Abstract
The global rise of the coronavirus disease 2019 pandemic resulted in an exponentially increasing demand for severe acute respiratory syndrome coronavirus 2 testing, which resulted in shortage of reagents worldwide. This shortage has been further worsened by screening of asymptomatic populations such as returning employees, students, and so on, as part of plans to reopen the economy. To optimize the utilization of testing reagents and human resources, pool testing of populations with low prevalence has emerged as a promising strategy. Although pooling is an effective solution to reduce the number of reagents used for testing, the process of pooling samples together and tracking them throughout the entire workflow is challenging. To be effective, samples must be tracked into each pool, pool-tested and reported individually. In this article, we address these challenges using robotics and informatics.
Keywords: coronavirus disease 2019, medical informatics, pathology, severe acute respiratory syndrome coronavirus 2, clinical laboratory information systems, pool testing
Introduction
With the rise in coronavirus disease 2019 (COVID-19) cases, 1,2 the need for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) testing resources is constantly increasing. Due to this increase and sudden demand for testing, there is a worldwide shortage of testing resources and delays in almost every aspect of the supply chain for SARS-CoV-2 testing reagents and consumables. 3 There are also huge delays in resulting COVID-19 tests. 3 Testing delays can increase the spread of the infection in the population and delayed patient care. With a demand for COVID-19 screening in a large asymptomatic populations 4,5 and the need to conserve testing materials, a general strategy is to pool samples. 6 -14 This could include screening of collegiate and employee populations and hospital admissions such as presurgical, nonrespiratory, and trauma cases. This also results in decreased turnaround times.
For example, if 100 samples are pooled with a pooling size of 10, then 10 tests would have to be run instead of 100 tests. If the prevalence of the disease in the population is 1%, one of these pools would be positive. The 10 samples making up this positive pool would then be run individually to identify the positive sample. In this scenario, a total of 20 tests would be run instead of 100, resulting in a saving of 80% of testing materials. Even at a 5% prevalence in the population, this same pooling strategy would save a minimum of 40% of testing materials. When there is a low positivity rate, most samples will be negative and can be reported. Obviously, the higher the positivity rate, the less effective this strategy will be. However, effectiveness can be improved by reducing the pooling size for higher percent positivity. A pool size of 10 would be more efficient for a 1% prevalence rate, whereas a pool size of 5 would be more efficient for a 5% prevalence. Table 1 illustrates a calculation of the number of tests that would be saved depending on the prevalence rate and the size of the pools being created. This table illustrates the most conservative estimate, which can be used in the laboratory when there is a rapidly fluctuating positivity rate. Probabilistic models suggest that it may be possible to save even more tests overtime. 12 For validation of the assay, the largest pool size was validated considering smaller pools would be more sensitive. The limit of detection (LOD) for a pool size of 10 was 5 copies/mL, which is comparable to the LOD for the Centers for Disease Control and Prevention (CDC) assay.
Table 1.
Approximate Number of Tests Saved Per 100 Samples for Various Combination of Pooling Factors and Percent Positives.
| Tests saved per 100 samples | Percent positives | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1% | 2% | 3% | 4% | 5% | 6% | 7% | 8% | 9% | 10% | 15% | 20% | 30% | ||
| Pooling factor | 2 | 48.0 | 46.0 | 44.0 | 42.0 | 40.0 | 38.0 | 36.0 | 34.0 | 32.0 | 30.0 | 20.0 | 10.0 | 0.0 |
| 3 | 63.7 | 60.7 | 57.7 | 54.7 | 51.7 | 48.7 | 45.7 | 42.7 | 39.7 | 36.7 | 21.7 | 6.7 | 0.0 | |
| 4 | 71.0 | 67.0 | 63.0 | 59.0 | 55.0 | 51.0 | 47.0 | 43.0 | 39.0 | 35.0 | 15.0 | 0.0 | 0.0 | |
| 5 | 75.0 | 70.0 | 65.0 | 60.0 | 55.0 | 50.0 | 45.0 | 40.0 | 35.0 | 30.0 | 5.0 | 0.0 | 0.0 | |
| 6 | 77.3 | 71.3 | 65.3 | 59.3 | 53.3 | 47.3 | 41.3 | 35.3 | 29.3 | 23.3 | 0.0 | 0.0 | 0.0 | |
| 7 | 78.7 | 71.7 | 64.7 | 57.7 | 50.7 | 43.7 | 36.7 | 29.7 | 22.7 | 15.7 | 0.0 | 0.0 | 0.0 | |
| 8 | 79.5 | 71.5 | 63.5 | 55.5 | 47.5 | 39.5 | 31.5 | 23.5 | 15.5 | 7.5 | 0.0 | 0.0 | 0.0 | |
| 9 | 79.9 | 70.9 | 61.9 | 52.9 | 43.9 | 34.9 | 25.9 | 16.9 | 7.9 | 0.0 | 0.0 | 0.0 | 0.0 | |
| 10 | 80.0 | 70.0 | 60.0 | 50.0 | 40.0 | 30.0 | 20.0 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | |
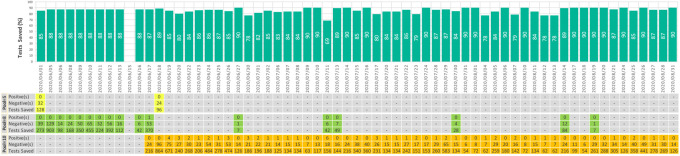
At our institution, about 2619 pooled tests were conducted during first 3 months (June-August 2020) with pool sizes 5, 8, and 10 (Figure 1). This practice resulted in testing of 22 659 specimens, which saved 19 215 (84.8%) tests that would otherwise have been needed. The pool testing strategy described in this article is now being implemented on students, faculty, and staff asymptomatic population screening only. Our expected turnaround time for pooled testing is 24 to 48 hours, with most results returned within 24 hours.
Figure 1.
Summary of total severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pool tests, their positive rates, and overall tests savings for the 3 months (June-August, 2020).
With traditional testing, the pathway from sample to result is straightforward. When pooling samples together, there are many workflow challenges to be addressed especially with large numbers of samples. In this article, we address 3 main challenges in pool testing—sample mapping and processing, storage tracking, and reporting.
Limitation of Native Laboratory Informatics Pooled Testing Workflows
To accomplish pool testing, there needs to be a way to track samples that are in a pool so that results can be returned for each sample tested. One way to do this is to manually track which samples are in which pool. This is time-consuming and prone to error. There are now several “out of the box” options available to handle this type of testing. For example, Epic Beaker laboratory information system provides a solution using an available workflow traditionally used for batch testing. With this method, users identify a batch of samples and then generate a new sample ID, or pool ID, for the batch. The samples in the batch are then pooled, and the assay is run under the new pool ID. When results are returned, if the pool is negative, all samples in that pool can be resulted at the same time.
One limitation to this method is that samples must be organized in advance so that individual samples are placed into the correct pool. Ideally, samples could be in any order and pooled together. The pool ID and individual sample IDs would then be scanned and mapped as the pooling occurs removing the need for up-front sample organization. This allows for samples to be pooled with minimal sample management. Our efforts to achieve this are described below.
Pooled Sample Mapping and Processing
We used robotic instrumentation, 15 to create pools, and to track sample information in a reliable manner to reduce sources of human error. By creating unique barcodes for the pool samples, a robot can correctly track pool identifiers. These barcodes can then be read by other instrumentation down the line. The testing results can be identified and linked to the pool sample and corresponding source samples. If a pool tests positive, the samples contained in the pool can not only be identified but also located for additional testing. Robotic tracking of each sample can efficiently locate each sample. This allows repeat testing of each sample to occur in a timely manner, both avoiding unnecessary increases in turnaround times and the potential for sample degradation. Programming of a robot also allows protocol changes as pooling size may need to change depending on changes in the frequency of positive cases in the population.
Our workflow was designed with specific instrumentation in mind, although it can be adapted to many platforms. The Hamilton Company NGS STAR system was chosen because it has the ability to read barcodes, create sample maps, and is programmable to accommodate different pool sizes. Individual barcoded patient samples are placed onto the deck of the Hamilton Company NGS STAR. To utilize the Hologic Panther system, pools need to be created in tubes containing lysis buffer. Each of these pool lysis tubes are assigned with unique barcodes (pool IDs). For example, pool sizes of 10 for 100 samples requires 10 pool IDs. The pool lysis tubes are labeled and placed onto the deck of the Hamilton Company NGS STAR. When the sample transfer program on Hamilton Company NGS STAR is initiated, equal amounts of sample is transferred from the source sample tube (patient sample) to the target pool lysis tube. When the transfer program is complete, an excel file is generated. This file acts as a map indicating which samples were transferred into which pool tube, all identified by their unique barcodes. The pool lysis tubes are transferred to the Hologic Panther system and run per the manufactures instructions. When completed, the results identified by the unique pool barcode ID are exported from the Hologic Panther system as a .LIS file (which is a tab delimited text file). These results can then be paired with the map from the Hamilton Company NGS STAR system for analysis.
One limitation with pooling is the possibility that a sample could be missed during the transfer and then resulted as not detected when the sample was never included in the pool. The Hamilton Company NGS STAR system is capable of liquid sensing, clot detection, and aspiration error detection. This will reduce the number of missed samples but does not completely eliminate the issue. One way to mitigate this would be to manually transfer the samples. However, the manual process of transferring and manually keeping track of all samples would make it impractical to use our testing approach. In our initial deliberations on this topic, we determined that the benefit of being able to routinely screen a large number of the campus population outweighed the limitations of pooling (decreased sensitivity and the possibility of a failed aliquot).
Sample Storage Tracking
When performing pool testing, sample storage tracking is required. If a pool is positive, each of the samples need to be located that made up the positive pool so they can be tested individually. By utilizing the Epic Beaker Storage Container function, we tracked samples (via barcode scanning) to a specific rack type, rack number, and position. A rack is first created in Epic Beaker with a unique name (Figure 2A). Each sample barcode is scanned and placed into the racks corresponding location (Figure 2B). This location is immediately available in the tracking information for each individual sample (Figure 2C). Once a positive pool is identified, Epic Beaker tracking information for each individual source sample can be used to locate the rack and rack position. We found that such sample tracking is an easy and effective solution for efficiently locating a small percentage of a large sample set.
Figure 2.
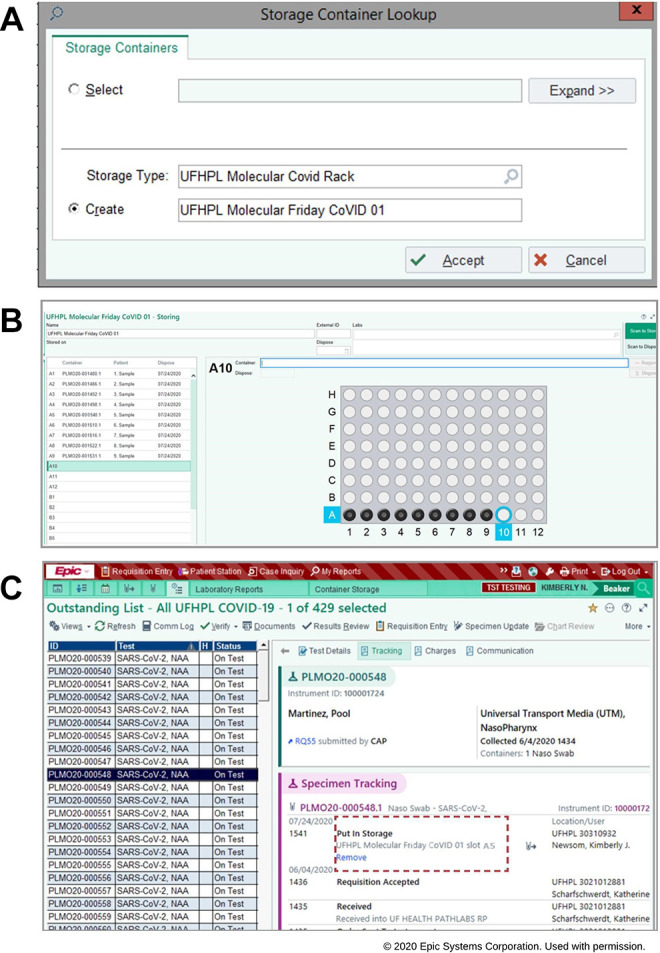
Sample storage tracking system in epic beaker (EPIC Systems Corporation) laboratory information system. (A) Creating the storage container tray. (B) Scanning the sample barcodes and placing it with corresponding slot in the storage tray. (C) Storage location is displayed under specimen tracing logs for the specimens.
Custom Middleware Solution for Reporting Pooled Testing Results
When a pool sample is negative, all samples in that pool should be resulted as negative. If a pool is positive, all the samples comprising that pool are located via storage container tracking and are retested individually.
Custom middleware was designed to link the sample pooling map from the Hamilton Company NGS STAR system with the results exported from the Hologic Panther system. In short, the middleware automates the interpretation of pool results to either result the negative samples or identify the component samples of the positive pools. Testing results are interfaced with Epic Beaker to the NextGen Connect Integration Engine (Formerly MirthConnect), which pushes the results into Epic Beaker. The custom middleware code repository is publicly available for download at https://bitbucket.org/srikarchamala/covid_ 19 _pool_testing_middleware. The repository contains a “README.md” file explaining the software dependencies, computational environment prerequisites, steps to run the software, and sample input/output files.
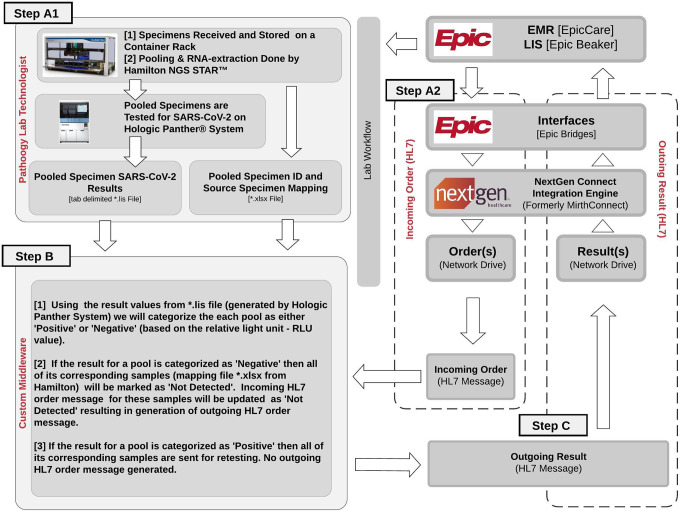
As shown in Figure 3, when a COVID-19 test is ordered and a specimen is collected, 2 events will happen simultaneously. One of them is that test order shows up on the pathology lab worklist in Epic Beaker laboratory information system (Figure 3, Step A1). We configured our electronic interface software Epic Bridges and NextGen Connect Integration Engine for simultaneously sending an order health level 7 (HL7) message to a network folder (Figure 3, Step A2). This HL7 file contains information related to the patient, test order, specimen ID, and empty test result value field. See example outgoing HL7 files in “Input_Files/Incoming_HL7” folder at the above code repository website.
Figure 3.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pool testing workflow. When a SARS-CoV-2 test order is placed by the health care provider, the order shows up on the molecular pathology work list (step A1) and a health level 7 international (HL7) message is generated (step A2) simultaneously. In step A1, specimens are pooled and processed through the SARS-CoV-2 pool assay. The outputs from step 1A and step 1B are processed by our custom middleware (step B), generating HL7 test result messages for each specimen, which are reported back to epic beaker (step C).
The samples are processed and resulted following the standard laboratory workflow using Hamilton Company NGS STAR and Hologic Panther systems (Figure 3, Step A1). Custom middleware (Figure 3, Step B) was designed to link the sample pooling map from the Hamilton Company NGS STAR system with the results exported from the Hologic Panther system and automatically resulting test values to Epic Beaker.
After reading the pool results file (tab delimited file) from the Hologic Panther system, we categorize the pool result as “Negative” if the following criteria are met (eg, “Input_Files/Panther_POOL_RESULTS.lis.xls” in the above code repository website)—column “Interpretation 1” is less than 350 (its relative light unit—RLU Score) AND column “Interpretation 2” is “valid” AND column “Interpretation 3” is “Negative.” Else, the pool result is categorized as “Positive.” The Hologic Panther system output file contains only the Pool ID and their corresponding results, and not the original Sample IDs. The next step retrieves all the original Sample IDs associated with each pool by linking “Specimen Barcode” column (which is a pool sample ID) of the Hologic Panther system output file with “Pooled Sample Barcode” column in the Hamilton Company NGS STAR output file (tab delimited file—eg, “Input_Files/Hamilton_SAMPLE_POOL_MAP.xlsx”). The samples associated with “Positive” pools are next retested individually. Each sample in the “Negative” pool is categorized as “Negative” and reported back to Epic Beaker. This is done by detecting the corresponding incoming HL7 order message file based on the sample ID, and inserting a result value (as “Not Detected” to OBX record) into the outgoing HL7 message. The outgoing HL7 message is appended with the test protocol description to the NTE record that appears in the comment box in Epic Beaker. This outgoing HL7 message is placed on the network drive (eg, outgoing HL7 files in “Output_Files/Result_HL7” folder at the above code repository website).
Samples corresponding to “Positive” pools (Figure 3, Step B) do not generate any outgoing HL7 files. Thus, the results for these samples are reported to Epic Beaker to be retested individually because at least one sample in that pool is positive. Outgoing HL7 messages generated by “Negative” samples are automatically picked up by NextGen Connect Integration Engine and pushed into Epic Beaker.
On average, we can result 4 samples per minute when processed manually. In this article, we reported testing of 22 659 specimens that resulted in time saving of about 94 hours. Another critical outcome of our informatics automation step is prevention of manual entry errors. Manual entry error estimation analysis of pathology data fields 16 yielded error rates ranging from 0.5% to 6.4%. This means that with our automation step, we potentially avoided COVID-19 test resulting errors in at least 113 (0.5%) of 22 659 tests.
Validation, Security, and Data Retention
Before deploying this informatics workflow in the production environment, our molecular pathology technologists and pathology informatics team created in silico cases for vigorous testing of the informatics pipeline using the development environment (including Epic Beaker). Following in silico validation, pools with known positive and known negative samples were created and were tested through our computational pipeline in the development environment. Additionally, when these custom clinical informatics pipelines were used, security concerns were taken into consideration. The custom middleware was deployed in our institutional approved Health Insurance Portability and Accountability Act (HIPPA) compliant computation infrastructure. Also, start to end of the informatics workflow was independently evaluated and approved by informational technology risk assessment team at our institution. All of our pool testing information was retained both as physical paper copies and in excel files on our institutional HIPPA complaint storage facility with regular storage backup.
Conclusions
We report robotic implementation and informatics solutions for pooling, resulting, subsequent sample mapping, and storage tracking. We designed this approach for meeting the unique challenges of COVID-19 testing, but it is generalizable to other types of testing. Also, although our strategy was designed for use with Hamilton Company NGS STAR, Hologic Panther, and Epic Beaker, it can easily be adapted for use with other platforms at other institutions. The use of robotics and tailored pathology informatics solutions can help conserve, reduce turnaround times, and remove sources of error.
Acknowledgments
The authors thank Ashley Chandler, Dawn Blood, and other members from the UF Health enterprise IT services for their contributions to some of the informatics components detailed in this paper. The authors are grateful for Hesamedin Hakimjavadi for providing assistance in data visualization and for UF Health Pathology members and leadership for supporting pooled testing work.
Authors’ Note: Balaji Balasubramani and Kimberly J. Newsom contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Srikar Chamala  https://orcid.org/0000-0001-6367-7615
https://orcid.org/0000-0001-6367-7615
References
- 1. Secon H, Woodward A, Mosher D. A comprehensive timeline of the new coronavirus pandemic, from China’s first COVID-19 case to the present. 2020. Accessed May 10, 2020. https://www.businessinsider.com/coronavirus-pandemic-timeline-history-major-events-2020-3
- 2. Florida Department of Health. Florida’s COVID-19 data and surveillance dashboard. 2020. Accessed March 29, 2020. https://experience.arcgis.com/experience/96dd742462124fa0b38ddedb9b25e429
- 3. Herper M, Branswell H. Shortage of crucial chemicals creates new obstacle to U.S. coronavirus testing. STAT. 2020. Accessed March 29, 2020. https://www.statnews.com/2020/03/10/shortage-crucial-chemicals-us-coronavirus-testing/
- 4. Ferenczi BA, Baliga C, Akl P, et al. Pre-procedural Covid-19 screening of asymptomatic patients: a model for protecting patients, community and staff during expansion of surgical care. Nejm Catal Innov Care Deliv. 2020. doi:10.1056/CAT.20.0261 [Google Scholar]
- 5. Krouse S. To get back to work, companies seek coronavirus tests for workers. 2020. Accessed May 10, 2020.https://www.wsj.com/articles/to-get-back-to-work-companies-seek-coronavirus-tests-for-workers-11587375003
- 6. Sunjaya AF, Sunjaya AP. Pooled testing for expanding COVID-19 mass surveillance. Disaster Med Public Health Prep. 2020;14:e42–e43. doi:10.1017/dmp.2020.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alcoba-Florez J, Gil-Campesino H, de Artola DG-M, et al. Increasing SARS-CoV-2 RT-qPCR testing capacity by sample pooling. Int J Infect Dis. 2020;103:19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borillo GA, Kagan RM, Baumann RE, et al. Pooling of upper respiratory specimens using a SARS-CoV-2 real-time RT-PCR assay authorized for emergency use in low-prevalence populations for high-throughput testing. Open Forum Infect Dis. 2020;7:ofaa466. doi:10.1093/ofid/ofaa466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denny TN, Andrews L, Bonsignori M, et al. Implementation of a pooled surveillance testing program for asymptomatic SARS-CoV-2 infections on a college campus—Duke University, Durham, North Carolina, August 2–October 11, 2020. Morb Mortal Wkly Rep. 2020;69:1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perchetti GA, Sullivan K-W, Pepper G, et al. Pooling of SARS-CoV-2 samples to increase molecular testing throughput. J Clin Virol. 2020;131:104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shental N, Levy S, Wuvshet V, et al. Efficient high-throughput SARS-CoV-2 testing to detect asymptomatic carriers. Sci Adv. 2020;6:eabc5961. doi:10.1126/sciadv.abc5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bilder CR, Iwen PC, Abdalhamid B. Pool size selection when testing for severe acute respiratory syndrome coronavirus 2. Clin Infect Dis. 2020;72:1104–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abdalhamid B, Bilder CR, Garrett JL, Iwen PC. Cost effectiveness of sample pooling to test for SARS-CoV-2. J Infect Dev Ctries. 2020;14:1136–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abdalhamid B, Bilder CR, McCutchen EL, Hinrichs SH, Koepsell SA, Iwen PC. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol. 2020;153:715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cresswell K, Ramalingam S, Sheikh A. Can robots improve testing capacity for SARS-CoV-2? J Med Internet Res. 2020;22:e20169. doi:10.2196/20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hong MKH, Yao HHI, Pedersen JS, et al. Error rates in a clinical data repository: lessons from the transition to electronic data transfer—a descriptive study. BMJ Open. 2013;3:e002406. [DOI] [PMC free article] [PubMed] [Google Scholar]