Abstract
The SARS‐CoV‐2 spike glycoprotein (spike) mediates viral entry by binding ACE2 receptors on host cell surfaces. Spike glycan processing and cleavage, which occur in the Golgi network, are important for fusion at the plasma membrane, promoting both virion infectivity and cell‐to‐cell viral spreading. We show that a KxHxx motif in the cytosolic tail of spike weakly binds the COPβ’ subunit of COPI coatomer, which facilitates some recycling of spike within the Golgi, while releasing the remainder to the cell surface. Although histidine (KxHxx) has been proposed to be equivalent to lysine within di‐lysine endoplasmic reticulum (ER) retrieval sequences, we show that histidine‐to‐lysine substitution (KxKxx) retains spike at the ER and prevents glycan processing, protease cleavage, and transport to the plasma membrane.
Keywords: COPI coatomer, COVID‐19 SARS‐CoV‐2, di‐lysine motif, ER retrieval signal, spike glycoprotein
Abbreviations
C‐tail, C‐terminal cytoplasmic tail
ER, endoplasmic reticulum
ERRS, ER retrieval signal
SARS‐CoV‐2, Severe Acute Respiratory Syndrome Coronavirus 2
The spike glycoprotein (spike) of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has garnered significant attention since the outbreak of a pandemic (COVID‐19) in early 2020 due to its importance in viral pathogenesis and immune response [1]. As of April 2021, all vaccines with emergency use authorization for COVID‐19 target spike by providing host cells with a genetic transcript (mRNA or adenovirus) that ribosomes translate into a mutated spike protein. However, how nascent spike protein gets processed and traffics within the cell remains unclear.
Spike is a type I transmembrane protein with a signal peptide that is initially synthesized in the cytoplasm. The rest of the protein, including the receptor binding domain, is then cotranslationally translocated into the lumen of the endoplasmic reticulum (ER) while leaving a 39 aa cytoplasmic tail (C‐tail) that is anchored by a C‐terminal transmembrane domain [2]. This inactive precursor polypeptide acquires up to 22 N‐linked high‐mannose glycan chains in the ER that are further processed to complex glycans in the Golgi [3]. Unlike spike from SARS‐CoV, CoV‐2 spike is further processed by cleavage at a multibasic site into S1 and S2 subunits by either furin at the trans‐Golgi network (TGN) [4, 5], or by another proprotein convertase [6]. Although the S1 and S2 subunits remain noncovalently bound, this first cleavage step likely primes them for binding with angiotensin I‐converting enzyme 2 (ACE2) at new host cell surfaces and for subsequent cleavage of S2 by transmembrane serine protease 2 (TMPRSS2) into an S2’ fragment [7]. During initial processing, after S1‐S2 cleavage, spike then either recycles back to the ER‐Golgi intermediate compartment (ERGIC) for assembly into SARS‐CoV‐2 virions or traffics to the plasma membrane of the host cell where it mediates cell–cell fusion, spreading the viral infection.
Much of our understanding of the Golgi recycling of SARS‐CoV‐2 spike is based on previous studies with spike protein from SARS‐CoV [8, 9, 10, 11]. There is only one amino acid difference between the C‐tails of spike from SARS‐CoV‐2 and SARS‐CoV, Cys1247 vs Ala1229, respectively. Faced with an ongoing pandemic, scientists are rapidly studying spike and a recent publication did partially address the trafficking itinerary of SARS‐CoV‐2 spike in the context of the viral envelope (E) and membrane (M) proteins [12]. Together, these previous studies identified an ER retrieval motif in the C‐tails of both SARS‐CoV and SARS‐CoV‐2, and for SARS‐CoV demonstrated interaction with whole COPI coatomer complex. In addition, the histidine reside within this motif has been proposed as functionally equivalent to lysine, which is present in the canonical KxKxx ER retrieval signals (ERRS) of many endogenous human ER type I transmembrane proteins [9, 13]. Given that SARS‐CoV‐2 spike alone is the basis for currently available COVID‐19 vaccines, we chose to study its trafficking in the absence of viral envelope and membrane proteins. Here, we present data showing that in the context of SARS‐CoV‐2 spike, a histidine‐to‐lysine substitution within the KxHxx motif changes the steady‐state distribution of spike from the Golgi to the ER and prevents protease cleavage, glycan processing, and transport to the plasma membrane. We also determine to which subunit within COPI coatomer SARS‐CoV‐2 spike binds, namely the COPβ’ subunit.
Materials and methods
Cell lines
HeLa and HEK 293 cell lines were maintained in Dulbecco’s modified Eagle medium (Life Technologies, Carlsbad, CA, USA) containing 0.11 g·L−1 sodium pyruvate and 4.5 g·L−1 glucose, supplemented with 10% (vol/vol) FBS (Atlanta Biologicals, Flowery Branch, GA, USA), 100 000 U·L−1 penicillin, 100 mg·L−1 streptomycin (Life Technologies, Carlsbad, CA, USA), and 2 mm l‐glutamine (Life Technologies, Carlsbad, CA, USA).
DNA constructs
Using the Tac‐TGN38 C‐tail chimera cDNA clone that was kindly provided by Ruben Aguilar (Purdue University), the native Tac C‐tail was restored by QuickChange mutagenesis. To make the Tac‐spike C‐tail chimera, a 138 nucleotide Ultamer Duplex (IDT, Coralville, IA, USA) was synthesized with a 5′ XbaI site and a 3′ BamHI site. TAC‐TGN38 cDNA was then cut with the same two enzymes as described [14], and the TGN38 C‐tail was replaced with the double‐digested Ultramer. QuickChange mutagenesis generated mutant Tac‐spike cDNA constructs. The various GST‐peptide cDNA constructs were made synthesizing Ultramers with a 5′ EcoRI site and a 3′ XhoI site and ligated into double‐digested pGEX5X3 vector cut with the same enzymes. The codon‐optimized full‐length spike cDNA (Plasmid #145032) in pCDNA3.1 was purchased from Addgene (Watertown, MA, USA). This cDNA encodes a 1D4 epitope at the C terminus of spike. To avoid any unnecessary perturbations to the trafficking itinerary of spike, this additional sequence was removed by swapping an approximately 900 bp BbvCI/XhoI PCR fragment into the cDNA to restore the untagged, native 3′ end. The C‐tail mutants in the full‐length spike were generated in a similar way using mutant reverse primers. The COPα‐myc and COPβ’‐myc cDNA clones have been described [15]. All cDNA constructs were verified by Sanger sequencing.
Antibodies and reagents
The following antibodies were used in this study: anti‐spike 1A9 mouse monoclonal antibody (Cat #GTX632604) from GeneTex (Irvine, CA, USA), anti‐giantin rabbit polyclonal antibody (Cat #924302) from BioLegend (San Diego, CA, USA), anti‐Tac 7G7/B6 mouse monoclonal antibody (Cat #05‐170) from Millipore‐Sigma (St. Louis, MO, USA), anti‐β‐COP rabbit polyclonal antibody (Cat #ab2899) from Abcam (Cambridge, MA, USA), anti‐δ‐COP rabbit polyclonal antibody (Cat #PA5‐21484) from Life Technologies, Carlsbad, CA, USA, anti‐GGA2 mouse monoclonal (Cat #612612) from BD Biosciences (San Jose, CA, USA), anti‐HA mouse monoclonal antibody (Cat #MMS‐101P) from Covance (Princeton NJ, USA), anti‐Flag mouse monoclonal antibody (Cat #200472‐21) from Agilent (Santa Clara, CA, USA), and anti‐myc 9E10 mouse monoclonal antibody from Development Studies Hybridoma Bank (Iowa, City, IA, USA). The anti‐µ1 polyclonal antibody RY/1 was kindly provided by the late Linton Traub (University of Pittsburgh School of Medicine, Pittsburgh, PA, USA). Endo H and PNGase F were purchased from New England BioLabs (Ipswich, MA, USA), while jetOPTIMUS transfection reagent was from Polyplus (Illkirch, France).
Protein expression, purification, and GST pull‐down assays
All GST‐fusion proteins were expressed in the Escherichia coli strain BL‐21 (RIL) (Agilent) and purified as described previously [16]. 50 µg of each fusion protein was immobilized on glutathione superflow agarose beads, and the binding assays were performed essentially as described [16] using either untransfected HEK 293 cell lysates, or cells transfected with the indicated cDNAs.
Immunoblotting
Proteins resolved by sodium dodecyl sulfate (SDS)/polyacrylamide gel under reducing conditions were transferred to nitrocellulose membrane and detected with antibodies as described in the figure legends. Equal amounts of whole‐cell extract were loaded on the gels.
Immunofluorescence microscopy
To determine the subcellular localization of the TAC chimeras or spike glycoprotein, the various constructs were transfected into HeLa cells on sterile glass coverslips using jetOPTIMUS transfection reagent according to the manufacturer’s protocol. Cells were fixed the following day with 4% formaldehyde (Sigma‐Aldrich) for 10min, permeabilized, and blocked with PBS containing 0.4% (v/v) Triton X‐100% and 2% immunoglobulinG‐free BSA (Jackson ImmunoResearch) for 1 hr, and then probed with the indicated antibodies in PBS containing 0.1% Triton X‐100% and 0.5% BSA. Following fluorophore‐conjugated secondary antibody treatment and washing, the processed cells were mounted in ProLong® Glass antifade mounting medium (Life Technologies, Carlsbad, CA, USA), and the images were acquired with an LSM880 confocal microscope (Carl Zeiss Inc., Peabody, MA, USA). Images were analyzed by imagej software (NIH, Bathesda, MD, USA).
Results
Replacement of the ER retrieval signal (ERRS) histidine with lysine prevents spike from being transported to the plasma membrane
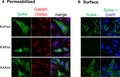
The 39 amino acid cytosolic tail of SARS‐CoV‐2 spike differs from that of SARS‐CoV at only one position, namely a cysteine for alanine substitution at amino acid 1247 (Fig. 1A, underlined cysteine). While others have studied the effect of mutating both the lysine and histidine residues to alanine in the C‐terminal 1251 KLHYT1255 of SARS‐CoV [9, 10, 11], or deleting the last 19 aa of SARS‐CoV‐2 [12], the consequence of a H1271K change, generating a canonical ERRS, has not been previously reported. In order to assess the behavior of mutant SARS‐CoV‐2 spike harboring a canonical 1269KLKYT1273 motif [17], we initially made a Tac chimera containing the 39 amino acid SARS‐CoV‐2 spike C‐tail, similar to constructs used to test SARS‐CoV spike [9]. The 1269 KLHYT1273 sequence within this chimera was then mutated to 1269KLKYT1273 or 1269 ALAYT1273 (Fig. 1A). A Tac‐Tac construct having the native C‐tail of Tac, which lacks an ERRS, is known to traffic to the cell surface [18]. Confocal immunofluorescence microscopy for Tac or the Golgi marker giantin reveals Tac‐Tac and Tac‐ALAYT to be predominantly on the plasma membrane with little overlap with giantin. Tac‐KLHYT showed both plasma membrane and Golgi localization (Fig. 1B), in agreement with published work for SARS‐CoV [9]. In contrast, Tac‐KLKYT displayed a cytoplasmic reticular pattern typical of the ER with no apparent surface staining (Fig. 1B). To detect Tac only on the cell surface, the immunofluorescence was repeated without permeabilizing cells. Cell surface localization was observed for Tac‐Tac, Tac‐KLHYT, and Tac‐ALAYT, but not Tac‐KLKYT (Fig. 1C). This indicates that the C‐tail of SARS‐CoV‐2 alone with a H1271K substitution is sufficient and very effective at retaining or recycling Tac to the ER.
Fig. 1.
Analysis of Tac‐SARS‐CoV‐2 spike C‐tail chimeric proteins in HeLa cells. (A) Schematic representation of full‐length Tac with its native C‐tail, or the C‐tail replaced with WT or mutant SARS‐CoV‐2 spike C‐tail. Sequence of SARS‐CoV‐2 spike C‐tail is shown with the one amino acid different from SARS‐CoV spike underlined. (B) Confocal immunofluorescence images of transfected HeLa cells, permeabilized and stained for Tac (green) and the Golgi marker protein giantin (red). (C) HeLa cells, treated as in (B), but without permeabilization to detect only surface localized Tac‐spike chimeras (green).
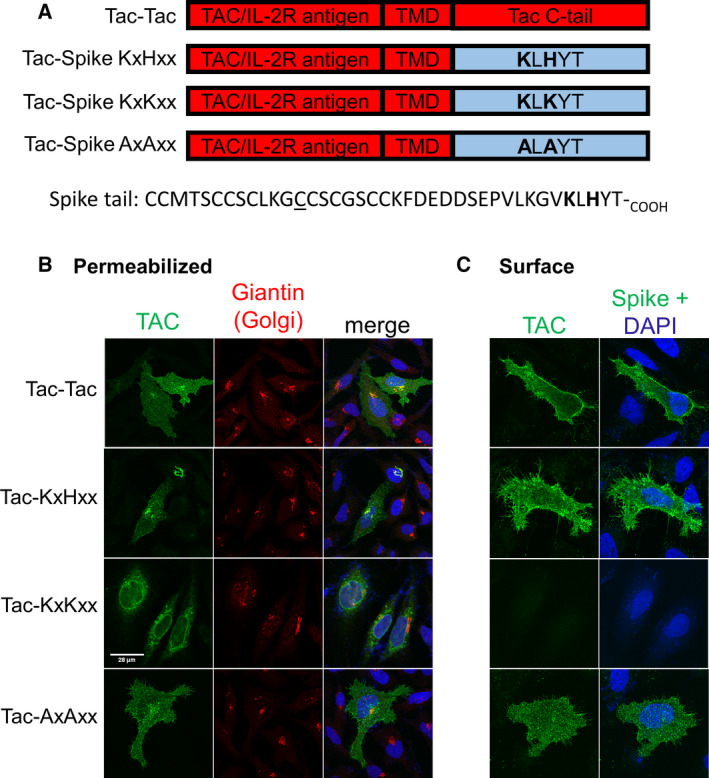
Since the histidine‐to‐lysine substitution elicited such a marked change in cellular localization of the Tac chimera, we next asked if a similar redistribution from plasma membrane to ER would also be seen with untagged, native, full‐length spike, which in HEK 293 cells has been shown to form a homotrimer [19], likely not present in our Tac chimeras. Confocal immunofluorescence microscopy of HeLa cells expressing untagged WT, 1269 KLKYT1273, or 1269 ALAYT1273 full‐length spike constructs showed only WT spike and the 1269 ALAYT1273 mutant on the cell surface (Fig. 2A,B), whereas the 1269 KLKYT1273 mutant was predominantly ER (Fig. 2A) with no plasma membrane staining in nonpermeabilized cells (Fig. 2B), similar to the Tac chimera.
Fig. 2.
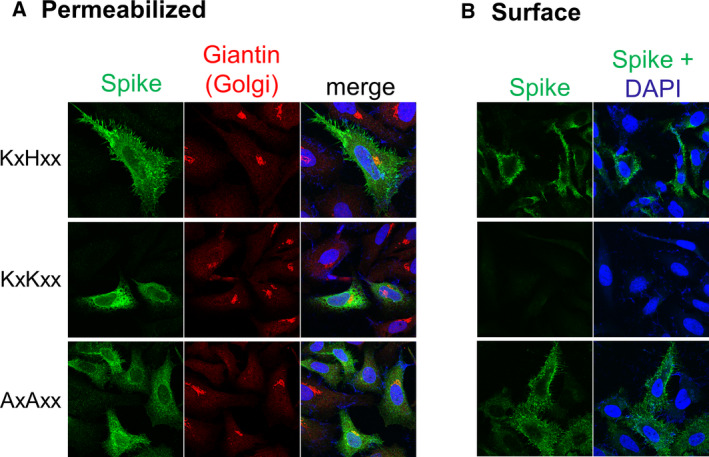
Subcellular distribution of untagged, full‐length WT SARS‐CoV‐2 spike and C‐tail mutants in HeLa cells. (A) HeLa cells were transfected with the indicated constructs. After 24 h, cells were fixed, permeabilized, and immunostained with anti‐spike (green) and for the Golgi marker protein giantin (red). (B) HeLa cells, treated as in (A), but without permeabilization to detect only surface localized full‐length spike constructs (green).
Replacement of the ERRS histidine with lysine enhances binding of spike C‐tail to COPα and COPβ’
The canonical KxKxx motif and its variant KxHxx have both been shown to immunoprecipitate COPI coatomer complex from whole‐cell lysates [9, 13, 20]. As reported for SARS‐CoV spike [9], the SARS‐CoV‐2 spike C‐tail fused to GST (Fig. 3A) also bound COPI coatomer complex (probed for endogenous COPIβ and COPIδ subunits) from untransfected whole‐cell lysate but not the clathrin adaptor GGA2 (Fig. 3B). Since di‐lysine‐based motifs are known to specifically interact with the COPα and/or COPβ’ subunits of COPI coatomer [21], we questioned whether the KxHxx motif of spike also bound these subunits. Individual myc‐tagged COPα and COPβ’ subunits of COPI coatomer were expressed in HEK293 cells, and cell lysates were used to performed pull‐down assays with various purified GST C‐tail fusion constructs (Fig. 3A). A control, GST UGT2B15 tail, with overlapping KxKxx and KKxx motifs (Fig. 3A) bound both coatomer subunits robustly (Fig. 3C, lane 6). In contrast, the GST SARS‐CoV‐2 spike WT C‐tail only weakly bound COPβ’ and not at all to COPα. As expected, the H1271K substitution within spike, which created a canonical KxKxx motif, resulted in strong binding of both COPβ’ and COPα (Fig. 3C). To rule out interaction with other tail regions, an AxAxx mutation within GST SARS‐CoV‐2 spike tail was tested and totally abrogated binding (Fig. 3C, KH→AA). In addition to the COPβ’/α/ε subcomplex, the seven subunits of COPI form two other subcomplexes: COPβ/δ and COPγ/ζ. Binding to these subcomplexes was tested by coexpressing epitope‐tagged subunits in Sf9 cells [15] and GST pull‐down. The GST SARS‐CoV‐2 spike C‐tail did not bind to either COPβ/δ or COPγ1/ζ1 hemicomplexes, or to COPε alone beyond background level (Fig. 3D).
Fig. 3.
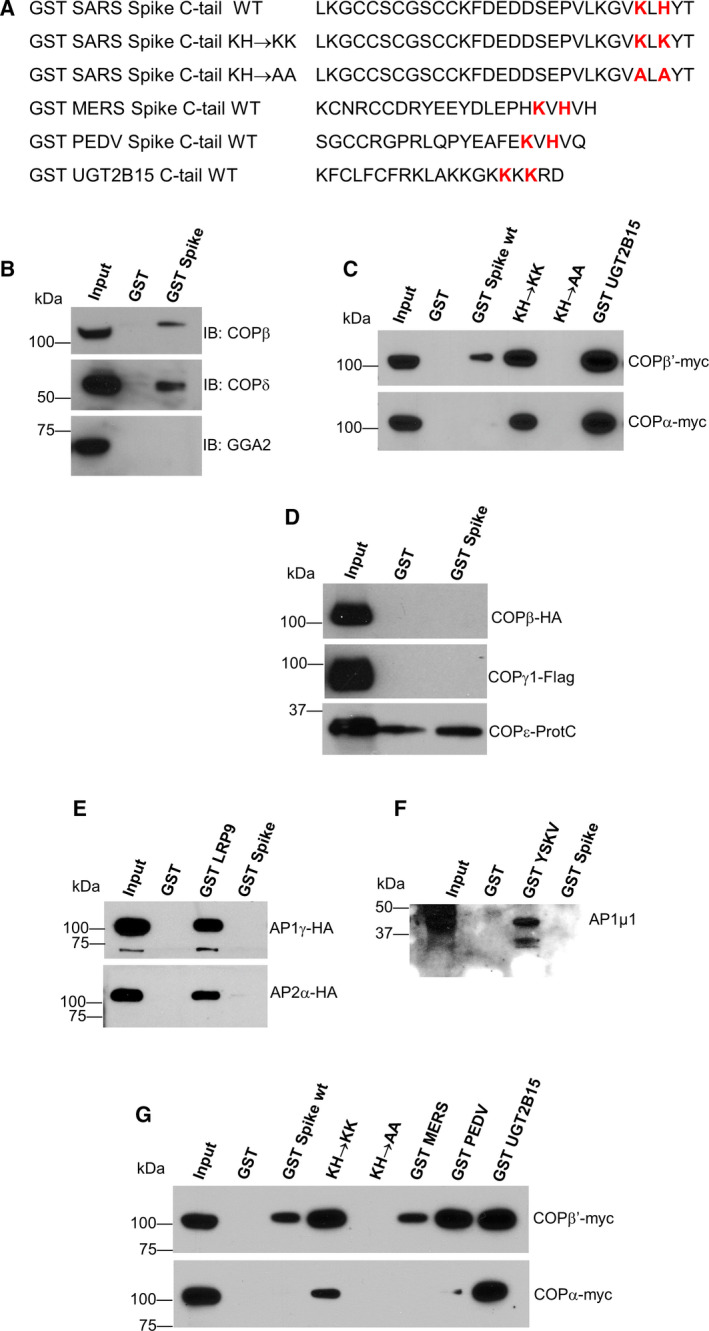
The SARS‐CoV‐2 spike C‐tail binds the COPI β’ subunit. (A) The indicated cytoplasmic C‐tails were cloned downstream of GST and expressed and purified from bacteria. (B) HEK293 whole‐cell lysate (input) was incubated with WT SARS‐CoV‐2 spike tail or GST, to control for nonspecific binding. Proteins in the pellet fraction were immunoblotted for endogenous COPβ and COPδ subunits and the monomeric clathrin adaptor GGA2. (C) GST pull‐downs were performed similar to (B) except HEK293 cells were transfected with either full‐length, human COPβ’‐myc, or COPα‐myc before preparing whole‐cell lysate. Blots were probed with anti‐myc antibody. (D) GST pull‐downs performed similar to (B) except using Sf9 insect cell lysates expressing COPI hemicomplexes β‐HA/δ (probed with anti‐HA) or γ1‐Flag/ζ1 (probed with anti‐Flag), or COPε‐ProtC alone. (E &F) GST pull‐downs performed using Sf9 insect cell lysates expressing the clathrin adaptor hemicomplexes AP‐1 γ1‐HA/σ1 (probed with anti‐HA), AP‐2 α‐HA/σ2 (probed with anti‐HA), or AP‐1 β1/µ1 (probed for µ1). (G) GST pull‐downs performed similar to C with other viral spike tails and probed with anti‐myc antibody.
Inspection of the SARS‐CoV‐2 spike C‐tail amino acids (Fig. 1A) revealed the presence of a DEDDSEPVL sequence reminiscent of the [D/E]XXXL[L/I] motif that binds the γ/σ1 and α/σ2 hemicomplexes of clathrin adaptors AP1 and AP2, respectively [22, 23, 24]. In addition, the acidic cluster could also potentially bind the µ subunit of AP‐1 [25]. However, pull‐down experiments with γ/σ1 and α/σ2 (Fig. 3E), or β1/µ1 (Fig. 3F) showed no binding of GST spike WT C‐tail to any of these subunits of AP‐1 and AP‐2, whereas the control GST‐LRP9 and GST‐YSKV peptide fusions bound as expected (Fig. 3E,F) [23].
Finally, we asked how the spike C‐tail from two other viruses compared with SARS‐CoV‐2. For this, we chose the spike C‐tails of Middle East Respiratory Syndrome (MERS) coronavirus and porcine epidemic diarrhea virus (PEDV). Pull‐down assays with GST MERS spike C‐tail and GST PEDV spike C‐tail showed that GST MERS spike C‐tail, like GST SARS‐CoV‐2 spike C‐tail, bound weakly to COPβ’ and not at all to COPα (Fig. 3G). GST PEDV spike C‐tail, on the other hand, bound COPβ’ as well as GST UGT2B15, with only trace binding to COPα.
Replacement of the ERRS histidine with lysine prevents spike cleavage and glycan processing
Our immunofluorescence data showing that spike with the ERRS histidine mutated to lysine does not traffic to the plasma membrane suggest that mutant spike may be efficiently recycled back to the ER via COPI vesicles without ever reaching the TGN, where the protease furin and other glycan processing enzymes reside. To test this possibility, HEK293 cells were transfected with either WT, KH→KK, or KH→AA full‐length spike, and cell lysates were tested for cleavage and glycan processing by immunoblotting with an anti‐spike S2 subunit antibody. WT spike (Fig. 4, lane 1) and KH→AA spike (lane 7) showed similar bands representing uncleaved, glycosylated precursor (S1S2Glyco) and cleaved S2 fragments (S2Glyco). In contrast, in cell lysates with KH→KK spike only the uncleaved, glycosylated precursor was detected (lane 4), suggesting that this mutant never reached the TGN for furin cleavage.
Fig. 4.
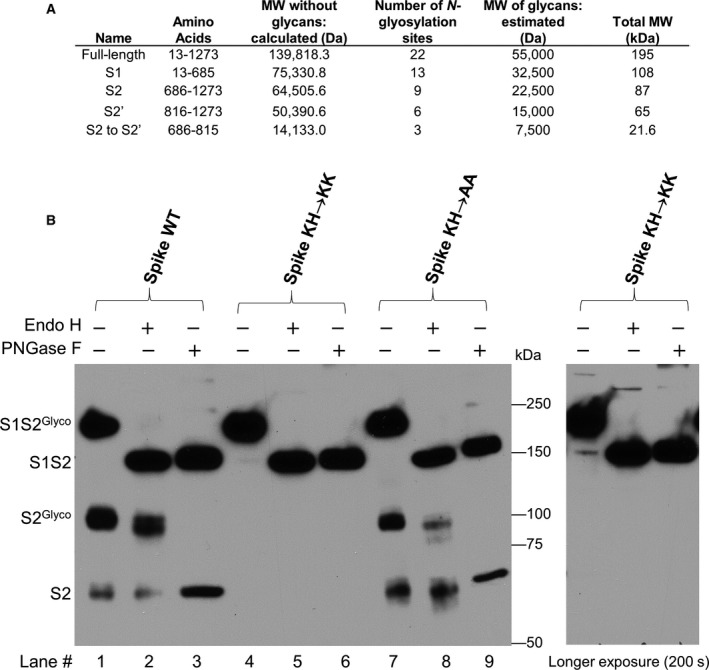
SARS‐CoV‐2 spike C‐tail requires KxHxx for cleavage and glycan processing. (A) Molecular weight estimates for various SARS‐CoV‐2 spike fragments after signal peptide removal were calculated. The number of glycan sites in spike has been published [3]. (B) Whole‐cell lysates from HEK293 cells expressing full‐length SARS‐CoV‐2 WT, KxKxx, or AxAxx spike proteins were either untreated, treated with Endo H, which removes high‐mannose but not complex N‐linked glycans, or treated with PNGase F, which removes all N‐linked glycans. Glyco indicates glycosylated bands. Immunoblots were probe with an antibody against the spike S2 fragment. Exposure on the left is 40 s, longer exposure is 200 s.
Endoglycosidase H (Endo H) only removes high‐mannose, N‐linked glycan side chains whereas peptide‐N‐glycosidase F (PNGase F) removes all N‐linked sugars. The glycan status of the three spike constructs was compared using these enzymes. The glycosylated precursors of all three spike constructs shifted similarly when treated with Endo H or PNGase F (compare lanes 2 & 3, 5 & 6, and 8 & 9), suggesting that all three precursors are similarly N‐glycosylated and contain high‐mannose glycans. However, the S2 fragment of WT spike is mostly Endo H resistant (lane 2, ratio S2Glyco to S2 bands) indicating complex glycan side chains processing within the Golgi. In contrast, the S2 fragment of KH→AA spike was only partially Endo H resistant (lane 8, ratio S2Glyco to S2 bands). This may reflect the ability of WT spike to recycle within the Golgi developing complex, Endo H‐resistant glycan side chains versus KH→AA spike which may traverse the Golgi once before being transported to the plasma membrane. Importantly, the S2 fragment was not detected with the KH→KK mutant, even with a long exposure, indicating the failure of this mutant to undergo cleavage.
Discussion
The data presented in this study provide an explanation for why SARS‐CoV‐2 uses a histidine instead of a lysine within the C‐tail of spike. Replacement of the histidine with a lysine not only prevented transport of spike to the plasma membrane but also prevented proteolytic cleavage of spike precursor into S1 and S2 fragments necessary for fusion [7]. This cleavage is, for the most part, performed by furin at the TGN [4, 5], although the requirement for furin is not absolute since some cleavage, presumably mediated by another protease, still occurs in a furin knockout cell line [6]. Although its identity is unknown, this secondary spike protease likely resides later in the Golgi trafficking pathway as the efficiently recycled KxKxx spike showed no cleavage even at long exposures (Fig. 4).
Cleavage at the multibasic site is critical not only for priming of SARS‐CoV‐2 spike on assembled virions to mediate viral entry, but also allows for the cell‐to‐cell spreading of infectivity in a virion‐independent manner [6]. In the presence of furin inhibitors, uncleaved spike can incorporate into virions but cannot serve as a substrate for cleavage by TMPRSS2 following attachment of spike to ACE2 at the plasma membrane [4]. We propose that SARS‐CoV‐2 spike includes a histidine instead of lysine to achieve TGN cleavage and glycan processing while maintaining dual‐targeting to both the ERGIC, by recycling back for virion assembly, and trafficking to the PM, for cell‐to‐cell infection. However, it is possible that in the presence of membrane and envelope viral proteins, spike assembly into a virion could also occur prior to cleavage and glycan processing, in which case, spike modification would need to happen as the preassembled virion traffics through the Golgi.
One open question that remains is why the KxHxx motif of SARS‐CoV‐2 only weakly pulled down COPβ’ whereas the corresponding KxHxx tail from PEDV strongly bound COPβ’ and even a little to COPα (Fig. 3G). This suggests that the histidine is not always suboptimal for binding COPI subunits. The identity of the x residues within the KxHxx motif, as well as upstream amino acids within these viral spike C‐tails, can likely enhance or reduce binding affinity. We have demonstrated that the terminal methionine residue within the cytosolic tail of cation‐dependent mannose phosphate receptor (CD‐MPR, …DDHLLPM) strongly inhibits binding of the DxxLLxx motif to Golgi‐localizing, γ‐adaptin ear homology, ARF‐binding (GGA) proteins [26], and CD‐MPR‐mediated lysosomal enzyme sorting [27]. Mutation of this terminal methionine to alanine (…DDHLLPA) relieves both inhibition to GGA binding and restores efficient sorting by the receptor.
In the study by Ma and Goldberg, [13], the structure of the PEDV KxHxx peptide bound to COPβ’ revealed a highly similar binding mode to that of KxKxx. Yet, the binding affinity of the PEDV peptide (FEKVHVQ) for COPβ’, as determined by isothermal calorimetry, was extremely weak (> 800 µm) as compared to COPα (82 µm). Our GST pull‐down assay showed the opposite specificity; namely that binding of the PEDV tail to COPβ’ is similar to a canonical KxKxx sequence (Fig. 3G). PEDV compared to KH→KK or UGT2B15, and only trace binding was detected to COPα. The reason for this discrepancy is not clear. One possibility is that Ma and Goldberg used fragments of yeast COPI subunits, whereas we used full‐length human subunits. Nevertheless, the weak binding of the SARS‐CoV‐2 KxHxx to COPβ’ when compared with that of PEDV suggests that the residues surrounding the lysine and histidine may in fact play a more important role than originally thought. A systematic investigation of the residues within and surrounding the KxHxx motifs of SARS‐CoV‐2 and PEDV spike could shed light on their binding differences to coatomer subunits. It could also aid in determining whether the twelve endogenous, human transmembrane proteins with cytoplasmic KxHxx motifs identified in our unpublished in silico search also bind COPI subunits. Current mRNA vaccines for COVID‐19 cause host cell production of spike antigen that to varying degrees activates both cell‐mediated immunity, through an ER‐based pathway involving MHC class I molecules, and antibody‐mediated immunity, through a plasma membrane secretion pathway and MHC class II molecules [28]. Understanding how residues surrounding the KxHxx motif of spike influence its recycling and trafficking to these pathways could allow for the design of new mRNA vaccines with tailored spike C‐tail sequences that cause a more desirable immune response. In the case of WT SARS‐CoV‐2 spike, it is reasonable to assume that the surrounding residues weaken binding to ensure decreased recycling to the ER and increased trafficking through the Golgi for proper cleavage, glycan processing, and plasma membrane display.
Note added in proof
As of the acceptance date of this Research Letter, we became aware of an unpublished, pre‐print manuscript describing related findings from the Munro group at the MRC Laboratory of Molecular Biology, Cambridge, United Kingdom [29].
Conflict of interest
The authors declare no conflict of interests.
Author contributions
BCJ and BD conceived and designed the project; BCJ and BD acquired the data; BCJ, SK, and BD analyzed and interpreted the data; BD drafted the initial manuscript, and BCJ edited it to produce the final version.
Dedication
This paper is dedicated to the memory of our former lab colleague, Dr. Linton M. Traub, who passed away on October 19, 2020, for his contribution to the field of protein trafficking.
Acknowledgements
This study was supported by National Institutes of Health Grant 2R01CA008759‐53 to SK.
Data accessibility
The data that support the findings of this study are available from the corresponding author [bd@wustl.edu] upon reasonable request.
References
- 1. Whittaker GR, Daniel S and Millet JK (2021) Coronavirus entry: how we arrived at SARS‐CoV‐2. Curr Opin Virol 47, 113–120. 10.1016/j.coviro.2021.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang Y, Yang C, Xu XF, Xu W and Liu SW (2020) Structural and functional properties of SARS‐CoV‐2 spike protein: potential antivirus drug development for COVID‐19. Acta Pharmacol Sin 41, 1141–1149. 10.1038/s41401-020-0485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Watanabe Y, Allen JD, Wrapp D, McLellan JS and Crispin M (2020) Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science 369, 330–333. 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bestle D, Heindl MR, Limburg H, Van Lam van T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O et al. (2020) TMPRSS2 and furin are both essential for proteolytic activation of SARS‐CoV‐2 in human airway cells. Life Sci Alliance 3, e202000786. 10.26508/lsa.202000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoffmann M, Kleine‐Weber H and Pohlmann S (2020) A Multibasic Cleavage Site in the Spike Protein of SARS‐CoV‐2 Is Essential for Infection of Human Lung Cells. Mol Cell 78, 779–784 e775. 10.1016/j.molcel.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Papa G, Mallery DL, Albecka A, Welch LG, Cattin‐Ortola J, Luptak J, James LC (2021) Furin cleavage of SARS‐CoV‐2 Spike promotes but is not essential for infection and cell‐cell fusion. PLoS Pathog 17, e1009246. 10.1371/journal.ppat.1009246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xia X (2021) Domains and Functions of Spike Protein in Sars‐Cov‐2 in the Context of Vaccine Design. Viruses 13, 109. 10.3390/v13010109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lontok E, Corse E and Machamer CE (2004) Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J Virol 78, 5913–5922. 10.1128/JVI.78.11.5913-5922.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McBride CE, Li J and Machamer CE (2007) The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J Virol 81, 2418–2428. 10.1128/JVI.02146-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sadasivan J, Singh M and Sarma JD (2017) Cytoplasmic tail of coronavirus spike protein has intracellular targeting signals. J Biosci 42, 231–244. 10.1007/s12038-017-9676-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ujike M, Huang C, Shirato K, Makino S and Taguchi F (2016) The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus‐like particles. J Gen Virol 97, 1853–1864. 10.1099/jgv.0.000494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boson B, Legros V, Zhou B, Siret E, Mathieu C, Cosset FL, Denolly S (2020) The SARS‐CoV‐2 Envelope and Membrane proteins modulate maturation and retention of the Spike protein, allowing assembly of virus‐like particles. J Biol Chem 296, 100111. 10.1074/jbc.RA120.016175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma W and Goldberg J (2013) Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J 32, 926–937. 10.1038/emboj.2013.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aguilar RC, Boehm M, Gorshkova I, Crouch RJ, Tomita K, Saito T, Bonifacino JS (2001) Signal‐binding specificity of the mu4 subunit of the adaptor protein complex AP‐4. J Biol Chem 276, 13145–13152. 10.1074/jbc.M010591200 [DOI] [PubMed] [Google Scholar]
- 15. Liu L, Doray B and Kornfeld S (2018) Recycling of Golgi glycosyltransferases requires direct binding to coatomer. Proc Natl Acad Sci USA 115, 8984–8989. 10.1073/pnas.1810291115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doray B and Kornfeld S (2001) Gamma subunit of the AP‐1 adaptor complex binds clathrin: implications for cooperative binding in coated vesicle assembly. Mol Biol Cell 12, 1925–1935. 10.1091/mbc.12.7.1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jackson MR, Nilsson T and Peterson PA (1990) Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J 9, 3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vergarajauregui S and Puertollano R (2006) Two di‐leucine motifs regulate trafficking of mucolipin‐1 to lysosomes. Traffic 7, 337–353. 10.1111/j.1600-0854.2006.00387.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Qian Z (2020) Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun 11, 1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cosson P and Letourneur F (1994) Coatomer interaction with di‐lysine endoplasmic reticulum retention motifs. Science 263, 1629–1631. 10.1126/science.8128252 [DOI] [PubMed] [Google Scholar]
- 21. Eugster A, Frigerio G, Dale M and Duden R (2004) The alpha‐ and beta'‐COP WD40 domains mediate cargo‐selective interactions with distinct di‐lysine motifs. Mol Biol Cell 15, 1011–1023. 10.1091/mbc.e03-10-0724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH and Bonifacino JS (2007) Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol 81, 3877–3890. 10.1128/JVI.02725-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doray B, Lee I, Knisely J, Bu G and Kornfeld S (2007) The gamma/sigma1 and alpha/sigma2 hemicomplexes of clathrin adaptors AP‐1 and AP‐2 harbor the dileucine recognition site. Mol Biol Cell 18, 1887–1896. 10.1091/mbc.e07-01-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Janvier K, Kato Y, Boehm M, Rose JR, Martina JA, Kim BY, Bonifacino JS (2003) Recognition of dileucine‐based sorting signals from HIV‐1 Nef and LIMP‐II by the AP‐1 gamma‐sigma1 and AP‐3 delta‐sigma3 hemicomplexes. J Cell Biol 163, 1281–1290. 10.1083/jcb.200307157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Navarro Negredo P, Edgar JR, Wrobel AG, Zaccai NR, Antrobus R, Owen DJ and Robinson MS (2017) Contribution of the clathrin adaptor AP‐1 subunit micro1 to acidic cluster protein sorting. J Cell Biol 216, 2927–2943. 10.1083/jcb.201602058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doray B, Bruns K, Ghosh P and Kornfeld S (2002) Interaction of the cation‐dependent mannose 6‐phosphate receptor with GGA proteins. J Biol Chem 277, 18477–18482. 10.1074/jbc.M201879200 [DOI] [PubMed] [Google Scholar]
- 27. Johnson KF and Kornfeld S (1992) A His‐Leu‐Leu sequence near the carboxyl terminus of the cytoplasmic domain of the cation‐dependent mannose 6‐phosphate receptor is necessary for the lysosomal enzyme sorting function. J Biol Chem 267, 17110–17115. [PubMed] [Google Scholar]
- 28. Shin MD, Shukla S, Chung YH, Beiss V, Chan SK, Ortega‐Rivera OA, Steinmetz NF (2020) COVID‐19 vaccine development and a potential nanomaterial path forward. Nat Nanotechnol 15, 646–655. 10.1038/s41565-020-0737-y [DOI] [PubMed] [Google Scholar]
- 29. Cattin‐Ortolá J, Welch L, Maslen SL, Skehel JM, Papa G, James LC and Munro S (2021) Sequences in the cytoplasmic tail of SARS‐CoV‐2 spike facilitate expression at the cell surface and syncytia formation. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.10.12.335562v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author [bd@wustl.edu] upon reasonable request.


