Highlights
-
•
Quantify the brain structural connectivity signature using DTI.
-
•
Reveal three types of structural connectivity alteration patterns in PD progression.
-
•
Identify brain dysconnectivity features of iRBD and PD by MDMR for the first time.
-
•
High accuracy performance for diagnosis of prodromal PD.
Keywords: DTI, Parkinson disease, Idiopathic rapid eye movement sleep behaviour disorder, Brain structural network, Multivariate distance matrix regression
Abstract
Pinpointing the brain dysconnectivity in idiopathic rapid eye movement sleep behaviour disorder (iRBD) can facilitate preventing the conversion of Parkinson's disease (PD) from prodromal phase. Recent neuroimage investigations reported disruptive brain white matter connectivity in both iRBD and PD, respectively. However, the intrinsic process of the human brain structural network evolving from iRBD to PD still remains largely unknown. To address this issue, 151 participants including iRBD, PD and age-matched normal controls were recruited to receive diffusion MRI scans and neuropsychological examinations. The connectome-wide association analysis was performed to detect reorganization of brain structural network along with PD progression. Eight brain seed regions in both cortical and subcortical areas demonstrated significant structural pattern changes along with the progression of PD. Applying machine learning on the key connectivity related to these seed regions demonstrated better classification accuracy compared to conventional network-based statistic. Our study shows that connectome-wide association analysis reveals the underlying structural connectivity patterns related to the progression of PD, and provide a promising distinct capability to predict prodromal PD patients.
1. Introduction
Over the past years, idiopathic rapid eye movement sleep behaviour disorder (iRBD) has been increasingly recognized as the heralding features of Parkinson’s disease (PD) and is characterized by a long incubation period (Boeve, 2010, Iranzo et al., 2014, Iranzo et al., 2006, Iranzo et al., 2013, Postuma et al., 2015a, Schenck et al., 2013, Schenck et al., 1996). According to a recent longitudinal study, 39.7% iRBD patients had converted to PD within four years (Fereshtehnejad et al., 2017). The update of Movement Disorder Society (MDS) criteria in 2019 demonstrates that polysomnogram-confirmed iRBD takes up the highest likelihood ratio of all the prodromal markers of the progression of PD (Heinzel et al., 2019). Therefore, identification of in-vivo brain structural abnormality in iRBD is crucial to promptly prevent the conversion of PD from prodromal phase to clinical stage in time.
Converging neurophysiological evidence suggested that progression of PD-related neurodegeneration is not prominent in terms of regional atrophy, but more associated with white matter integrity due to axonopathy and synaptic dysfunction (Burke and O'Malley, 2013, O'malley 2010, Tinaz et al., 2017). According to the Braak staging scheme of PD, the primary pathology patterns emerge in brainstem and progressively propagate to the neocortex, through the related white matter fibre pathway (Braak et al., 2003, Braak et al., 2004). In particular, this pathway could be directly damaged by the misfolded α-synuclein through regulating membrane levels of acidic phospholipids (Schechter et al., 2020). On a macroscopic scale, widespread diffusion MRI studies confirmed decreased white matter connectivity in PD patients, including but not limited to frontoparietal-striatal pathway (Tinaz et al., 2017), nigro-pallidal pathway (Barbagallo et al., 2017), and other white matter tracts linking brainstem such as the longitudinal fasciculus and the internal capsule (Haghshomar et al., 2018, Scherfler et al., 2011). Notably, these white matter damages could be a preceding sign of surrounding grey matter neuronal loss (Rektor et al., 2018). Therefore, it is intuitive to hypothesize the reorganization of brain structural network may occur in iRBD stage. Another study using graph-theory based analysis showed disrupted topological disorganization of the global brain network and hub reorganization in iRBD (Park et al., 2019). However, the intrinsic process of the human brain structural network evolving from iRBD to PD remains largely unknown.
On the other hand, studies exploring the brain network reorganization due to neurodegeneration have often been hampered by the subjective analytical design on manipulating massive brain connectivity, such as the seed-based strategies or within a restricted subnetwork. To avoid this bias, the connectome-wide association study (CWAS) paradigms allow one to systematically quantify topological properties of brain networks as a whole continuum (Milham, 2012, Shehzad et al., 2014). However, the conventional analytical strategies in CWAS, such as dimensionality reduction (Zhan et al., 2015) and graph analysis (Park et al., 2019), have been criticized for substantial local information loss (Poldrack et al., 2017). To address this issue, the multivariate distance matrix regression (MDMR) was recently introduced to statistically characterize the local pattern of brain connectome without sacrificing connectome dimension (Shehzad et al., 2014). This multivariate analytical framework allows to clarify the substrate of the specific structural connectivity patterns associated with various clinical phenotypes, such as anhedonia (Sharma et al., 2017), and psychosis-spectrum symptoms (Satterthwaite et al., 2015). Besides, our previous investigation by using MDMR also revealed widespread brain network reorganization caused by AD pathology (Ye et al., 2019), indicating that CWAS framework could open a new avenue to uncover the signature of brain structural network changes along with PD progression.
Here, we exploited the MDMR framework to provide insights into the relationship between brain connectome and PD progression from prodromal to clinical stage. In a cohort of 151 participants including iRBD, PD and age-matched normal controls (NC), the anatomical and diffusion MRIs were scanned for each individual in Xuanwu Hospital of Capital Medical University, Beijing, China. We anticipated that the signature of brain white matter structural connectivity would explicitly describe a trajectory of early PD. To evaluate its potential value in clinical use, we also adopted machine learning classification methods to examine the effectiveness of brain connectivity patterns revealed by the CWAS strategy.
2. Methods
2.1. Study participants and clinical assessment
The subjects in this study were recruited from the Movement Disorders Clinic of the Xuanwu Hospital of Capital medical University and were all approved by the Institutional Review Board of Xuanwu Hospital, including 61 normal controls (NC), 35 iRBDs and 55 PDs. All the participants were given written informed consent to the experiment. In order to remove the potential handedness effects, all the patients were all right-handed measured according to the Edinburgh Inventory (Tinelli et al., 2013). In particular, the iRBD patients were screened by the International Classification of Sleep Disorders-Third Edition (ICSD 3) diagnostic criteria and confirmed by polysomnography (Sateia, 2014). The NCs were all older than 40 years, with no family history of movement disorders and no obvious cerebral lesions found in MR images. The PDs were diagnosed according to the MDS Clinical Diagnostic Criteria for Parkinson's disease (Postuma et al., 2015b). Multiple assessments were applied to evaluate the iRBDs and PDs, such as the Rapid Eye Movement Sleep Behaviour Disorder Questionnaire–Hong Kong (RBDQ-HK), Pittsburgh sleep quality index (PSQI), Montreal Cognitive Assessment (MoCA), the Movement Disorder Society Unified Parkinson’s Disease Rating Scale, part III (UPDRS III) and Epworth sleepiness score (ESS). Table 1 summarizes the demographic details.
Table 1.
Characteristics of participants.
| NC n = 61 Mean (SD) |
iRBD n = 35 Mean (SD) |
PD n = 55 Mean (SD) |
F statistic | ANOVA p |
Post-hoc comparisons |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F statistic | NC VS iRBD adjusted p |
F statistic | iRBD VS PD adjusted p |
F statistic | NC VS PD adjusted p |
||||||
| Sex (M/F) b | 27/34 | 19/16 | 31/24 | 0.8532 | 0.428 | 0.432 | 0.513 | 0.161 | 0.689 | 1.69 | 0.196 |
| Age (years) a | 60.1 (10.1) | 61.5 (7.83) | 59.6 (7.98) | 0.4404 | 0.646 | 0.446 | 0.506 | 1.092 | 0.299 | 0.08 | 0.776 |
| Duration (years) a | – | 8.19 (8.53) | 6.64 (11.0) | – | – | – | – | 0.4387 | 0.5096 | – | – |
| UPDRS III a | – | 4.97 (4.56) | 26.16 (13.4) | – | – | – | – | 9.5332 | 0.002** | – | – |
| H&Y | – | – | 1.79 (0.74) | – | – | – | – | – | – | – | – |
| RBDQ-HK a | 9 (6.57) | 40.2 (17.4) | 21.3 (15.4) | 56.703 | <0.001*** | 150.2 | <0.001*** | 26.047 | <0.001*** | 31.99 | <0.001*** |
| MoCA a | 25.5 (2.91) | 24.2 (3.67) | 24.0 (3.88) | 3.108 | 0.047 * | 3.1568 | 0.079 | 0.088 | 0.767 | 5.8313 | 0.017** |
| ESS a | 4.61 (3.38) | 5.57 (4.79) | 6.6 (4.84) | 3.0267 | 0.051 | 1.1907 | 0.278 | 0.8159 | 0.369 | 6.4805 | 0.0123** |
| PSQI a | 4.2 (3.25) | 9.12 (5.75) | 6.74 (4.34) | 13.062 | <0.001*** | 25 | <0.001*** | 3.7161 | 0.0582 | 11.583 | <0.001*** |
Abbreviations: PD = Parkinson’s Desease; NC = Normal Control; iRBD = Idiopathic Rapid eye movement sleep Behavior Disorder; RBDQ-HK = Rapid Eye Movement Sleep Behavior Disorder Questionnaire–Hong Kong; PSQI = Pittsburgh sleep quality index; MoCA = Montreal Cognitive Assessment; ESS = Epworth Sleepiness Score.
Main effect of group in ANOVA.
Chi-squared test.
2.2. Imaging protocol
MR images were collected on a 3.0 T MR system (MAGNETOM Skyra, Siemens, Germany) including the axial diffusion scan with echo-planar imaging sequence, the T1 weighted scan with sagittal magnetization-prepared rapid gradient-echo sequence, as well as quantitative susceptibility mapping (QSM) scan with axial 3-D single-echo gradient echo sequence. The T1 weighted MRI data was acquired on with the following parameters: inversion time (TI) = 1100 ms; echo time (TE) = 2.98 ms; repetition time (TR) = 2530 ms; field of view (FOV) = 224 × 256 mm2; isotropic voxel size = 1 mm; flip angle = 7°. A spin echo EPI sequence was performed to acquire diffusion weighted images with the following parameters: TE, 105 ms; TR, 5000 ms; flip angle, 90°; isotropic voxel size, 2 mm; anterior to posterior phase encoding direction, 60 gradient directions with b = 1000/2000 s/mm2, and two additional b0 image were acquired with reversal of the acquisition direction along the phase-encode axis. A 3-D single-echo gradient echo sequence: slice thickness = 1.5 mm; TE = 17.5 ms; TR = 25 ms; echo number = 1; flip angle = 15°; display field of view (DFOV) = 256 × 256 × 80 pixels; voxel size = 0.667 × 0.667 × 1.5 mm3; scanning time = 306 s.
2.3. Image preprocessing and tractography based on diffusion MRI
The anatomical scans were segmented with a parcellation scheme with n = 87 regions. After bias filed correction, each individual T1 weighted image was processed by FreeSurfer V6 (Fischl, 2012) to separate all 77 cortical brain regions according to the Desikan-Killiany-Tourville (DKT) protocol. QSM reconstruction was performed using MATLAB-based susceptibility imaging software STI Suite (Li et al., 2014) (refer details to our previous work (Sun et al., 2020)). Given that QSM images exhibit better signal contrast on certain subcortical nuclei as compared to T1 weighted images (Deistung et al., 2017), ten subcortical regions of interest, including the substantia nigra (SN), the globus pallidus, the red nucleus, the caudate nucleus, and the putamen on both hemispheres, were manually drawn on the QSM images by an experienced neuroradiologist (J. S.). Given that the low intensity voxels of QSM where SN locates don't precisely coincide with voxels including pars compacta, we basically followed the previous robust principle (He et al., 2015) to maximally avoid bias. 1) SN was drawn on all successive axial slices where the boundary with hyperintensity signal were visible and clear; 2) the first and last layers, as well as the boundary voxels were excluded to avoid partial volume effects; 3) obvious vascular structures were removed. All these parcellation labels were further mapped to diffusion MRI space by linear image registration from the susceptibility weighted magnitude image to T1 weighted image using Advanced Normalization Tools (http://picsl.upenn.edu/software/ants/) (Avants et al., 2009), and served as nodes for brain connectivity extraction and brain structural network construction. All the MR images and parcellation results have been visually checked by the neuroradiologist (J. S.).
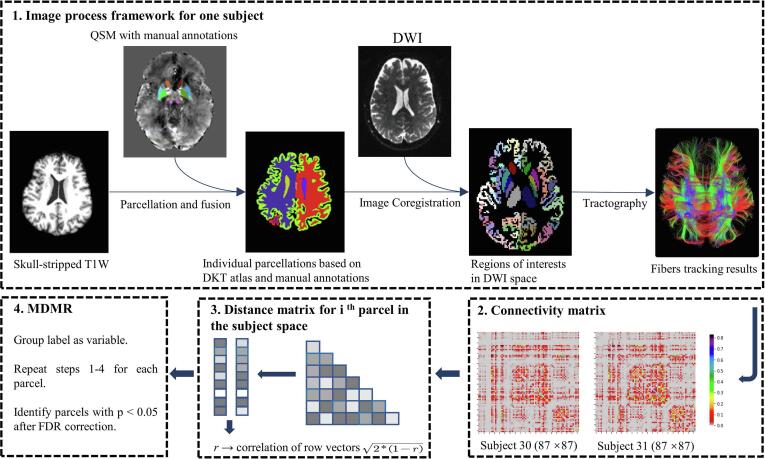
The preprocessing of diffusion weighted images includes image denoising (Veraart et al., 2016), head motion, eddy-current and susceptibility distortion correction (dwifslpreproc command), and field inhomogeneity correction (Tustison et al., 2010), which were all performed by MRtrix3 (www.mrtrix.org). After preprocessing, the fiber orientation distributions were estimated by constrained spherical deconvolution (Jeurissen et al., 2011, Tournier et al., 2008) with a maximum spherical harmonic degree lmax of 8. Then in conjunction with the anatomical-constrained tractography protocol (Smith et al., 2012), we performed the 2nd order Integration over Fiber Orientation Distributions (iFOD2) algorithm (Tournier et al., 2010) on each voxel to reconstruct ten million probabilistic streamlines within a brain mask, with the default parameters: FOD amplitude threshold = 0.05, step size = 0.5 × voxel size, maximum curvature per step = 45°. The probabilistic fiber tracking algorithm was adopted here because it exhibits higher anatomical reproducibility than other approaches in terms of connectivity calculation (Bonilha et al., 2015). Furthermore, Spherical-deconvolution Informed Filtering of Tractograms (SIFT) algorithm (Smith et al., 2013, Smith et al., 2015) was applied to reduce the overall streamline count into five million, meanwhile provides more biologically meaningful estimates of structural connection density and improve biological accuracy and reproducibility. From these data, a brain white matter structural connectivity matrix was derived for each subject, for which each element of this matrix represents the number of streamlines connecting the parcellated brain regions (Hermundstad et al., 2013). An overview of processing steps is given in Fig. 1.
Fig. 1.
Schematic flowchart of multivariate distance matrix regression analysis based on the process framework, including 3D T1 weighted images parcellation, tractography of diffusion tensor images and registration. DWI: Diffusion weighted imaging; FDR, false discovery rate; DKT, Desikan–Killiany–Tourville atlas.
2.4. Multivariate distance matrix regression
MDMR was applied here to test the variation of distance in brain structural connectivity patterns among different cohorts. First, a distance matrix in the subject space was calculated for each region. Within each distance matrix, the distance between connectivity patterns for every possible subject pair among all groups related to region was calculated by
where represents the Pearson correlation coefficient between connectivity vectors of subject and . The connectivity vector, referring to the connection of a brain region to the rest 86 regions, demonstrates the connectivity strength from the given to others.
The MDMR could exhibit excellent test level accuracy, yielding a pseudo F-statistic analogous to an F-statistic from a standard ANOVA model (Zalesky et al., 2010). Specifically, the total sum of squares for region was calculated by
where , the total number of subjects. Meanwhile, the within group sum of squares was calculated by
where and denote the number of each group respectively. represents the belonging of the subject and , which equals to one when and within the same group. And then the F-statistic score of the region would be obtained by
A random permutation was applied to subjects with 1000 times to simulate the null distribution and the pseudo F-statistic score was re-computed each time. Finally, p value was calculated by counting the pseudo F-statistics from permutated values greater than those derived from the original data. Age and sex were incorporated in this model as covariates. Similarly, the procedure was repeated for all the brain regions. False discovery rate correction was further applied to control the false positive rate. Essentially, MDMR could discriminate each voxel whether patterns of whole brain connectivity would be more similar in individuals with similar phenotypes than those with dissimilar phenotypes (Ye et al., 2019). Additionally, p < 0.05 after false discovery rates correction was determined significance within the above experiments.
2.5. Post-hoc analysis
To explicitly measure which specific connectivity pattern is primarily driving the association between brain network changes and disease progression, the alteration in connectivity pattern within these pair connections would be also examined. Specially, the -statistic was applied here to identify the top five connected network nodes with the greatest effect size from each seed brain region among the three subject groups (McArtor et al., 2017). The alterations of the connectivity strengths are further examined in those pairwise connections. In other words, the post-hoc analysis was conducted descriptively, according to those seed regions selected based on the significance from MDMR results.
2.6. Classification performance
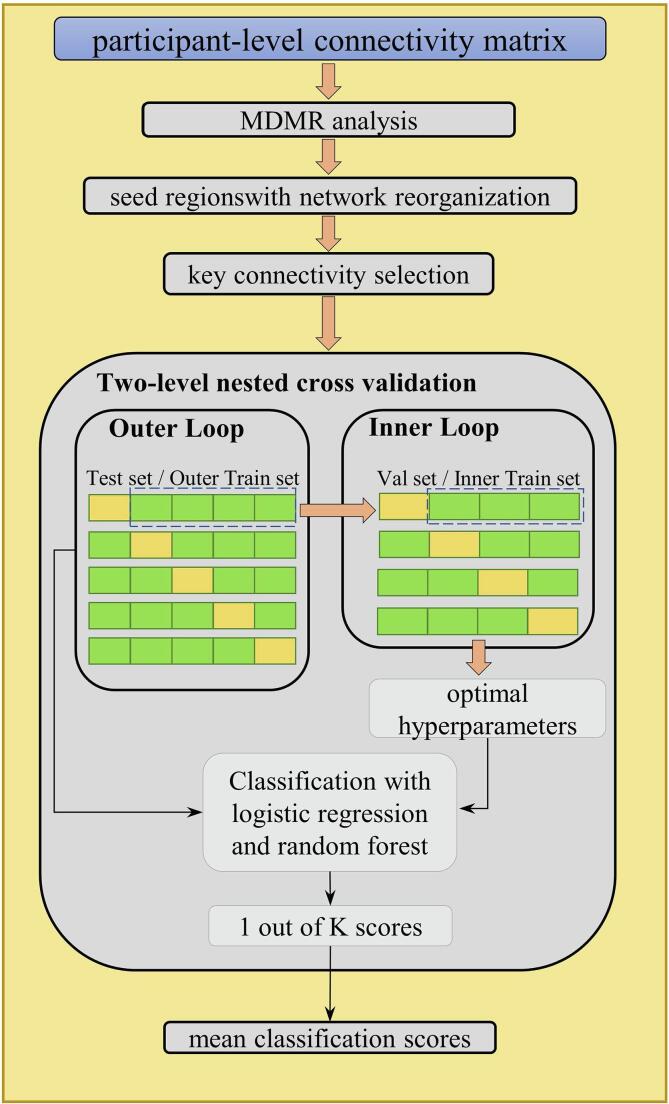
Aiming to examine the discriminative ability of the key structural connectivity returned by MDMR and post-hoc analysis, we conducted the supervised learning experiment to classify the different subject groups. Briefly, we applied two classification models (i.e. logistic regression with L2 regularization and random forest) to detect iRBD or PD patients based on the connectivity features with the greatest effect size. A two-level nested cross validation was implemented to reduce potential overfitting, as illustrated in Fig. 2. In this nested cross-validation design, the outer cross validation holds out 20% of the samples as test set within 5 folds and an additional inner 4-fold cross validation was performed on the remaining 80% samples to select the optimal model by hyper-parameter grid searching. This process was repeated 5 times with leaving 20% samples out and finally the classification performance was measured from all the samples. The two-level nested cross validation strategy helps to reduce the bias and gives an error estimate which is very close to the testing set. Dichotomous classification was applied for NC vs. RBD, and for NC vs. PD. In contrast, we also tested the discriminative performance of the whole brain pairwise connectivity features (87 × 86 / 2 = 3741 pairs), and of the features extracted from network-based statistic (NBS) (Zalesky et al., 2010). The NBS algorithm was first proposed to exploit the extent to which the connections comprising the contrast or effect of interest are interconnected, and reflects the advanced superiority of data-driven statistical method. Given that the NBS parameter threshold is difficult to choose, we empirically determined this value to make the number of significant connectivity from NBS approximate the number returned from MDMR analysis. The classification performance was evaluated in terms of sensitivity, specificity, accuracy, as well as the area under curve of receiver operating characteristic on the test set. All the statistical analysis was performed in R (https://www.r-project.org). The classification model was built and evaluated with python package scikit-learn (Pedregosa et al., 2011).
Fig. 2.
The supervised learning pipeline with nested cross validation design, where logistic regression and random forest are implemented for distinguishing iRBD and PD.
3. Results
3.1. Demographic and clinical information
The demographic characteristics of the participants in this study was shown in Table 1. No significant group differences were found in the sex (p = 0.351) and age (p = 0.646). Both iRBD and PD patients showed significantly worse sleep quality than NC, reflected by RBDQ-HK and PSQ (p < 0.001). MoCA assessment also demonstrated PD patients with significantly worse cognitive function compared with NC variations (p < 0.001).
3.2. Brain regions with altered connectivity
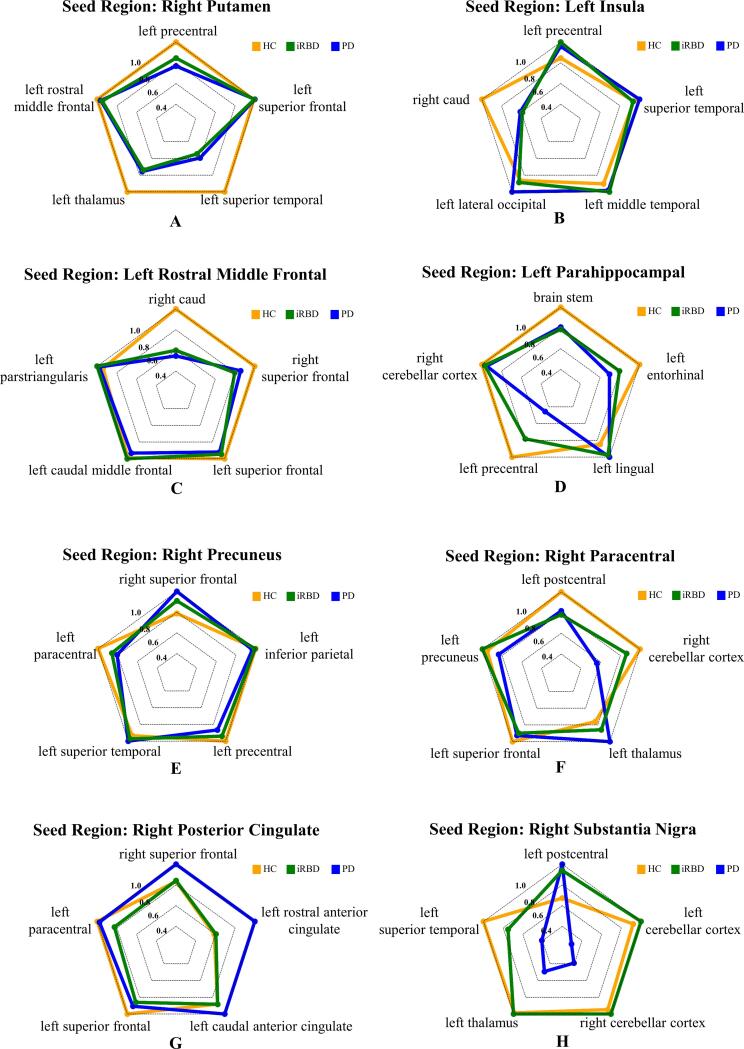
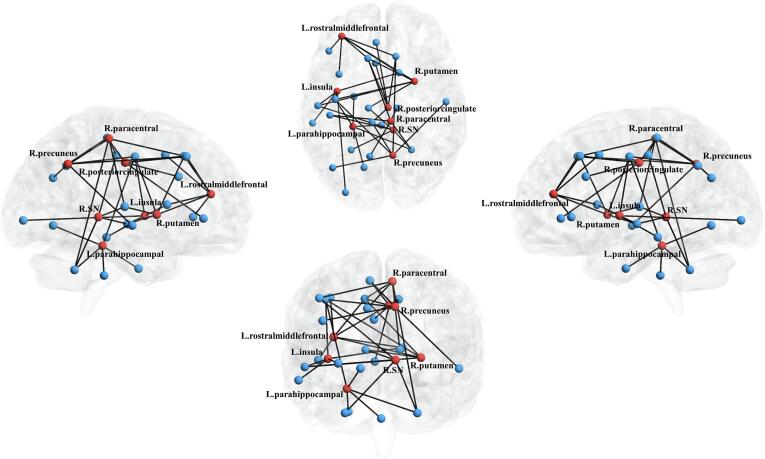
Eight brain seed regions with significant network reorganizations were observed from the MDMR analysis, including the right putamen, the right substantia nigra, the left parahippocampal gyrus, the left rostral middle frontal cortex, the left insula, the right paracentral, the right posterior cingulate, and the right precuneus (see Table S1 for statistical results of all brain regions). We further empirically selected five pair-wise connectivity with greatest effect size for each seed region as the key structural connectivity. Radar charts in Fig. 3 characterize the comprehensive connectivity patterns for each seed region, where each axis represents scaled connectivity strength. In Fig. 4, seed regions with significant network reorganizations were visualized in red, and the linked key connectivity were displayed in black. Based on the above findings, the brain connectivity signature associated with PD progression can be integrated into three types: Type A) remarkably disrupted connectivity in both iRBD and PD (the right putamen, the left rostral middle frontal cortex and the left insula); Type B) connectivity incrementally decreasing from iRBD to PD (the left parahippocampal gyrus, the right precuneus, and the right paracentral lobule); and Type C) preserved connectivity in iRBD and remarkably disrupted connectivity in PD (the right substantia nigra and the right posterior cingulate). Note that this final determination of seed regions clustering mainly depends on the reshaping pattern of the most typical key connectivity, rather than all key connectivity for each seed region (please refer to the supplementary univariate test shown in Table S2 and Fig. S2). For comparison, we also calculated the brain structural connectivity with significant groupwise difference based on NBS, as shown in Fig. S1.
Fig. 3.
Post-hoc analysis of connectivity patterns for the seed regions subsequent to multivariate distance matrix regression (MDMR). Eight nodes with significant differences in connectivity patterns in the three-group comparison were selected for the post-hoc analysis. For each seed region returned by MDMR, the top five connections with the greatest effect size are represented as axes in the radar chart.
Fig. 4.
Comprehensive connectivity patterns for all seed regions identified by the post-hoc analysis. Network nodes representing each brain region selected in this paper are shown, where seed regions with significant difference by MDMR and post-hoc analysis are represented in red and others in blue. The edges with top five greatest connectivity strength for each seed region are displayed in black. SN: substantia nigra; L, left; R, right. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Evaluation of connectivity feature numbers
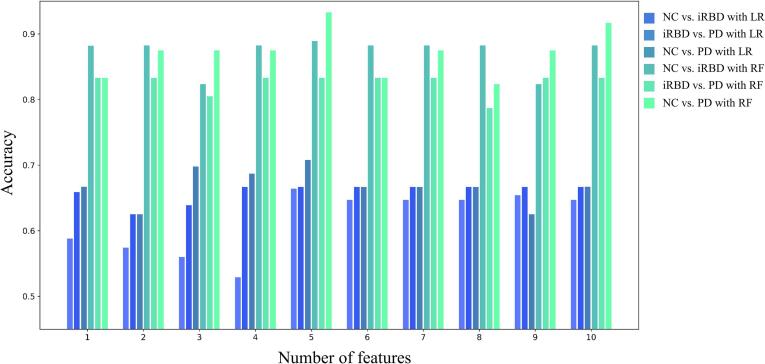
In order to evaluate the optimal number of key connectivity (features) related to each seed region, we compared the corresponding classification accuracy based on various numbers of features from n = 1 to n = 10. The bar plot in Fig. 5 confirmed that key connectivity with n = 5 could achieve optimal classification accuracy across different group comparisons and different classification strategies. Therefore, the key connectivity regarding all eight seed regions (5 × 8 = 40) for each participant were collected to constitute the full feature set for the following evaluation.
Fig. 5.
Classification performance with the number of features, which were selected from the regions with the greatest effect size.
3.4. Classification performance
To comprehensively evaluate the disease discriminating capability underlying the key structural connectivity patterns, the classification performance based on machine learning was compared among the three different feature selection approaches (i.e. whole brain connectivity, connectivity pairs returned from MDMR and NBS). As demonstrated in Fig. 6, MDMR with random forest achieved optimal accuracy performance in discriminating NC and PD (accuracy = 0.933), compared to the other two approaches (whole brain: accuracy (RF) = 0.833, accuracy (LR) = 0.658; NBS: accuracy (RF) = 0.875, accuracy (LR) = 0.683). As expected, the random forest method prevails over the logistic regression in terms of accuracy for all feature selection approaches, probably due to advantage of ensemble strategy. Even for prodromal stage, the accuracy of discrimination between iRBDs and NCs was 0.833 (sensitivity = 0.886; specificity = 0.815), suggesting that the MDMR analysis could offer effective brain feature sets for PD detection and early warning (see Table 2).
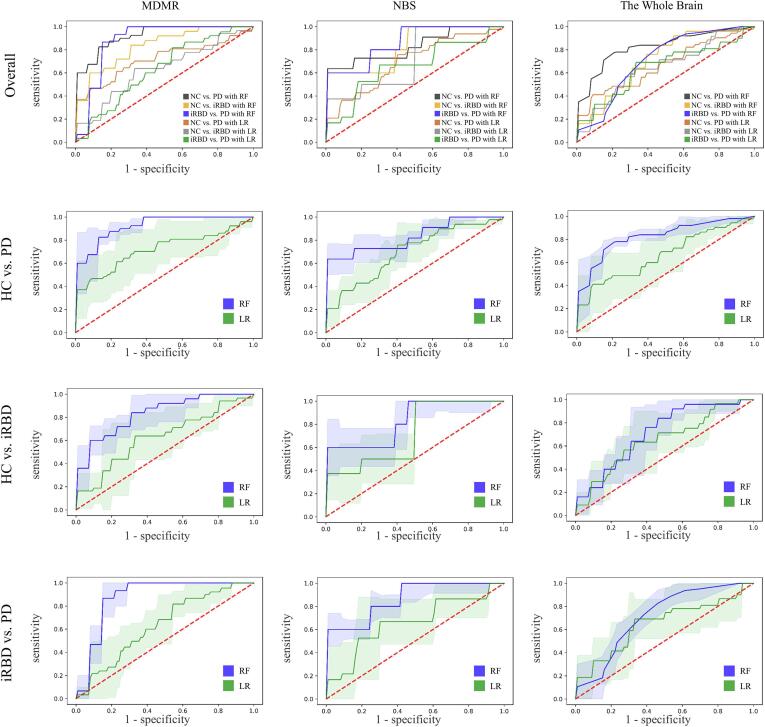
Fig. 6.
Receiver operating characteristic (ROC) curves show the brain connectivity patterns returned from MDMR (first column), NBS (second column), whole brain (third column) briefly in the first row, distinguishing PD from NC (second row), iRBD from NC (third row) and PD from iRBD by random forest (blue line) and logistic regression (green line) in detail. The shaded areas represent the confidence intervals of the ROC curves. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Classification performance between groups (NC vs. iRBD and NC vs. PD) with two classification algorithm, logistic regression, and random forest. ACC = accuracy; SEN = sensibility; SPE = specificity, MDMR = multivariate distance matrix regression; NBS = network based statistics.
| Connectivity features | MDMR |
NBS |
Whole Brain |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NC vs. PD | NC vs. iRBD | iRBD vs. PD | NC vs. PD | NC vs. iRBD | iRBD vs. PD | NC vs. PD | NC vs. iRBD | iRBD vs. PD | ||
| Logistic Regression | ACC | 0.708 | 0.667 | 0.667 | 0.683 | 0.724 | 0.635 | 0.658 | 0.625 | 0.588 |
| SEN | 0.750 | 0.667 | 0.701 | 0.634 | 0.725 | 0.548 | 0.607 | 0.705 | 0.550 | |
| SPE | 0.667 | 0.527 | 0.519 | 0.772 | 0.650 | 0.646 | 0.738 | 0.551 | 0.625 | |
| Random Forest | ACC | 0.933 | 0.833 | 0.889 | 0.875 | 0.833 | 0.824 | 0.833 | 0.778 | 0.824 |
| SEN | 0.967 | 0.886 | 0.925 | 0.917 | 0.875 | 0.869 | 0.857 | 0.765 | 0.812 | |
| SPE | 0.917 | 0.815 | 0.920 | 0.833 | 0.750 | 0.900 | 0.800 | 0.667 | 0.900 | |
4. Discussion
Increasing evidences suggest iRBD could be one of the initial manifestation and a prodromal symptom of PD (Fereshtehnejad et al., 2015, Zhang et al., 2017). In this study, we endeavoured to quantitatively characterize PD progression in early and late stages. To this end, we used diffusion MRI to establish individual-based brain structural network in NC, iRBD and PD groups, and performed multiple data-driven multivariate statistical approaches to identify potential brain connectivity signatures related to PD pathology. In our findings, while extensive decreased cortico-striatal structural connectivity was observed in iRBD patients, the incremental alterations of connectivity between cortical regions were associated with the progression of PD. More importantly, these connectivity features underlying this PD-related network reorganization demonstrated excellent performance to distinguish iRBD and PD from NC. In the following discussion, we first summarize the PD-related structural connectivity patterns.
Briefly, three distinct types of structural network reorganization patterns in PD progression were revealed related to PD progression. The first type is the disrupted cortico-striatal connectivity observed in both iRBD and PD (namely Type A connectivity pattern), including the ipsilateral putamen-temporal connectivity (see Fig. 3A), the contralateral caudate-frontal connectivity (see Fig. 3B), and the contralateral caudate-insula connectivity (see Fig. 3C). Consistently, post mortem and nuclear imaging studies have reported that the nigro-striatal dopamine depletion, one hallmark of PD, will trigger decreased connectivity between the striatum and the telencephalon (Brooks et al., 1990, Brück et al., 2006, Guttman et al., 1997, Kish et al., 1988, Nurmi et al., 2001). Given pathological evidences showing that prominent motor symptoms would not emerge until about half dopaminergic nerves were irreversibly damaged (Fearnley and Lees, 1991), it’s no surprise to observe early decrease of cortico-striatal structural connectivity in prodromal PD stage. Congruently, compensatory reactivation due to dopaminergic deafferentation was also confirmed in brain functional network in patients with RBD (Rolinski et al., 2016). Taken together, our findings comprehensively reflected extensive decreased cortico-striatal structural connectivity occurred in iRBD patients, which may further induce significant motor symptoms in PD patients.
Besides, the incremental alterations of connectivity between cortical regions along with the progression of PD were also extensively observed in our study, including connectivity related to the parahippocampus (see Fig. 3D), the precuneus (see Fig. 3E) and the paracentral lobule (see Fig. 3F). This connectivity pattern (namely type B connectivity pattern) supports the hypothesis that patients with iRBD are on the path to synucleinopathy development (Bauckneht et al., 2018). Previous studies have found that extensive patterns of reduced connectivity in PD were observed within and between the temporal, parietal and occipital areas (Shah et al., 2017a) and precuneus degeneration might be related to this non-motor symptom in PD (Shin et al., 2017). The gradually decreased connectivity linked to paracentral lobule has also been reported on PD patients in previous studies (Hepp et al., 2017) (Kim et al., 2014).
Contrary to our hypothesis, the contralateral cingulate connectivity (see Fig. 3G) and the SN-related connectivity (see Fig. 3H) significantly reconfigured on clinical PD stage, as compared to preserved connectivity strength in prodromal stage (namely type C connectivity pattern). Although the interpretation of the significant increased contralateral cingulate connectivity needs more evidence, many observations regarding structural compensation of commissural fibers in later stage have been reported (see literature review in (Sanjari Moghaddam et al., 2020)), possibly due to improved interhemispheric information transfer to counterbalance pathological changes in one hemisphere. As a hallmark brain region of PD, SN has been widely reported in terms of fiber damages in clinical stage (Kamagata et al., 2018, Menke et al., 2009, Shah et al., 2017). However, no MRI study has ever investigated SN-related structural connectivity integrity in iRBD patients. Although our previous study reported excessive iron deposition accumulated in the SN at the iRBD stage (Sun et al., 2020), the related connectivity in iRBD remains controversial. While a previous study with a limited sample size of participants found that the brain functional connectivity of iRBD was different from that of NC and PD, which shows the progression from iRBD to PD involves increases and decreases in different nigra subcortical-cortical connections correlations (Ellmore et al., 2013), another recent study reported no significant difference in functional connectivity between iRBD and PD (Rolinski et al., 2016). Overall, these findings imply that the SN-related structural connectivity is more likely to be relatively intact at prodromal PD and altered after prodromal PD. Longitudinal evidence is further needed to test this hypothesis.
Furthermore, the brain dysconnectivity features identified by MDMR demonstrated remarkably performance in detecting iRBD and PD from NC (NC vs. iRBD accuracy: 83.3%, NC vs PD accuracy: 93.3%), compared with the connectivity features detected by the conventional NBS approach (NC vs. iRBD accuracy: 83.3%, NC vs PD accuracy: 87.5%), or from the whole brain (NC vs. iRBD accuracy:77.8%, NC vs PD accuracy: 83.3%). MDMR evidently exhibits excellent sensitivity and specificity to identify the driving components that leads to brain network reorganization, and to localize the brain connectivity signature unique to particular disease states in a data-driven manner. Overall, our findings coincide with the recent paradigm shift in investigating etiological mechanism of PD from regional abnormalities to an integrated outlook. Studies have shown that the misfolding of α-synuclein propagation occurs along the structural connection network of the human brain (Zeighami et al., 2015, Zheng et al., 2019). Therefore, clarifying the nature of structural connectivity signature of PD progression is crucial for searching new intervention targets to alter disease trajectory in early PD patients.
This study was not without limitations. First, the optimal fiber tracking method in structural brain network construction is still controversial, for regions with more frequent fiber disposition are more likely to obtain potential errors during fiber tract reconstruction. To maximally avoid such effects, anatomical-constrained tractography method for tractography estimation with anatomical priors was performed to improve the biological plausibility of the generated streamlines (Smith et al., 2012). Second, it’s challenging to biologically interpret the substrate underlying the strength changes of brain structural connectivity. The multi-shell high spatial and angular diffusion MR data combined with anatomical-specific post-processing analysis, such as voxel-based analysis (Mito et al., 2018), can possibly explain whether the degeneration of WM tracts in iRBD and PD patients is mainly due to axonal loss or demyelination. Finally, the sample size of our study was relatively small. Although nested cross validation strategy has been implemented to overcome overfitting, larger cohorts are needed to validate the observed brain network signature related to PD progression.
In conclusion, we established the brain structural network of iRBD and PD using diffusion MRI data, and identified unique patterns of network reorganization along with the progression of PD. Three types of brain structural connectivity reorganizations of PD progression were detected in eight brain regions. These progressive alterations support the hypothesis that iRBD patients are on the path to developing a synucleinopathy (Bauckneht et al., 2018). The extracted specific structural connectivity patterns associated with various clinical phenotypes obtained high accuracy performance to predict iRBD and PD. Cheap, safe and reliable means of identifying iRBD with the highest risk of PD will promote the targeted use of novel disease modification therapies and revolutionize clinical trials in this field.
5. Contributors
YY, and CY did statistical analyses, and drafted the manuscript and figures. JS collected and supervised data collection. YY, CY, TW and TM conceived and supervised the study. HL, and LL collected and cleaned data.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This study is supported by the National Key Research and Development Program of China (2018YFC1312000) and the National Nature Science Foundation of China (82071423). The funding sources of this study did not play any role in the study design, the collection, analysis, and interpretation of the data, in the writing of the manuscript and in the decision to submit the paper for publication.
Data sharing statement
This database is available from the authors upon request. Contact: wutao69@gmail.com. The code for the computational pipeline is publicly available at https://github.com/podismine/MDMR-based-Brain-Connectome-Analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2021.102715.
Contributor Information
Ting Ma, Email: tma@hit.edu.cn.
Tao Wu, Email: wutao69@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Avants B.B., Tustison N., Song G. Advanced normalization tools (ANTS) Insight J. 2009;2(365):1–35. [Google Scholar]
- Barbagallo G., Caligiuri M.E., Arabia G., Cherubini A., Lupo A., Nisticò R., Salsone M., Novellino F., Morelli M., Cascini G.L., Galea D., Quattrone A. Structural connectivity differences in motor network between tremor-dominant and nontremor Parkinson's disease. Hum. Brain Mapp. 2017;38(9):4716–4729. doi: 10.1002/hbm.23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauckneht M., Chincarini A., De Carli F., Terzaghi M., Morbelli S., Nobili F., Arnaldi D. Presynaptic dopaminergic neuroimaging in REM sleep behavior disorder: A systematic review and meta-analysis. Sleep Med. Rev. 2018;41:266–274. doi: 10.1016/j.smrv.2018.04.001. [DOI] [PubMed] [Google Scholar]
- Boeve B.F. REM sleep behavior disorder: updated review of the core features, the REM sleep behavior disorder-neurodegenerative disease association, evolving concepts, controversies, and future directions. Ann. N. Y. Acad. Sci. 2010;1184(1):15–54. doi: 10.1111/j.1749-6632.2009.05115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L., Gleichgerrcht E., Fridriksson J., Rorden C., Breedlove J.L., Nesland T., Paulus W., Helms G., Focke N.K., Hayasaka S. Reproducibility of the structural brain connectome derived from diffusion tensor imaging. PLoS One. 2015;10(9):e0135247. doi: 10.1371/journal.pone.0135247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H., Tredici K.D., Rüb U., de Vos R.A.I., Jansen Steur E.N.H., Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging. 2003;24(2):197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Braak H., Ghebremedhin E., Rüb U., Bratzke H., Del Tredici K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004;318(1):121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- Brooks D.J., Ibanez V., Sawle G.V., Quinn N., Lees A.J., Mathias C.J., Bannister R., Marsden C.D., Frackowiak R.S.J. Differing patterns of striatal 18F-dopa uptake in Parkinson's disease, multiple system atrophy, and progressive supranuclear palsy. Ann. Neurol.: Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 1990;28(4):547–555. doi: 10.1002/ana.410280412. [DOI] [PubMed] [Google Scholar]
- Brück A., Aalto S., Nurmi E., Vahlberg T., Bergman J., Rinne J.O. Striatal subregional 6-[18F] fluoro-L-dopa uptake in early Parkinson's disease: A two-year follow-up study. Movement Disord.: Off. J. Movement Disorder Soc. 2006;21(7):958–963. doi: 10.1002/mds.20855. [DOI] [PubMed] [Google Scholar]
- Burke R.E., O'Malley K. Axon degeneration in Parkinson's disease. Exp. Neurol. 2013;246:72–83. doi: 10.1016/j.expneurol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deistung A., Schweser F., Reichenbach J.R. Overview of quantitative susceptibility mapping. NMR Biomed. 2017;30(4) doi: 10.1002/nbm.3569. [DOI] [PubMed] [Google Scholar]
- Ellmore T.M., Castriotta R.J., Hendley K.L., Aalbers B.M., Furr-Stimming E., Hood A.J., Schiess M.C. Altered nigrostriatal and nigrocortical functional connectivity in rapid eye movement sleep behavior disorder. Sleep. 2013;36(12):1885–1892. doi: 10.5665/sleep.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnley J.M., Lees A.J. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain. 1991;114(Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- Fereshtehnejad S.-M., Romenets S.R., Anang J.B., Latreille V., Gagnon J.-F., Postuma R.B. New clinical subtypes of Parkinson disease and their longitudinal progression: a prospective cohort comparison with other phenotypes. JAMA neurology. 2015;72(8):863–873. doi: 10.1001/jamaneurol.2015.0703. [DOI] [PubMed] [Google Scholar]
- Fereshtehnejad S.-M., Montplaisir J.Y., Pelletier A., Gagnon J.-F., Berg D., Postuma R.B. Validation of the MDS research criteria for prodromal Parkinson's disease: longitudinal assessment in a REM sleep behavior disorder (RBD) cohort. Mov. Disord. 2017;32(6):865–873. doi: 10.1002/mds.26989. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Burkholder J., Kish S.J., Hussey D., Wilson A., DaSilva J., Houle S. [11C] RTI-32 PET studies of the dopamine transporter in early dopa-naive Parkinson's disease: implications for the symptomatic threshold. Neurology. 1997;48(6):1578–1583. doi: 10.1212/wnl.48.6.1578. [DOI] [PubMed] [Google Scholar]
- Haghshomar M., Dolatshahi M., Ghazi Sherbaf F., Sanjari Moghaddam H., Shirin Shandiz M., Aarabi M.H. Disruption of Inferior Longitudinal Fasciculus Microstructure in Parkinson's Disease: A Systematic Review of Diffusion Tensor Imaging Studies. Front. Neurol. 2018;9:598. doi: 10.3389/fneur.2018.00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N., Ling H., Ding B., Huang J., Zhang Y., Zhang Z., Liu C., Chen K., Yan F. Region-specific disturbed iron distribution in early idiopathic Parkinson's disease measured by quantitative susceptibility mapping. Hum. Brain Mapp. 2015;36(11):4407–4420. doi: 10.1002/hbm.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel S., Berg D., Gasser T., Chen H., Yao C., Postuma R.B., Disease M.T.F. Update of the MDS research criteria for prodromal Parkinson's disease. Mov. Disord. 2019;34(10):1464–1470. doi: 10.1002/mds.27802. [DOI] [PubMed] [Google Scholar]
- Hepp D.H., Foncke E.M.J., Olde Dubbelink K.T.E., van de Berg W.D.J., Berendse H.W., Schoonheim M.M. Loss of functional connectivity in patients with Parkinson disease and visual hallucinations. Radiology. 2017;285(3):896–903. doi: 10.1148/radiol.2017170438. [DOI] [PubMed] [Google Scholar]
- Hermundstad A.M., Bassett D.S., Brown K.S., Aminoff E.M., Clewett D., Freeman S., Frithsen A., Johnson A., Tipper C.M., Miller M.B., Grafton S.T., Carlson J.M. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proc. Natl. Acad. Sci. U.S.A. 2013;110(15):6169–6174. doi: 10.1073/pnas.1219562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo Alex, Fernández-Arcos Ana, Tolosa Eduard, Serradell Mónica, Molinuevo José Luis, Valldeoriola Francesc, Gelpi Ellen, Vilaseca Isabel, Sánchez-Valle Raquel, Lladó Albert, Gaig Carles, Santamaría Joan, Toft Mathias. Neurodegenerative disorder risk in idiopathic REM sleep behavior disorder: study in 174 patients. PLoS One. 2014;9(2):e89741. doi: 10.1371/journal.pone.008974110.1371/journal.pone.0089741.g00110.1371/journal.pone.0089741.g00210.1371/journal.pone.0089741.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo Alex, Molinuevo José Luis, Santamaría Joan, Serradell Mónica, Martí María José, Valldeoriola Francesc, Tolosa Eduard. Rapid-eye-movement sleep behaviour disorder as an early marker for a neurodegenerative disorder: a descriptive study. Lancet Neurol. 2006;5(7):572–577. doi: 10.1016/S1474-4422(06)70476-8. [DOI] [PubMed] [Google Scholar]
- Iranzo Alex, Tolosa Eduard, Gelpi Ellen, Molinuevo José Luis, Valldeoriola Francesc, Serradell Mónica, Sanchez-Valle Raquel, Vilaseca Isabel, Lomeña Francisco, Vilas Dolores, LLadó Albert, Gaig Carles, Santamaria Joan. Neurodegenerative disease status and post-mortem pathology in idiopathic rapid-eye-movement sleep behaviour disorder: an observational cohort study. Lancet Neurol. 2013;12(5):443–453. doi: 10.1016/S1474-4422(13)70056-5. [DOI] [PubMed] [Google Scholar]
- Jeurissen B., Leemans A., Jones D.K., Tournier J.D., Sijbers J. Probabilistic fiber tracking using the residual bootstrap with constrained spherical deconvolution. Hum. Brain Mapp. 2011;32(3):461–479. doi: 10.1002/hbm.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamagata Koji, Zalesky Andrew, Hatano Taku, Di Biase Maria Angelique, El Samad Omar, Saiki Shinji, Shimoji Keigo, Kumamaru Kanako K., Kamiya Kouhei, Hori Masaaki, Hattori Nobutaka, Aoki Shigeki, Pantelis Christos. Connectome analysis with diffusion MRI in idiopathic Parkinson's disease: Evaluation using multi-shell, multi-tissue, constrained spherical deconvolution. Neuroimage Clin. 2018;17:518–529. doi: 10.1016/j.nicl.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Ji Sun, Yang Jin-ju, Lee Jong-min, Youn Jinyoung, Kim Ju-min, Cho Jin Whan. Topographic pattern of cortical thinning with consideration of motor laterality in Parkinson disease. Parkinsonism Related Disord. 2014;20(11):1186–1190. doi: 10.1016/j.parkreldis.2014.08.021. [DOI] [PubMed] [Google Scholar]
- Kish Stephen J., Shannak Kathleen, Hornykiewicz Oleh. Uneven pattern of dopamine loss in the striatum of patients with idiopathic Parkinson's disease. N. Engl. J. Med. 1988;318(14):876–880. doi: 10.1056/NEJM198804073181402. [DOI] [PubMed] [Google Scholar]
- Li W., Wu B., Liu C. STI Suite: a Software Package for Quantitative Susceptibility Imaging. Proc. Intl. Soc. Mg. Reson. Med. 2014:22. [Google Scholar]
- McArtor Daniel B., Lubke Gitta H., Bergeman C.S. Extending multivariate distance matrix regression with an effect size measure and the asymptotic null distribution of the test statistic. Psychometrika. 2017;82(4):1052–1077. doi: 10.1007/s11336-016-9527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke R.A., Scholz J., Miller K.L., Deoni S., Jbabdi S., Matthews P.M., Zarei M. MRI characteristics of the substantia nigra in Parkinson's disease: a combined quantitative T1 and DTI study. Neuroimage. 2009;47(2):435–441. doi: 10.1016/j.neuroimage.2009.05.017. [DOI] [PubMed] [Google Scholar]
- Milham Michael Peter. Open neuroscience solutions for the connectome-wide association era. Neuron. 2012;73(2):214–218. doi: 10.1016/j.neuron.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Mito R., Raffelt D., Dhollander T., Vaughan D.N., Tournier J.D., Salvado O., Connelly A. Fibre-specific white matter reductions in Alzheimer's disease and mild cognitive impairment. Brain. 2018;141(3):888–902. doi: 10.1093/brain/awx355. [DOI] [PubMed] [Google Scholar]
- Nurmi Elina, Ruottinen Hanna M., Bergman Jrgen, Haaparanta Merja, Solin Olof, Sonninen Pirkko, Rinne Juha O. Rate of progression in Parkinson's disease: a 6-[18F] fluoro-L-dopa PET study. Movement Disord.: Off. J. Movement Disord. Soc. 2001;16(4):608–615. doi: 10.1002/mds.1139. [DOI] [PubMed] [Google Scholar]
- O'malley K.L. The role of axonopathy in Parkinson's disease. Exp. Neurobiol. 2010;19(3):115–119. doi: 10.5607/en.2010.19.3.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Kang Min, Lee Ho-Joon, Lee Byung In, Kim Sung Eun. Alterations of the brain network in idiopathic rapid eye movement sleep behavior disorder: structural connectivity analysis. Sleep Breathing. 2019;23(2):587–593. doi: 10.1007/s11325-018-1737-0. [DOI] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Dubourg V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- Poldrack Russell A., Baker Chris I., Durnez Joke, Gorgolewski Krzysztof J., Matthews Paul M., Munafò Marcus R., Nichols Thomas E., Poline Jean-Baptiste, Vul Edward, Yarkoni Tal. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci. 2017;18(2):115–126. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postuma R.B., Berg D., Stern M., Poewe W., Olanow C.W., Oertel W., Lang A.E. MDS clinical diagnostic criteria for Parkinson's disease. Mov. Disord. 2015;30(12):1591–1601. doi: 10.1002/mds.26424. [DOI] [PubMed] [Google Scholar]
- Postuma R.B., Iranzo A., Hogl B., Arnulf I., Ferini-Strambi L., Manni R., Montplaisir J.Y. Risk factors for neurodegeneration in idiopathic rapid eye movement sleep behavior disorder: A multicenter study. Ann. Neurol. 2015;77(5):830–839. doi: 10.1002/ana.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rektor Ivan, Svátková Alena, Vojtíšek Lubomir, Zikmundová Iva, Vaníček Jirí, Király András, Szabó Nikoletta, Najbauer Joseph. White matter alterations in Parkinson's disease with normal cognition precede grey matter atrophy. PLoS One. 2018;13(1):e0187939. doi: 10.1371/journal.pone.0187939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolinski Michal, Griffanti Ludovica, Piccini Paola, Roussakis Andreas A., Szewczyk-Krolikowski Konrad, Menke Ricarda A., Quinnell Timothy, Zaiwalla Zenobia, Klein Johannes C., Mackay Clare E., Hu Michele T.M. Basal ganglia dysfunction in idiopathic REM sleep behaviour disorder parallels that in early Parkinson's disease. Brain. 2016;139(8):2224–2234. doi: 10.1093/brain/aww124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjari Moghaddam Hossein, Dolatshahi Mahsa, Mohebi Farnam, Aarabi Mohammad Hadi. Structural white matter alterations as compensatory mechanisms in Parkinson's disease: A systematic review of diffusion tensor imaging studies. J. Neurosci. Res. 2020;98(7):1398–1416. doi: 10.1002/jnr.v98.710.1002/jnr.24617. [DOI] [PubMed] [Google Scholar]
- Sateia M.J. International classification of sleep disorders. Chest. 2014;146(5):1387–1394. doi: 10.1378/chest.14-0970. [DOI] [PubMed] [Google Scholar]
- Satterthwaite T.D., Vandekar S.N., Wolf D.H., Bassett D.S., Ruparel K., Shehzad Z., Gennatas E.D. Connectome-wide network analysis of youth with Psychosis-Spectrum symptoms. Mol. Psychiatry. 2015;20(12):1508–1515. doi: 10.1038/mp.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter M., Grigoletto J., Abd-Elhadi S., Glickstein H., Friedman A., Serrano G.E., Sharon R. A role for alpha-Synuclein in axon growth and its implications in corticostriatal glutamatergic plasticity in Parkinson's disease. Mol. Neurodegener. 2020;15(1):24. doi: 10.1186/s13024-020-00370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck C.H., Boeve B.F., Mahowald M.W. Delayed emergence of a parkinsonian disorder or dementia in 81% of older men initially diagnosed with idiopathic rapid eye movement sleep behavior disorder: a 16-year update on a previously reported series. Sleep Med. 2013;14(8):744–748. doi: 10.1016/j.sleep.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Schenck C.H., Bundlie S.R., Mahowald M.W. Delayed emergence of a parkinsonian disorder in 38% of 29 older men initially diagnosed with idiopathic rapid eye movement sleep behavior disorder. Neurology. 1996;46(2):388–393. doi: 10.1212/wnl.46.2.388. [DOI] [PubMed] [Google Scholar]
- Scherfler Christoph, Frauscher Birgit, Schocke Michael, Iranzo Alex, Gschliesser Viola, Seppi Klaus, Santamaria Joan, Tolosa Eduardo, Högl Birgit, Poewe Werner. White and gray matter abnormalities in idiopathic rapid eye movement sleep behavior disorder: a diffusion-tensor imaging and voxel-based morphometry study. Ann. Neurol. 2011;69(2):400–407. doi: 10.1002/ana.22245. [DOI] [PubMed] [Google Scholar]
- Shah Apurva, Lenka Abhishek, Saini Jitender, Wagle Shivali, Naduthota Rajini M., Yadav Ravi, Pal Pramod Kumar, Ingalhalikar Madhura. Altered brain wiring in Parkinson's disease: a structural connectome-based analysis. Brain Connect. 2017;7(6):347–356. doi: 10.1089/brain.2017.0506. [DOI] [PubMed] [Google Scholar]
- Sharma Anup, Wolf Daniel H., Ciric Rastko, Kable Joseph W., Moore Tyler M., Vandekar Simon N., Katchmar Natalie, Daldal Aylin, Ruparel Kosha, Davatzikos Christos, Elliott Mark A., Calkins Monica E., Shinohara Russell T., Bassett Danielle S., Satterthwaite Theodore D. Common dimensional reward deficits across mood and psychotic disorders: a connectome-wide association study. Am. J. Psychiatry. 2017;174(7):657–666. doi: 10.1176/appi.ajp.2016.16070774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Zarrar, Kelly Clare, Reiss Philip T., Cameron Craddock R., Emerson John W., McMahon Katie, Copland David A., Xavier Castellanos F., Milham Michael P. A multivariate distance-based analytic framework for connectome-wide association studies. Neuroimage. 2014;93:74–94. doi: 10.1016/j.neuroimage.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Jung Hwan, Shin Seong A, Lee Jee-Young, Nam Hyunwoo, Lim Jae-Sung, Kim Yu Kyeong. Precuneus degeneration and isolated apathy in patients with Parkinson’s disease. Neurosci. Lett. 2017;653:250–257. doi: 10.1016/j.neulet.2017.05.061. [DOI] [PubMed] [Google Scholar]
- Smith Robert E., Tournier Jacques-Donald, Calamante Fernando, Connelly Alan. Anatomically-constrained tractography: improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage. 2012;62(3):1924–1938. doi: 10.1016/j.neuroimage.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Smith Robert E., Tournier Jacques-Donald, Calamante Fernando, Connelly Alan. SIFT: Spherical-deconvolution informed filtering of tractograms. Neuroimage. 2013;67:298–312. doi: 10.1016/j.neuroimage.2012.11.049. [DOI] [PubMed] [Google Scholar]
- Smith Robert E., Tournier Jacques-Donald, Calamante Fernando, Connelly Alan. SIFT2: Enabling dense quantitative assessment of brain white matter connectivity using streamlines tractography. Neuroimage. 2015;119:338–351. doi: 10.1016/j.neuroimage.2015.06.092. [DOI] [PubMed] [Google Scholar]
- Sun Junyan, Lai Zhaoyu, Ma Jinghong, Gao Linlin, Chen Meijie, Chen Jie, Fang Jiliang, Fan Yangyang, Bao Yan, Zhang Dongling, Chan Piu, Yang Qi, Ye Chenfei, Wu Tao, Ma Ting. Quantitative evaluation of iron content in idiopathic rapid eye movement sleep behavior disorder. Mov. Disord. 2020;35(3):478–485. doi: 10.1002/mds.27929. [DOI] [PubMed] [Google Scholar]
- Tinaz Sule, Lauro Peter M., Ghosh Pritha, Lungu Codrin, Horovitz Silvina G. Changes in functional organization and white matter integrity in the connectome in Parkinson's disease. Neuroimage: Clinical. 2017;13:395–404. doi: 10.1016/j.nicl.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinelli E., Francia A., Quartuccio E.M., Morreale M., Contessa G.M., Pascucci S., Sbardella E., Pozzilli C., Pantano P. Structural brain MR imaging changes associated with obsessive-compulsive disorder in patients with multiple sclerosis. AJNR Am. J. Neuroradiol. 2013;34(2):305–309. doi: 10.3174/ajnr.A3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier J.-Donald, Yeh Chun-Hung, Calamante Fernando, Cho Kuan-Hung, Connelly Alan, Lin Ching-Po. Resolving crossing fibres using constrained spherical deconvolution: validation using diffusion-weighted imaging phantom data. Neuroimage. 2008;42(2):617–625. doi: 10.1016/j.neuroimage.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Tournier J.D., Calamante F., Connelly A. Paper presented at the Proceedings of the international society for magnetic resonance in medicine. 2010. Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions. [Google Scholar]
- Tustison N.J., Avants B.B., Cook P.A., Zheng Y., Egan A., Yushkevich P.A., Gee J.C. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraart Jelle, Fieremans Els, Novikov Dmitry S. Diffusion MRI noise mapping using random matrix theory. Magn. Reson. Med. 2016;76(5):1582–1593. doi: 10.1002/mrm.26059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Mori S., Chan P., Ma T. Connectome-wide network analysis of white matter connectivity in Alzheimer's disease. Neuroimage: Clinical. 2019;22 doi: 10.1016/j.nicl.2019.101690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky Andrew, Fornito Alex, Bullmore Edward T. Network-based statistic: identifying differences in brain networks. Neuroimage. 2010;53(4):1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Zeighami Y., Ulla M., Iturria-Medina Y., Dadar M., Zhang Y., Larcher K.M.-H., Dagher A. Network structure of brain atrophy in de novo Parkinson's disease. ELife. 2015;4 doi: 10.7554/eLife.08440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L., Zhou J., Wang Y., Jin Y., Jahanshad N., Prasad G., Thompson P.M. Comparison of nine tractography algorithms for detecting abnormal structural brain networks in Alzheimer’s disease. Front. Aging Neurosci. 2015;7 doi: 10.3389/fnagi.2015.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Xiaona, Sun Xiaoxuan, Wang Junhong, Tang Liou, Xie Anmu. Prevalence of rapid eye movement sleep behavior disorder (RBD) in Parkinson’s disease: a meta and meta-regression analysis. Neurol. Sci. 2017;38(1):163–170. doi: 10.1007/s10072-016-2744-1. [DOI] [PubMed] [Google Scholar]
- Zheng Ying-Qiu, Zhang Yu, Yau Yvonne, Zeighami Yashar, Larcher Kevin, Misic Bratislav, Dagher Alain, Kennedy Henry. Local vulnerability and global connectivity jointly shape neurodegenerative disease propagation. PLoS Biol. 2019;17(11):e3000495. doi: 10.1371/journal.pbio.3000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.