Abstract
Background
NDM-producing Enterobacteriaceae are a major clinical concern worldwide. We characterized NDM-positive pathogens isolated from patients and assessed the dissemination patterns of the blaNDM genes in a hospital setting.
Methods
Eleven NDM-positive Enterobacteriaceae (three Enterobacter hormaechei, six Klebsiella pneumoniae and two Escherichia coli) were isolated from nine patients over a 1 year period. Antimicrobial susceptibility was assessed by MICs. A combination of short- and long-read WGS was used for genome analysis. Clinical treatment history of patients was linked with genetic features of individual isolates to investigate the dissemination patterns of the blaNDM genes and NDM-positive strains.
Results
bla NDM in clonal K. pneumoniae were transmitted between two patients. In other instances, an identical IncC plasmid encoding NDM-1 was transmitted between E. coli and K. pneumoniae isolated from the same patient, and an IncX3 plasmid, carrying blaNDM-1 or blaNDM-5, was harboured in non-clonal E. hormaechei. Varying patterns of IS elements were identified as a critical transmission mechanism in association with blaNDM genes.
Conclusions
Multiple transmission patterns were identified in hospitalized patients, including dissemination of clonal bacterial strains carrying resistance genes and horizontal transfer of resistance genes among divergent bacterial strains. Controlling spread of NDM is complex: while attention to standard infection control practices is critically important, this needs to be matched by aggressive efforts to limit unnecessary antimicrobial use, to minimize the selection for and risk of transfer of ‘high mobility’ resistance genes among Enterobacteriaceae.
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) were identified as an ‘urgent threat’ in the 2019 Antibiotic Resistance Threats Report by CDC.1 In 2017, CDC estimated that there were 13 100 cases, 1100 deaths and $130 million in healthcare costs caused by CRE.1 CRE have two subsets: carbapenemase-producing Enterobacteriaceae (CPE) and non-carbapenemase-producing Enterobacteriaceae. As CPE spread quickly in hospital settings, they are a rising clinical concern globally.1–4 Carbapenemases produced by Enterobacteriaceae are divided into three classes of β-lactamase (class A, B and D), based on their molecular structure.5,6 The five common carbapenemases include Klebsiella pneumoniae carbapenemase (KPC) belonging to class A, New Delhi metallo-β-lactamase (NDM), Verona integron-encoded metallo-β-lactamase (VIM) and imipenemase (IMP) belonging to class B, and oxacillinase-48 (OXA-48) belonging to class D.6
Due to its unprecedented speed of spread, NDM has drawn global attention in the last decades.7 NDM-1 was first identified in a K. pneumoniae strain isolated from a patient from New Delhi, India, who was hospitalized in Sweden in 2008.8 By 2019, a total of 24 NDM variants (NDM-1 to NDM-24) were identified in various bacterial species globally.9 Among the NDM-positive Enterobacteriaceae, K. pneumoniae account for more than half of the total number, followed by Escherichia coli and Enterobacter cloacae complex (which includes Enterobacter hormaechei isolated in this study).9 Given the ability to hydrolyse most β-lactams (with the exception of monobactams), infections with NDM-producing strains have very limited therapeutic options, with a high associated mortality rate among infected patients.10,11 However, limited data are available on the treatment of infections caused by NDM-bearing microorganisms using existing antibiotic options. In addition, much of the newer antimicrobials and experimental pipeline is unable to inhibit this group of carbapenemases.
The NDM-encoding genes, blaNDM-1 to blaNDM-24, have been reported in conjugative plasmids belonging to several incompatibility groups (Inc).9 In addition to plasmids, other mobile genetic elements (MGEs), such as transposons, IS elements, and integrons, have also been associated with NDM mobilization.12,13 However, lower resolution techniques such as PCR-based targeted partial genotyping or short-read WGS based unclosed genome often fail to provide a comprehensive picture of mechanisms underlying NDM transmission.14,15 Here, we focus on routes of NDM transmission over time in a major, tertiary-care referral hospital. By using a combination of short- and long-read WGS we provide insights into transmission mechanisms, highlighting areas of concern in trying to control nosocomial spread of this potentially dangerous gene complex.
Methods
MICs and bacterial isolation
Presence of CRE and associated MICs were identified using the Vitek®2 Microbial Identification System (bioMérieux) in the University of Florida (UF) Health Shands Hospital Clinical Microbiology Laboratory. MICs were subsequently confirmed via Etest (bioMérieux). Organisms were further analysed via the Xpert® Carba-R (Cepheid) to detect the presence of carbapenemase genes.16 As part of standard daily hospital infection control activities, all Gram-negative bacterial isolates found by the hospital microbiology laboratory to have resistance to carbapenem were identified through an automated infection control/laboratory surveillance system, and basic data on patient location and movement during hospitalization were collected. This resulted in identification of 11 NDM-carrying strains across a 1 year time period; sequence data were obtained for all 11 strains, as described below. A subset of patients infected with NDM strains were enrolled in an institutional review board-approved study at the University of Florida that permitted collection of additional clinical and epidemiological data from patients infected with antimicrobial-resistant bacterial strains. Informed consent was not required for the use of de-identified samples.
WGS and assembly
For Illumina WGS and assembly, DNA of the 11 strains identified as carrying NDM was extracted using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA) following the protocol for Gram-negative bacteria. DNA libraries were constructed using the Nextera XT sample preparation kit (Illumina, San Diego, CA, USA) following the protocol from manufacturer. The sequencing steps and assembly settings were performed as described previously.17
For PacBio sequencing, DNA was extracted from six selected isolates using the Puregene Yeast/Bact. Kit B (Qiagen) and was cleaned up using the DNeasy PowerClean Cleanup Kit (Qiagen), following the protocols for Gram-negative bacteria. Then, the DNA samples were sent to the Interdisciplinary Center for Biotechnology Research (ICBR) of University of Florida for PacBio Sequel sequencing. The raw reads generated from the PacBio Sequel I sequencing system were demultiplexed with the PacBio SMRT Analysis (7.0.1.66974). The sub-reads for each sample were assembled by the HGAP4 (hierarchical genome assembly process) and Canu v2.0 with optimized parameters to generate de novo genome chromosomes and plasmids.18 Both assemblers filtered the sequencing data to remove SMRTbell adapter sequences and recover high-quality genomic content. The initial assemblies were further checked and validated with the samtools, FASTX-toolkit and R-based scripts developed in house at ICBR. The validated assemblies were imported into SMRT Link for subsequent polishing with the Resequencing Analysis to attain a higher base quality. The finalized genomes and plasmids were circularized using the Circlator tool.19 Assembled genomes were deposited in NCBI (Table S1, available as Supplementary data at JAC-AMR Online).
Phylogenetic tree analysis and antibiotic resistance gene (ARG) identification
Phylogenetic trees were generated using Illumina sequencing results for each species separately, using Parsnp (https://harvest.readthedocs.io/en/latest/) with default settings, based on the core-genome SNPs. The reference strains for each species were chosen by Parsnp randomly, and KCJ3K13, KCJ3K293 and KCJ3K291 were selected as the reference strains for E. hormaechei, K. pneumoniae and E. coli, respectively. SNPs in each phylogenetic clade were calculated using NCBI Pathogen Detection (https://www.ncbi.nlm.nih.gov/pathogens/).
To identify the ARGs, the genomic sequences were compared with the reference sequences in the Comprehensive Antibiotic Resistance Database (CARD 3.0.8) using the Resistance Gene Identifier (RGI 5.1.0).20 Bacterial DNA sequences were submitted to the web portal of CARD with the selected parameters of ‘Perfect and Strict hits only’, ‘Include nudge’ and ‘Low quality/coverage’.
Plasmid typing and NDM genetic environment
Plasmids were assigned to incompatibility groups using PlasmidFinder 2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/). The completed circular genome maps of chromosomal DNA and plasmids were generated using BLAST Ring Image Generator (BRIG).21 The annotations for the genome mapping and generic environments of blaNDM flanking genes were acquired from Prokka and the NCBI Prokaryotic Genome Annotation Pipeline (PGAP).22
Bacterial conjugation
The conjugation experiment was performed between K. pneumoniae KCJ3K292 and E. coli XL1-Blue (tetracycline-resistant strain). Overnight cultures of K. pneumoniae KCJ3K292, harbouring NDM-encoding-plasmid pKC148K (donor) and E. coli XL1-Blue (recipient) were combined (3:1 ratio). The combined bacteria were conjugated for 20 h on tryptic soy agar (TSA) at 25 °C and E. coli transconjugants were selected on TSA plates containing tetracycline (10 mg/L) and meropenem (16 mg/L). Total recipients used for conjugation were counted by selecting on TSA containing only tetracycline (10 mg/L). Conjugation frequency was calculated by dividing the number of transconjugants by the number of recipients used.
Results
The 11 NDM-producing Enterobacteriaceae strains (three E. hormaechei, six K. pneumoniae and two E. coli), were isolated over a 1 year period from nine patients from UF Health Shands hospital. All nine patients were discharged alive after the admission during which the NDM-bearing organism was identified. The median age was 52 years (range 2–67), five were male (56%) and four were female (44%). Four patients (44%) were admitted from a congregate setting or another hospital. Two patients were admitted in septic shock, and four were admitted with necrotizing fasciitis. Patients were relocated between North Tower (NT), South Tower (ST), and East Tower (ET) of the hospital for treatment. Length of hospitalization ranged from 11 days to more than 9 months. The average number of days that elapsed between admission and isolation of the NDM-bearing organism was 14 (range 0–38). Five of the nine (56%) had been hospitalized in the year prior to isolation of the NDM-bearing organism, and of these the number of prior admissions ranged from one to five. Of the 11 isolates, 6 were obtained from wound cultures from four patients, 3 isolates were from urine, 1 was from ascitic fluid and 1 was from blood. All patients were initially treated with cefepime, and therapy was changed to agents active against the NDM-bearing organism(s) once they were identified in eight of the nine patients (one result was obtained after the patient was discharged to the sending facility) (Table S2).
To investigate the antibiotic resistance profile, the MIC was obtained using standard Vitek®2 susceptibility cards (Table 1). Twenty-three antibiotics and combinations, belonging to 12 antibiotic classes, including aminoglycoside, penicillin, penicillin/β-lactamase inhibitor, cephalosporin, fluoroquinolone, carbapenem, cephalosporin/β-lactamase inhibitor, sulphonamide, glycylcycline, polymyxin, tetracycline and nitrofurantoin (urine isolates only) were tested (Table 1). All strains were MDR in which K. pneumoniae KCJ3K307 was resistant to 14 antibiotics, while K. pneumoniae KCJ3K65 was resistant to the least number of antibiotics (n = 12). All isolates were resistant to piperacillin/tazobactam, cephalosporin class drugs and meropenem. All isolates were susceptible to amikacin.
Table 1.
MIC (mg/L) of NDM strains isolated from hospitalized patients
| Strains |
E. hormaechei
|
K. pneumoniae
|
E. coli
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KCJ3K13 | KCJ3K19 | KCJ3K22 | KCJ3K53 | KCJ3K65 | KCJ3K270 | KCJ3K292 | KCJ3K293 | KCJ3K307 | KCJ3K291 | KCJ3K426 | |
| Patients | A | B | G | E | F | H | C | C | D | C | I |
| Specimen source | wound | wound | urine | blood | urine | urine | wound | wound | wound | wound | ascitic fluid |
| Isolated date | 3 Apr 2019 | 6 Mar 2019 | 2 Nov 2018 | 4 Jul 2018 | 8 Aug 2018 | 15 Apr 2019 | 6 May 2019 | 6 May 2019 | 4 Jun 2019 | 6 May 2019 | 13 Jul 2019 |
| Antimicrobial | |||||||||||
| amikacin | ≤2 (S) | 4 (S) | ≤2 (S) | ≤2 (S) | ≤2 (S) | ≤2 (S) | ≤2 (S) | ≤2 (S) | ≤2 (S) | 4 (S) | ≤2 (S) |
| gentamicin | ≤1 (S) | 8 (I) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) |
| tobramycin | ≤1 (S) | ≥16 (R) | 8 (I) | 8 (I) | 8 (I) | ≥16 (R) | 8 (I) | 8 (I) | ≥16 (R) | ≥16 (R) | 8 (I) |
| ampicillin | NA | NA | NA | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) |
| ampicillin/ sulbactam | NA | NA | NA | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) | ≥32 (R) |
| piperacillin/ tazobactam | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) | ≥128 (R) |
| cefazolin | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) |
| cefepime | 16 (R) | ≥64 (R) | ≥64 (R) | 8 (SDD) | 2 (SDD) | 8 (SDD) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | 16 (R) |
| cefoxitin | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) |
| ceftazidime | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) |
| ceftriaxone | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) | ≥64 (R) |
| ciprofloxacin | 1 (R) | 2 (R) | 1 (R) | ≤0.25 (S) | 1 (R) | 1 (R) | ≤0.25 (S) | ≤0.25 (S) | ≤0.25 (S) | ≤0.25 (S) | ≤0.25 (S) |
| levofloxacin | 4 (R) | 4 (R) | 1 (R) | ≤0.12 (S) | 1 (R) | 1 (R) | ≤0.12 (S) | ≤0.12 (S) | ≤0.12 (S) | ≤0.12 (S) | ≤0.12 (S) |
| meropenem | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) | ≥16 (R) |
| aztreonam | 0.19 (S) | NA | NA | 0.064 (S) | NA | NA | 0.094(S) | 0.125 (S) | 0.19 (S) | 0.016 (S) | NA |
| trimethoprim/ sulphonamide | ≤20 (S) | ≥320 (R) | ≥320 (R) | ≥320 (R) | ≥320 (R) | ≥320 (R) | ≥320 (R) | ≥320 (R) | ≥320 (R) | ≥320 (R) | ≥320 (R) |
| tigecycline | ≥8 (R) | NA | NA | NA | NA | 1 (S) | ≤0.5 (S) | ≤0.5 (S) | NA | ≤0.5 (S) | NA |
| ceftazidime/ avibactam | >256 (R) | >256 (R) | NA | NA | NA | >256 (R) | >256 (R) | >256 (R) | ≥256 (R) | >256 (R) | NA |
| ceftolozane/ tazobactam | >256 (R) | >256 (R) | NA | NA | NA | NA | NA | NA | ≥256 (R) | NA | NA |
| colistin | 0.38 | NA | 0.125 | 0.38 | 0.38 | 0.75 | 0.5 | 0.5 | 0.5 | 0.125 | 0.38 |
| polymyxin B | 0.5 | NA | 0.38 | 0.38 | 0.5 | 0.5 | 0.5 | 0.75 | 1.5 | 0.25 | NA |
| minocycline | NA | NA | NA | 4 (S) | NA | NA | NA | NA | NA | NA | NA |
| nitrofurantoin | NA | NA | 64 (I) | NA | ≤16 (S) | 64 (I) | NA | NA | NA | NA | NA |
NA, not applicable; S, susceptible; SDD, susceptible dose-dependent; I, intermediate; R, resistant.
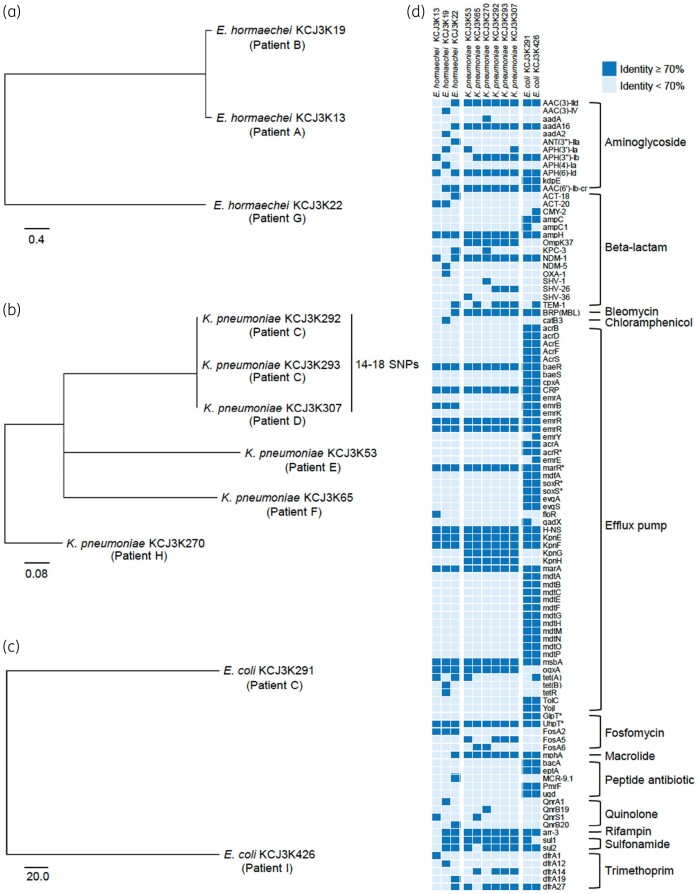
To investigate the genetic relatedness of the NDM isolates, WGS was conducted with the Illumina platform and subjected to a phylogenetic analysis. The phylogenetic trees were constructed for E. hormaechei, K. pneumoniae and E. coli (Figure 1a–c). Most isolates were genetically distinct. However, three Klebsiella strains (K. pneumoniae KCJ3K292, KCJ3K293, and KCJ3K307), isolated from patients C and D, were clonal variants with a difference of less than 18 SNPs in the genome. These data are consistent with transmission of this particular NDM-bearing bacterial strain among hospitalized patients.
Figure 1.
Genetic relatedness and ARG profile of the NDM isolates. The maximum-likelihood phylogenetic trees were constructed based on core-genome SNPs of Illumina sequencing data. Diverse E. hormaechei (a), K. pneumoniae (b) and E. coli (c) were isolated from patients. Clonal variants were isolated from different patients. (d) ARG profile. The ARGs of 11 strains were identified by comparing their genomic DNA sequences with CARD. The identified ARGs with more than 70% identity are shown in dark blue, while the ones with less than 70% identity are shown in light blue. The asterisks indicate the genes with mutations conferring antimicrobial resistance.
With the CARD analysis, we identified 102 ARGs that are related to resistance to 12 classes of antibiotic. E. coli carried the highest number of ARGs (n = 63 and 65), while E. hormaechei carried 23 to 34 ARGs and K. pneumoniae carried 30 to 33 ARGs. All isolates carried genes conferring resistance to aminoglycoside, β-lactam, fosfomycin, quinolone and trimethoprim, as well as genes encoding efflux pumps that confer MDR. Most of the identified ARGs were functional based on MIC (Table 1). Additionally, we identified genes conferring resistance to bleomycin, chloramphenicol, macrolide, peptide antibiotic, rifampicin and sulphonamide. Interestingly, mcr-9.1, the novel colistin resistance gene, was identified in E. hormaechei KCJ3K22, eliminating a key ‘last resort’ antimicrobial for these highly resistant strains, but this strain was susceptible to colistin (Table 1), likely due to low expression levels. Fortunately, this particular strain retained some activity to fluoroquinolones, which were used in combination in the treatment regimen for this patient.
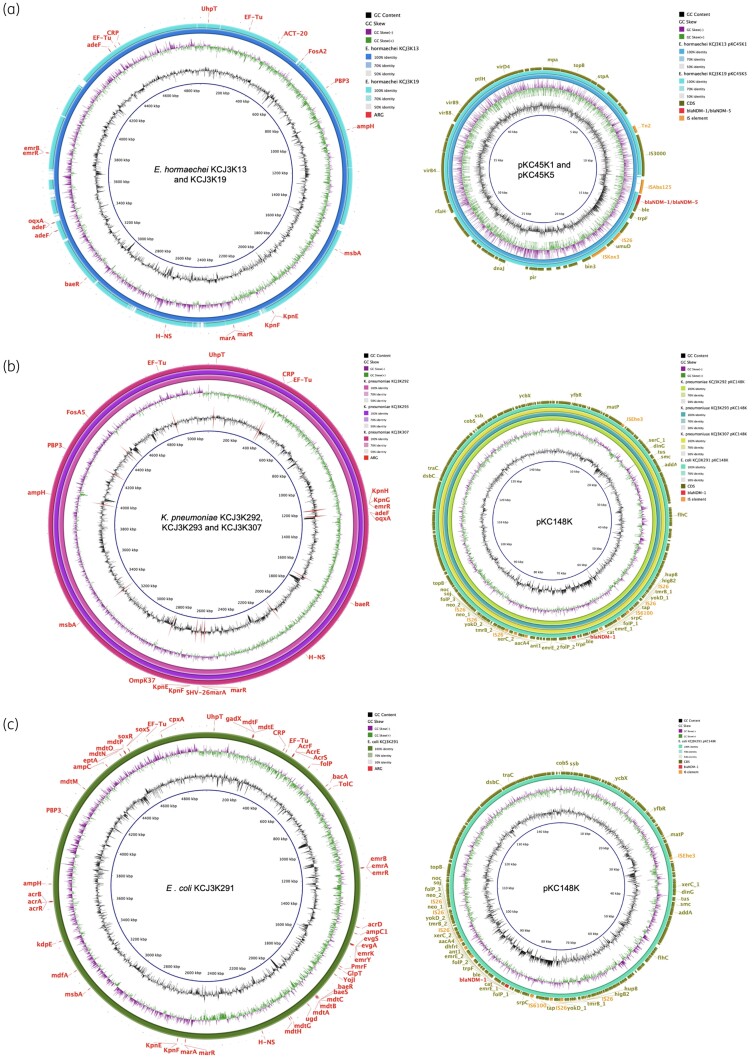
To further dissect the dissemination mechanisms of NDM-bearing strains among hospitalized patients, we used the PacBio sequencing platform to close the genome for six isolates (E. hormaechei KCJ3K13 from patient A, E. hormaechei KCJ3K19 from patient B, K. pneumoniae KCJ3K292 from patient C, K. pneumoniae KCJ3K293 from patient C, K. pneumoniae KCJ3K307 from patient D, and E. coli KCJ3K291 from patient C); strains were selected based on close genetic distance to further characterize genetic variations and transmission patterns of NDM genes and NDM-bearing strains between patients. The six strains carried multiple plasmid types, including IncF family, IncR and IncX3 plasmids with a narrow host range of Enterobacteriaceae as well as IncHI2 and IncC plasmids with a broad host range (Table S3). The IncX3 plasmid carrying blaNDM-1 and blaNDM-5 was found in E. hormaechei strains, and the IncC plasmids carrying blaNDM-1 were identified in K. pneumoniae and E. coli strains (Table S3).
The chromosomes of E. hormaechei KCJ3K13 and KCJ3K19 had a similar genetic backbone, albeit with multiple insertion-deletions (indels) which clearly differentiated the two genomes. However, these two chromosomes carried the same 22 ARGs, such as β-lactamase-encoding genes ACT-20 and ampH (Figure 2a). At the plasmid level, pKC45K1 carrying blaNDM-1 was identified in E. hormaechei KCJ3K13, while pKC45K5 carrying blaNDM-5 was identified in E. hormaechei KCJ3K19. These plasmids were identical except for a two amino acid difference between blaNDM-1 and blaNDM-5 and two IS elements, ISAba125 and IS5, which were located upstream of the blaNDM genes (Figure 2a). This non-homologous IS region indicates that different IS-mediated transposition events occurred at the same loci of a plasmid carried by two different E. hormaechei strains. These two plasmids encode the VirB type IV secretion system that may transfer plasmids by conjugation.
Figure 2.
Chromosomal and plasmid genome maps of the NDM pathogens. Circular genome maps of chromosomal DNA and plasmids were generated using PacBio sequencing data. (a) E. hormaechei. (b) K. pneumoniae. (c) E. coli. The IncX3 plasmids, pKC45K1 and pKC45K5, were identical except for the blaNDM genes and IS elements. The IncC plasmid, pKC148K, was harboured by three K. pneumoniae strains and one E. coli strain.
The clonal variant K. pneumoniae strains isolated from two patients (KCJ3K292/KCJ3K293 from patient C; KCJ3K307 from patient D) carried 21 ARGs in the chromosome, including the ESBL-encoding gene blaSHV-26 and the AmpC-encoding blaAmpH (Figure 2b). Based on the genetic identity, the K. pneumoniae KCJ3K307 might have originated from patient C or vice versa. Patient C was also infected with E. coli KCJ3K291, which carried 53 ARGs on the chromosome, including the β-lactamase-encoding genes blaAmpC1 and blaAmpH (Figure 2c). Interestingly, the same IncC plasmid, pKC148K, encoding blaNDM, was harboured by E. coli KCJ3K291, K. pneumoniae KCJ3K292, K. pneumoniae KCJ3K293 and K. pneumoniae KCJ3K307, indicating that plasmid transmission probably occurred between two different genera in patient C, possibly mediated by the type IV conjugation system. pKC148K in the four strains was identical (≥99.99% identify, with 100% query coverage). The bacterial conjugation experiment confirmed that pKC148K could be transferred from K. pneumoniae KCJ3K292 to E. coli strain at frequencies ranging from 3 × 10−4 to 7.5 × 10−4 per recipient cell. The blaNDM-1 gene was located in a resistance island, carrying multiple resistance genes, tmrB, ant1, neo and cat. The resistance island carried five IS26 elements and one IS6100, suggesting that multiple insertion events happened on this island. Additional plasmid, pKC141K was harboured by these Klebsiella isolates (Figure S1). Overall, we found that the blaNDM gene was inserted into a broad host range conjugative plasmid that mediated transmission into different genera, indicating NDM transmission occurred within and between patients in the hospital.
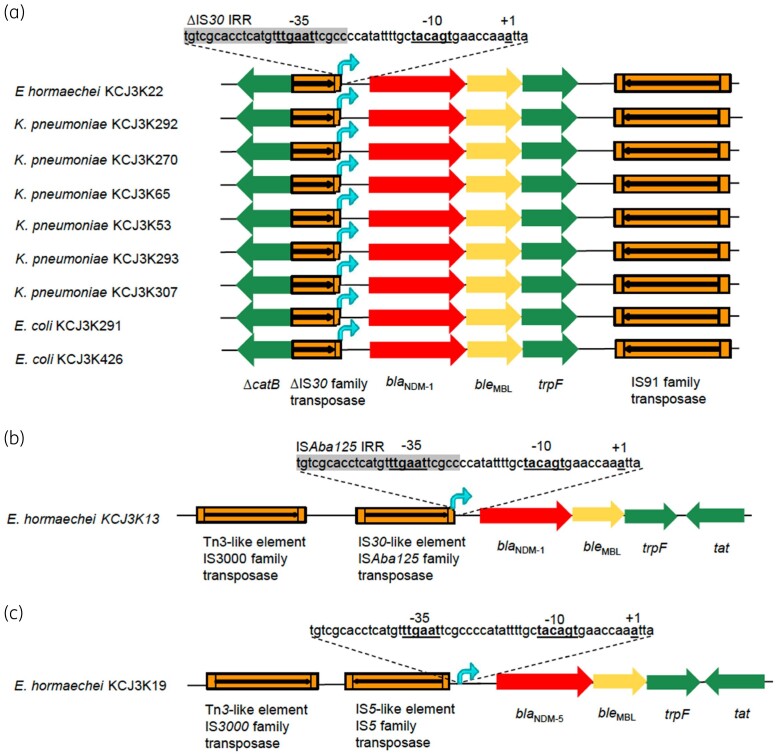
To further understand the role of IS elements on NDM expression, we investigated transcription mechanisms of the NDM genes. Three different transcription mechanisms were found in 11 NDM strains (Figure 3). Mechanism 1 was found in nine strains. A truncated IS30 element provided the −35 region for the transcription of the blaNDM-1 gene, and the −10 region was provided by the NDM gene sequences (Figure 3a). The truncated IS30 element was flanking a truncated catB gene, suggesting that an IS30 element inserted into the catB gene. Similar to mechanism 1, mechanism 2 also used a hybrid promoter region: the −35 region was provided by the ISAba125 element, and the −10 box was provided by the NDM gene sequences (Figure 3b). In mechanism 3, IS5 was located upstream of the blaNDM-5 gene, but the transcription promoter regions were fully coded by the NDM gene sequences (Figure 3c). Regardless of the genetic environment, bleMBL and trpF were always identified downstream of the blaNDM gene, indicating that these two genes were transferred together with the blaNDM gene. Therefore, besides the blaNDM gene mobilization, IS elements would appear to regulate the transcription of the gene.
Figure 3.
Transposition of blaNDM genes by IS elements. Transposition types of blaNDM were constructed by using Illumina sequencing data. (a) Mechanism 1 transposition. (b) Mechanism 2 transposition. (c) Mechanism 3 transposition. Three different ISs were identified upstream of the blaNDM genes. bleMBL and trpF were identified downstream of blaNDM in all 11 strains. Truncated IS30 family transposase and intact ISAba125 provided the −35 box for blaNDM-1 transcription promoters. IS5 was identified upstream of blaNDM-5 without involving in the promoter formation. Δ, truncated gene; IRR, right inverted repeats of IS element.
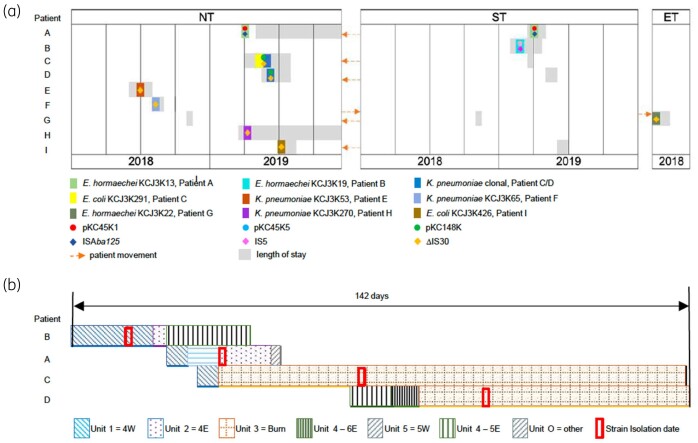
To better illustrate the potential dissemination events related to the blaNDM-carrying strains, a patient movement map was built, based on the clinical records and genetic characteristics of the isolates (Figure 4). Clonal variants—K. pneumoniae KCJ3K292, K. pneumoniae KCJ3K293 and K. pneumoniae KCJ3K307—were isolated from patients C and D who had a similar hospitalization history. Both of them stayed in the hospital ST for a short period, and then moved to the NT where they were present at the same time (Figure 4a). Patient C was negative for NDM-bearing organisms from initial wound cultures when the patient was admitted to Unit 1. However, 33 days after transfer to Unit 3, the patient was positive for NDM-bearing pathogens—K. pneumoniae KCJ3K292, K. pneumoniae KCJ3K293 and E. coli KCJ3K291—suggesting that patient C acquired these strains from Unit 3. To understand potential transmission routes of these strains, we conducted rectal screening and carbapenem resistance of all patients on Unit 3, but none of the patients was positive for NDM-1. However, 28 days later, the clonal variant strain K. pneumoniae KCJ3K307 was isolated from patient D in Unit 3 (Figure 4b). Patients C and D were in rooms next to one another, and the clonal variant strain was isolated 16 days after patient D admitted into Unit 3, suggesting NDM-producing K. pneumoniae was transmitted between the two patients during their hospital stay indirectly. However, environmental cultures obtained to understand possible transmission routes, including surfaces inside and outside the patient rooms, sink, the nurses’ station, equipment and operating room, were negative for any NDM-bearing or carbapenem-resistant organism. Besides K. pneumoniae, NDM-positive E. coli KCJ3K291 was also isolated from patient C. The blaNDM-1-carrying IncC plasmid, pKC148K, was harboured by K. pneumoniae clonal variants and E. coli KCJ3K291, suggesting the plasmid was transferred between different genera in patient C. However, E. coli bearing NDM-1 was not isolated in patient D. In addition, the same plasmids, pKC45K1 and pKC45K5, were isolated from patients A and B but we could not identify an epidemiological connection that might explain plasmid transmission between two patients.
Figure 4.
Schematic diagrams of patient movement and probable transmission of NDM strains, plasmids and IS elements in the hospital. Patient movement history in the hospital related to genetic features of the NDM-positive strains (a). Patients moved frequently between different facilities. Arrows indicate patient movement directions. The size of grey rectangle represents the length of hospital stay of individual patient. Coloured rectangles represent 12 NDM-positive strains isolated from the patients indicated. K. pneumoniae clonal variant was isolated from patient C and D. Coloured circles represent plasmids identified in the six strains shown as the rectangles. The identical plasmids, pKC45K1 and pKC45K5, were harboured in E. hormaechei KCJ3K13 and KCJ3K15, respectively. Plasmid pKC148K was identified in both K. pneumoniae clonal variant and E. coli KCJ3K291. Coloured diamonds represent different insertion sequences located upstream of blaNDM. NT, North Tower; ST, South Tower; ET, East Tower. (b). Schematic diagram of overlapped hospital stays at the units while patients B, A, C and D were hospitalized. Each patterned rectangle represents the length of stay and units. Red boxes indicate the dates of isolation of each NDM strain.
Discussion
In this study we characterized 11 MDR NDM-bearing pathogens from nine patients hospitalized in a single hospital during a 1 year time period. We found multiple genetic mechanisms involved in NDM transmission including clonal strain transmission, horizontal gene transfer mediated by plasmid conjugation and transposition by IS elements. With multiple antibiotic resistance determinants and various dissemination mechanisms, selection and transmission of NDM is complicated in the hospital setting. Connecting the genetic features of NDM-bearing strains with patient-movement history helped us link possible transmission mechanisms of the NDM genes.
Plasmids can contribute to the spread and evolution of ARGs.12 The result of plasmid typing showed that the blaNDM genes were carried by IncX3 or IncC type plasmids (Table S3). As the most common blaNDM gene carrier, IncX3 plasmids have been reported carrying multiple blaNDM variants, raising the possibility that the blaNDM gene evolved on IncX3 plasmids.9 The NDM-encoding plasmids, pKC45K1 and pKC45K5, harboured by E. hormaechei KCJ3K13 and KCJ3K19, respectively, are identical, except for the blaNDM variants, blaNDM-1 and blaNDM-5 and IS elements. According to previous studies, NDM-5 confer greater resistance than NDM-1.23 In addition to the blaNDM gene, the IncX3 plasmid is associated with blaSHV-12, blaKPC and blaOXA-181.24,25 As a conjugative plasmid, IncX3 contributes to the horizontal gene transfer of the blaNDM gene and other ARGs, and thus helps them spread among the Enterobacteriaceae. IncC is another common type of the blaNDM gene-carrying plasmids.9 It has a broad host range, not only among Enterobacteriaceae, but also Morganellaceae and Vibrionaceae, contributing to the worldwide spread of blaNDM.9 Currently, a total of 20 replicon types have been reported in NDM-carrying plasmids in Enterobacteriaceae, suggesting blaNDM can be acquired and transferred by multiple plasmids among strains, which makes it difficult to restrain the spread of blaNDM.9 In this study we found that the IncC plasmid could transfer to another patient and between K. pneumoniae and E. coli.
CRE usually carry multiple IS elements that affect neighbouring gene expression.26 The IncC plasmid, pKC148K, found in K. pneumoniae and E. coli carried five IS26 and one IS6100 on the resistance islands (Figure 2b and c), resulting in an MDR phenotype. Consistently, both IS26 and IS6100 belong to the IS6 family and IS26 has been reported associated with the blaNDM gene transfer.27 There are two features commonly found next to the blaNDM gene, IS elements and the bleomycin resistance gene bleMBL.9 IS elements were systematically found upstream of the blaNDM gene providing the −35 box for transcription.28 Consistently, we identified IS elements in the upstream of the blaNDM-1 gene that initiate transcription. The bleMBL gene is systematically found downstream of blaNDM, and expression of the bleMBL gene is regulated by the same promoter regions. Bleomycin has been mostly used as an anticancer agent in clinical therapy, suggesting blaNDM-carrying strains might be selected by antibiotics or co-selected by bleomycin.29
Interestingly, all isolates tested against aztreonam displayed very low MIC with retained susceptibility. Though aztreonam is the only available β-lactam antibiotic stable to MBLs, it is susceptible to enzymatic degradation by ESBL and AmpC enzymes. However, all isolates from patients C and D retained in vitro susceptibility to aztreonam, despite encoding both ESBL and AmpC β-lactamases.
Our paper demonstrates the spread, persistence and complexity involved in movement of NDM resistance among patients in a hospital setting; it also highlights the utility of molecular analysis in understanding transmission mechanisms, and, in turn, in developing optimal control strategies for these pathogens. The demonstration of movement of resistant clonal strains among patients underscores the importance of optimizing ‘standard’ infection control procedures, particularly in high-risk settings where there are seriously ill patients with a history of prolonged and/or multiple recent hospital stays. Care should also be taken with use of empirical antimicrobial regimens, which have the potential for selecting out strains carrying complex resistance genes, and also encouraging the movement of mobile resistance elements (plasmids and IS elements) among strains. With the number of resistance determinants we have demonstrated in these NDM clinical strains, it is paramount to have robust antimicrobial stewardship efforts in place that control all classes of antimicrobials and the duration of their use.
Supplementary Material
Acknowledgements
This project is part of the University of Florida (UF)’s ‘Creating the Healthiest Generation’ Moonshot initiative, which is supported by the UF Office of the Provost, UF Office of Research, UF Health, UF College of Medicine and UF Clinical and Translational Science Institute.
Funding
This work was supported, in part, by an internal grant (Moonshot Initiative) from the University of Florida.
Transparency declarations
None to declare.
Disclaimer
UF had no role in the design of the study, collection, analysis and interpretation of the data. The authors are solely responsible for the contents.
Supplementary data
Tables S1 to S3 and Figure S1 are available as Supplementary data at JAC-AMR Online.
References
- 1. CDC. Antibiotic Resistance Threats in the United States, 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- 2. Gandra S, Burnham CD.. Carbapenem-resistant Enterobacterales in the USA. Lancet Infect Dis 2020; 20: 637–9. [DOI] [PubMed] [Google Scholar]
- 3. Gupta N, Limbago BM, Patel JB. et al. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 2011; 53: 60–7. [DOI] [PubMed] [Google Scholar]
- 4. Haverkate MR, Bootsma MC, Weiner S. et al. Modeling spread of KPC-producing bacteria in long-term acute care hospitals in the Chicago region, USA. Infect Control Hosp Epidemiol 2015; 36: 1148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ambler RP. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci 1980; 289: 321–31. [DOI] [PubMed] [Google Scholar]
- 6. Bush K, Fisher JF.. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from Gram-negative bacteria. Annu Rev Microbiol 2011; 65: 455–78. [DOI] [PubMed] [Google Scholar]
- 7. Walsh TR. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents 2010; 36 Suppl 3: S8–14. [DOI] [PubMed] [Google Scholar]
- 8. Yong D, Toleman MA, Giske CG. et al. Characterization of a new metallo-β-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 2009; 53: 5046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu W, Feng Y, Tang G. et al. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev 2019; 32: e00115–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Logan LK, Weinstein RA.. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 2017; 215: S28–S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guducuoglu H, Gursoy NC, Yakupogullari Y. et al. Hospital outbreak of a colistin-resistant, NDM-1- and OXA-48-producing Klebsiella pneumoniae: high mortality from pandrug resistance. Microb Drug Resist 2018; 24: 966–72. [DOI] [PubMed] [Google Scholar]
- 12. Stokes HW, Gillings MR.. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol Rev 2011; 35: 790–819. [DOI] [PubMed] [Google Scholar]
- 13. Giufre M, Errico G, Accogli M. et al. Emergence of NDM-5-producing Escherichia coli sequence type 167 clone in Italy. Int J Antimicrob Agents 2018; 52: 76–81. [DOI] [PubMed] [Google Scholar]
- 14. Conlan S, Thomas PJ, Deming C. et al. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 2014; 6: 254ra126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheppard AE, Stoesser N, Wilson DJ. et al. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 2016; 60: 3767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cepheid. Xpert® Carba-R. https://www.cepheid.com/Package%20Insert%20Files/Xpert-Carba-R-Rx-Only-US-IVD-ENGLISH-Package-Insert-301-2438-Rev-F.pdf.
- 17. Teng L, Lee S, Ginn A. et al. Genomic comparison reveals natural occurrence of clinically relevant multidrug-resistant extended-spectrum-β-lactamase-producing Escherichia coli strains. Appl Environ Microbiol 2019; 85: e03030–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koren S, Walenz BP, Berlin K. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 2017; 27: 722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hunt M, De Silva N, Otto TD. et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 2015; 16: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jia B, Raphenya AR, Alcock B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017; 45: D566–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alikhan NF, Petty NK, Ben Zakour NL. et al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011; 12: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–9. [DOI] [PubMed] [Google Scholar]
- 23. Hornsey M, Phee L, Wareham DW.. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 2011; 55: 5952–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li P, Lin Y, Hu X. et al. Characterization of blaNDM-1- and blaSHV-12-positive IncX3 plasmid in an Enterobacter hormaechei new sequence type 1000 from China. Infect Drug Resist 2020; 13: 145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mouftah SF, Pal T, Darwish D. et al. Epidemic IncX3 plasmids spreading carbapenemase genes in the United Arab Emirates and worldwide. Infect Drug Resist 2019; 12: 1729–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siguier P, Gourbeyre E, Chandler M.. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 2014; 38: 865–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahillon J, Chandler M.. Insertion sequences. Microbiol Mol Biol Rev 1998; 62: 725–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mussi MA, Limansky AS, Viale AM.. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob Agents Chemother 2005; 49: 1432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dortet L, Nordmann P, Poirel L.. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob Agents Chemother 2012; 56: 1693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.