Consequently, the metagenomic assembly from WMS data now clustered together with isolates and assembled Brucella -specific reads (Fig. 4b). Hence, cgMLST results clearly depend on the assembly algorithm, which was also proven for the WGS data of isolates that clustered differently (Fig. 4a, b).
Fig. 4.
Genomic typing of Brucella from Egyptian raw milk samples by cgMLST (a, b) and SNP analysis (c) using WGS and WMS datasets. (a) Excerpt of a minimum spanning tree (Fig. S4 gives an overview) based on cgMLST allelic profiles of 336 publicly available genomes, and assemblies of WGS and WMS data from raw milk sample number 151. Assemblies from WMS data were generated either by metagenomic assembly (metagenome) or after extraction of Brucella -specific reads (metagenome Brucella reads). Numbers at the branches stand for allelic distances, and branches presenting less than 10 alleles difference are collapsed. Assemblies of WGS data of isolates and Brucella -specific reads were generated either with Shovill and SPAdes, respectively (a), or with megahit (b). (c) Phylogenetic relationship of four closely related publicly available draft genomes and WGS and WMS data obtained from raw milk sample number 151 based on core SNP distances illustrated in a maximum-likelihood tree.
A SNP analysis using the closest complete reference genome available in NCBI, previously identified by RefSNPer, was performed. Besides sequencing data of sample number 151, including genomic sequences of isolates, WMS data and Brucella -specific reads extracted from WMS data, we analysed publicly available genomes (GCF_000370845, GCF_006507065, GCF_001608355 and GCF_001608425) found in the same cgMLST cluster. The B. melitensis isolates from goat's milk revealed 190 SNPs and the metagenomic data 179 SNPs in comparison with the reference genome. Relationships based on SNP analysis were depicted in a maximum-likelihood tree with B. melitensis strain 2008724259 (GCF_001715485.1) as an outgroup (Fig. 4c). The three isolates and the metagenomic samples shared the same SNPs at 179 positions and revealed a distance of 11 SNPs, while the publicly available genomes in the same cgMLST cluster showed a SNP distance of 21–72 to the isolates in our study. In contrast to cgMLST analysis, the relationship of the WGS data of isolates was much closer with the WMS data gained from our experiments than with the four publicly available genomes.
In summary, a close relationship between isolates and Brucella sequences from WMS data was proven, but they were not found in the same subclade using high-resolution SNP analysis. There was no difference between the total WMS dataset and the extracted Brucella -specific reads in the SNP analysis. In contrast, our cgMLST analysis was less robust and dependent on the assembler used.
Milk microbiota and associated zoonotic bacterial pathogens
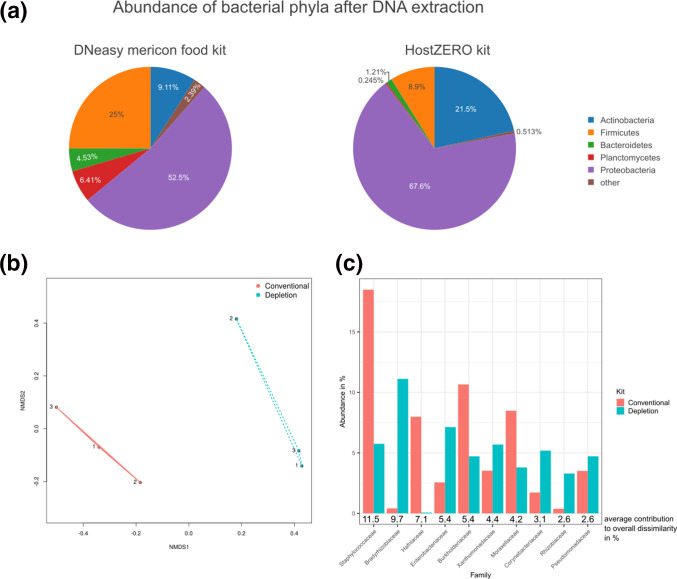
In addition to zoonotic pathogens, we investigated the composition of the prokaryotic community in raw milk in order to assess the effects of eukaryotic cell depletion. We first analysed the taxonomic composition of bacterial phyla in the pure raw milk used for inoculation experiments (Fig. 5a). Regardless of the extraction method applied, the major phylum was Proteobacteria . The fractions of Firmicutes , Planctomycetes and Bacteroidetes were larger in total DNA extracts than in DNA extracted after eukaryotic cell depletion, and vice versa for the fractions of Proteobacteria and Actinobacteria . The differences in the relative abundance of bacterial phyla in milk, depending on the DNA extraction method used, could be confirmed at family level by using a non-metric multidimensional scaling analysis (NMDS), which revealed two different clusters (Fig. 5b). While analysis of similarities (ANOSIM) showed a high within-group similarity and dissimilarity between groups (R=1), the difference between the groups was not statistically significant (P=0.1). The analysis of similarity percentages (SIMPER) showed that these dissimilarities were mainly caused by Staphylococcaceae , Hafniaceae , Burkholderiaceae and Moraxellaceae , which were more abundant when conventional DNA extraction was applied, and Bradyrhizobiaceae , Enterobacteriaceae , Xanthomonadaceae , Corynebacteriaceae , Rhizobiaceae and Pseudomonadaceae , which were more abundant after eukaryotic cell depletion (Fig. 5c).
Fig. 5.
Analysis of the effects of different DNA extraction strategies on the abundance of bacterial phyla in pure cow’s milk. The prokaryotic composition of raw milk used for inoculation experiments was determined by KrakenUniq after conventional DNA extraction with the DNeasy mericon food kit, and after eukaryotic cell depletion and DNA extraction with the HostZERO kit. (a) Mean abundances (%) of the bacterial phyla most prevalent in three replicates are presented in a pie chart for each extraction kit. (b) Non-metric multidimensional scaling (NMDS) analysis based on Bray–Curtis dissimilarity of the detected bacterial families in all samples after conventional DNA extraction (red) and DNA extraction with eukaryotic cell depletion (blue). (c) Mean abundances (%) of the families that were most discriminating between the two DNA extraction kits obtained by analysis of similarity percentage (SIMPER) using Bray–Curtis dissimilarity are presented in a bar chart together with the mean contribution to the overall dissimilarity.
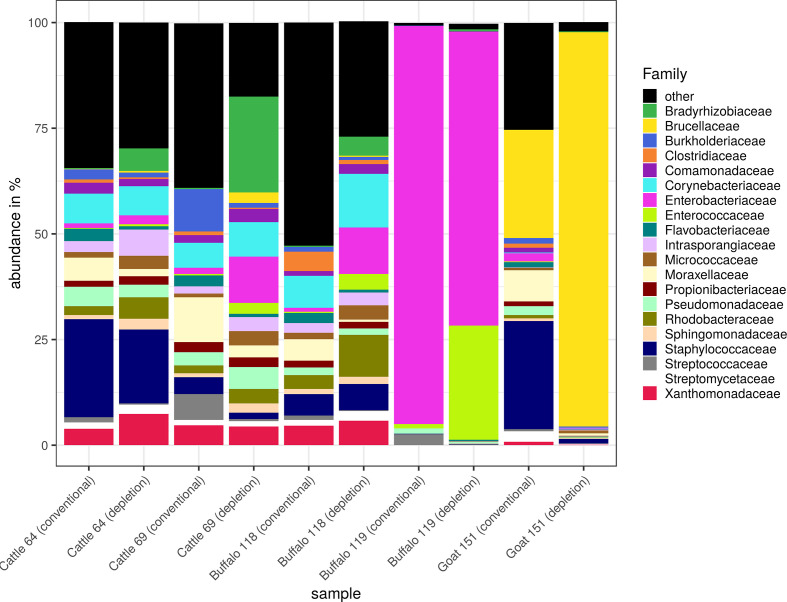
We also analysed raw milk samples from different dairy animals for the most abundant bacterial families (Fig. 6). In general, the profiles found in sample numbers 64, 69 and 118 were quite similar. In these samples, the fractions of Bradyrhizobiaceae , Enterobacteriaceae , Rhodobacteraceae and Micrococcaceae were larger after DNA extraction with eukaryotic cell depletion than without, whereas the fractions of Flavobacteriaceae and Moraxellaceae were smaller. In sample number 119, the fraction of Enterococcaceae was strongly increased after depletion of eukaryotic cells, and the fraction of Streptococcaceae was slightly decreased. In general, we observed a smaller fraction of low abundance families after eukaryotic cell depletion together with decreased species richness (data not shown). In sample number 151, the fraction of Staphylococcaceae and Moraxellaceae was strongly decreased, while the fraction of Brucellaceae was enriched after eukaryotic cell depletion. Furthermore, we observed a high prevalence of Enterobacteriaceae in sample number 119, usually present in the gut of mammals and, therefore, indicating a faecal contamination of the milk.
Fig. 6.
Taxonomic composition of raw milk samples from different dairy animals. The prokaryotic composition in raw milk after conventional DNA extraction with DNeasy mericon food kit and after eukaryotic cell depletion and DNA extraction with HostZERO kit was determined using KrakenUniq. Relative abundances (%) of bacterial families in different milk samples are presented as stacked bar graphs, with both DNA extraction kits next to each other for comparison.
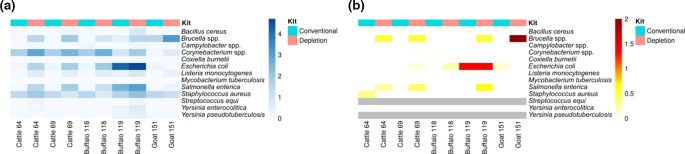
Additionally, we looked for hazardous bacteria [40] in the milk samples from Egypt that can be transmitted through the consumption of raw milk and products thereof using KrakenUniq. Besides Brucella spp., other potentially pathogenic species such as E. coli , Salmonella enterica and Staphylococcus aureus were detected in almost every sample (Fig. 7a). A few samples contained Corynebacterium spp., Yersinia spp., Listeria monocytogenes and Bacillus cereus in low abundance. For many of the pathogenic species (namely Bacillus cereus , Brucella spp., Corynebacterium spp., E. coli , Salmonella enterica , Staphylococcus aureus and Yersinia spp.), we observed an improved detection when eukaryotic cell depletion was applied prior to DNA extraction, showing the applicability of the method not only to Gram-negative Brucella spp., but also to Gram-positive bacteria such as Bacillus and Staphylococcus .
Fig. 7.
Hazardous bacterial species and their virulence factors in raw milk samples from different dairy animals. Detection of potentially hazardous bacterial species and their virulence factors in raw milk after conventional DNA extraction and after DNA extraction with eukaryotic cell depletion. (a) Classified species-specific reads per million total reads depicted in a heatmap. (b) Percentage of virulence factors detected in a species in relation to the total number of known virulence factors in the respective species according to VFDB was calculated. Grey colour indicates the absence of virulence factors in these species in the VFDB. To improve data presentation in the heatmaps, values were transformed by 1+log10.
Although we detected many potentially pathogenic bacterial species, we only identified virulence factors of E. coli , Brucella spp., Salmonella enterica and Staphylococcus aureus (Fig. 7b, Table S2). A reason for that might be the low genome coverage for most species. In sample number 119, E. coli was highly abundant, and was found together with typical indicators of faecal contamination such as Enterococcus faecalis and Enterococcus faecium . Seventy E. coli -specific virulence factors could be identified in this sample. No virulence genes typical for enteropathogenic E. coli encoding Shiga toxins, heat-stable or heat-labile enterotoxins, or other pathogenicity factors such as intimin (eae) and invasin plasmid antigen (ipaH), were detected. However, six type III secretion effectors were found, which are encoded on the pathogenicity island locus of enterocyte effacement (LEE), mediating the formation of attaching and effacing lesions in the intestinal epithelium.
In summary, there are differences in the composition of bacterial families depending on the DNA extraction kit applied. If eukaryotic cells were depleted, the detection of several pathogenic bacterial species could be improved.
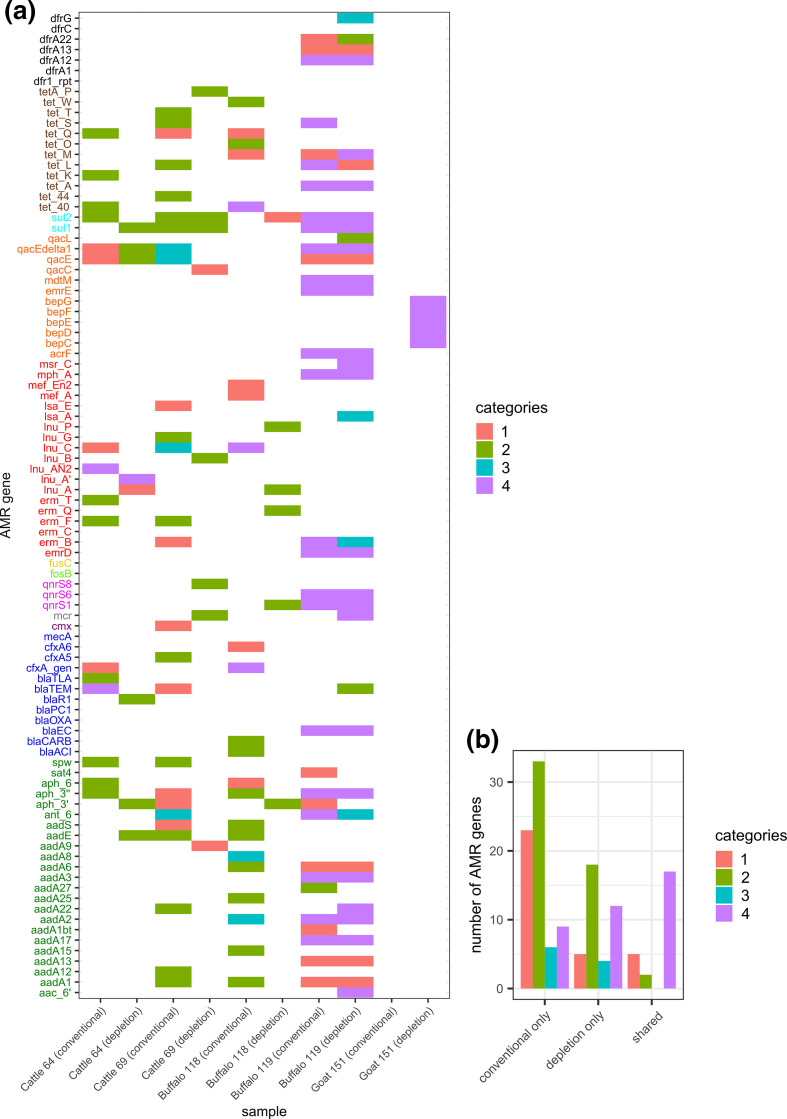
Detection of AMR genes in raw milk
The screening of metagenomic datasets for AMR genes revealed 94 genes with a coverage >30 %, distributed across twelve different antimicrobial classes (Fig. 8a). Several of these genes confer resistance to last-resort antibiotics. The detected AMR genes were categorized by certainty of evidence considering gene coverage and coverage depth. In general, DNA extraction methods again affected results. The depletion of eukaryotic cells prior to DNA extraction by differential lysis resulted in lower retrieval rates of AMR genes (Fig. 8b), especially in case of a lower certainty of evidence. Across all samples, only 38 genes revealed high certainty (coverage >90). Seventeen out of these were detected independent of the DNA extraction method applied. The detection of 12 genes could be improved by eukaryotic cell depletion. The highest prevalence of AMR genes (n=17) belonging to category 4 [high evidence (coverage >90)] were found in sample number 119, which was from a buffalo. This sample also contained a high proportion of Enterobacteriaceae (>60 %) that might be the reservoir of verified AMR genes. Sample number 151, from a goat, contained several components of the multidrug efflux RND (Resistance–Nodulation–Division) transporter, which is assumed to confer resistance to various antibiotics such as tetracycline, doxycycline and fluoroquinolones [41]. These genes can be also assigned to the Brucella genome, and were found in the isolates obtained from the milk sample (data not shown).
Fig. 8.
Presence of antibiotic resistance genes in raw milk. WMS data of raw milk samples were screened for AMR genes after conventional DNA extraction with a DNeasy mericon food kit or after eukaryotic cell depletion and DNA extraction with a HostZERO kit. The probability of gene presence was categorized into four classes according to coverage and depth: category 1, very low evidence – truncated gene (coverage <90, depth >1.5); category 2, low evidence – very low abundance (coverage <60, depth <1.5); category 3, medium evidence – low abundance (coverage >60, depth <1.5); category 4, high evidence (coverage >90). (a) Colours of gene names refer to AMR classes: aminoglycoside (green), β-lactam (dark blue), chloramphenicol (violet), colistin (grey), fluoroquinolone (pink), fosfomycin (light green), fusidic acid (yellow), macrolide (red), multidrug resistance (orange), sulfonamide (cyan), tetracycline (brown), trimethoprim (black). (b) Number of AMR genes exclusively detected after conventional DNA extraction (conventional only) and after eukaryotic cell depletion (depletion only) or by both DNA extraction methods (shared).
Discussion
Our study demonstrates that metagenomic sequencing can be used for direct pathogen detection and characterization in raw milk without pre-enrichment culture. For a proof of concept, we chose Brucella as the ideal model pathogen because it is naturally shed in milk of infected ruminants, is difficult to isolate from complex samples due to its fastidious growth, exhibits a high homology among its species and has a low infectious dose, which is why it is considered as highly pathogenic. In our study, we determined the detection limits of metagenomic sequencing and qPCR to correctly identify Brucella in artificially contaminated raw milk. Metagenomic sequencing of DNA after eukaryotic cell depletion decreased the detection limit for Brucella in raw milk to <20 c.f.u. ml−1. Hence, metagenomics showed a higher sensitivity than the gold standard in molecular diagnostics of brucellosis, which is a genus-specific qPCR after conventional DNA extraction (>1000 cells ml−1). Our results are comparable to previously reported detection limits of metagenomics for E. coli in spinach with 10 c.f.u. g−1 after 5 h of enrichment [7]. Eukaryotic cell depletion also performed quite well in naturally contaminated raw milk, although most brucellae are known to reside in milk macrophages [42]. Furthermore, ethanol pre-treatment of the milk to inactivate pathogens before DNA extraction did not impair the performance of eukaryotic cell depletion. In a recent metabarcoding study, different kits used for the depletion of host signals in infected human tissue samples were compared, and bacterial DNA could be enriched more than 10-fold by the HostZERO kit [19]. After eukaryotic cell depletion with the same method, we proved a similar enrichment of Brucella -specific reads in raw milk samples, which makes this approach suitable for pathogen detection in animal food products.
Since Brucella genomes display a strong sequence homology among species, a monophyletic genus has been assumed [43, 44]. The close relationship of Brucella spp. makes it challenging to determine the species when only a few sequence reads are available. In our study, we compared two different bioinformatics approaches for species and strain prediction: Mash Screen and RefSNPer. At low read numbers (<8000 reads) Brucella species prediction is much more reliable with RefSNPer than with Mash Screen. However, RefSNPer also needs more than 1000 reads to reliably identify the species. Mash Screen was not able to predict Brucella strains, whereas RefSNPer enabled strain prediction but at least 2000 genus-specific reads were needed. Of course, these results might be different for other genera that exhibit less homology among species and strains. Since Brucella is shed in milk in very low numbers, strain prediction remains a challenge that can be successfully tackled by reducing matrix background through the depletion of eukaryotic cells, as demonstrated in our study.
Metagenomic sequencing of sample number 151 resulted in >100 000 Brucella -specific reads after eukaryotic cell depletion. In this way, we could define the closest species with a similarity of >99 % using RefSNPer. Strain prediction with RefSNPer was even feasible with fewer reads (n=2787) without preceding eukaryotic cell depletion, in contrast to Mash Screen, which actually predicted the wrong species. We did not find any indications of a second Brucella strain in the milk sample after bioinformatics analysis with ConFindr [45] and sigma [46] (data not shown). Since RefSNPer might be unsuitable for distinguishing strains of the same species or genus in a mixed sample, we propose read extraction, for instance, with sparse and sigma as recently reported [47], before applying RefSNPer. We also tested two different assembly strategies for metagenomic sequence reads. Brucella -specific read extraction after Kraken2 classification, as previously described [5], and assembly with SPAdes yielded fewer contigs and a higher recovery of the genome fraction than metagenomic assembly by megahit. In a cgMLST analysis, the assembly after genus-specific read extraction (pre-assembly binning method) clustered closer to the isolates from the same milk sample than the metagenomic assembly of complete WMS data. When the megahit assembler was used for all data, the isolates clustered together with the metagenomic assembly and with the assembly of Brucella -specific reads. The dependence of cgMLST results on the assembler has been described previously [48]. The cgMLST analysis, which was performed with all B. melitensis assemblies available from the NCBI at that time, revealed four closely related strains, of which three also originated from Egypt. Since no further metadata of samples and strains were available, an epidemiological context could not be clarified. In the cgMLST analysis, the isolates in our study were more closely related to four publicly available genomes than to the WMS data obtained from the same milk sample. In contrast, SNP analysis revealed a closer association between isolates and WMS data, and is, therefore, more robust when WMS datasets shall be included. The metagenome was located in the same clade as the isolates, although WGS and WMS data differed in 11 nucleotide positions. Nevertheless, isolate sequences and WMS data can be clearly matched due to the majority of overlapping SNPs. SNP differences between isolates and metagenome might either be a consequence of insufficient sequence coverage at respective positions or due to multiple alleles in the sample. The latter is supported by the fact that the three isolates from the same milk sample differed at three nucleotide positions when directly compared to each other. Briefly, the striking resemblance of genomic and metagenomic data indicates the applicability of WMS in outbreak investigations.
In consistency with Lim and colleagues [13], we noticed differences in the composition of the milk microbiome depending on the DNA extraction method applied. These differences, however, were not statistically significant. Nevertheless, some families such as Staphylococcaceae and Moraxellaceae were underrepresented after eukaryotic cell depletion. This phenomenon was also observed in the Egyptian raw milk samples. Since Staphylococcaceae are Gram-positive bacteria and depletion of eukaryotic cells is in fact based on the selective lysis of mammalian cells due to specific membrane properties, we doubt that bacterial cells were lysed together with mammalian cells. In addition, insufficient lysis of Gram-positive bacteria can probably be ruled out by the slightly improved detection of Corynebacteriaceae after eukaryotic cell depletion. It is conceivable that the detection of Staphylococcaceae by metagenomics after conventional DNA extraction was based on the detection of soluble DNA, which was not detected when eukaryotic cell depletion was applied, because the selective lysis step also eliminates extracellular DNA. The presence of extracellular DNA is well known for members of the family Staphylococcaceae during biofilm formation. In addition, the depletion of extracellular DNA that distorts the actual microbial composition might be a desired effect if the aim is for only intact bacteria to be detected. The absence of several low-abundance families in naturally contaminated raw milk samples after eukaryotic cell depletion might also be a consequence of the methodological loss of extracellular DNA. By analysing negative controls (data not shown), we noticed that most of these differences in the abundance of Bradyrhizobiaceae , Burkholderiaceae , Rhizobiaceae , Corynebacteriaceae and Pseudomonadaceae originated from DNA contaminations within extraction kits known as the kitome [49].
Various AMR genes could be identified in the raw milk samples. Most of these genes showed a coverage of <90 %, which might emerge from low abundance, from partial similarities to other genes or from truncation. We observed more partial genes after conventional DNA extraction. Interestingly, there were also some high-certainty AMR genes (n=9), which were detected only after conventional DNA extraction. As mentioned above, this might be a consequence of the depletion of extracellular DNA along with eukaryotic DNA.
Source attribution studies applying metagenomics in foodstuffs are still rare. Most of these studies reported problems with the detection of low-abundance pathogens due to a high matrix background. While enrichment culture takes time and is not always a good alternative for fastidious pathogens, we could show that selective lysis of mammalian cells improved the detection of various pathogens in the complex milk matrix, including Brucella spp., Salmonella enterica and E. coli . In addition, our study has provided the proof-of-concept that metagenomics is a highly sensitive tool for microbial risk assessment of food, particularly when pathogens are difficult to isolate. In this way, we provide a simple approach for the detection and characterization of pathogenic bacteria in milk without pre-enrichment. Since purchasable kits and open-source software were used, our tool can be easily applied to other scientific questions and also to routine food microbiology.
Supplementary Data
Funding information
This study was conducted within the framework of the Ess-B.A.R. project funded by the German Federal Ministry of Education and Research (FKZ 13N13982). M. G. was funded by the Science and Technology Development Fund (Egypt) (grant no. 25425).
Acknowledgements
We thank Anne Stephan and Beatrice Baumann for excellent technical assistance.
Author contributions
The study was conceptualized by J. G., C. D., H. B., D. H., B. M. and S. A. D. Laboratory work was performed by J. G., A. S., M. G. and M. P. Dairy animals were clinically examined and milk was sampled by M. G. and M. E.-A. Data were analysed and visualized by J. G. and C. D. Bioinformatics software was developed by C. D. The manuscript was written by J. G., and reviewed and edited by S. A. D., B. M., C. D., M. P., H. B., M. G., M. E.-A., D. H. and A. S.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Samples were collected according to Directive 2010/63/EU on the protection of animals used for scientific purposes. The study design only included non-invasive procedures. The farmers were informed about the purpose of the study and participated on a voluntary basis. The use of genetic resources was compliant with the Nagoya Protocol.
Footnotes
Abbreviations: AMR, antimicrobial resistance; cgMLST, core-genome multilocus sequence typing; NA, nucleic acid; NCBI, National Center for Biotechnology Information; qPCR, quantitative PCR; VFDB, Virulence Factor Database; WGS, whole-genome sequencing; WMS, whole-metagenome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary figures and two supplementary tables are available with the online version of this article.
References
- 1.Corbel M J. Brucellosis in Humans and Animals. Geneva: World Health Organization; 2006. [Google Scholar]
- 2.Capparelli R, Parlato M, Iannaccone M, Roperto S, Marabelli R, et al. Heterogeneous shedding of Brucella abortus in milk and its effect on the control of animal brucellosis. J Appl Microbiol. 2009;106:2041–2047. doi: 10.1111/j.1365-2672.2009.04177.x. [DOI] [PubMed] [Google Scholar]
- 3.Jansen W, Linard C, Noll M, Nöckler K, Al Dahouk S. Brucella-positive raw milk cheese sold on the inner European market: a public health threat due to illegal import? Food Control. 2019;100:130–137. doi: 10.1016/j.foodcont.2019.01.022. [DOI] [Google Scholar]
- 4.Cocolin L, Mataragas M, Bourdichon F, Doulgeraki A, Pilet M-F, et al. Next generation microbiological risk assessment meta-omics: the next need for integration. Int J Food Microbiol. 2018;287:10–17. doi: 10.1016/j.ijfoodmicro.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Grützke J, Malorny B, Hammerl JA, Busch A, Tausch SH, et al. Fishing in the soup – pathogen detection in food safety using metabarcoding and metagenomic sequencing. Front Microbiol. 2019;10:1805. doi: 10.3389/fmicb.2019.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EFSA Panel on Biological Hazards (EFSA BIOHAZ Panel) Koutsoumanis K, Allende A, Alvarez-Ordóñez A, Bolton D, et al. Whole genome sequencing and metagenomics for outbreak investigation, source attribution and risk assessment of food-borne microorganisms. EFSA J. 2019;17:e05898. doi: 10.2903/j.efsa.2019.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard SR, Mammel MK, Lacher DW, Elkins CA. Application of metagenomic sequencing to food safety: detection of Shiga toxin-producing Escherichia coli on fresh bagged spinach. Appl Environ Microbiol. 2015;81:8183–8191. doi: 10.1128/AEM.02601-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartsch C, Hoper D, Made D, Johne R. Analysis of frozen strawberries involved in a large norovirus gastroenteritis outbreak using next generation sequencing and digital PCR. Food Microbiol. 2018;76:390–395. doi: 10.1016/j.fm.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 9.Ottesen AR, Gonzalez A, Bell R, Arce C, Rideout S, et al. Co-enriching microflora associated with culture based methods to detect Salmonella from tomato phyllosphere. PLoS One. 2013;8:e73079. doi: 10.1371/journal.pone.0073079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis KG, White JR, Grim CJ, Ewing L, Ottesen AR, et al. Cilantro microbiome before and after nonselective pre-enrichment for Salmonella using 16S rRNA and metagenomic sequencing. BMC Microbiol. 2015;15:160. doi: 10.1186/s12866-015-0497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feehery GR, Yigit E, Oyola SO, Langhorst BW, Schmidt VT, et al. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLoS One. 2013;8:e76096. doi: 10.1371/journal.pone.0076096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macher J-N, Speksnijder A, Choo LQ, van der Hoorn B, Renema W. Uncovering bacterial and functional diversity in macroinvertebrate mitochondrial-metagenomic datasets by differential centrifugation. Sci Rep. 2019;9:10257. doi: 10.1038/s41598-019-46717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim YW, Haynes M, Furlan M, Robertson CE, Harris JK, et al. Purifying the impure: sequencing metagenomes and metatranscriptomes from complex animal-associated samples. J Vis Exp. 2014;94:52117. doi: 10.3791/52117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thoendel M, Jeraldo PR, Greenwood-Quaintance KE, Yao JZ, Chia N, et al. Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. J Microbiol Methods. 2016;127:141–145. doi: 10.1016/j.mimet.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasan MR, Rawat A, Tang P, Jithesh PV, Thomas E, et al. Depletion of human DNA in spiked clinical specimens for improvement of sensitivity of pathogen detection by next-generation sequencing. J Clin Microbiol. 2016;54:919–927. doi: 10.1128/JCM.03050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marotz CA, Sanders JG, Zuniga C, Zaramela LS, Knight R, et al. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome. 2018;6:42. doi: 10.1186/s40168-018-0426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson MT, Pope CE, Marsh RL, Wolter DJ, Weiss EJ, et al. Human and extracellular DNA depletion for metagenomic analysis of complex clinical infection samples yields optimized viable microbiome profiles. Cell Rep. 2019;26:2227–2240. doi: 10.1016/j.celrep.2019.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong W, Rockett R, Timms V, Sintchenko V. Optimization of sample preparation for culture-independent sequencing of Bordetella pertussis . Microb Genom. 2020;6:e000332. doi: 10.1099/mgen.0.000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heravi FS, Zakrzewski M, Vickery K, Hu H. Host DNA depletion efficiency of microbiome DNA enrichment methods in infected tissue samples. J Microbiol Methods. 2020;170:105856. doi: 10.1016/j.mimet.2020.105856. [DOI] [PubMed] [Google Scholar]
- 20.Farrell ID. The development of a new selective medium for the isolation of Brucella abortus from contaminated sources. Res Vet Sci. 1974;16:280–286. doi: 10.1016/S0034-5288(18)33726-3. [DOI] [PubMed] [Google Scholar]
- 21.Karger A, Melzer F, Timke M, Bettin B, Kostrzewa M, et al. Interlaboratory comparison of intact-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry results for identification and differentiation of Brucella spp. J Clin Microbiol. 2013;51:3123–3126. doi: 10.1128/JCM.01720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alton GG, Jones LM, Angus RD, Verger JM. Techniques for the Brucellosis Laboratory. Paris: Institut National de la Recherche Agronomique; 1988.. [DOI] [Google Scholar]
- 23.Al Dahouk S, Nöckler K, Scholz HC, Pfeffer M, Neubauer H, et al. Evaluation of genus-specific and species-specific real-time PCR assays for the identification of Brucella spp. Clin Chem Lab Med. 2007;45:1464–1470. doi: 10.1515/CCLM.2007.305. [DOI] [PubMed] [Google Scholar]
- 24.Cloeckaert A, Grayon M, Grepinet O. An IS711 element downstream of the bp26 gene is a specific marker of Brucella spp. isolated from marine mammals. Clin Diagn Lab Immunol. 2000;7:835–839. doi: 10.1128/CDLI.7.5.835-839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breitwieser FP, Baker DN, Salzberg SL. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018;19:198. doi: 10.1186/s13059-018-1568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ondov BD, Starrett GJ, Sappington A, Kostic A, Koren S, et al. Mash Screen: high-throughput sequence containment estimation for genome discovery. Genome Biol. 2019;20:232. doi: 10.1186/s13059-019-1841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 34.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva M, Machado MP, Silva DN, Rossi M, Moran-Gilad J, et al. chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb Genom. 2018;4:e000166. doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janowicz A, De Massis F, Ancora M, Cammà C, Patavino C, et al. Core genome multilocus sequence typing and single nucleotide polymorphism analysis in the epidemiology of Brucella melitensis infections. J Clin Microbiol. 2018;56:e00517-18. doi: 10.1128/JCM.00517-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.EFSA Panel on Biological Hazards (BIOHAZ) Scientific opinion on the public health risks related to the consumption of raw drinking milk. EFSA J. 2015;13:3940. doi: 10.2903/j.efsa.2015.3940. [DOI] [Google Scholar]
- 41.Martin FA, Posadas DM, Carrica MC, Cravero SL, O'Callaghan D, et al. Interplay between two RND systems mediating antimicrobial resistance in Brucella suis . J Bacteriol. 2009;191:2530–2540. doi: 10.1128/JB.01198-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harmon BG, Adams LG, Frey M. Survival of rough and smooth strains of Brucella abortus in bovine mammary gland macrophages. Am J Vet Res. 1988;49:1092–1097. [PubMed] [Google Scholar]
- 43.Al Dahouk S, Scholz HC, Tomaso H, Bahn P, Göllner C, et al. Differential phenotyping of Brucella species using a newly developed semi-automated metabolic system. BMC Microbiol. 2010;10:269. doi: 10.1186/1471-2180-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leclercq SO, Cloeckaert A, Zygmunt MS. Taxonomic organization of the family Brucellaceae based on a phylogenomic approach. Front Microbiol. 2019;10:3083. doi: 10.3389/fmicb.2019.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Low AJ, Koziol AG, Manninger PA, Blais B, Carrillo CD. ConFindr: rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ. 2019;7:e6995. doi: 10.7717/peerj.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn TH, Chai J, Pan C. Sigma: strain-level inference of genomes from metagenomic analysis for biosurveillance. Bioinformatics. 2015;31:170–177. doi: 10.1093/bioinformatics/btu641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saltykova A, Buytaers FE, Denayer S, Verhaegen B, Piérard D, et al. Strain-level metagenomic data analysis of enriched in vitro and in silico spiked food samples: paving the way towards a culture-free foodborne outbreak investigation using STEC as a case study. Int J Mol Sci. 2020;21:5688. doi: 10.3390/ijms21165688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uelze L, Grützke J, Borowiak M, Hammerl JA, Juraschek K, et al. Typing methods based on whole genome sequencing data. One Health Outlook. 2020;2:3. doi: 10.1186/s42522-020-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.