Abstract
Background
The emerging resistance to the last-resort antimicrobial colistin is being reported globally. Underestimation of the burden of colistin resistance and misinterpretation of colistin susceptibility test results, using suboptimal testing methods, may be causing unexplained treatment failures and even mortality among critically ill patients. Thus, this study was conducted at an apex trauma centre to assess the performance of Vitek®2 for colistin susceptibility testing.
Methods
A total of 910 clinical isolates of Gram-negative bacteria (GNB), including Enterobacterales, Acinetobacter baumannii and Pseudomonas aeruginosa, were tested and analysed for colistin resistance using Vitek®2. Broth microdilution (BMD) was taken as the reference method. The essential (EA) and categorical (CA) agreements and very major error (VME) and major error (ME) rates were calculated. An MIC correlation was taken to be positive with EA ≥ 90%, CA ≥ 90%, VME ≤ 1.5% and ME ≤ 3.0% rates. Spearman’s coefficient was calculated and P < 0.05 was considered statistically significant.
Results
A total of 64% of isolates were MDR. Overall, 196 (21.5%) and 110 (12%) of isolates were resistant to colistin by BMD and Vitek®2, respectively. The automated Vitek®2 method failed to detect the resistance in up to 48.5% of GNB tested. When comparing Vitek®2 colistin interpretive results with reference BMD for all 910 isolates, the CA was 88% (798/910) with 10% (95/910) VMEs and 1% (9/910) MEs.
Conclusions
The Vitek®2 method for colistin susceptibility testing, still in use in some settings; is a suboptimal and unreliable method.
Introduction
Polymyxins (polymyxin B and colistin), penta-cationic antibiotics that selectively bind to the LPS of Gram-negative bacteria (GNB), were introduced in the late 1950s. Resistance to these last-resort antimicrobials is facilitated by a cationic modification of LPS, impermeability and other efflux mechanisms. As well as the intrinsically resistant organisms such as Morganella spp., Proteus spp. and Providencia spp., acquired resistance to polymyxins can be both plasmid-mediated and chromosomal. While chromosomal resistance is attributable to the cross transmission of resistant isolates and to the previous use of colistin, plasmid-mediated resistance by the mcr gene was first described in China and it is now being reported all over the world.1
In 2016, CLSI and EUCAST jointly recommended only the broth microdilution (BMD) methodology to perform the colistin susceptibility tests.2,3
It is now well established that disc diffusion methodology is unreliable to detect colistin resistance.4–7 Error rates up to 41.5% have been reported for colistin Etests, which now are not recommended as a testing method.5–9
Automated systems have become the backbone of diagnostic microbiology labs even in developing countries. In smaller labs, it is difficult to ensure quality in disc diffusion and BMD. There is also a scarcity of trained technical staff; all of these drive the use of semi- or fully automated identification/antimicrobial susceptibility testing (ID/AST) systems. A very recent study reported that with a very major error (VME) rate of 36% for colistin testing, Vitek®2 may not be that reliable.10 This study mainly evaluated Gram-negative pathogens of family Enterobacterales. In recent times, non-fermenters such as Acinetobacter spp. and Pseudomonas spp. have become the most common pathogens causing healthcare-associated infections at many centres.11–13 The increasing use of colistin for treatment of suspected sepsis in ICUs makes it necessary to evaluate the automated methods for testing and reporting colistin susceptibility, which is essential for any successful antimicrobial stewardship programme.14
Due to the lack of trained personnel in India, BMD is not an attractive option for AST. Automated methods such as Vitek®2 that provide more objective results and are less prone to operator error are preferred.
The aim of this study was to evaluate the performance of Vitek®2 to detect colistin resistance and to determine the prevalence of colistin resistance among GNB isolated from diagnostic specimens from January to August 2019.
Methods
This prospective study was conducted at the Microbiology Laboratory of the JPNA Trauma Centre of the All India Institute of Medical Sciences, New Delhi, India. We tested and analysed a total of 910 sequential, non-duplicate GNB isolates collected from various clinical specimens of patients admitted to our centre from January to August 2019. The isolates were identified using the Vitek®2 automated system (Vitek®2 GN-card). The intrinsically colistin-resistant isolates—Morganella morganii, Proteus mirabilis, Proteus penneri, Proteus vulgaris, Providencia rettgeri, Providencia stuartii, Serratia marcescens and Burkholderia cepacia—were excluded from the study. Colistin MICs (range: 0.50–16 mg/L) were determined using the commercial Vitek®2 AST system [Vitek®2 AST-N280 (for lactose fermenters) and AST-N281 (for non-lactose fermenters)] (bioMérieux, Marcy-l’Étoile, France) as per the manufacturer’s instructions.
The colistin MICs were also determined using the reference BMD method (MIC range: 0.25–16 mg/L) colistin sulfate salt (Sigma, St. Louis, MO, USA) dissolved in CAMHB (BD, Franklin Lakes, NJ, USA), and according to CLSI recommendations in untreated 96-well polystyrene microplates (Greiner, Frickenhausen, Germany).15
EUCAST MIC breakpoints of > 2 mg/L for resistance and ≤ 2 mg/L for susceptibility were used for Enterobacterales, Pseudomonas aeruginosa and Acinetobacter baumannii.2 The same breakpoints were used for all the other organisms (including Moraxella group, Bordetella hinzii, Comamonas testosteroni, Myroides spp., Sphingomonas paucimobilis) tested in this study, since currently no CLSI and EUCAST MIC-interpretation criteria for defining susceptibility are available.
The MICs for mcr-1-positive Escherichia coli NCTC 13846 (range: 2–8 mg/L), E. coli ATCC® 25922 (range: 0.25–2 mg/L) and P. aeruginosa ATCC® 27853 (range: 0.5–4 mg/L) and a clinical isolate of P. mirabilis (colistin MIC >16 mg/L) were used as quality-control strains for each run of the colistin MIC tests. Each isolate was tested in duplicate, and for all discordant results repeat testing was performed. All resistant strains were tested twice by both the methods. Sensitivity, specificity and positive and negative predictive values (PPV and NPV) were determined, since the prevalence of colistin resistance impacts the PPV and NPV of tests. To assess performance of Vitek®2 as compared with BMD for MIC testing of colistin, essential agreement (EA) and categorical agreement (CA) were evaluated. EA was defined as the percentage of Vitek®2 MIC results that were within ± 1 log2 dilution of reference BMD MIC results. CA was the percentage of Vitek®2 interpretive results (susceptible or resistant) that agreed with reference BMD interpretive results.
Categorical disagreements were classified as VMEs and major errors (MEs). A VME for Vitek®2 was defined as a colistin-susceptible isolate determined using BMD interpreted as a colistin-resistant isolate (false susceptibility result). VME rates were calculated using the number of isolates resistant by BMD as the denominator. An ME for Vitek®2 was defined as a colistin-resistant isolate determined using BMD interpreted as a colistin-susceptible isolate (false resistant result). ME rates were calculated using the number of isolates susceptible by BMD as the denominator.
Acceptable agreement for Vitek®2 compared with BMD was defined as EA ≥ 90%, CA ≥ 90%, VME ≤ 1.5% and ME ≤ 3% as described by CLSI.16 Spearman’s coefficient was calculated to determine the concordance of Vitek®2 MICs with those of BMD. A P value <0.05 was considered statistically significant. Since the clinical isolates included in the study were sent for routine AST testing to the laboratory, no ethical clearance was obtained for this study.
Results
The clinical isolates (n = 910) belonging to order Enterobacterales included Klebsiella pneumoniae (n = 245), E. coli (n = 158), Enterobacter cloacae (n = 42), Citrobacter freundii (n = 8), Salmonella Typhi (n = 7), Raoultella planticola (n = 4), Cronobacter spp. (n = 2) and Pantoea spp. (n = 2). The non-Enterobacterales isolates included A. baumannii (n = 273), P. aeruginosa (n = 139), Aeromonas hydrophila (n = 10), Stenotrophomonas maltophilia (n = 9), S. paucimobilis (n = 5), B. hinzii (n = 2), Moraxella group (n = 2), C. testosteroni (n = 1) and Myroides spp. (n = 1).
Pus and wound swab (n = 322, 35%) were the most common source of isolates, followed by endotracheal aspirates (n = 119, 13%), blood (n = 183, 20%), bronchoalveolar lavage fluid (n = 89, 10%), sterile body fluids (n = 60, 7%), tissue (n = 52, 6%), urine (n = 49, 5%), pleural fluid (n = 27, 3%), and miscellaneous samples (n = 9, 1%).
The sensitivity of Vitek®2 compared with BMD ranged from 12.5% to 72%, and specificity was ≥ 94% (Table 1). The PPV ranged from 83% to 100% while NPV ranged from 10% to 95.5%. Despite 100% specificity of Vitek®2 for isolates for which no colistin breakpoints are available (A. hydrophila, S. maltophilia and other non-fermenters), the sensitivity was very low (0%–33%).
Table 1.
Sensitivity, specificity, and predictive values of detecting colistin resistance using Vitek®2 with BMD as reference method
| Organism (n) | Vitek®2 | BMD |
Sensitivity (false S) |
Specificity (false R) |
PPV | NPV | |
|---|---|---|---|---|---|---|---|
| R | S | ||||||
| Enterobacterales (n = 468) | |||||||
| K. pneumoniae | R | 73 | 5 | 72% | 96.5% | 94% | 83% |
| S | 29 | 138 | |||||
| E. coli | R | 1 | 0 | 12.5% | 100% | 100% | 95.5% |
| S | 7 | 150 | |||||
| E. cloacae | R | 6 | 0 | 35% | 100 % | 100% | 69% |
| S | 11 | 25 | |||||
| othersa | R | 0 | 1 | 0% | 94% | 0 | 77% |
| S | 5 | 17 | |||||
| Non-Enterobacterales (n = 442) | |||||||
| A. baumannii | R | 15 | 3 | 47% | 99% | 83% | 93% |
| S | 17 | 238 | |||||
| P. aeruginosa | R | 5 | 0 | 42% | 100% | 100% | 95% |
| S | 7 | 127 | |||||
| A. hydrophila | R | 0 | 0 | 0% | 100% | NA | 10% |
| S | 9 | 1 | |||||
| S. maltophilia | R | 0 | 0 | 0% | 100% | NA | 10% |
| S | 8 | 1 | |||||
| othersb | R | 1 | 0 | 33% | 100% | 100% | 80% |
| S | 2 | 8 | |||||
R, resistant; S, susceptible; NA, not applicable.
Citrobacter spp.(n = 8), Cronobacter spp. (n = 2), Pantoea spp. (n = 2), R. planticola (n = 4), Salmonella Typhi (n = 7).
B. hinzii (n = 2), C. testosteroni (n = 1), Moraxella group (n = 2), Myroides spp. (n = 1), S. paucimobilis (n = 5).
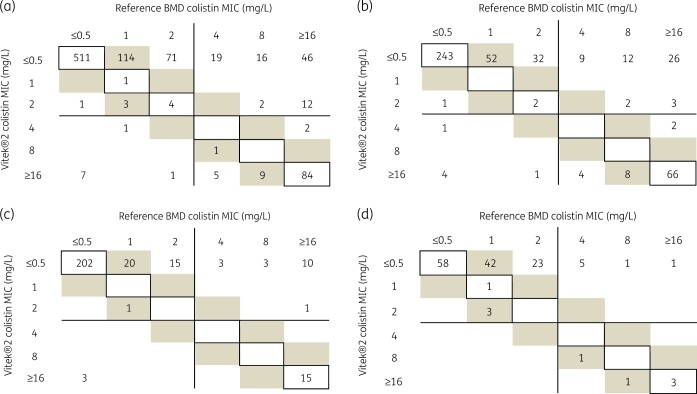
The correlation with reference MICs was poor for Vitek®2 and a 45-degree correlation could not be obtained. Vitek®2 tended to underestimate MICs for resistant isolates (Figure 1).
Figure 1.
Correlation between Vitek®2 and reference BMD for (a) all Gram-negative bacterial isolates studied (n = 910), (b) Enterobacterales (n = 468), (c) A. baumannii (n = 273) and (d) P. aeruginosa (n = 139). MICs within EA (within ± 1 dilution of reference MICs) are shaded and MICs identical to reference MICs are within boxes. EUCAST breakpoints (resistant > 2 mg/L) are shown as lines.
The performance of Vitek®2 and reference BMD to determine colistin MICs is presented in Table 2. Generally, for all the isolates tested, the BMD MIC values were higher than those obtained by Vitek®2. Figure 1 shows the correlation between both the tests for all Gram-negative bacterial isolates studied (n = 910), Enterobacterales (n = 468), A. baumannii and P. aeruginosa (n = 139).
Table 2.
Performance characteristics of the reference BMD method and Vitek®2
| Organism (n) | Method | No. (%) of isolates exhibiting |
Spearman’s coefficient | |||||
|---|---|---|---|---|---|---|---|---|
| R | S | EA | CA | VME | ME | |||
| Enterobacterales (n = 468) | ||||||||
| K. pneumoniae (n = 245) | BMD | 102 | 143 | 187 (76) | 211 (86) | 29 (28) | 5 (3.5) | ρ = 0.69596 (P = 0.000)* |
| Vitek®2 | 78 | 167 | ||||||
| E. coli (n = 158) | BMD | 8 | 150 | 141 (89) | 151 (96) | 7 (86) | 0 | ρ = 0.15382 (P = 0.054) |
| Vitek®2 | 1 | 157 | ||||||
| E. cloacae (n = 42) | BMD | 17 | 25 | 29 (69) | 31 (74) | 11 (65) | 0 | ρ = 0.48858 (P = 0.001)* |
| Vitek®2 | 6 | 36 | ||||||
| othersa (n = 23) | BMD | 5 | 18 | 14 (61) | 9 (39) | 5 (100) | 1 (6) | ρ = −0.01672 (P = 0.100) |
| Vitek®2 | 1 | 22 | ||||||
| Non-Enterobacterales (n = 442) | ||||||||
| A. baumannii (n = 273) | BMD | 32 | 241 | 239 (88) | 253 (93) | 17 (53) | 3 (1) | ρ = 0.36258 (P = 0.000)* |
| Vitek®2 | 18 | 255 | ||||||
| P. aeruginosa (n = 139) | BMD | 12 | 127 | 109 (78) | 132 (95) | 7 (58) | 0 | ρ = 0.28633 (P = 0.007)* |
| Vitek®2 | 5 | 134 | ||||||
| A. hydrophila (n = 10) | BMD | 9 | 1 | 1 (10) | 1 (11) | 9 (100) | 0 | ρ = 0.66667 (P = 0.035)* |
| Vitek®2 | 0 | 10 | ||||||
| S. maltophilia (n = 9) | BMD | 8 | 1 | 0 | 1 (11) | 8 (100) | 0 | ρ = 0.7333 (P = 0.023)* |
| Vitek®2 | 0 | 9 | ||||||
| othersb (n = 11) | BMD | 3 | 8 | 8 (73) | 9 (82) | 2 (67) | 0 | ρ = 0.45707 (P = 0.158) |
| Vitek®2 | 1 | 10 | ||||||
Significant differences are highlighted in bold (*P < 0.05).
R, resistant; S, susceptible.
Citrobacter spp.(n = 8), Cronobacter spp. (n = 2), Pantoea spp. (n = 2), R. planticola (n = 4), Salmonella Typhi (n = 7).
B. hinzii (n = 2), C. testosteroni (n = 1), Moraxella group (n = 2), Myroides spp. (n = 1), S. paucimobilis (n = 5).
Overall, 714 (78.5%) and 800 (88%) of isolates were colistin susceptible by BMD and Vitek®2, respectively. The EA was found to be highest in E. coli (89%) followed by A. baumannii (88%). This could be because there were only 8/158 resistant isolates of E. coli. Poor CA (≤ 90%) was found in all the isolates except for E. coli (among Enterobacterales), A. baumannii and P. aeruginosa (non-Enterobacterales). Very high VME (28%–100%) and up to 6% ME were observed among all the tested isolates. Except for K. pneumoniae, A. hydrophila and S. maltophilia, strong MIC correlation (Spearman’s ρ > 0.8) was not seen for any of the isolates.
Discussion
As the burden of acquired resistance to colistin is rising, accurate detection and reporting is essential to roll out a diagnostic stewardship programme, especially for counties like India.17
The semi-automated Vitek®2 has been reported as a reliable colistin testing method.6,8,9,18 However, we found that the automated Vitek®2 method failed to detect the resistance in 87.5% (n = 7) E. coli, 65% (n = 11) E. cloacae, 58% (n = 7) P. aeruginosa, 53% (n = 17) A. baumannii and 28% (n = 29) K. pneumoniae colistin-resistant isolates. The acceptable EA ≥ 90% was observed in none of the isolates. Although an acceptable CA (> 90%) among E. coli, A. baumannii and P. aeruginosa was observed, VME rates of 86%, 53% and 58% were also observed, respectively. Despite the strong positive MIC correlation among K. pneumoniae, A. hydrophila, and S. maltophilia, the VME rates were found to be well in excess of the 1.5% rate recommended by the CLSI.
In a recent study, colistin MICs determined by Vitek®2 were reported to be unreliable, especially for E. cloacae and A. baumannii complex isolates.19 Another study showed that semi-automated systems including Vitek®2 performed poorly, with 31 VMEs.20 Our study highlights that Vitek®2 is not reliable for colistin susceptibility testing, especially for Enterobacterales, A. baumannii and P. aeruginosa, for which CLSI and EUCAST MIC interpretation criteria for defining susceptibility have been published.2
However, the lower numbers of isolates (and resistant isolates) for some species is a limitation of this study. This often happens when clinical isolates routinely tested are studied, rather than picking specific resistant isolates to study.
It is noteworthy to mention that misinterpreting colistin susceptibility test results may lead to inexplicable treatment failures and even mortality, as isolates identified as susceptible may rather resist antibiotic therapy owing to colistin heteroresistance.17,21–23
Although performing AST methods such as BMD for clinical testing is technically demanding, laboratories need to train their staff to perform BMD and overcome common difficulties including making initial dilutions, multiple skipped wells, contamination, or other quality control problems, none of which are involved in automated systems. Further detection of mcr genes amongst these bacteria would also provide molecular epidemiological data.
Most studies on colistin resistance have included one or a few species of Enterobacterales. This is one of few studies that report colistin resistance amongst a large collection of GNB, for many of which a breakpoint is also not available.
Acknowledgements
We thank Ms Manisha Kaim and Ms Manisha Chaurasiya for data collection and technical assistance.
Funding
This study was carried out as part of the routine work in the microbiology division at the JPNA Trauma Centre, All India Institute of Medical Sciences, New Delhi.
Transparency declarations
None to declare.
References
- 1. Liu Y-Y, Wang Y, Walsh TR et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 2. EUCAST. Recommendations for MIC Determination of Colistin (Polymyxin E) as Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. 2016. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
- 3. Matuschek E, Åhman J, Kahlmeter G et al. Antimicrobial susceptibility testing of Kingella kingae with broth microdilution and disk diffusion using EUCAST recommended media. Clin Microbiol Infect 2018; 24: 396–401. [DOI] [PubMed] [Google Scholar]
- 4. Hindler JA, Humphries RM. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 2013; 51: 1678–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maalej SM, Meziou MR, Rhimi FM et al. Comparison of disc diffusion, Etest and agar dilution for susceptibility testing of colistin against Enterobacteriaceae. Lett Appl Microbiol 2011; 53: 546–51. [DOI] [PubMed] [Google Scholar]
- 6. Lee SY, Shin JH, Lee K et al. Comparison of the Vitek 2, MicroScan, and Etest methods with the agar dilution method in assessing colistin susceptibility of bloodstream isolates of Acinetobacter species from a Korean University Hospital. J Clin Microbiol 2013; 51: 1924–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moskowitz SM, Garber E, Chen Y et al. Colistin susceptibility testing: evaluation of reliability for cystic fibrosis isolates of Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J Antimicrob Chemother 2010; 65: 1416–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dafopoulou K, Zarkotou O, Dimitroulia E et al. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother 2015; 59: 4625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lo-Ten-Foe JR, de Smet AMGA, Diederen BMW et al. Comparative evaluation of the VITEK 2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob Agents Chemother 2007; 51: 3726–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chew KL, La M-V, Lin RTP et al. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive Enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol 2017; 55: 2609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mehrad B, Clark NM, Zhanel GG et al. Antimicrobial resistance in hospital-acquired Gram-negative bacterial infections. Chest 2015; 147: 1413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khurana S, Mathur P, Batra P et al. Does infection with multidrug resistant bacteria necessarily lead to adverse patient outcome?: A prospective study. J Patient Saf Infect Control 2015; 3: 56. [Google Scholar]
- 13. Khurana S, Mathur P, Kapil A et al. Molecular epidemiology of β-lactamase producing nosocomial Gram-negative pathogens from North and South Indian hospitals. J Med Microbiol 2017; 66: 999–1004. [DOI] [PubMed] [Google Scholar]
- 14. Despotovic A, Milosevic B, Milosevic I et al. Hospital-acquired infections in the adult intensive care unit—Epidemiology, antimicrobial resistance patterns, and risk factors for acquisition and mortality. Am J Infect Control 2020; 48: 1211–15. [DOI] [PubMed] [Google Scholar]
- 15. CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Thirtieth Edition: M100. 2020. [Google Scholar]
- 16. CLSI. Verification of Commercial Microbial Identification and Antimicrobial Susceptibility Testing Systems—First Edition: M52. 2015. [Google Scholar]
- 17. Mathur P, Khurana S, De Man TJB et al. Multiple importations and transmission of colistin-resistant Klebsiella pneumoniae in a hospital in northern India. Infect Control Hosp Epidemiol 2019; 40: 1387–93. [DOI] [PubMed] [Google Scholar]
- 18. La M-V, Lee B, Hong BZM et al. Prevalence and antibiotic susceptibility of colistin-resistance gene (mcr-1) positive Enterobacteriaceae in stool specimens of patients attending a tertiary care hospital in Singapore. Int J Infect Dis 2019; 85: 124–6. [DOI] [PubMed] [Google Scholar]
- 19. Lai CC,, Chen YS, Lee NY et al. Susceptibility rates of clinically important bacteria collected from intensive care units against colistin, carbapenems, and other comparative agents: Results from surveillance of multicenter antimicrobial resistance in Taiwan (SMART). Infect Drug Resist 2019; 12: 627–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pfennigwerth N, Kaminski A, Korte-Berwanger M et al. Evaluation of six commercial products for colistin susceptibility testing in Enterobacterales. Clin Microbiol Infect 2019; 25: 1385–9. [DOI] [PubMed] [Google Scholar]
- 21. Rojas LJ, Salim M, Cober E et al. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis 2017; 64: 711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Machuca I, Gutiérrez-Gutiérrez B, Gracia-Ahufinger I et al. Mortality associated with bacteremia due to colistin-resistant Klebsiella pneumoniae with high-level meropenem resistance: importance of combination therapy without colistin and carbapenems. Antimicrob Agents Chemother 2017; 61: e00406-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Band VI, Satola SW, Burd EM et al. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 2018; 9: e02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]