Abstract
Background
Genomic epidemiology of antibiotic resistance is not sufficiently studied in low-income countries.
Objectives
To determine prevalence of ESBL production, and resistome and virulome profiles, of Klebsiella pneumoniae isolated at Jimma Medical Center, Ethiopia.
Methods
Strains isolated from patients with suspected infections between June and November 2016 were characterized by MALDI-TOF for species identification and disc diffusion for antimicrobial susceptibility testing. All K. pneumoniae isolates were characterized by double disc diffusion for ESBL production and all ESBL-producing strains (ESBL-KP) were subjected to WGS on the Illumina (HiSeq 2500) platform. DNA was extracted by automated systems (MagNA Pure 96). Genome assembly was performed using SPAdes (v. 3.9) and draft genomes were used for analysing molecular features of the strains. Maximum likelihood trees were generated using FastTree/2.1.8 based on SNPs in shared genomic regions to identify transmission clusters.
Results
Of the 146 K. pneumoniae strains isolated, 76% were ESBL-KP; 93% of the ESBL-KP strains showed resistance to multiple antimicrobial classes. blaCTX-M-15 (84.4%) was the most prevalent ESBL gene. Resistance genes for aminoglycosides and/or fluoroquinolones [aac(6′)-Ib-cr (65.1%)], phenicols [catB3 (28.4%)], sulphonamides [sul1 (61.2%) and sul2 (60.5%)], trimethoprim [dfrA27 (32.1%)], macrolides [mph(A) (12.8%)] and rifampicin [arr2/arr3 (39.4%)] were prevalent. Plasmids of the IncF and IncR families were prevalent among ST218, ST147, ST15 and ST39. KL64 and KL57 capsular types and O1 and O2 LPSs were prevalent. A high-risk clone, ST218-KL57 encoding rmpA1/rmpA2 and iutA, was detected. Phylogenetic analysis showed a cluster of clonally related strains from different units of the hospital.
Conclusions
Prevalence of ESBL-KP was high and blaCTX-M-15 was the predominant ESBL gene. ESBL genes had spread through both clonal and polyclonal expansion of high-risk and hypervirulent clones. Nosocomial transmission of MDR strains between different units of the hospital was observed.
Introduction
Enterobacteriaceae are widely distributed in nature and are frequently isolated from infected patients, animals and the environment. In concordance with their abundance in nature, Escherichia coli and Klebsiella pneumoniae are frequently isolated from clinical samples. K. pneumoniae is most prevalent in hospitals among admitted patients and is often identified with more resistance to antimicrobials than E. coli. K. pneumoniae causes different types of infection among immunocompromised individuals, children and elderly people.1,2
Though K. pneumoniae is largely recognized as a cause of hospital-acquired infections, hypervirulent strains can also cause serious infections among apparently healthy individuals.3 These hypervirulent strains are often resistant to antimicrobials commonly prescribed for infections caused by Gram-negative bacteria. These antimicrobials include extended-spectrum cephalosporins, carbapenems, aminoglycosides, fluoroquinolones and polymyxins. Currently, ESBL-producing K. pneumoniae (ESBL-KP) has become a challenge in healthcare in every country in the world (https://www.who.int/foodsafety/publications/antimicrobials-sixth/en/), and high-risk and hypervirulent clones of K. pneumoniae are increasingly prevalent.4,5
The presence of a large accessory genome and the genomic plasticity of K. pneumoniae to acquire both resistance and virulence genes have been fairly well understood. The increased prevalence of antimicrobial resistance (AMR) and virulence in K. pneumoniae strains is to a large extent due to acquisition of mobile genetic elements.6 Furthermore, an increased fitness of successful clones and continuous exchange of genetic determinants for AMR may lead to pan-resistant clones of K. pneumoniae.3 Global efforts of tackling AMR consider antimicrobial surveillance strategies that cover screening for high-risk clones, however, genomic epidemiology and surveillance studies from low-income countries are limited. On top of a compromised economy, an increased burden of AMR in low-income countries is a real threat to healthcare systems.
While few phenotypic studies show high prevalence of AMR, genomic epidemiology of ESBL-producing Enterobacterales was not previously studied in Ethiopia. We here characterized the prevalence of ESBLs, molecular features, STs and virulence genes in K. pneumoniae strains isolated from suspected infections at Jimma Medical Center, Ethiopia. Moreover, we analysed SNPs between strains isolated at different units of the hospital to determine potential nosocomial transmission of strains within the hospital.
Methods
A cross-sectional study was conducted among patients seeking medical care at Jimma Medical Center in Jimma, Ethiopia; a 600 bed hospital with 13 units serving a total population of 15 million people. Non-repeat single specimens were collected from each patient with suspected infection (pneumonia, diarrhoea, wound infection and urinary tract infection) from June to October 2016. Data regarding sociodemographic variables and putative risk factors (presence of underlying chronic illness, admission, and current usage of antibiotics) were collected using a structured questionnaire. Strains were isolated by conventional biochemical methods at the study site.
All K. pneumoniae isolates (n = 146) were tested for ESBL production phenotypically by the double disc diffusion test (DDST) using cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg) and amoxicillin/clavulanic acid (30 μg, 20/10). Species identification was further validated by MALDI-TOF, and antimicrobial susceptibility testing was performed by the Kirby–Bauer disc diffusion technique according to EUCAST guidelines (https://www.eucast.org/ast_of_bacteria/). All ESBL-KP strains (n = 111) were subjected to WGS. After overnight aerobic incubation on cystine lactose electrolyte deficient agar at 37°C, 2–5 pure colonies were taken with inoculating loop and were then suspended in BSA (0.5 mg/mL) in PBS (pH 7.4). Then, the cell suspensions were aliquotted in a 96-well plate and genomic DNA was extracted using an automated DNA extraction system (MagNA Pure 96, Roche Life Sciences, Sweden). DNA concentrations were measured with QubitTM3.0 (Thermo scientific, MA, USA). Sequencing libraries were generated using Nextera XT (Illumina kits) and short-read sequencing was run on Illumina (HiSeq 2500) systems with a 150 bp insert size paired end sequencing protocol at Science for Life Laboratory, Stockholm, Sweden.
SPAdes (version 3.9) was used for genome assembly. The assembled draft genomes were used for analysis of resistome and virulome, as well as plasmid replicon and sequence typing using CGE tools (http://www.genomicepidemiology.org/). For virulence genes and other genomic analysis BIGsdb (https://bigsdb.pasteur.fr/index.html), a K. pneumoniae genome analysis pipeline hosted at Institut Pasteur, was used. Kaptive/Holt Lab (https://kaptive-web.erc.monash.edu) was used to predict capsular types and O-LPSs. For all databases, the putative genes were considered positive when the coverage and the percentage of identity of the nucleotides was 100%. For statistical significance, Fisher’s exact test was used as test of association and a P value ≤0.05 was applied as a cut-off for statistical significance.
Variant calling, mapping and de novo assembly was done using CLC Assembly Cell Version: 4.4.2.133896 (Qiagen Bioinformatics). Minimum coverage was set to 10× and SNP ratio cut-off for SNP support was set to 90%. Gubbins version 2.3.1, with standard settings, was used to remove SNPs from recombinant regions.7 Maximum likelihood trees were inferred with FastTree/2.1.88 using GTR nucleotide substitution model and bootstrapping (1000 replicates). Visualization of trees and metadata was done using iTOL (https://itol.embl.de/).9
The study was ethically approved by AAU-ERB, AHRI-ERC and NERC-Ministry of Science and Technology, Ethiopia. Written consent was obtained from study participants, and the findings for each patient were communicated to the physician in charge of care of each patient.
Results
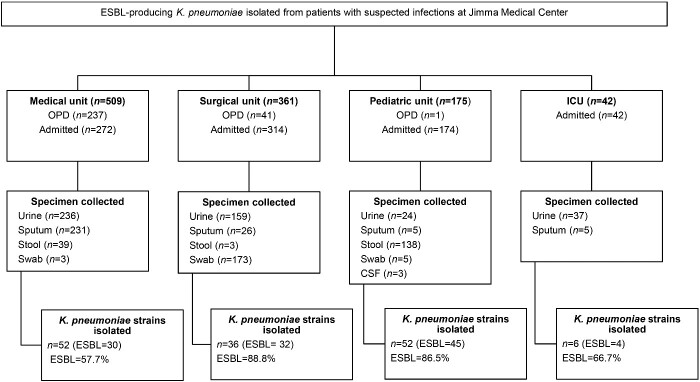
A total of 1087 patients (n = 802 admitted to the hospital and n = 285 seeking care at the outpatient department) with suspected infections (pneumonia, diarrhoea, wound infections and urinary tract infections) from four units of the hospital—the medical unit (n = 509), the surgical unit (n = 361), the paediatric unit (n = 175) and the ICU (n = 45)—were included in the study. Based on the specific condition of each patient, sputum, wound swab, urine and stool samples were collected. The prevalence of ESBL was higher at the surgical unit (88.8%) and the paediatric unit (86.5%) compared with the ICU (66.7%) and the medical unit (57.7%) (Figure 1).
Figure 1.
Flow chart showing prevalence of ESBL-KP strains at different units of the hospital. OPD, outpatient department.
A total of 146 K. pneumoniae strains were isolated and 111 (76%) were ESBL producing. Thirty-three (22.6%) of the ESBL-KP were non-susceptible to carbapenems, but only one strain encoded a carbapenemase gene (blaNDM-1). The blaCTX-M gene was the most prevalent ESBL-type and variants included: blaCTX-M-15 (84%, 92/109), blaCTX-M-11 (3.6%, 4/109), blaCTX-M-14 (1.8%, 2/109), as well as blaCTX-M-3 and blaCTX-M-9 (each accounting for 0.9%, 1/109). Only one strain carried two blaCTX-M genes (blaCTX-M-14 and blaCTX-M-15). Multiple variants of the blaSHV-genes were detected, but they were less prevalent than other ESBL genes: blaSHV-11 (21.1%, 23/109), blaSHV-1 (12.8%, 14/109), blaSHV-28 (6.3%, 7/109), blaSHV-27 (3.6%, 4/109) and blaSHV-26 (2.7%, 3/109). Among other β-lactamase genes, the blaTEM gene blaTEM-1b (69.7%, 76/109) was the most prevalent, the blaOXA, blaCMY, and blaTEM-1A genes were also detected, but with lower prevalence (Table 1).
Table 1.
List of AMR genes and their prevalence in K. pneumoniae strains isolated at Jimma Medical Center, Ethiopia
| Serial no. | Class of antimicrobial | Gene description and resistance mechanism |
Genes detected | Prevalence | ||
|---|---|---|---|---|---|---|
| sub-class genes | putative mechanisms | drugs targeted | ||||
| 1 | Aminoglycosides | acetylation genes | acetyl transferase | aminoglycosides | aac (3)-IIa | 18 (16.5%) |
| acetyl transferase | aminoglycosides | aac (3)-IId | 1 (0.9%) | |||
| acetyl transferase | aminoglycosides and fluoroquinolones | aac (6′) Ib-cr | 71 (65.1%) | |||
| acetyl transferase | aminoglycosides | aac (6′)-IIc | 2 (1.8%) | |||
| acetyl transferase | aminoglycosides | aacA4 | 2 (1.8%) | |||
| adenylation | adenyl transferase | aminoglycosides | aadA1 | 4 (3.6%) | ||
| tetracycline efflux | aminoglycosides | aadA2 | 14 (12.8%) | |||
| adenyl transferase | aminoglycosides | aadA5 | 1 (0.9%) | |||
| adenyl transferase | aminoglycosides | aadB | 3 (2.7%) | |||
| streptomycin | streptomycin phosphotransferase | aminoglycosides | strA | 45 (41.3%) | ||
| streptomycin phosphotransferase | aminoglycosides | strB | 45 (41.3%) | |||
| phosphorylation | aminoglycosides phosphotransferase | aminoglycosides | aph (3′)-Ia | 10 (9.2%) | ||
| 2 | β-lactams | bla CMY | β-lactamase | cephamycins | bla CMY-6 | 1 (0.9%) |
| β-lactamase | cephamycins | bla CMY-42 | 2 (1.8%) | |||
| bla CTX-M | ESBL | aminopenicillins and cephalosporins | bla CTX-M-11 | 4 (3.6%) | ||
| ESBL | aminopenicillins and cephalosporins | bla CTX-M-14 | 2 (1.8%) | |||
| ESBL | aminopenicillins and cephalosporins | bla CTX-M-15 | 92 (84.4%) | |||
| ESBL | aminopenicillins and cephalosporins | bla CTX-M-3 | 1 (0.9%) | |||
| ESBL | aminopenicillins and cephalosporins | bla CTX-M-9 | 1 (0.9%) | |||
| bla OXA | β-lactamase | aminopenicillins | bla OXA-1 | 38 (34.8%) | ||
| ESBL | aminopenicillins and cephalosporins | bla OXA-10 | 12 (11.0%) | |||
| bla TEM | β-lactamase | aminopenicillins | bla TEM-1B | 76 (69.7%) | ||
| β-lactamase | aminopenicillins | bla TEM-1A | 2 (1.8%) | |||
| β-lactamase | aminopenicillins | bla SHV-1 | 14 (12.8%) | |||
| bla SHV | β-lactamase | aminopenicillins | bla SHV-11 | 23 (21.1%) | ||
| β-lactamase | aminopenicillins | bla SHV-26 | 3 (2.7%) | |||
| ESBL | aminopenicillins and cephalosporins | bla SHV-27 | 4 (3.6%) | |||
| β-lactamase | aminopenicillins | bla SHV-28 | 7 (6.3%) | |||
| 3 | Carbapenem | carbapenemase | metallo-β-lactamase | aminopenicillins, cephalosporins and carbapenems | bla NDM-1 | 1 (0.9%) |
| 4 | Phenicols | acetyl transferase | chloramphenicol | catB3 | 31 (28.4%) | |
| acetyl transferase | chloramphenicol | catB4 | 6 (5.4%) | |||
| 5 | Trimethoprim | dihydrofolate reductase | sulfamethoxazole/co-trimoxazole | dfrA1 | 5 (4.5%) | |
| dihydrofolate reductase | sulfamethoxazole/co-trimoxazole | dfrA12 | 10 (9.2%) | |||
| dihydrofolate reductase | sulfamethoxazole/co-trimoxazole | dfrA17 | 2 (1.8%) | |||
| dihydrofolate reductase | sulfamethoxazole/co-trimoxazole | dfrA27 | 35 (32.1%) | |||
| dihydrofolate reductase | sulfamethoxazole/co-trimoxazole | dfrA5 | 1 (0.9%) | |||
| dihydrofolate reductase | sulfamethoxazole/co-trimoxazole | dfrA7 | 14 (12.8%) | |||
| 6 | Fosfomycin | Glutathione transferase | fosfomycin | fosA | 11 (10.1%) | |
| 7 | Macrolides | macrolide phosphotransferase | chloramphenicol | mph(A) | 14 (12.8%) | |
| 8 | Quinolones | QepA | efflux pump | fluoroquinolones | qepA | 1 (0.9%) |
| QnrB | plasmid mediated quinolone resistance | fluoroquinolones | qnrB6 | 13 (11.9%) | ||
| QnrS | plasmid mediated quinolone resistance | fluoroquinolones | qnrS1 | 18 (16.5%) | ||
| 8 | Tetracycline | Tet | efflux pump | tetracyclines | tet(A) | 41 (37.6%) |
| efflux pump | tetracyclines | tet(c) | 4 (3.6%) | |||
| efflux pump | tetracyclines | tet(D) | 20 (18.3%) | |||
| 10 | Sulphonamides | Sul | dihydropteroate synthase | sulphonamides and co-trimoxazole | sul1 | 67 (61.5%) |
| dihydropteroate synthase | sulphonamides and co-trimoxazole | sul2 | 66 (60.5%) | |||
| 11 | Rifampicin | ARR | ADP-ribosylation catalysing enzyme | rifampicin | arr-2 | 8 (7.3%) |
| ADP-ribosylation catalysing enzyme | rifampicin | arr-3 | 35 (32.1%) | |||
Bold indicates the most prevalent gene for a particular class of antimicrobial.
Phenotypic analysis showed a high prevalence of resistance to non-β-lactam antimicrobials like aminoglycosides [81.8% (90/109) for gentamicin and 5.5% (6/109) for amikacin], ciprofloxacin (63.3%, 69/109), sulfamethoxazole (95.4%, 104/109), and piperacillin/tazobactam (50%). Based on phenotypic analysis, 93% (102/109) of the strains were resistant to three or more classes of antimicrobials, 76% (83/109) were resistant to four or more classes of antimicrobials, and 15% (17/109) were resistant to six classes of antimicrobials (Table 2). Similarly, the prevalence of genetic determinants of resistance to non-β-lactam antimicrobials was high among these ESBL strains. Aminoglycoside resistance genes were present in all ESBL-producing strains, and other commonly encountered resistance gene encoded resistance to fluoroquinolones (72.4%), sulphonamides (84.4%), trimethoprim (61.3%) and phenicols (33.8%). The aac(6′)-Ib-cr gene was the most prevalent aminoglycoside and fluoroquinolone resistance gene (65.1%) (Table 1) and 95.7% of the aac(6′)-Ib-cr co-existed with blaCTX-M-15. The coexistence of blaCTX-M-15 and aac(6′)-Ib-cr genes in the same strain was statistically significant (P < 0.0001).
Table 2.
Prevalence of MDR, type of antimicrobial and the combined resistance of the K. pneumoniae strains
| Strains, n (%) | Antibiogram of ESBL strains of K. pneumoniae (%) | Number of drugs | Number of antimicrobial classes |
|---|---|---|---|
| 8 (7%) | CTX-CAZ-SXT (62.5%) | 3 | 2 |
| CTX-CAZ-GEN (12.5%) | |||
| CTX-CAZ-GEN (25%) | |||
| 19 (17%) | CTX-CAZ-SXT-GEN (57.9%) | 4 | 3 |
| CTX-CAZ-CIP-SXT (26.3%) | |||
| CTX-CAZ-MEM-GEN (5.2%) | |||
| CTX-CAZ-SXT-MEM-GEN (10.5%) | 5 | ||
| 40 (36%) | CTX-CIP-SXT-GEN (2.5%) | 4 | 4 |
| CTX-CAZ-CIP-SXT-GEN (35%) | 5 | ||
| CTX-CAZ-TZP-SXT-GEN (32.5%) | |||
| CTX-CAZ-CIP-SXT-MEM (12.5%) | |||
| CTX-CAZ-SXT-MEM-GEN (10%) | |||
| CTX-CAZ-TZP-CIP-SXT (5%) | |||
| CTX-CAZ-TZP-CIP-AMK (2.5%) | |||
| 26 (24%) | CTX-TZP-CIP-SXT-GEN (3.8%) | 5 | 5 |
| CTX-CAZ-TZP-CIP-SXT-GEN (73%) | 6 | ||
| CTX-CAZ-CIP-SXT-MEM-GEN (19.2%) | |||
| CTX-CAZ-TZP-SXT-MEM-GEN (3.8%) | |||
| 17 (16%) | CTX-CAZ-TZP-CIP-SXT-MEM-GEN (94.2%) | 7 | 6 |
| CTX-CAZ-TZP-CIP-SXT-ETP-MEM-GEN-AMK (5.8%) | 9 |
CTX, cefotaxime; TZP, piperacillin/tazobactam; CIP, ciprofloxacin, CAZ, ceftazidime, AMK, amikacin; SXT, trimethoprim/sulfamethoxazole; ETP, ertapenem; MEM, meropenem; IPM, imipenem; GEN, gentamicin.
Bold indicates the most prevalent resistance patterns.
MLST revealed more than 40 different STs. Some of the STs were novel, whereas ST218 (13%, n = 7), ST15 (11.1%, n = 6), ST17 (11.1%, n = 6), ST147 (11.1%, n = 6) and ST39 (9.3%, n = 5), were most prevalent. ST14, ST15, ST17, ST20, ST101, ST147, ST218 and ST340 were among the epidemic clones detected in this study and overall epidemic clones accounted for 22.0% of the isolates (Table S1, available as Supplementary data at JAC-AMR Online).
At least one plasmid replicon type was identified in 88.9% (97/109) of the strains. IncFIB (IncFIBK, IncFIBMar, IncFIBpQil, 32.9%, 32/97), IncR (24.7%, 24/109) and IncQ1 (12.4%, 12/109) were the most prevalent plasmid replicon types (Table S2).
Genetic determinants of virulence, including capsules, O-LPSs, siderophores and adhesins (fimH-1 and fimH-3), were analysed. KL62 (9.3%, 10/108), KL57 (9.3%, 10/108), KL25 (6.5%, 8/108) and KL64 (6.5%, 8/108) were the most prevalent capsular types. Regulators of mucoid phenotype (rmpA1/rmpA2) were detected among epidemiologically important strains. Both Type 1 and Type 3 adhesive fimbrial antigens were encoded by all strains. The O-LPSs O1 (53.7%, 57/108) and O2 (23.15%, 25/108) were the most prevalent types (Table S3). Several genetic determinants of siderophores were identified, the prevalence varied from 6.5% to 42.5%. Yersiniabactin irp1 and irp2 (6.5%, 7/108), and ybtQ (42.6%, 46/108), salmochelin iroBCDE (7.4%, 8/108), and aerobactin iucABCD/iutA (9.2%, 10/108) were the most prevalent siderophores (Table S4).
An SNP-based maximum likelihood tree showed several clusters of clonally related high-risk clones that were isolated from different units in the hospital (Figure 2). Multiple clonally related strains of ST15, ST17, ST39, ST48, ST147 and ST218, were isolated from clinical samples collected from surgical, paediatric, medical and intensive care units. The findings demonstrate nosocomial transmission of K. pneumoniae featuring MDR and hypervirulence.
Figure 2.
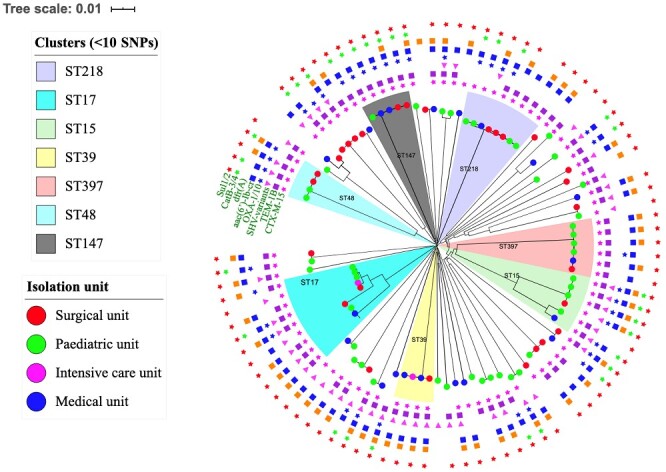
Clusters of epidemic clones isolated from the surgical unit, paediatric unit, ICU and medical unit of the hospital.
Discussion
The very high prevalence of ESBL-KP in this study is higher than a number of previous reports from Africa (Tanzania, South Africa, Gabon, Guinea-Bissau, and many other parts of the continent).10,11 However, it might be comparable to the finding of a study conducted in 2014 in Bamako, Mali, which reported 76.9% among K. pneumoniae.12 The fact that 93% of the ESBL-KP strains were resistant to at least three classes of antimicrobials and 15% to at least six classes of antimicrobials may indicate the tip of the iceberg of a high prevalence of MDR K. pneumoniae strains disseminating in hospitals in Ethiopia. In most African countries, such evidence might have been obscured because of the lack of sufficient technically standardized studies. Similarly, a recent review on AMR from Africa reported that there is a lack of updated AMR data for 40% of the African countries.13 The same review noted that the available evidence is neither sufficient nor has optimum microbiological quality. On the other hand, the low prevalence of carbapenem resistance is important for patients with severe infections from ESBL-producing strains. This low prevalence might be due to the limited availability of carbapenems for clinical use in Ethiopia. Similarly, a study from Kenya reported a low prevalence of blaNDM-1 among K. pneumoniae strains at a tertiary hospital in Nairobi.14 Low prevalence of blaNDM-1 has also been reported from other East African countries.14–17 Infections caused by strains resistant to both aminoglycosides and fluoroquinolones, as well as being ESBL-producing, are increasingly compromising the therapeutic options for serious/invasive infections. Moreover, when such strains cause infection in low-income countries, where diagnostic capacity and therapeutic options are limited, the problem might be worse.
Most of the epidemiological clones identified were MDR and may acquire further antimicrobial resistance through selection, and also encoded virulence genes. The ST15 strain, second most prevalent in this study, encoded both blaCTX-M-15 and blaSHV-28. A similar clone was previously reported from an outbreak in Copenhagen.18 An ST101 clone detected in this study was also previously reported from an outbreak in a Hungarian teaching hospital together with other epidemic clones (ST15 and ST147).19 One of the ST45 strains identified in this study encoded blaNDM-1. Though ST45 has previously been reported from an outbreak in Portugal, the clone from Portugal encoded blaKPC-3 and mcr-1, but not blaNDM-1.20 To the knowledge of these investigators, blaNDM-1 has not been previously reported in K. pneumoniae ST45. The strain found in this study encoded resistance genes for nine different classes of antimicrobials and carried a broad-range IncA/C2 plasmid. The blaNDM-1 encoding IncA/C2 plasmid has previously been reported in K. pneumoniae strains and was able to capture other resistance genes and transfer to other species.16 Considering that this strain is MDR and a high-risk clone carrying an epidemic plasmid, the risk of spreading the blaNDM-1 gene locally may be substantial.
The IncF family plasmids (IncFIB, IncFII and IncFIA) are usually regarded as epidemic plasmids and are prevalent among epidemic clones. A high prevalence of IncF plasmids in this study might have contributed to the emergence and spread of resistance to multiple antimicrobial classes in this setting. Most of the plasmids detected in this study, except IncQ1 and ColE, were conjugative plasmids that can spread between several genera/species. The IncR plasmids, another important family of plasmids in the epidemiology AMR, were identified among ST15 and ST147, which are also epidemic clones.
Some of the K. pneumoniae isolates identified in this study were characterized as hypervirulent. Globally, the prevalence of hypervirulent strains of K. pneumoniae causing community-acquired infections is on the rise.21,22 Generally, hypervirulent strains encode a unique profile of virulence genes and employ heterogenous strategies to protect themselves from the host immune response to cause infections among apparently healthy individuals. Though not every virulence factor plays the same role in all strains, profiling of the virulence genes encoded is important to understand the virulence and pathogenesis of K. pneumoniae strains.22 Such in-depth studies are rare from low-income countries. The hypervirulent strains identified in this study encoded several virulence genes categorized into four groups of virulence factors: protectins (capsule, LPSs), siderophores/iron scavenging molecules (enterobactin, yersiniabactin, salmochelin, aerobactin), fimbriae adhesins (Fim-1, and Fim-3) and miscellaneous (Table S4). Among these virulence genes, the regulators of mucoid phenotype genes (rmpA and rmpA2) for hypermucoviscous property of strains were identified exclusively with KL57, one of the most prevalent capsular types, and the KL57 was predominantly detected in ST218 strains. Besides the hypercapsular phenotype, these clones encoded multiple groups of siderophores (molecules for scavenging iron) (iroBCDN, iucABCD, iutA, entA, ybtA), and FimH-3 (mrkABCD clusters). On the other hand, an experimental study from Russia characterized strains with similar genotype (K. pneumoniae-ST218-KL57) as hypermucoviscous/hypervirulent clones.23 Other epidemiological studies from Russia and China showed that KL57 capsular types were associated with diarrhoea, pyogenic liver abscess, meningitis and bloodstream infections.24,25
With regard to LPS structures, the O2V2 types were predominant among the strains of ST218-KL57 clone. As described earlier, all of them encoded KL-57 capsular type, the similar patterns of surface molecules (KL-locus and O-LPSs) on the surface of high-risk and hypervirulent K. pneumoniae strain might be important information for intervention strategies targeting surface molecules. The O-LPSs were rarely reported from epidemiological studies, however we reported here the unique pattern of KL57-O2V2 surface molecules identified on a high-risk clone (ST218), which might be important epidemiological knowledge. However, large scale studies are required to consolidate evidence for prevention strategies to target these structures.
Furthermore, analysis of a population structure of the strains showed nosocomial transmission between the different units of the hospital. Seven independent outbreak clusters of high-risk clones were detected by a phylogenetic tree constructed using SNP analysis of the shared core genome. A clonal cluster of strains isolated from different clinical samples collected from surgical, paediatric, intensive care and medical units of the hospital indicates a likely occurrence of transmission clusters. Most importantly, the detection of multiple nosocomial transmission clusters of high-risk clones of K. pneumoniae can severely compromise the standard of care and patient safety at this hospital, where therapeutic options are limited.
The high prevalence and nosocomial transmission of multiple ESBL-producing high-risk clones of K. pneumoniae strains from a low-income country is a severe problem. In a setting where the microbiology laboratory capacity is compromised, supply of antimicrobials is limited and there is a lack of personnel with specialty training to consult prescribers, patient care and safety can be significantly affected. Moreover, lack of stringent infection control practices and functional antimicrobial stewardship programme may warrant further spread of more stringent high-risk clones.
Conclusions
The prevalence of ESBL-KP was very high and blaCTX-M-15 was the predominant ESBL gene. The spread of resistance genes was mediated by multiple high-risk clones that also encoded epidemic plasmids. The prevalence of carbapenemases was low, however, the detection of blaNDM-1 carbapenemase in a hypervirulent high-risk clone is a worrisome challenge for the future. We were able to document nosocomial transmission of multiple high-risk clones between different units in the hospital.
Supplementary Material
Acknowledgements
We acknowledge Addis Ababa University, Armauer Hansen Research Institute, Karolinska Institutet. We acknowledge Marat Murzabekov for technical assistance in genome assembly.
Funding
Addis Ababa University, Addis Ababa University-AHRI collaborative project through BSPP grant from Swedish International Development Co-operation Agency (Sida), Åke Wibergs stiftelse (F.W.) and Bill and Melinda Gates Foundation (F.W.).
Transparency declarations
None to declare.
Author contributions
T.S. contributed to the design and conception of the study, data collection, data analysis and interpretation, and writing the manuscript. C.G.G. contributed to the design and conception of the study, data acquisition and analysis, and writing the manuscript. D.A. and Y.W. contributed to the design and conception of the study and revision of the manuscript. S.N. contributed to data analysis and drafting and revision of the manuscript. F.W. contributed to the study design and revision of the manuscript. A.A. contributed to the study design, data acquisition and revision of the manuscript. All authors read and approved the manuscript for submission.
Supplementary data
Tables S1 to S6 are available as Supplementary data at JAC-AMR Online.
References
- 1. Gorrie CL, Mirceta M, Wick RR et al. Antimicrobial-resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clin Infect Dis 2018; 67: 161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moradigaravand D, Martin V, Peacock SJ. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. mBio 2017; 8: e01976–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cubero M, Grau I, Tubau F et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013. ). Clin Microbiol Infect 2016; 22: 154–60. [DOI] [PubMed] [Google Scholar]
- 4. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 2017; 41: 252–75. [DOI] [PubMed] [Google Scholar]
- 5. Ko KS, Lee J-Y, Baek JY et al. Predominance of an ST11 extended-spectrum β-lactamase-producing Klebsiella pneumoniae clone causing bacteraemia and urinary tract infections in Korea. J Med Microbiol 2010; 59: 822–8. [DOI] [PubMed] [Google Scholar]
- 6. Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 2018; 8: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Croucher NJ, Page AJ, Connor TR et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Price MN, Dehal PS, Arkin AP. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 2016; 44: W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saravanan M, Ramachandran B, Barabadi H. The prevalence and drug resistance pattern of extended spectrum β–lactamases (ESBLs) producing Enterobacteriaceae in Africa. Microb Pathog 2018; 114: 180–92. [DOI] [PubMed] [Google Scholar]
- 11. Storberg V. ESBL-producing Enterobacteriaceae in Africa a non-systematic literature review of research published 2008–2012. Infect Ecol Epidemiol 2014; 4: doi: 10.3402/iee.v4.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sangare SA, Rondinaud E, Maataoui N et al. Very high prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in bacteriemic patients hospitalized in teaching hospitals in Bamako, Mali. PLoS One 2017; 12: e0172652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tadesse BT, Ashley EA, Ongarello S et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis 2017; 17: 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poirel L, Revathi G, Bernabeu S, Nordmann P. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother 2011; 55: 934–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pritsch M, Zeynudin A, Messerer M et al. First report on bla NDM-1-producing Acinetobacter baumannii in three clinical isolates from Ethiopia. BMC Infect Dis 2017; 17: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carattoli A, Villa L, Poirel L et al. Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob Agents Chemother 2012; 56: 783–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holman AM, Allyn J, Miltgen G et al. Surveillance of carbapenemase-producing Enterobacteriaceae in the Indian Ocean Region between January 2010 and December 2015. Med Mal Infect 2017; 47: 333–9. [DOI] [PubMed] [Google Scholar]
- 18. Nielsen JB, Skov MN, Jorgensen RL et al. Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur J Clin Microbiol Infect Dis 2011; 30: 773–8. [DOI] [PubMed] [Google Scholar]
- 19. Melegh S, Schneider G, Horvath M et al. Identification and characterization of CTX-M-15 producing Klebsiella pneumoniae clone ST101 in a Hungarian university teaching hospital. Acta Microbiol Immunol Hung 2015; 62: 233–45. [DOI] [PubMed] [Google Scholar]
- 20. Mendes AC, Novais Â, Campos J et al. mcr-1 in carbapenemase-producing Klebsiella pneumoniae in hospitalized patients, Portugal, 2016–2017. Emerg Infect Dis 2018; 24: 762–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marr CM, Russo TA. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev anti Infect Ther 2019; 17: 71–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol Mol Biol Rev 2016; 80: 629–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lev AI, Astashkin EI, Kislichkina AA et al. Comparative analysis of Klebsiella pneumoniae strains isolated in 2012-2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog Glob Health 2018; 112: 142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liao CH, Huang YT, Chang CY et al. Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur J Clin Microbiol Infect Dis 2014; 33: 365–9. [DOI] [PubMed] [Google Scholar]
- 25. Zhang X, Wang L, Li R et al. Presence and characterization of Klebsiella pneumoniae from the intestinal tract of diarrhoea patients. Lett Appl Microbiol 2018; 66: 514–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.