Abstract
Recent epitranscriptomics studies unravelled that ribosomal RNA (rRNA) 2′O-methylation is an additional layer of gene expression regulation highlighting the ribosome as a novel actor of translation control. However, this major finding lies on evidences coming mainly, if not exclusively, from cellular models. Using the innovative next-generation RiboMeth-seq technology, we established the first rRNA 2′O-methylation landscape in 195 primary human breast tumours. We uncovered the existence of compulsory/stable sites, which show limited inter-patient variability in their 2′O-methylation level, which map on functionally important sites of the human ribosome structure and which are surrounded by variable sites found from the second nucleotide layers. Our data demonstrate that some positions within the rRNA molecules can tolerate absence of 2′O-methylation in tumoral and healthy tissues. We also reveal that rRNA 2′O-methylation exhibits intra- and inter-patient variability in breast tumours. Its level is indeed differentially associated with breast cancer subtype and tumour grade. Altogether, our rRNA 2′O-methylation profiling of a large-scale human sample collection provides the first compelling evidence that ribosome variability occurs in humans and suggests that rRNA 2′O-methylation might represent a relevant element of tumour biology useful in clinic. This novel variability at molecular level offers an additional layer to capture the cancer heterogeneity and associates with specific features of tumour biology thus offering a novel targetable molecular signature in cancer.
INTRODUCTION
Emerging evidence suggests that changes in ribosomal RNA 2′O-ribose methylation (rRNA 2′OMe) in the human ribosome play a key role in regulating translation thereby contributing in setting particular phenotypes such as hallmarks of cancer cells (1–5). Using cancer cellular models, we and others indeed reported an association between variation in rRNA 2′O-methylation and modulation of translation efficiency of particular mRNA subsets in cancer (1,6–9). In particular, we demonstrated that inactivation of the tumour suppressor p53 protein in mammary epithelial cells is associated with an increased expression of the 2′O-methyltransferase FBL, an alteration of the 2′O-methylation level at some of the 106 rRNA-methylated sites and an increased translation of some cellular Internal Ribosome Entry Sites (IRES)-containing mRNAs such as those encoding the oncogenic IGF1R and cMyc proteins (1). Moreover, we have shown that FBL overexpression promotes IGF1R-dependent proliferation, colony formation and resistance to chemotherapy in breast cancer cell lines, suggesting a role of rRNA 2′O-methylation in mammary tumorigenesis and breast cancer progression (1). Importantly, using cell-free in vitro translation assays, we recently demonstrated that modulation of rRNA 2′O-methylation directly affects translation initiation of IRES-containing mRNAs, the IGF1R mRNA and the Cricket paralysis virus mRNA (2). This new layer of gene expression regulation recently uncovered, joins the numerous descriptions of chemical modifications of both coding and non-coding RNAs that regulates post-transcriptional processes, a field known as epitranscriptomics (3,4,10). However at present, it is unclear whether rRNA 2′O-methylation varies in humans.
Although genetics, genomics, epigenetics, transcriptomics and proteomics revealed the heterogeneity of breast tumours at the molecular level, identification of novel molecular layers are required to fully capture all the inter-patient variability, essential for developing clinical applications (11). Amongst them, epigenetics appears as promising additional molecular fingerprints to improve patient classification, prognosis or treatment, including in breast cancer (11–13). In these molecular portraits of breast cancers, RNA epitranscriptomics and more specifically rRNA 2′O-methylation, has never been investigated. Thanks to the recent resolution of technological issues, including the development of RNA-seq-based approaches dedicated to the profiling of rRNA 2′O-methylation (4,5,14,15) and the recent developments of high-resolution cryo-EM that has provided insights into the detailed structure of the human ribosome including rRNA modifications (16–18), rRNA 2′O-methylation profiling can now be achieved in human samples to determine whether it can provide a molecular fingerprint in breast cancer.
Here, we present the first rRNA 2′O-methylation landscape of human breast tumours. We used the next-generation RiboMeth-seq technology that we remodelled to perform high-throughput analyses of human clinical samples. Our data reveal the unexpected existence of specific modulation of 2′O-methylation at only particular rRNA positions between human breast tumours that are associated with clinical outcome and biological features.
MATERIALS AND METHODS
Human samples
A series of 195 female primary breast tumours were collected from 1997 to 2011 and maintained by the Tayside Tissue Bank (TTB, Dundee, Scotland, UK) under ethical approval (REC Reference 07/S1402/90) (Supplementary Table ST1) (19). This retrospective series of primary breast tumours is composed of both non-invasive (45%) and invasive (55%) lymph node tumours (Supplementary Table ST1). Total RNA was extracted from frozen tissues by TTB services as already described (19,20). A second series of 7 mammoplasties (i.e. non-transformed, non-proliferative but mostly adipocyte cells) derived from healthy donors was sequenced independently (Institut Curie, France) (21). A third series composed of 10 mammary malignant tumours and 8 mammary benign tumours (i.e. non-transformed but hyper-proliferative epithelial cells) were collected between 2006 and 2007 (Institut Gustave Roussy, France) from consenting patients using fine-needle aspiration after suspicion of breast lesion (Supplementary Table ST2) (22). A human RNA reference sample (i.e. RNA reference) was used as a calibrated source of rRNA (Human XpressRef Universal Total RNA, Qiagen) prepared from 20 different human adult and foetal normal major organs.
RiboMeth-seq
Levels of rRNA 2′O-methylation at the 106 rRNA 2′O-methylated sites were determined by RiboMeth-seq (14,15,23–25). Presence of 2′O-methylation protects the phosphodiester bond located at the 3′ of the 2′O-methylated nucleotide from alkaline hydrolysis. Thus, the presence of 2′O-methylation at the given nucleotide n induces under-representation of RNA fragments starting at the nucleotide n + 1 and ending at position n allowing to calculate a 2′O-methylation level at the corresponding nucleotide position (or C-score) varying from 0 to 1 (15). RiboMeth-seq was performed using the Illumina sequencing technology and raw data were processed as previously described (14,25). The median number of total reads reaches 7.2 millions after trimming, these reads being aligned on the 7.2 kb-long rRNA sequences, that corresponds to the optimal sequencing depth (14,26).
Analysis of the breast cancer series
Amongst an initial series of 214 primary breast tumours, RiboMeth-seq data of 195 primary breast tumour samples passed the QC criteria (representing 91% of the initial series) and were thus retained for the downstream analyses (Supplementary Table ST1). Unsupervised data analysis was performed (hierarchical clustering and principal component analysis (PCA)) using the C-scores at the 106 individual rRNA 2′O-methylated sites of the 195 samples. rRNA 2′O-methylation profiles were shown as either box-and-whiskers plots, line charts or barcharts. Classification of the rRNA 2′O-methylation sites into ‘stable’ and ‘variable’ classes was done empirically based on the variability of the sites (in terms of interquartile range, IQR), the cut-off site being the site from which the IQR values no longer lie on this straight line (see Supplementary Materials and Methods for details). Between-group comparisons were performed using Fisher's exact test for categorical data or Mann–Whitney test for quantitative data. Bonferroni or false discovery rate correction methods were applied for multiple comparisons (P.adj). All P-values corresponded to two-tailed P-values. A P-value < 0.05 was considered to be statistically significant. Statistical analyses and graphical representations were performed using either R v3.6.3 or GraphPad Prism v7.0a software (GraphPad Software, Inc).
rRNA 2′O-methylation evolution and mapping
To further characterize the two classes of human rRNA 2′O-methylated sites, their conservation during evolution was compared using box C/D snoRNA guides as surrogates of rRNA 2′O-methylated sites, since to date availability of rRNA 2′O-methylation profiles is limited to only few organisms. Three evolution groups were established using the snoRNA-LBME-db database (www-snorna.biotoul.fr). Using Fisher's exact test, enrichment analysis was performed to compare the proportion of stable versus variable sites amongst the three groups of rRNA 2′O-methylated sites. The rRNA 2′O-methylated sites were mapped on the structure of the HeLa cancer cell human ribosome determined by cryo-EM (16–18) and images were drawn using the PyMol software. Observations were based on previous reported three-dimensional molecular docking analysis of tRNAs or ribosome-associated factors, as already discussed in (ref 16–18).
RESULTS
Optimization of RiboMeth-seq technology for human samples
To profile rRNA 2′O-methylation in human primary breast tumours, we used the novel RiboMeth-seq technology. The measurement of 2′O-methylation frequency by RiboMeth-seq technology relies on a partial alkaline hydrolysis of the rRNA phosphodiester bonds, which become refractory to hydrolysis when adjacent riboses are methylated in position 2′ (14,15,25). RiboMeth-seq processing yields a score (i.e. C-score) at each of the 106 rRNA 2′O-methylated positions, reflecting the level of 2′O-methylation.
Since rRNA 2′O-methylation profiling has never been performed on large series of human samples, we developed RiboMeth-seq-dedicated extensive quality controls based on numerous metrics (see Supplementary Materials and Methods). Using human RNA reference samples composed of a mix of 20 different human adult and foetal normal major organs and a test set of 20 primary breast tumour samples, we show a strong correlation in the C-scores amongst the technical replicates (Supplementary Figures S1 and 2). Overall, our data demonstrate that the high-throughput RiboMeth-seq technology is a reproducible and robust technology even when sequencing delicate biological material such as frozen tumour biopsies.
Identification of two classes of rRNA 2′O-methylation sites
We then profiled rRNA 2′O-methylation in a series of 195 primary breast cancers composed of a mix of invasive and non-invasive tumours using RiboMeth-seq (Supplementary Table S1) (20). Unsupervised statistical analysis revealed variability in rRNA 2′O-methylation levels (Figure 1). It first showed that, for a given tumour, the 2′O-methylation level varies between 0 and 0.98 amongst the 106 rRNA 2′O-methylation sites (Figure 1 and Supplementary Figure S3a). This demonstrates that in a single human tumour sample, all rRNA molecules are not fully and equally 2′O-methylated at the 106 sites.
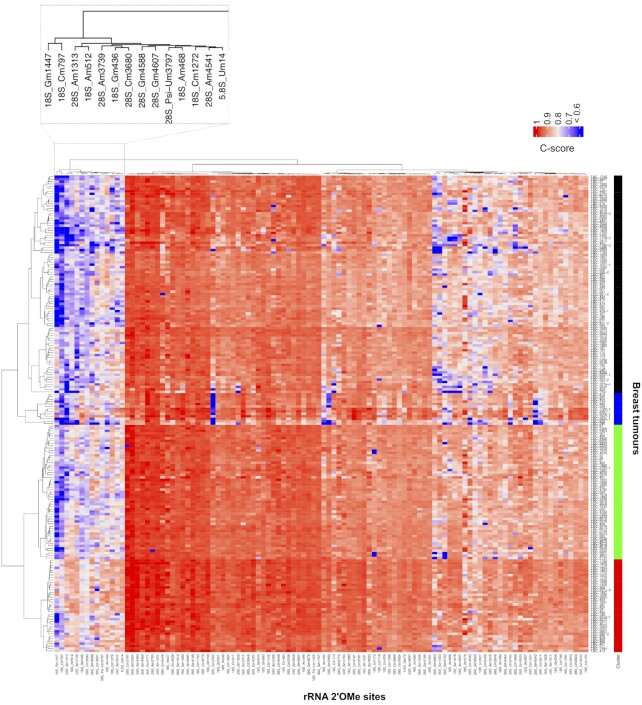
Figure 1.
Stability and variability of rRNA 2′O-methylation levels between breast tumours. Levels of rRNA 2′O-methylation (i.e. C-score) were determined at the 106 rRNA 2′O-methylated sites using RiboMeth-seq in a series of 195 primary human breast tumours. Data are presented as a hierarchical clustering where the C-score of each position (column) for each tumour (row) is presented (colour scale). Dendrograms represent relationships of similarity between breast tumours on the basis of their rRNA 2′O-methylation profiles (left panel) that identify four groups of breast tumour samples (right colour panel), or in the rRNA 2′O-methylation level at a given site between tumours (top panel). An enlarged view of a group of clustered rRNA 2′O-methylated sites is presented.
Hierarchical clustering also revealed that, for a given rRNA site, 2′O-methylation levels differ when comparing the 195 human tumours (i.e. inter-patient variability) (Figure 1; Supplementary Figures S3a and 4). We defined two classes of rRNA 2′O-methylated sites based on the variability of their 2′O-methylation level amongst patients (Supplementary Figure S3c). The classification into these two classes was done empirically based on the intra-variability of the sites (in terms of interquartile range, IQR, at a given site). The first class contains 60 rRNA 2′O-methylated sites (56.6% of all the rRNA 2′O-methylated positions), the levels of which exhibit low inter-patient variability between the 195 tumour samples, despite these series representing the full spectrum of breast cancer subtypes (Figure 2a and Supplementary Figure S4, red). Thereafter, they are termed ‘stable’ sites since they display the most stable C-scores between the 195 tumours. The second class corresponds to a limited number of rRNA 2′O-methylated sites (46 sites, around 43.4% of the rRNA 2′O-methylated positions) that exhibit high inter-patient variability in their 2′O-methylation level (‘variable’ sites, blue). Amongst them, the 28S-Am1313, 28S-Cm2352 and 28S-Gm3723 sites displayed important inter-patient variability, the C-scores ranging from 0 to >0.90 and a null C-score being observed in different tumours (Figure 2a and Supplementary Figure S3a). Position-by-position comparison of the ∂C-score [(mean C-score + 2sd) – (mean C-score – 2sd)] between the 195 breast tumours and the statistical analysis based on five technical replicates of normal human RNA reference samples reveals that the high inter-patient variability in C-scores corresponds to a biological variability rather than to technical variability (Supplementary Figure S3b). Similarly, these two classes of most stable and most variable sites derived from the 195 breast tumours were observed in a series of 7 mammoplasties derived from healthy female donors, most of the sites being classified in the same classes with the except of few ones (Supplementary Figure S5). The difference might come either from the small size of this series or from the fact that variability in rRNA 2′O-methylation might be tissue-specific, mammoplasties mostly corresponding to adipocyte cells. These data show that rRNA 2′O-methylation varies between human samples and demonstrate the co-existence of stable and variable classes of rRNA 2′O-methylated sites within human samples.
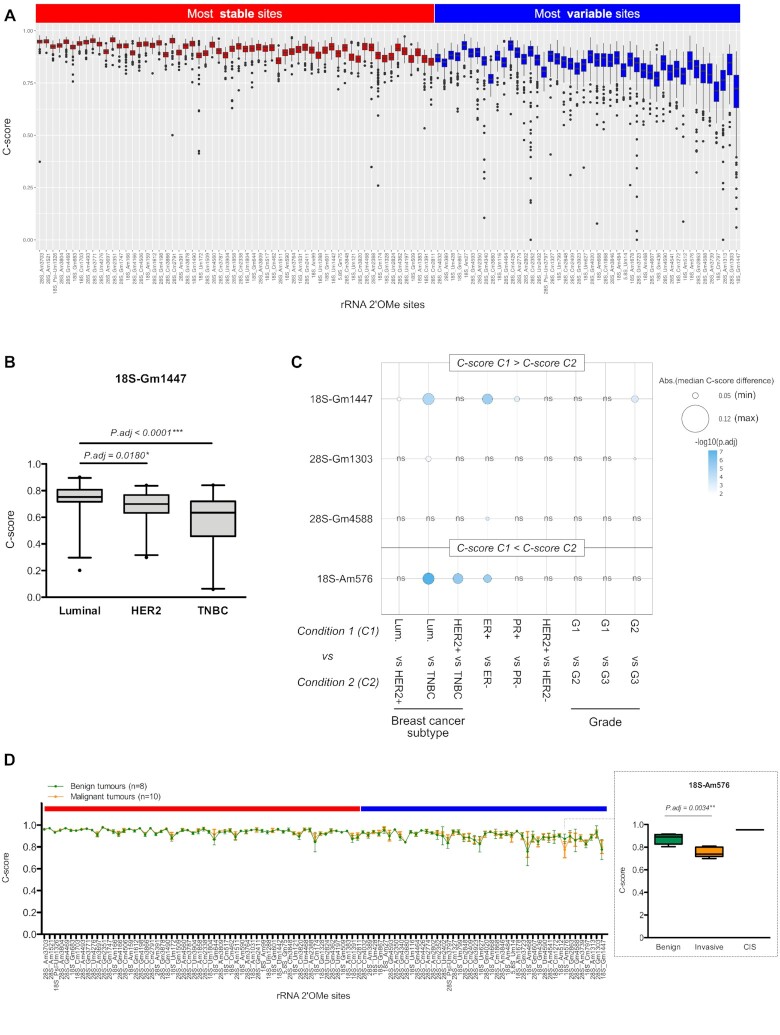
Figure 2.
Biological relevance of variable rRNA 2′O-methylation levels in breast cancer. (A) C-score variation amongst the 195 human primary breast tumours at each of the 106 rRNA 2′O-methylated sites was ranked by increasing interquartile range (IQR). Based on IQR divergence, two classes of rRNA 2′O-methylated sites were defined: sites with the most stable C-scores (red); and sites with the most variable C-scores (blue). (B) Example of significant change in rRNA 2′O-methylation levels at the 18S-Gm1447 site between luminal, HER2+ and TNBC breast tumours. (C) Summary of the four variable rRNA sites, whose 2′O-methylation levels were significantly different between breast cancer subtypes, hormonal and HER2 receptors, and tumour grade. ER: oestrogen receptor; PR: progesterone receptor; Luminal: ER+ PR± HER2−; HER2+: ER+/− PR+/− HER2+; TNBC: ER− PR− HER2-; G: grade. (D) Comparison of rRNA 2′O-methylation profiles in a series of 8 benign (green) and 10 malignant (orange) tumours. An enlarged view presents the significant change in rRNA 2′O-methylation levels at the variable 18S-Am576 site in malignant compared to benign mammary tumours, with carcinoma in situ (CIS) presenting the highest rRNA 2′O-methylation level. *: adjusted P-value < 0.05; **: P.adj < 0.01; ***: P.adj < 0.0001.
Association of rRNA 2′O-methylation level with biological and clinical characteristics
To assess the biological significance of the co-existence of two classes of rRNA 2′O-methylated positions displaying either the most ‘stable’ 2′O-methylation level between human tumour samples or the most ‘variable’ 2′O-methylation level, we evaluated the association between variations in C-score at the 106 rRNA 2′O-methylated positions and biological or clinical characteristics of primary breast tumours (Figure 2B and C). Remarkably, amongst the stable rRNA 2′O-methylated sites, no association between change in C-score and well-known characteristics of breast cancers was observed (i.e. tumour grade, tumour size, lymph node involvement, hormonal/HER2 receptor status, breast cancer subtype or TP53 mutation). In contrast, we identified four variable rRNA 2′O-methylated sites (18S-Gm1447, 28S-Gm1303, 28S-Gm4588 and 18S-Am576), the levels of which are significantly different between breast cancer subtypes, oestrogen and progesterone statuses, as well as tumour grades (Figure 2B and C; Supplementary Table S3). In both human tumour samples and cell lines, the 2′O-methylation level of the 18S-Gm1447 site decreased in triple-negative breast cancers (TNBCs) compared to the luminal tumours, whilst that of the 18S-Am576 site increased in TNBCs (Figure 2B and C; Supplementary Figure S6). Furthermore, we compared rRNA 2′O-methylation profile in a small series of eight benign (i.e. hyperproliferative, but non-transformed epithelial cells) and 10 malignant tumours, the former do not represent the full breast cancer inter-patient heterogeneity. In this series, 2′O-methylation level at the 18S-Am576 site distinguished benign from malignant mammary tumours (Figure 2D).
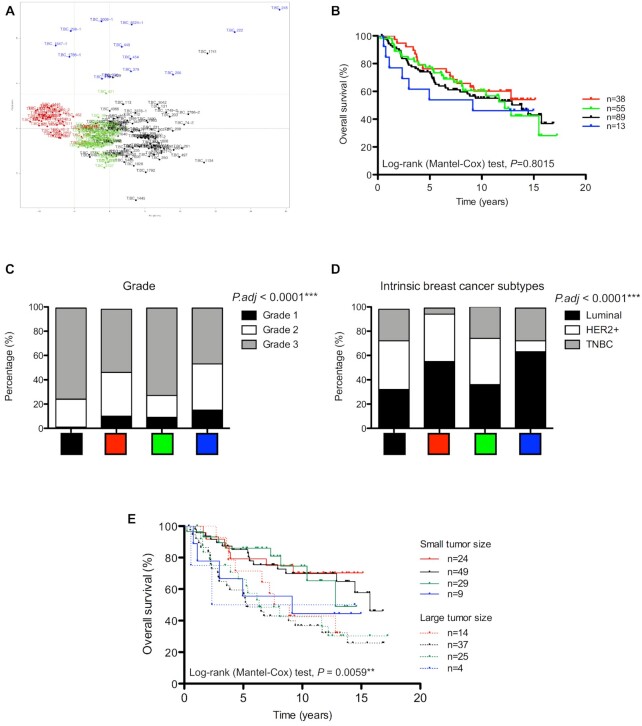
Secondly, we evaluated the association between breast cancer patients harbouring tumours exhibiting similar rRNA 2′O-methylation profiles and biological/clinical characteristics of breast tumours. Hierarchical clustering and PCA methods identified 4 clusters of breast cancer tumours based on their rRNA 2′O-methylation profiles (Figures 1 and 3A). Analysis of survival indicates that, although no significant association was observed, the patients carrying breast tumours characterized by distinct rRNA 2′O-methylation profile display distinguishable overall survival (Figure 3B). In particular the ‘blue’ group displays the poorest overall survival. This blue group was enriched in TNBC and in tumours of high grade compared with the other groups (Figure 3C and D; Supplementary Figure S7). Regarding the fact that tumours in the blue groups exhibited the lowest C-score at most of the rRNA 2′O-methylated sites (Figure 1), these data suggest that global decrease in rRNA 2′O-methylation might be associated with breast tumour aggressiveness. Finally, clustering breast cancer tumours on the basis of their 2′O-methylation level at the 106 rRNA sites highlights at diagnosis, patients carrying small tumours size who display prognosis as poor as patients carrying large tumours size (Figure 3E, blue group). Indeed, patients carrying small tumour size exhibit different survival rates depending on the rRNA 2′O-methylation profile. Overall, these data indicate that 2′O-methylation levels at variable rRNA sites are associated with breast cancer subtypes and tumour grade.
Figure 3.
Association of rRNA 2′O-methylation profiles with biological and clinical breast cancer characteristics. (A) PCA showed four clusters of breast tumours based on their rRNA 2′O-methylation profiles that correspond to the ones identified using the hierarchical clustering approach (Figure 1). (B) Kaplan–Meier curve suggested the patients carrying breast tumours characterized by a ‘blue’ rRNA 2′O-methylation profile display the poorest overall survival. (C andD) Significant differences in repartition amongst the four clusters were observed regarding tumour grade (C) and breast cancer subtype (D). Luminal: ER+ PR± HER2−; HER2+: ER+/− PR+/− HER2+; TNBC: ER− PR− HER2−. (E) Kaplan–Meier curve depicts the overall survival of patients grouped on the basis of both their tumour size at diagnosis and their rRNA 2′O-methylation profiles. **: P < 0.01; ***: P < 0.0001.
Evaluation of the structural impact of the two classes of rRNA 2′O-methylation sites
To further characterize the two classes of human rRNA 2′O-methylated positions identified in the 195 breast tumours, we compared their conservation during evolution. Amongst the 106 rRNA 2′O-methylation sites described in humans, some sites have also been identified in yeast and bacteria (snoRNA-LBME-db database, www-snorna.biotoul.fr). We thus compared the proportion of either ‘stable’ or ‘variable’ sites classified on the basis of our data, amongst three groups: the group of rRNA 2′O-methylated sites only detected in humans (‘humans’, n = 68); the group of rRNA 2′O-methylated sites detected both in humans and yeast (‘eukaryotes’, n = 35); and the group of rRNA 2′O-methylated sites conserved in humans, yeast and bacteria (‘universal’, n = 3; see also annotation on the human ribosome structure in ref 17). Enrichment analysis showed an increased proportion of the variable rRNA 2′O-methylated sites in humans compared to other organisms (Figure 4A), suggesting a link between the appearance of variable rRNA 2′O-methylated sites and evolution, characterized by an increasing number of fine-tuning gene expression mechanisms.
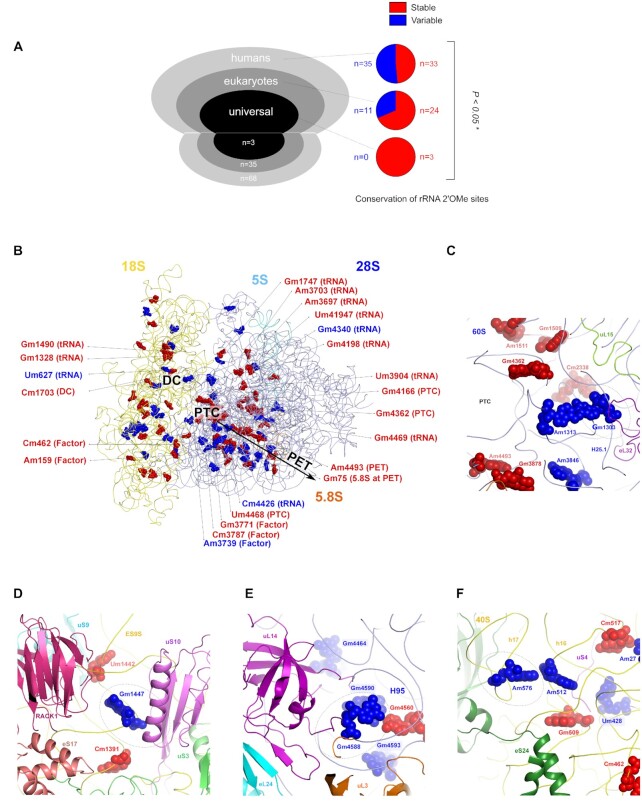
Figure 4.
Structural properties of stable and variable rRNA 2′O-methylated sites. (A) Amongst the 106 rRNA 2′O-methylated sites in humans, 68 are specific to humans (humans), 35 are found across eukaryotes (eukaryotes) and 3 are conserved from bacteria to humans (universal) (left panel). The proportion of stable and variable sites amongst these three groups is represented using pie charts (right panel). (B andC), Mapping of the two classes of rRNA 2′O-methylated sites on the human ribosome (B) and at the PTC (C). Only rRNA 2′O-methylated positions directly involved in function domains or in interaction with components of the translational machinery have been annotated. (D–F) Structural environment of the variable rRNA 2′O-methylated sites significantly associated with breast cancer features. Red: stable sites; blue: variable sites; dotted circles: rRNA 2′O-methylation sites of interest; DC: decoding centre; PTC: peptidyl-transferase centre; PET: peptide exit tunnel. *: P < 0.05.
We then mapped the stable and variable rRNA 2′O-methylated positions on the human ribosome structure (16–18). It revealed an interesting distribution in space, where the decoding centre (DC), the peptidyl-transferase centre (PTC) and the polypeptide exit tunnel are mostly decorated with stable 2′O-methylated positions, whilst some variable positions are found in 1 or 2 nt layers away from the functional sites (i.e. maximum 4.5 Å away from the functional sites) (Figure 4B and C, annotated sites). However, 2′O-methylated sites are also found in functional sites, methyl carbon in very few 2′O-methylated sites are directly involved in van der Waals contacts with tRNAs, mRNA and other ribosome functional large ligands, as can be predicted from modelling these into the human ribosome structure. Most tRNAs binding regions in the large and small subunits are decorated with stable 2′O-methylated positions with the exception of 4 variable 2′O-methylated sites (18S-Um627, 28S-Am3739, 28S-Gm4340 and 28S-Cm4426) (Figure 4B). In particular, the 28S-Am3739 nucleotide is located at the hairpin loop of helix H69, which is located right next to the inter-subunit rotation centre describing pre- and post-translocation rotational states of the 40S with respect to the 60S ribosomal subunit and whose 2′O-methylation may influence P-site tRNA binding. The variable 28S-Gm4340 site is located at the CCA end region of the E-site tRNA that provides favourable van der Waals contacts to the E-site tRNA as observed experimentally in the human 80S structure. Finally, the four rRNA sites displaying changes in 2′O-methylation associated with biological and clinical features of breast cancer (28S-Gm1303, 18S-Gm1447, 28S-Gm4588 and 18S-Am576), are located at the periphery of the PTC of the ribosomes and are accessible for tRNAs, mRNA and other ribosome functional large ligands such as GTPases that associate transiently during the translation process (Figure 4C–F). Various factor binding pockets and their vicinity are mostly decorated with variable sites. Molecular docking tRNA and ribosomal factors suggest that stable rRNA 2′O-methylated sites are mostly located at the catalytic sites of the ribosomes, whilst the variable sites are found at the regulatory sites of the ribosomes.
DISCUSSION
Taken together, we show here for the first time that rRNA molecules exist naturally in humans as molecules not fully and equally 2′O-methylated at all the 106 positions. Indeed, our structure/function and evolution enrichment analyses as well as the differential association with biological characteristics, demonstrates that at least two classes of rRNA 2′O-methylated sites concur in human breast cancers, depending on their ability to tolerate (i.e. variable sites) or not (i.e. stable sites) the absence of chemical modifications. This unique discovery made by comparing human samples demonstrates the co-existence of stable and variable 2′O-methylation at specific rRNA positions.
Historically, plethora of studies mainly performed in both yeast and Xenopus laevis models, which have been the historical prototypic, eukaryotic models used during many years finely decipher the highly complex molecular mechanisms of rRNA transcription, chemical modifications and processing (27–31). These studies allowed the mapping of rRNA 2′O-methylation sites but also the demonstration of the importance of 2′O-methylation in the conserved intrinsic activity of the ribosome, on which life relies. Indeed, it appeared over the past years that loss of 2′O-methylation at a few number of rRNA sites in yeast is critical for ribosome functions and induces cell death (27,32). However, more recent findings indicate that in yeast, loss of rRNA 2′O-methylation at other sites have only little or no effect.
Our finding sustain the emerging notion that rRNA chemical modifications provide plasticity to the ribosome structure on which its regulatory effects rely, thus extending the modulation and activity of the ribosome to translational regulation in humans, including in cancer (4,33). In a single human sample, it appears that rRNA 2′O-methylation varies between the 106 rRNA 2′O-methylated positions (0 < C-score < 0.98). It indicates that even within a single human sample, rRNA molecules exist naturally as molecules not fully and equally 2′O-methylated at all the 106 positions. This observation is consistent with previous studies that were performed, albeit in limited number of models, either in cell lines (i.e. HeLa, PA1, HCT-116 and 293) (2,8,18,34–37), in cellular models describing tumour initiation and progression of solid and haematological cancers (1,6,38) or in few number of adult/embryonic organs (39). In addition to these previous studies, our findings also demonstrate that difference in rRNA 2′O-methylation occur between different human samples and reflects inter-patient variability. Variation in rRNA 2′O-methylation level between tumours might result from different mechanisms. Change in 2′O-methylation levels can result from alteration in expression of each individual component of the rRNA 2′O-methylation complex (i.e. the rRNA methyl-transferase FBL, the structural proteins Nop56, Nop58 or NHP2L1, or the small nucleolar snoRNA, which hybridizes in a sequence-specific manner the rRNA and thus specifically guides the enzyme) (1,7,9,28). Expression of these different components has been correlated with genomic alteration, loss of tumour suppressor or oncogene activation (1,7,28,40), suggesting that the 106 rRNA 2′O-methylation sites can reflect numerous alterations of the tumour. Moreover, changes in the ratio substrate:enzyme (rRNA:rRNA 2′O-methylation complex) might be sufficient to alter rRNA 2′O-methylation level, the rRNA level being also dependent of particular tumour suppressors and oncogenes (41). Interestingly, the rRNA 2′O-methylated positions that are not fully 2′O-methylated in a single human sample overlap with those displaying strong inter-variability between breast cancer samples. Development of statistical and methodological tools for future meta-analyses will be required to identify the complete list of rRNA sites accepting lack of 2′O-methylation, either commonly or specifically to a particular physio/pathological context.
The diversity of rRNA 2′O-methylation profiles naturally occurring in humans reinforces the altered translational control induced by experimental modulation of rRNA 2′O-methylation (1,2,28). Amongst the variable rRNA 2′O-methylation positions, the 18S-Gm1447 displays the highest variability in rRNA 2′O-methylation amongst tumours with the identification of human tumours and breast cancer cell lines that lack 2′O-methylation at this particular nucleotide. 18S-Gm1447 is in close vicinity of the uS10 ribosomal protein, which has been shown to bind to HCV IRES, regulates translation initiation of short 5′UTR containing mRNAs and contributes in resolving stalled ribosomes (42–46). Thus, absence or presence of rRNA 2′O-methylation at 18S-Gm1447 might finely modulate uS10 activity, which applies to the intrinsic ribosome function in general depending on its plasticity. Overall, our data reinforce, in humans, both the existence of rRNA 2′O-methylated compulsory/stable sites, which display no or low inter-patient variability and on which ribosome activities might rely as suggested by structural observations, and of rRNA 2′O-methylated plastic sites, which provides novel properties to the ribosome that can now be seen as a direct actor of translational regulation.
In addition to 2′O-methylation, rRNA exhibits other chemical modifications that affect translational regulation and contribute in the acquisition of cancer hallmarks. rRNA pseudouridylation (Ψ) remains the prototypic example. Alteration in Ψ rRNA profile induced by mutant DKC1, the enzyme-directed rRNA pseudouridine synthesis, has been shown to inhibit translation of the tumour suppressor p53 protein (47,48). More recently, the 18S-U609/U863Ψ has been shown to display tumour suppressive activity in response to KRAS-induced oncogenic stress in liver cancer (49). Similarly, loss of 18S-m1acp3U1248Ψ and 28S-m6A4220 rRNA are associated with colorectal and hepatocellular carcinoma, respectively (50,51). Thus, like other rRNA chemical modifications, rRNA 2′O-methylation can regulate translation and might provide novel molecular understanding of cancer biology, as it has been previously suggested, alteration of rRNA 2′O-methylation profile impairing cell proliferation and resistance to chemotherapy (1). Future studies will thus be required to finely identify rRNA 2′O-methylation sites involved in tumorigenesis by comparing series of normal and tumour breast tissues. Here, using benign tissue corresponding to non-transformed epithelial, we identify that in hyper-proliferative cells 2′O-methylation level of the 18S-Am576 site differs between benign and malignant tumours, suggesting that the rRNA 2′O-methylation profile is different between transformed and non-transformed tissue. It has to be noted that due to the cycling evolution of breast cancer through life (i.e. menstruation, pregnancy, menopause…) and the discrepancies between benign tumours (i.e. hyper-proliferative), mammoplasty (i.e. astrocyte enrichment) or stromal tissue (i.e. molecular traits) and breast cancer tissues (52), it remains difficult to build matched collection of normal mammary tissues.
Our data also demonstrate that rRNA 2′O-methylation corresponds to an additional layer of molecular characterization of breast cancer patient heterogeneity. Indeed, we show for the first time that rRNA 2′O-methylation levels differ between breast cancer subtypes and tumour grades. rRNA 2′O-methylation corresponds to an additional layer of molecular characterization that may help in the near future to improve breast cancer patient classification but also patients management. First, rRNA 2′O-methylation might be useful to improve the refinement of breast cancer patients stratification required in precision medicine. For example, the rRNA 2′O-methylation profile could help in identifying patients with the poorest outcome although they carry small tumours that are generally associated with a low risk factor in breast cancer. Such patients may thus benefit from a more aggressive treatment protocol usually reserved to treat largest breast tumours. Second, cancer-specific alterations of ribosome structure induced by naturally occurring variation in rRNA 2′O-methylation might be the source of powerful and specific anti-cancer molecules (18,53). Indeed, presence or absence of 2′O-methylation at a particular position might provide formation of specific ligand pockets to specifically target cancer-related ribosomes. In cancer, ribosome indeed appears as an emerging therapeutic target, inhibitors of RNA polymerase I activity specifically killing cancer cells being currently evaluated in clinical trials (54) and small molecules such as eukaryote-specific antibiotics directly targeting the human ribosome reducing proliferation of cancer cells (18,53,55–57). However, future studies will be required to determine whether diversity in rRNA 2′O-methylation profiles result from the intra-tumoral heterogeneity and/or heterogeneity in chemically modified rRNA molecules between and/or within a single cell.
The discovery of rRNA chemical modifications promoting ribosome plasticity will undoubtedly lead to a deeper and more complete understanding of rRNA chemical modifications in ribosome function and of cell fate determination providing a novel layer of cancer-related molecular fingerprinting.
DATA AVAILABILITY
Deposit FastQ and/or raw counts as dataset in GEO Profile (GSE143415).
To review GEO accession GSE143415:
Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143415.
Enter token wpknugomppoxlmh into the box.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Brigitte Manship for English editing.
Author contributions: V.Marcel, J.K., V.Marchand, H.P., F.N.V.L., L.A., V.B.I., P.L.M., D.M. and L.T. performed and analysed experiments. V.Marcel, J.K., H.P., L.T., Y.M. and A.V. performed statistical analyses. F.N.V.L., V.S., S.E.B., A.D., L.B.J., A.M.T., J.C.B., T.D. and F.A. provide human samples, clinical data and cellular models. V.Marcel, J.K., S.K.N., F.C., A.P., B.O.K. and J.J.D. analysed and interpreted the data. V.Marcel and J.J.D. shaped the research question and coordinated the project. V.Marcel, J.K., F.C. and A.V. supervised the daily experiments. V.Marcel, A.P. and J.J.D. raised funding. V.Marcel, J.K., S.K.N., B.P.K. and J.J.D. wrote the first draft of the manuscript.
Contributor Information
Virginie Marcel, Univ Lyon, Université Claude Bernard Lyon 1, Inserm 1052, CNRS 5286, Centre Léon Bérard, Centre de Recherche en Cancérologie de Lyon, 69008 Lyon, France.
Janice Kielbassa, Synergie Lyon Cancer, Gilles Thomas Bioinformatics Platform, Centre Léon Bérard, 69008 Lyon, France.
Virginie Marchand, UMS2008 IBSLor CNRS-INSERM-Lorraine University, Biopôle, 9 avenue de la forêt de haye, 54505 Vandoeuvre-les-Nancy, France.
Kundhavai S Natchiar, Centre for Integrative Biology (CBI), Department of Integrated Structural Biology, IGBMC, CNRS, Inserm, Université de Strasbourg, 1 rue Laurent Fries, 67404 Illkirch, France; Institut of Genetics and of Molecular and Cellular Biology (IGBMC), 1 rue Laurent Fries, 67404 Illkirch, France; Centre National de la Recherche Scientifique (CNRS), UMR 7104, 67404 Illkirch, France; Institut National de la Santé et de la Recherche Médicale (Inserm), U964, 67404 Illkirch, France; Université de Strasbourg, 67404 Illkirch, France.
Hermes Paraqindes, Univ Lyon, Université Claude Bernard Lyon 1, Inserm 1052, CNRS 5286, Centre Léon Bérard, Centre de Recherche en Cancérologie de Lyon, 69008 Lyon, France; Synergie Lyon Cancer, Gilles Thomas Bioinformatics Platform, Centre Léon Bérard, 69008 Lyon, France.
Flora Nguyen Van Long, Univ Lyon, Université Claude Bernard Lyon 1, Inserm 1052, CNRS 5286, Centre Léon Bérard, Centre de Recherche en Cancérologie de Lyon, 69008 Lyon, France.
Lilia Ayadi, UMS2008 IBSLor CNRS-INSERM-Lorraine University, Biopôle, 9 avenue de la forêt de haye, 54505 Vandoeuvre-les-Nancy, France; IMoPA, UMR 7365 CNRS-UL, Biopole UL, 54505 Vandoeuvre-les-Nancy, France.
Valérie Bourguignon-Igel, UMS2008 IBSLor CNRS-INSERM-Lorraine University, Biopôle, 9 avenue de la forêt de haye, 54505 Vandoeuvre-les-Nancy, France; IMoPA, UMR 7365 CNRS-UL, Biopole UL, 54505 Vandoeuvre-les-Nancy, France.
Piero Lo Monaco, Univ Lyon, Université Claude Bernard Lyon 1, Inserm 1052, CNRS 5286, Centre Léon Bérard, Centre de Recherche en Cancérologie de Lyon, 69008 Lyon, France.
Déborah Monchiet, Univ Lyon, Université Claude Bernard Lyon 1, Inserm 1052, CNRS 5286, Centre Léon Bérard, Centre de Recherche en Cancérologie de Lyon, 69008 Lyon, France.
Véronique Scott, Predictive biomarkers and novel therapeutic strategies Group, Institut Gustave Roussy, University of Paris Sud, INSERM 981, Université Paris Saclay, 114 rue Edouard Vaillant, 94800 Villejuif, France.
Laurie Tonon, Synergie Lyon Cancer, Gilles Thomas Bioinformatics Platform, Centre Léon Bérard, 69008 Lyon, France.
Susan E Bray, Tayside Tissue Bank, Ninewells Hospital and Medical School, NHS Tayside, Dundee DD1 9SY, Scotland, UK.
Alexandra Diot, Division of Cancer Research, University of Dundee, Ninewells Hospital and Medical School, Dundee DD1 9SY, Scotland, UK.
Lee B Jordan, Department of Pathology, University of Dundee, Ninewells Hospital and Medical School, Dundee DD1 9SY, UK.
Alastair M Thompson, Division of Cancer Research, University of Dundee, Ninewells Hospital and Medical School, Dundee DD1 9SY, Scotland, UK; Olga Keith Wiess Chair of Surgery, Dan L Duncan Breast Center, Division of Surgical Oncology, Baylor College of Medicine, Houston, TX 77030, USA.
Jean-Christophe Bourdon, Division of Cancer Research, University of Dundee, Ninewells Hospital and Medical School, Dundee DD1 9SY, Scotland, UK.
Thierry Dubois, Breast Cancer Biology Group, Translational Research Department, Institut Curie-PSL Research University, 26 rue d’Ulm, 75005 Paris, France.
Fabrice André, Predictive biomarkers and novel therapeutic strategies Group, Institut Gustave Roussy, University of Paris Sud, INSERM 981, Université Paris Saclay, 114 rue Edouard Vaillant, 94800 Villejuif, France.
Frédéric Catez, Univ Lyon, Université Claude Bernard Lyon 1, Inserm 1052, CNRS 5286, Centre Léon Bérard, Centre de Recherche en Cancérologie de Lyon, 69008 Lyon, France.
Alain Puisieux, Univ Lyon, Université Claude Bernard Lyon 1, Inserm 1052, CNRS 5286, Centre Léon Bérard, Centre de Recherche en Cancérologie de Lyon, 69008 Lyon, France.
Yuri Motorin, UMS2008 IBSLor CNRS-INSERM-Lorraine University, Biopôle, 9 avenue de la forêt de haye, 54505 Vandoeuvre-les-Nancy, France; IMoPA, UMR 7365 CNRS-UL, Biopole UL, 54505 Vandoeuvre-les-Nancy, France.
Bruno P Klaholz, Centre for Integrative Biology (CBI), Department of Integrated Structural Biology, IGBMC, CNRS, Inserm, Université de Strasbourg, 1 rue Laurent Fries, 67404 Illkirch, France; Institut of Genetics and of Molecular and Cellular Biology (IGBMC), 1 rue Laurent Fries, 67404 Illkirch, France; Centre National de la Recherche Scientifique (CNRS), UMR 7104, 67404 Illkirch, France; Institut National de la Santé et de la Recherche Médicale (Inserm), U964, 67404 Illkirch, France; Université de Strasbourg, 67404 Illkirch, France.
Alain Viari, Synergie Lyon Cancer, Gilles Thomas Bioinformatics Platform, Centre Léon Bérard, 69008 Lyon, France; INRIA Grenoble Rhône-Alpes, 38330 Montbonnot-Saint-Martin, France.
Jean-Jacques Diaz, Univ Lyon, Université Claude Bernard Lyon 1, Inserm 1052, CNRS 5286, Centre Léon Bérard, Centre de Recherche en Cancérologie de Lyon, 69008 Lyon, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Cancer Online.
FUNDING
Institut National du Cancer (INCa) [PLBIO program n°2019-138 MARACAS, PRT-K program n°2018-024 EMT-CoNCEPT, PAIR Sein program ARC_INCa_LNCC_7625 RiboTEM, and to B.P.K.]; Fondation ARC pour la recherche sur le cancer [20161204686 MARACAS.v0, and to B.P.K.]; ITMO Cancer [18CN044-00 RESIST]; SIRIC program [INCa-DGOS-Inserm_12563 LyRICAN]; Labex program [DEVCAN2UMAN]; Synergie Lyon Cancer Foundation; Fondation pour la Recherche Médicale (FRM) (to B.P.K.); Ligue Nationale Contre le Cancer (LNCC) (to B.P.K.); Agence Nationale pour la Recherche (ANR) (to B.P.K.); USIAS of the University of Strasbourg [USIAS-2018-012 to B.P.K.]; LNCC Fellowship (to H.P., F.N.V.L.); FRM Fellowship (to F.N.V.L.). French Research Minister Fellowship (to P.L.M.); Centre National de la Recherche Scientifique (CNRS) (to S.K.N., F.C., B.K.); Institut National de la Santé et de la Recherche Médicale (Inserm) (to J-.J.D., V.Marcel); Breast Cancer Now Fellowship [2012MaySF127 to A.D., J-,C.B,]; Chief Scientist Office; NHS Scotland; University of Dundee.
Conflict of interest statement. None declared.
This paper is linked to: 10.1093/narcan/zcaa035.
REFERENCES
- 1. Marcel V., Ghayad S.E., Belin S., Therizols G., Morel A.P., Solano-Gonzàlez E., Vendrell J.A., Hacot S., Mertani H.C., Albaret M.A. et al. P53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013; 24:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Erales J., Marchand V., Panthu B., Gillotf S., Belin S., Ghayad S.E., Garcia M., Laforets F., Marcel V. et al. Evidence for rRNA 2 {’}-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shi Z., Fujii K., Kovary K.M., Genuth N.R., Röst H.L., Teruel M.N., Barna M. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell. 2017; 67:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Genuth N.R., Barna M. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet. 2018; 19:431–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Helm M., Motorin Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017; 18:275–291. [DOI] [PubMed] [Google Scholar]
- 6. Belin S., Beghin A., Solano-Gonzàlez E., Bezin L., Brunet-Manquat S., Textoris J., Prats A.-C., Mertani H.C., Dumontet C., Diaz J.-J. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One. 2009; 4:e7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gachet S., El-Chaar T., Avran D., Genesca E., Catez F., Quentin S., Delord M., Therizols G., Briot D. et al. Deletion 6q drives T-cell leukemia progression by ribosome modulation. Cancer Discov. 2018; 8:1614–1631. [DOI] [PubMed] [Google Scholar]
- 8. D’Souza M.N., Gowda N.K.C., Tiwari V., Babu R.O., Anand P., Dastidar S.G., Singh R., Selvaraja B., Pal R., Ramesh A. et al. FMRP interacts with C/D box snoRNA in the nucleus and regulates ribosomal RNA methylation. iScience. 2018; 9:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su H., Xu T., Ganapathy S., Shadfan M., Long M., Huang T.H.-M., Thompson I., Yuan Z.-M. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2014; 33:1348–1358. [DOI] [PubMed] [Google Scholar]
- 10. Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011; 12:861–874. [DOI] [PubMed] [Google Scholar]
- 11. Harbeck N., Penault-Llorca F., Cortes J., Gnant M., Houssami N., Poortmans P., Ruddy K., Tsang J., Cardoso F. Breast cancer. Nat. Rev. Dis. Prim. 2019; 5:66–97. [DOI] [PubMed] [Google Scholar]
- 12. Beekman R., Chapaprieta V., Russiñol N., Vilarrasa-Blasi R., Verdaguer-Dot N., Martens J.H.A., Duran-Ferrer M., Kulis M., Serra F., Javierre B.M. et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat. Med. 2018; 24:868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E. et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018; 555:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marchand V., Blanloeil-Oillo F., Helm M., Motorin Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. NucleicAcids Res. 2016; 44:135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. 2015; 54:451–455. [DOI] [PubMed] [Google Scholar]
- 16. Khatter H., Myasnikov A.G., Natchiar S.K., Klaholz B.P. Structure of the human 80S ribosome. Nature. 2015; 520:640–645. [DOI] [PubMed] [Google Scholar]
- 17. Natchiar S.K., Myasnikov A.G., Kratzat H., Hazemann I., Klaholz B.P. Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017; 551:472–477. [DOI] [PubMed] [Google Scholar]
- 18. Natchiar S., Myasnikov A., Hazemann I., Klaholz B. Visualizing the role of 2′-OH rRNA methylations in the human ribosome structure. Biomolecules. 2018; 8:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bourdon J.-C., Khoury M.P., Diot A., Baker L., Fernandes K., Aoubala M., Quinlan P., Purdie C.A., Jordan L.B., Prats A.-C. et al. p53 mutant breast cancer patients expressing p53γ have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res. 2011; 13:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nguyen Van Long F., Lardy-Cleaud A., Bray S., Chabaud S., Dubois T., Diot A., Jordan L.B., Thompson A.M., Bourdon J.-C. et al. Druggable Nucleolin identifies breast tumours associated with poor prognosis that exhibit different biological processes. Cancers (Basel). 2018; 10:390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maire V., Baldeyron C., Richardson M., Tesson B., Vincent-Salomon A., Gravier E., Marty-Prouvost B., De Koning L., Rigaill G., Dumont A. et al. TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS One. 2013; 8:e63712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. André F., Michiels S., Dessen P., Scott V., Suciu V., Uzan C., Lazar V., Lacroix L., Vassal G., Spielmann M. et al. Exonic expression profiling of breast cancer and benignlesions: a retrospective analysis. Lancet Oncol. 2009; 10:381–390. [DOI] [PubMed] [Google Scholar]
- 23. Krogh N., Nielsen H. Sequencing-based methods for detection and quantitation of ribose methylations in RNA. Methods. 2018; 156:5–15. [DOI] [PubMed] [Google Scholar]
- 24. Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. Engl. 2015; 54:451–455. [DOI] [PubMed] [Google Scholar]
- 25. Marchand V., Ayadi L., El HajjA., Blanloeil-Oillo F., Helm M., Motorin Y.. Lusser A. High-throughput mapping of 2′-O-Me residues in RNA using next-generation sequencing (Illumina RiboMethSeq Protocol). RNA Methylation: Methods and Protocols, Methods in Molecular Biology. 2017; NY: Springer; 171–187. [DOI] [PubMed] [Google Scholar]
- 26. Pichot F., Marchand V., Ayadi L., Bourguignon-Igel V., Helm M., Motorin Y. Holistic optimization of bioinformatic analysis pipeline for detection and quantification of 2′-O-Methylations in RNA by RiboMethSeq. Front. Genet. 2020; 11:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Decatur W.A., Fournier M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002; 27:344–351. [DOI] [PubMed] [Google Scholar]
- 28. Lo Monaco P., Marcel V., Diaz J.J., Catez F. 2′-O-methylation of ribosomal RNA: towards an epitranscriptomic control of translation?. Biomolecules. 2018; 8:106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maden B.E.H., Corbett M.E., Heeney P.A., Pugh K., Ajuh P.M. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995; 77:22–29. [DOI] [PubMed] [Google Scholar]
- 30. Maden B.E.H. Methylation map of xenopus laevis ribosomal RNA. Nature. 1980; 288:293–296. [DOI] [PubMed] [Google Scholar]
- 31. Sirum-Connolly K., Mason T.L. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993; 262:1886–1889. [DOI] [PubMed] [Google Scholar]
- 32. Baudin-Baillieu A., Fabret C., Liang X.H., Piekna-Przybylska D., Fournier M.J., Rousset J.P. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009; 37:7665–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Truitt M.L., Ruggero D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2016; 16:288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma S., Marchand V., Motorin Y., Lafontaine D.L.J. Identification of sites of 2′-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 2017; 7:11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krogh N., Jansson M.D., Häfner S.J., Tehler D., Birkedal U., Christensen-Dalsgaard M., Lund A.H., Nielsen H. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016; 44:7884–7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Incarnato D., Anselmi F., Morandi E., Neri F., Maldotti M., Rapelli S., Parlato C., Basile G., Oliviero S. High-throughput single-base resolution mapping of RNA 2′-O-methylated residues. NucleicAcids Res. 2016; 45:1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu Y., Pirnie S.P., Carmichael G.G. High-throughput and site-specific identification of 2′-O-methylation sites using ribose oxidation sequencing (RibOxi-seq). RNA. 2017; 23:1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou F., Liu Y., Rohde C., Pauli C., Gerloff D., Kohn M., Misiak D., Baumer N., Cui C., Gollner S. et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 2017; 19:844–855. [DOI] [PubMed] [Google Scholar]
- 39. Hebras J., Krogh N., Marty V., Nielsen H., Cavaillé J. Developmental changes of rRNA ribose methylations in the mouse. RNA Biol. 2020; 17:150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mourksi N.-E.-H., Morin C., Fenouil T., Diaz J.-J., Marcel V. snoRNAs offer novel insight and promising perspectives for lung cancer understanding and management. Cells. 2020; 9:541–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bywater M.J., Pearson R.B., McArthur G.A., Hannan R.D. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat. Rev. Cancer. 2013; 13:299–314. [DOI] [PubMed] [Google Scholar]
- 42. Haimov O., Sinvani H., Martin F., Ulitsky I., Emmanuel R., Tamarkin-Ben-Harush A., Vardy A., Dikstein R. Efficient and accurate translation initiation directed by TISU involves RPS3 and RPS10e binding and differential eukaryotic initiation factor 1A regulation. Mol. Cell. Biol. 2017; 37:e00150-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malygin A.A., Kossinova O.A., Shatsky I.N., Karpova G.G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. NucleicAcids Res. 2013; 41:8706–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sundaramoorthy E., Leonard M., Mak R., Liao J., Fulzele A., Bennett E.J. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell. 2017; 65:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsuo Y., Ikeuchi K., Saeki Y., Iwasaki S., Schmidt C., Udagawa T., Sato F., Tsuchiya H., Becker T., Tanaka K. et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017; 8:159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. DiGiuseppe S., Rollins M.G., Bartom E.T., Walsh D. ZNF598 plays distinct roles in interferon-stimulated gene expression and poxvirus protein synthesis. Cell Rep. 2018; 23:1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bellodi C., Krasnykh O., Haynes N., Theodoropoulou M., Peng G., Montanaro L., Ruggero D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010; 70:6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruggero D. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science (80-.). 2003; 299:259–262. [DOI] [PubMed] [Google Scholar]
- 49. McMahon M., Contreras A., Holm M., Uechi T., Forester C.M., Pang X., Jackson C., Calvert M.E., Chen B., Quigley D.A. et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife. 2019; 8:e48847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Babaian A., Rothe K., Girodat D., Minia I., Djondovic S., Milek M., Spencer Miko S., Wieden H.-J., Landthaler M., Morin G. et al. Loss of m 1 acp 3 Ψ ribosomal RNA modification is a major feature of cancer. Cell Rep. 2019; 31:107611. [DOI] [PubMed] [Google Scholar]
- 51. Ma H., Wang X., Cai J., Dai Q., Natchiar S.K., Lv R., Chen K., Lu Z., Chen H., Shi Y.G. et al. Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019; 15:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aran D., Camarda R., Odegaard J., Paik H., Oskotsky B., Krings G., Goga A., Sirota M., Butte A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017; 8:1077–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Myasnikov A.G., Kundhavai Natchiar S., Nebout M., Hazemann I., Imbert V., Khatter H., Peyron J.-F.F., Klaholz B.P. Structure-function insights reveal the human ribosome as a cancer target for antibiotics. Nat. Commun. 2016; 7:12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Catez F., Dalla Venezia N., Marcel V., Zorbas C., Lafontaine D.L.J., Diaz J.-J. Ribosome biogenesis: an emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 2019; 159:74–81. [DOI] [PubMed] [Google Scholar]
- 55. Boer R.E., Schneekloth J.S. Targeting mammalian translational inhibition with tetracyclines. Cell Chem. Biol. 2018; 25:1437–1438. [DOI] [PubMed] [Google Scholar]
- 56. Gilles A., Frechin L., Natchiar K., Biondani G., von Loeffelholz O., Holvec S., Malaval J.-L., Winum J.-Y., Klaholz B.P., Peyron J.-F. Targeting the human 80S ribosome in cancer: from structure to function and drug design for innovative adjuvant therapeutic strategies. Cells. 2020; 9:629–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pellegrino S., Meyer M., Zorbas C., Bouchta S.A., Saraf K., Pelly S.C., Yusupova G., Evidente A., Mathieu V., Kornienko A. et al. The amaryllidaceae alkaloid haemanthamine binds the eukaryotic ribosome to repress cancer cell growth. Structure. 2018; 26:416–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deposit FastQ and/or raw counts as dataset in GEO Profile (GSE143415).
To review GEO accession GSE143415:
Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143415.
Enter token wpknugomppoxlmh into the box.