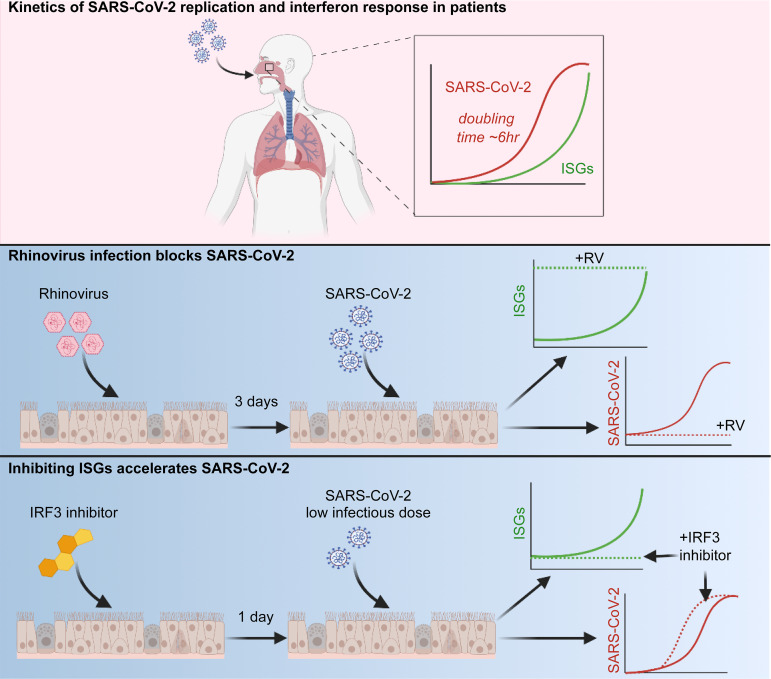
SARS-CoV-2 replication in the upper respiratory tract initiates infection and leads to viral transmission. Cheemarla et al. show that a dynamic innate immune response can curtail viral replication early in infection, and that recent exposure to rhinovirus accelerates innate defenses and blocks viral replication.
Abstract
Initial replication of SARS-CoV-2 in the upper respiratory tract is required to establish infection, and the replication level correlates with the likelihood of viral transmission. Here, we examined the role of host innate immune defenses in restricting early SARS-CoV-2 infection using transcriptomics and biomarker-based tracking in serial patient nasopharyngeal samples and experiments with airway epithelial organoids. SARS-CoV-2 initially replicated exponentially, with a doubling time of ∼6 h, and induced interferon-stimulated genes (ISGs) in the upper respiratory tract, which rose with viral replication and peaked just as viral load began to decline. Rhinovirus infection before SARS-CoV-2 exposure accelerated ISG responses and prevented SARS-CoV-2 replication. Conversely, blocking ISG induction during SARS-CoV-2 infection enhanced viral replication from a low infectious dose. These results show that the activity of ISG-mediated defenses at the time of SARS-CoV-2 exposure impacts infection progression and that the heterologous antiviral response induced by a different virus can protect against SARS-CoV-2.
Graphical Abstract
Introduction
SARS-CoV-2 enters the body and first replicates in the upper respiratory tract, usually peaking within the first week of infection. The level of viral replication in the nasopharynx correlates strongly with the likelihood of transmission, with most transmission occurring in the first week (Cevik et al., 2021; He et al., 2020). Furthermore, significant viral replication following SARS-CoV-2 exposure is likely a prerequisite (although certainly not the only factor) for COVID-19 progression. Thus, successful host responses at this early stage could reduce viral transmission and progression to COVID-19 disease.
Innate immune responses such as the antiviral IFN response are often critical for curtailing early viral replication at mucosal surfaces; however, recent work shows that SARS-CoV-2 antagonizes and delays this response (Banerjee et al., 2020b; Blanco-Melo et al., 2020; Konno et al., 2020; Xia et al., 2020; Zhou et al., 2020; Martin-Sancho et al., 2021; Ravindra et al., 2021). Nonetheless, there is strong evidence that the IFN response protects against severe COVID-19, from both genetic and autoantibody studies, and from in vitro studies showing protection of cells using recombinant IFNs. However, it is unclear how early in infection these defenses function (Lee and Shin, 2020; Meffre and Iwasaki, 2020; Zhang et al., 2020; Bastard et al., 2020; Vanderheiden et al., 2020; Pairo-Castineira et al., 2021; Lokugamage et al., 2020). A recent study linked rapid induction of virus-specific T cells to early viral clearance (Tan et al., 2021). However, the timing and role of the IFN response during initial SARS-CoV-2 replication in the upper respiratory tract has not been reported. This is due in part to the difficulty of obtaining samples from the start of infection, when the infection is often asymptomatic, and also due to the lack of established, practical methods to track these responses in human subjects.
The mucosal IFN response is initiated when pattern recognition receptors within epithelial cells and immune cells sense general features shared by many viruses, such as common structural features of viral RNA. This recognition event triggers expression of type I and type III IFNs and IFN-stimulated genes (ISGs). Secreted IFNs, in turn, bind to cell surface receptors on nearby cells, amplifying ISG expression and creating an antiviral state in the mucosal barrier (Schneider et al., 2014; Odendall and Kagan, 2015; Iwasaki, 2012). Most respiratory viruses induce an IFN response in the airway, although the kinetics and magnitude vary, and most are also highly susceptible to this host defense mechanism if it is sufficiently activated during active viral replication.
Recent work on the host response to SARS-CoV-2 in the airway epithelium or model cell lines has shown that SARS-CoV-2 can trigger IFN and ISG expression via the viral RNA sensor MDA5, albeit with delayed kinetics, but that this response in some cases was ineffective at curtailing viral replication; furthermore, while pretreatment with exogenous IFN blocked SARS-CoV-2 infection, IFN was much less effective if added after the infection was established (Yin et al., 2021; Rebendenne et al., 2021; Hsin et al., 2021 Preprint). Single-cell analysis of epithelial cells from the nasopharynx of COVID-19 patients showed ISG induction in epithelial cells primarily from patients with mild-to-moderate disease, but not severe disease, consistent with the observation that patients with severe disease are more likely to have genetic or acquired deficiencies in IFN signaling pathways (Ziegler et al., 2021 Preprint). Together, these findings indicate that epithelial cells are capable of mounting an IFN response to SARS-CoV-2 but that the timing and magnitude of the IFN response relative to viral replication may be critical for determining the course of infection.
In this study, we sought to capture early host–virus dynamics in the human nasopharynx using serial patient samples and study the functional consequences of modulating the host innate immune response in primary human airway epithelial air–liquid interface (ALI; organoid) cultures. While SARS-CoV-2 is known to antagonize and delay the IFN response, we hypothesized that this defense mechanism might still be critical for restricting initial SARS-CoV-2 replication under some circumstances, depending on the relative kinetics of viral replication and ISG induction.
Due to SARS-CoV-2 screening and testing practices at our hospital at the start of the pandemic in March 2020, we were able to obtain nasopharyngeal (NP) swab samples from patients at multiple time points after infection, including serial samples collected close to the start of infection in asymptomatic subjects. Using transcriptomic and biomarker-based analysis of these samples, we observed robust ISG induction in the airway mucosa in diverse COVID-19 patients. Longitudinal analysis revealed a dynamic mucosal response, with a rise and fall in ISG expression tracking with the rise and fall in viral load. Using the organoid model, we found that prior infection with rhinovirus (RV) accelerated ISG activity early in SARS-CoV-2 infection and completely prevented SARS-CoV-2 replication, an effect that was reversed by preventing ISG induction. Conversely, blocking homologous ISG induction during SARS-CoV-2 infection enhanced the replication rate in the setting of a low infectious dose. Together, these results demonstrate that innate immunity in the airway of human subjects is highly dynamic and demonstrate that innate immune responses profoundly influence initial infection with SARS-CoV-2.
Results
SARS-CoV-2 induces an IFN response in the nasopharynx across diverse patient groups
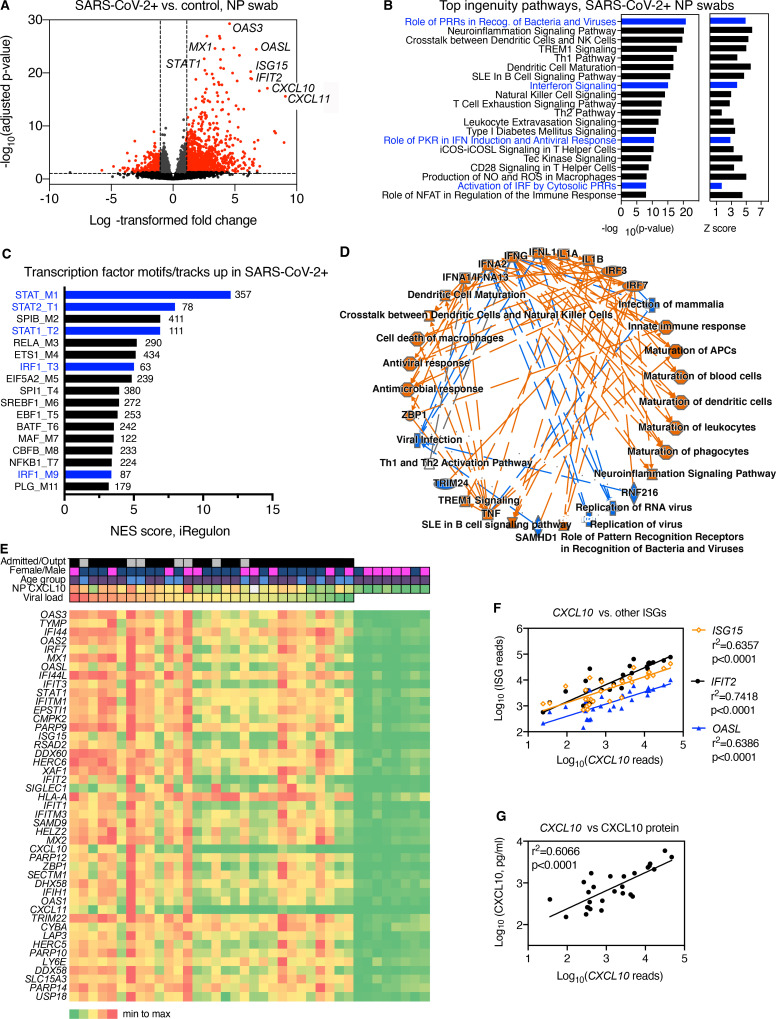
The host response to SARS-CoV-2 in the upper respiratory tract and its relationship to viral replication at the start of infection are not well defined. To initially characterize host responses during SARS-CoV-2 infection in vivo, we performed RNA sequencing (RNA-seq) on NP swab RNA from SARS-CoV-2–positive patients (n = 30) and SARS-CoV-2–negative healthcare worker controls (n = 8). Patients included outpatients and patients admitted to the hospital of both sexes, who ranged in age from 20 to 90 yr, with the majority being >60 yr of age (Table S1). Samples varied in viral load over five orders of magnitude as assessed by quantitative RT-PCR (RT-qPCR) for the SARS-CoV-2 N1 gene or by read mapping of NP RNA to the SARS-CoV-2 genome, with a strong correlation seen between PCR Ct value for the SARS-CoV-2 N1 gene and viral RNA reads by RNA-seq (r2 = 0.8380, P < 0.0001).
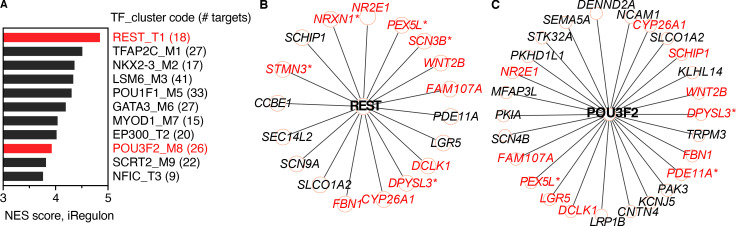
Of reads mapping to the human genome, 1,770 RNAs differed significantly between SARS-CoV-2–positive patients and control subjects (Fig. 1 A). These included 1,567 protein-coding genes, of which 1,245 (79.4%) were enriched and 322 reduced in patients relative to controls. The most significantly enriched genes in the nasopharynx of SARS-CoV-2 patients were known ISGs, including three NP ISG transcripts previously shown by our group to accurately identify patients with viral respiratory infection (OASL, IFIT2, and CXCL10; Fig. 1 A and Table S2; Landry and Foxman, 2018). Analysis of Ingenuity pathways and transcription factor–binding sites associated with enriched transcripts demonstrated activation of multiple pathways related to ISG induction in SARS-CoV-2 patients compared with controls, as well as other pathways linked to innate immunity, leukocyte recruitment, and initiation of mucosal inflammatory responses (Fig. 1, B–D; and Table S3). Notably, transcripts regulated by neuronal tissue–associated transcription factors were enriched in control subjects compared with SARS-CoV-2–positive patients (Fig. S1). Many of these transcripts were previously shown to be more highly expressed in the human olfactory epithelium and/or olfactory sensory neurons compared with the respiratory epithelium, consistent with reports that SARS-CoV-2 targets the olfactory epithelial cells and possibly neurons in the nasopharynx (Olender et al., 2016; Fodoulian et al., 2020).
Figure 1.
Transcriptome analysis of RNA isolated from SARS-CoV-2–positive NP swabs. Related to Fig. S1 and Table S1, Table S2, Table S3, Table S4, Table S5, and Table S6. (A) Volcano plot showing significantly differentially expressed protein-coding genes based on RNA-seq of NP swab RNA from SARS-CoV-2 patients (n = 30) compared with control SARS-CoV-2–negative subjects (n = 8). Transcripts with FC > 2 and adjusted P value < 0.05 are highlighted in red. (B) Top 20 Ingenuity pathways enriched in SARS-CoV-2–positive patients compared with controls based on 1,770 differentially expressed RNAs. P value and Z score for each pathway are indicated on the x axis. Pathways related to IFN and IFN regulatory factor signaling are highlighted in blue. NK, natural killer; Th, T helper; NO, nitric oxide; IRF, IFN regulatory factor. (C) Transcription factor–binding sites associated with NP transcripts enriched in SARS-CoV-2–positive patients compared with controls. Bars show strength of association of motifs/tracks with enriched transcripts, indicated by normalized enrichment score (NES). The y axis label indicates top transcription factor associated with each cluster of motifs (M) or tracks (T) and the cluster code. The number of enriched transcripts associated with each track/motif is indicated to the right of each bar. Transcription factors associated with the IFN response are highlighted in blue. (D) Graphical summary of pathways and regulators enriched based on Ingenuity Pathway Analysis of differentially expressed genes enriched in NP RNA of SARS-CoV-2–positive patients compared with controls. Colored lines indicate relationship between nodes, with orange lines showing enhancement and blue arrows showing suppression of a biological process by an upstream regulator (e.g., IFNλ1→suppression of viral infection). (E) Heatmap showing relative expression level of the top 45 most significant differentially expressed genes in patients (left) or SARS-CoV-2–negative controls (right). Clinical characteristics of each patient are indicated by color: viral load (red = highest viral load/lowest Ct value, green = lowest viral load/highest Ct value); NP CXCL10 protein level (red = highest, green = lowest, white = data not available). Heatmap colors represent values from highest (red) to lowest (green) for viral load (based on Ct value), CXCL10 concentration (pg/ml), or gene expression level, scaled from minimum to maximum (green = 0; yellow = 0.5, red = 1) Patient characteristics indicated at the top of the graph include admission status (gray, outpatient; black, admitted), sex (blue, male; pink, female), and age (blue, <55 yr; purple, >60 yr); white, data not available. Outpt., outpatient. (F) Correlation between reads mapping to CXCL10 and reads mapping to other ISGs (IFIT2, OASL, and ISG15). (G) Correlation between reads mapping to CXCL10 and CXCL10 protein measured by ELISA in NP swab–associated viral transport medium. PRR, pattern recognition receptor; SLE, systemic lupus erythematosus; PKR, protein kinase R; iCOS-iCOSL, inducible T cell costimulator and inducible T cell costimulator ligand.
Figure S1.
iRegulon analysis of transcription factor–binding sites enriched in RNA from control subjects compared with SARS-CoV-2–positive patients. Related to Fig 1. (A) Transcription factor (TF) motifs/track–binding sites enriched in NP RNA from control subjects. Bars show strength of association (normalized enrichment score [NES]) with motifs/tracks and associated transcription factors. Transcription factors associated with regulation of neuronal tissue–specific genes are highlighted in red. (B and C) Targets of transcription factors REST (B) and POU3F2 (C) enriched in NP RNA from control subjects. Red text indicates targets previously shown to be more highly expressed in olfactory compared with respiratory epithelium. Asterisks indicate transcripts also enriched in olfactory sensory neurons (olfactory epithelium, respiratory epithelium, and olfactory sensory neuron expression data are from Olender et al., 2016).
Examination of gene expression across patient samples revealed several patterns (Fig. 1, E–G). First, the 45 most significantly enriched genes were all ISGs, according to the Interferome database (Rusinova et al., 2013) and consistent with previous reports (Mick et al., 2020; Lieberman et al., 2020). ISGs appeared to be coregulated within individual patients (i.e., patients with high expression of one ISG tended to have high expression of other ISGs; Fig. 1 E). This was also demonstrated by analysis of the correlation between reads for different ISGs across samples (Fig. 1 F). Second, ISG expression appeared to be loosely correlated with viral load, with those patients with the highest viral load (Fig. 1 E, left) tending to have higher ISG expression than those with the lowest viral loads (Fig. 1 E, right). However, while all SARS-CoV-2–positive samples showed enrichment of ISG expression compared with controls, there was no clear relationship between ISG expression level and sex, age, or outpatient/admitted status in this diverse set of 30 samples. Notably, IFNλ1 and IFNγ, but not other IFN subtypes, showed significant enrichment in COVID-19 patients compared with controls, although expression levels were relatively low compared with many ISGs, similar to findings in a single-cell RNA-seq (scRNA-seq) study of epithelial cells from the nasopharynx of COVID-19 patients (Table S2; Ziegler et al., 2021 Preprint). Direct comparison of DEGs in patient groups with distinct clinical characteristics (male versus female, older versus younger, and inpatient versus outpatient) showed no significant differences in ISG expression, although there were some other notable biological differences in NP gene expression that have not been reported previously. Male and female patients showed differential NP expression of a subset of sex-linked genes, including the innate immune associated RNA helicase DDX3Y (Table S4; Szappanos et al., 2018). Also, younger patients (<55 yr) showed enrichment for mRNAs expressed by the repair-associated Hillock cells of the airway epithelium (KRT4 and KRT13) compared with older patients (>70 yr), and a similar pattern was seen in outpatients compared with admitted patients (Table S5 and Table S6) In addition, some previously reported patterns were also noted, such as increased expression of a subset of airway mucins in COVID-19 patients compared with controls (Table S2; Biering et al., 2021 Preprint).
NP CXCL10 protein correlates with ISG RNA in NP swabs
Next, we sought a quantitative biomarker of ISG induction in order to evaluate this response across a larger set of samples and over time in individual patients. We previously showed that NP CXCL10 is detected in the viral transport medium during other acute viral respiratory infections and correlates with expression of ISGs at the mRNA level (Landry and Foxman, 2018). Therefore, we measured the level of CXCL10 protein in the NP swab–associated viral transport medium using ELISA. We observed a significant positive correlation between NP CXCL10 protein level with the NP mRNA level of CXCL10 in SARS-CoV-2–positive patient NP samples (Fig. 1, E and G). Together, these results indicated that across subjects with diverse clinical presentations, SARS-CoV-2 induced a robust IFN response in the nasopharynx and that the NP CXCL10 protein level correlated with ISG expression at the RNA level.
NP host response correlates with SARS-CoV-2 viral load across patient groups
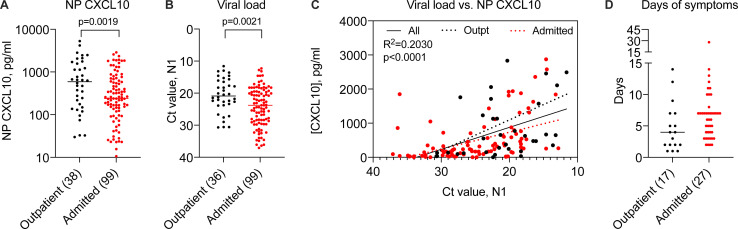
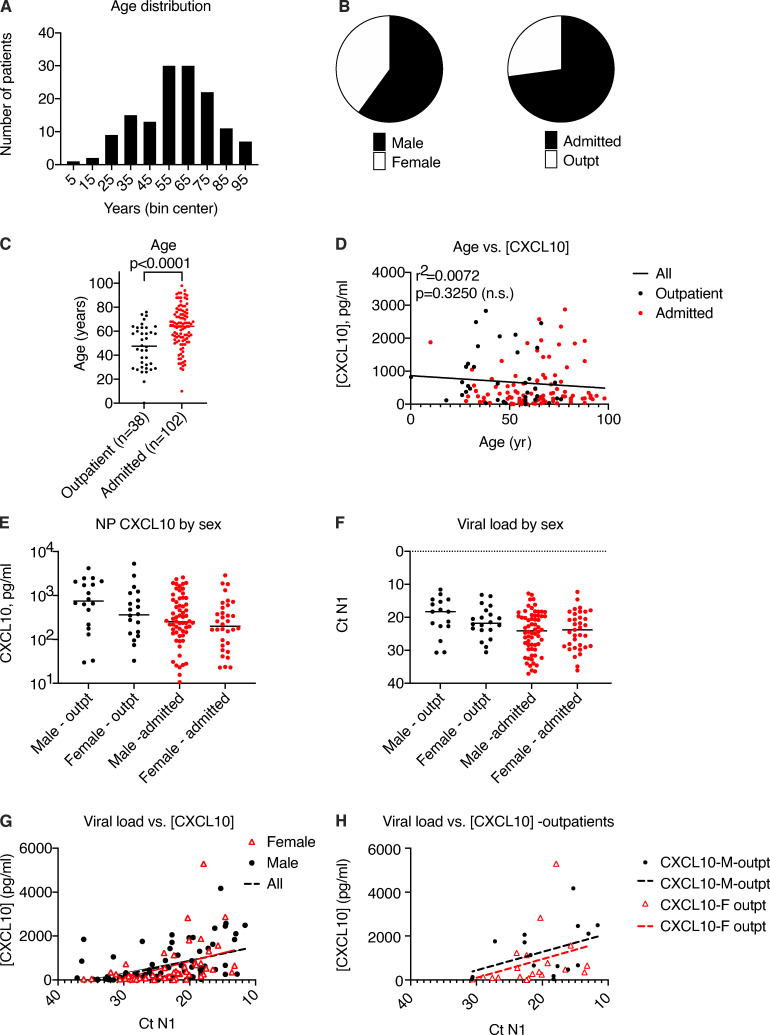
Using CXCL10 as a biomarker, we evaluated a larger set of samples from SARS-CoV-2–positive patients tested in our healthcare system in March and April of 2020 (n = 140) in order to gain further insight into the relationships among viral replication, disease status, and host response to infection (Fig. 2; and Fig. S2, A and B). First, we examined NP CXCL10 level in SARS-CoV-2–positive individuals who were tested as outpatients and not admitted to the hospital compared with those who were admitted. Notably, we observed significantly higher NP CXCL10 levels in outpatients compared with admitted patients (Fig. 2 A; P = 0.0019). To understand the reason for this, we first examined patient age, since as a group, admitted patients were significantly older than outpatients (16 yr older on average; Fig. S2 C). However, there was no correlation between age and CXCL10 level (Fig. S2 D). Next, we examined viral load. We were initially surprised to find that admitted patients had significantly lower viral loads than outpatients (Fig. 2 B). This suggested that the main factor driving CXCL10 level was viral load. Supporting this idea, correlation analysis showed a significant positive correlation between NP CXCL10 level and viral load by RT-qPCR (for all patients, r2 = 0.2030, P < 0.0001; Fig. 2 C). This correlation was also seen in separate analyses of outpatients and admitted patients, but there was no significant difference in the slope of the CXCL10 vs. viral load correlation between these groups, although there was a trend toward a higher slope in outpatients (Fig. 2 C). We also observed no significant relationship between sex and NP CXCL10 or sex and viral load in this sample set (Fig. S2, E–H).
Figure 2.
Relationship between NP CXCL10 and viral load in 140 SARS-CoV-2–positive patients. Related to Fig. S2. (A) NP CXCL10 level in patients testing positive for SARS-CoV-2 by RT-qPCR at Yale New Haven Hospital in March 2020. Black symbols indicate patients tested as outpatients or in the emergency department and not admitted to the hospital, and red symbols indicate patients admitted to the hospital. (B) Ct value for SARS-CoV-2 N1 gene in outpatients and admitted patients. Number in parentheses (A and B) indicates the number of outpatient (total n = 38) and admitted (total n = 102) samples for which data were available. Difference between groups was evaluated using an unpaired Student’s t test, with P values shown. (C) Regression analysis showing relationship between viral load and NP CXCL10 protein for all samples (black solid line, r2 and P value indicated) or only outpatients (dashed black line) or admitted patients (dotted red line). (D) Days of symptoms reported before testing for samples with this information available (number indicated in parentheses). Difference was not significant by a Student’s t test.
Figure S2.
Relationship between age or sex and and NP CXCL10 in 140 SARS-CoV-2–positive patients. Related to Fig 2. (A) Age distribution of 140 patients studied. (B) Sex distribution of 140 patients studied. (B) Sex distribution and inpatient/outpatient status of 140 patients studied. (C) Comparison of age in outpatients versus admitted patients. Number of patients shown in parentheses. Groups were compared by unpaired t test, with P value shown. (D) Correlation analysis showing lack of significant correlation between age and NP CXCL10 protein level, based on 140 patients. (E and F) Comparison of NP CXCL10 protein level (E) and viral load (F) based on age and inpatient/outpatient status. No significant differences were detected between groups by unpaired t test. (G and H) Correlation between viral load and NP CXCL10 in all patients (G) and outpatients (H) in all patients or patients segregated by sex. For all patients, according to simple linear regression analysis, the slope of all samples is significantly nonzero (r2 = 0.2030, P < 0.0001), but slopes are not significantly different for each sex. For outpatients only, the slope of the regression line is not significantly nonzero, likely due to lower numbers (male [M] outpatient, n = 18; female [F] outpatient, n = 20).
Prior work on SARS-CoV-2 has shown that the NP viral load is highest in the first few days of infection and that the more severe symptoms of COVID-19 requiring hospitalization occur in the second or third week of infection (Cevik et al., 2021). Therefore, we hypothesized that admitted patients may have had lower viral loads at the time of testing than outpatients because they presented later in infection, after peak viral replication in the nasopharynx. Consistently, outpatients tended to report fewer days of symptoms before testing compared with admitted patients, although this information was only available for a subset of patients (approximately one third, n = 44; Fig. 2 D).
NP CXCL10 reveals kinetics of host response to SARS-CoV-2 at the start of infection
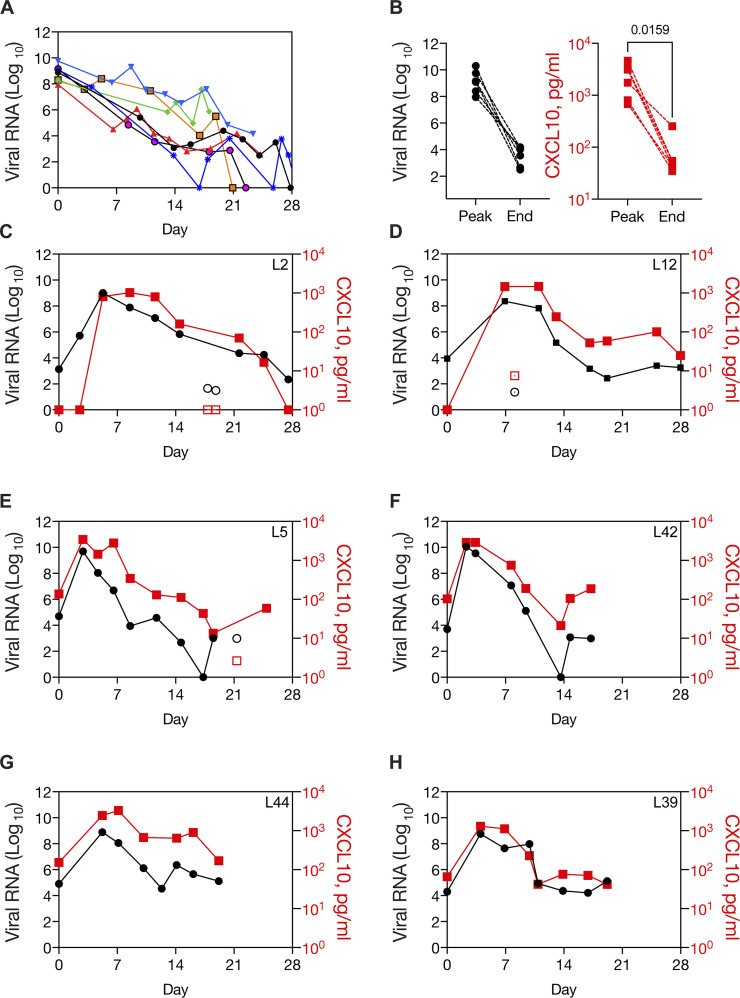
To further evaluate the relationship between viral replication and the innate antiviral response in the nasopharynx, we examined viral load and NP CXCL10 data in longitudinal patient samples. First, we examined viral load from 29 inpatients from March 12 and April 30, 2020 for whom we had at least eight sequential test results for SARS-CoV-2 with at least the first sample tested using the CDC assay (our clinical laboratory also had other testing platforms; Fig. 3 and Table S7). In March and April 2020, most serial testing in our healthcare system was aimed at patient clearance for discharge. Consistently, the majority of patients (15/29) showed low viral loads (Ct N1 > 21), which remained low throughout the time course. Another common pattern was high viral load in the first sample (Ct N1 < 20) followed by a decline in viral load over time, similar to patterns reported in the literature for patients who presented close to the start of symptomatic illness (7/29 patients; Fig. 3 A). These patients showed high CXCL10 level in the sample with peak viral load and a decline in NP CXCL10 after the viral load had decreased (Fig. 3 B). One patient had consistently high NP viral loads for 20 d and did not survive (not shown). Finally, a third pattern was seen in several patients (6/29), in which the first sample had a low viral load that subsequently increased to a high peak level (Ct N1 <20) and then decreased over time (Fig. 3, C–H). This pattern is consistent with patient presentation close to the start of infection. Two of these patients had no symptoms of SARS-CoV-2 at presentation, and the virus was detected incidentally on screening during hospitalization for other reasons (Fig. 3, C and D; and Table S7). One of these patients had an inconclusive test result and a positive test result on the same day, 12 h later (first test, N1 not detected, N2 Ct = 38.4; second test, N1 Ct = 34.6, N2 Ct = 35.4), suggesting that this might have been the first day of infection for this patient (L2, Fig. 3 C). The other four patients presented with acute symptoms, including fever and, in some cases, cough and/or shortness of breath (Fig. 3, E–H; and Table S7). NP CXCL10 in these patients was undetectable or low in the first positive sample with low viral load, rose with viral RNA, and then subsequently declined as viral load declined. Together, the longitudinal data show a correlation with NP CXCL10 and viral load in individual patients over time, similar to what we observed across 140 patients tested at a single time point (Fig. 2 C). Notably, for patient L2, the only patient for whom three samples were available before peak viral load, there appeared to be a delay between CXCL10 production relative to viral replication during the first few days of infection, suggesting that initially, viral replication outpaced the host innate immune response in the nasopharynx.
Figure 3.
Dynamic innate immune response to SARS-CoV-2 in the nasopharynx in patients diagnosed before peak viral load. Related to Fig. S3 and Table S7. (A) Viral load over time in longitudinal samples from seven patients with high viral load in the first sample (Ct N1 > 20). (B) Paired viral RNA and NP CXCL10 measurements at the peak viral load and at the end viral load, defined as the first sample with Ct N1 > 30, for six patients shown in G (data not available for one sample). CXCL10 level was significantly different in peak and end samples by paired t test. (C–H) Viral load and NP CXCL10 level in longitudinal samples from SARS-CoV-2–positive patients who presented with a low viral load (Ct N1 > 28) that increased to a high viral load (Ct N1 < 20). Viral load is expressed as FC from the limit of detection for the SARS-CoV-2 N1 gene (black circles) and CXCL10 is expressed as picograms per milliliter in the NP swab–associated viral transport medium (red squares). Samples with low levels of RNaseP, an indicator of sample quality, are shown with open symbols. Patient characteristics are described in Table S7.
SARS-CoV-2 RNA increases exponentially, with a doubling time of ∼6 h, and infection induces a robust ISG response in organoids by day 4 after infection
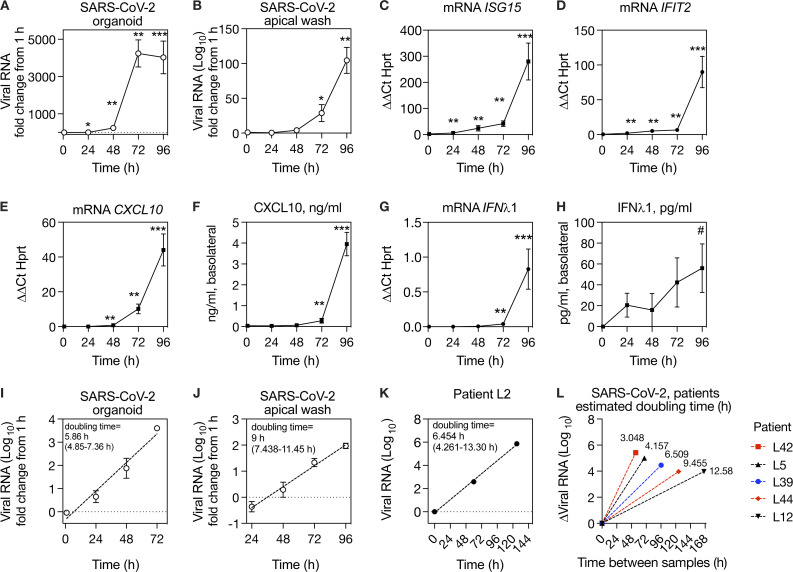
To further evaluate the kinetics of SARS-CoV-2 replication and host response, we performed a time course of SARS-CoV-2 infection using primary human airway epithelial cells grown at ALI, which differentiate into organoids with beating cilia and mucus production, recapitulating the airway mucosal surface in vivo. Reminiscent of what we observed during SARS-CoV-2 infection in vivo (Fig. 3, C–H), intracellular viral load in the organoids increased rapidly between 1 and 72 h and then plateaued between 72 and 96 h; in the apical wash, viral load increased exponentially starting slightly later, from 24 to 96 h after infection (Fig. 4, A and B). ISGs were also induced, with ISG mRNA levels and CXCL10 protein level in the basolateral medium increasing markedly from 72 to 96 h (Fig. 4, C–F). Notably, a very high level of CXCL10 protein was produced by infected epithelia (∼4 ng/ml by 96 h), consistent with the strong NP CXCL10 signal observed in the nasopharynx in vivo (Fig. 4 F). While in theory the CXCL10 measured in frozen NP swabs could reflect intracellular CXCL10, this result indicates that epithelial cells likely secrete CXCL10, consistent with our prior study in which CXCL10 was detected in the NP swab–associated media from virus-positive patients when cells were first removed from fresh samples by centrifugation (Landry and Foxman, 2018). Next, we checked for IFNλ1 at the mRNA and protein level, since the RNA-seq data showed significant induction of IFNλ1 and IFNγ in the nasopharynx of COVID-19 patients and IFNλ1 is known to be highly produced by epithelial cells, whereas IFNγ is usually produced by immune cells. We observed robust induction of IFNλ1 mRNA and a small but significant increase in IFNλ1 protein by 96 h after infection (Fig. 4, G and H).
Figure 4.
Kinetics of SARS-CoV-2 replication in organoids and in vivo.(A and B) Time course of SARS-CoV-2 replication in human primary airway epithelial organoids, expressed as fold increase from 1 h (postinoculation time point). (C–E) ISG mRNA level relative to HPRT mRNA in organoids during SARS-CoV-2 infection. (F) CXCL10 protein in the basolateral medium during SARS-CoV-2 replication. (G) IFNλ1 mRNA level relative to HPRT in organoids during SARS-CoV-2 infection. (H) IFNλ1 protein in the basolateral medium during SARS-CoV-2 infection. (I) Exponential curve fit for increase in SARS-CoV-2 RNA in organoids from 1 to 72 h and calculated doubling time for exponential growth, based on data also shown in A, with 95% CI. (J) Exponential curve fit for increase in SARS-CoV-2 RNA shed from apical surface from 24 to 96 h and calculated doubling time for exponential growth, based on data also shown in B, with 95% CI. (K) Exponential curve fit for increase in viral RNA during SARS-CoV-2 replication for first three virus-positive samples from patient L2 and calculated doubling time for exponential growth with 95% CI. (L) Estimated doubling times for increase in viral RNA during SARS-CoV-2 replication in patients with one SARS-CoV-2–positive sample before peak viral load, shown in Fig 3, D–H. The y axis shows change in viral RNA, and the x axis shows time interval between samples. Doubling time calculation assumes exponential growth between first and peak viral load samples. For A–J, symbols show mean and SEM of four to six biological replicates per condition, and data are representative of two independent experiments with primary epithelial cells from different donors. For K and L, data from six individual patients are shown (one patient in K and five patients in L). Symbols indicate significant difference from the t = 1 h after inoculation time point by Mann–Whitney test (*, P = 0.0317; #, P = 0.0152; **, P = 0.0079; ***, P = 0.0043).
Viral replication in organoid cultures appeared to follow exponential growth during the first few days of infection. Although viruses do not replicate by doubling per se, an exponential increase in viral load indicates that during this phase of growth, the increase in virus at a given time is proportional to the current amount. In this situation, the doubling time, which describes the rate of exponential increase, is a useful mathematical descriptor for viral replication rate. Therefore, we used curve fitting to exponential growth to estimate the doubling time of SARS-CoV-2 in organoids, which was 5.858 h (95% confidence interval [CI], 4.85–7.357 h; Fig. 4 I) and in the apical wash, which was 9 h (95% CI, 7.438–11.45 h; Fig. 4 J).
Although we did not evaluate viral load by plaque assay, other studies that measured SARS-CoV-2 titer during replication in similar culture systems also observed a period of exponential replication at the start of infection, consistent with our results. However, not unexpectedly, the reported kinetics vary with experimental conditions, such as viral load in the inoculum and incubation temperature (Hou et al., 2020; Zhu et al., 2020; V’kovski et al., 2021). Thus, we sought to estimate the replication kinetics in vivo using serial patient samples. For patient L2, viral load from the first three SARS-CoV-2–positive time points also appeared to follow exponential growth; therefore, we used the same method to estimate the SARS-CoV-2 doubling time in vivo from these data, which was 6.454 h (95% CI, 4.261–13.30 h based on 3 y-values; Fig. 4 K). For all other patients for whom viral load increased in serial samples (Fig. 3), we had only one sample before peak viral load. We asked what the doubling times for SARS-CoV-2 would be in these samples if we assumed exponential replication between the first and peak viral RNA values. The calculated doubling times across patients ranged from 3.048 to 6.509 h for samples ≤5 d apart (Fig. 4 L). For the two patients with a larger sampling interval (L44, L12), calculated doubling times were 9.455 and 12.58 h, although these calculations would be expected to overestimate doubling time if the second sample was taken after viral replication had plateaued or began to decline (Fig. 4 L). Together, the in vitro and in vivo results indicate that SARS-CoV-2 replicates exponentially during the first few days after infection before the peak host antiviral response, with an average doubling time of ∼6 h.
RV infection increases mRNA level of dACE2, but not full-length ACE2, in an IRF3-dependent manner
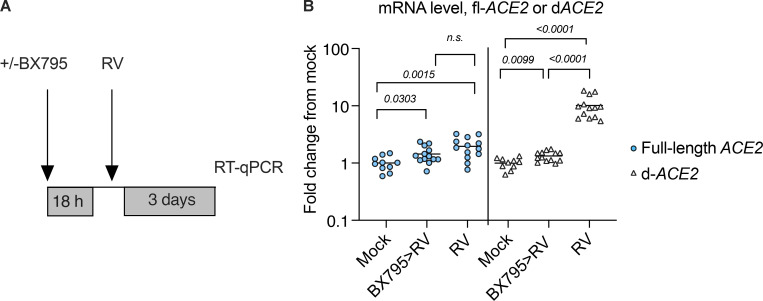
In organoid cultures, SARS-CoV-2 appeared to induce ISGs with delayed timing relative to viral replication, consistent with other cellular infection models (Yin et al., 2021). We considered that other physiological exposures, such as a prior infection with a different virus, could potentially accelerate these ISG responses. We focused on RV, since it is the most frequently detected virus in the human upper respiratory tract (Turner, 2007). Based on previous studies, we expected that RV might curtail infection by enhancing antiviral responses but also potentially promote infection by increasing expression of SARS-CoV-2 entry receptors (Wu et al., 2020; Ziegler et al., 2020). Using conditions previously established to produce robust viral amplification, we infected organoids with RV (HRV-01A, multiplicity of infection [MOI] 0.05) and evaluated the effect on expression of ACE2, the SARS-CoV-2 entry receptor. ACE2 was originally reported to be an ISG, but a subsequent study reported that full-length ACE2, which functions as an entry receptor, is not an ISG, and that a truncated form, dACE2, is an ISG but is not a functional SARS-CoV-2 entry receptor (Onabajo et al., 2020; Ziegler et al., 2020). Consistent with this finding, we observed that dACE2 was significantly induced by RV infection (∼10-fold). Induction was prevented by BX795, a signaling inhibitor that prevents IRF3 nuclear translocation and that we previously showed prevents ISG induction by RV in airway epithelial cells (Clark et al., 2009; Wu et al., 2020; Fig. S3). Notably, full-length ACE2 expression was also significantly increased by RV infection, albeit to a much lesser extent than dACE2 (approximately twofold), and this change was not abrogated by BX795 (Fig. S3) RV infection had no effect on expression of TMPRSS2, a cofactor for SARS-CoV-2 entry (not shown). Together, these results indicate that at the mRNA level, RV induces mRNA for dACE2 ∼10-fold by an IRF3-dependent mechanism, as expected for an ISG (Onabajo et al., 2020), and induces a small increase in mRNA for the SARS-CoV-2 entry receptor, full-length ACE2, approximately twofold by a different mechanism.
Figure S3.
Expression level of full-length ACE2 and the truncated variant dACE2 in mock-treated airway epithelial organoids following RV infection (HRV-01A). Related to Fig 5. (A) Organoids were mock infected or infected with RV. Some RV-infected cultures were pretreated for 18 h with 6 μM BX795 before infection with RV. (B) RT-qPCR was performed to quantify transcripts for full-length ACE2 (blue circles) or dACE2 (white triangles). Plot shows compiled data from three independent experiments, each with three to five biological replicates per condition. Individual values are shown (10–13 per condition). Numbers indicate P values of the Mann–Whitney test for significant differences between conditions. The primers used for full-length ACE2 target the junction of exons 8 and 9 are unique to the full-length ACE2, and the primers used for dACE2 target the junction of exons 1 and 2 are unique to dACE2, as previously described by Onabajo et al., 2020.
Prior RV infection blocks SARS-CoV-2 replication
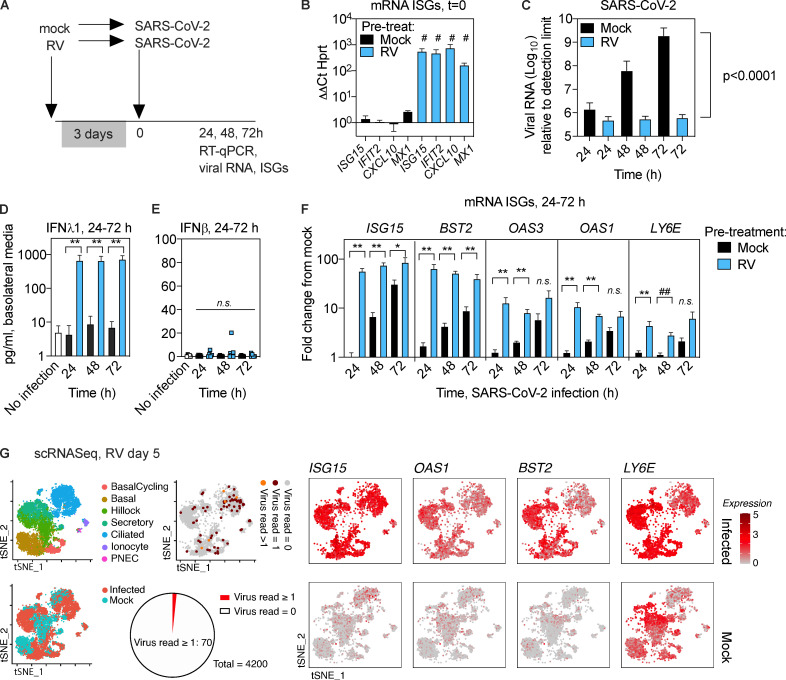
Next, we evaluated SARS-CoV-2 replication and ISG induction following infection of airway epithelial organoids, with or without prior RV infection (Fig. 5 A). RV induced expression of ISGs by day 3 after infection (Fig. 5 B). Importantly, RV-infected organoids appeared to be healthy at 3 d after infection, with cultures showing beating cilia similar to uninfected cultures (Video 1 and Video 2). SARS-CoV-2 viral load increased exponentially in infected cultures without prior RV infection, as observed previously (Fig. 4), but showed essentially no increase between 24 and 72 h when cultures had been exposed to RV 3 d prior (Fig. 5 C). Nonlinear regression analysis was consistent with exponential replication of SARS-CoV-2 from 24–72 h after infection in mock-pretreated cultures, a significant difference from RV-preinfected cultures (Fig. 5 C, P value). These results indicate that despite the slight increase in mRNA for full-length ACE2 triggered by RV (Fig. S3), the net result of RV infection 3 d before SARS-CoV-2 infection is to block replication of SARS-CoV-2. Next, we performed IFNλ1 and IFNβ ELISA on the basolateral media from organoid cultures infected with SARS-CoV-2 only or RV followed by SARS-CoV-2. For IFNλ1, we observed minimal IFNλ1 production in cultures infected with SARS-CoV-2 only but high levels of IFNλ1 from organoids sequentially infected with RV followed by SARS-CoV-2 (Fig. 5 D). There was no significant increase in IFNβ in SARS-CoV-2–infected or RV/SARS-CoV-2–coinfected cultures (Fig. 5 E), consistent with the literature showing that type III IFNs, not type I, are the major IFNs produced by respiratory epithelial cells in response to viral infection (Khaitov et al., 2009; Okabayashi et al., 2011). Consistently, evaluation of ISG expression over the course of infection showed that at early time points of SARS-CoV-2 infection, ISGs were significantly more highly expressed in RV-preinfected cultures that in cultures infected with SARS-CoV-2 without prior RV exposure (Fig. 5 F). This included several ISGs that have been previously reported to limit coronavirus replication or for which polymorphisms are linked to disease severity of SARS-CoV or SARS-CoV-2, including ISG15, BST2 (tetherin), LY6E, and OAS1-3 (Hamano et al., 2005; He et al., 2006; Ma et al., 2014; Taylor et al., 2015; Pfaender et al., 2020; Martin-Sancho et al., 2021; Pairo-Castineira et al., 2021; Fig. 5 F).
Figure 5.
Effect of prior RV infection on ISG induction and SARS-CoV-2 replication in human airway epithelial organoids.(A) Timing of infection of epithelial organoids with RV followed by SARS-CoV-2. (B) Expression of ISGs in airway epithelial organoids 3 d after RV infection, relative to mRNA for the housekeeping gene HPRT. (C) SARS-CoV-2 viral RNA at 24, 48, and 72 h after infection, with or without RV preinfection. P value represents difference between exponential growth curves fit to the data. (D) ELISA for IFNλ1 in basolateral media from uninfected organoid cultures or cultures infected with SARS-CoV-2 with or without RV preinfection. (E) ELISA for IFNβ in basolateral media from uninfected organoid cultures or cultures infected with SARS-CoV-2 with or without RV preinfection. (F) Expression of ISGs at 24, 48, and 72 h after SARS-CoV-2 infection, with or without RV preinfection, expressed as FC from uninfected cells. (G) Single-cell sequencing of human airway epithelial cell organoids, mock or 5 d after RV infection. Red and orange dots indicate 70/4,200 cells with detectable viral RNA at this time point in RV-infected cultures. tSNE plots show expression of mRNA for ISGs in mock and infected cultures at the same time point. Both conditions are from a single experiment using organoids from the same donor, with data from 8,711 cells (mock, n = 4,200 cells; infected, n = 4,511 cells). PNEC, pulmonary neuroendocrine cell. For B, bars show mean and SEM of four replicates per condition. Data are representative of three independent experiments with organoids from different primary cell donors. For C–F, bars show mean and SEM of four to six biological replicates per condition. Data are representative of two independent experiments with organoids from different primary cell donors. For B–F, significant differences between conditions were evaluated by the Mann–Whitney test (*, P = 0.0317; #, P = 0.0286; ##, P = 0.0159; **, P = 0.0079).
Video 1.
Cilia movement in mock-treated differentiated bronchial epithelial culture at ALI, day 3 after mock inoculation. Recorded at 30 frames per second, Olympus CKX53 light microscope, 20× magnification.
Video 2.
Cilia movement in RV-infected differentiated bronchial epithelial cells at ALI, day 3 after infection. Recorded at 30 frames per second, Olympus CKX53 light microscope, 20× magnification.
RV induces an IFN response in bystander cells
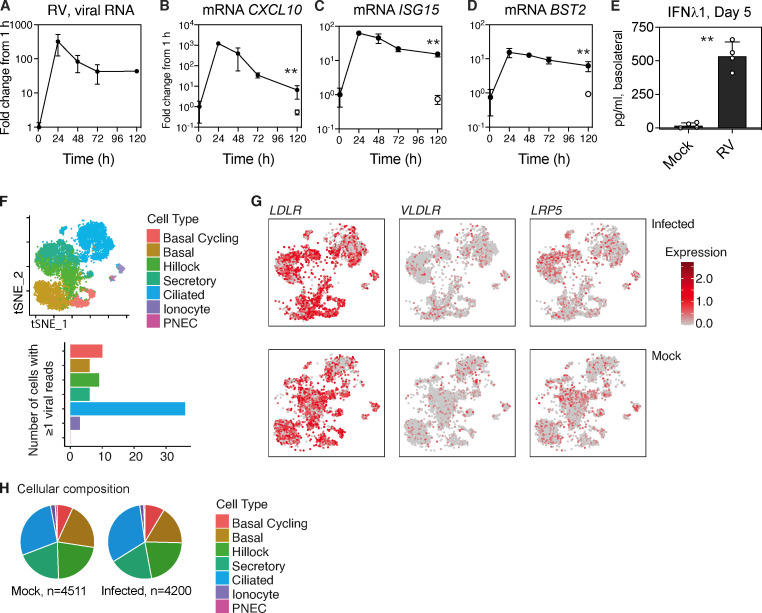
To better understand the timing and breadth of the epithelial host response to RV that appeared to limit SARS-CoV-2 replication, we evaluated ISG expression over time for 5 d and examined ISG expression and viral infection at the single-cell level. Time-course analysis showed that following inoculation, RV replicated robustly, peaking at 24 h after infection, and then declined significantly but was still detectable by RT-qPCR at day 5, a time point corresponding to 48 h after SARS-CoV-2 infection in the sequential infection experiment (Fig. S4 A). ISG expression increased and decreased in parallel with viral replication but was still significantly higher than in mock-treated cells at day 5 after RV infection (Fig. S4, B–D). In addition, a high level of IFNλ1 protein was detected in the basolateral media of RV-infected cultures at day 5 after infection (Fig. S4 E). Next, we performed scRNA-seq of mock-treated and RV-infected organoid cultures to evaluate the host ISG response at single-cell resolution at day 5 after infection At this time point, RV viral RNA reads were detected in only 70 out of 4,200 cells (Fig. 5 G). The infected cells were predominantly ciliated cells but included all major cell types, consistent with the HRV-01A entry receptor low-density lipoprotein receptor being ubiquitously highly expressed throughout the culture (Fig. S4, F and G). Although RV RNA was only detected in a small subset of cells in infected cultures at day 5 (1.67%), ISGs were elevated in all cells compared with mock-treated cultures (Fig. 5 G), indicating that RV infection induces a robust antiviral response in uninfected cells and consistent with detection of a high level of IFNλ1 in the basolateral media (Fig. S4 E). Although it is possible that some of the ISG-expressing cells in the day 5 culture had been previously been infected with RV, RV is generally considered to be a lytic infection and to infect only a small proportion of cells within the airway mucosa; thus, it is likely that the decline in viral load from day 1 to day 5 reflects death of infected cells and the majority of cells in the culture at day 5 are bystander cells (Jacobs et al., 2013). Consistent with this idea, RV infection did not significantly alter the cellular composition of organoid cultures on day 5 (Fig. S4 H). Together, these lines of evidence indicate that RV infection induces robust ISG expression in all cells of infected cultures, many of which are bystander cells.
Figure S4.
Effects of RV1A replication in differentiated primary human bronchial epithelial cultures. Related to Fig 5. (A–E) HRV-01A replication in bronchial epithelial organoids and host response over 5 d. (A–D) Viral RNA level and ISG mRNA levels are graphed as FC from ISG level at t = 1 h (postinoculation time point). Open symbols represent ISG levels in mock-infected cultures at day 5. Each point shows mean and SD of four biological replicates per condition. Results are representative of two independent experiments. Significant difference between mock and infected at day 5 is indicated by asterisks (**, P = 0.0286, Mann–Whitney test). (E) Protein level of IFNλ1 in the basolateral media at day 5 for mock- and RV-infected cultures. Graphs show mean and SD of four biological replicates per condition (A–E). Significant difference between mock and infected at day 5 is indicated by asterisks (**, P = 0.0286, Mann–Whitney test). (F) Cell type–specific RV infection in human bronchial epithelial organoid cultures 5 d after infection with RV (HRV-01A) or mock infection. Top panel shows cell types present, and bottom panel shows cell type–specific distribution of 70 cells containing at least one viral read. (G) Expression level of entry receptors for HRV-01A (LDLR, VLDLR, and LRP5) in RV-infected (top) or in mock-treated cultures (bottom panels). (H) Cellular composition of mock- or RV-1A–infected organoids at day 5 after infection.
RV blocks SARS-CoV-2 replication through heterologous ISG induction
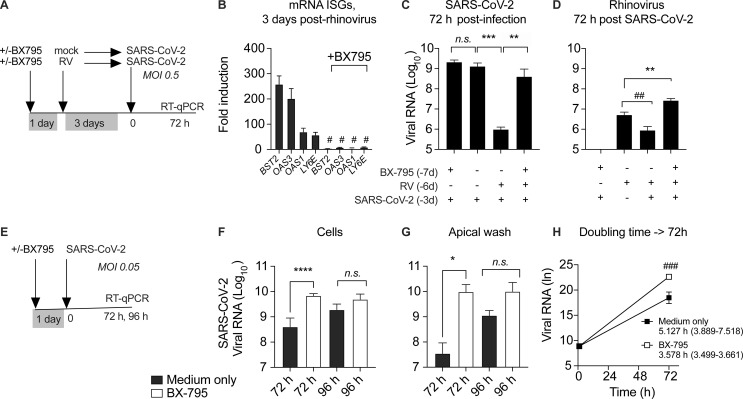
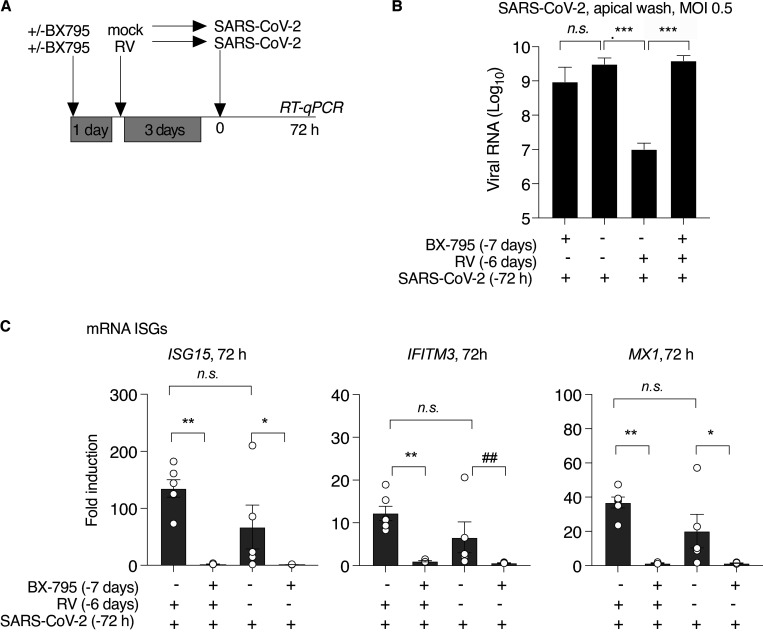
Next, to test whether suppression of SARS-CoV-2 replication in the presence of RV was dependent upon the host innate immune response, we blocked ISG induction by including the signaling inhibitor BX795 throughout single or sequential infections (Fig. 6 A and Fig. S5). Notably, under these conditions (MOI 0.5, 72 h), the homologous ISG response to SARS-CoV-2 did not appear to be limiting viral replication, since BX795 suppressed ISG induction by SARS-CoV-2 but did not alter SARS-CoV-2 viral load within the culture or apical wash (Figs. 6 C and S5). However, BX795 dramatically altered SARS-CoV-2 replication in the context of RV infection. As seen previously (Fig. 5), prior infection with RV suppressed SARS-CoV-2 replication by >1,000-fold (Fig. 6 C). However, following BX795 pretreatment, SARS-CoV-2 replication was restored (Fig. 6 C). These results indicate that ISG induction is required for the inhibition of SARS-CoV-2 replication by prior RV infection.
Figure 6.
Effect of pretreatment with BX795 during sequential RV, SARS-CoV-2 infection, and low-MOI SARS-COV-2 infection. (A) Organoid cultures were pretreated with or without BX795 for 18 h, mock infected or infected with HRV-01A, incubated for 3 d, and then infected with SARS-CoV-2 MOI 0.5. BX795 was present throughout the experiment. (B) Effect of BX795 pretreatment on ISG induction 3 d after RV infection. Bars show FC in ISG mRNA level in RV infected cultures compared with mock without (left) or with (right) BX795 pretreatment. Bars shown mean and SEM of four replicates per condition and are representative of three independent experiments using primary cells from different donors. Significant difference in ISG level by Mann–Whitney is indicated (#, P = 0.0303). (C) SARS-CoV-2 viral RNA level relative to the limit of detection in organoid cultures 72 h after SARS-CoV-2 infection, with and without BX795 and/or RV pretreatment. (D) HRV-01A viral RNA level relative to the limit of detection in organoid cultures 72 h after SARS-CoV-2 infection, with and without BX795 and/or RV pretreatment. This graph also includes cultures infected with RV, but not subsequently infected with SARS-CoV-2. (E) Cultures were pretreated with 6 µM BX795 or medium only for 18 h and then inoculated with SARS-CoV-2, MOI 0.05, at t = 0 (F–H). (F) SARS-CoV-2 viral RNA level relative to the limit of detection in organoid cultures 72 h after infection, with and without BX795 pretreatment (black or white bars, respectively. (G) SARS-CoV-2 viral RNA level relative to the limit of detection in apical wash 72 h after infection, with and without BX795 pretreatment (black or white bars, respectively). (H) Doubling time calculations for SARS-CoV-2 in organoids with and without BX795 pretreatment, assuming exponential growth between 1 and 72 h. Exponential growth curves were compared by the extra sum-of-squares F-test and found to be significantly different (###, P = 0.0011). For C, D, F, and G, bars show mean and SEM of four to six biological replicates per condition. P values indicate significant differences in ISG or viral RNA by Mann–Whitney test (*, P = 0.0449; ##, P = 0.0286; **, P = 0.0095; ***, P = 0.0043; ****, P < 0.0001). Data are representative of two independent experiments using primary cells from different donors.
Figure S5.
SARS-CoV-2 viral load in apical wash, and ISG expression in organoids following pretreatment with BX795 in sequential RV, SARS-CoV-2 infection. Related to Fig 6, A–D. (A–C) Organoid cultures were pretreated with or without BX795 for 18 h, mock infected or infected with HRV-01A, incubated for 3 d, and then infected with SARS-CoV-2, MOI 0.5 (from the experiment shown in Fig 6, A–D). SARS-CoV-2 viral RNA level relative to the limit of detection in RNA isolated from the apical wash collected 72 h after SARS-CoV-2 infection, with and without BX795 and/or RV pretreatment. ISG mRNA levels relative to levels in mock-treated cultures. Bars show mean, and symbols show individual replicates. Bars show mean and SEM of five or six replicates per condition. Symbols indicate significant difference by Mann–Whitney test (***, P = 0.0043; ##, P = 0.0079; **, P = 0.0095; *, P = 0.0159). Data are representative of two independent experiments each with at four to six biological replicates per condition.
RV RNA was detected at much lower levels than SARS-CoV-2 RNA at this time point (72 h after SARS-CoV-2 infection, 6 d after RV infection), and showed a slight reduction during SARS-CoV-2 coinfection without BX795, but significantly higher levels when both viruses were present in the presence of BX795 (Fig. 6 D). This result indicates that the antiviral response also limits RV replication in the setting of SARS-CoV-2 coinfection at this time point. In other words, in the presence of an intact antiviral response, the viral load of both viruses was reduced, but if ISG induction was inhibited, the viral load was equal (for SARS-CoV-2) or higher (for RV) during coinfection. This experiment models sequential viral infections in a host with an intact IFN response (both viruses decrease during coinfection) compared with a host with a deficient IFN response (equal or greater replication of both viruses during coinfection).
Homologous ISG response limits SARS-CoV-2 replication in a low-MOI infection
Next, to probe whether there are any conditions under which the homologous host response to SARS-CoV-2 might limit early viral replication, we tested the effect of BX795 using a 10-fold-lower level of virus in the inoculum (MOI 0.05), since under these conditions, SARS-CoV-2 replication would potentially be more sensitive to suppression by ISGs (Fig. 6, E–H). In this experiment, BX795 treatment led to an ∼10-fold increase in organoid-associated virus and an ∼300-fold increase in virus shedding into the apical wash at 72 h after infection. A trend toward an increase (5–10 times) was also seen at 96 h after infection, although due to variability among replicates, this difference was not statistically significant (Fig. 6, F and G). Next, based on the increase in viral RNA from 1 to 72 h during the low-MOI infection, we estimated the effect of BX795 on SARS-CoV-2 doubling time in organoid culture. Assuming exponential growth between 1 and 72 h, the SARS-CoV-2 doubling time was 5.127 h without BX795 (95% CI, 3.889–7.518 h) and 3.578 h with BX795 (95% CI, 3.499–3.661 h; Fig. 6 H). These results indicate that homologous ISG induction during the first 3 d of infection limits SARS-CoV-2 replication rate in the setting of a low infectious dose.
Taken together, the data from patient samples and organoid culture experiments show that SARS-CoV-2 undergoes exponential replication at the start of infection and induces ISGs that peak just before the fall in NP viral load. The heterologous response to a previous RV infection accelerated ISG induction and inhibited SARS-CoV-2 replication in its target tissue, the airway epithelium. Furthermore, the homologous ISG response induced by the virus itself significantly curtailed viral replication from a low starting viral load. These results demonstrate that ISG-mediated defenses in the airway epithelium are dynamic and define specific instances in which this host defense profoundly limits SARS-CoV-2 replication at the start of infection.
Discussion
SARS-CoV-2 transmission fuels the continuation of the COVID-19 pandemic. Peak viral replication in the upper respiratory tract occurs during the first few days of infection and correlates directly with transmission (Cevik et al., 2021; He et al., 2020). However, host–virus interactions at this stage of disease are not fully understood, in part due to the difficulties in capturing samples from this early stage of the infection and in part due to lack of straightforward methods to measure such responses. Here, we establish such methods and show that SARS-CoV-2 replicates exponentially at the start of infection and robustly induces ISGs in the nasopharynx within the first week, but not from the start, of infection. Through follow-on mechanistic studies in an organoid model, we show that varying the timing of ISG induction profoundly impacts the extent of viral replication (Figs. 5, 6, and S5).
Due to the nature of exponential growth, even a small change in the replication rate, as we observed with inhibition of ISG induction during low-MOI infection (Fig. 6, E–H), could have a profound impact on peak viral load. For example, if conditions allowed the viral doubling time to decrease by 2 h from 6 h to 4 h, this would lead to a 64-fold greater NP viral load after 72 h. NP viral load has been shown to correlate with viral transmission, an issue that has come into focus recently due to the emergence of SARS-CoV-2 variants with enhanced transmission (Cevik et al., 2021; He et al., 2020; Singanayagam et al., 2020; Centers for Disease Control and Prevention, 2021a). Our results suggest that viral mutations that enhance IFN/ISG antagonism would enable a faster doubling time and compel studying at this aspect of the biology as a possible mechanism for increased transmissibility in emerging viral variants.
Conversely, factors that enhance IFN-mediated defenses would be expected to reduce peak viral load, viral transmission, and host susceptibility. In this study, we focused on heterologous innate immunity induced by RV, the most frequent cause of common colds (Turner, 2007). Community studies have revealed that RV is much more prevalent in the upper respiratory tract than previously appreciated, detected in ∼15% of asymptomatic subjects at a given time and often at much higher rates in young children (Foxman and Iwasaki, 2011; Jartti et al., 2008; Byington et al., 2015). RVs and other respiratory viruses have also been shown to induce ISGs in the nasopharynx in vivo, in both the presence and absence of symptoms (Landry and Foxman, 2018; Wolsk et al., 2016; Yahya et al., 2017; Yu et al., 2019). Therefore, heterologous ISG induction may be important in defense against closely spaced viral infections, as we observed in a previous study of RV and influenza A (Wu et al., 2020). However, the outcome of closely spaced infections could also be influenced by virus-specific features, including ISG antagonism and entry receptor usage. The predicted effect of prior RV infection on SARS-CoV-2 replication was unclear, since RV induces transcription of the SARS-CoV-2 entry receptor, full-length ACE2, in the airway epithelium (Fig. S3) Also, SARS-CoV-2 has been shown to antagonize both ISG induction and activity at multiple levels (Banerjee et al., 2020a; Sa Ribero et al., 2020; Xia et al., 2020; Martin-Sancho et al., 2021). However, our experiments showed that prior RV infection protected against replication of SARS-CoV-2, that this protection was dependent upon ISG induction, and that a significant bystander IFN response was detected in cells throughout the epithelium for at least 5 d after RV infection, even after the RV itself was largely cleared (Figs. 5, 6, and S4). These results indicate that the protective effect of heterologous ISG induction throughout the epithelium by RV predominated over other effects, potently suppressing SARS-CoV-2 replication.
While host–pathogen interactions are often studied one at a time in experimental models, in the human upper respiratory tract, exposure to multiple microbes simultaneously or in series is a frequent occurrence and likely an important influence on innate immunity. The idea that viral interference could be shaping broad patterns of respiratory virus transmission has only recently begun to gain traction, in part due to epidemiological data suggesting interference among RNA respiratory viruses (Isaacs and Lindenmann, 1957; Greer et al., 2009; Linde et al., 2009; Casalegno et al., 2010; Ånestad and Nordbø, 2011; Schultz-Cherry, 2015; Karppinen et al., 2016; Nickbakhsh et al., 2019; Wu et al., 2020). Heterologous ISG induction could be particularly important for host defense against a virus like SARS-CoV-2, since it would preempt mechanisms SARS-CoV-2 has in place to antagonize and delay IFN and ISG induction in response to its own replication.
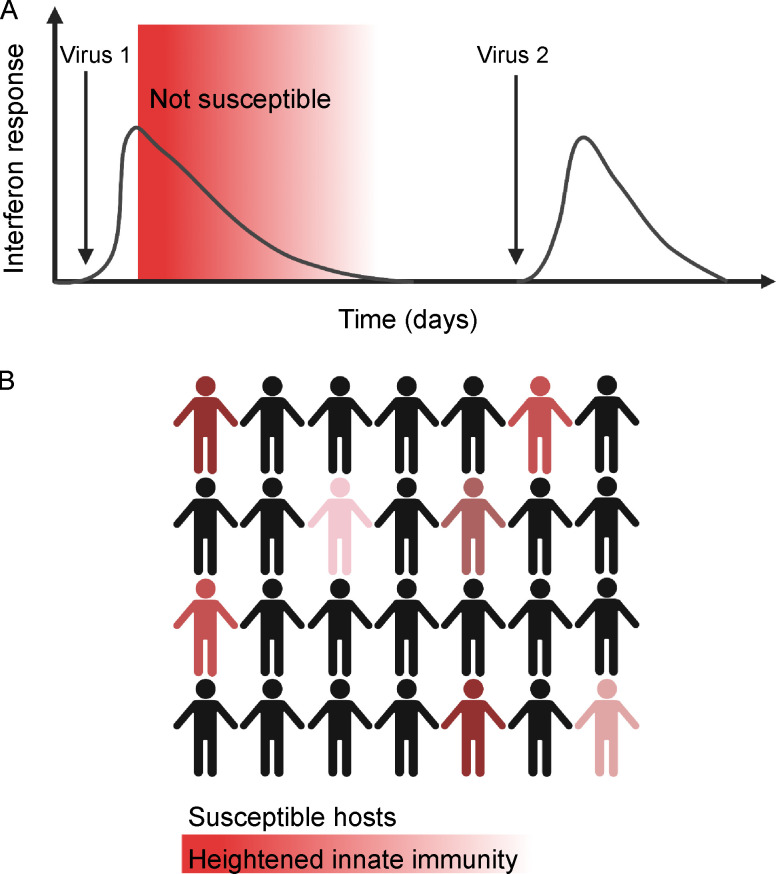
The extent to which heterologous ISG responses have affected and will affect SARS-CoV-2 transmission during the pandemic remains unclear. The SARS-CoV-2 pandemic and the public health response have disrupted the status quo in many ways, including dramatically reducing the circulation of common respiratory viruses (Olsen et al., 2020; Jones, 2020; Cowling et al., 2020; Yeoh et al., 2020; Sullivan et al., 2020; Centers for Disease Control and Prevention, 2021b). This change has led to speculation that diminished population adaptive immunity will lead to a surge of infections when these viruses again recirculate, and thus far, early data from school reopenings indicate a likely robust resurgence of RV (Baker et al., 2020; Fong et al., 2021; Poole et al., 2020). Our findings suggest that heterologous innate immune responses could be a mitigating factor, since the model predicts an upper limit in the extent to which IFN-sensitive viruses can simultaneously or sequentially infect the same host (see model, Fig. 7 A). This would also potentially slow viral transmission in the population by reducing the number of susceptible hosts at any given time (Fig. 7 B). Importantly, we are proposing this model based on our data to highlight the importance of considering viral interference in epidemiological patterns that may emerge in the coming year, but we are not arguing that potential effects of heterologous innate immunity outweigh the proven benefits of public health measures for directly preventing SARS-CoV-2 transmission.
Figure 7.
Model. Heterologous innate immunity creates a subset of individuals refractory to infection during periods of high respiratory virus circulation. (A) Virus 1 induces a mucosal IFN response, which creates a refractory period following infection during which ISGs are elevated and the host is protected from a second viral infection (red shading). After ISGs return to baseline, the host is again susceptible and can be infected with virus 2. (B) During periods of high respiratory virus circulation, a fraction of the population is refractory to infection at any given time due to ISG activation from a recent infection (red shaded figures). Thus, heterologous innate immune protection could mitigate against viral transmission at times of high respiratory virus circulation. Figure created with BioRender.com.
There are several important caveats to our study. First, the organoid model contains only epithelial cells, but homologous IFN responses induced by SARS-CoV-2 during initial viral replication are likely amplified by immune cells during natural infection. Based on the early and robust induction of CXCL10, we would expect leukocytes that express the cognate receptor CXCR3, such as T cells and natural killer cells, to be recruited and participate in antiviral defense; thus, in vivo innate responses could be significantly amplified compared with what we observe in vitro. Second, attenuation of the second infection is not the only possible outcome of a viral coinfection. We studied RV, which in most individuals results in asymptomatic infection or common colds. However, for viruses that damage the lung, such as influenza, residual tissue damage could exacerbate rather than protect against a subsequent viral respiratory illness. Studying virus–host–virus interactions for different virus pairs in physiological models is an important future direction of this study. Finally, the concept of protection based on heterologous induction of IFN responses assumes a host with intact innate immune defenses, but this is not always the case. Both genetic and environmental variables can prevent robust ISG induction in response to RV and other respiratory viruses (Lamborn et al., 2017; Asgari et al., 2017; Zhang et al., 2020; Foxman et al., 2015; Mihaylova et al., 2018; Kudo et al., 2019). In our experimental model, there was a profound difference in the outcome of RV-SARS-CoV-2 sequential infection in the presence and absence of an intact host-cell IFN response. With an intact host response, viral loads of both viruses decreased, but when the IFN response was blocked, viral loads of both viruses were equal to or higher than in single infections (Figs. 6 and S5) This result illustrates that the expected outcome of sequential viral infections is likely to be very dependent upon host innate immune status. Other factors include the extent to which coinfecting viruses trigger or block ISG induction and the kinetics of ISG induction relative to the interval between infections.
In sum, our results demonstrate an important role for IFN-mediated defenses in curtailing SARS-CoV-2 replication at the start of infection, including heterologous innate immune responses induced by prior RV infection. These results, and our findings in longitudinal patient samples, support the concept that airway innate immunity is dynamic, with innate immune defense rapidly changing in response to current and recent viral infections. Our findings also demonstrate that ISG-mediated defenses can profoundly curtail SARS-CoV-2 replication under certain conditions and compel further studies of the role of heterologous innate immunity in protecting against SARS-CoV-2 and other respiratory viruses.
Materials and methods
Experimental model and subject details
Human experimental guidelines approval statement
The use of clinical samples and data in this study was approved by the Yale University’s Human Investigation Committee (institutional review board protocol ID #200002765). Procedures for testing residual clinical samples and recording linked patient data, followed by sample and data deidentification, were evaluated, and the requirement for specific patient consent was waived. In vitro experiments used primary human cells obtained from Lonza Bioscience. Lonza Bioscience guarantees that all tissue used for human cell products is ethically obtained with donor informed consent in accordance with processes approved by an institutional review board or comparable independent review body.
Clinical samples
We used residual NP samples remaining after clinical testing for CXCL10 measurements and transcriptome analysis. Swab-associated viral transport medium was stored at −80°C following clinical testing and thawed just before ELISA assay or RNA isolation for RNA-seq. Clinical information, including age, sex, virology results, and specific features of clinical course (including presenting symptoms, hospital admission, and length of stay), was extracted from the electronic medical record and recorded, after which samples were assigned a study code and deidentified. In the clinical laboratory, SARS-CoV-2 was detected in most samples using an Emergency Use Authorization–approved TaqMan assay detecting the CDC targets N1, N2, and RNaseP (Centers for Disease Control and Prevention, 2020). In some longitudinal samples, SARS-CoV-2 was diagnosed with the commercial Cepheid assay (Cepheid, 2021); in this case, RT-qPCR for the CDC N1 gene was repeated using RT-qPCR TaqMan assay for the CDC N1 gene as described previously (catalog no. 10006600; Integrated DNA Technologies; Vogels et al., 2020).
Primary human bronchial epithelial cells
Primary human bronchial epithelial cells from healthy adult donors were obtained commercially (Lonza Bioscience) and cultured at ALI according to the manufacturer’s instructions using reduced hydrocortisone (Stem Cell Technologies). Cells were allowed to differentiate for 4 wk, by which time they displayed beating cilia and mucus production. This project used cells from eight different healthy adult donors (four male and four female). Each result was confirmed with independent experiments using organoids grown from at least two different cell donors.
Viruses
RV 1A (HRV-01A; VR-481; ATCC) was amplified in H1-HeLa cells (CRL-1985; ATCC) and used to generate a high-titer filtered cell lysate. Viral titer was determined by plaque assay as reported previously (Foxman et al., 2015). SARS-CoV-2 (USA-WA1/2020; BEI Resources) was generously provided by the Wilen laboratory (Yale University, New Haven, CT). Virus was cultured on Vero E6 cells, a filtered supernatant was used as the virus stock, and titer was determined by plaque assay as described previously (Ravindra et al., 2021). We confirmed that the cell lysate/supernatant did not contain significant levels of IFN or other molecules that activate ISG expression by mock-inoculating BEAS2B cells with UV-inactivated stocks. This did not induce ISG expression after 24-h incubation, in contrast to robust ISG induction in response to recombinant IFNβ.
RNA isolation from clinical samples
At the time of accessioning, the residual viral transport medium from clinical samples was stored at −80°C. Upon thawing, RNA was isolated from 140 µl transport medium using the Qiagen Viral RNA isolation kit per the manufacturer’s instructions (reference 52904; Qiagen), and one aliquot was reserved for ELISA.
Library preparation and RNA-seq
RNA samples were quantified and checked for quality using the Agilent 2100 Bioanalyzer Pico RNA Assay. Library preparation was performed using Kapa Biosystems Kapa HyperPrep Kit with RiboErase (human, rat, mouse) in which samples were normalized with a total RNA input of 25 ng. Libraries were amplified using 15 PCR cycles. Libraries were validated using Agilent TapeStation 4200 D1000 assay and quantified using the Kapa Library Quantification Kit for Illumina Platforms kit. Libraries were diluted to 1.3 nM and pooled at 1.25% each of an Illumina NovaSeq 6000 S4 flowcell using the XP workflow to generate 25 million read pairs/sample.
RNA-seq data analysis
Low-quality reads were trimmed and adaptor contamination was removed using Trim Galore (v0.5.0, https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Trimmed reads were mapped to the human reference genome (hg38) using HISAT2 (v2.1.0; Kim et al., 2019). Gene expression levels were quantified using StringTie (v1.3.3b) with gene models (v27) from the GENCODE project (Pertea et al., 2015). Differentially expressed genes (adjusted P value < 0.05, fold-change [FC] cutoff = 2) were identified using DESeq2 (v1.22.1; Love et al., 2014). To avoid the unexpected outlier replacement for sex-linked genes, we turned off the outlier replacement option in the male versus female comparison by setting minReplicatesForReplace = Inf for the DESeq() function in the DESeq2 package.
Visualization of RNA-seq data
Protein-coding genes differentially expressed in SARS-CoV-2–positive versus negative control were visualized on a volcano plot, with an x axis cutoff l log2FC = 10. All differentially expressed RNAs are included in Table S1 (n = 1,770). Significantly differentially expressed transcripts were defined as those with log2FC > 1 and adjusted P value < 0.05. Heatmap shows gene expression levels of top 45 most significant DEGs using minimum-to-maximum scaling of normalized read counts. Pathway analysis was performed using Ingenuity Pathway Analysis (version 01–16). Transcription factor motif enrichment analysis was performed using Cytoscape (version 3.8.1) with the iRegulon plug-in (version 1.3; Janky et al., 2014).
In vitro infections
We infected primary human bronchial epithelial cells differentiated at ALI with HRV-01A, SARS-CoV-2, or both. For SARS-CoV-2, high-MOI infection was MOI 0.5, and low-MOI infection was MOI 0.05. For HRV-01A, MOI 0.1 was used, as this was the minimum viral inoculum that reproducibly led to robust HRV-01A viral replication in ALI cultures based on prior studies.
To evaluate the effect of RV on subsequent infection with SARS-CoV-2, we infected with each virus individually or sequentially and examined the time course of viral amplification and ISG induction. To formally test whether prior exposure to RV inhibits SARS-CoV-2 replication through activation of the host-cell IFN response, we performed sequential infection studies in the presence of BX795. For BX795 inhibitor experiments, 6 µM BX795 (Sigma-Aldrich) was added to the media 1 d before the first infection. On the day of the first infection, the medium was replaced with fresh media with or without 6 µM BX795. On the day of the second infection, 150 μl additional media was added to the basolateral chamber, with or without 6 µM BX795. Results shown are representative of at least two independent experiments using primary bronchial epithelial cells from different donors, with four to six biological replicates per condition in each experiment.
RT-qPCR
For RT-qPCR, RNA was isolated from each well of differentiated epithelial cells using the QIAGEN RNeasy kit by incubating each 24-well insert with 350 µl lysis buffer at room temperature for 5 min, followed by RNA isolation and cDNA synthesis using iScript cDNA synthesis kit (Bio-Rad). To quantify viral RNA and mRNA levels for ISGs and the housekeeping gene HPRT, qPCR was performed using SYBR green iTaq universal (Bio-Rad) per the manufacturer’s instructions. Viral RNA was quantified using primers to the RV genome. Viral RNA per ng total RNA is graphed as FC from the limit of detection (40 cycles of PCR) as 240-Ct. ISG mRNA levels are graphed as FC from mock-treated cells or are presented relative to the level of mRNA for the housekeeping gene HRPT (2−ΔΔCT). RT-qPCR for SARS-CoV-2 within cultures was performed using the previously described TaqMan assay for the CDC N1 gene with primers and probes provided by Integrated DNA Technologies (catalog no. 10006600; Integrated DNA Technologies). RT-qPCR for SARS-CoV-2 in apical was performed using a combined reverse transcription and qPCR reaction using the Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs). RT-qPCR for ISG expression during SARS-CoV-2 infection was assessed with both HPRT and β-actin housekeeping gene controls to confirm that the HPRT level was stable following viral infection.
The following primers were used for RT-qPCR with SYBR green: HPRT (forward: 5′-TGGTCAGGCAGTATAATCCAAAG-3′; reverse: 5′-TTTCAAATCCAACAAAGTCTGGC-3′), β-actin (forward: 5′-CCTGGCACCCAGCACAAT-3′; reverse: 5′-GCCGATCCACACGGAGTACT-3′), ISG15 (forward: 5′-CATCTTTGCCAGTACAGGAGC-3′; reverse: 5′-GGGACACCTGGAATTCGTTG-3′), RSAD2 (forward: 5′-TCGCTATCTCCTGTGACAGC-3′; reverse: 5′-CACCACCTCCTCAGCTTTTG-3′), MX1 (forward: 5′-AGAGAAGGTGAGAAGCTGATCC-3′; reverse: 5′-TTCTTCCAGCTCCTTCTCTCTG-3′), IFITM3 (forward: 5′-ATCGTCATCCCAGTGCTGAT-3′; reverse: 5′-ATGGAAGTTGGAGTACGTGG-3′), IFIT2 (forward: 5′-CCTCAAAGGGCAAAACGAGG-3′; reverse: 5′-CTGATTTCTGCCTGGTCAGC-3′), CXCL10 (forward: 5′-CCTGCAAGCCAATTTTGTCC-3′; reverse: 5′-ATGGCCTTCGATTCTGGATTC-3′), LY6E (forward: 5′-GCATTGGGAATCTCGTGACA-3′; reverse: 5′-ATGGAAGCCACACCAACATT-3′), BST2 (forward: 5′-CACACTGTGATGGCCCTAAT-3′; reverse: 5′-TGTAGTGATCTCTCCCTCAAGC-3′), IFITM3 (forward: 5′-ATCGTCATCCCAGTGCTGAT-3′; reverse: 5′-ATGGAAGTTGGAGTACGTGG-3′), OAS1 (forward: 5′-GCTCCTACCCTGTGTGTGTGT-3′; reverse: 5′-TGGTGAGAGGACTGAGGAAGA-3′), OAS3 (forward: 5′-GAAAACTGTCAAGGGAGGCTC-3′; OAS3 reverse: 5′-CCCTCTGGTCCACATAGCTC-3′), HRV-01A (forward: 5′-CAGGCCAAATTAAAGTCAATAAGC-3′; reverse: 5′-AGGCTGAAGTTTGGTTTTGC-3′), Fl-ACE2 (forward: 5′-GGGCGACTTCAGGATCCTTAT-3′; reverse: 5′-GGATATGCCCCATCTCATGATG-3′), and d-ACE2 (forward: 5′-GGAAGCAGGCTGGGACAAA-3′; reverse: 5′-AGCTGTCAGGAAGTCGTCCATT-3′).
Quantification and statistical analysis and RT-qPCR data
GraphPad Prism (version 9.0.0) was used for data analysis. Data are shown as mean ± SEM. Statistical significance of differences between conditions was determined by t tests (two tailed) for patient data and by Mann–Whitney tests for in vitro experiments. Linear regression analysis was used to determine association between clinical parameters, such as viral load and NP CXCL10 in clinical samples, and test the null hypothesis that the slope of the association was significantly different from zero. Nonlinear regression analysis was used to fit viral growth to an exponential curve (exponential growth with log(population)) to determine virus doubling times and test the null hypothesis that one curve fit both datasets for the SARS-CoV-2 growth curve with and without RV preinfection. P < 0.05 was considered statistically significant.
ELISA
CXCL10 levels in cell-free residual NP swab samples or basolateral media of organoid cultures were quantified using a solid-phase sandwich ELISA (catalog no. DY266; R&D Systems). Briefly, frozen viral transport medium from residual NP swab samples or basolateral medium from organoid cultures was thawed on ice and centrifuged to remove cell debris. ELISA was performed according to the manufacturer’s instructions. IFNλ1 protein concentration in culture basolateral media was measured using a commercial IFNλ1 ELISA kit (catalog no. DY7246; R&D Systems) or IFNβ ELISA kit (DY814-05; R&D Systems). Basolateral media from mock-infected and infected cultures was collected from the bottom chamber of the ALI culture, UV inactivated, and stored at −80°C. The ELISA assay was performed on neat and diluted media per the manufacturer’s instructions.
scRNA-seq of ALI organoid cultures
Library preparation and sequencing
Organoid cultures were digested with trypsin/EDTA to form a single-cell suspension. The 10X Genomics single-cell 3′ protocol was used to produce Illumina-ready sequencing libraries with standard Illumina paired-end constructs according to the manufacturer’s instructions.
Analysis of scRNA-seq data
All downstream analyses were implemented using R version 3.6.3 and the package Seurat v3.1.4 (Stuart et al., 2019). Gene expression matrix of mock-infected and RV1A-infected samples was first individually analyzed in this procedure. Genes expressed in fewer than three cells and cells expressing fewer than 200 genes were discarded. A distribution histogram of unique molecular identifier (UMI) count in all cells was made, and cells with <8,000 UMI counts were discarded. This resulted in a matrix of 21,086 genes expressing in 4,511 cells in the mock sample and a matrix of 21,195 genes expressing in 4,200 cells in the RV1A-infected sample. The raw counts were normalized using the Seurat function NormalizeData with normalization.method = “LogNormalize” and scale.facto = 10,000. All genes were scaled using Seurat function ScaleData with default parameters. Variable features were determined using method “vst.” The top 2,000 variable features were used for principal-component analysis. Graph-based clustering was performed individually on mock- and RV1A-infected samples. The K-nearest neighbor graph was built by FindNeighbors function with first 20 principal components and k.param = 10. Louvain clustering was done using FindCluster function with resolution = 0.8 for both samples. Clusters were separately annotated in the mock- and RV1A-infected samples using the following markers of the major cell groups in the airway epithelium: basal cycling (MKI67 and HIST1H4C), basal (KRT14, KRT15, and KRT5), Hillock (SPINK5 and KRT13), secretory (SCGB1A1, BPIFA1, and LYPD2), ciliated (CAPS, PIFO, and MORN2), ionocyte (FOXI1), and pulmonary neuroendocrine cells (markers include AZGP1 and AVIL; Plasschaert et al., 2018). Each cluster found by clustering was assigned to one of the above seven major groups. A group of developing ciliated cells with marker cyclin-O was found in the mock sample and was merged into the ciliated cell cluster. After cluster annotation, mock and infected samples were merged together to produce t-distributed stochastic neighbor embedding (tSNE) maps and make comparisons. Both samples used same normalization method and the gene expression level was rescaled. Dimensionality reduction was performed by latent semantic analysis using Seurat function RunLSI with the first 50 singular values. The tSNE maps were then produced with the first 50 dimensions and perplexity 30. The color coding of tSNE plots used cell type, sample source, viral read per cell and expression levels of genes of interest.
Online supplemental material
Fig. S1 shows neuron-associated transcripts that are enriched in the nasopharynx of healthy controls compared with SARS-CoV-2–positive patients. Fig. S2 shows the relationship between NP CXCL10 levels in SARS-CoV-2–positive patients and age, sex, and inpatient/outpatient status, showing a significant association between NP CXCL10 and outpatient versus admitted status but no significant associations between NP CXCL10 and age or sex within these groups. Fig. S3 shows that mRNA for dACE2, a truncated form of ACE2, is induced ∼10-fold by RV in an IRF3-dependent manner and that full-length ACE2, the SARS-CoV-2 entry receptor, is also induced, but to a lesser extent (approximately twofold), and the induction is not IRF3 dependent. Fig. S4 shows the time course of RV infection and associated IFN and ISG induction in organoid cultures over the course of 5 d. Fig. S5 shows the viral load shed from the apical surface of organoid cultures during high-MOI SARS-CoV-2 infection and ISG expression with and without BX795. Table S1 and Table S7 describe clinical characteristics of patients described in Fig. 1 and Fig. 3, respectively. Table S2, Table S3, Table S4, Table S5, and Table S6 provide information about differentially expressed genes in SARS-CoV-2 patients compared with controls, as shown in Fig. 1. Video 1 and Video 2 show cilia movement in organoid cultures at day 3 after mock inoculation or infection with RV.
Supplementary Material
lists characteristics of 30 SARS-CoV-2–positive patient samples used for transcriptome analysis.
lists differentially expressed genes in SARS-CoV-2–positive patients versus negative controls.
lists Ingenuity pathways differentially enriched in SARS-CoV-2–positive NP samples compared to controls.
lists differentially expressed genes in SARS-CoV-2–positive female versus male patients.
lists differentially expressed genes in SARS-CoV-2–positive older versus younger patients.
lists differentially expressed genes in SARS-CoV-2–positive admitted patients versus outpatients.
lists clinical descriptions of longitudinal samples.
Acknowledgments
We thank Maureen Owen, Greta Edelman, the entire staff of the Yale New Haven Hospital Clinical Virology laboratory, and Amy Likens for their dedicated assistance. We thank Craig Wilen and Wilen laboratory members for valuable help and advice and providing SARS-CoV-2 virus stock. We also thank Bryan Pasqualucci and Christopher Castaldi at the Yale Center for Genomic Analysis.
Funding was provided by Fast Grants for COVID-19 from Emergent Ventures at the Mercatus Center of George Mason University (E.F. Foxman); the Yale Department of Laboratory Medicine and COVID-19 Dean’s Fund (E.F. Foxman); the China Scholarship Council Yale World Scholars Fellowship (B. Wang); the Gruber Foundation Fellowship (T.A. Watkins); and the National Institutes of Health (grant T32AI007019 to T.A. Watkins).
Author contributions: Conceptualization, N.R. Cheemarla and E.F. Foxman; Data Curation, N.R. Cheemarla, T.A. Watkins, and B. Wang; Methodology, N.R. Cheemarla, T.A. Watkins, V.T. Mihaylova, B. Wang, D. Zhao, and E.F. Foxman; Investigation, N.R. Cheemarla, T.A. Watkins, V.T. Mihaylova, and B. Wang; Formal Analysis, B. Wang, D. Zhao, and G. Wang; Resources, M.L. Landry; Supervision, E.F. Foxman; Visualization: N.R. Cheemarla, B. Wang, and E.F. Foxman; Validation, N.R. Cheemarla and E.F. Foxman; Writing – Original Draft, E.F. Foxman; Writing – Review & Editing, N.R. Cheemarla, T.A. Watkins, V.T. Mihaylova, B. Wang, D. Zhao, G. Wang, M.L. Landry, and E.F. Foxman; Funding Acquisition, E.F. Foxman.
Data availability
scRNA-seq data are available in the GEO database under accession no. GSE164982. RNA-seq data derived from patient samples have been deposited to dbGaP under accession no. phs002433.v1.p1. To access these data, users may apply for access to the dbGaP data repository (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?login=&page=login).
References
- Ånestad, G., and Nordbø S.A.. 2011. Virus interference. Did rhinoviruses activity hamper the progress of the 2009 influenza A (H1N1) pandemic in Norway? Med. Hypotheses. 77:1132–1134. 10.1016/j.mehy.2011.09.021 [DOI] [PubMed] [Google Scholar]
- Asgari, S., Schlapbach L.J., Anchisi S., Hammer C., Bartha I., Junier T., Mottet-Osman G., Posfay-Barbe K.M., Longchamp D., Stocker M., et al. 2017. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc. Natl. Acad. Sci. USA. 114:8342–8347. 10.1073/pnas.1704259114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R.E., Park S.W., Yang W., Vecchi G.A., Metcalf C.J.E., and Grenfell B.T.. 2020. The impact of COVID-19 nonpharmaceutical interventions on the future dynamics of endemic infections. Proc. Natl. Acad. Sci. USA. 117:30547–30553. 10.1073/pnas.2013182117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., et al. 2020a. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell. 183:1325–1339.e21. 10.1016/j.cell.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., et al. 2020b. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell. 183:1325–1339.e21. 10.1016/j.cell.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard, P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. COVID Human Genetic Effort . 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 370:eabd4585. 10.1126/science.abd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biering, S.B., Sarnik S.A., Wang E., Zengel J.R., Sathyan V., Nguyen X., Van Dis E., Catamura C., Yamashiro L.H., Begeman A., et al. 2021. Genome-wide, bidirectional CRISPR screens identify mucins as critical host factors modulating SARS-CoV-2 infection. bioRxiv. 10.1101/2021.04.22.440848 (Preprint posted April 23, 2021) [DOI]
- Blanco-Melo, D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. 2020. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 181:1036–1045.e9. 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byington, C.L., Ampofo K., Stockmann C., Adler F.R., Herbener A., Miller T., Sheng X., Blaschke A.J., Crisp R., and Pavia A.T.. 2015. Community Surveillance of Respiratory Viruses Among Families in the Utah Better Identification of Germs-Longitudinal Viral Epidemiology (BIG-LoVE) Study. Clin. Infect. Dis. 61:1217–1224. 10.1093/cid/civ486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalegno, J.S., Ottmann M., Duchamp M.B., Escuret V., Billaud G., Frobert E., Morfin F., and Lina B.. 2010. Rhinoviruses delayed the circulation of the pandemic influenza A (H1N1) 2009 virus in France. Clin. Microbiol. Infect. 16:326–329. 10.1111/j.1469-0691.2010.03167.x [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel https://www.fda.gov/media/134922/download (accessed May 20, 2020).
- Centers for Disease Control and Prevention . 2021a. Emerging SARS-CoV-2 Variants. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html(accessed January 8, 2021). [PubMed]
- Centers for Disease Control and Prevention . 2021b. The National Respiratory and Enteric Virus Surveillance System. https://www.cdc.gov/surveillance/nrevss/index.html(accessed February 1, 2021).
- Cevik, M., Tate M., Lloyd O., Maraolo A.E., Schafers J., and Ho A.. 2021. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2:e13–e22. 10.1016/S2666-5247(20)30172-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K., Plater L., Peggie M., and Cohen P.. 2009. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 284:14136–14146. 10.1074/jbc.M109.000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling, B.J., Ali S.T., Ng T.W.Y., Tsang T.K., Li J.C.M., Fong M.W., Liao Q., Kwan M.Y., Lee S.L., Chiu S.S., et al. 2020. Impact assessment of non-pharmaceutical interventions against coronavirus disease 2019 and influenza in Hong Kong: an observational study. Lancet Public Health. 5:e279–e288. 10.1016/S2468-2667(20)30090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodoulian, L., Tuberosa J., Rossier D., Boillat M., Kan C., Pauli V., Egervari K., Lobrinus J.A., Landis B.N., Carleton A., and Rodriguez I.. 2020. SARS-CoV-2 Receptors and Entry Genes Are Expressed in the Human Olfactory Neuroepithelium and Brain. iScience. 23:101839. 10.1016/j.isci.2020.101839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, M.W., Leung N.H.L., Cowling B.J., and Wu P.. 2021. Upper Respiratory Infections in Schools and Childcare Centers Reopening after COVID-19 Dismissals, Hong Kong. Emerg. Infect. Dis. 27:1525–1527. 10.3201/eid2705.210277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman, E.F., and Iwasaki A.. 2011. Genome-virome interactions: examining the role of common viral infections in complex disease. Nat. Rev. Microbiol. 9:254–264. 10.1038/nrmicro2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman, E.F., Storer J.A., Fitzgerald M.E., Wasik B.R., Hou L., Zhao H., Turner P.E., Pyle A.M., and Iwasaki A.. 2015. Temperature-dependent innate defense against the common cold virus limits viral replication at warm temperature in mouse airway cells. Proc. Natl. Acad. Sci. USA. 112:827–832. 10.1073/pnas.1411030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer, R.M., McErlean P., Arden K.E., Faux C.E., Nitsche A., Lambert S.B., Nissen M.D., Sloots T.P., and Mackay I.M.. 2009. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J. Clin. Virol. 45:10–15. 10.1016/j.jcv.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano, E., Hijikata M., Itoyama S., Quy T., Phi N.C., Long H.T., Ha L.D., Ban V.V., Matsushita I., Yanai H., et al. 2005. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem. Biophys. Res. Commun. 329:1234–1239. 10.1016/j.bbrc.2005.02.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J., Feng D., de Vlas S.J., Wang H., Fontanet A., Zhang P., Plancoulaine S., Tang F., Zhan L., Yang H., et al. 2006. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case-control study. BMC Infect. Dis. 6:106. 10.1186/1471-2334-6-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., et al. 2020. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 26:672–675. 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- Hou, Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H. III, Kato T., Lee R.E., Yount B.L., Mascenik T.M., et al. 2020. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 182:429–446.e14. 10.1016/j.cell.2020.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin, F., Chao T., Chan Y., Kao H., Liu W., Wang J., Pang Y., Lin C., Tsai Y., Lin J., and Chang S.. 2021. Distinct Inductions of and Responses to Type I and Type III Interferons Promote Infections in Two SARS-CoV-2 Isolates. bioRXiv 10.1101/2020.04.30.071357 (Preprint posted May 1, 2020) [DOI]
- Isaacs, A., and Lindenmann J.. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:258–267. 10.1098/rspb.1957.0048 [DOI] [PubMed] [Google Scholar]
- Iwasaki, A.2012. A virological view of innate immune recognition. Annu. Rev. Microbiol. 66:177–196. 10.1146/annurev-micro-092611-150203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, S.E., Lamson D.M., St George K., and Walsh T.J.. 2013. Human rhinoviruses. Clin. Microbiol. Rev. 26:135–162. 10.1128/CMR.00077-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janky, R., Verfaillie A., Imrichová H., Van de Sande B., Standaert L., Christiaens V., Hulselmans G., Herten K., Naval Sanchez M., Potier D., et al. 2014. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLOS Comput. Biol. 10:e1003731. 10.1371/journal.pcbi.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti, T., Jartti L., Peltola V., Waris M., and Ruuskanen O.. 2008. Identification of respiratory viruses in asymptomatic subjects: asymptomatic respiratory viral infections. Pediatr. Infect. Dis. J. 27:1103–1107. 10.1097/INF.0b013e31817e695d [DOI] [PubMed] [Google Scholar]
- Jones, N.2020. How COVID-19 is changing the cold and flu season. Nature. 588:388–390. 10.1038/d41586-020-03519-3 [DOI] [PubMed] [Google Scholar]
- Karppinen, S., Toivonen L., Schuez-Havupalo L., Waris M., and Peltola V.. 2016. Interference between respiratory syncytial virus and rhinovirus in respiratory tract infections in children. Clin. Microbiol. Infect. 22:208.e1–208.e6. 10.1016/j.cmi.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Khaitov, M.R., Laza-Stanca V., Edwards M.R., Walton R.P., Rohde G., Contoli M., Papi A., Stanciu L.A., Kotenko S.V., and Johnston S.L.. 2009. Respiratory virus induction of alpha-, beta- and lambda-interferons in bronchial epithelial cells and peripheral blood mononuclear cells. Allergy. 64:375–386. 10.1111/j.1398-9995.2008.01826.x [DOI] [PubMed] [Google Scholar]
- Kim, D., Paggi J.M., Park C., Bennett C., and Salzberg S.L.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37:907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno, Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Nakagawa S., and Sato K.. USFQ-COVID19 Consortium . 2020. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 32:108185. 10.1016/j.celrep.2020.108185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo, E., Song E., Yockey L.J., Rakib T., Wong P.W., Homer R.J., and Iwasaki A.. 2019. Low ambient humidity impairs barrier function and innate resistance against influenza infection. Proc. Natl. Acad. Sci. USA. 116:10905–10910. 10.1073/pnas.1902840116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamborn, I.T., Jing H., Zhang Y., Drutman S.B., Abbott J.K., Munir S., Bade S., Murdock H.M., Santos C.P., Brock L.G., et al. 2017. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J. Exp. Med. 214:1949–1972. 10.1084/jem.20161759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, M.L., and Foxman E.F.. 2018. Antiviral Response in the Nasopharynx Identifies Patients With Respiratory Virus Infection. J. Infect. Dis. 217:897–905. 10.1093/infdis/jix648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S., and Shin E.C.. 2020. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 20:585–586. 10.1038/s41577-020-00429-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman, N.A.P., Peddu V., Xie H., Shrestha L., Huang M.L., Mears M.C., Cajimat M.N., Bente D.A., Shi P.Y., Bovier F., et al. 2020. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 18:e3000849. 10.1371/journal.pbio.3000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde, A., Rotzén-Ostlund M., Zweygberg-Wirgart B., Rubinova S., and Brytting M.. 2009. Does viral interference affect spread of influenza? Euro Surveill. 14:14. 10.2807/ese.14.40.19354-en [DOI] [PubMed] [Google Scholar]
- Lokugamage, K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., and Menachery V.D.. 2020. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 94:e01410–e01420. 10.1128/JVI.01410-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X.Z., Bartczak A., Zhang J., He W., Shalev I., Smil D., Chen L., Phillips J., Feld J.J., Selzner N., et al. 2014. Protein interferon-stimulated gene 15 conjugation delays but does not overcome coronavirus proliferation in a model of fulminant hepatitis. J. Virol. 88:6195–6204. 10.1128/JVI.03801-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Sancho, L., Lewinski M.K., Pache L., Stoneham C.A., Yin X., Becker M.E., Pratt D., Churas C., Rosenthal S.B., Liu S., et al. 2021. Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell. 10.1016/j.molcel.2021.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre, E., and Iwasaki A.. 2020. Interferon deficiency can lead to severe COVID. Nature. 587:374–376. 10.1038/d41586-020-03070-1 [DOI] [PubMed] [Google Scholar]
- Mick, E., Kamm J., Pisco A.O., Ratnasiri K., Babik J.M., Castañeda G., DeRisi J.L., Detweiler A.M., Hao S.L., Kangelaris K.N., et al. 2020. Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses. Nat. Commun. 11:5854. 10.1038/s41467-020-19587-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova, V.T., Kong Y., Fedorova O., Sharma L., Dela Cruz C.S., Pyle A.M., Iwasaki A., and Foxman E.F.. 2018. Regional Differences in Airway Epithelial Cells Reveal Tradeoff between Defense against Oxidative Stress and Defense against Rhinovirus. Cell Rep. 24:3000–3007.e3. 10.1016/j.celrep.2018.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickbakhsh, S., Mair C., Matthews L., Reeve R., Johnson P.C.D., Thorburn F., von Wissmann B., Reynolds A., McMenamin J., Gunson R.N., and Murcia P.R.. 2019. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. USA. 116:201911083. 10.1073/pnas.1911083116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendall, C., and Kagan J.C.. 2015. The unique regulation and functions of type III interferons in antiviral immunity. Curr. Opin. Virol. 12:47–52. 10.1016/j.coviro.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabayashi, T., Kojima T., Masaki T., Yokota S., Imaizumi T., Tsutsumi H., Himi T., Fujii N., and Sawada N.. 2011. Type-III interferon, not type-I, is the predominant interferon induced by respiratory viruses in nasal epithelial cells. Virus Res. 160:360–366. 10.1016/j.virusres.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Olender, T., Keydar I., Pinto J.M., Tatarskyy P., Alkelai A., Chien M.S., Fishilevich S., Restrepo D., Matsunami H., Gilad Y., and Lancet D.. 2016. The human olfactory transcriptome. BMC Genomics. 17:619. 10.1186/s12864-016-2960-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, S.J., Azziz-Baumgartner E., Budd A.P., Brammer L., Sullivan S., Pineda R.F., Cohen C., and Fry A.M.. 2020. Decreased Influenza Activity During the COVID-19 Pandemic - United States, Australia, Chile, and South Africa, 2020. MMWR Morb. Mortal. Wkly. Rep. 69:1305–1309. 10.15585/mmwr.mm6937a6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onabajo, O.O., Banday A.R., Stanifer M.L., Yan W., Obajemu A., Santer D.M., Florez-Vargas O., Piontkivska H., Vargas J.M., Ring T.J., et al. 2020. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 52:1283–1293. 10.1038/s41588-020-00731-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira, E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. Gen-COVID Investigators . 2021. Genetic mechanisms of critical illness in COVID-19. Nature. 591:92–98. 10.1038/s41586-020-03065-y [DOI] [PubMed] [Google Scholar]
- Pertea, M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., and Salzberg S.L.. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33:290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaender, S., Mar K.B., Michailidis E., Kratzel A., Boys I.N., V’kovski P., Fan W., Kelly J.N., Hirt D., Ebert N., et al. 2020. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat. Microbiol. 5:1330–1339. 10.1038/s41564-020-0769-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert, L.W., Žilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A.M., and Jaffe A.B.. 2018. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 560:377–381. 10.1038/s41586-018-0394-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, S., Brendish N.J., Tanner A.R., and Clark T.W.. 2020. Physical distancing in schools for SARS-CoV-2 and the resurgence of rhinovirus. Lancet Respir. Med. 8:e92–e93. 10.1016/S2213-2600(20)30502-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra, N.G., Alfajaro M.M., Gasque V., Huston N.C., Wan H., Szigeti-Buck K., Yasumoto Y., Greaney A.M., Habet V., Chow R.D., et al. 2021. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 19:e3001143. 10.1371/journal.pbio.3001143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebendenne, A., Valadão A.L.C., Tauziet M., Maarifi G., Bonaventure B., McKellar J., Planès R., Nisole S., Arnaud-Arnould M., Moncorgé O., and Goujon C.. 2021. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. 95:e02415–e02420. 10.1128/JVI.02415-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova, I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., and Hertzog P.J.. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41(Database issue, D1):D1040–D1046. 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa Ribero, M., Jouvenet N., Dreux M., and Nisole S.. 2020. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 16:e1008737. 10.1371/journal.ppat.1008737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, W.M., Chevillotte M.D., and Rice C.M.. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32:513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz-Cherry, S.2015. Viral Interference: The Case of Influenza Viruses. J. Infect. Dis. 212:1690–1691. 10.1093/infdis/jiv261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singanayagam, A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., Ladhani S., Zambon M., and Gopal R.. 2020. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 25:25. 10.2807/1560-7917.ES.2020.25.32.2001483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive Integration of Single-Cell Data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, S.G., Carlson S., Cheng A.C., Chilver M.B.N., Dwyer D.E., Irwin M., Kok J., Macartney K., MacLachlan J., Minney-Smith C., et al. 2020. Where has all the influenza gone? The impact of COVID-19 on the circulation of influenza and other respiratory viruses, Australia, March to September 2020. Euro Surveill. 25:26. 10.2807/1560-7917.ES.2020.25.47.2001847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szappanos, D., Tschismarov R., Perlot T., Westermayer S., Fischer K., Platanitis E., Kallinger F., Novatchkova M., Lassnig C., Müller M., et al. 2018. The RNA helicase DDX3X is an essential mediator of innate antimicrobial immunity. PLoS Pathog. 14:e1007397. 10.1371/journal.ppat.1007397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. 2021. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 34:108728. 10.1016/j.celrep.2021.108728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J.K., Coleman C.M., Postel S., Sisk J.M., Bernbaum J.G., Venkataraman T., Sundberg E.J., and Frieman M.B.. 2015. Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference. J. Virol. 89:11820–11833. 10.1128/JVI.02274-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, R.B.2007. Rhinovirus: more than just a common cold virus. J. Infect. Dis. 195:765–766. 10.1086/511829 [DOI] [PubMed] [Google Scholar]
- Vanderheiden, A., Ralfs P., Chirkova T., Upadhyay A.A., Zimmerman M.G., Bedoya S., Aoued H., Tharp G.M., Pellegrini K.L., Manfredi C., et al. 2020. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 94:e00985–e20. 10.1128/JVI.00985-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski, P., Gultom M., Kelly J.N., Steiner S., Russeil J., Mangeat B., Cora E., Pezoldt J., Holwerda M., Kratzel A., et al. 2021. Disparate temperature-dependent virus-host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium. PLoS Biol. 19:e3001158. 10.1371/journal.pbio.3001158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels, C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., et al. 2020. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 5:1299–1305. 10.1038/s41564-020-0761-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolsk, H.M., Følsgaard N.V., Birch S., Brix S., Hansel T.T., Johnston S.L., Kebadze T., Chawes B.L., Bønnelykke K., and Bisgaard H.. 2016. Picornavirus-Induced Airway Mucosa Immune Profile in Asymptomatic Neonates. J. Infect. Dis. 213:1262–1270. 10.1093/infdis/jiv594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, A., Mihaylova V.T., Landry M.L., and Foxman E.F.. 2020. Interference between rhinovirus and influenza A virus: a clinical data analysis and experimental infection study. Lancet Microbe. 1:e254–e262. 10.1016/S2666-5247(20)30114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., and Shi P.Y.. 2020. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 33:108234. 10.1016/j.celrep.2020.108234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahya, M., Rulli M., Toivonen L., Waris M., and Peltola V.. 2017. Detection of Host Response to Viral Respiratory Infection by Measurement of Messenger RNA for MxA, TRIM21, and Viperin in Nasal Swabs. J. Infect. Dis. 216:1099–1103. 10.1093/infdis/jix458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh, D.K., Foley D.A., Minney-Smith C.A., Martin A.C., Mace A.O., Sikazwe C.T., Le H., Levy A., Blyth C.C., and Moore H.C.. 2020. The impact of COVID-19 public health measures on detections of influenza and respiratory syncytial virus in children during the 2020 Australian winter. Clin. Infect. Dis. ciaa1475. 10.1093/cid/ciaa1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., Sakai K., Gotoh S., Miorin L., De Jesus P.D., et al. 2021. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 34:108628. 10.1016/j.celrep.2020.108628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., Peterson D.R., Baran A.M., Bhattacharya S., Wylie T.N., Falsey A.R., Mariani T.J., and Storch G.A.. 2019. Host Gene Expression in Nose and Blood for the Diagnosis of Viral Respiratory Infection. J. Infect. Dis. 219:1151–1161. 10.1093/infdis/jiy608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. NIAID-USUHS/TAGC COVID Immunity Group . 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 370:eabd4570. 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., et al. 2020. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 27:883–890.e2. 10.1016/j.chom.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Wang W., Liu Z., Liang C., Wang W., Ye F., Huang B., Zhao L., Wang H., Zhou W., et al. 2020. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 11:3910. 10.1038/s41467-020-17796-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. HCA Lung Biological Network . 2020. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 181:1016–1035.e19. 10.1016/j.cell.2020.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, C.G.K., Miao V.N., Owings A.H., Navia A.W., Tang Y., Bromley J.D., Lotfy P., Sloan M., Laird H., Williams H.B., et al. 2021. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. bioRxiv. 10.1101/2021.02.20.431155 (Preprint posted February 20, 2021) [DOI] [PMC free article] [PubMed]
- Cepheid. 2021. Xpert Express SARS-CoV-2. https://www.fda.gov/media/136314/download (accessed June 8, 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists characteristics of 30 SARS-CoV-2–positive patient samples used for transcriptome analysis.
lists differentially expressed genes in SARS-CoV-2–positive patients versus negative controls.
lists Ingenuity pathways differentially enriched in SARS-CoV-2–positive NP samples compared to controls.
lists differentially expressed genes in SARS-CoV-2–positive female versus male patients.
lists differentially expressed genes in SARS-CoV-2–positive older versus younger patients.
lists differentially expressed genes in SARS-CoV-2–positive admitted patients versus outpatients.
lists clinical descriptions of longitudinal samples.
Data Availability Statement
scRNA-seq data are available in the GEO database under accession no. GSE164982. RNA-seq data derived from patient samples have been deposited to dbGaP under accession no. phs002433.v1.p1. To access these data, users may apply for access to the dbGaP data repository (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?login=&page=login).