Abstract
An Italian male with no link to China Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) epidemic presented at Emergency Room (ER) with severe respiratory impairment. The RT‐PCR on 20 February 2020, nasopharyngeal swab revealed SARS‐CoV‐2 infection, confirmed with viral culture and sequencing. This was the first identified autochthonous SARS‐CoV‐2 transmission in Italy, that unveiled global pathogen diffusion. This clinical case highlights an underestimation of SARS‐CoV‐2 circulation, making initial containment measures unfit to face the real situation and delaying the management of potentially affected SARS‐CoV‐2 patients.
Keywords: pandemic, RT‐PCR, SARS‐CoV‐2, serology, viral culture
1. INTRODUCTION
The pandemic of SARS‐CoV‐2 originated in Wuhan, China, in December 2019. 1 Chinese health authorities put in place containment measures, apparently limiting diffusion inside national borders and occasionally involving travelers abroad: 2 in Italy, on 5 February 2020, only three cases were reported coming from Hubei province, two Chinese tourists and an Italian repatriate. 3 However, the first identification of local transmission in Italy here reported revealed that global diffusion was a fact and no more a hypothesis.
2. CASE PRESENTATION
A 38 years old male presented at ER of Codogno General Hospital (Lodi province, Lombardy, Northern Italy) on 18 February 2020, with fever and nonproductive cough which had arisen on 10 February 2020; however, despite pneumonia diagnosis, no epidemiological link with Coronavirus Disease 2109 (COVID‐19). Chinese epidemic was found and the patient was discharged with prescription of levofloxacin therapy. On 19 February 2020, he was presented at ER again due to worsened conditions and the situation negatively evolved in few hours: the patient was dysphonic, with a body temperature of 40.8°C, a P/F = 80, normal WBC, negative PCT and CRP 80, while the chest CT showed multiple foci bilateral pneumonia and ground glass areas (Figure 1); considering the clinical picture, the subject was moved to Intensive Care Unit (ICU), where after CPAP trail failure, he was intubated and pronated. During anesthetist visit, the patient's wife disclosed a possible exposure to the new Coronavirus: in the previous days they found out that the patient's colleague had fever and respiratory symptoms after a business meeting with Chinese co‐workers.
FIGURE 1.
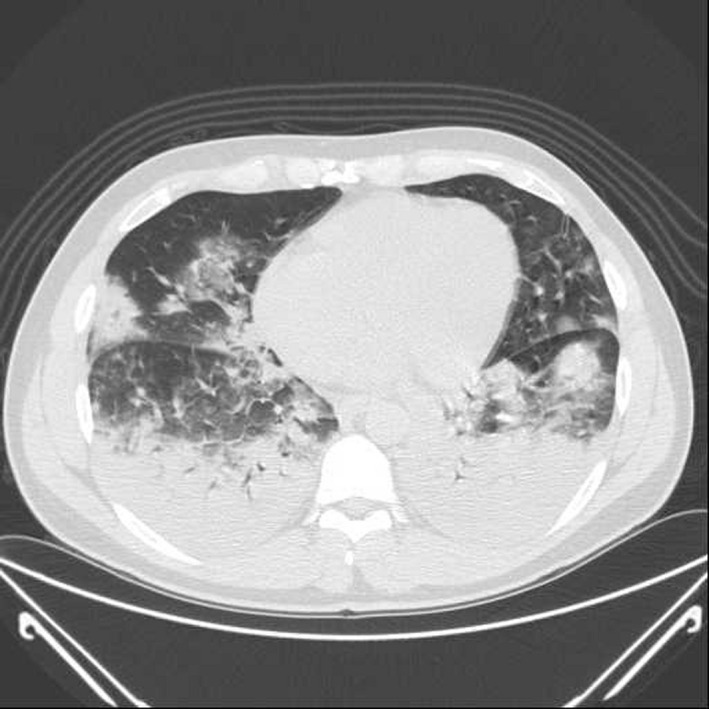
Chest Computer Tomography Scan. Computer Tomography scan that shows some areas of ground glass and consolidations in the lower lobes
A nasopharyngeal swab, collected on 20 February 2020, was sent to Sacco Hospital Laboratory of Clinical Microbiology, Virology and Bioemergencies (CLIMVIB) for SARS‐CoV‐2 diagnosis. According to internal procedure, the presence of respiratory pathogens was investigated using the multiplex BioFire® FilmArray® Respiratory 2 Panel (Biomerieux) assay: all targets resulted negative, excluding infection with most common bacteria and viruses and confirming the negative results of legionella and pneumococcal urine antigen tests. At the same time, a sample aliquot of 500 μL was used for RNA extraction by means of NUCLISENS® easyMAG® (Biomerieux) instrument; RNA was then processed using the SARS‐CoV‐2 Hong‐Kong University (HKU) detection protocol, 4 including SARS‐CoV RNA from laboratory biological repository as positive control, as indicated in the HKU protocol: both Nucleoprotein (N) and Orf‐1ab targets (for screening and confirmation, respectively) were detected, putting the last piece into the diagnostic puzzle. According to Italian guidelines, a swab aliquot was immediately delivered to the Istituto Superiore di Sanità (ISS, National Institute of Health), Department of Infectious Diseases for diagnosis confirmation. However, considering that no other cases had been previously found, in order to further support these preliminary data the sample was analyzed using reagents provided as trial kits by different companies: Allplex™ 2019‐nCoV Assay (Seegene Inc), Liferiver Novel Coronavirus (2019‐nCoV) Real Time Multiplex RT‐PCR Kit (Liferiver™, Shanghai ZJ Bio‐Tech Co., Ltd.), and TaqMan 2019‐nCoV Assay Kit v1 (Applied Biosystems, Thermo Fisher Scientific Inc). All commercial assays returned positive results for all the gene targets, as confirmed in the following days also by ISS.
Further investigations were conducted during the following months to better depict the case. A plasma sample collected on 21 February 2020, was tested for SARS‐CoV‐2‐specific antibodies, using anti‐SARS‐CoV‐2 IgG and IgM chemiluminescence immunoassay kits on fully automated iFlash1800 analyzer (Shenzen YHLO Biotech Co., Ltd.,): 5 both IgM (0.30 AU/mL, cut‐off = 10.00 AU/mL) and IgG (0.57 AU/mL, cut‐off = 10.00 AU/mL) resulted negative, in accordance with the time spanning between symptoms onset and blood collection (10 days). Given the patient's disease severity and considering that the presence of viral genome in blood was recognized as a negative prognostic factor, 6 , 7 , 8 a RT‐PCR was performed on the same sample using ELITe InGenius® system and the GeneFinder™ COVID‐19 Plus RealAmp Kit assay (ELITechGroup): not surprisingly, a positivity to N gene (Ct value = 33) was found. Viral culture on VERO E6 cells (ATCC® CRL‐1586™) was also performed from both nasopharyngeal swab and Bronchoalveolar Lavage Fluid (BALF). The first one permitted whole genome sequencing by means of Illumina Miseq Reagent Nano kit: the sequence was deposited in the Global Initiative on Sharing All Influenza Data (GISAID; accession number EPI_ISL_412973) and phylogenetic analysis placed the viral strain in a cluster close to the German isolated one, but distant from the three first imported Italian cases. 9 Interestingly, a cytopathic effect was observed after only 24 h of incubation of BALF sample, suggesting a high viral concentration in the patient's low respiratory tract, likely linked to severe clinical conditions.
After the notification of positivity, Codogno Hospital promptly activated isolation and containment measures and the case was notified to Italian Ministry of Health and competing authorities. Patient's close contacts were tested for SARS‐CoV‐2: several individuals resulted positive, including his pregnant wife; on the contrary, both nasopharyngeal swab and serum sample from the putative transmitting subject (promptly collected on February 21st, 6 weeks after his return from China) were negative, thus increasing uncertainty on the possible infection acquisition. The patient was moved to San Matteo Hospital in Pavia on 22 February 2020, and he completely recovered after several weeks of hospitalization.
3. DISCUSSION
The identification of this first local transmission paved the way to pandemic recognition. CLIMVIB had processed 68 samples in the period January 22nd‐February 20th, all from people with a direct link to Chinese epidemic (ie: travelers from China) and all of them tested negative for SARS‐CoV‐2 and some positive for other respiratory pathogens (Flu A = 7, Flu B = 11, RSV = 5, HCoV‐NL63 = 1, EV/RV/hBoV = 1). On 21 February 2020 only, the received nasopharyngeal swabs were more than 50, becoming thousands in few days in all regional referral centers, with plenty of symptomatic individuals, probable contacts and frightened people rushing to ERs. In only 2 weeks SARS‐CoV‐2 epidemiology radically changed, as depicted by the European situation: accordingly to WHO reports, on 20 February 2020 confirmed cases were 3, 12, 16, 2, and 9 in Italy (IT), France (FR), Germany (DE), Spain (SP), and the United Kingdom (UK), respectively, rising on 4 March 2020 to IT = 2502, FR = 212, DE = 196, FR = 151, and UK = 51; 10 , 11 these data, however, were just the tip of the iceberg of the real situation, as demonstrated by more than three million infections and 217 thousand deaths by the end of April, 2020 worldwide. 12
This first unexpected autochthonous case increased incertitude on when and how the virus started its global circulation. Zhou et al 1 stated that the initial SARS‐CoV‐2 outbreak started on 12 December 2019 in Wuhan, based on the admission date of seven patients with severe pneumonia to ICU of Jin Yin‐Tan Hospital: therefore, it is reasonable that these subjects acquired the infection at least in late November from an unknown source. Considering that Chinese authorities declared the Wuhan quarantine on 22 January 2020, a virus unrestrained circulation of approximately 2 months can be hypothesized, during which SARS‐CoV‐2 spread due to human mobility, also reaching travelers moving around the world: the World Bank estimated more than 611 million passengers in China airports for 2018, each of them representing a potential virus carrier during an epidemic. 13 It is relevant that the first local transmission in Europe was detected in January 2020: a Chinese woman from Shanghai, who had attended a business meeting in Munich on January 20th and 21st, with mild symptoms wrongly attributed to jetlag, fell ill on the flight back home and tested positive on January 26th; the company as well as German health authorities were immediately informed and the contact tracing was started: a total of 20 cases out of more than 200 contacts were included in this epidemic cluster (four as first generation cases, the other 16 belonging to a subsequent group) and the outbreak was considered contained, with no further expansion in the country; notably only two patients (n 1 and n 2) had direct contact with the index patient (the Chinese woman), the others had direct or secondary contact only with German patient n 1. 14 , 15 Aside from the outbreak description, the German studies made evident that SARS‐CoV‐2 can be transmitted by asymptomatic individuals too: the Chinese woman developed symptoms only after contact with her colleague, who then infected relatives and co‐workers before becoming sick. The keystone of the unrestricted circulation of SARS‐CoV‐2 at least at the beginning of the outbreak is that asymptomatic people tended not to get tested despite their capacity to spread the infection. Interestingly, as above mentioned, phylogenetic studies revealed that SARS‐CoV‐2 likely entered Italy with at least two transmission events, probably in late January, and that the patient one virus mapped in a cluster containing also German sequences, but not the other Italian ones. 9 , 16 Such evidences suggest that SARS‐CoV‐2 was spreading undisturbed everywhere via asymptomatic subjects, thus making epidemic estimations and containment measures unfit to face the real situation.
Following Italian Patient one identification and the increase in confirmed cases, on March 9th, the Italian Government declared the “lockdown” status, limiting citizens’ activities and transfers, in order to control the epidemic spread; with the same purpose, other countries prohibited access to Italian travelers, even in absence of local containment measures or lockdown policies.
But it was too late: Pandora's box was already wide open.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Conceptualization, VM, AM, AM; Investigation, AM, PGV, CP, OA; Data curation, VM, AM, DM, AR; Writing ‐ Original Draft, AM, DM, AR; Writing ‐ Review & Editing, VM, AM; Supervision, VM, MRG.
CONSENT STATEMENT
Published with written consent of the patient.
ETHICAL APPROVAL
All data used in the study were previously made anonymous, according to the requirements set by Italian Data Protection Code (Legislative Decree 196/2003) and the general authorizations issued by the Data Protection Authority. Under Italian law, all sensitive data were deleted and only age, gender and sampling date were collected making Ethics Committee approval unnecessary (Art. 6 and Art. 9 of Legislative Decree 211/2003).
AKNOWLEDGMENTS
We thank all the staff of the Laboratory of Clinical Microbiology, Virology and Bioemergencies ASST Fatebenefratelli Sacco and of Anesthesia and ICU ASST Lodi for the strong effort in the pandemic response, especially during the very next days after identification of this first case. We also thank Istituto Superiore di Sanità for sequence data generation and sharing.
Micheli V, Mancon A, Malara A, et al. What was behind the first recognition and characterization of autochthonous SARS‐CoV‐2 transmission in Italy: The impact on European scenario. Clin Case Rep. 2021;9:e04154. 10.1002/ccr3.4154
Valeria Micheli, Alessandro Mancon are equally contributed to this work.
REFERENCES
- 1. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270‐273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Italian Ministry of Health . COVID‐19: nuove indicazioni e chiarimenti, 2020. http://www.trovanorme.salute.gov.it/norme/renderNormsanPdf?anno=2020&codLeg=73195&parte=1+&serie=null. Accessed June 24, 2020.
- 3. Capobianchi MR, Rueca M, Messina F, et al. Molecular characterization of SARS‐CoV‐2 from the first case of COVID‐19 in Italy. Clinical Microb Inf. 2020;26:954‐956. 10.1016/j.cmi.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. School of Public Health, The University of Hong Kong, Hong Kong . Detection of 2019 novel coronavirus (2019‐cCoV) in suspected human cases by RT‐PCR, 2020. https://www.who.int/docs/default‐source/coronaviruse/peiris‐protocol‐16‐1‐20.pdf?sfvrsn=af1aac73_4. Accessed June 24, 2020.
- 5. Infantino M, Grossi V, Lari B, et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti‐SARS‐CoV‐2 IgM and IgG antibodies: an Italian experience. J Med Virol. 2020;92(9):1671–1675. 10.1002/jmv.25932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiaohua C, Binghong Z, Yueming Q, et al. Detectable serum SARS‐CoV‐2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL‐6) level in critically ill COVID‐19 patients. Clin Infect Dis. 2020;17:ciaa449. 10.1093/cid/ciaa449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fajnzylber J, Regan J, Coxen K, et al. SARS‐CoV‐2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11(1):5493. 10.1038/s41467-020-19057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Veyer D, Kernéis S, Poulet G, et al. Highly sensitive quantification of plasma SARS‐CoV‐2 RNA shelds light on its potential clinical value. Clin Infect Dis, 2020;17:ciaa1196. 10.1093/cid/ciaa1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stefanelli P, Faggioni G, Lo Presti A, et al. Whole genome and phylogenetic analysis of two SARS‐CoV‐2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020;25:2000305. 10.2807/1560-7917.ES.2020.25.13.2000305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. World Health Organization . Coronavirus disease 2019 (COVID‐19): situation report, 31, 2020. https://apps.who.int/iris/handle/10665/331120. Accessed June 24, 2020.
- 11. World Health Organization . Coronavirus disease 2019 (COVID‐19): situation report, 44, 2020. https://apps.who.int/iris/handle/10665/331355. Accessed June 26, 2020.
- 12. World Health Organization . Coronavirus disease 2019 (COVID‐19): situation report, 101, 2020. https://apps.who.int/iris/handle/10665/332054. Accessed June 24, 2020.
- 13. The World Bank . Air transport, passengers carried, 2020. https://data.worldbank.org/indicator/IS.AIR.PSGR. Accessed June 26, 2020.
- 14. Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019‐nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970‐971. 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Böhmer MM, Buchholz U, Corman VM, et al. Investigation of a COVID‐19 outbreak in Germany resulting from a single travel‐associated primary case: a case series. Lancet Infect Dis. 2020;20(8):920‐928. 10.1016/S1473-3099(20)30314-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Micheli V, Rimoldi SG, Romeri F, et al. Geographical reconstruction of the SARS‐CoV‐2 outbreak in Lombardy (Italy) during the early phase. J Med Virol. 2021;93(3):1752‐1757. 10.1002/jmv.26447 [DOI] [PMC free article] [PubMed] [Google Scholar]


