Abstract
Objectives
To evaluate the current prevalence status of mecC MRSA among dairy farms in England and Wales 5 years after a previous survey conducted in 2011–12.
Methods
A convenience sample of 697 dairy farms in England and Wales was used for the study, conducted in 2017–18, testing bulk tank milk samples for the presence of mecC MRSA using high salt broth enrichment and chromogenic MRSA agar selection. All putative MRSA isolates were screened by PCR for the presence of mecA and mecC genes and subjected to antimicrobial susceptibility testing using both the disc diffusion method and VITEK® 2. MRSA isolates were also sequenced for genomic characterization.
Results
mecC MRSA were detected on 4 out of 697 dairy farms in England and Wales (prevalence 0.57%, 95% CI 0.16%–1.46%). Three of the mecC isolates were ST425 and one was ST4652 (in the CC130 lineage). Two mecA MRSA were also isolated: one ST5 and one ST398.
Conclusions
These results indicate that there has been a substantial reduction in the prevalence of mecC MRSA in England and Wales with a 72% reduction (2.15% to 0.57%) compared with a previous study. While the levels of mecA MRSA remain very low the continued presence of ST398, a livestock-associated MRSA, suggests that this lineage is established in the UK.
Introduction
MRSA represent a significant healthcare problem causing infections in both people and animals. MRSA typically have high-level resistance to β-lactams. The resistance to β-lactams in staphylococci most commonly results from expression of a blaZ gene (encoding a β-lactamase) and/or a mec gene (mecA, mecC or mecB encoding an alternative PBP2, the target of β-lactams).1
The mecC gene was initially found among bovine and human isolates from the UK and Denmark in 2011.2 Although mecC MRSA was first isolated from cattle and humans, it has a wide range of host species, including other livestock species, companion animals and wildlife.3
The mecC gene has 69% nucleotide identity with mecA. As a result, mecC MRSA test negative when using molecular detection methods based on the mecA gene. For example, agglutination assays for mecA encoded PBP2a (using latex beads coated with monoclonal antibodies) will misidentify mecC MRSA as being susceptible.4 A number of PCR assays5 have been developed or adapted to include detection of the mecC gene. Phenotypic testing for MRSA based on antimicrobial susceptibility fails to discriminate between mecA and mecC MRSA and severe and fatal cases of mecC MRSA infections have been described in the literature.6
Zoonotic transmission of mecC MRSA from livestock to humans has been reported,7,8 highlighting the importance of monitoring its prevalence in both human and livestock populations. Paterson et al.9,10 reported a 0.45% prevalence rate among 2010 MRSA isolates collected from individual human patients from six microbiology laboratories in England and a 2.15% prevalence rate in bulk tank milk samples collected from 465 dairy farms in England and Wales in 2011–12. In order to provide an updated prevalence of mecC MRSA on dairy farms in England and Wales this survey was undertaken using bulk tank milk samples collected in 2017–18.
Methods
Bulk tank milk samples and processing
Milk samples (n = 697) from England and Wales were supplied by a commercial milk testing company responsible for more than 95% of quality assurance testing of bulk tank milk from GB dairy farms. The samples were being used for another study between January 2017 and April 2018 and were exploited as a convenience sample for this study. The testing laboratory was asked to provide randomly selected samples although no formal randomization protocol was followed. No farms were sampled more than once, and the date of sampling and the location of each farm were the only metadata available.
The bulk tank milk samples were collected aseptically and stored at 4 °C for up to 5 days before freezing at −20°C prior to testing. The processing of the samples was carried out as described previously.10 Briefly, samples were thawed at 37°C and 1 mL of milk was added to 10 mL of 6% NaCl broth (E&O Laboratories, UK). After static incubation at 37°C for 24 h, 50 μL of culture was spread onto MRSA Brilliance 2 plates (Oxoid) and incubated at 37°C for 24 h. Potential MRSA colonies (blue colour) were subcultured on Staph Brilliance 24 plates (Oxoid) and subsequently screened for mecA, mecC and femB by multiplex PCR as described previously.11
Antimicrobial susceptibility testing
Disc diffusion method susceptibility testing for penicillin (Oxoid, 1 unit) and cefoxitin (Oxoid, 30 μg) was performed according to the EUCAST standard (Version 7.1, March 2017). As EUCAST does not define a disc diffusion breakpoint for oxacillin, we also performed oxacillin (Oxoid, 1 μg) disc diffusion according to BSAC (Version 14, January 2015). The VITEK® 2 system was used to validate the antimicrobial susceptibility testing results using an AST-GP79 card with the EUCAST breakpoint parameter set configuration, in accordance with the manufacturer’s instructions.
Genome sequencing and bioinformatic analysis
Genomic DNA of MRSA isolates was extracted from overnight cultures grown in TSB at 37°C with 200 rpm shaking using the MasterPure Gram-Positive DNA Purification Kit (Cambio, UK). Illumina library preparation, HiSeq sequencing, and genome assembly were carried out as previously described.7 The presence of mecA or mecC genes in all MRSA isolates was confirmed and the multilocus ST was also determined using the sequencing data. The presence of antimicrobial resistance genes in these isolates were determined using the Resfinder gene database to search the assembled genomes using ≥80% identity over ≥60% of the length of the target gene as a threshold for a positive identification. A maximum likelihood core genome phylogenetic tree was produced using an alignment generated by Panaroo12 and IQ-TREE 213 to create the tree.
Ethical review
The study was subject to ethical review at the University of Cambridge, Department of Veterinary Medicine (ref. 2017/012).
Data availability
The sequencing data for the six isolates described in this paper have been deposited in the European Short Read Archive (ebi.ac.uk) with the following sample accession numbers: QM059, ERS2607810; QM212, ERS2607839; QM312, ERS2607851; QM355, ERS2607859; QM518, ERS2607880; QM652, ERS2607892. A list of accession numbers for other sequence data used in analyses is provided in the Supplementary data (available at JAC-AMR Online).
Results
Prevalence of mecC MRSA among dairy farm bulk milk samples
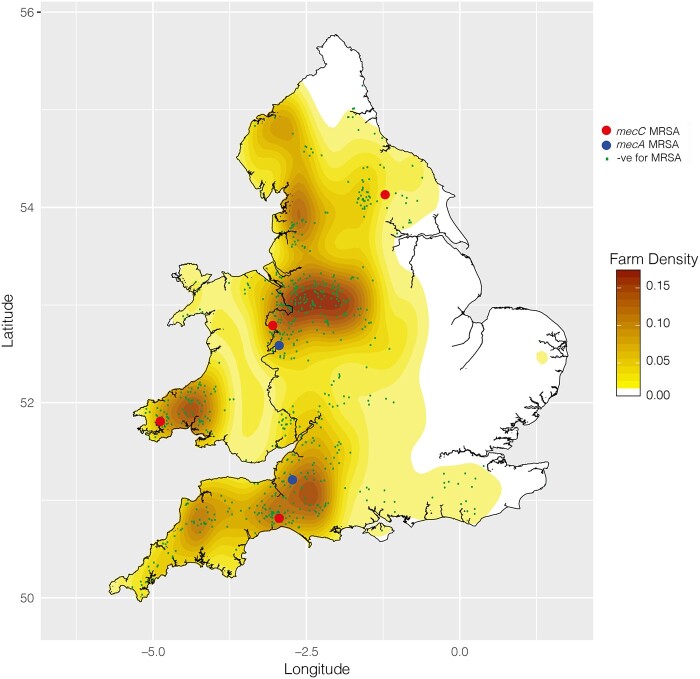
Four dairy farms from a total of 697 were positive for mecC MRSA, which represents a farm level prevalence rate of 0.57% (95% CI 0.16%–1.46%) in England and Wales. Three of the positive farms were in England and one was in Wales. Figure 1 maps the locations of farms sampled, the locations of the positive farms, and a heat map of the density of dairy farms across England and Wales. A comparison of the distribution of the sampled farms with the average density of dairy farms shows that the sampling distribution matches that of the farms being sampled with no obvious deviant clustering.
Figure 1.
Geographical distribution of farms included in the survey with results. Map of England and Wales showing the location of farms tested and their results. Farms testing negative are shown as green dots, mecC MRSA-positive farms are shown with larger red circles and mecA MRSA-positive farms are shown with larger blue circles. A heatmap showing the density of dairy farms per square kilometre is also displayed on the map.
Characterization of bovine mecC MRSA
All of the four mecC MRSA isolates were resistant to penicillin, oxacillin and cefoxitin according to disc diffusion antimicrobial susceptibility testing (Table 1). Further antimicrobial susceptibility testing on these isolates using the VITEK® 2 system confirmed the resistance to these three antimicrobials and also reported that all isolates were resistant to cefalotin, a first-generation cephalosporin antibiotic. All mecC isolates had blaZ and mecC genes identical to those described in the original mecC paper,2 but no other antimicrobial resistance genes.
Table 1.
Phenotypic and genotypic characteristics of MRSA isolates in this study
| Isolate | County | mec gene | ST | spa type | Oxacillin |
Cefoxitin |
Penicillin |
Cefalotin | Human immune evasion cluster |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IZD (mm) | VITEK | IZD (mm) | VITEK | IZD (mm) | VITEK | VITEK | chp | scn | sak | |||||
| QM059 | North Yorkshire | mecC | 4652 | t843 | 7 | R | 18 | R | 15 | R | R | − | − | − |
| QM212 | Somerset | mecA | 5 | t002 | 6 | R | 10 | R | 6 | R | R | + | + | + |
| QM312 | Shropshire | mecA | 398 | t034 | 6 | R | 15 | R | 6 | R | R | + | + | + |
| QM355 | Shropshire | mecC | 425 | t10855 | 11 | R | 20 | R | 14 | R | R | − | − | − |
| QM518 | Devon | mecC | 425 | t6292 | 6 | R | 18 | R | 10 | R | R | − | − | − |
| QM652 | Pembrokeshire | mecC | 425 | t6292 | 6 | R | 17 | R | 10 | R | R | − | − | − |
IZD, inhibition zone diameter; R, resistant.
6 mm in disc diffusion indicates no inhibition zone.
For penicillin, resistant breakpoint for disc diffusion is <26 mm diameter.
For cefoxitin, resistant breakpoint for disc diffusion is <22 mm diameter.
For oxacillin, resistant breakpoint for disc diffusion is ≤14 mm diameter (BSAC).
spa types were determined from the sequencing data using spaTyper 1.0 (https://cge.cbs.dtu.dk/services/spaTyper-1.0/).
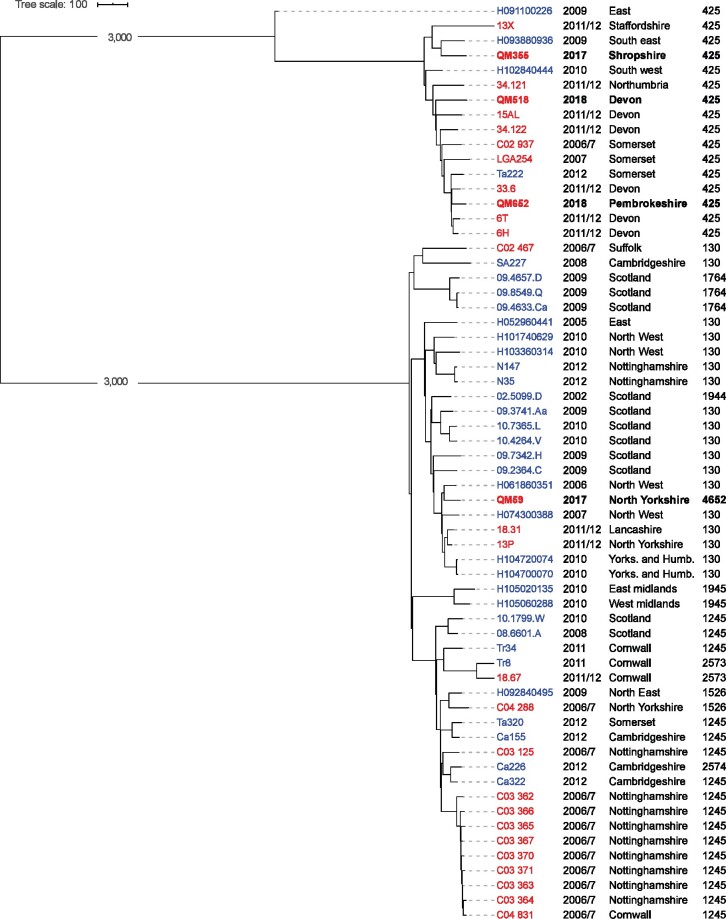
A phylogenetic tree showing the four positive isolates in the context of previous mecC isolates found in the UK is shown in Figure 2. Three of the isolates were ST425 and one was ST4625 (a single locus variant of ST130) representing the two main lineages found in the UK.
Figure 2.
Core-genome phylogenetic tree showing the mecC MRSA isolates found in this study (indicated by bold text) together with other published mecC isolates from the UK.2,10,16 The core genome alignment was produced using Panaroo12 and the maximum likelihood tree generated using IQ-TREE 2.13 The year of isolation, geographical region MLSTs are provided in the text next to the isolate names. Isolates with names in blue text came from human samples. Isolates with names in red text were of bovine origin. The branches of the trees separating the two different lineages (CC425 and CC130) are not shown to scale in order to make the relationships within the two clades easier to see. Accession numbers for sequencing data for all isolates in the tree are provided in the Supplementary data.
Prevalence and characteristics of mecA MRSA from GB bulk milk
Two mecA MRSA were also identified in the survey giving a prevalence of 0.29% (95% CI 0.08%–1.04%). The location of the positive farms is shown in Figure 1. Both isolates were resistant to penicillin, oxacillin and cefoxitin based on the results of disc diffusion and VITEK® 2 (Table 1). The VITEK® 2 results also showed that both isolates were resistant to cefalotin, erythromycin and clindamycin and isolate QM312 was also resistant to ceftiofur and cefquinome. Isolate QM212 belongs to ST5 and possessed mecA, blaZ and ermA antimicrobial resistance genes. Isolate QM312 was an ST398 MRSA associated with livestock-associated MRSA epidemiology. This isolate possessed all three canonical SNPs associated with the livestock-independent clade of this lineage and, along with the ST5 MRSA, possessed the immune evasion cluster genes scn, chp and sak.14 QM312 possessed an ermC gene in addition to mecA.
Discussion
In this study, we conducted a follow-up prevalence study of mecC MRSA on dairy farms in England and Wales 5 years after the previous survey.10 We were unable to include Scotland in this survey although in the previous study no MRSA were found in Scottish dairy herds. Four out of 697 dairy farms were found to have mecC MRSA, which gives an estimated prevalence of 0.57%, compared with a 2.15% prevalence reported for 2011–12, a reduction of 72%.10 The original mecC prevalence study employed formal methodology to ensure random selection of dairy farms whereas this study used a convenience sample. Nonetheless an examination of the distribution of farms sampled does not indicate any obvious bias in this sample. Furthermore, the results from this study are similar to another study, published recently, which sampled 363 farms in 2015–16 and reported two mecC MRSA positives (0.5%).15
Although the frequency of mecC MRSA remains low, its presence in dairy cattle poses a zoonotic risk to humans. Two case studies have demonstrated the transmission of mecC MRSA from livestock to people evidenced using molecular epidemiology.7,8 Three human prevalence surveys have been performed in the UK. A study performed in 2011–12 reported that mecC was responsible for 0.45% of MRSA9 in six hospitals across the UK and a 2012–13 study in the East of England reported a mecC MRSA rate of 0.52%.16 A more recent study of human MRSA isolates from North-West England, collected in 2015, reported a mecC MRSA prevalence of 0.07%,17 a reduction in the human prevalence that appears to parallel the fall seen in livestock. Any reduction in livestock of this MRSA is to be welcomed and will reduce the potential for zoonotic transmission.
We can only speculate on what may have caused a reduction in prevalence of mecC MRSA in UK dairy farms. The samples in this prevalence study were collected in 2017–18, which coincides with the end of the ‘UK five year AMR strategy 2013 to 2018’ published by the UK Department of Health18 and it seems reasonable to attribute the observed decrease to the successful implementation of this strategy, although many other events or phenomena may be responsible. According to the UK One Health Report published in early 2019,19 an overall reduction of 19% in antibiotic usage was achieved in people and animals between 2013 and 2017 with a drop of 35% in animals (436 to 282 tonnes). Antibiotic use is an obvious driver of antibiotic resistance and the use of β-lactams, particularly cephalosporins, on dairy farms is likely to select for mecC MRSA due to the differential affinity of the mecC PBP2a for cephalosporins.20 The 2018 UK Veterinary Antibiotic Resistance and Sales Surveillance Report21 reports a 50% reduction in third/fourth-generation cephalosporin use on dairy farms from 2017 to 2018.
Apart from mecC MRSA, this study has also identified two mecA MRSA-positive farms in England, at similar levels to those seen in 2011–12. One mecA MRSA isolate belongs to ST5, a typical human S. aureus lineage and the other to the livestock-associated MRSA lineage ST398. As the ST398 is a member of the livestock independent lineage, with the human immune evasion cluster, it is possible that both these mecA MRSA were of recent human origin. These findings are notable but not alarming at this time.
Conclusions
This study has found a welcome decrease in mecC MRSA prevalence on the dairy farms in England in comparison to that of 2011–12 that may be the result of improved antibiotic stewardship in the UK. In light of the zoonotic potential of this MRSA from livestock to humans, it is important to continue to monitor levels of mecC MRSA in agriculture and the environment.
Funding
The research was funded by UK Medical Research Council awards MR/P007201/1 and MR/S013660/1. Cheng Cui was supported by a British Council fellowship.
Transparency declarations
None to declare.
Supplementary data
A list of accession numbers is available as Supplementary data at JAC-AMR Online.
Supplementary Material
References
- 1. Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev 2017; 41: 430–49. [DOI] [PubMed] [Google Scholar]
- 2. García-Álvarez L, Holden MT, Lindsay H et al. A descriptive study of MRSA harbouring a novel mecA homologue emerging in human and bovine populations in the United Kingdom and Denmark. Lancet Infect Dis 2011; 11: 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paterson GK, Harrison EM, Holmes MA. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol 2014; 22: 42–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dupieux C, Bouchiat C, Larsen AR et al. Detection of mecC-positive Staphylococcus aureus: what to expect from immunological tests targeting PBP2a? J Clin Microbiol 2017; 55: 1961–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Becker K, Larsen AR, Skov RL et al. Evaluation of a modular multiplex-PCR methicillin-resistant Staphylococcus aureus detection assay adapted for mecC detection. J Clin Microbiol 2013; 51: 1917–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia-Garrote F, Cercenado E, Marin M et al. Methicillin-resistant Staphylococcus aureus carrying the mecC gene: emergence in Spain and report of a fatal case of bacteraemia. J Antimicrob Chemother 2014; 69: 45–50. [DOI] [PubMed] [Google Scholar]
- 7. Harrison EM, Paterson GK, Holden MT et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. Embo Mol Med 2013; 5: 509–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Petersen A, Stegger M, Heltberg O et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect 2013; 19: E16–22. [DOI] [PubMed] [Google Scholar]
- 9. Paterson GK, Morgan FJE, Harrison EM et al. Prevalence and characterization of human mecC methicillin-resistant Staphylococcus aureus isolates in England. J Antimicrob Chemother 2014; 69: 907–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paterson GK, Morgan FJE, Harrison EM et al. Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (MRSA) in bovine bulk tank milk in Great Britain. J Antimicrob Chemother 2014; 69: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paterson GK, Larsen AR, Robb A et al. The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J Antimicrob Chemother 2012; 67: 2809–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tonkin-Hill G, MacAlasdair N, Ruis C et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol 2020; 21: 180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Minh BQ, Schmidt HA, Chernomor O et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 2020; 37: 1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stegger M, Liu CM, Larsen J et al. Rapid differentiation between livestock-associated and livestock-independent Staphylococcus aureus CC398 clades. PLoS One 2013; 8: e79645.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fisher EA, Paterson GK. Prevalence and characterisation of methicillin-resistant staphylococci from bovine bulk tank milk in England and Wales. J Glob Antimicrob Resist 2020; 22: 139–44. [DOI] [PubMed] [Google Scholar]
- 16. Harrison EM, Coll F, Toleman MS et al. Genomic surveillance reveals low prevalence of livestock-associated methicillin-resistant Staphylococcus aureus in the East of England. Sci Rep 2017; 7: 7406.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paterson GK. Low prevalence of livestock-associated methicillin-resistant Staphylococcus aureus clonal complex 398 and mecC MRSA among human isolates in North-West England. J Appl Microbiol 2020; 128: 1785–92. [DOI] [PubMed] [Google Scholar]
- 18. UK Five Year Antimicrobial Resistance Strategy 2013 to 2018. Department of Health, 2013. https://www.gov.uk/government/publications/uk-5-year-antimicrobial-resistance-strategy-2013-to-2018.
- 19. Veterinary Medicines Directorate. UK One Health Report: Joint Report on Antibiotic Use and Antibiotic Resistance, 2013–2017. 2019. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/921039/Ted_Final_version__1318703-v45-One_Health_Report_2019_FINAL-accessible.pdf.
- 20. Kim C, Milheirico C, Gardete S et al. Properties of a novel PBP2A protein homologue from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam resistant phenotype. J Biol Chem 2012; 287: 36854–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Veterinary Medicines Directorate. UK Veterinary Antibiotic Resistance and Sales Surveillance Report (UK-VARSS 2018). 2019. https://www.gov.uk/government/publications/veterinary-antimicrobial-resistance-and-sales-surveillance-2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data for the six isolates described in this paper have been deposited in the European Short Read Archive (ebi.ac.uk) with the following sample accession numbers: QM059, ERS2607810; QM212, ERS2607839; QM312, ERS2607851; QM355, ERS2607859; QM518, ERS2607880; QM652, ERS2607892. A list of accession numbers for other sequence data used in analyses is provided in the Supplementary data (available at JAC-AMR Online).