Abstract
The broad-spectrum herbicide, glyphosate, is considered safe for animals because it selectively affects the shikimate pathway that is specific to plants and microorganisms. We sought a previously unknown mechanism to explain the concerns that glyphosate exposure can negatively affect animals, including humans. Computer modeling showed a probable interaction between glyphosate and eukaryotic translation elongation factor 1 subunit alpha 1 (eEF1α1), which was confirmed by microcalorimetry. Only restricted, nondisrupted spermatogenesis in rats was observed after chronic glyphosate treatments (0.7 and 7 mg/L). Cytostatic and antiproliferative effects of glyphosate in GC-1 and SUP-B15 cells were indicated. Meta-analysis of public health data suggested a possible effect of glyphosate use on sperm count. The in silico, in vitro, and in vivo experimental results as well as the metastatistics indicate side effects of chronic glyphosate exposure. Together, these findings indicate that glyphosate delays protein synthesis through an interaction with eEF1α1, thereby suppressing spermatogenesis and cell growth.
1. Introduction
Glyphosate, N-(phosphonomethyl)glycine, is the most frequently used herbicide globally, and its increasing popularity for agricultural and nonagricultural use has been documented.1 The glyphosate active substance acts as a broad-spectrum nonselective herbicide that specifically and exclusively inhibits 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS). EPSPS, which is involved in the shikimate pathway, is present in plants and microorganisms but not in animals; thus, glyphosate has been considered to be safe for animals.2 However, in the last decade, concerns that glyphosate can impact animals, including humans, as nontargeted organisms have increased.3 There is a controversy on whether glyphosate is a carcinogen to humans. In March 2015, the World Health Organization International Agency for Research on Cancer (IARC) classified glyphosate as a probable group 2A carcinogen to humans, but this classification was based on limited evidence of cancer in humans.4 In 2017, the European Chemicals Agency (ECHA) Committee for Risk Assessment (RAC)5 and the United States Environmental Protection Agency (EPA)6 claimed that glyphosate is not likely to be carcinogenic. Overall, authorities principally involved in the regulation of pesticides have conducted an intensified assessment of glyphosate safety, but the different outputs have resulted in a controversy.7−9 Indeed, additional research is required to ascertain whether previously unknown mechanisms exist in traditional toxicological studies.10 An indirect adverse effect of glyphosate may be involved in affecting the gut microbiome, which contains EPSPS, and the possibility of dysbiosis.11,12 It is also possible that glyphosate exposure may affect the biochemistry of nontargets in animals and humans by unexpected side mechanisms through interactions with molecules derived from their own genome.
Some studies have hypothesized that glyphosate affects amino acid metabolism/protein synthesis in mammals, but there is disunity in studies regarding the supposed cancer effect. It is important to consider not only glyphosate as the parent compound but also its relevant metabolites. In animals and humans, similar to the case in microorganisms, glyphosate is metabolized not only to aminomethylphosphonic acid (AMPA) but also to other metabolites, such as glyoxylate.13 Although Ford et al.14 showed that glyphosate is metabolized in mouse liver to glyoxylate, they used high doses (200 mg/kg) that were intraperitoneally administered once a day for 7 days. Thus, the reactive metabolite may affect cysteines in proteins and suppress fatty acid oxidation only at an excessive exposure.14 Further, it has been suggested that glyphosate is a substitute for glycine in protein polypeptide chains,15 but another study has negated this assumption.16 Molecular modeling has shown that glyphosate binding to glycyl-tRNA synthetase is unlikely,16 but the same study did not indicate any significant changes in human breast cancer cells via proteomic analysis, which was similar to another previous study by Mesnage et al.,17 who did not observe changes in MDA-MB-231 cell growth characteristics after treatment with 100 mg/L glyphosate.16 However, Mesnage et al.17 observed that ≥10 mg/L glyphosate treatment promotes proliferation in MCF-7 cells because these cells are estrogen receptor (ER)-positive. Importantly, the MDA-MB-231 cell line used by Antoniou et al.16 is ER-negative (triple negative) and insensitive to antiestrogen treatments.18,19 Furthermore, a previous study hypothesized that glyphosate and AMPA, as glycine analogues, inhibit serine hydroxymethyltransferase (SHMT), which catalyzes serine to glycine and vice versa.20 Indeed, Li et al.20 provided evidence that glyphosate and AMPA inhibit proliferation and promote apoptosis in certain cancer cell lines but not in two immortalized normal cell lines, and they indicated that hormone sensitivity of the cells is the likely factor for this phenomenon.20 The link between the hormone/estrogen sensitivity of certain cancer cell lines toward glyphosate/AMPA treatment may be the involved mechanism.17,20
Studies have indicated that glyphosate and AMPA affect the cell cycle in certain cell lines. Li et al.20 indicated that the effect is cell cycle-dependent, suggesting that glyphosate/AMPA are more effective against rapidly proliferating cells. In addition, the glyphosate metabolite, AMPA, has been shown to arrest cancer cells at the G1/G0 phase.20 Lin et al.21 observed cell cycle-specific eukaryotic translation elongation factor 1 subunit alpha 1 (eEF1α1) expression in breast carcinomas, and they demonstrated that eEF1α1 mRNA levels are high in G1 and low in proliferating cells. However, the eEF1α1 transcript level is underexpressed, while the eEF1α1 protein level is overexpressed in ductal breast carcinomas, including ER-positive tumors. These researchers also suggested a link between estrogen signaling and the eEF1α1 mRNA level because estrogen promotes proliferation.21 Moreover, depletion of eEF1α1 impairs cell vitality and cell growth but arrests cells in the G1/G0 phase.22 Indeed, eEF1α1 influenced HCC cell proliferation via regulation of the G1 phase.23 Thus, there may be a potential link between glyphosate/AMPA treatment and impairment of the cell cycle, and the target may be eEF1α1.
We do not reject the hypothesis that glyphosate alters protein synthesis in nontargets.15 In agreement with Antoniou et al.,16 we did not consider that glyphosate binds the active site of aminoacyl-tRNA synthetase, which is responsible for the covalent amino acid linking to tRNA.24 Instead, we sought to investigate whether glyphosate treatment alters the different phases of protein synthesis. As a target for our study, we selected the stage in which the aminoacyl-tRNA is delivered to the ribosome, an event mediated by eEF1α,25−27 which is present in two isoforms, and both isoforms are oncogenes.28 Importantly, while eEF1α1 has been established to be proapoptotic, an inverse antiapoptotic effect has been suggested for eEF1α2.29 Another difference between the two isoforms is the different expression in tissues and their replacement during cellular differentiation, i.e., eEF1α1 expressed in embryonic and postnatal development is later replaced by eEF1α2 expressed in long-lasting terminally differentiated cells.25,30,31 Considering the observed misexpression of eEF1α1 in tumor cells21 and the proapoptotic effects of glyphosate and AMPA on cancer cell lines,20 we hypothesize that glyphosate and AMPA are associated with the proapoptotic eEF1α1 functions.29
Previous studies have reported that the contraceptive drug, gamendazole, interacts with eEF1α1.32,33 Thus, if glyphosate treatment suppresses eEF1α1 similarly to gamendazole,32,33 then an analogous role of glyphosate treatment in spermatogenesis may be suggested. Therefore, we additionally hypothesized that there may be a link between glyphosate use and the increased problems in human fertility and testicular cancer over the last few decades.34,35 Several studies have investigated the adverse effect of glyphosate on cancer and reproduction, but the results of the studies are controversial. Some of these studies used high nonrealistic doses and/or formulated glyphosate.10,36 However, a low glyphosate effect has been observed on rat male reproductive organs,37 and a meta-analysis has indicated a potential effect of glyphosate on sperm counts in rodents.38 Thus, it is necessary to further investigate the adverse potential of glyphosate, especially from chronic exposure.10 The realistic contents of glyphosate and AMPA in the environment can be considered up to hundreds of micrograms or a few milligrams parts per million (ppm). Incidentally, the U.S. Environmental Protection Agency has set a drinking water maximum contaminant level (MCL) of 0.7 mg/L for glyphosate.39,40
In this study, we sought to determine whether glyphosate and/or AMPA impact the functions of eEF1α1. In addition, we investigated the potential involvement of glyphosate treatment in spermatogenesis and its cytostatic effects, which may be affected by the interaction of glyphosate with eEF1α1.
2. Experimental Section
2.1. Molecular Modeling
The structure comparison of glyphosate and AMPA 3D was performed using the QSAR Toolbox.41 The eEF1α1 3D protein structure was obtained as described previously.32 Briefly, human eEF1α1 was identified in UniProt (Accession No. P68104), and the BLAST tool42 on this server was used to identify template structures. The 1F60, 1G7C, 1IJE, 1IJF, 2B7B, 2B7C, and 4C0S crystallographic template structures available from the RCSB PDB were selected for comparative modeling using MODELLER.43−45 The template structures were from different yeasts and rabbits. Yeast eEF1α1 showed 80.7% identity with human eEF1α1, and rabbit eEF1α1 had an identity of 92.6%. The first step in comparative modeling was the 3D alignment of the templates and target sequences performed using the SALIGN 3D module of MODELLER.43−45 This automodel module generated 100 protein structures. The generated structures were verified using VERIFY3D46 and PROCHECK,47 and the best 10 models were selected for docking experiments as targeted protein structures. Scripts from AutoDockTools48 were used to create input files for the ligands and the protein for submission to AutoDock Vina.48 Each optimized ligand structure was docked 10 times in each protein model using both standard B3LYP (Becke, 3-parameter, Lee–Yang–Parr) and RHF (restricted Hartree–Fock) methods,49 which resulted in 100 docked structures for each compound, from which only the best pose was selected for further investigation. With regard to gamendazole, the 10 best docking results were used to make the statistical sampling more reliable. The structures of glyphosate and AMPA were obtained from the ChemSpider database.50,51 The molecular docking experiment was performed using AutoDock Vina software version 1.1.2.48
2.2. Isothermal Titration Calorimetry (ITC) Experiments
The interaction of glyphosate with elongation factor eEF1α1 (ProSpec, Rehovot, Israel) was studied using ITC. ITC measures the changes in heat during an interaction and provides thermodynamic information about the binding affinity of a ligand to a protein. ITC is a straightforward method for determining the binding affinity constant (K) and binding stoichiometry (n), and the enthalpy of binding (ΔH) that occurs over the course of a reaction in solution and the entropy changes (ΔS) are calculated from the following equation: ΔG = −RT ln Ka = ΔH – TΔS.
The ITC experiment was performed at 25 °C with a Nano ITC Low Volume instrument (TA Instruments, New Castle, DE). During all measurements, 20 injections of 4 μM ligand (2.5 μL each) were titrated into 250 μL of protein (1 μM) with time intervals of 300 s and a stirring speed of 250 rpm. All ITC experiments were conducted with degassed 100 mM phosphate buffer solutions (pH 7.4). Control experiments included the titration of each complex solution into buffer. Corrected data refer to the experimental data after subtraction of the compounds from the buffer control data. The resulting thermograms were analyzed using the “Independent” model within NanoAnalyze software (TA Instruments, New Castle, DE).
2.3. Animal Experiment
The animal experiment was performed on the premises of the Institute of Pharmacology and Pharmacy (Building No. 22, Room 222) following the approved Rules of Conduct. The animal experiment was approved by the Ethics Committee, Ministry of Education, Youth and Sports of the Czech Republic, Czechia, and was in accordance with the Czech Animal Protection Act No. 246/1992 Coll. - VFU (18020).
Sixty Wistar rats aged approximately 10 weeks and weighing approximately 225 g were used in the experiment. The rats were kept in a controlled temperature environment (25 °C) on a normal photoperiod (12 h light, 12 h dark) and provided regular daily surveillance and health care. The animals were divided into three groups with 10 males and 10 females in each group. The first group was untreated throughout the experimental period and received water ad libitum without glyphosate (control group). The remaining two groups received water ad libitum with 0.7 or 7 mg/L glyphosate (Cat No. 89432, TraceCERT, Supelco, Sigma-Aldrich, St. Louis, MO) for 100 days.52 All animals received standard nutrition. Water consumption was monitored in each experimental group every day. At the end of the experimental period, the rats were starved overnight and then painlessly sacrificed. Tissue samples were then collected and used for histological examination. To verify that the source of drinking water did not influence our experiment, we confirmed that the drinking water did not contain pesticides or metabolite residues. This analysis included validation methods for 301 pesticide residue compounds performed as a service in an accredited laboratory of ALS Czech Republic (Part of ALS Limited). Thus, we verified that the water source did not significantly influence our results because it did not contain any pyrethroids, quaternary ammonium salts, glyphosate, or AMPA. In addition, two compounds were detected in trace amounts from a wide range of screening, namely, 0.087 μg/L chloridazon-desphenyl (experimental uncertainty analysis ∼ ±35%) and 0.069 μg/L alachlor ESA (experimental uncertainty analysis ∼ ±35%), which were present in levels that were approximately 10 000- and 100 000-fold lower than that of the tested glyphosate, respectively.
2.4. Histology of Rat Testicles
Rat testicles from the above-described experiment were fixed in 10% neutral buffered formalin. Each testicle was embedded in paraffin wax and cross-sectioned at a thickness of 4 μm. Sections were made through the center of the testicle, and the tissue was stained with hematoxylin and eosin. In one cross-section per animal, the tubules were evaluated for the presence of spermatogonia, spermatocytes, and spermatids. One testicle was rated for its spermatogenic potential (spermatogenic index) on a scale of 1–6 from 10 circular sections of tubules, each from a different testicular region that was homogeneous throughout with respect to cell association and spermiogenesis. The spermatogenic index was based on the appearance of spermatogenic cells throughout the testicle and included the number of cell layers, the types of cells, and the presence of late spermatids in the seminiferous tubules. The index and criteria were as follows: (1) only spermatogonia present; (2) spermatogonia and spermatocytes present; (3) spermatogonia, spermatocytes, and round (early) spermatids present with <5 late spermatids per tubule; (4) spermatogonia, spermatocytes, and round spermatids present with up to 25 late spermatids per tubule; (5) all cell types present and 50–75 late spermatids per tubule; and (6) all cell types present and >100 late spermatids per tubule.
The criteria (Table S1) for the assessment of the spermatogenic index was based on the testicular morphology from Whitsett et al.53 The area covered by ripe spermatic cells in tubules was determined using Fiji, a standard open-source platform for biological image analysis.54
2.5. Inhibition of Cancer Cells
2.5.1. Cell Culture
The spermatogonia GC-1 cell line (ATCC CRL-2053)55 derived from BALB/c mouse testes and the human B-lymphoblastic leukemia cell line SUP-B15 (ATCC CRL-1929)56 were obtained from the American Type Cell Culture Collection (ATCC). The GC-1 cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich), while the SUP-B15 cell line was grown in RPMI-1640 complete medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Gibco, Dublin, Ireland) and 1% penicillin–streptomycin (Sigma-Aldrich). Cells were incubated in 5% CO2 at 37 °C.
2.5.2. Cytotoxicity Assays
GC-1 cells were seeded in 96-well plates (2500 cells per well), and SUP-B15 cells were seeded in 24-well plates (2.5 × 105 cells per well). In our experiments, SUP-B15 cells grew better in flat bottom, low adherent 24-well plates than in 96-well plates. On the next day, cells were treated for 24, 48, or 72 h with glyphosate (Cat No. 89432, TraceCERT, Supelco, Sigma-Aldrich) using four different concentrations (25 mM, 4.17 mM, 694 μM, and 116 μM). Further, glyphosate cytotoxicity was evaluated using two different assays.
A Cytotoxicity Detection KitPLUS (Roche, Basel, Switzerland) was used to evaluate the cytotoxicity based on lactate dehydrogenase (LDH) activity. The absorbance of the supernatants was measured with an Infinite 200 PRO reader (Tecan, Mannedorf, Switzerland). The reference wavelength was set at 680 nm, and the samples were measured at 490 nm. Each LDH activity measurement involved wells with growth medium without cells as a background control, a positive control with maximum LDH release (obtained by the addition of 5 μL of lysis solution from the kit at the end of 30 min of incubation at 37 °C), and nontreated cells.
The second cytotoxicity analysis was performed using a WST-1 Cell Proliferation Assay Kit (Roche). This assay is used for cell viability, cell proliferation, and cytotoxicity analysis by measuring the level of formazan, which is a cleavage product of the WST-1 tetrazolium salt. Absorbance was measured in the proliferating cells at 450 nm using an Infinite 200 PRO reader (Tecan). The reference absorbance was measured at 630 nm. The results were normalized by comparing each value to the negative controls.
2.6. Public Health Statistics
Because we did not find any publicly available statistical data on the total sperm count, we performed a meta-analysis of published data. For this purpose, we closely followed a previously described procedure.57 The glyphosate use data were obtained from the U.S. Department of the Interior National Water-Quality Assessment (NAWQA) Project58 and from other published articles.1,59 Data on cancer incidence were obtained from the U.S. Department of Health and Human Services, National Institutes of Health, and National Cancer Institute.60 For all statistical computing, we used the R suite.61
2.7. Role of the Funding Source
The funder had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all data in the studies and had the final responsibility for the decision to submit these data for publication.
3. Results and Discussion
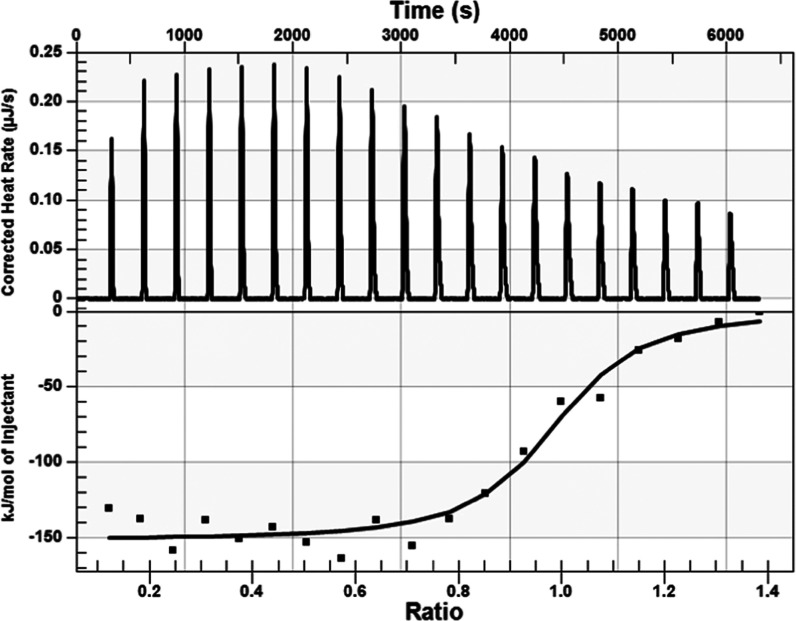
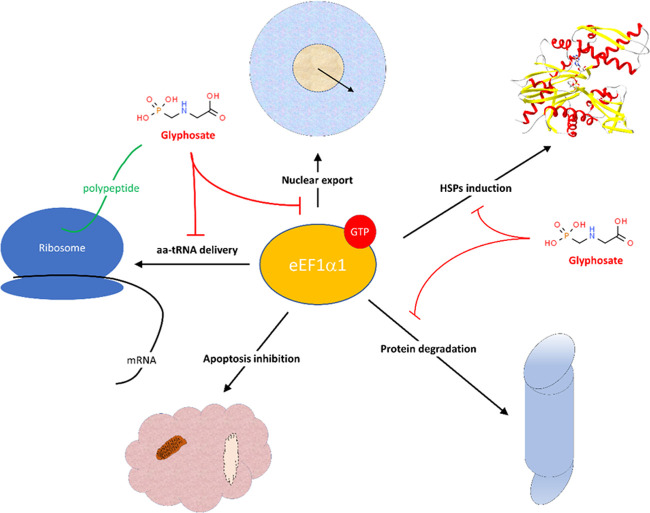
The results of our in silico experiment in Figure 1 show that glyphosate interacted with eEF1α1 in a similar manner as previously identified for the eEF1α1 inhibitor, gamendazole;32,33 however, the interaction energy was less favorable than that of gamendazole (Table 1). We also verified whether the AMPA metabolite interacts with eEF1α1, but the interaction sites of AMPA with eEF1α1 were different (Figure 1), and the interaction energy for AMPA was less favorable than that of glyphosate (Table 1). Because our results indicated that it is not likely that AMPA, in contrast to glyphosate, affects the reaction center in eEF1α1, we used only glyphosate for the microcalometric experiment. Importantly, the microcalorimetric estimation of the dissociation constant (Figure 2) confirmed the in silico analyses. The resulting dissociation constant value of 6.5 × 10–9 kJ/mol indicated the existence of a stable complex between glyphosate and eEF1α1. Thus, our results indicated that glyphosate may affect the delivery of aminoacyl-tRNAs to ribosomes.25−27 However, the eEF1α1 function altered by glyphosate may have various possible consequences affecting cell signal transduction, nuclear export, proliferation/apoptosis, cell vitality, and heat shock protein (HSP) response.21,22,28,62−67 Thus, based on our finding that eEF1α1 interacts with glyphosate, the proposed effects that glyphosate may exert may be due to this interaction (Figure 3). For instance, a change of the eEF1α1 conformation due to interaction with glyphosate affects the phosphorylation at Ser300 induced by the type I transforming growth factor β receptor (TβR-I), resulting in inhibition of cell proliferation.62 This is an example of a potential glyphosate effect on eukaryotic translation through affecting the aminoacyl-tRNA interaction site of eEF1α1.62 Thus, despite the unlikely direct binding of glyphosate to aminoacyl-tRNA synthetase,16 our results indicated that glyphosate affects protein synthesis through the aminoacyl-tRNA delivery to ribosomes.
Figure 1.
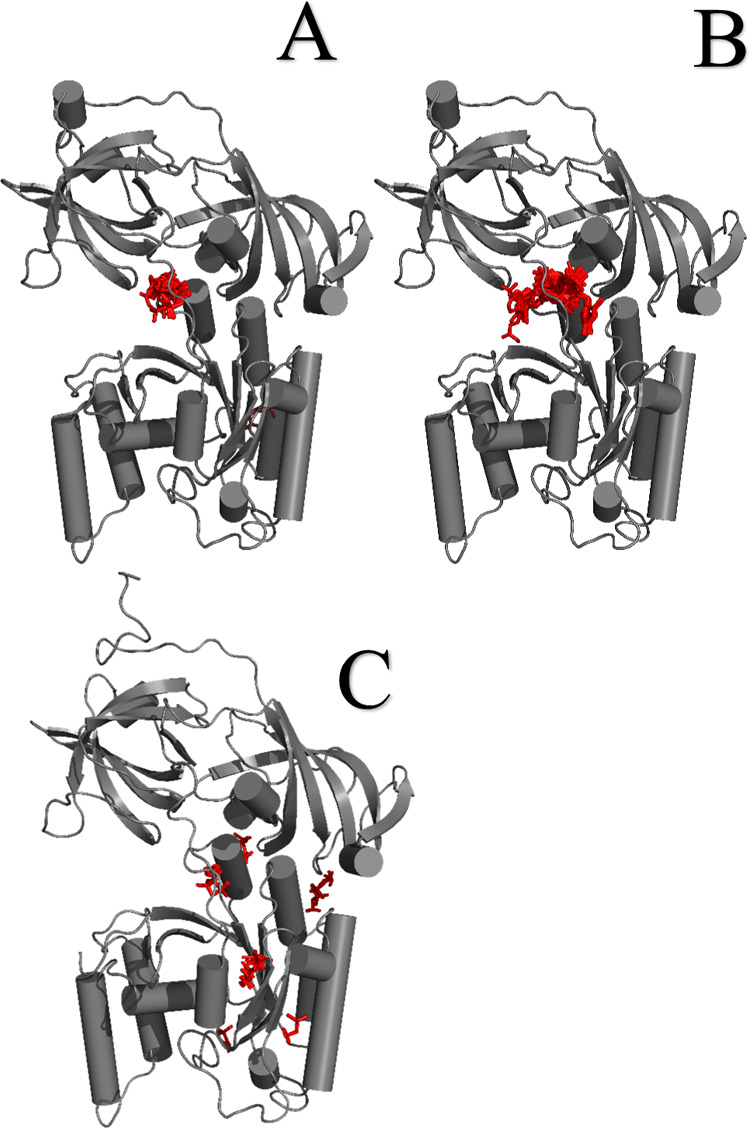
3D Structure of eEF1α1 obtained by computer modeling together with docked glyphosate (A), a male contraceptive gamendazole (B), and AMPA (C). These visualizations show that the glyphosate interaction site with eEF1α1 is the same as that for gamendazole; however, the interaction sites of AMPA with eEF1α1 are different.
Table 1. Comparison of the Interaction Energy of Gamendazole, Glyphosate, and AMPA with eEF1α1a.
| method | compound | docking energy (kcal/mol) |
|---|---|---|
| B3LYP | gamendazole | –9.1 |
| glyphosate | –5.3 | |
| AMPA | –4.1 | |
| RHF | gamendazole | –9.1 |
| glyphosate | –5.3 | |
| AMPA | –4.1 |
These results were obtained by docking the ligand structures computed by the B3LYP (Becke, 3-parameter, Lee–Yang–Parr) and RHF (restricted Hartree–Fock) methods49 into the protein structure obtained by homology modeling. The negative docking energy indicates the possible interaction of the three tested compounds with eEF1α1.
Figure 2.
Microcalorimetric estimation results of the dissociation constant of the glyphosate and eEF1α1 interaction. The dissociation constant of Kd = 6.494 × 10–9 (ΔH = −150.8 kJ/mol, ΔS = −349.0 J/(mol·K)) indicates a strong interaction of glyphosate with eEF1α1.
Figure 3.
Proposed consequences of glyphosate interaction with eEF1α1 due to known eEF1α1 function. The modulated function of eEF1α1 affects the aminoacyl-tRNA delivery to ribosome, HSP response, and apoptosis/proliferation.
Because tRNA is composed of ribonucleosides,68,69 we investigated whether glyphosate and AMPA mimic ribonucleotides. Although an exhaustive quantitative structure–activity relationship (QSAR) search41 has shown high similarity of both glyphosate and AMPA with the four ribonucleotide monophosphates (Table 2), we consider this effect unlikely because otherwise the effect of glyphosate and AMPA would be obvious and destructive at low concentrations.
Table 2. Results of the Exhaustive Quantitative Structure–Activity Relationship (QSAR) Searcha.
| compound | nucleotide | QSAR similarity (Yule, PubChem features) |
|---|---|---|
| glyphosate | AMP | 74.359% |
| CMP | 76.923% | |
| GMP | 87.179% | |
| UMP | 82.051% | |
| AMPA | AMP | 88.235% |
| CMP | 88.235% | |
| GMP | 88.235% | |
| UMP | 88.235% |
These results indicate high similarity of glyphosate and AMPA to all four nucleotides, but it is not likely that they substitute glyphosate or AMPA.
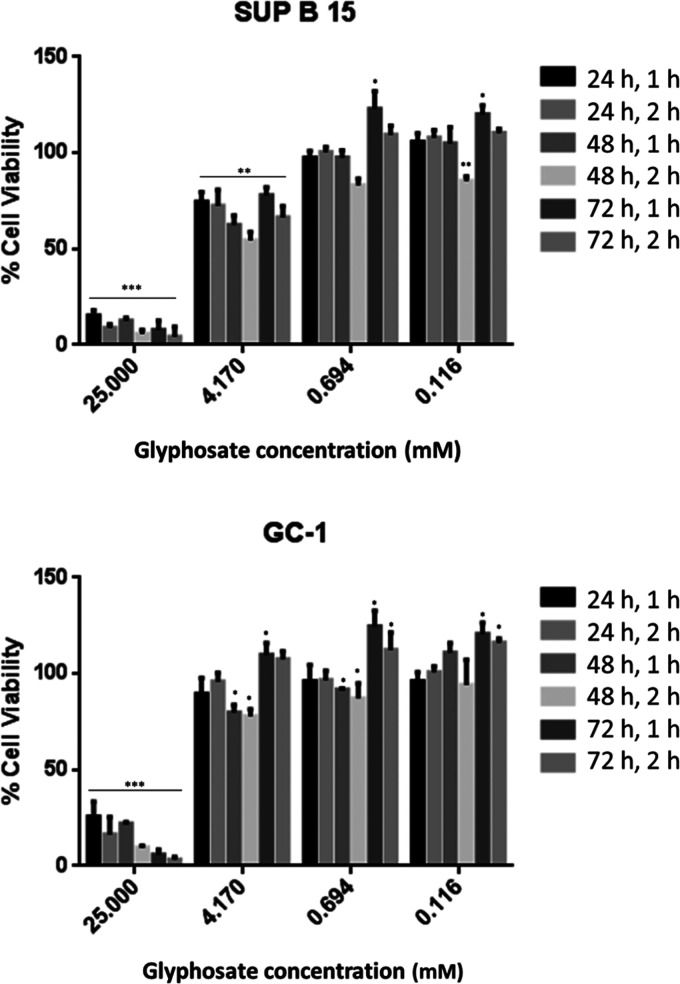
Our results showed the cytostatic effect of glyphosate using two cell lines, namely, GC-1 and SUP-B15, which are both ER-positive.70−73 The use of LDH (Figure 4) and the WST-1 assay (Figure S2) provided evidence for cytostatic and antiproliferative effects of glyphosate in both tested cell lines. These in vitro studies demonstrated that glyphosate treatment decreased the number of viable cells. Thus, our results support the previously observed cytostatic effect on ER-positive cancer cell lines.17,20 We were unable to elucidate the exact mechanism to explain the observed higher susceptibility of ER-positive cancer cells to glyphosate; however, our results suggested an association between estrogen signaling and the eEF1α1 mRNA level.21 Future studies will be performed to identify whether glyphosate treatment alters eEF1α1 levels at the transcript or protein level in certain cells and whether the expression is cell cycle-dependent.
Figure 4.
Vitality graphs determined by lactate dehydrogenase assay using SUP-B15 and GC-1 cell lines. p-values = 0 < (***) < 0.001 < (**) < 0.01 < (*) < 0.05. The controls were set as 100% vitality. For the proliferation assays, only the results compared to the controls are shown (100% proliferation). These results clearly indicate the cytostatic effect of glyphosate, which is attributed to eEF1α1 inhibition.
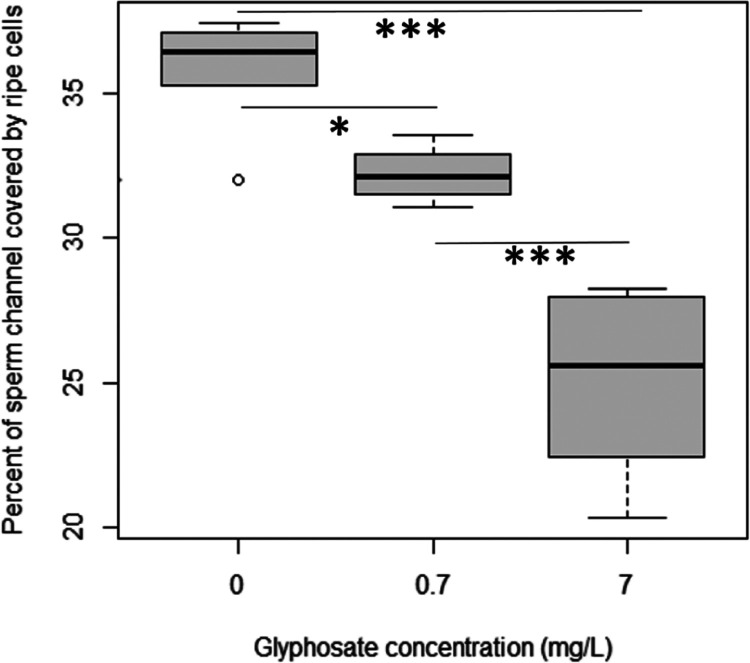
According to our results, glyphosate interaction with eEF1α1 was similar to that of gamendazole (Figure 1), thereby prompting us to investigate whether glyphosate exhibits an antispermatogenic effect.32,33 To verify whether glyphosate treatment affects spermatogenesis in vivo, we determined the total amount of sperm cells in the tubules of male rats. We did not observe any unripe sperm cells in the tubules, which was similar to a previous study37 but contradictory to another study reporting abnormal sperm cells and degenerative testicular lesions.74 We identified a significant decrease in the space covered by ripe sperm cells (ANOVA; p = 1.65 × 10–6), suggesting possible antispermatogenic effects of glyphosate (Figures 5 and S1), which agrees with the decreased sperm count observed in previous studies.37,74 In addition to the observed effect on spermatogenesis, rats in glyphosate-treated groups were smaller at the end of the 100-day experiment, but only the female rats were significantly smaller (p = 0.005) after the 7 mg/L treatment (Figure S3). The decrease in body weight has been previously observed in chronic/subchronic glyphosate exposure in mice.75 The decrease of body weight due to glyphosate interaction with eEF1α1 is likely due to the effect on protein synthesis.
Figure 5.
Percentage of sperm channel area covered by ripe sperm cells (p = 1.65 × 10–6). Oral glyphosate treatment of rats results in decreased spermatogenesis. Both 0.7 and 7 mg/L glyphosate treatments for 100 days in drinking water significantly (p < 0.05) decreased the sperm channel coverage by ripe cells. The negative effect of glyphosate on spermatogenesis increased with increasing glyphosate concentration.
Because our results indicated spermatogenesis reduction in rats and a cytostatic effect on cells (see above) and a previous meta-analysis of published studies has indicated a potential effect of glyphosate on sperm counts in rodents,38 we investigated whether this effect occurs at a population level in humans. Our meta-analysis using the published yearly glyphosate use1,58 showed a small negative correlation for glyphosate use. Although the resulting Spearman correlation coefficient (R = −0.2781609) was low, the correlation was still significant (p = 1.511 × 10–5). Thus, the results in Figure 6 do not rule out the possible negative impact of glyphosate on spermatogenesis, which may be attributed to the glyphosate interaction with eEF1α1.
Figure 6.
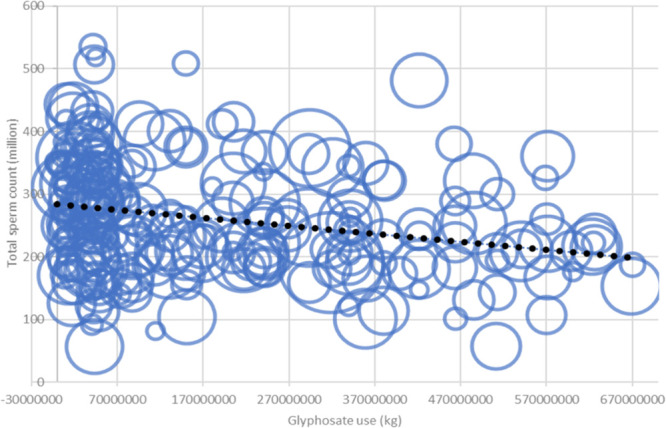
Meta-analysis results of average sperm count related to total glyphosate use in the USA (Spearman correlation R = −0.2781609 and p = 1.511 × 10–5).
Because studies have implicated an association between aberrant eEF1α1 and cancer,21,23,76−78 we analyzed several cancer incidences and their correlations with glyphosate use. The most striking observed correlation was the incidence of testicular cancer (Figure 7A) with a Spearman correlation of R = 0.9149815 and p = 3.885 × 10–10. We then compared this result with the previously identified correlation with non-Hodgkin lymphoma (Figure 7B),79−81 which resulted in a Spearman correlation of R = 0.5582609 and p = 0.005253. Based on these results, glyphosate may impact tumorigenesis.
Figure 7.
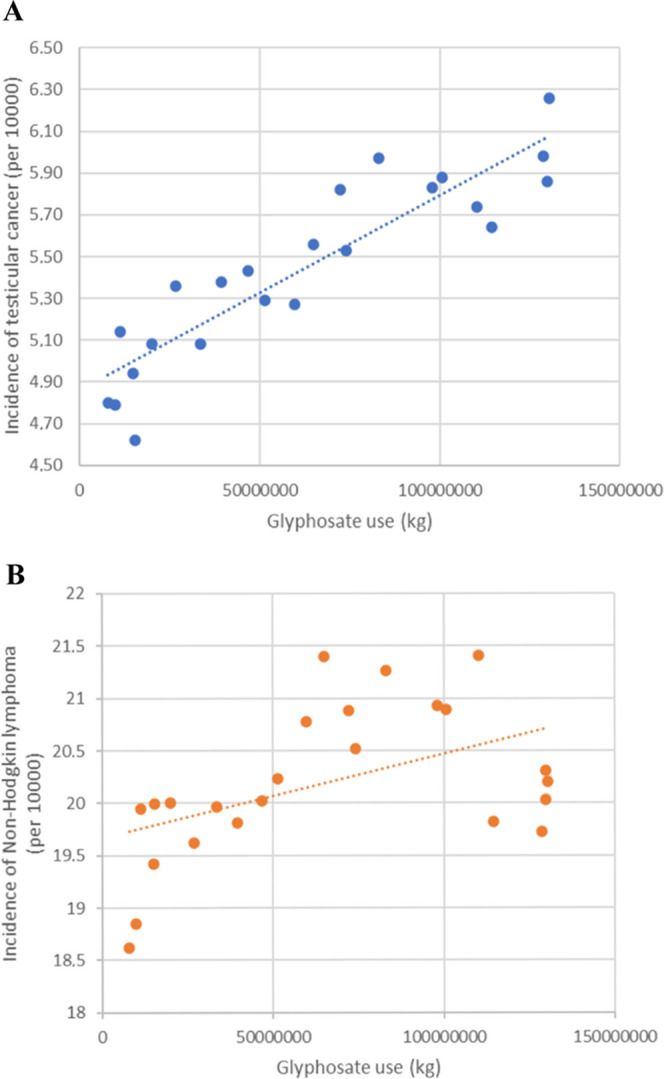
Graphs of selected cancer incidences plotted against glyphosate use in the USA. The resulting Spearman correlations were (A) R = 0.9149815 and p = 3.885 × 10–10 for testicular cancer and (B) R = 0.5582609 and p = 0.005253 for non-Hodgkin lymphoma.
Even though the negative correlation coefficient is relatively low, it is still significant (p < 0.01). Together with the other results in this study, these data indicate that the effect of glyphosate on human spermatogenesis due to alteration of eEF1α1 function cannot be ruled out.
Acknowledgments
We would like to thank Eva Filova and Martin Markovic for their valuable help.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c00449.
Details of the criteria for the assessment of the spermatogenic index; representative microphotographs of rat testes tissue; the average weight gain differences in female and male rats among glyphosate treatments; and the proliferation of GC-1 and SUP-B15 cells analyzed by WST-1 assay (PDF)
Author Contributions
B.S. and T.E. were involved in the conception of the study. B.S. was involved in the study design. G.T., E.A., and B.S. performed the modeling experiments. B.S. was the study statistician. M.M. and M.H. were responsible for the microcalorimetric estimations. J.C., J.F., and M.S. performed the animal experiment and subsequent histological analysis. G.B. and M.A. were involved in the cell culture analysis. B.S. and T.E. were involved in interpreting the data. T.E. and B.S. wrote the main manuscript. All authors have read and approved the final manuscript.
This research was supported by funding no. RO0418 from the Ministry of Agriculture of the Czech Republic (http://eagri.cz/). Computational resources were provided by the CESNET LM2015042 and the CERIT Scientific Cloud LM2015085 under the “Projects of Large Research, Development, and Innovations Infrastructures” program.
The authors declare no competing financial interest.
Supplementary Material
References
- Benbrook C. M. Trends in Glyphosate Herbicide Use in the United States and Globally. Environ. Sci. Eur. 2016, 28, 3 10.1186/s12302-016-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann K. M.; Weaver L. M. The Shikimate Pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 473–503. 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- Gill J. P. K.; Sethi N.; Mohan A.; Datta S.; Girdhar M. Glyphosate Toxicity for Animals. Environ. Chem. Lett. 2018, 16, 401–426. 10.1007/s10311-017-0689-0. [DOI] [Google Scholar]
- International Agency for Research on Cancer (IARC). Q&A on Glyphosate, 2016. https://www.iarc.fr/wp-content/uploads/2018/11/QA_Glyphosate.pdf (accessed July 11, 2019).
- European Chemicals Agency (ECHA). Glyphosate Not Classified as a Carcinogen by ECHA: ECHA/PR/17/06, 2017. https://echa.europa.eu/-/glyphosate-not-classified-as-a-carcinogen-by-echa (accessed April 30, 2021).
- U.S. Environmental Protection Agency (US EPA). EPA Releases Draft Risk Assessments for Glyphosate: For Release: December 18, 2017, 2017. https://www.epa.gov/pesticides/epa-releases-draft-risk-assessments-glyphosate (accessed April 30, 2021).
- Evaluation of the Impact of Glyphosate and Its Residues in Feed on Animal Health. EFSA J. 2018, 16, e05283 10.2903/j.efsa.2018.5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C. J.; Armstrong B. K.; Baguley B. C.; Baur X.; Belyaev I.; Belle R.; Belpoggi F.; Biggeri A.; Bosland M. C.; Bruzzi P.; Budnik L. T.; Bugge M. D.; Burns K.; Calaf G. M.; Carpenter D. O.; Carpenter H. M.; Lopez-Carrillo L.; Clapp R.; Cocco P.; Consonni D.; Comba P.; Craft E.; Dalvie M. A.; Davis D.; Demers P. A.; De Roos A. J.; DeWitt J.; Forastiere F.; Freedman J. H.; Fritschi L.; Gaus C.; Gohlke J. M.; Goldberg M.; Greiser E.; Hansen J.; Hardell L.; Hauptmann M.; Huang W.; Huff J.; James M. O.; Jameson C. W.; Kortenkamp A.; Kopp-Schneider A.; Kromhout H.; Larramendy M. L.; Landrigan P. J.; Lash L. H.; Leszczynski D.; Lynch C. F.; Magnani C.; Mandrioli D.; Martin F. L.; Merler E.; Michelozzi P.; Miligi L.; Miller A. B.; Mirabelli D.; Mirer F. E.; Naidoo S.; Perry M. J.; Petronio M. G.; Pirastu R.; Portier R. J.; Ramos K. S.; Robertson L. W.; Rodriguez T.; Roosli M.; Ross M. K.; Roy D.; Rusyn I.; Saldiva P.; Sass J.; Savolainen K.; Scheepers P. T. J.; Sergi C.; Silbergeld E. K.; Smith M. T.; Stewart B. W.; Sutton P.; Tateo F.; Terracini B.; Thielmann H. W.; Thomas D. B.; Vainio H.; Vena J. E.; Vineis P.; Weiderpass E.; Weisenburger D. D.; Woodruff T. J.; Yorifuji T.; Yu I. J.; Zambon P.; Zeeb H.; Zhou S.-F. Differences in the Carcinogenic Evaluation of Glyphosate between the International Agency for Research on Cancer (IARC) and the European Food Safety Authority (EFSA). J. Epidemiol. Community Health 2016, 70, 741–745. 10.1136/jech-2015-207005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Environmental Protection Agency (EPA). EPA Takes Next Step in Review Process for Herbicide Glyphosate, Reaffirms No Risk to Public Health, 2019. https://www.epa.gov/newsreleases/epa-takes-next-step-review-process-herbicide-glyphosate-reaffirms-no-risk-public-health (accessed Jul 11, 2019).
- Davoren M. J.; Schiestl R. H. Glyphosate-Based Herbicides and Cancer Risk: A Post-IARC Decision Review of Potential Mechanisms, Policy and Avenues of Research. Carcinogenesis 2018, 39, 1207–1215. 10.1093/carcin/bgy105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda-Ruzafa L.; Cruz F.; Roman P.; Cardona D. Gut Microbiota and Neurological Effects of Glyphosate. NeuroToxicology 2019, 75, 1–8. 10.1016/j.neuro.2019.08.006. [DOI] [PubMed] [Google Scholar]
- Motta E. V. S.; Raymann K.; Moran N. A. Glyphosate Perturbs the Gut Microbiota of Honey Bees. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 10305–10310. 10.1073/pnas.1803880115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsel A.; Seneff S. Glyphosate, Pathways to Modern Diseases IV: Cancer and Related Pathologies. J. Biol. Phys. Chem. 2015, 15, 121–159. 10.4024/11SA15R.jbpc.15.03. [DOI] [Google Scholar]
- Ford B.; Bateman L. A.; Gutierrez-Palominos L.; Park R.; Nomura D. K. Mapping Proteome-Wide Targets of Glyphosate in Mice. Cell Chem. Biol. 2017, 24, 133–140. 10.1016/j.chembiol.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Samsel A.; Seneff S. Glyphosate Pathways to Modern Diseases V: Amino Acid Analogue of Glycine in Diverse Proteins. J. Biol. Phys. Chem. 2016, 16, 9–46. 10.4024/03SA16A.jbpc.16.01. [DOI] [Google Scholar]
- Antoniou M. N.; Nicolas A.; Mesnage R.; Biserni M.; Rao F. V.; Martin C. V. Glyphosate Does Not Substitute for Glycine in Proteins of Actively Dividing Mammalian Cells. BMC Res. Notes 2019, 12, 494 10.1186/s13104-019-4534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R.; Phedonos A.; Biserni M.; Arno M.; Balu S.; Corton J. C.; Ugarte R.; Antoniou M. N. Evaluation of Estrogen Receptor alpha Activation by Glyphosate-Based Herbicide Constituents. Food Chem. Toxicol. 2017, 108, 30–42. 10.1016/j.fct.2017.07.025. [DOI] [PubMed] [Google Scholar]
- Theodossiou T. A.; Ali M.; Grigalavicius M.; Grallert B.; Dillard P.; Schink K. O.; Olsen C. E.; Walchli S.; Inderberg E. M.; Kubin A.; Peng Q.; Berg K. Simultaneous Defeat of MCF7 and MDA-MB-231 Resistances by a Hypericin PDT-Tamoxifen Hybrid Therapy. npj Breast Cancer 2019, 5, 13 10.1038/s41523-019-0108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C. K. Tamoxifen in the Treatment of Breast Cancer. N. Engl. J. Med. 1998, 339, 1609–1618. 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- Li Q.; Lambrechts M. J.; Zhang Q.; Liu S.; Ge D.; Yin R.; Xi M.; You Z. Glyphosate and AMPA Inhibit Cancer Cell Growth through Inhibiting Intracellular Glycine Synthesis. Drug Des., Dev. Ther. 2013, 7, 635–643. 10.2147/DDDT.S49197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-Y.; Beattie A.; Baradaran B.; Dray E.; Duijf P. H. G. Contradictory mRNA and Protein Misexpression of EEF1A1 in Ductal Breast Carcinoma Due to Cell Cycle Regulation and Cellular Stress. Sci. Rep. 2018, 8, 13904 10.1038/s41598-018-32272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farra R.; Scaggiante B.; Guerra C.; Pozzato G.; Grassi M.; Zanconati F.; Perrone F.; Ferrari C.; Trotta F.; Grassi G.; Dapas B. Dissecting the Role of the Elongation Factor 1A Isoforms in Hepatocellular Carcinoma Cells by Liposome-Mediated Delivery of siRNAs. Int. J. Pharm. 2017, 525, 367–376. 10.1016/j.ijpharm.2017.02.031. [DOI] [PubMed] [Google Scholar]
- Huang J.; Zheng C.; Shao J.; Chen L.; Liu X.; Shao J. Overexpression of eEF1A1 Regulates G1-Phase Progression to Promote HCC Proliferation through the STAT1-Cyclin D1 Pathway. Biochem. Biophys. Res. Commun. 2017, 494, 542–549. 10.1016/j.bbrc.2017.10.116. [DOI] [PubMed] [Google Scholar]
- Rajendran V.; Kalita P.; Shukla H.; Kumar A.; Tripathi T. Aminoacyl-tRNA Synthetases: Structure, Function, and Drug Discovery. Int. J. Biol. Macromol. 2018, 111, 400–414. 10.1016/j.ijbiomac.2017.12.157. [DOI] [PubMed] [Google Scholar]
- Sasikumar A. N.; Perez W. B.; Kinzy T. G. The Many Roles of the Eukaryotic Elongation Factor 1 Complex. Wiley Interdiscip. Rev.: RNA 2012, 3, 543–555. 10.1002/wrna.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H.; Hurt E.; Simos G. An Aminoacylation-Dependent Nuclear tRNA Export Pathway in Yeast. Genes Dev. 2000, 14, 830–840. [PMC free article] [PubMed] [Google Scholar]
- Lee J. S.; Park S. G.; Park H.; Seol W.; Lee S.; Kim S. Interaction Network of Human Aminoacyl-tRNA Synthetases and Subunits of Elongation Factor 1 Complex. Biochem. Biophys. Res. Commun. 2002, 291, 158–164. 10.1006/bbrc.2002.6398. [DOI] [PubMed] [Google Scholar]
- Thornton S.; Anand N.; Purcell D.; Lee J. Not Just for Housekeeping: Protein Initiation and Elongation Factors in Cell Growth and Tumorigenesis. J. Mol. Med. 2003, 81, 536–548. 10.1007/s00109-003-0461-8. [DOI] [PubMed] [Google Scholar]
- Ruest L.-B.; Marcotte R.; Wang E. Peptide Elongation Factor eEF1A-2/S1 Expression in Cultured Differentiated Myotubes and Its Protective Effect against Caspase-3-Mediated Apoptosis. J. Biol. Chem. 2002, 277, 5418–5425. 10.1074/jbc.M110685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.; Francoeur A.-M.; Liu S.; Wang E. Tissue-Specific Expression in Mammalian Brain, Heart, and Muscle of S1, a Member of the Elongation Factor-1α Gene Family. J. Biol. Chem. 1992, 267, 24064–24068. 10.1016/S0021-9258(18)35946-5. [DOI] [PubMed] [Google Scholar]
- Kahns S.; Lund A.; Kristensen P.; Knudsen C. R.; Clark B. F. C.; Cavallius J.; Merrick W. C. The Elongation Factor 1 A-2 Isoform from Rabbit: Cloning of the cDNA and Characterization of the Protein. Nucleic Acids Res. 1998, 26, 1884–1890. 10.1093/nar/26.8.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglová K.; Rylova G.; Markos A.; Prichystalova H.; Soural M.; Petracek M.; Medvedikova M.; Tejral G.; Sopko B.; Hradil P.; Dzubak P.; Hajduch M.; Hlavac J. Identification of Eukaryotic Translation Elongation Factor 1-α 1 Gamendazole-Binding Site for Binding of 3-Hydroxy-4(1H)-Quinolinones as Novel Ligands with Anticancer Activity. J. Med. Chem. 2018, 61, 3027–3036. 10.1021/acs.jmedchem.8b00078. [DOI] [PubMed] [Google Scholar]
- Tash J. S.; Chakrasali R.; Jakkaraj S. R.; Hughes J.; Smith S. K.; Hornbaker K.; Heckert L. L.; Ozturk S. B.; Hadden M. K.; Kinzy T. G.; Blagg B. S. J.; Georg G. I. Gamendazole, an Orally Active Indazole Carboxylic Acid Male Contraceptive Agent, Targets HSP90AB1 (HSP90BETA) and EEF1A1 (eEF1A), and Stimulates Il1a Transcription in Rat Sertoli Cells. Biol. Reprod. 2008, 78, 1139–1152. 10.1095/biolreprod.107.062679. [DOI] [PubMed] [Google Scholar]
- Joffe M. What Has Happened to Human Fertility?. Hum. Reprod. 2010, 25, 295–307. 10.1093/humrep/dep390. [DOI] [PubMed] [Google Scholar]
- Le Cornet C.; Lortet-Tieulent J.; Forman D.; Beranger R.; Flechon A.; Fervers B.; Schuz J.; Bray F. Testicular Cancer Incidence to Rise by 25% by 2025 in Europe? Model-Based Predictions in 40 Countries Using Population-Based Registry Data. Eur. J. Cancer 2014, 50, 831–839. 10.1016/j.ejca.2013.11.035. [DOI] [PubMed] [Google Scholar]
- Williams A. L.; Watson R. E.; DeSesso J. M. Developmental and Reproductive Outcomes in Humans and Animals after Glyphosate Exposure: A Critical Analysis. J. Toxicol. Environ. Health, Part B 2012, 15, 39–96. 10.1080/10937404.2012.632361. [DOI] [PubMed] [Google Scholar]
- Dai P.; Hu P.; Tang J.; Li Y.; Li C. Effect of Glyphosate on Reproductive Organs in Male Rat. Acta Histochem. 2016, 118, 519–526. 10.1016/j.acthis.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Cai W.; Ji Y.; Song X.; Guo H.; Han L.; Zhang F.; Liu X.; Zhang H.; Zhu B.; Xu M. Effects of Glyphosate Exposure on Sperm Concentration in Rodents: A Systematic Review and Meta-Analysis. Environ. Toxicol. Pharmacol. 2017, 55, 148–155. 10.1016/j.etap.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Lupi L.; Bedmar F.; Puricelli M.; Marino D.; Aparicio V. C.; Wunderlin D.; Miglioranza K. S. B. Glyphosate Runoff and Its Occurrence in Rainwater and Subsurface Soil in the Nearby Area of Agricultural Fields in Argentina. Chemosphere 2019, 225, 906–914. 10.1016/j.chemosphere.2019.03.090. [DOI] [Google Scholar]
- Gunarathna S.; Gunawardana B.; Jayaweera M.; Manatunge J.; Zoysa K. Glyphosate and AMPA of Agricultural Soil, Surface Water, Groundwater and Sediments in Areas Prevalent with Chronic Kidney Disease of Unknown Etiology, Sri Lanka. J. Environ. Sci. Health, Part B 2018, 53, 729–737. 10.1080/03601234.2018.1480157. [DOI] [PubMed] [Google Scholar]
- Dimitrov S. D.; Diderich R.; Sobanski T.; Pavlov T. S.; Chankov G. V.; Chapkanov A. S.; Karakolev Y. H.; Temelkov S. G.; Vasilev R. A.; Gerova K. D.; Kuseva C. D.; Todorova N. D.; Mehmed A. M.; Rasenberg M.; Mekenyan O. G. QSAR Toolbox – Workflow and Major Functionalities. SAR QSAR Environ. Res. 2016, 27, 203–219. 10.1080/1062936X.2015.1136680. [DOI] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Šali A.; Blundell T. L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Martí-Renom M. A.; Stuart A. C.; Fiser A.; Sanchez R.; Melo F.; Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Fiser A.; Do R. K. G.; Sali A. Modeling of Loops in Protein Structures. Protein Sci. 2000, 9, 1753–1773. 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D.; Luthy R.; Bowie J. U. VERIFY3D: Assessment of Protein Models with Three-Dimensional Profiles. Methods Enzymol. 1997, 277, 396–404. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; MacArthur M. W.; Moss D. S.; Thornton J. M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Petersson G. A.; Nakatsuji H.; Li X.; Caricato M.; Marenich A. V.; Bloino J.; Janesko B. G.; Gomperts R.; Mennucci B.; Hratchian H. P.; Ortiz J. V.; Izmaylov A. F.; Sonnenberg J. L.; Williams-Young D.; Ding F.; Lipparini F.; Egidi F.; Goings J.; Peng B.; Petrone A.; Henderson T.; Ranasinghe D.; Zakrzewski V. G.; Gao J.; Rega N.; Zheng G.; Liang W.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Throssell K.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M. J.; Heyd J. J.; Brothers E. N.; Kudin K. N.; Staroverov V. N.; Keith T. A.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A. P.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Millam J. M.; Klene M.; Adamo C.; Cammi R.; Ochterski J. W.; Martin R. L.; Morokuma K.; Farkas O.; Foresman J. B.; Fox D. J.. Gaussian 16, Revision B.01; Gaussian, Inc.: Wallingford, CT, 2016.
- ChemSpider. N-(phosphonomethyl)glycine, 2019. http://www.chemspider.com/Chemical-Structure.3376.html (accessed July 11, 2019).
- ChemSpider. (Aminomethyl)phosphonate, 2019. http://www.chemspider.com/Chemical-Structure.13399.html (accessed July 11, 2019).
- Larsen K.; Najle R.; Lifschitz A.; Virkel G. Effects of Sub-Lethal Exposure of Rats to the Herbicide Glyphosate in Drinking Water: Glutathione Transferase Enzyme Activities, Levels of Reduced Glutathione and Lipid Peroxidation in Liver, Kidneys and Small Intestine. Environ. Toxicol. Pharmacol. 2012, 34, 811–818. 10.1016/j.etap.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Whitsett J. M.; Noden P. F.; Cherry J.; Lawton A. D. Effect of Transitional Photoperiods on Testicular Development and Puberty in Male Deer Mice (Peromyscus maniculatus). Reproduction 1984, 72, 277–286. 10.1530/jrf.0.0720277. [DOI] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J.-Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M.-C.; Narisawa S.; Hess R. A.; Millan J. L. Immortalization of Germ Cells and Somatic Testicular Cells Using the SV40 Large T Antigen. Exp. Cell Res. 1992, 201, 417–435. 10.1016/0014-4827(92)90291-F. [DOI] [PubMed] [Google Scholar]
- Fainstein E.; Marcelle C.; Rosner A.; Canaani E.; Gale R. P.; Dreazen O.; Smith S. D.; Croce C. M. A New Fused Transcript in Philadelphia Chromosome Positive Acute Lymphocytic Leukaemia. Nature 1987, 330, 386–388. 10.1038/330386a0. [DOI] [PubMed] [Google Scholar]
- Levine H.; Jorgensen N.; Martino-Andrade A.; Mendiola J.; Weksler-Derri D.; Mindlis I.; Pinotti R.; Swan S. H. Temporal Trends in Sperm Count: A Systematic Review and Meta-Regression Analysis. Hum. Reprod. Update 2017, 23, 646–659. 10.1093/humupd/dmx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of the Interior/U.S. Geological Survey. National Water-Quality Assessment (NAWQA) Project, 2018. http://water.usgs.gov/nawqa/pnsp/usage/maps/county-level/ (accessed Nov 11, 2018).
- Myers J. P.; Antoniou M. N.; Blumberg B.; Carroll L.; Colborn T.; Everett L. G.; Hansen M.; Landrigan P. J.; Lanphear B. P.; Mesnage R.; Vandenberg L. N.; vom Saal F. S.; Welshons W. V.; Benbrook C. M. Concerns over Use of Glyphosate-Based Herbicides and Risks Associated with Exposures: A Consensus Statement. Environ. Health 2016, 15, 19 10.1186/s12940-016-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services/National Institutes of Health/National Cancer Institute. Cancer Stat Facts: Testicular Cancer, 2018. https://seer.cancer.gov/statfacts/html/testis.html (accessed Nov 11, 2018).
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, 2016. https://www.r-project.org/ (accessed Nov 11, 2018).
- Lin K. W.; Yakymovych I.; Jia M.; Yakymovych M.; Souchelnytskyi S. Phosphorylation of eEF1A1 at Ser300 by TβR-I Results in Inhibition of mRNA Translation. Curr. Biol. 2010, 20, 1615–1625. 10.1016/j.cub.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Liu X.; Chen L.; Ge J.; Yan C.; Huang Z.; Hu J.; Wen C.; Li M.; Huang D.; Qiu Y.; Hao H.; Yuan R.; Lei J.; Yu X.; Shao J. The Ubiquitin-Like Protein FAT10 Stabilizes eEF1A1 Expression to Promote Tumor Proliferation in a Complex Manner. Cancer Res. 2016, 76, 4897–4907. 10.1158/0008-5472.CAN-15-3118. [DOI] [PubMed] [Google Scholar]
- Mateyak M. K.; Kinzy T. G. eEF1A: Thinking Outside the Ribosome. J. Biol. Chem. 2010, 285, 21209–21213. 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanch A.; Robinson F.; Watson I. R.; Cheng L. S.; Irwin M. S. Eukaryotic Translation Elongation Factor 1-Alpha 1 Inhibits p53 and p73 Dependent Apoptosis and Chemotherapy Sensitivity. PLoS One 2013, 8, e66436 10.1371/journal.pone.0066436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M.; Pani B.; Griffiths L. A.; Muchardt C.; Abbott C. M.; Singer R. H.; Nudler E. The Translation Elongation Factor eEF1A1 Couples Transcription to Translation During Heat Shock Response. eLife 2014, 3, e03164 10.7554/eLife.03164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H.; Ning L.; Yu Z.; Dou G.; Li L. Proteomic Identification of eEF1A1 as a Molecular Target of Curcumol for Suppressing Metastasis of MDA-MB-231 Cells. J. Agric. Food Chem. 2017, 65, 3074–3082. 10.1021/acs.jafc.7b00573. [DOI] [PubMed] [Google Scholar]
- Fujishima K.; Kanai A. tRNA Gene Diversity in the Three Domains of Life. Front. Genet. 2014, 5, 142 10.3389/fgene.2014.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S.; Tomita M.; Suzuki H.; Kanai A. Systematic Analysis of the Binding Surfaces between tRNAs and Their Respective Aminoacyl tRNA Synthetase Based on Structural and Evolutionary Data. Front. Genet. 2018, 8, 227 10.3389/fgene.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicini E.; Loiarro M.; Di Agostino S.; Corallini S.; Capolunghi F.; Carsetti R.; Chieffi P.; Geremia R.; Stefanini M.; Sette C. 17-β-Estradiol Elicits Genomic and Non-Genomic Responses in Mouse Male Germ Cells. J. Cell. Physiol. 2006, 206, 238–245. 10.1002/jcp.20454. [DOI] [PubMed] [Google Scholar]
- Sirianni R.; Chimento A.; Ruggiero C.; De Luca A.; Lappano R.; Ando S.; Maggiolini M.; Pezzi V. The Novel Estrogen Receptor, G Protein-Coupled Receptor 30, Mediates the Proliferative Effects Induced by 17β-Estradiol on Mouse Spermatogonial GC-1 Cell Line. Endocrinology 2008, 149, 5043–5051. 10.1210/en.2007-1593. [DOI] [PubMed] [Google Scholar]
- Zhou W.; Wang S.; Ying Y.; Zhou R.; Mao P. miR-196b/miR-1290 Participate in the Antitumor Effect of Resveratrol via Regulation of IGFBP3 Expression in Acute Lymphoblastic Leukemia. Oncol. Rep. 2017, 37, 1075–1083. 10.3892/or.2016.5321. [DOI] [PubMed] [Google Scholar]
- Iacobucci I.; Di Rora A. G.; Falzacappa M. V.; Agostinelli C.; Derenzini E.; Ferrari A.; Papayannidis C.; Lonetti A.; Righi S.; Imbrogno E.; Pomella S.; Venturi C.; Guadagnuolo V.; Cattina F.; Ottaviani E.; Abbenante M. C.; Vitale A.; Elia L.; Russo D.; Zinzani P. L.; Pileri S.; Pelicci P. G.; Martinelli G. In Vitro and in Vivo Single-Agent Efficacy of Checkpoint Kinase Inhibition in Acute Lymphoblastic Leukemia. J. Hematol. Oncol. 2015, 8, 125 10.1186/s13045-015-0206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owagboriaye F. O.; Dedeke G. A.; Ademolu K. O.; Olujimi O. O.; Ashidi J. S.; Adeyinka A. A. Reproductive Toxicity of Roundup Herbicide Exposure in Male Albino Rat. Exp. Toxicol. Pathol. 2017, 69, 461–468. 10.1016/j.etp.2017.04.007. [DOI] [PubMed] [Google Scholar]
- Ait Bali Y.; Ba-Mhamed S.; Bennis M. Behavioral and Immunohistochemical Study of the Effects of Subchronic and Chronic Exposure to Glyphosate in Mice. Front. Behav. Neurosci. 2017, 11, 146 10.3389/fnbeh.2017.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas W.; Kumar A.; Herbein G. The eEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front. Oncol. 2015, 5, 75 10.3389/fonc.2015.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.-L.; Lu S.-X.; Liu L.-L.; Wang C.-H.; Yang X.; Zhang Z.-Y.; Zhang H.-Z.; Yun J.-P. eEF1A1 Overexpression Enhances Tumor Progression and Indicates Poor Prognosis in Hepatocellular Carcinoma. Transl. Oncol. 2018, 11, 125–131. 10.1016/j.tranon.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaggiante B.; Dapas B.; Bonin S.; Grassi M.; Zennaro C.; Farra R.; Cristiano L.; Siracusano S.; Zanconati F.; Giansante C.; Grassi G. Dissecting the Expression of EEF1A1/2 Genes in Human Prostate Cancer Cells: The Potential of EEF1A2 as a Hallmark for Prostate Transformation and Progression. Br. J. Cancer 2012, 106, 166–173. 10.1038/bjc.2011.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinasi L.; Leon M. E. Non-Hodgkin Lymphoma and Occupational Exposure to Agricultural Pesticide Chemical Groups and Active Ingredients: A Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2014, 11, 4449–4527. 10.3390/ijerph110404449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E. T.; Delzell E. Systematic Review and Meta-Analysis of Glyphosate Exposure and Risk of Lymphohematopoietic Cancers. J. Environ. Sci. Health, Part B 2016, 51, 402–434. 10.1080/03601234.2016.1142748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Rana I.; Shaffer R. M.; Taioli E.; Sheppard L. Exposure to Glyphosate-Based Herbicides and Risk for Non-Hodgkin Lymphoma: A Meta-Analysis and Supporting Evidence. Mutat. Res., Rev. Mutat. Res. 2019, 781, 186–206. 10.1016/j.mrrev.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.