Abstract
Intrinsic resistance to current therapies, leading to dismal clinical outcomes, is a hallmark of glioblastoma multiforme (GBM), the most common and aggressive brain tumor. Understanding the underlying mechanisms of such malignancy is, therefore, an urgent medical need. Deregulation of the protein translation machinery has been shown to contribute to cancer initiation and progression, in part by driving selective translational control of specific mRNA transcripts involved in distinct cancer cell behaviors. Here, we focus on eIF3, a multimeric complex with a known role in the initiation of translation and that is frequently deregulated in cancer. Our results show that the deregulated expression of eIF3e, the e subunit of eIF3, in specific GBM regions could impinge on selective protein synthesis impacting the GBM outcome. In particular, eIF3e restricts the expression of proteins involved in the response to cellular stress and increases the expression of key functional regulators of cell stemness. Such a translation program can therefore serve as a double-edged sword promoting GBM tumor growth and resistance to radiation.
INTRODUCTION
Glioblastomas (or glioblastoma multiformes, GBMs) are one of the most common brain tumors in adults and represent the second most common cause of death in children and the third in adults in the world (1). Despite the standard treatments, which combine surgical resection followed by radiotherapy and chemotherapy, the median survival does not exceed 14–15 months (2,3). The main reason for this dismal course is the high rate of tumor recurrence, which is associated with both the extremely infiltrative nature of glioblastoma growth and a frequently occurring resistance to conventional treatments, in part due to tumor heterogeneity. Indeed, aside from sharing common characteristics, such as high vascularization, pseudopalisading necrosis and infiltrating growth, these tumors are also very heterogeneous at the genetic, molecular and cellular levels. Among the sources of this heterogeneity are (i) the presence of a wide spectrum of driver mutations (4), (ii) distinct molecular subtypes (5) and (iii) cellular plasticity relying on cancer stem cells (or glioma stem cells, GSCs) responsible for tumor recurrence and resistance to therapy (6). In this context, in-depth study of gene expression programs in GBM cells and tumors, their mechanisms of regulation and functional consequences provide an opportunity to identify new targets and develop new therapeutics that will fill a major unmet clinical need in oncology.
Translation is the most energetically demanding process in the cell and its deregulation, often associated with the modification of the expression/activity of certain translation factors, contributes to development, tumor progression and the response to therapeutic treatments (7–10). Beyond an increase in global translation rates supporting cellular proliferation and growth, selective mRNA translation plays key roles in rapidly and precisely regulating gene expression steering distinct aspects of the transformed phenotype. Recent work pointed out the heterogeneity and specialized functions of individual components of the ribosome and of the translation initiation factor complexes eIF3 and eIF4F, which are required for recruitment of the ribosome to the mRNA 5′ untranslated region (5′UTR) (11).
Here, we focus on the expression and function of eIF3, a multimeric complex, composed of 13 subunits (a–m), connecting the 43S pre-initiation and the eIF4F initiation complexes. The expression of the different subunits is altered in cancers, but the function associated with this deregulation is not fully understood (9,10). Novel insights into the link between eIF3 and cancer have been provided by recent studies showing that eIF3 specifically recognizes mRNA structures (12), mRNA modifications (m6A) or, in the case of the eIF3d subunit, the 5′ cap of mRNAs (13,14), suggesting that eIF3 subunits drive selective mRNA translation in cancer. Our previous work showing that eIF3e is essential for proliferation and survival of GBM cells (15) prompted us to investigate whether, where and how this factor impacts GBM patient outcome by regulating mRNA translation. Our results support a model whereby the heterogeneous expression of eIF3e in GBM is linked to its specialized function in mRNA translational regulation and steers functional pathways involved in GBM progression and response to treatments.
MATERIALS AND METHODS
Cell lines and culture conditions
Human glioblastoma cell lines U87 (ATCC HTB-14, obtained from the American Type Culture Collection, Manassas, VA, USA), U251, LN18 and SF767 (obtained from C. Simon’s laboratory, UPENN, Philadelphia, PA, USA) were used and routinely maintained in Dulbecco’s Modified Eagle Medium (Lonza, Portsmouth, NH, USA) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in 5% CO2-humidified incubators as previously described. Cells were tested for mycoplasma contamination by PCR. When indicated, cells were treated with cobalt chloride (CoCl2; Sigma, St Louis, MO, USA) at 100 μM (2 × 50 μM at 24 h and 48 h after transfection) or with MG132 (5 μM for 2 h; Sigma, St Louis, MO, USA) or irradiated at 5 Gy using the Gammacell 40 Exactor irradiator (Nordion, Ottawa, ON, Canada).
Cell transfection
For siRNA transfections, human glioblastoma cells were transfected with 20 nmol/l of siScramble (siScr: Qiagen, Venlo, Limburg, The Netherlands) or 20 nmol/l of siRNA against human Int6/eIF3e (si3e: SI02662499, FlexiTube eIF3E siRNA, 20 nmol, 5′-CCCAAAGGUCGCGAUAAUAUU-3′, Qiagen; or si3e#2: Dharmacon ON-TARGET plus SMART pool eIF3E siRNA 3646, 10 nmol) or against DDX3X (siDDX3X: SIGMA, 50 nmol, 5′-CCUAGACCUGAACUCUUCAGAUAAU-3′), combined with Lipofectamine™ RNAiMAX as recommended by the manufacturer (Invitrogen Life Technologies, Carlsbad, CA, USA) as previously described (15). For luciferase mRNA transfections, 250 ng of reporter mRNA was transfected 48h after siRNA transfection in 48-well plates using Lipofectamine 2000 reagent according to the manufacturer’s instructions. Cells were subsequently incubated at 37°C for 16 h following mRNA reporter transfections before harvesting and analysis.
Western blot analysis
Cells were lysed in lysis buffer [50 mM HEPES, pH 7, 150 mM NaCl, 10% glycerol, 1% Triton, 100 mM NaF, 1 mM EGTA, 1.5 mM MgCl2, 10 mM sodium pyrophosphate complemented by protease inhibitors (Sigma-Aldrich, St Louis, MO, USA) at 1/100]. For immunoblotting analysis, proteins were resolved on 10% denaturing polyacrylamide gels and were transferred to nitrocellulose membranes. The blots were blocked for 30 min with TBST–5% milk and then probed overnight with primary antibodies against Int6/eIF3e (Bethyl, A302-985A), HIF-2α (Novus Biologicals, NB100-122), HIF-1α (Cayman Chemical, 10006421), GADD45α (Cell Signaling, CS4362), CDC45 (Abcam, ab-108350), FAS (Santa Cruz, sc-4233), β-actin (Santa Cruz, sc-8432), eIF3c (Cell Signaling, CS2068), eIF3a (Abcam, ab-128996), eIF3b (Santa Cruz, sc-16377), eIF3d (Bethyl, A301-758A), eIF3f (Bethyl, A303-005A), GAPDH (Santa Cruz, sc-32233), DDX3X (Santa Cruz, sc-365768), eIF4E (Cell Signaling, CS9742S), hnRNPU (Santa Cruz, sc-32315), eIF4A (Santa Cruz, sc-50354), eIF4G (Santa Cruz, sc-11373), phospho-ser209-eIF4E (Cell Signaling, CS9741S) and β-tubulin (Cell Signaling, CS2146). Gel quantification was performed using ImageJ (Research Services Branch, NIH, Bethesda, MD, USA). The secondary antibody was either HRP-conjugated anti-rabbit or HRP-conjugated anti-mouse. The blots were developed using the ECL system (Amersham Pharmacia Biotech) according to the manufacturer’s instructions.
RT-qPCR
Reverse Transcription (RT) was performed on 1 μg total RNA using the RevertAidH Minus First (Thermo Fisher) according to the manufacturer’s instructions. A total of 12.5 ng of cDNA was analyzed by qPCR with the SybrGreen (KAPA KK4605) and a StepOnePlus (Applied Biosystems/Life Technologies, Carlsbad, CA, USA). Expression of eIF3e, HIF-1α, HIF-2α, GADD45α, FAS, UBE2V1, CDC45 and GAS5 was standardized using B2M as a reference. Relative levels of expression were quantified by calculating 2^ΔΔCT, where ΔΔCT is the difference in CT (cycle number at which the amount of amplified target reaches a fixed threshold) between target and reference. All primer sequences are available in Table 1.
Table 1.
qPCR primer sequences
| Targets | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| B2M | ACCCCCACTGAAAAAGATGA | ATCTTCAAACCTCCATGATG |
| Int6/eIF3e | TTCTTCAATCACCCCAAAGG | TAGAACCTGCCGACGTTTTC |
| GADD45α | GAGAGCAGAAGACCGAAAGGA | CACAACACCACGTTATCGGG |
| UBE2V1 | CGGGCTCGGGAGTAAAAGTC | CCCCAGCTAACTGTGCCATC |
| HIF-1α | TTTACCATGCCCCAGATTCAG | GGTGGACTTTGTCTAGTGCTTCCA |
| HIF-2α | CCACCAGCTTCACTCTCTCC | TCAGAAAAAGGCCACTGCTT |
| FAS | TCTGGTTCTTACGTCTGTTGC | CTGTGCAGTCCCTAGCTTTCC |
| CDC45 | GCCATGGTGATGTTTGAGCT | TGTGTTCTCCTCATCCTCGT |
| GAS5 | TGAAGTCCTAAAGAGCAAGCC | ACCAGGAGCAGAACCATTAAG |
Flow cytometry
72 h after transfection with siRNA (siScr or si3e) and 48 h after ionizing radiation (IR) treatment (5 Gy) or CoCl2 treatment (100 μM), cells were collected, washed in Phosphate-Buffered Saline (PBS) and incubated in 100 μl of Annexin-binding buffer 5X (10 mM HEPES, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2), containing 5 μl of Annexin V-FITC antibody and 1 μl of Propidium Iodide (PI; Invitrogen Life Technologies, Carlsbad, CA, USA) solution at 100 μg/ml (FITC Annexin V/Dead Cell Apoptosis Kit with FITC Annexin V and PI; Invitrogen Life Technologies, Carlsbad, CA, USA) during 15 min at room temperature in the dark. 400 µl of Annexin-binding buffer 5X were then added after washes with PBS–bovine serum albumin 1%. Labeled cells were preserved on ice and run on a flow cytometer (BD Accuri™ C6, BD Biosciences, Franklin Lakes, NJ, USA).
Polysome profiling and isolation of total, non-polysomal, light polysomal and heavy polysomal mRNA fractions
Cells were incubated for 15 min at 37°C with 100 μg/ml cycloheximide (CHX), washed in PBS, lysed in hypotonic polysome lysis buffer (5 mM Tris–HCl, pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 100 μg/ml CHX, 2 mM DTT, 40 U/μl RNAse OUT, 10% Triton X-100, 10% sodium deoxycholate) on ice, mixed by vortexing and centrifuged at 1400 rpm for 5 min. Lysates were layered onto 15–50% sucrose gradients for fractionation (2 h at 39 000 rpm in a Beckman SW40.1 rotor and L8 ultracentrifuge at 4°C). After ultracentrifugation, fractions were harvested with a gradient fractionator and polysome profiles were continuously visualized with a monitoring at A254 nm. The fractions were collected and pooled before mRNA extraction; non-polysomal (NP), light polysomal (LP) and heavy polysomal (HP) mRNAs. Pooled fraction mRNAs and cytoplasmic mRNAs were extracted using TRIzol® reagent (Invitrogen). TRIzol® reagent was added on samples (3 volumes) and incubated for 5 min at room temperature. Chloroform was added (0.2 ml per 1 ml of TRIzol®) and samples were shacked vigorously by hand for 15 s and incubated for 2–5 min at room temperature. After centrifugation at 12 000 × g for 15 min at 4°C, the aqueous phase was recovered and submitted to the RNA isolation with isopropanol (0.5 ml per 1 ml of TRIzol®) overnight at −20°C. The samples were then centrifugated at 12 000 × g for 30 min at 4°C and RNAs were washed by adding 75% ethanol (1 ml 75% ethanol per 1 ml of TRIzol®), vortexing briefly and centrifugating samples (at 7500 × g for 5 min at 4°C). RNAs were then dried and resuspended in RNase-free water.
Microarray analysis
Before labeling, RNA concentration and integrity were measured using a Nanodrop™ spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and controlled on ‘Lab-on-Chip’ by a Bioanalyzer (Agilent Technologies, Stockport, UK), according to the manufacturer’s instructions. From each pool, 100 ng of RNA was converted into labeled cRNA with nucleotides coupled to a Cy3 fluorescent dye using the Low Input Quick Amp Labeling kit (Agilent Technologies, Palo Alto, CA, USA) following the manufacturer’s instructions. The quality and quantity of the resulting labeled cRNA were assessed using a NanoDrop™ spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). We have done two labelings/sample to obtain the quantity of Cy3 -labeled cRNA necessary for the hybridization (600ng).Each Cy3-labeled cRNA was hybridized to human microarray ship (Agilent Human V2 Microarrays, 8*60k, G4858A) for 17 h at 65°C, 10 rpm as described in the One-Color Microarray-Based Gene ExpressionAnalysis Protocol (Agilent). The hybridized arrays were then washed and scanned using an TECAN MS200 scanner. Data were extracted from the scanned image using Feature Extraction V11.5.1.1 (Agilent Technologies, Redwood City, CA, USA) and analyzed using GeneSpring v.7.2 software (Agilent Technologies). Differential translation analysis was assessed with ANOTA analysis as described previously (16).
5′UTR sequence analysis
Sequence motifs were discovered using DREME (17). Where more than one transcript from one gene was present, only the longest UTR sequence was used; 156 sequences were used in the analysis. Motifs up to 8 nt long were sought in the 5′UTR sequences against the same sequences randomized using DREME. We considered the predicted consensus sequences with P < 1 × 10−4 as significant motifs. Minimum free energies (ΔG) of 5′UTR secondary structures were calculated using the Vienna RNA package (version 1.8.5) (18). The Kolmogorov–Smirnov test was performed to investigate whether there were significant differences between eIF3e-dependent and eIF3e-independent groups.
Plating efficiency clonogenicity assay
U251 and LN18 glioblastoma cells were transfected with siRNA (siScr or si3e); after 48 h, cells were harvested and plated in six-well plates at different concentrations (10, 15 or 20 cells/cm2 for siScr and 20, 30 or 40 cells/cm2 for si3e) in duplicate. 48 h later, cells were irradiated with an IR scale (from 0 to 4 Gy) using the Gammacell 40 Exactor irradiator (Nordion, Ottawa, ON, Canada). Cells were then incubated for ∼10 days until colonies were visible with the naked eye without any joining between colonies. Then, plates were washed and cells were fixed with 10% formalin for 10 min, the formalin was removed and cells were covered with 10% crystal violet oxalate (RAL Diagnostics, Martillac, France) for 10 min; plates were rinsed with water until no additional color comes off the plate. Colonies were then counted to calculate the dose enhancement factor (DEF). DEF is measured as follows: for the same biological effect, the dose on the curve radiation alone (here Scr) is divided by the dose on the curve radiation + treatment (here 3e). A DEF >1 means that the treatment is functioning as a radiosensitizer.
Immunohistochemistry and scoring
Formalin-fixed, paraffin-embedded glioblastoma tissue sections were immersed in a Tris–EDTA buffer (pH 9) provided in the EnVision™ FLEX High pH kit K8000 (Dako, Ely, UK) and incubated for 20 min at 96°C for epitope retrieval. EnVision™ FLEX Peroxidase-Blocking Reagent was used for blocking (provided in the K8000 kit, Dako, Ely, UK) for 5 min. Sections were incubated for 1 h with diluted 1/50 (diluted in EnVision™ FLEX Antibody Diluent) human eIF3e-specific primary antibody, HPA 023973 (Sigma, St Louis, MO, USA), and washed with PBS–Tween 0.3%. Sections were incubated with secondary antibody-HRP (EnVision™ FLEX/HRP, K8000 kit, Dako, Ely, UK) for 20 min and after washes with PBS–Tween 0.3%, EnVision™ FLEX system (DAB+ Chromogen, K8000 kit, Dako, Ely, UK) was used for signal visualization according to the manufacturer’s instructions and we counterstained sections with hematoxylin (EnVision™ FLEX Hematoxylin, provided in the K8000 kit, Dako, Ely, UK). After dehydration, slides were mounted with EUKITT® mounting medium. The levels of cytoplasmic eIF3e staining were scored by two anatomopathologists using a 4-point score intensity to distinguish between ‘high’ (2 or 3 points) and ‘low’ (0 or 1 point) eIF3e staining levels. Patient survival between high and low eIF3e-stained glioblastomas was measured using the log-rank test and the Gehan–Breslow–Wilcoxon test.
Immunoprecipitation
U251 cells were lysed in lysis buffer [20 mM Tris–HCl, pH 8, 0.5% NP-40, 450 mM NaCl, 0.5% Triton, 2% glycerol complemented by protease inhibitors (Sigma-Aldrich, St Louis, MO, USA) at 1/100)]. Cell extracts were incubated for 1 h at room temperature with Benzonase (Millipore E1014) and DNase I (Thermo Scientific EN0521; see Supplementary Figure S3) and precleared with protein-Sepharose beads for 1 h at 4°C. DDX3X (Santa Cruz, sc-365768) and IgG (Invitrogen, 026502) antibodies were incubated with 20 μl of slurry beads (washed and equilibrated in cell lysis buffer) for 1 h at 4°C. Beads were incubated with 1 mg of cell extracts overnight at 4°C. Beads were washed four times with cell lysis buffer and co-immunoprecipitated proteins were analyzed by western blot.
RNA immunoprecipitation
U251 cells were lysed in lysis buffer [20 mM Tris–HCl, pH 8, 0.5% NP-40, 450 mM NaCl, 0.5% Triton, 2% glycerol complemented by protease inhibitors (Sigma-Aldrich, St Louis, MO, USA) at 1/100)]. Cell extracts were incubated for 1 h at room temperature with DNase I (Thermo Scientific EN0521) and precleared with protein-Sepharose beads for 1 h at 4°C. DDX3X (Santa Cruz, sc-365768) and IgG (Invitrogen, 026502) antibodies were incubated with 20 μl of slurry beads (washed and equilibrated in cell lysis buffer) for 1 h at 4°C. Beads were incubated with 1 mg of cell extracts overnight at 4°C. Beads were washed four times with cell lysis buffer and eluted in NT2 buffer [50 mM Tris, pH 7.4, 1 mM MgCl2, 0.05% NP-40, 0.1% SDS and 30 μg proteinase K (Qiagen, Venlo, Limburg, The Netherlands)]. RNAs are then extracted by phenol/chloroform and analyzed by RT-qPCR.
In vitro transcription
RNAs used in RNA chromatography experiments were transcribed using the MEGAscript Kit (Invitrogen AM1333) as per manufacturer’s instructions. 7.5 mM ATP/CTP/GTP, 6.75 mM UTP and 0.75 mM biotinylated UTP (Biotin-16-UTP, Lucigen BU6105H) were used. For luciferase reporter mRNAs, m7G-cap was added using the Vaccinia capping system kit (M208S NEB) according to the manufacturer’s instructions. To generate the DNA templates to synthetize the luciferase reporter mRNAs, oligonucleotides CDC45, FAS and GADD45α were amplified by PCR using the following primers. For CDC45, PCR products were used as template for in vitro transcription. For GADD45α and FAS, PCR products were cloned in the pSC-B-amp/kan plasmid from the Strataclone Blunt PCR Cloning Kit, and then digested by, respectively, BamHI and XhoI (restriction sites for transcription run-off) and purified. All oligonucleotide sequences are available in the following table (bold: T7 promoter). For GADD45α, T7 sequence used was in pSC-B-amp/kan plasmid.
| Name | Sequence |
|---|---|
| GADD45α | FW: GGAGAGCGGGGCCCTTTGTC RV: CTGGATCATAAACTTTCGAAGTCATATTGCAAACTGCAGGTCGCCC |
| CDC45 | FW: TAATACGACTCACTATAGGGAAAGAAAGGAAGGCTGGGAACTAA RV: CTGGATCATAAACTTTCGAAGTCATAGCCACGGCGGCCGGACG |
| FAS | FW: TAATACGACTCACTATAGGGATCAATGGAGCCCTCCCCAAC RV: CTGGATCATAAACTTTCGAAGTCATGGTTGTTGAGCAATCCTCCGAA |
In cellulo luciferase activity analysis
U251 cells transfected with luciferase Renilla and Firefly reporter mRNAs were harvested in 100 μl of Passive Lysis Buffer (Promega). 10 µl of this extract were analyzed with the luciferase assay.
RNA chromatography
200 µg of the U251 cells were lysed in 500 μl of lysis buffer [20 mM Tris–HCl, pH 8, 1% NP-40, 150 mM NaCl, 2 mM EDTA complemented by protease inhibitors (Sigma-Aldrich, St Louis, MO, USA) at 1/100]. Cells extracts were precleared with 20 μl of streptavidin acrylamide beads (Thermo Fisher Scientific) in the binding buffer containing 20 mM Tris, pH 8, 1 mM DTT, 100 mM KCl, 0.2 mM EDTA and 0.05% NP-40 for 1 h at 4°C. 3 µg of in vitro‐transcribed biotinylated RNAs were fixed on 10 μl of streptavidin acrylamide beads by incubation in the binding buffer for 1 h at 4°C. The RNA fixed on beads was then combined with the precleared extracts for 3 h at 4°C. The beads were collected by centrifugation, washed five times with 1 ml of the binding buffer, resuspended in 30 μl of Laemmli buffer, boiled for 5 min, loaded onto an SDS-PAGE gel and analyzed by western blot.
GBM tumor samples
For Figure 5C(i), total protein lysates in RIPA buffer (Sigma) [50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 5 mM sodium fluoride, 0.5 mM sodium vanadate and 1× protease inhibitor cocktail (Roche)] were extracted from three GBM (grade IV) and four diffuse low-grade gliomas (grade II: two astrocytomas and two oligodendrogliomas). Tumors samples were obtained from the Montpellier Hospital (‘Biological Resource Centre’, Collection NEUROLOGIE, DC-2013-2027/DC-2010-1185/Authorization AC-2017–3055/Research Protocol P487) with patient consent. All the methods used were carried out in accordance with relevant guidelines and regulations of French Institut National de la Santé et de la Recherche Médicale (INSERM). All experimental protocols were approved by INSERM. The tissues were obtained from patients, who underwent surgical resection. The tissues were processed, classified and graded as previously described. The clinicopathological parameters of the patients and tumors are described in the following table.
Figure 5.
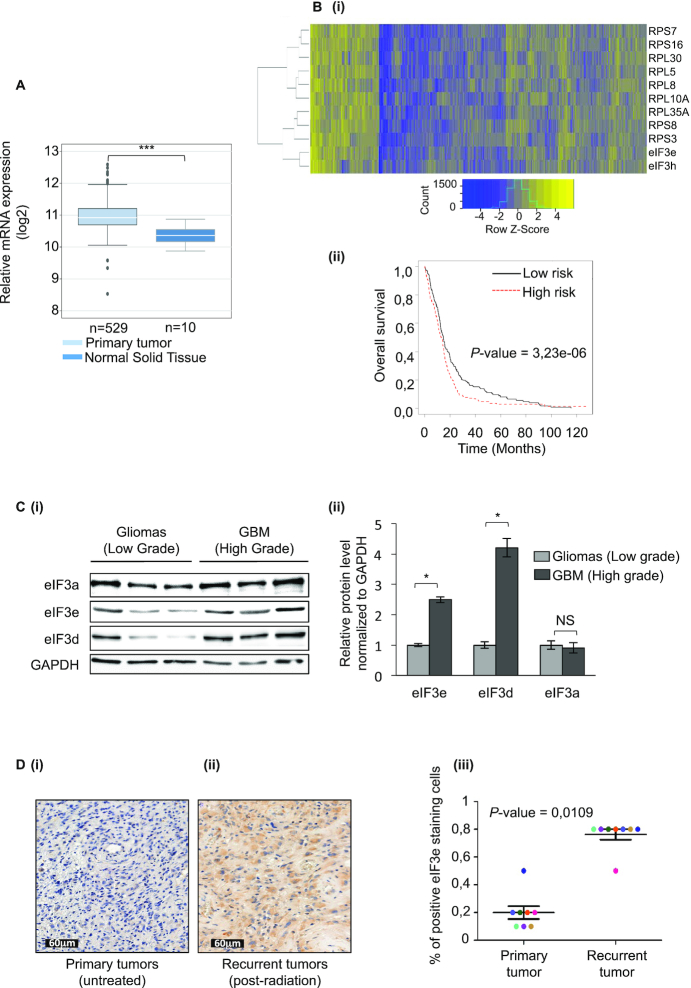
High eIF3e expression in GBM impacts GBM patient outcome. (A)eIF3e mRNA expression using TCGA data in normal tissue (n = 10) and GBM primary tumor (n = 529). The band inside the box shows the median and the whiskers show the upper and lower extremes (two-sided non-parametric Mann–Whitney test and Benjamini–Hochberg procedure was used for multiple comparisons). (B) GBM patient clustering with the top-ranked eIF3e-correlated genes (see Supplementary Table S2) (i). Kaplan–Meyer survival analysis from (ii). (C) Western blot analysis of eIF3a, eIF3e and eIF3d levels in protein extracts from diffuse low-grade gliomas (grade II) and high-grade GBMs (grade IV) (i). Protein levels in (i) from high-grade GBMs were normalized to GAPDH protein levels and plotted relatively to the levels of protein from the low-grade gliomas (ii). Data are presented as mean values ± SEM of two independent experiments; *P < 0.05, NS: non-significant (Wilcoxon test). (D) Representative IHC images for eIF3e expression in primary untreated tumors (i) and post-radiation recurrent tumors (ii) from matched GBM patients. Percentage of eIF3e positive-stained cells in primary untreated tumors and post-radiation recurrent tumors from matched GBM patients (n = 8; P < 0.05, Wilcoxon test) (iii).
| Designation | Tumor type | 1p/19q codeletion | IDH1 mutation | ATRX |
|---|---|---|---|---|
| LGG112 | Oligodendroglioma II | Deleted | R132H | Preserved |
| LGG110 | Astrocytoma II | No deletion | R132H | Loss |
| LGG141 | Astrocytoma II | No deletion | R132H | Loss |
| Gli22b | GBM | |||
| Gli25 | GBM | |||
| Gli26 | GBM |
Patient characteristics for the tumors studied in Figure 5D(iii):
| % of positive cells | ||||||
|---|---|---|---|---|---|---|
| Patient number | Gender | Age | Delay for recurrence (months) | Survival (months) | Initial diagnosis | Recurrence |
| 6 | F | 57 | 6 | 13 | 20 | 50 |
| 9 | F | 43 | 16 | 26 | 20 | 80 |
| 12 | M | 56 | 5 | 24 | 20 | 80 |
| 17 | M | 68 | 18 | 24 | 20 | 80 |
| 18 | F | 63 | 42 | 24 | 10 | 80 |
| 19 | M | 58 | 4 | 54 | 10 | 80 |
| 20 | M | 50 | 15 | 10 | 10 | 80 |
| 22 | M | 79 | 19 | 26 | 50 | 80 |
Ivy GAP
Gene expression in the various anatomical regions of glioblastoma tumors was analyzed using the Ivy Glioblastoma Atlas Project (GAP; http://glioblastoma.alleninstitute.org/). Gene expression in major anatomic structures of glioblastoma (leading edge, infiltrating tumor, cellular tumor, perinecrotic zone, pseudopalisading, hyperplastic blood vessels and microvascular area) was quantified by RNA sequencing in the Anatomic Structures RNA-Seq Study. The Ivy GAP cohort is comprised of 42 tumors.
TCGA GBM
Gene expression from 529 patients in the glioblastoma database of The Cancer Genome Atlas (TCGA; http://xena.ucsc.edu/) and 10 normal tissues was used to compare GBM subtypes and assess correlations between genes among different subgroups.
Statistical analysis
Correlations between quantitative data were assessed using the Spearman’s rank correlation coefficient. Differences between subgroups were assessed with the Mann–Whitney U test or t-test. The signed-rank test was used to assess relationships between paired data. Dunn’s pairwise comparisons were adjusted for multiple testing with Benjamini–Hochberg correction. For the TCGA cohort, overall survival was defined as the time from initial diagnosis to death from any cause (event) or the last follow-up (censored data). Univariable and multivariable analyses were performed using the log-rank test and Cox proportional hazards model; hazard ratios (HR) were estimated with 95% confidence intervals. Univariable analyses were performed using the log-rank test. Two-sided P < 0.05 was considered statistically significant. Significance was represented by asterisks, where *P < 0.05, **P < 0.01 and ***P < 0.001.
RESULTS
eIF3e regulates HIF mRNA translation
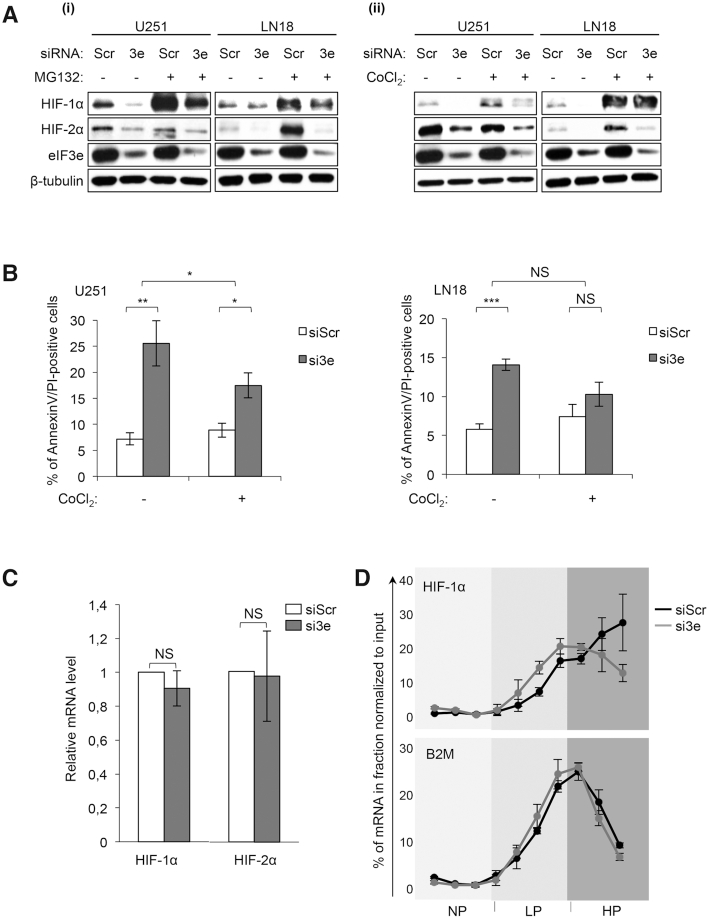
We previously demonstrated that eIF3e inhibition resulted in decreased expression of the hypoxia-inducible factors (HIFs) in GBM cells associated with a decreased proliferation and increased apoptosis (15). HIFs are transcription factors, composed of two subunits: HIF-α and HIF-β, and the two major HIF-α subunits are HIF-1α and HIF-2α. These factors control the transcription of >150 genes important for GBM cells, mainly involved in angiogenesis, metabolism, proliferation and cell migration (19). HIFs and hypoxia are known to compromise the cytotoxic effects of IR and limit the success of radiotherapy (20). To further investigate the eIF3e–HIF interplay, we defined whether eIF3e depletion in U251 and LN18 GBM cell lines downregulated HIF proteins by transcriptional or post-transcriptional mechanisms. As shown in Figure 1A, knockdown of eIF3e in GBM cells decreased HIF protein levels and this effect was maintained in conditions that induce HIF protein accumulation as a result of protein stabilization by the proteasome inhibitor MG132. Consistent with this result, treating GBM cell lines with CoCl2, a hypoxia mimetic agent that stabilizes HIF proteins, only partially reverses HIF protein loss (Figure 1A) or apoptosis (Figure 1B) induced by eIF3e silencing. The percentage of apoptotic cells (increasing from 7.2% to 25.5% in U251 and from 5.8% to 14% in LN18) following eIF3e depletion is indicative of biological significance. These results demonstrated that the effect of eIF3e modulation on HIF protein levels was related neither to GBM cell type nor solely to a change in protein stability. In addition, eIF3e silencing in U251 cells did not alter HIF mRNA levels (Figure 1C).
Figure 1.
eIF3e regulates HIF translation in glioblastoma cells. (A) Western blot analysis of HIF-1α and HIF-2α in GBM cell lines treated with control (siScr) and eIF3e (si3e) siRNAs, with or without a proteasome inhibitor treatment (MG132 at 5 μM for 2 h) (i) and with or without a hypoxia mimetic treatment (CoCl2 at 150 μM for 12 h) (ii). Shown is a representative result from two to four experiments. (B) Annexin V/PI staining and flow cytometry analysis for GBM cells treated as in (A). Data are presented as mean values ± SEM of four independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001 (paired t-test). (C) Quantitative RT-qPCR using specific primers for HIF-1αand HIF-2α mRNAs extracted from U251 cells treated with control (siScr) or eIF3e (si3e) siRNAs. Data are plotted relatively to B2M mRNA amounts and are presented as mean values ± SEM of four to six independent experiments. NS: non-significant (Wilcoxon test). (D) RT-qPCR analysis using specific primers for HIF-1α and B2M mRNAs extracted from translationally inactive (NP) or active (including LP and HP) fractions obtained after fractionation on a sucrose gradient of cytoplasmic extracts of U251 cells treated with control (siScr) or eIF3e (si3e) siRNAs. Shown is a representative distribution of two independent experiments.
To explore whether the amount of HIF mRNA engaged in translation was affected by the eIF3e knockdown, we transfected U251 cells with eIF3e-specific (si3e) or control siRNAs (siScr) and then fractionated cytoplasmic extracts of these cells by sucrose density gradient centrifugation. RNA was isolated from translationally inactive (NP) or active (including LP and HP) fractions, and the distribution of HIF mRNA (or the control B2M mRNA) along these fractions was determined by RT-qPCR. We observed that eIF3e loss induced a change in the distribution of the HIF-1α mRNA (Figure 1D), but not the control B2M mRNA or the HIF-2α mRNA (data not shown), along the sucrose gradient. Indeed, HIF-1α mRNAs shifted from the highest to the lowest polysome fractions, suggesting that eIF3e inhibits HIF-1α mRNA translation.
eIF3e affects the translation of mRNAs relevant to GBM progression and response to treatments
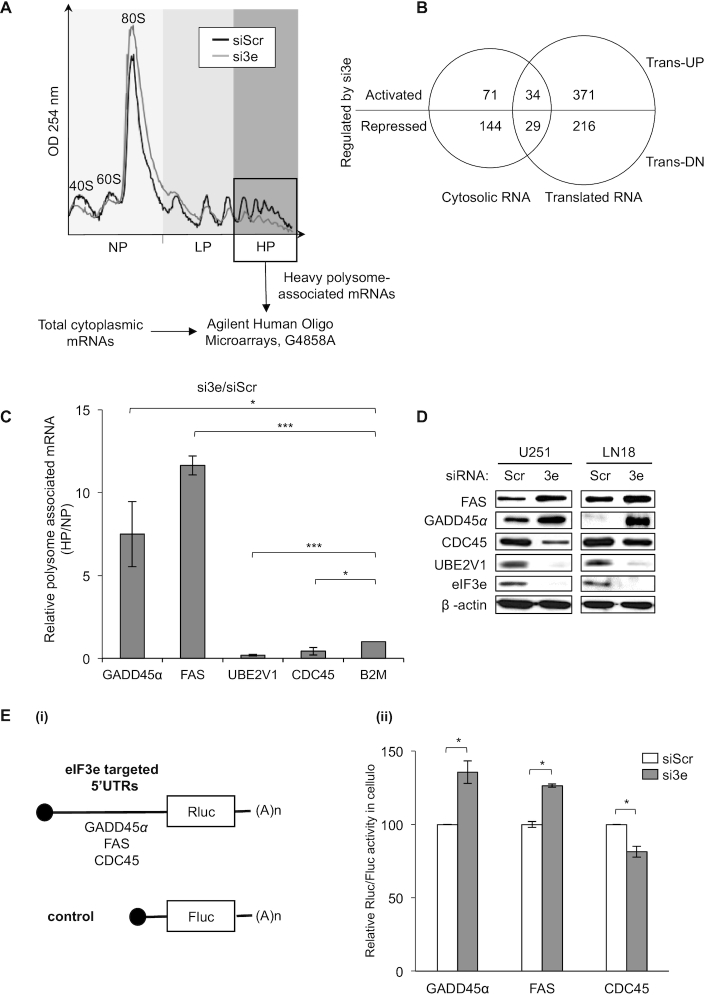
Next, based on eIF3 functions in the regulation of both global and mRNA-specific translation (11), we further investigated the potential involvement of eIF3e in protein synthesis. We previously reported that eIF3e knockdown had no significant impact on bulk protein synthesis in GBM cells (15). Therefore, we pushed further our analysis to identify mRNAs whose translation is mediated by eIF3e by performing genome-wide analysis of mRNA polysomal profiles in U251 GBM cells. Interestingly, sucrose density gradient separation revealed no major differences in polysome distribution in U251 cells where eIF3e is silenced (Figure 2A and Supplementary Figure S1A). These data show that eIF3e modulation does not significantly affect global mRNA translation, which corroborates our previous 35S-methionine metabolic labeling of protein synthesis (15). However, combining polysome profiling and microarray hybridization, we identified 278 differentially expressed mRNAs between control cells and cells transfected with si3e in total cytosolic fraction, and 650 differentially translated mRNAs associated with polysomes (Supplementary Table S1). Interestingly, the abundance and/or polysome association of 405 mRNAs were increased upon eIF3e knockdown, whereas those of 245 mRNAs were decreased (thereafter called trans-UP and trans-DN, respectively; Figure 2B).
Figure 2.
eIF3e drives selective mRNA translation of mRNAs involved in GBM tumor progression and response to treatments. (A) Polysome profile of U251 cells treated with control (siScr) or eIF3e (si3e) siRNAs. mRNA was isolated from fractions containing three ribosomes (box) and analyzed in parallel with cytoplasmic mRNA using microarrays (see the ‘Materials and Methods’ section). The peaks corresponding to the 40S, 60S and 80S ribosomal subunits and polysomes are indicated, as well as NP, LP and HP fractions. (B) Microarray data analysis using a Venn diagram showing the overlap between cytosolic mRNAs and translationally regulated mRNAs. The number of eIF3e activated or repressed mRNAs in each group is indicated. (C) Validation of the genome-wide polysome profiling in (A) by RT-qPCR analysis from pooled NP and HP fractions using specific primers for the indicated mRNAs and quantification by analyzing the ratio HP/NP [error bars ± SEM of three experiments; *P < 0.05, ***P < 0.0005 (paired t-test)]. (D) Western blot analysis of the indicated proteins in U251 and LN18 cells treated with control (siScr) or eIF3e (si3e) siRNAs. (E) Ratio of Renilla/Firefly luciferase activities (Rluc/Fluc) (ii) determined using U251 cells treated with control (siScr) and eIF3e (si3e) siRNAs, followed by co-transfection with Rluc and Fluc mRNA reporters depicted in (i) [error bars ± SEM of three independent experiments; *P < 0.05 (paired t-test)].
Within the mRNAs affected by eIF3e deletion in GBM cells, we focused our attention on the altered expression of those that are relevant to cancer progression and response to treatments. Translation of two major mRNAs of the p53 pathway, GADD45α and FAS, was increased, respectively, by 6- and 3.5-fold, and translation of the survival and DNA replication/repair response mRNAs FGF11, UBE2V1, JAK3 and CDC45 was decreased, respectively, by 5-, 4-, 2- and 2-fold, in eIF3e-depleted U251 cells (Supplementary Table S1). In order to confirm our analysis, we validated some of these targets, including HIF-1α, by RT-qPCR on polysomal fractions and total mRNAs from U251 cells (Figure 2C and Supplementary Figure S1B). The expression of the eIF3e-regulated targets identified above followed the same pattern as observed in the microarray data analysis. More specifically, we observed an increased translation of GADD45α and FAS and a decreased translation of UBE2V1 and CDC45 mRNAs (Figure 2C). Consistent with Figure 1D, HIF-1α mRNA translation was significantly decreased by 2-fold in eIF3e-depleted U251 cells (Supplementary Figure S1B). Additionally, we were able to confirm these changes of mRNA levels in polysomes at the protein level by western blot analysis in several GBM cell lines (Figure 2D and Supplementary Figure S1C and D). Although the effects at the translation and protein levels for all genes were qualitatively consistent, we noted a quantitative difference, in particular for GADD45α and FAS (Figure 2D and Supplementary Figure S1C). While this could be explained by previous observations suggesting a rapid turnover of these two proteins (21,22), it remains to be determined in future studies whether this discrepancy is related to eIF3e.
To directly investigate whether the 5′UTR drives eIF3e-specific translational regulation, we focused on validating eIF3e targets that were highly regulated, either positively or negatively. To this end, we transfected RNA reporters containing the luciferase open reading frame fused to the 5′UTRs of GADD45α, FAS or CDC45 mRNAs into U251 cells (Figure 2E). In agreement with both polysomal profiling and western blot analysis, we found that the activity of 5′UTR–luciferase fusions was either upregulated (for GADD45α or FAS reporters) or downregulated (for CDC45 reporters) upon knockdown of eIF3e relative to control cells (Figure 2E). Differences in the magnitude of the effects (Figure 2C–E and Supplementary Figure S1C and D) indicate the possibility of complex translational and possibly post-translational regulation, which is quite consistent with the multivalent function of eIF3e in translation, and more generally in gene expression. Overall, these results suggest that eIF3e affects the selective translation of mRNAs with opposite effects on protein synthesis and regardless of the GBM cell line. Of note, mRNAs were not the only type of transcripts whose association with polysomes was modulated by eIF3e depletion. Indeed, among the RNAs targeted for translation by eIF3e, we found several other RNA types, including long non-coding RNAs (lncRNAs; Supplementary Figure S2A). While the regulation of lncRNAs by eIF3e remains to date unappreciated, this is consistent with the work of Lee et al. who showed a direct interaction between eIF3 and several ncRNAs (12). To validate this result, we focused on GAS5 since this lncRNA is translationally regulated by eIF3e (Supplementary Table S1) and directly targeted by eIF3 subunits in living cells (12). Importantly, GAS5 plays a role in glioma cell growth, invasion and apoptosis (23). Consistent with polysome profiling data (Supplementary Table S1), we found that eIF3e silencing increased the association of GAS5 with polysomal fractions (Supplementary Figure S2A).
Next, we sought to go further in the investigation of the translatome of eIF3e. Recent transcriptome-wide analysis of in cellulo eIF3–RNA interactions revealed that eIF3 subunits preferentially interacted with specific subsets of mRNAs, predominantly in 5′UTRs (12). Further structure–function investigation of eIF3 target mRNAs uncovered the existence of specific stem–loop structures with opposite effects on translation (12). These results are consistent with our eIF3e translation data (Figure 2) and led us to explore and compare 5′UTR features of both eIF3e translationally repressed and activated mRNAs. We found that trans-UP mRNAs had longer 5′UTRs with greater predicted secondary structure stability [i.e. lower minimum free folding energy (ΔG)] compared to the trans-DN mRNAs. However, there was no significant relationship with the G/C content (Supplementary Figure S2B). In the trans-UP group of mRNAs, but not in the trans-DN mRNAs, the DREME software showed the enrichment of partially self-complementary GC-rich sequences, possibly forming secondary structures (Supplementary Figure S2C). The lack of motif enrichment in the trans-DN mRNAs suggested that this effect is not related to a particular sequence or structure.
Cooperation of eIF3e, eIF3d and DDX3X drives selective translational regulation
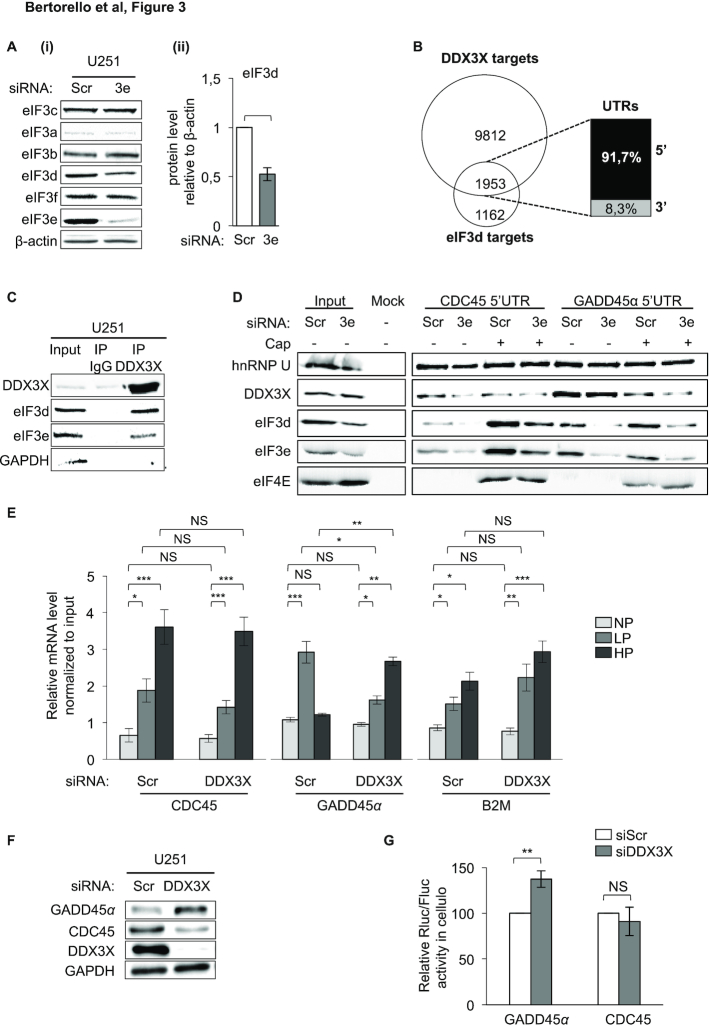
Previous works demonstrated that eIF3e forms a binary complex with the d subunit, eIF3d, of the eIF3 multimeric assembly. Moreover, this interaction occurs independently of the whole eIF3 complex and requires the C-terminal region of eIF3e for assembly into eIF3 (24). Based on these observations, it has been proposed that eIF3e forms a module with eIF3d (25). Since eIF3e could lead to co-depletion of eIF3d (24,26,27), we tested whether this occurs in GBM cells. We found that eIF3e silencing reduced eIF3d expression while leaving the other subunits unaffected (Figure 3A). This suggests that, as proposed by Shah et al. (27), eIF3e could function in synergy with eIF3d to orchestrate an mRNA-specific translational program in GBM cells. Since, unlike eIF3d, eIF3e does not have RNA interaction domains, its binding to the RNA could be mediated by eIF3d. Therefore, to get insights into the molecular mechanism underlying the function of eIF3e in mRNA-specific translation, we studied the network of RNA-binding proteins (RBPs) associated with the 5′UTRs bound by eIF3d (derived by available CLIP datasets) (12) using the AURA database (28). The co-regulation analysis revealed that the most enriched factor bound to eIF3d-associated 5′UTRs (Supplementary Figure S4A) is DDX3X, an RNA helicase that plays opposite roles in mRNA translation (29–31). eIF3d and DDX3X (32) shared 1953 sites (62.7% of eIF3d hits, 16.6% of DDX3X hits) that map mainly to 5′UTRs (Figure 3B). Collectively, eIF3d and DDX3X share many target mRNAs, suggesting a possible co-regulation of common target mRNAs by these two RBPs.
Figure 3.
eIF3e’s mechanism of regulation involves eIF3d and DDX3X. (A) Western blot analysis of the indicated eIF3 subunits in U251 cells treated with control (Scr) or eIF3e (3e) siRNAs (i). eIF3d protein levels in (i) were normalized to β-actin protein levels and plotted relatively to the Scr condition (ii). Data are presented as mean values ± SEM of nine independent experiments; *P < 0.05 (Wilcoxon test). (B) Venn diagram showing unique or common 5′UTR CLIP-derived binding sites between DDX3X (32) and eIF3d (12). (C) Immunoprecipitation of U251 total extracts, followed by western blot analysis and probing with the indicated antibodies. Shown is a representative result from two independent experiments. (D) RNA chromatography using the 5′UTR of the CDC45 and GADD45α RNAs either capped or not and total cell extracts from U251 cells treated with control (Scr) or eIF3e (3e) siRNAs, followed by western blot analysis. Shown is a representative result from two independent experiments. (E) Quantitative RT-qPCR analysis from pooled NP, LP and HP fractions, using specific primers for indicated mRNAs extracted from U251 cells treated with control (Scr) or DDX3X siRNAs. Data are plotted relatively to input mRNA amounts and are presented as mean values ± SEM of three independent experiments; NS: non-significant, *P < 0.05, **P < 0.005, ***P < 0.0005 (paired t-test). (F) Western blot analysis of the indicated proteins in U251 cells treated with control (Scr) or DDX3X siRNAs. Shown is a representative image of three independent experiments. (G) Ratio of Renilla/Firefly luciferase activities (Rluc/Fluc) determined using U251 cells treated with control (siScr) or DDX3X siRNAs, followed by co-transfection with Rluc and Fluc mRNA reporters. Data are presented as mean values ± SEM of three independent experiments; NS: non-significant, **P < 0.005 (paired t-test).
The possibility that eIF3e, eIF3d and DDX3X may work in concert on the same targets to regulate their translation was first tested by immunoprecipitating DDX3X using GBM cytoplasmic extracts. In agreement with (33), we found that DDX3X co-immunoprecipitated both eIF3 subunits (but not GAPDH that also displays RNA-binding activity) in an RNA/DNA-independent manner (Figure 3C and Supplementary Figure S3). Then, we analyzed whether DDX3X interacted with mRNAs translationally regulated by eIF3e. The intersection of datasets from eIF3e polysome profiling (Supplementary Table S1) and DDX3X CLIP data (32) revealed a significant enrichment of DDX3X with the mRNAs whose translation is increased after eIF3e silencing (Supplementary Figure S4A). Then, we performed RNA chromatography to verify the binding status of eIF3e, eIF3d and DDX3X on two previously validated eIF3e mRNA translational targets, CDC45 and GADD45α, which we showed to be down- and upregulated by eIF3e, respectively (Figure 2C–E). Consistent with the enrichment of DDX3X binding sites on the trans-UP mRNAs (Figure 3B and Supplementary Figure S4A), we found that DDX3X preferentially bound the GADD45α 5′UTR compared to the CDC45 5′UTR (Figure 3D). This differential binding was most evident when comparing uncapped RNAs, and a similar pattern was shared by eIF3e/eIF3d, suggesting that the binding of the three factors to the GADD45α 5′UTR is cap independent and relies on a specific sequence/structure. Another important difference between the two RNAs was the differential binding of eIF3e/eIF3d to capped and uncapped CDC45 or GADD45α RNAs. Indeed, while the recruitment of eIF3e/eIF3d on the CDC45 5′UTR occurred mainly following the addition of the cap, their binding to the GADD45α was mainly cap independent with only a limited change after cap addition (Figure 3D). Consistent with these in vitro results, immunoprecipitation of DDX3X bound to mRNAs from U251 GBM cells resulted in preferential binding of DDX3X to the GADD45α mRNA compared to the CDC45 mRNA (Supplementary Figure S4B). In addition, similarly to Figure 3D, loss of eIF3e decreased the recruitment of DDX3X on GADD45α mRNA but not on CDC45 mRNA (Supplementary Figure S4B), indicating an RNA-specific RBP-helicase cooperative recruitment. To further support this model, we performed polysome analysis of DDX3X-depleted U251 cells (Supplementary Figure S4C). We observed that DDX3X silencing did not significantly modify global polysomal profiles (Supplementary Figure S4C), yet the translation of the GADD45α mRNA was significantly increased compared to that of CDC45 or the B2M control mRNA (Figure 3E). This conclusion was further supported by the analysis of the effect of DDX3X silencing on protein accumulation (Figure 3F and Supplementary Figure S4D) and expression from RNA reporters (Figure 3G). Overall, these results show that the activating and repressive effects of eIF3e on mRNA translation might be explained in part by a distinct binding pattern of eIF3e, eIF3d and DDX3X on the target mRNAs, suggesting the existence of an RBP-helicase cooperative interaction mechanism (34).
In addition to these differential protein/RNA interactions, and in agreement with previous results (35), eIF3e depletion induced a decrease in the phosphorylation of eIF4E (Supplementary Figure S4E). MNK-mediated phosphorylation of eIF4E affects eIF4F activity and is associated with cancer progression, likely by modulating the translation of specific mRNAs (36,37), suggesting that this modification might contribute to the mechanisms underlying eIF3e-driven selective mRNA translation.
eIF3e modulation impact on GBM cell radiosensitization
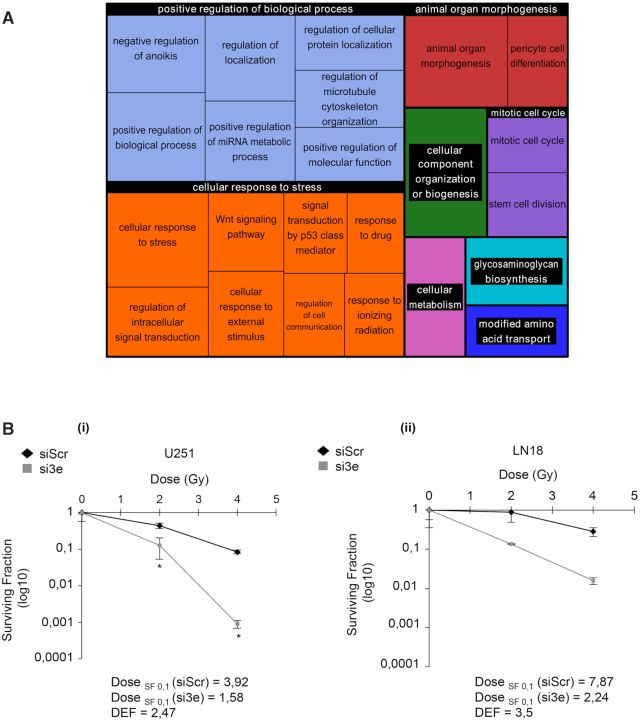
To explore the biological consequences associated with eIF3e modulation, positively and negatively eIF3e-regulated mRNAs were subjected to gene ontology (GO) enrichment analysis. The 405 genes that are preferentially translated after eIF3e silencing were largely enriched for genes involved in the cellular response to stress, including response to external stimulus, drugs or IR (Figure 4A). Strikingly, proteins encoded by the trans-DN mRNAs may also influence these pathways. Indeed, among these genes we found ALDH1A (Supplementary Table S1), a GSC marker that is associated with tumor growth and resistance to chemoradiotherapy (38). Radiation is a fundamental component of GBM therapeutic treatments, though radiation-surviving cells can evolve toward more aggressive and invasive phenotypes, constituting one major resistance mechanism (39,40). Tumors respond to radiation-induced DNA lesions by triggering a sophisticated network of DNA damage signaling and repair pathways that determine cellular fate, including survival, death and genomic stability. Based on our previous findings (15), we wondered whether eIF3e silencing could enhance the cytotoxic effects of radiation. To assess the radiosensitizer potential of eIF3e inhibition, we depleted eIF3e in U251 and LN18 GBM cells (that are radiosensitive and radioresistant, respectively) and performed a plating efficiency clonogenic assay. Interestingly, we observed that eIF3e silencing significantly decreased the survival fraction after irradiation of U251 cells with a mean DEF >1 (Figure 4B). A similar trend was observed for LN18 cells (Figure 4B). This suggests that eIF3e inhibition leads to a radiosensitization phenomenon in both U251 and LN18 cells. Overall, these results suggest that eIF3e knockdown could impact GBM cell response to treatments.
Figure 4.
eIF3e silencing increases GBM cell radiosensitization. (A) GO enrichment analysis of the mRNAs that are upregulated after eIF3e silencing and plotted with REVIGO. Size of the rectangles reflects the P-value (log10). (B, C) Plating efficiency assays measuring the surviving fraction in LN18 (B) or U251 (C) cells treated with control (siScr) or eIF3e (si3e) siRNAs and submitted to a radiation dose scale. Data are presented as mean values ± SEM of three independent experiments; **P < 0.005, ***P < 0.0005 (paired t-test). DEF for U251 and LN18 cells is measured as follows: for the same biological effect (here surviving fraction of log 0.1), the dose on the curve radiation alone (here siScr) is divided by the dose on the curve radiation + treatment (here si3e)). A DEF >1 means that the treatment is functioning as a radiosensitizer.
Deregulated expression of eIF3e in specific GBM regions could impinge on selective protein synthesis impacting GBM outcome
The murine Int6 gene, encoding eIF3e, was originally identified as a site of mouse mammary tumor virus (MMTV) integration in MMTV-induced tumors and a neoplastic lesion (41). Since then, the role of eIF3e in tumorigenesis has been controversial and various reports have implicated differing roles for this protein, both as an oncogene and as a tumor suppressor depending on cancer type, tumor stage or the baseline level of eIF3e expression (42,43). To examine the expression pattern of eIF3e in GBM patients, we analyzed eIF3e mRNA expression on a large cohort of GBM cancer patients (Cancer Genome Atlas Network) made available by the TCGA (https://www.cancer.gov/tcga). Strikingly, eIF3e mRNA expression was significantly increased in the primary GBM tumors compared to normal tissue (Figure 5A). This result was reinforced by data extracted from the Oncomine database (http://www.oncomine.org), where several sets that compare expression in GBM with normal brain show a highly significant increase in eIF3e mRNA levels (Supplementary Figure S5A). A Pearson’s correlation analysis was performed using the TCGA database, and the results indicated that eIF3e expression was highly correlated with genes with functions in protein synthesis and translational regulation (Supplementary Table S2). As shown in Figure 5B, stratification of samples along low and high expression (median threshold) of the top-ranked eIF3e-correlated genes showed significant differences in overall survival, with high expression associated with poor prognosis. The risk score based on eIF3e and top-ranked correlated genes (linear predictor) was significantly associated with overall survival (HR = 2.72 [1.78; 4.14], P < 0.0001) and remains a good prognostic factor when adjusting on clinical parameters (P = 0.0249; Supplementary Figure S5B). Thus, multivariate analysis supports the clinical relevance of the eIF3e signature. Additionally, eIF3e overexpression in human GBM samples was confirmed at the protein level by western blot analysis of graded human glioma specimens (Figure 5C). The expression of eIF3e correlated with morphologically determined tumor grades, with low levels in low-grade tumors and high levels in high-grade tumors. To investigate whether these results were clinically relevant, we collected eight matched GBM tissues from the initial surgery for untreated tumors and from the second surgery after treatment failure with radiation and chemotherapy (temozolomide) for eIF3e immunohistochemistry (IHC) analysis. We observed an overall trend of increased eIF3e expression in recurrent tumors compared with primary untreated tumor sections (Figure 5D). Similarly, the proportions of eIF3e-positive tumor cells were generally increased in recurrent tumors compared with matched primary tumors (Figure 5D). Collectively, these results show that eIF3e is upregulated in post-treatment GBM tissues, indicating that eIF3e may play an important role in the development of chemo- and radioresistance.
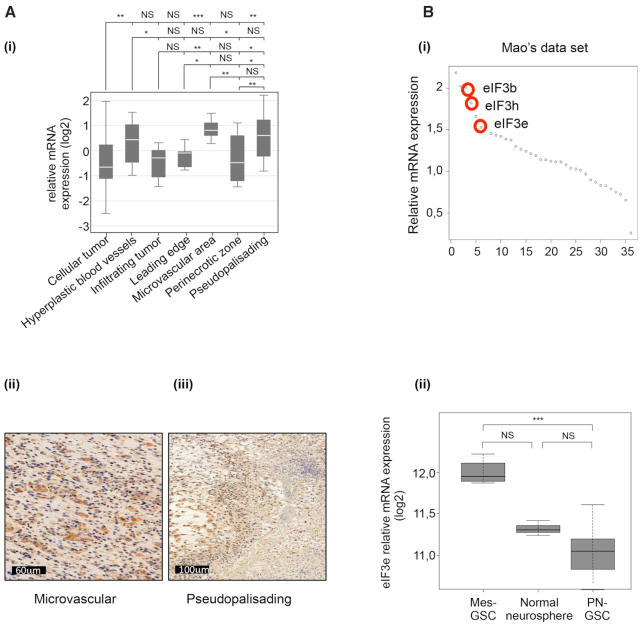
GBMs are highly heterogeneous malignancies at histological, genetic and gene expression levels (5,44). With the aim to characterize eIF3e mRNA distribution in different GBM histological regions, we explored the Ivy GBM database (http://glioblastoma.alleninstitute.org/) that provides RNA sequencing data from seven laser-microdissected GBM regions: leading edge, infiltrating tumor, cellular tumor, perinecrotic zone, pseudopalisading cells around necrosis, hyperplastic blood vessels and microvascular proliferation. Data analysis indicated that the expression of eIF3e was highly enriched at the microvascular proliferation and pseudopalisading areas of tumors compared with other regions within the tumor (Figure 6A). To validate these results, we performed IHC and observed that eIF3e staining colocalized with both GBM regions. However, eIF3e subunits showed differential expression over distinct regions of GBM (Supplementary Figure S6A). For instance, the eIF3c mRNA did not show significant enrichment in a specific area of the tumor, while eIF3a expression was significantly increased in hyperplastic blood vessels and microvascular proliferation area but not in pseudopalisading region (Supplementary Figure S6B). It is important to note that the expression of eIF3e in both regions is correlated with gene groups having different functions (Supplementary Figure S7 and Supplementary Table S2). Specifically, in pseudopalisading we found several terms associated with translational regulation. Among the best-correlated genes, we noticed several mRNAs containing an oligopyrimidine tract at their 5′ termini (5′ TOP). These mRNAs encode components of the translational machinery whose expression at the transcriptional and translational levels is sensitive to mTOR regulation. This observation, together with previous findings showing that phosphorylated mTOR displayed a predilection for pseudopalisading perinecrotic cells in GBM (45), suggests that eIF3e is part of a specific gene expression program requiring coordinated activation of the translational machinery, possibly under control of mTOR. By contrast, the expression of eIF3e in the microvascular area was correlated with genes involved in chromosome duplication (Supplementary Figure S7), in accordance with the notion that endothelial cells of tumor blood vessels have high frequencies of excess centrosomes (46).
Figure 6.
eIF3e expression in different GBM histological regions and in GSCs. (A) Relative mRNA expression for eIF3e in different tumor regions from the Ivy GBM database. Data are presented as mean values ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001, NS: non-significant (Benjamini–Hochberg test) (i). Representative IHC images for eIF3e expression in microvascular proliferating (ii) and pseudopalisading regions (iii). (B) Genome-wide transcriptome microarray analysis (GSE67089) showing that eIF3e was among the most upregulated translation factors in glioma spheres (i). mRNA expression analysis (GSE67089) of eIF3e in the mesenchymal (Mes) and the proneural (PN) subtypes of GSCs compared with normal neurosphere (ii). *P < 0.05 and ***P < 0.001, by one-way ANOVA followed by Dunnett’s post-hoc test. n = 6 for PN, n = 3 for Mes and n = 1 for normal neurosphere.
Pseudopalisading necrosis and microvascular proliferation are unique histological features that differentiate GBM from lower grade gliomas and make GBM one of the most hypoxic and angiogenic of all tumors (47). Pseudopalisading cells express high nuclear levels of HIF-1α that in turn activates an adaptive transcriptional response to hypoxia, which includes activation of glycolytic metabolism, secretion of proangiogenic factors and increased cell migration. Importantly, the pseudopalisading areas with a necrotic core create the hypoxic niche for GSCs. Indeed, hypoxia promotes stemness through the activation of genes implicated in self-renewal and dedifferentiation (including HIF-1α), and protects tumor cells and GSCs from chemo- and radiotherapy (48). Since we observed that eIF3e regulates the expression of HIFs (Figure 1) and GSC markers (ALDH1A1; Supplementary Table S1), we hypothesized that a deregulation of its expression in GSCs could contribute to cell stemness by selectively modifying translation of key GSC regulators. To investigate the expression of eIF3e in GSCs, we analyzed the microarray data from Mao’s dataset (GSE67089) including 11 GSC and 5 normal astrocyte samples (38). We found that eIF3e was one of the most upregulated translation factor-encoding gene in GSCs compared with normal human astrocytes (Figure 6B). Then, we analyzed whether eIF3e expression changed between the Mes and the PN subtypes. These are characterized by striking genetic and phenotypic differences, including aberrantly high expression of ALDH1A, marked resistance to radiation and more aggressive phenotype in Mes GSCs (38). Interestingly, eIF3e expression was specifically higher in the Mes subtype than PN one (Figure 6B), indicating that eIF3e might be involved in GBM heterogeneity through directing specific gene expression programs associated with therapeutic resistance in GSCs.
DISCUSSION
The eIF complex is a paradigm of the complexity of the translational apparatus, linked to the deregulation of the expression of its components, their modulability and heterogeneity in cancer cells (7–10), whose understanding is key for elaborating novel and effective targeting strategies. More recently, this notion of complexity has been broadened by the, as yet poorly understood, ability of some factors, including eIF3 (12,13), to influence both the basic functional mechanisms and those guiding their activity to shape mRNA translation programs allowing cells to respond and adapt to environmental conditions. By focusing on the e subunit of the eIF3 complex, this work revealed additional facets of this complexity associated with both intratumoral heterogeneity of eIF3 expression and the combinatorial activation and repression of eIF3e translational programs linked to GBM resistance to radiation therapy.
Underlying the reduced sensitivity to radiation is the ability of cells to repair damaged DNA, limit apoptosis, evolve to a stem-like phenotype and cope with hypoxic stress and metabolic reprogramming, among others. Our results support the notion that eIF3e is a hub that coordinates these different pathways through combinatorial activation and repression of synthesis of key protein factors at the crossroad of different resistance mechanisms. Among the genes positively regulated by eIF3e, HIF-1α stands out because its enhanced expression after irradiation enables adaptation to hypoxic stress through gene expression reprogramming impacting angiogenesis, apoptosis, migration and, as reported more recently, metabolism and is a potential target for the development of future GBM therapeutics (49). A possible link between eIF3e and reprogramming of GBM metabolism is also suggested by our polysome profiling analysis showing selective translation of genes involved in glycolysis/gluconeogenesis, including the stem cell marker ALDH1 (Supplementary Table S1). Considering that (i) eIF3e, HIF-1α and ALDH1 are strongly expressed in high-grade glioma (Figure 5) (50), particularly in hypoxic pseudopalisading regions including niches for GSC and conferring survival advantage in the tumor microenvironment (Figure 6) (51), (ii) the expression of eIF3e is correlated in these areas with a translational signature of poor prognostic possibly related to the mTOR control (Figure 5), (iii) eIF3e increases the translation (but not the accumulation) of HIF-1α (Figure 1) and ALDH1 mRNAs (Supplementary Table S1) and finally (iv) a link between eIF3e and glucose metabolism has recently been established (27,52,53), we propose that eIF3e could activate selective translational programs related to energy metabolism contributing to the mechanisms underlying resistance to treatments in specific GBM regions.
Our polysome profiling results, which were validated at the mRNA and protein levels as well as using RNA reporters (Figure 2), suggest that eIF3 could concomitantly modulate these mechanisms by repressing the translation of mRNAs involved in the stress response, including GADD45α. The activation of this gene with multiple functions in DNA repair, cell cycle, senescence and induction of apoptosis is essential for mediating anticancer activity of multiple genotoxic treatments and its absence might abrogate their effects in cancer cells (54). In GBMs and GSCs, radiation modifies gene expression primarily at the level of translation and GADD45α has been found targeted for increased protein synthesis (55–57). Our study suggests that translational repression modulated by eIF3e high expression in GBM adds to the post-transcriptional regulatory mechanisms targeting GADD45α for both selective repression in unstressed cells and activation upon DNA damage (58,59).
In addition to the alteration of expression and opposite translational effects, this work highlights that the complexity of eIF3e-mediated translational regulation involves multiple overlapping mechanisms. First, the abundance of eIF3e impacts the accumulation and/or activity of other translation factors, including other eIF3 subunits, notably eIF3d [in agreement with (24,27) and Figure 3] with which eIF3e interacts, and eIF4E at both the phosphorylation [in agreement with (35)] and mRNA translation levels (Figure 3 and Supplementary Table S1). Second, the eIF3e-dependent regulatory mechanisms rely on sequence/structure features and protein partners (Figure 3) whose combination may determine the translational outcome. Among the trans-regulators, we identified DDX3X enriched on mRNAs repressed by eIF3e (Figure 3), including GADD45α mRNAs (Figure 3). These results are consistent with the observation that these transcripts have longer and more stable 5′UTRs. While the role of DDX3X bound to these RNAs requires further investigation, the pattern of protein interaction to the CDC45 and GADD45α mRNAs (which are activated and repressed by eIF3e, respectively) displayed a different requirement of eIF3e/eIF3d/DDX3X for the presence of the 5′ cap (Figure 3). The ability of eIF3e/eIF3d to similarly bind uncapped and capped GADD45α mRNAs strongly supports the possibility that eIF3e/eIF3d selectively inhibit translation through binding cis-regulatory elements/structures within the 5′UTR (12). Cap addition resulted in a decreased binding of DDX3X to GADD45α 5′UTR, but not to CDC45 5′UTR, which could be explained by its displacement from direct binding to RNA and indirect recruitment by eIF4E, thus participating in the translational repression of GADD45α through the formation of a DDX3X–eIF4E inhibitory complex (30). Based on the observation that DDX3X is abnormally increased in high-grade gliomas (60), that this increase in expression induces nucleation of stress granules (SGs) depending on the DDX3X–eIF4E interaction (61,62) and that SGs associated with translational inhibition also contain eIF3 subunits (63), it is possible that the translational regulation of eIF3e targets involves these cellular compartments (63). However, it seems unlikely that eIF3e/DDX3X regulation would involve upstream open reading frames, as proposed for some (but not all) eIF3 subunits (31), since we did not find ATF4 mRNA as a translational target for eIF3e.
Taken together, these data indicate that eIF3e acts as an important component of the initiation translation machinery in GBM cells. Although the molecular mechanisms underlying the anti-GBM activity of eIF3e silencing are not clear yet, we could think that by directly regulating the translation of specific subsets of mRNAs involved in tumorigenic pathways, eIF3e seems to support GBM malignant phenotype and progression. In addition to its potential prognostic value, eIF3e inhibition, through the modulation of the HIFs, could also represent an effective adjuvant therapy to the standard radio/chemotherapies. In conclusion, our work shows that eIF3e could become a promising target for GBM patients and it might be of special interest to develop specific new therapeutic agents to reverse eIF3e-regulated changes in the protein synthesis profile.
DATA AVAILABILITY
The polysome profiling has been deposited in the Gene Expression Omnibus (GEO) under accession GSE157608.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marie Tosolini and Frederic Pont from CRCT Technology Cluster for initial bioinformatic analysis, GeT-Biopuces core of INSA Toulouse for assistance with the polysome profiling microarray technology, D. da Mota and F. Arnauduc for assistance with IHC and J.P. Hugnot for providing GBM samples. C. Oertlin and O. Larsson at Karolinska Institutet helped with anota2seq analysis.
Author contributions: S.M. and N.S. conceived the project. S.M., N.S., C.S. and A.C. designed and supervised the experiments. J.B. and J.S. performed most of the experiments, together with A.C. M.A. prepared the GBM protein extracts. A.C., J.S. and J.B. made the figures. J.G. performed statistical analysis. S.E., M.C.-S. and E.U.-C. performed and analyzed ISH. E.C.J.-M., C.T., C.S. and E.U.-C. contributed with helpful discussion. E.D. performed bioinformatics analysis. S.M. and N.S. wrote the manuscript with input from all the authors.
Contributor Information
Juliette Bertorello, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France.
Julie Sesen, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France.
Julia Gilhodes, Institut Universitaire du Cancer de Toulouse-Oncopole, 31100 Toulouse, France.
Solène Evrard, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France; Institut Universitaire du Cancer de Toulouse-Oncopole, 31100 Toulouse, France.
Monique Courtade-Saïdi, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France; Institut Universitaire du Cancer de Toulouse-Oncopole, 31100 Toulouse, France.
Meera Augustus, INSERM U1051, Institute for Neurosciences, Hôpital Saint Eloi, 80 Avenue Augustin Fliche, 34091 Montpellier Cedex 5, France.
Emmanuelle Uro-Coste, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France; Institut Universitaire du Cancer de Toulouse-Oncopole, 31100 Toulouse, France.
Christine Toulas, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France; Institut Universitaire du Cancer de Toulouse-Oncopole, 31100 Toulouse, France.
Elizabeth Cohen-Jonathan Moyal, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France; Institut Universitaire du Cancer de Toulouse-Oncopole, 31100 Toulouse, France.
Catherine Seva, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France.
Erik Dassi, Department of Cellular, Computational and Integrative Biology (CIBIO), University of Trento, Via Sommarive 9, 38123 Trento (TN), Italy.
Anne Cammas, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France.
Nicolas Skuli, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France.
Stefania Millevoi, Cancer Research Center of Toulouse, INSERM UMR 1037, 31037 Toulouse, France; Université Toulouse III—Paul Sabatier, 2 Avenue Hubert Curien, 31100 Toulouse, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Cancer Online.
FUNDING
Institut National de la Santé et de la Recherche Médicale (INSERM); Université Toulouse III—Paul Sabatier; Centre National de la Recherche Scientifique (CNRS); Ligue Contre le Cancer (LNCC) [to J.S.]; Labex TOUCAN [ANR11-LABX]; Ministère de l’Enseignement supérieur, de la Recherche et de l’Innovation (MENRT) [to J.B.].
Conflict of Interest statement. None declared.
REFERENCES
- 1. Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K., Burger P.C., Jouvet A., Scheithauer B.W., Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007; 114:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K.et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009; 10:459–466. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U.et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005; 352:987–996. [DOI] [PubMed] [Google Scholar]
- 4. Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S.M.et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016; 164:550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P.et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010; 17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dirkse A., Golebiewska A., Buder T., Nazarov P.V., Muller A., Poovathingal S., Brons N.H.C., Leite S., Sauvageot N., Sarkisjan D.et al. Stem cell-associated heterogeneity in glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 2019; 10:1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015; 14:261–278. [DOI] [PubMed] [Google Scholar]
- 8. Truitt M.L., Ruggero D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2017; 17:332. [DOI] [PubMed] [Google Scholar]
- 9. de la Parra C., Walters B.A., Geter P., Schneider R.J. Translation initiation factors and their relevance in cancer. Curr. Opin. Genet. Dev. 2018; 48:82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu J., Cargnello M., Topisirovic I., Pelletier J. Translation initiation factors: reprogramming protein synthesis in cancer. Trends Cell Biol. 2016; 26:918–933. [DOI] [PubMed] [Google Scholar]
- 11. Genuth N.R., Barna M. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet. 2018; 19:431–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee A.S., Kranzusch P.J., Cate J.H. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015; 522:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee A.S., Kranzusch P.J., Doudna J.A., Cate J.H. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016; 536:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R. 5′ UTR m6A promotes cap-independent translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sesen J., Cammas A., Scotland S.J., Elefterion B., Lemarie A., Millevoi S., Mathew L.K., Seva C., Toulas C., Moyal E.C.et al. Int6/eIF3e is essential for proliferation and survival of human glioblastoma cells. Int. J. Mol. Sci. 2014; 15:2172–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larsson O., Sonenberg N., Nadon R. anota: analysis of differential translation in genome-wide studies. Bioinformatics. 2011; 27:1440–1441. [DOI] [PubMed] [Google Scholar]
- 17. Bailey T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011; 27:1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Majmundar A.J., Wong W.J., Simon M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010; 40:294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barker H.E., Paget J.T., Khan A.A., Harrington K.J. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat. Rev. Cancer. 2015; 15:409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wright D.A., Futcher B., Ghosh P., Geha R.S. Association of human fas (CD95) with a ubiquitin-conjugating enzyme (UBC-FAP). J. Biol. Chem. 1996; 271:31037–31043. [DOI] [PubMed] [Google Scholar]
- 22. Jiang H.Y., Jiang L., Wek R.C. The eukaryotic initiation factor-2 kinase pathway facilitates differential GADD45a expression in response to environmental stress. J. Biol. Chem. 2007; 282:3755–3765. [DOI] [PubMed] [Google Scholar]
- 23. Zhao X., Wang P., Liu J., Zheng J., Liu Y., Chen J., Xue Y. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Mol. Ther. 2015; 23:1899–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wagner S., Herrmannova A., Sikrova D., Valasek L.S. Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic Acids Res. 2016; 44:10772–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith M.D., Arake-Tacca L., Nitido A., Montabana E., Park A., Cate J.H. Assembly of eIF3 mediated by mutually dependent subunit insertion. Structure. 2016; 24:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sha Z., Brill L.M., Cabrera R., Kleifeld O., Scheliga J.S., Glickman M.H., Chang E.C., Wolf D.A. The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol. Cell. 2009; 36:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shah M., Su D., Scheliga J.S., Pluskal T., Boronat S., Motamedchaboki K., Campos A.R., Qi F., Hidalgo E., Yanagida M.et al. A transcript-specific eIF3 complex mediates global translational control of energy metabolism. Cell Rep. 2016; 16:1891–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dassi E., Re A., Leo S., Tebaldi T., Pasini L., Peroni D., Quattrone A. AURA 2: empowering discovery of post-transcriptional networks. Translation (Austin). 2014; 2:e27738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soto-Rifo R., Rubilar P.S., Limousin T., de Breyne S., Decimo D., Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012; 31:3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shih J.W., Tsai T.Y., Chao C.H., Wu Lee Y.H. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008; 27:700–714. [DOI] [PubMed] [Google Scholar]
- 31. Chen H.H., Yu H.I., Yang M.H., Tarn W.Y. DDX3 activates CBC-eIF3-mediated translation of uORF-containing oncogenic mRNAs to promote metastasis in HNSCC. Cancer Res. 2018; 78:4512–4523. [DOI] [PubMed] [Google Scholar]
- 32. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K.et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee C.S., Dias A.P., Jedrychowski M., Patel A.H., Hsu J.L., Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008; 36:4708–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dassi E. Handshakes and fights: the regulatory interplay of RNA-binding proteins. Front. Mol. Biosci. 2017; 4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walsh D., Mohr I. Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev. 2014; 28:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Furic L., Rong L., Larsson O., Koumakpayi I.H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L.A.et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl Acad. Sci. U.S.A. 2010; 107:14134–14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Proud C.G. Mnks, eIF4E phosphorylation and cancer. Biochim. Biophys. Acta. 2015; 1849:766–773. [DOI] [PubMed] [Google Scholar]
- 38. Mao P., Joshi K., Li J., Kim S.H., Li P., Santana-Santos L., Luthra S., Chandran U.R., Benos P.V., Smith L.et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl Acad. Sci. U.S.A. 2013; 110:8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wild-Bode C., Weller M., Rimner A., Dichgans J., Wick W. Sublethal irradiation promotes migration and invasiveness of glioma cells: implications for radiotherapy of human glioblastoma. Cancer Res. 2001; 61:2744–2750. [PubMed] [Google Scholar]
- 40. Kargiotis O., Geka A., Rao J.S., Kyritsis A.P. Effects of irradiation on tumor cell survival, invasion and angiogenesis. J. Neurooncol. 2010; 100:323–338. [DOI] [PubMed] [Google Scholar]
- 41. Miyazaki S., Imatani A., Ballard L., Marchetti A., Buttitta F., Albertsen H., Nevanlinna H.A., Gallahan D., Callahan R. The chromosome location of the human homolog of the mouse mammary tumor-associated gene INT6 and its status in human breast carcinomas. Genomics. 1997; 46:155–158. [DOI] [PubMed] [Google Scholar]
- 42. Hershey J.W. The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta. 2015; 1849:792–800. [DOI] [PubMed] [Google Scholar]
- 43. Sesen J., Casaos J., Scotland S.J., Seva C., Eisinger-Mathason T.S., Skuli N. The bad, the good and eIF3e/INT6. Front. Biosci. (Landmark Ed.). 2017; 22:1–20. [DOI] [PubMed] [Google Scholar]
- 44. Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L.et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006; 9:157–173. [DOI] [PubMed] [Google Scholar]
- 45. Korkolopoulou P., Levidou G., El-Habr E.A., Piperi C., Adamopoulos C., Samaras V., Boviatsis E., Thymara I., Trigka E.A., Sakellariou S.et al. Phosphorylated 4E-binding protein 1 (p-4E-BP1): a novel prognostic marker in human astrocytomas. Histopathology. 2012; 61:293–305. [DOI] [PubMed] [Google Scholar]
- 46. Hida K., Hida Y., Amin D.N., Flint A.F., Panigrahy D., Morton C.C., Klagsbrun M. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 2004; 64:8249–8255. [DOI] [PubMed] [Google Scholar]
- 47. Rong Y., Durden D.L., Van Meir E.G., Brat D.J. ‘Pseudopalisading’ necrosis in glioblastoma: a familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J. Neuropathol. Exp. Neurol. 2006; 65:529–539. [DOI] [PubMed] [Google Scholar]
- 48. Hambardzumyan D., Bergers G. Glioblastoma: defining tumor niches. Trends Cancer. 2015; 1:252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Semenza G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003; 3:721–732. [DOI] [PubMed] [Google Scholar]
- 50. Chen Z., Wang H.W., Wang S., Fan L., Feng S., Cai X., Peng C., Wu X., Lu J., Chen D.et al. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J. Clin. Invest. 2019; 129:2043–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brat D.J., Castellano-Sanchez A.A., Hunter S.B., Pecot M., Cohen C., Hammond E.H., Devi S.N., Kaur B., Van Meir E.G. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 2004; 64:920–927. [DOI] [PubMed] [Google Scholar]
- 52. Guan B.J., van Hoef V., Jobava R., Elroy-Stein O., Valasek L.S., Cargnello M., Gao X.H., Krokowski D., Merrick W.C., Kimball S.R.et al. A unique ISR program determines cellular responses to chronic stress. Mol. Cell. 2017; 68:885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miao B., Wei C., Qiao Z., Han W., Chai X., Lu J., Gao C., Dong R., Gao D., Huang C.et al. eIF3a mediates HIF1alpha-dependent glycolytic metabolism in hepatocellular carcinoma cells through translational regulation. Am. J. Cancer Res. 2019; 9:1079–1090. [PMC free article] [PubMed] [Google Scholar]
- 54. Tamura R.E., de Vasconcellos J.F., Sarkar D., Libermann T.A., Fisher P.B., Zerbini L.F. GADD45 proteins: central players in tumorigenesis. Curr. Mol. Med. 2012; 12:634–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lu X., de la Pena L., Barker C., Camphausen K., Tofilon P.J. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006; 66:1052–1061. [DOI] [PubMed] [Google Scholar]
- 56. Wahba A., Rath B.H., Bisht K., Camphausen K., Tofilon P.J. Polysome profiling links translational control to the radioresponse of glioblastoma stem-like cells. Cancer Res. 2016; 76:3078–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wahba A., Lehman S.L., Tofilon P.J. Radiation-induced translational control of gene expression. Translation (Austin). 2017; 5:e1265703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lal A., Abdelmohsen K., Pullmann R., Kawai T., Galban S., Yang X., Brewer G., Gorospe M. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol. Cell. 2006; 22:117–128. [DOI] [PubMed] [Google Scholar]
- 59. Badura M., Braunstein S., Zavadil J., Schneider R.J. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc. Natl Acad. Sci. U.S.A. 2012; 109:18767–18772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hueng D.Y., Tsai W.C., Chiou H.Y., Feng S.W., Lin C., Li Y.F., Huang L.C., Lin M.H. DDX3X biomarker correlates with poor survival in human gliomas. Int. J. Mol. Sci. 2015; 16:15578–15591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shih J.W., Wang W.T., Tsai T.Y., Kuo C.Y., Li H.K., Wu Lee Y.H. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 2012; 441:119–129. [DOI] [PubMed] [Google Scholar]
- 62. Hilliker A., Gao Z., Jankowsky E., Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F–mRNA complex. Mol. Cell. 2011; 43:962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009; 36:932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The polysome profiling has been deposited in the Gene Expression Omnibus (GEO) under accession GSE157608.